Abstract
Objective.
Exosomes are lipid bilayer-bound microvesicles containing various macromolecules including numerous microRNA (miRNA). Exosomes mediate intercellular communication by fusing and releasing their macromolecular content into target cells. Here, we analysed the content of profibrotic and antifibrotic miRNAs in exosomes isolated from the serum of systemic sclerosis (SSc) patients and tested their ability to induce a profibrotic pheno-type in normal human dermal fibroblasts in vitro.
Methods.
Exosomes were isolated from serum from patients with limited cutaneous or diffuse cutaneous SSc and were characterised by Nanosight Particle Tracking Analysis, exosome antibody arrays, and transmission electron microscopy. The content of nine profibrotic and eighteen antifibrotic miRNA was assessed in the isolated exosomes by semiquantitative real time PCR. The effects of the isolated exosomes on cultured normal human dermal fibroblasts were assessed by real time PCR and Western blotting.
Results.
The isolated serum exosomes displayed the expected exosome size and morphology and contained characteristic exosome proteins. Six profibrotic miRNAs were increased and ten antifibrotic miRNAs were decreased in SSc serum exosomes compared to normal serum exosomes. The levels of eight miRNA were significantly different between exosomes from limited and diffuse SSc. Exosomes isolated from both limited or diffuse SSc patients caused dose-dependent stimulation of profibrotic gene expression and type I collagen and fibronectin production and secretion in normal human dermal fibroblasts in vitro.
Conclusion.
Serum exosomes from SSc patients contain miRNA displaying a markedly profibrotic profile and induce a profibrotic phenotype in target normal fibroblasts in vitro suggesting a plausible mechanism for the extension of the fibrotic SSc process to non-affected tissues.
Keywords: systemic sclerosis, exosomes, miRNA, fibrosis, SSc biomarkers, cell-cell communication
Introduction
Systemic sclerosis (SSc) is a systemic autoimmune disease of unknown aetiology characterised by progressive tissue fibrosis, cellular and humoral immunity abnormalities, and prominent microvascular alterations (1, 2). The mechanisms involved in SSc pathogenesis are complex and have not been fully elucidated (3–5), although it has been recognised that most of the clinical and pathologic SSc manifestations result from progressive skin and internal organ fibrosis and a severe fibroproliferative vasculopathy (6, 7). A unique SSc feature is the progressive extension of the fibrotic process to non-affected tissues. Despite the crucial importance of this process to SSc clinical manifestations, therapeutic approaches and prognosis, the mechanisms responsible have remained elusive, although recent studies of circulating exosomes have suggested novel potential mechanisms. Exosomes are 30–150 nm microvesicles surrounded by lipid bilayer membranes that are released from all human cells into the extracellular space (8–10). Exosomes contain numerous macromolecules including mRNAs, microRNAs (miRNA), small non-coding RNAs, cytokines, chemokines, and growth factors (11–13). Exosomes play an important role in intercellular communications owing to their ability to alter the phenotype of distant target cells (14–17). This pathogenetic mechanism may play a crucial role in the paracrine modulation of molecular programs in neighboring and distant normal cells and could explain the extension of the SSc-associated fibrotic process to unaffected cells and tissues.
Activated myofibroblasts are the cells ultimately responsible for the severe fibroproliferative process in SSc (18–21). Myofibroblasts display a remarkable profibrotic phenotype characterised by the increased production of fibrillar type l and type lll collagens, initiation of expression of α-smooth muscle actin (α-SMA), and reduction of ECM-degradative enzymes (22, 23). In response to injury tissue resident fibroblasts become activated and transdifferentiate into myofibroblasts (24–26), thus, an analysis of the mechanisms involved in the generation of activated myofibroblasts may provide insights into the establishment and spread of the fibroproliferative process in SSc.
Recently, much interest has been focused on the participation of miRNA in the epigenetic regulation of gene expression (27–29) and on their role in the pathogenesis of tissue fibrosis and fibrotic diseases including SSc (30–33). Indeed, crucial intracellular transduction pathways implicated in the SSc fibrotic process, including TGF-β signaling, are modulated and targeted by miRNA (34–39). Furthermore, several recent studies have identified miRNAs whose levels were altered in the serum of different clinical subsets of SSc patients or that played a role in SSc pathogenesis (40–43). However, comparison of miRNA content of exosomes isolated from serum of normal individuals and from SSc patients has not been performed. These studies have the potential to identify signaling miRNAs that regulate novel molecular pathways involved in the pathogenesis and progression of SSc, and may provide valuable biomarkers to predict the response of individual SSc patients to disease modifying treatment modalities.
Materials and methods
Patients
Serum samples from three donors with limited cutaneous SSc (lcSSc), and three donors with diffuse cutaneous SSc (dcSSc) were obtained following IRB approval from the Thomas Jefferson Scleroderma Center Serum Bank. These donors fulfilled the American College of Rheumatology classification criteria for SSc (44) and the criteria for clinical subset classification described by Le Roy et al. (45). The demographic and disease characteristics of the patients included in the study are summarised in Table I. Two samples of normal serum were utilised as controls. One sample consisted of serum from one normal donor whereas a second sample was a pool of serum from twelve normal blood donors. This pooled serum sample was included to minimise possible heterogeneity introduced by normal variability. The blood samples were allowed to clot at room temperature for 30 min and then they were centrifuged at 5000 rpm for 10 min at 4°C. The serum supernatants were stored at −80°C before analysis.
Table I.
Selected features of systemic sclerosis patients studied*.
| lcSSc 1 | lcSSc 2 | lcSSc 3 | dcSSc 1 | dcSSc 2 | dcSSc 3 | |
|---|---|---|---|---|---|---|
| Age | 58 | 49 | 43 | 60 | 32 | 61 |
| Sex | F | F | F | F | F | F |
| Disease Duration, (years)† | 4 | 2 | 1 | 1 | 1 | 2 |
| Extent of maximal skin induration,‡ | 3% | 2% | 2% | 27% | 30% | 35% |
| mRSS | 7 | 4 | 5 | 25 | 26 | 28 |
| Organ Involvement | ||||||
| Raynaud’s Phenomenon | YES | YES | YES | YES | YES | YES |
| G-I | YES | NO | YES | YES | NO | YES |
| Pulmonary | NO | NO | NO | NO | YES | YES |
| Cardiac | NO | NO | NO | NO | NO | NO |
| Renal | NO | NO | NO | NO | NO | NO |
| ANA Test | ||||||
| ANATiter | 1:640 | 1:320 | 1:5120 | 1:2560 | 1:360 | 1:640 |
| Scl-70 | NEG | NEG | NEG | NEG | POS | POS |
| ACA | POS | POS | POS | NEG | NEG | NEG |
| Treatment | ||||||
| Antifibrotic | NO | NO | NO | D-Pen | D-Pen | D-Pen |
| Immunosupressive | NO | NO | NO | NO | NO | NO |
lcSSc: limited cutaneous SSc; dcSSc: diffuse cutaneous SSc.
Determined from onset of skin induration or first non-Raynaud’s phenomenon manifestation.
Extent of affected body surface (% of total body surface). mRSS: modified Rodman Skin Score.
D-Pen: D-penicillamine.
Exosome isolation and electron microscopy
Exosomes were isolated from 250 μL of serum by a one-step polymer precipitation procedure utilising ExoQuick Exosome Precipitation Solution (System Biosciences, Palo Alto, CA), according to the manufacturer’s instructions, followed by incubation at 4°C for 30 min and pelleting by centrifugation at 13000 rpm for 2 min (46, 47). For transmission electron microscopy (TEM), exosome pellets were resuspended in 200 μL 0.2M phosphate buffer and then mixed with an equal volume (1:1) of 4% paraformaldehyde. A 5 μL aliquot from each sample was pipetted onto UV-treated 200 mesh formvar/carbon-coated nickel electron microscopy grids and allowed to adsorb for 20 min. Exosomes were further fixed for 5 min in 2.5% glutaraldehyde, contrasted for 10 min in 2% uranyl acetate, and analysed using a Hitachi H-7600 TEM microscope at 80 kV.
Exosome antibody array analysis
Exosome protein content was quantified using the bicinchoninic acid (BCA) assay (Pierce Biotechnology, Woburn, MA). For exosome array binding assays, an aliquot of isolated exosomes from each patient containing 500 μg exosome proteins was lysed in Exosome Lysis buffer. Each lysate was combined with Exosome Array Binding buffer and incubated with a separate Exo-Check antibody membrane array overnight at 4°C. Each membrane array has 12 pre-printed spots. Eight spots contain one of eight antibodies for known exosome marker proteins (CD63, CD81, ALIX, FLOT1, ICAM1, EpCam, ANXA5 or TSG101), one spot contains an antibody that detects GM130, a cis-Golgi protein marker, two spots are positive controls for HRP-mediated reagents, and the last spot is empty and serves as a blank control. The membranes were then washed using Array Wash buffer and incubated at room temperature with infrared dye-labeled secondary anti-rabbit antibody (1:10000) in Odyssey (Li-Cor Biosciences, Lincoln, NE) blocking buffer. Signals were detected using an Odyssey Infrared Imaging System (Li-Cor).
Nanosight particle tracking analysis
Freshly isolated serum exosome samples were diluted 1:50 with sterile PBS and injected into the Nanosight NS300 unit (Malvern Instruments, Westborough, MA). Capture and analysis settings were manually set according to the manufacturer’s protocol. Particles were visualised by laser light scattering and their Brownian motion captured on digital video. Three separate runs were conducted for each sample. The recorded videotapes were analysed utilising Nanoparticle Tracking Analysis (NTA) 2.3 software based on tracking at least two hundred individual particles per run. The software generates high resolution size distribution profiles and concentration measurements of tracked particles using the properties of both light scattering and Brownian motion as they move through the flow chamber.
RT-PCR assessment of exosome miRNA
For RT-PCR assessment the isolated exosome pellets were lysed with lysis buffer and RNA was purified using SeraMir RNA spin columns (System Biosciences). The concentration of RNA was quantified using the Nano-Drop ND-1000 system (NanoDrop, Wilmington, DE). Isolated exosome RNA was polyadenylated and cDNA was synthesised by reverse transcription utilising a SeraMir 3’ Adaptor primer according to the manufacturers’ instructions. Real-time quantitative PCR was performed using standard protocols on an Applied Biosystems 7900HT Sequence Detection System equipped with a 384-well reaction plate using 2 μL of exosome cDNA in 2× SYBR green PCR master mix (Applied Biosystems, Foster City, CA), 800 nM each of the miRNA specific primer and the SeraMir Reverse primer. The primers utilised for miRNA amplification are shown in Table II. To normalise for the amounts of loaded cDNA, the small nucleolar RNA SNORD25 was used as an internal control. Differences were calculated employing the comparative Ct method.
Table II.
Primers used in real-time PCR studies.
| MicroRNA | Sequence (5’−3’) |
|---|---|
| let-7a-5p | TGAGGTAGTAGGTTGTATAGTT |
| let-7d-5p | AGAGGTAGTAGGTTGCATAGTT |
| let-7g-5p | TGAGGTAGTAGTTTGTACAGTT |
| miR-7–5p | TGGAAGACTAGTGATTTTGTTGT |
| miR-17–5p | CAAAGTGCTTACAGTGCAGGTAG |
| miR-21–5p | TAGCTTATCAGACTGATGTTGA |
| miR-23b-5p | TGGGTTCCTGGCATGCTGATTT |
| miR-26a-5p | TTCAAGTAATCCAGGATAGGCT |
| miR-26b-5p | TTCAAGTAATTCAGGATAGGT |
| miR-29a-3p | TAGCACCATCTGAAATCGGTTA |
| miR-29b-3p | TAGCACCATTTGAAATCAGTGTT |
| miR-92a-3p | TATTGCACTTGTCCCGGCCTGT |
| miR-125b-5p | TCCCTGAGACCCTAACTTGTGA |
| miR-129–5p | CTTTTTGCGGTCTGGGCTTGC |
| miR-132–3p | TAACAGTCTACAGCCATGGTCG |
| miR-133a-3p | TTTGGTCCCCTTCAACCAGCTG |
| miR-140–5p | CAGTGGTTTTACCCTATGGTAG |
| miR-145–5p | GTCCAGTTTTCCCAGGAATCCCT |
| miR-146a-5p | TGAGAACTGAATTCCATGGGTT |
| miR-150–5p | TCTCCCAACCCTTGTACCAGTG |
| miR-155–5p | TTAATGCTAATCGTGATAGGGGT |
| miR-196a-5p | TAGGTAGTTTCATGTTGTTGGG |
| miR-200a-3p | TAACACTGTCTGGTAACGATGT |
| miR-200b-3p | TAATACTGCCTGGTAATGATGA |
| miR-206–5p | TGGATGTAAGGAAGTGTGTGG |
| miR-215–5p | ATGACCTATGAATTGACAGAC |
| miR-223–3p | TGTCAGTTTGTCAAATACCCCA |
| miR-503–5p | TAGCAGCGGGAACAGTTCTGCAG |
Treatment of normal dermal fibroblasts with normal andSSc serum exosomes
To examine the effects of the isolated serum exosomes on the gene expression and biosynthetic profile of normal human dermal fibroblasts the cells were exposed to various concentrations of isolated exosomes. A normal human dermal fibroblast cell line, established from a full-thickness skin biopsy from a normal donor was obtained from the Thomas Jefferson University Scleroderma Center Tissue Bank. The Institutional Review Board of Thomas Jefferson University approved the use of the tissues from normal donors or from patients remaining after diagnostic histopathologic studies for in vitro analyses. The normal human dermal fibroblast cell line (passage 4) was cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, antibiotics, 50 mM HEPES, and glutamine in 12 well plates until confluent. Prior to exposure to isolated exosomes, media were removed and cells were extensively washed in PBS followed by incubation in serum-free medium for 24h. Exosomes isolated from the sample of pooled normal serum and from the serum of two patients with limited SSc and from two patients with diffuse SSc were examined. After 24h, fresh serum-free media was added to the cells along with three replicates of each isolated exosome preparation to yield a final exosome concentration of either 2.5, 5.0, or 10.0 μg/mL based on total protein concentration. Control wells received only normal saline. After 72h, culture supernatants were removed, the cells were lysed and mRNA and cellular proteins were isolated for subsequent studies.
Gene expression levels and Western blot analysis of normal samples from human dermal fibroblasts following treatment with normal and SSc serum exosomes
For gene expression assessment, total RNA was isolated from dermal fibroblasts utilising Trizol extraction and cDNA was synthesised by reverse transcription. The expression levels of genes for various extracellular matrix components, myofibroblast-specific proteins, and profibrotic growth factors were examined. The differences in the number of mRNA copies in each PCR were corrected for human GAPDH endogenous control transcript levels; levels in control experiments were set at 100 and all other values expressed as multiples of control values. For the detection of type I collagen and total fibronectin, equal volumes of culture medium containing proteins secreted by exosome-treated fibroblasts were processed for Western blotting under denaturing conditions. Media from cell cultures were collected after 72h and 35 μL of culture media were heated to 95°C for 5 min. The proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred to nitrocellulose membranes (Invitrogen, Carlsbad, CA). The blots were blocked for 1 h in Odyssey blocking buffer (Li-Cor, Lincoln, NE). The membranes were incubated overnight at 4° C with either polyclonal anti-COL1 antibody (Southern Biotech, Birmingham, AL), polyclonal anti-COL3 antibody (Sigma-Aldrich, St. Louis, MO), or monoclonal anti-fibronectin 1 (anti-FN1; Santa Cruz Biotechnology, Santa Cruz, CA) in the same blocking buffer. The membranes were then washed with PBS-0.2% Tween 20 and incubated for 1 h with the appropriate infrared dye-labeled secondary antibodies (Li-Cor), diluted 10,000 fold in the blocking buffer. Signals were detected and quantitated using an Odyssey Infrared Imaging System (Li-Cor).
Statistical analysis
Data are presented as the mean ± standard deviation of three replicates from each serum exosome sample. The statistical significance of the real-time PCR data was assessed by Student’s two-tailed t test. P-values less than 0.05 were considered statistically significant.
Results
Quantitative analysis, characterisation, and TEM of exosomes isolated from normal serum and from the serum of patients with SSc
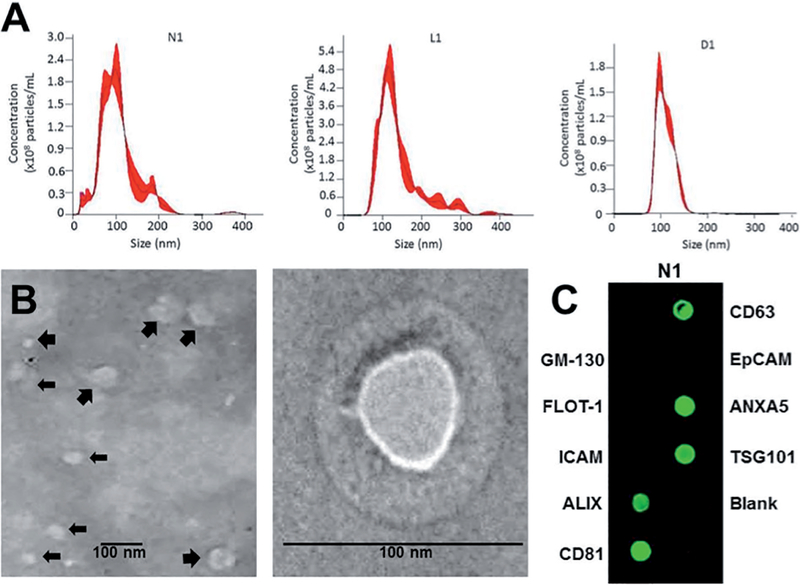
Exosomes were isolated from the blood serum of three donors with limited cutaneous SSc (lcSSc), and three donors with diffuse cutaneous SSc (dc-SSc) as well as from a single normal donor and from a sample comprised of a pool of serum from twelve normal individuals that was included to mini-mise possible heterogeneity introduced by normal variability. Nanosight particle tracking analysis of isolated exosomes demonstrated the presence of particles ranging in size from ~75 nm to 200 nm consistent with the expected size range of exosomes in all samples of serum studied (Fig. 1A). Although exosomes from sera of limited SSc and diffuse SSc donors also displayed the size range observed from normal sera, smaller exosomes in the range of 40–60 nm were predominant in these samples. The average particle concentration and the average RNA content were higher in serum exosomes from limited SSc and diffuse SSc patients compared to normal serum exosomes (Table III). TEM was performed on exosomes isolated from serum of one normal, one limited SSc and two diffuse SSc donors. TEM showed that exosomes isolated from normal and SSc serum appeared as spherical vesicles surrounded by well-defined membranes ranging in size from 40 nm to ~120 nm (Fig. 1B). The isolated exosomes from all samples were also analysed by exosome antibody membrane arrays. All serum exosome samples examined were positive for the presence of CD63, CD81, ALIX, TSG101, and ANXA5 exosome-associated proteins (Fig. 1C). No signal was detected with the GM-130 cis-Golgi marker indicating that the samples were not contaminated with cellular proteins.
Fig. 1.
Characterisation of isolated serum exosomes. A. Light scattering (Nanosight) measurements of particle size and concentration. Purified exosomes preparations were injected into the Nanosight NS300 instrument and the particles motion was captured and video-recorded. Particle size and concentrations were calculated by NTA software. Size distribution graphs represent the merged profiles generated from three separate runs and analyses per sample. Particle diameter sizes were between 75 and 200 nm, and the particle concentrations ranged between 1.3×1010 and 7.2×1010 particles per mL. N1: normal; L1: Limited SSc; D1: Diffuse SSc. B. Transmission electron microscopy. Analysis of isolated exosomes showed the presence of two populations of vesicles in the size range of 40–120 nm at 25000× magnification (left panel). Larger vesicles of ~100–120 nm size are interspersed (large arrows) with a population of smaller vesicles of ~40–60 nm (small arrows). Enlarged electron micrograph of a representative 90 nm exosome. Scale bars = 100 nm (right panel). C. Antibody membrane arrays. Exosome antibody membrane arrays incubated with exosome lysates confirming the presence of the exosome-specific proteins CD63, CD81, ALIX, ANXA5 and TSG101. Samples were negative for the cis-Golgi protein GM130, an indicator of cytosolic protein contamination. A representative array obtained with exosomes isolated from sample N1 is displayed.
Table III.
Comparison of the numbers, size, and RNA and protein content of isolated exosomes from normal and SSc patient serum.
| Donor | Total concentration (Particles/ml) | Particle size (nm) | RNA concentration (ng/ml) | Protein concentration (μg) |
|---|---|---|---|---|
| Average for N | 1.5 × 1010 | 113.3 ± 11.8 | 840 ± 148.5 | 1078 ± 26.2 |
| Average for L | 2.9 × 1010 ± 1.4×1010 | 158.2 ± 14.9 | 960 ± 240 | 1210 ± 105.1 |
| Average for D | 4.3 × 1010 ± 2.7×1010 | 164.2 ± 70.8 | 1720 ± 615.8 | 1220 ± 26.9 |
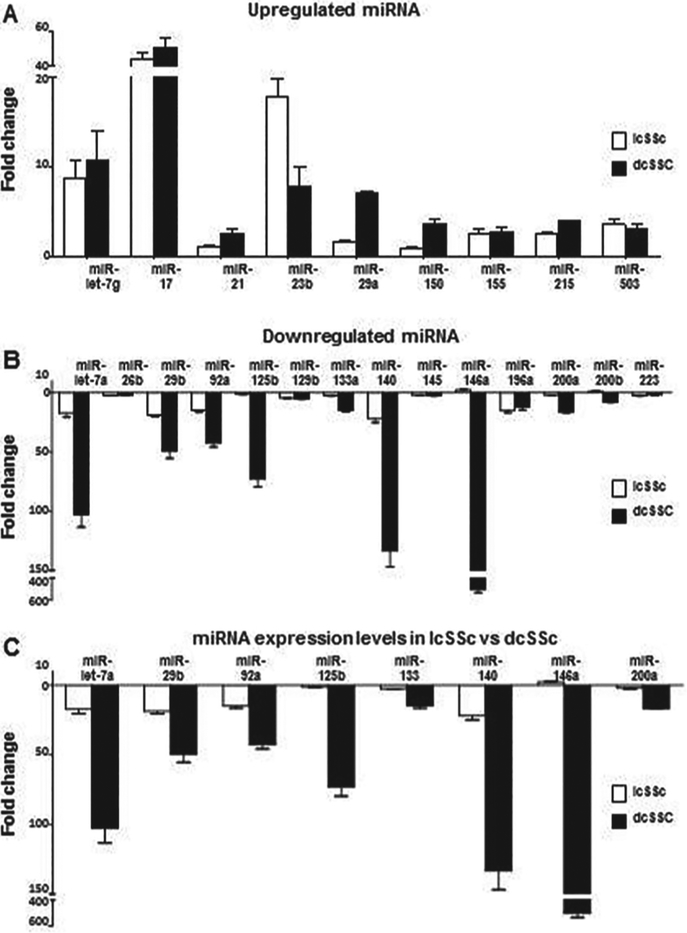
Content of Profibrotic and Antifibrotic miRNA in serum exosomes from patients with limited and diffuse cutaneous SSc compared to exosomes isolated from normal serum
These studies were performed in order to assess differences in the miRNA content of exosomes isolated from the SSc patient subgroups from the mi-RNA content of exosomes isolated from the serum of normal donors. The exosome content of twenty eight miRNA that have been previously implicated in the pathogenesis of the fibrotic process in SSc (32, 33, 32) was assessed. Nine of these were considered to be profibrotic and eighteen were considered to be antifibrotic. Exosomal miRNA levels were normalised to levels of the small nucleolar RNA SNORD25. The levels of each miRNA present in exosomes isolated from normal serum were averaged and arbitrarily set at the 1 fold content level for comparison. The relative levels of all the miRNAs examined are shown in Table IV, and the miRNA that displayed levels that were either greater than two-fold higher or two-fold less or lower compared to the values obtained from the normal serum exosomes or that differed by greater than two-fold between the exosomes from the two groups of SSc patients are shown in Figure 2. The content levels of six profibrotic miRNAs were significantly elevated in exosomes isolated from both limited SSc and diffuse SSc serum compared to the levels in exosomes from normal serum. The levels of three other profibrotic miRNA were elevated significantly only in exosomes isolated from diffuse SSc serum (Fig. 2A). The exosome content of twelve antifibrotic miRNAs was lower by greater than two-fold in exosomes from both limited SSc and diffuse SSc groups compared to exosomes from normal serum, and two other miRNA displayed lower levels only in diffuse SSc serum samples (Fig. 2B). The levels of five antifibrotic miRNA did not show significant differences between SSc and normal exosomes (Table IV).
Table IV.
Mean content of Profibrotic and antifibrotic miRNAs in serum exosomes from SSc patients compared to the average levels in exosomes from normal serum.
| miRNA | Expression Level | Expected Effect | Limited SSc Fold Change (avg)1 | Diffuse SSc Fold Change (avg)1 |
|---|---|---|---|---|
| let-7g-5p | Increased | Profibrotic | 8.67*** | 10.71*** |
| miR-17–5p | Increased | Proflbrotic | 35.79*** | 46.66*** |
| miR-21–5p | Increased | Profibrotic | 1.16 | 2.62** |
| miR-23b-5p | Increased | Proflbrotic | 17.83*** | 7.85*** |
| miR-29a-3p | Increased | Profibrotic | 1.61 | 7.17*** |
| miR-150–5p | Increased | Profibrotic | 1.04 | 3.65*** |
| miR-155–5p | Increased | Profibrotic | 2.62** | 2.80** |
| miR-215–5p | Increased | Profibrotic | 2.53** | 4.01** |
| miR-503–5p | Increased | Profibrotic | 3.62*** | 3.20** |
| let-7a-5p | Decreased | Antifibrotic | −17.45*** | −102.80*** |
| miR-26b-5p | Decreased | Antifibrotic | −2.33** | −1.98* |
| miR-29b-3p | Decreased | Antifibrotic | −18.93*** | −49.49*** |
| miR-92a-3p | Decreased | Antifibrotic | −15.01*** | −42.66*** |
| miR-125b-5p | Decreased | Antifibrotic | −1.33 | −73.03*** |
| miR-129–5p | Decreased | Antifibrotic | −4.53*** | −5.19*** |
| miR-133a-3p | Decreased | Antifibrotic | −2.6** | −14.8*** |
| miR-140–5p | Decreased | Antifibrotic | −22.1*** | −133.5*** |
| miR-145–5p | Decreased | Antifibrotic | −2.16* | −2.65** |
| miR-146a-5p | Decreased | Antifibrotic | 2.26** | −502.5*** |
| miR-196a-5p | Decreased | Antifibrotic | −15.12*** | −12.04*** |
| miR-200a-3p | Decreased | Antifibrotic | −2.27* | −16.67*** |
| miR-200b-3p | Decreased | Antifibrotic | 1.0 | −8.02*** |
| miR-223–3p | Decreased | Antifibrotic | −2.24** | −2.10* |
| let-7d-5p | Unchanged | Antifibrotic | 1.01 | 1.11 |
| miR-7–5p | Unchanged | Antifibrotic | −1.31 | −1.79 |
| miR-26a-5p | Unchanged | Antifibrotic | 1.04 | 1.14 |
| miR-132–3p | Unchanged | Antifibrotic | −1.11 | 1.37 |
| miR-206–5p | Unchanged | Antifibrotic | 1.27 | −1.79 |
Fold change was calculated by comparison of the averaged miRNA values measured in exosome isolated from the serum of SSc patients following correction employing the small nucleolar RNA SNORD25 compared to the average values of each miRNA obtained in the exosomes isolated from normal serum.
p<0.05,
p<0.01,
p<0.001.
Fig. 2.
Content levels of miRNA present in exosomes isolated from serum of donors with limited and diffuse SSc as assessed by real time PCR. Values represent the mean (± SD) fold change levels of three replicates of exosomes isolated from three patients per group. Content levels in the two normal serum samples were averaged and arbitrarily set at 1 fold content level. A. Profibrotic miRNAs with a 2-fold or greater difference in content level in one or both disease subgroups. B. Antifibrotic miRNAs with a 2-fold or greater difference in content level in one or both disease subgroups. C. Antifibrotic miRNAs displaying content levels that were significantly different between lcSSc and dcSSc subgroups.
Differences in profibrotic and antifibrotic miRNA content of exosomes from sera of patients with limited and diffuse cutaneous SSc
Comparison of the miRNA content levels between exosomes isolated from serum of the two SSc subsets showed that the levels of eight antifibrotic mi-RNAs differed significantly between exosomes from the limited SSc group and the diffuse SSc group (Fig. 2C and Table IV), whereas there were no significant differences in any of the profibrotic miRNA.
SSc serum exosomes induce the expression of genes associated with fibrosis and myofibroblast activation in normal human dermal fibroblastsin vitro.
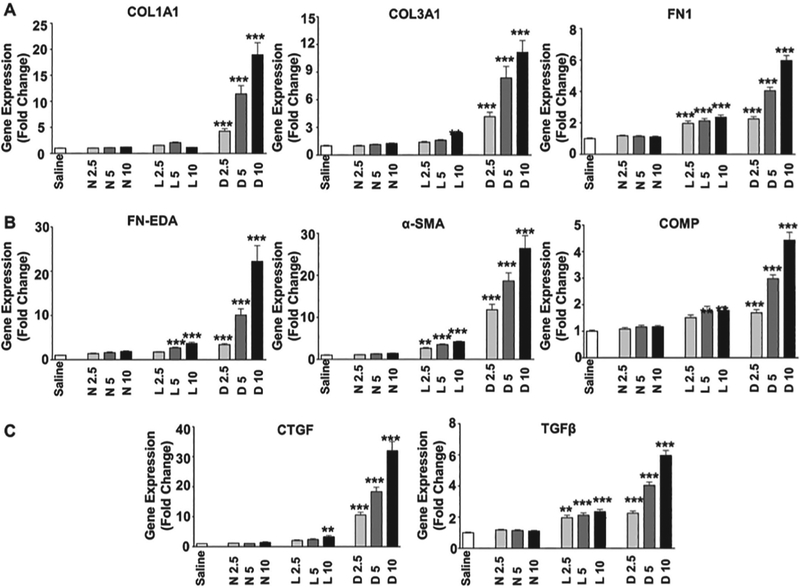
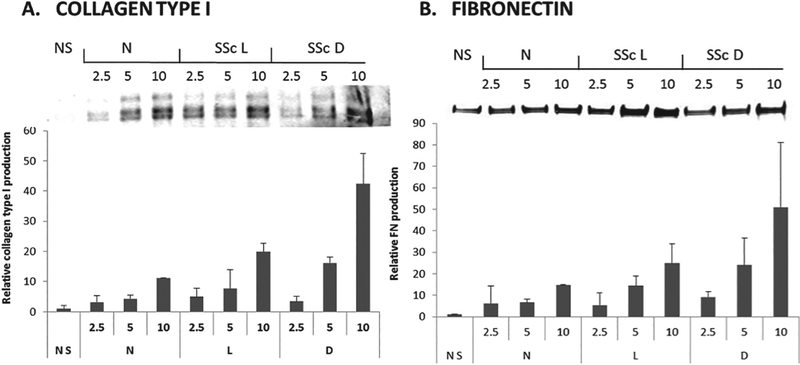
To determine whether serum exosomes from SSc patients could induce phenotypic changes in target cells, normal dermal human fibroblasts that had been cultured in FBS-free culture media were exposed to three concentrations (2.5, 5 or 10 μg) of exosomes isolated from the serum of normal individuals or from patients with limited SSc or diffuse SSc. The concentration of exosomes added to the fibroblast cultures was normalised by adding equivalent amounts of exosomes based on their total protein concentrations. Changes in gene expression in response to treatment with the added exosomes was performed by RT-PCR and C(t) levels were normalised to the GAPDH endogenous control. The results showed that exosomes isolated from the serum of SSc patients stimulated the expression of genes encoding extracellular matrix components COL1A1, COL3A1, and FN1 when compared to the levels measured in saline-treated control fibroblasts or in fibroblasts treated with exosomes isolated from the serum of a normal individual (Fig. 3A). Quantitative analysis revealed that the strongest upregulation was mediated by serum exosomes from diffuse SSc patients and moderate upregulation was mediated by exosomes isolated from limited SSc donor sera. Serum exosomes from both subsets of SSc patients also induced dose-dependent increases in the expression of three genes associated with myofibroblast activation, COMP, ACTA2 (α-SMA) and the FN1 alternative splice variant FN-EDA (Fig. 3B). Finally, exosomes from serum of patients with either of the clinical SSc subgroups induced dose-dependent increases in expression of TGF-β and CTGF genes (Fig. 3C). Notably, exosomes isolated from the serum of diffuse SSc patients induced higher levels of expression of all the genes examined than exosomes isolated from limited SSc patient serum. Western blots validated at the protein level the results of gene expression analyses showing increased levels of type I collagen (Fig. 4A), total FN1 (Fig. 4B), and type III collagen (not shown) in culture supernatants of fibroblasts treated with exosomes isolated from diffuse SSc and limited SSc patient sera compared to supernatants from fibroblasts exposed to exosomes isolated from normal donor sera.
Fig. 3.
Effects of exosomes isolated from the serum of SSc patients on the expression of profibrotic genes in cultured normal human dermal fibroblasts. A. COL1A1, COL3A1, and FN. B. FN-EDA, α-SMA (ACTA2) and COMP. C. CTGF, and TGF-β. Three concentrations of added exosomes (2.5, 5 or 10 μg) determined by protein concentration of the isolated exosomes were examined. Each concentration was added to triplicate cultures of confluent normal human dermal fibroblasts incubated with 1 ml of culture media. Each value shown represents the mean (± SD) fold change levels of gene expression from each treatment group. Triplicate wells of fibroblasts treated with individual exosome preparations for each treatment group (Normal: 2 samples; Limited and Diffuse SSc: 3 samples each). C(t) levels were normalised to GAPDH. Content levels from the saline-treated fibroblasts were arbitrarily set at the 100% expression level. Values for other samples are expressed relative to this control. Significance was determined by Student’s T-test. Statistical significance: **p<0.01; ***p<0.001.
Fig. 4.
Increased production of extracellular matrix proteins by exosome-treated cultured normal human dermal fibroblasts. Representative Western blots from secreted proteins present in the culture medium of exosome-treated normal human dermal fibroblasts. A. type I collagen, B. FN1. Quantitative analysis of the results of Western blotting was performed employing a fluorescence imaging system and the values are displayed below the corresponding protein and represent the mean fluorescence units from fibroblasts treated with each exosome concentration as fold change compared to the mean fluorescence of fibroblasts cultured without exosomes (saline).
Discussion
Numerous studies have examined the differences in secreted and intracellular proteins between various cell types obtained from normal individuals and homologous cells obtained from patients with SSc [reviewed in refs. 2–4]. However, differences in the macromolecular content of exosomes released by various cells or present in the serum have not been studied. We report here the comparative analysis of the miRNA content of exosomes isolated from sera from normal individuals and from sera from three patients with limited SSc and three patients with diffuse SSc. Nanoparticle tracking analysis and TEM confirmed that the isolated microvesicles were of the sizes expected for exosomes. Nanoparticle analysis revealed differences in particle number and size between exosomes isolated from sera of the two SSc subsets compared to exosomes isolated from the sera of normal individuals. Exosome antibody arrays further validated that the isolated particles were indeed exosomes based on the detection of specific exosome-associate proteins and the lack of cellular protein contamination of the samples.
Analysis of miRNAs previously reported to be associated either with tissue fibrosis or SSc pathogenesis (3, 32, 33) demonstrated remarkable differences between exosomes isolated from sera from SSc patients compared to exosomes isolated from normal serum. The levels of several miRNAs that are predicted to stimulate profibrotic target genes or downregulate antifibrotic genes were significantly increased in the SSc exosome samples and, conversely, several anti-fibrotic miRNAs that can potentially down-regulate fibrosis-promoting molecular targets were decreased in the exosomes isolated from the serum of SSc patients. Four profibrotic miRNAs (let-7g, miR17, miR23b and miR29a) were increased greater than 5-fold and four antifibrotic miRNAs (let-7a, miR125b, miR140, and miR146a) were decreased by greater than 70-fold in the exosomes isolated from diffuse SSc serum. There were also significant differences in the content of antifibrotic miRNAs in exosomes isolated from diffuse SSc patients serum compared to exosomes from limited SSc patients. The most notable differences were that three antifibrotic miRNAs (let-7a, miR-125b, and miR-140), displayed greater than 6-fold decrease, and that, remarkably, miR-146a displayed greater than 200-fold decrease in the diffuse SSc exosomes compared to the limited SSc exosomes. These results are in partial agreement with recent studies that examined differences in plasma extracellular miRNA in diffuse SSc patients compared with limited SSc patients (40–43). In agreement with our observations, a recently published study (48) reported that expression of exosomespecific CD63 protein was increased in the skin of SSc patients compared to normal individuals and that exosomes isolated from cultured SSc dermal fibroblasts displayed changes in the expression of profibrotic miRNAs. Interestingly, they found that TGF-β1 treatment of normal dermal fibroblasts did not trigger increased production of exosomes. However, in contrast with our results they found significantly lower numbers of circulating exosomes in SSc patients with vascular involvement, possibly due to inability of the exosomes to migrate to the blood.
The marked differences in the content of profibrotic and antifibrotic miRNAs in exosomes isolated from the serum of SSc patients compared to exosomes in normal serum described here suggest a novel pathogenetic mechanism for SSc progression. This mechanism postulates that cells induced to express a profibrotic phenotype in response to the SSc initiating event or etiologic factor package profibrotic macromolecules including miRNA into exosomes that are shed into their extracellular milieu. The released exosomes enter the circulation and fuse with distant target cells, releasing their macromolecular contents and as a result, inducing a profibrotic molecular program in the target cells. In this way, the response to the initial SSc-etiologic factor would be sustained and its effects would progressively spread to normal cells and tissues at sites distant from the originating site. In support of this pathogenetic mechanism we found here that exosomes isolated from serum from patients with both clinical SSc subsets induced increased expression of various genes associated with a profibrotic phenotype in cultured normal human dermal fibroblasts including the increased expression of genes encoding interstitial collagens (Type I and III) and of fibronectin. A remarkable observation was that treatment of normal fibroblasts with equivalent amounts of SSc serum exosomes induced the expression of genes associated with the transdifferentiation of these cells to activated myofibroblasts including α-SMA, COMP, and FN-EDA as well as the increased expression of genes for the crucial profibrotic growth factors, TGF-β and CTGF. We further found that the exosomes isolated from the serum of diffuse SSc patients demonstrated greater upregulation of the expression of these genes compared to the exosomes isolated from the serum of patients with limited SSc. The induction of a profibrotic phenotype in the normal fibroblasts was further confirmed by the demonstration of increased production of type I and type III interstitial collagens and FN1 by Western blots of culture media from the normal dermal fibroblasts following incubation with exosomes isolated from the serum of patients with Limited and Diffuse SSc. This mechanism of exosome-induced alteration of target cell phenotype could affect a variety of target cells important in the initiation, establishment, and progression of the fibrotic process such as endothelial cells, macrophages and other immune cells.
Collectively, these data support a model whereby exosomes released from affected profibrotic cells transmit information that establishes a profibrotic phenotype in normal or unaffected target fibroblasts, thus resulting in extension and propagation of the fibrotic process to distant normal sites. These results further indicate that besides being a potential source of diagnostic and prognostic biomarkers for SSc, serum exosomes may represent attractive targets for the development of treatment modalities designed to prevent the extension of the profibrotic SSc phenotype to unaffected areas of the body thus, halting the progression of the fibrotic component of this devastating disease.
We are aware that the small number of samples analysed represents a potential limitation of this study, particularly in the identification of specific miRNA capable of discriminating normal individuals from patients with SSc and we recognise that additional studies will be needed, including the validation of the profibrotic or antifibrotic role of the exosome miRNA employing SSc-derived cells and tissues, as well as, the inclusion of greater numbers of samples to increase the statistical power and eliminate the risk of potential false positives. However, the results demonstrate that there are substantial differences in the miRNA profiles of serum exosomes from normal individuals and from patients with limited and diffuse SSc. Furthermore, the results indicate that regulatory molecules contained in exosomes present in the sera of SSc patients can affect and modify the phenotype of target cells inducing them to acquire a profibrotic phenotype.
Acknowledgements
We thank the expert assistance of Nancy Peltier at Villanova University, Biology Department Electron Microscopy facility in the acquisition of TEM images. We also thank Ruth M. Johnson and Alana Pagano for their assistance in the preparation of the manuscript. Design of the study and manuscript preparation by PJW and SAJ. Data acquisition by PJW and SP-V. Analysis of data by PJW, SP-V and SAJ. Final manuscript revised and approved by PJW and SAJ.
Funding: Supported by NIH grant AR 19616 to S.A. Jimenez.
Footnotes
Competing interests: none declared.
References
- 1.GABRIELLI A, AVVEDIMENTO EV, KRIEG T: Scleroderma. N Engl J Med 2009; 360: 1989–2003. [DOI] [PubMed] [Google Scholar]
- 2.VARGA J, ABRAHAM D: Systemic sclerosis: a prototypic multisystem fibrotic disorder. J Clin Invest 2007; 117: 557–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.PATTANAIK D, BROWN M, POSTLETHWAITE BC, POSTLETHWAITE AE: Pathogenesis of systemic sclerosis. Front Immunol 2015; 6: 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.STERN EP, DENTON CP: The pathogenesis of systemic sclerosis. Rheum Dis Clin North Am 2015; 41: 367–82. [DOI] [PubMed] [Google Scholar]
- 5.BARSOTTI S, STAGNARO C, D’ASCANIO A, DELLA ROSSA A: One year in review 2016: systemic sclerosis. Clin Exp Rheumatol 2016; 34: (Suppl. 100): S3–13. [PubMed] [Google Scholar]
- 6.HO YY, LAGARES D, TAGER AM, KAPOOR M: Fibrosis–a lethal component of systemic sclerosis. Nat Rev Rheumatol 2014; 10: 390–402. [DOI] [PubMed] [Google Scholar]
- 7.MATUCCI-CERINIC M, KAHALEH B, WIGLEY FM: Review: evidence that systemic sclerosis is a vascular disease. Arthritis Rheum 2013; 65: 1953–62. [DOI] [PubMed] [Google Scholar]
- 8.VLASSOV AV, MAGDALENO S, SETTERQUIST R, CONRAD R: Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochem Biophys Acta 2012; 1820: 940–8. [DOI] [PubMed] [Google Scholar]
- 9.PANT S, HILTON H, BURCZYNSKI ME: The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol 2012; 83: 1484–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.COLOMBO M, RAPOSO G, THÉRY C: Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 2014; 30: 255–89. [DOI] [PubMed] [Google Scholar]
- 11.RAPOSO G, STOORVOGEL W : Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013; 200: 373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.SIMPSON RJ, LIM JW, MORITZ RL, MATHIVANAN S: Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics 2009; 6: 267–83. [DOI] [PubMed] [Google Scholar]
- 13.GUSACHENKO ON, ZENKOVA MA, VLASSOV VV: Nucleic acids in exosomes: disease markers and intercellular communication molecules. Biochemistry (Mosc) 2013; 78: 1–7. [DOI] [PubMed] [Google Scholar]
- 14.THÉRY C, OSTROWSKY M, SEGURA E: Membrane vesicles as conveyors of immune responses. Nat Rev Immunol 2009; 9: 581–93. [DOI] [PubMed] [Google Scholar]
- 15.YUANA Y, STURK A, NIEUWLAND R Extra-cellular vesicles as an emerging mechanism of cell-to-cell communication. Biochem Pharmacol 2012; 83: 1484–94.22230477 [Google Scholar]
- 16.CHRISTIANSON HC, SVENSSON KJ, BELTING M: Exosomes and microvesicles mediated phene transfer in mammalian cells. Semin Cancer Biol 2014; 28: 31–8. [DOI] [PubMed] [Google Scholar]
- 17.LO CICERO A, STAHL PD, RAPOSO G: Extra-cellular vesicles shuffling intercellular messages: for good or for bad. Curr Opin Cell Biol 2015; 35: 69–77. [DOI] [PubMed] [Google Scholar]
- 18.KRIEG T, ABRAHAM D, LAFYATIS R: Fibrosis in connective tissue disease: the role of the myofibroblast and fibroblast-epithelial cell interactions. Arthritis Res Ther 2007; 9 (Suppl. 2): S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ABRAHAM DJ, ECKES B, RAJKUMAR V, KRIEG T: New developments in fibroblast and myofibroblast biology: implications for fibrosis and scleroderma. Curr Rheumatol Rep 2007; 9: 136–43. [DOI] [PubMed] [Google Scholar]
- 20.HINZ B, GABBIANI G: Fibrosis: recent advances in myofibroblast biology and new therapeutic perspectives. F1000 Biol Rep 2010; 2: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.GILBANE AJ, DENTON CP, HOLMES AM: Scleroderma pathogenesis: a pivotal role for fibroblasts as effector cells. Arthritis Res Ther 2013; 15: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.HINZ B, PHAN SH, THANNICKAL VJ et al. : Recent developments in myofibroblast biology: paradigms for connective tissue remodeling. Am J Pathol 2012; 180: 1340–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.HINZ B, PHAN SH, THANNICKAL VJ et al. : The myofibroblast: one function, multiple origins. Am J Pathol 2007; 170: 1807–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.KISSIN E, KORN JH: Apoptosis and myofibroblasts in the pathogenesis of systemic sclerosis. Curr Rheumatol Rep 2002; 4: 129–35. [DOI] [PubMed] [Google Scholar]
- 25.BEON M, HARLEY RA, WESSELS A, SILVER RM, LUDWICKA-BRADLEY A: Myofibroblast induction and microvascular alteration in scleroderma lung fibrosis. Clin Exp Rheumatol 2004; 22: 733–42. [PubMed] [Google Scholar]
- 26.WATSKY MA, WEBER KT, SUN Y, POSTLETH-WAITE A: New insights into the mechanism of fibroblast to myofibroblast transformation and associated pathologies. Int Rev Cell Mol Biol 2010; 282: 165–92. [DOI] [PubMed] [Google Scholar]
- 27.BARTEL DP: MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–97. [DOI] [PubMed] [Google Scholar]
- 28.EULALIO A, HUNTZINGER E, IZAURRALDE E: Getting to the root of miRNA-mediated gene silencing. Cell 2008; 132: 9–14. [DOI] [PubMed] [Google Scholar]
- 29.OLIVE V, MINELLA AC, HE L: Outside the coding genome, mammalian microRNAs confer structural and functional complexity. Sci Signal 2015; 8: re2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.JIANG X, TSITSIOU E, HERRICK SE, LINDSAY MA: MicroRNAs and the regulation of fibrosis. FEBS J 2010; 277: 2015–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.VETTORI S, GAY S, DISTLER O: Role of MicroRNAs in Fibrosis. Open Rheumatol J 2012; 6: 130–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LI H, YANG R, FAN X et al. : MicroRNA array analysis of microRNAs related to systemic scleroderma. Rheumatol Int 2012; 32: 307–13. [DOI] [PubMed] [Google Scholar]
- 33.ZHU H, LUO H, ZUO X: MicroRNAs: their involvement in fibrosis pathogenesis and use as diagnostic biomarkers in scleroderma. Exp Mol Med 2013; 45: e41.doi: 10:10.1038/emm.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.BOWEN T, JENKINS RH, FRASER DJ: MicroRNAs, transforming growth factor beta-1, and tissue fibrosis. J Pathol 2013; 229: 274–85. [DOI] [PubMed] [Google Scholar]
- 35.PETROCCA F, VECCHIONE A, CROCE CM: Emerging role of miR-106b/miR-17–92 clusters in the control of transforming growth factor beta signaling. Cancer Res 2008; 68: 8191–4. [DOI] [PubMed] [Google Scholar]
- 36.INUI M, MARTELLO G, PICCOLO S: MicroRNA control of signal transduction. Nat Reve Mol Cell Biol 2010; 11: 252–63. [DOI] [PubMed] [Google Scholar]
- 37.ALTOROK N, ALMESHAL N, WANG Y, KAHALEH B: Epigenetics, the holy grail in the pathogenesis of systemic sclerosis. Rheumatology 2015; 54: 1759–70. [DOI] [PubMed] [Google Scholar]
- 38.LI Y, HUANG J, GUO M, ZUO X: MicroRNAs regulating signaling pathways: potential biomarkers in systemic sclerosis. Genomics Proteomics Bioinformatics 2015; 13: 234–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LUI PY, JIN DY, STEVENSON NJ: MicroRNA: master controllers of intracellular signaling pathways. Cell Mol Life Sci 2015; 72: 3531–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.TANAKA S, SUTO A, IKEDA K et al. : Alteration of circulating miRNAs in SSc: miR-30b regulates the expression of PDGF receptor β. Rheumatology (Oxford) 2013; 52: 1963–72. [DOI] [PubMed] [Google Scholar]
- 41.MAKINO K, JINNIN M, KAJIHARA I et al. : Circulating miR-142–3p levels in patients with systemic sclerosis. Clin Exp Dermatol 2012; 37: 34–9. [DOI] [PubMed] [Google Scholar]
- 42.WUTTGE DM, CARLSEN AL, TEKU G et al. : Specific autoantibody profiles and disease subgroups correlate with circulating micro-RNA in systemic sclerosis. Rheumatology (Oxford) 2015; 54: 2100–07. [DOI] [PubMed] [Google Scholar]
- 43.STEEN SO, IVERSEN LV, CARLSEN AL et al. : The circulating cell-free microRNA profile in systemic sclerosis is distinct from both healthy controls and systemic lupus erythematosus. J Rheumatol 2015; 42: 214–21. [DOI] [PubMed] [Google Scholar]
- 44.VAN DEN HOOGEN F, KANNA D, FRANSEN J et al. : 2013 classification criteria for systemic sclerosis: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Ann Rheum Dis 2013; 72: 1747–55. [DOI] [PubMed] [Google Scholar]
- 45.LEROY EC, BLACK C, FLEISHMAJER R et al. : Scleroderma (systemic sclerosis): classification, subsets and pathogenesis. J Rheumatol 1988; 15: 202–5. [PubMed] [Google Scholar]
- 46.REKKER K, SAARE M, ROOST AM et al. : Comparison of serum exosome isolation methods for microRNA profiling. Clin Biochem 2014; 47: 135–38. [DOI] [PubMed] [Google Scholar]
- 47.PETERSON MF, OTOC N, SETHI JK, GUPTA A, ANTES TJ: Integrated systems for exosome investigation. Methods 2015; 87: 31–45. [DOI] [PubMed] [Google Scholar]
- 48.NAKAMURA K, JINNIN M, HARADA M et al. : Altered expression of CD63 and exosomes in scleroderma dermal fibroblasts. J Dermatol Sci 2015; 84: 30–39. [DOI] [PubMed] [Google Scholar]