Abstract
Optogenetics provides promising tools for the precise control of receptor-mediated cell behaviors in a spatiotemporal manner. Most photoreceptors, yet, require extensive genetic manipulation and respond only to ultraviolet or visible light, which are suboptimal for in vivo applications because they do not penetrate thick tissues. Here we report a novel near-infrared light-activated DNA agonist (NIR-DA) nanodevice for nongenetic manipulation of cell signaling and phenotype in deep tissues. This nanodevice is prepared by conjugating a pre-inactivated DNA agonist onto the gold nanorods (AuNRs). Upon NIR light treatment, the DNA agonist is released through the localized surface plasmon resonance (LSPR)-based photothermal effect of AuNRs and becomes active. The active DNA agonist dimerizes the DNA-modified chimeric or native receptor tyrosine kinase (RTK) on cell surfaces and activates downstream signal transduction in live cells. Such NIR-DA activation of RTK signaling enables the control of cytoskeletal remodeling, cell polarization, and directional migration. Furthermore, we demonstrate that the NIR-DA system can be used in vivo to mediate RTK signaling and skeletal muscle satellite cell migration and myogenesis, which are critical cellular behaviors in the process of skeletal muscle regeneration. Thus, the NIR-DA system offers a powerful and versatile platform for exogenous modulation of deep tissues for purposes such as regenerative medicine.
Keywords: Au nanorods (AuNRs), aptamer, DNA nanodevice, dimerization, receptor tyrosine kinase
A cell’s signaling network is wired in ways that coordinately links organellar functions to regulate a wide variety of essential physiological processes including development, homeostasis, and tissue repair.1 In response to extracellular changes, ligand-mediated activation of the cell surface receptor subsequently transduces and amplifies the signals intracellularly by acting on specific effector molecules for specific cell behaviors, such as secretion, movement, growth, division, or death.2 To precisely modulate a cell behavior through a specific signaling pathway, photoactivation is highly desired because it is a noninvasive approach that can be precisely deployed with a high degree spatiotemporal control.3–6 Many optogenetic tools have been developed to achieve precise spatiotemporal control over cellular signals and to impact diverse cellular phenotypes, of which many involve the introduction of optogenetically modified receptors into cells.7–13 However, the process of genetic manipulation is time-consuming, is unpredictable and inefficient, which may introduce artifacts because of overexpression and tagging related artifacts.14 Thus, the need for methods that enable exogenous precision control of cell signaling and behaviors with light (non-invasive) without genetic manipulation remains unmet.
Deoxyribonucleic acid (DNA) nanotechnology has been recently applied to nongenetically modulated cell functions via engineering DNA-tailored surface receptor due to several advantageous properties of DNA, i.e., its aptamer-based targeting, predictable hybridization, and programmable assembly.15–17 We and others have developed unique DNA nanodevices, that noncovalently bind the extracellular domains of receptor tyrosine kinases (RTKs) to generate a chimeric DNA-receptor.18 Using the functional DNAs (aptamers or DNAzymes) as a sensory module, the chimeric DNA-receptor enables cells to respond to different external stimuli including various small-molecular cues,18 proteins,19 or oligonucleotides.20 Applying the recently developed valuable optochemical tools,21, 22 the DNA nanodevices could be reprogrammed to achieve the light-controlled receptor-mediated cell signaling and cellular responses; this is an avenue that has remained largely unexploited. A recent study reported the optical control over cell responses via the light-triggered DNA assembly using a photocleavable caging chemical linker.23 This photocleavage-based uncaging process requires visible or ultraviolet light, which has the shallow depth of tissue penetration and has phototoxic effects on living systems, both limiting the in vivo and future clinical application.24
Compared with commonly used visible or ultraviolet light, near-infrared (NIR) light (700 to 1100 nm) enables deeper tissue penetration than visible or ultraviolet light, noninvasive manipulation, accurate remote control, and minimal photodamage for the living system.25–27 For the NIR light-based activation, plasmonic nanomaterial is an ideal mediator owing to the photothermal response to NIR light.28 For example, Au nanorods (AuNRs), with the adjustable size, facile surface modification, and unique localized surface plasmon resonance (LSPR) properties, have been used for numerous biomedical applications, including the controlled release of biomolecules,29, 30 hyperthermia therapy,31 and biological sensing.32 Therefore, AuNRs provide promising and versatile materials as the NIR light-responsible module to fabricate the light-activatable DNA nanodevice for deep tissue applications.
Here, we propose a novel NIR light-activatable DNA agonist system (NIR-DA) based on the plasmonic AuNRs for the optical control of the activation of cellular signaling and cell behaviors in vitro and in vivo. We use NIR-DA to target RTKs as example because this family of proteins, comprised of more than fifty cell-surface receptors with intrinsic tyrosine kinase activity, is known to regulate various cellular functions, whose functions are frequently deregulated in diseased states.2, 33 Upon activation, the natural agonist binds and promotes homo or hetero-dimerization of RTKs, leading to the tyrosine autophosphorylation and activation of downstream pathways for specific cell responses.33 According to this activation mode, we first designed a DNA agonist capable of dimerization with DNA-modified chimeric or natural RTK at cell surfaces. We functionalize the DNA agonist on the AuNRs as a latent agonist complex, which enables the NIR light-triggered release and activation of DNA agonist via the LSPR-based photothermal effect. Upon NIR light irradiation, functionalized AuNRs release and activate the DNA agonist, which triggers binding-induced dimerization of RTK and subsequent autophosphorylation, activation of downstream signal transduction and enhancement of cell migration and proliferation. We further demonstrate that the NIR-DA system is capable of initiating NIR light-activated RTK signaling, which we used to modulate the behavior of myogenic skeletal muscle cells in mice during muscle regeneration after an acute injury.
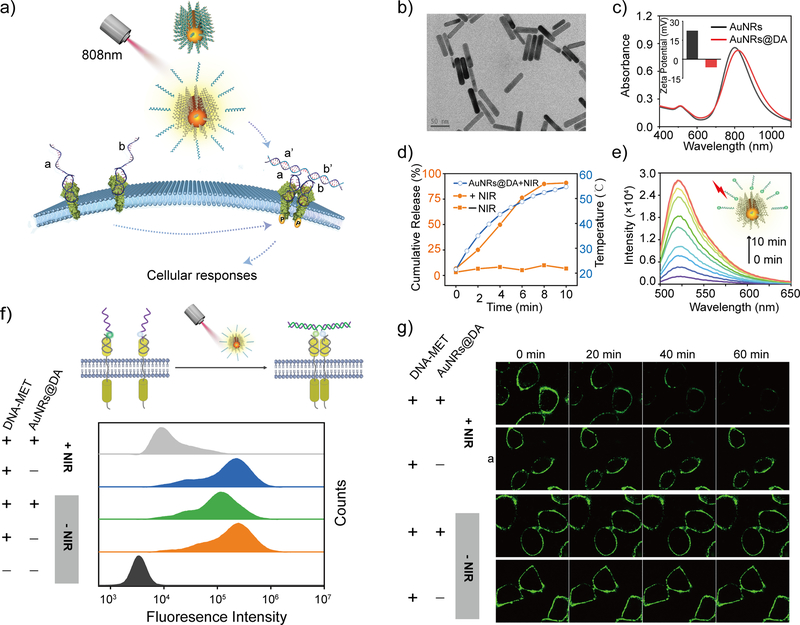
As a proof-of-principle experiment, we first developed a NIR-DA system to control signaling by the tyrosine-protein kinase MET or hepatocyte growth factor receptor (HGFR). MET is the RTK for a native agonist hepatic growth factor (HGF), and HGF-induced MET activation is a critical signaling event for embryonic development, angiogenesis, and regeneration.34 The optical modulation of MET signaling at deep-tissue-penetrable levels is highly desirable for cell-based therapy. In order to reprogram the target specificity of MET from the native agonist HGF to a DNA agonist, we designed a DNA-protein chimeric receptor system by nongenetically engineered MET with a pair of DNA-based receptors. Each DNA-based receptor contains the same MET-anchored module with a MET-targeting aptamer (Figures S1 and S2) and a different sensory module (a and b), which can respectively recognize two segments (a’ and b’) of the DNA agonist (Table S1), thus allowing binding-induced dimerization (Figure S3). To make the DNA agonists responsive to NIR light, we subsequently prepared the AuNRs according to a seed-mediated protocol and inactivated the DNA agonist via hybridization through the partially complementary blocking of DNA strands (b-DA), which were conjugated to the AuNRs’ surfaces using thiol-gold chemistry (Table S1).35 The DNA-harbored AuNRs were further modified with thiolated PEG-5000 to improve the biocompatibility and stability.32 The DNA agonist functionalized AuNRs (AuNRs@DA) had an average length of 61.3 ± 0.3 nm and width of 14.3 ± 0.08 nm, and the aspect ratio was about 4.3 (Figures 1b and S4). A UV-vis spectroscopy analysis showed a longitudinal plasmon resonance peak ~810 nm (Figure 1c). Furthermore, the Zeta potential switched from +22.4 mV to −6.37 mV due to the increased negative charges associated with the phosphate backbone of the DNA (Figure 1c insert) and the hydrodynamic size of length increased by ~20 nm as determined by the dynamic light scattering (DLS) analysis (Figure S5), indicating the successful immobilization of the hybridized DNA onto the AuNRs. The amount of DNA agonist on AuNRs@DA was measured and normalized by the amount of AuNRs (Figure S6)35, 36 and it was estimated that each AuNR was loaded with 199 ± 23 molecules of DNA agonist.
Figure 1. Rationale design and characterization of a NIR-DA system.
(a) Scheme illustration of NIR light-induced RTK dimerization using the NIR-DA system. (b) TEM image of AuNRs (Scale bar, 50 nm), (c) Absorption spectra analysis and the insert is Zeta potential of AuNR@DA. (d) A time-dependent increase of temperature (blue cycle) and the releasing ratio of DNA agonist from AuNR@DA with (orange cycle) or without (orange square) NIR light treatment (laser irradiation: 808 nm, 1.00 W/cm2). (e) Time-dependent release of FAM-labeled DNA agonist from AuNRs@DA induced by NIR light. (f) Upper: scheme of fluorophore / quencher labeled DNA-MET chimeric receptors for demonstrating NIR light-induced dimerization on the cell surface; lower: Fluorescence decay on the cells with DNA-MET chimeric receptors (200 nM) in the presence of AuNRs@DA (0.5 nM) with or without NIR light treatment (808 nm laser irradiation for 4 min) were monitored by flow cytometry. (g) Time-lapse confocal fluorescence microscope images of decayed fluorescence on the cell surface by NIR light treatment.
Next, we assessed the NIR light responsiveness of AuNRs@DA by monitoring the temperature of the solution with and without NIR laser irradiation (808 nm laser, 1 W/cm2). A rapid increase in temperature of the AuNRs solution was observed upon a NIR light (Figures 1d and S7), indicating that the AuNRs transformed the energy of the NIR light into local heat. No increase of temperature was observed during the continuous irradiation of the solution without AuNRs over a period of 20 minutes due to the minimal absorption by water molecules (Figure S7), indicating that the increase we observe is due to AuNRs. NIR laser irradiation in the presence of AuNRs increased the temperature to 43.6 °C in less than 4 minutes (Figure 1d), which is higher than the melting temperature (42 °C) of the hybridized DNA agonist and the blocking strand, and could induce the de-hybridization and release of the DNA agonist. Consequently, we observed that NIR laser irradiation triggered a time-dependent release of the DNA agonist from AuNRs@DA (Figure 1e). Approximately 50% of the DNA agonist was released within 4 minutes of NIR laser irradiation (Figure 1d), and the concentration of released DNA agonist was calculated to ~50 nM from 0.5 nM of AuNRs@DA. Moreover, NIR light-triggered DNA agonist released from AuNRs@DA hybridized with both DNA strands for DNA-MET chimeric receptors and assembled a DNA complex as determined by a non-denaturing (native) gel electrophoresis (Figure S8). Based on these in vitro characterizations, we used 0.5 nM AuNRs@DA and a 4-min period NIR laser irradiation as a standard procedure in subsequent experiments for NIR light activation.
Next, we tested the function of the NIR-DA system on the dynamics of DNA-MET chimeric receptors at the cell surface. To this end, we chose to study the A549 cells because it is known to overexpress MET. We used one DNA-MET chimeric receptor tagged with a fluorophore (FAM) and the other one with a quencher (BHQ1) (Table S2). Upon NIR light-activation, the two DNA-based receptors dimerize to close proximity, and quench the fluorescence signals (Figures 1f and S9). Time-lapse experiments (Figure 1g) showed that upon NIR light-activation, the fluorescence signal at the cell surface was rapidly quenched, ~40% in 20 minutes and more than 90% in 60 minutes, indicating the NIR light triggered the dimerization of both chimeric DNA-MET receptors at the cell surface. Fluorescence remained unchanged in the presence of the AuNRs@DA when cells were not exposed to NIR light. Findings were further validated using flow cytometry (Figure 1f). Furthermore, using two different MET-positive cell lines, A549 cell and DU145, we confirmed that the AuNRs@DAs, NIR light treatment and the pre-modifications with DNA-based receptors did not affect the cell viability (Figure S10), indicating that the NIR-DA system is biocompatible. These findings suggested that the newly developed NIR-DA system may serve as a precise and non-toxic approach for controlling MET receptor activation on the cell surface.
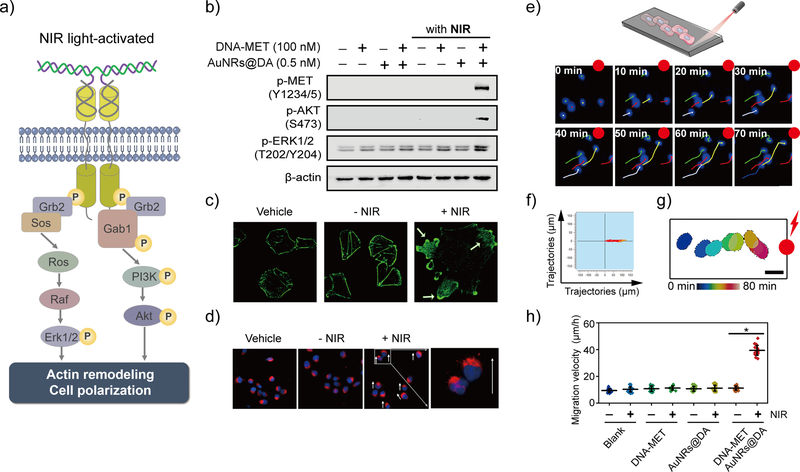
Next, we investigated how the NIR-DA system induced MET activation impacts intracellular signaling pathways (Figure 2a). The first hallmark of MET receptor dimerization and activation is cross-phosphorylation of the cytoplasmic tails of two dimerized receptors on tyrosines (Y1234/5) within the activation loop. We analyzed these phosphorylation events by western blotting. The NIR light significantly induced these MET tyrosine phosphorylation in the cells exclusively in the presence of both the AuNRs@DA and the pre-modified DNA-MET chimeric receptors but not in control cells (Figures 2b and S11), indicating that the NIR-DA system was necessary for NIR light-induced MET activation. We also found that MET autophosphorylation in the NIR-treated cells was associated also significant enhancement of the two major signaling pathways immediately downstream of MET (Figure 2a), i.e., Ras/mitogen-activated protein kinase (Ras/MAPK) and phosphoinositide 3-kinase/protein kinase B (PI3K/AKT).34 NIR light-responsive activation of ERK1/2 (as determined by phosphorylation of T202/Y204 residues) and Akt (as determined by the degree of its phosphorylation at S473 residue) were significantly increased in the NIR-DA-treated cells compared with that in the control cells (Figure 2b). These results confirm that the NIR-DA system could trigger MET activation and major downstream signaling events in live cells.
Figure 2. NIR light-activated MET signaling and cellular responses.
(a) Scheme illustration of NIR light-induced MET dimerization and downstream signaling. (b) Activation of MET signaling triggered by NIR light on A549 cells was investigated by western blot analysis. (c) NIR light-induced actin-cytoskeletal remodeling. The DNA-MET chimeric receptors (100 nM) modified A549 cells were incubated with AuNRs@DA (0.5 nM) and were treated without or with 4-min NIR light-irradiation, then were incubated at 37 °C for 1 hour followed by fixation and staining with FITC-phalloidin. (d) NIR light-directed cell polarization. DU145 cells were pre-modified with DNA-MET chimeric receptors and then incubated with AuNRs@DA. The opposite terminal of the µ-slide was irradiated with laser 808 nm for 4 min and was incubated at 37 °C for 30 min. The cells were fixed and stained using the fluorescent probe for Golgi (red) and nuclei (blue); the white arrow indicates the moving direction. (e) Upper: scheme illustration of NIR light-activated directional migration using ibidi μ-slide, lower: NIR light-induced directed migration of cells population. The cell movement was tracked under fluorescence microscopy, the migration trajectories of the cell were segmented from fluorescence images every 10 min. The scale bar: 10 μm. (f) Directional migration trajectories of cells. The DU145 cells were treated with DNA-MET chimeric receptors and AuNRs@DA, and the opposite terminal of the µ-slide was irradiated with NIR laser 808 nm for 4 min. The migration trajectories of cells (n=10) were illustrated. (g) Time-dependent tracking of the single cell toward the NIR light. The cell movement was tracked under fluorescence microscopy, the outer border of the cell was segmented from fluorescence images every 10 min, and the contours of the cell are illustrated for increasing time points (from blue to red) over 70 min. Scale bar: 20 μm. (h). Moving velocities of cells induced by NIR light (n=20). The average velocity of directional migration of DU145 cells during the duration (2.5 h) was quantified and analyzed. Data are represented as means ± SD, *P<0.05.
Next, we investigated how NIR-DA-activated MET signaling impacts cellular behavior. To this end, we analyzed the most prominent cellular responses triggered by MET signaling, i.e., the actin-cytoskeletal remodeling, cell migration, and cell proliferation. When we visualized the actin-cytoskeleton with FITC-phalloidin (specially stained filament actin, F-actin) and analyzed them by confocal microscopy, we found that NIR light triggered the formation of the lamellipodia sheets and actin-cytoskeletal remodeling exclusively in the NIR-DA-treated cells (Figure 2c), whereas the actin stress fibers remained unaltered in control cells (Figure S12). Such morphological changes were associated with increased cell motility, as determined by cell scattering assays which showed that random migratory distances were accentuated in cells treated with the NIR light (Figure S13). In addition, NIR light significantly promoted the cell proliferation of NIR-DA-treated cells by 1.45-fold compared to the control cells (Figure S14).
Next, we analyzed the spatiotemporal precision of the NIR-DA system. More specifically, we asked if the direction of cell migration could be controlled optically using the NIR-DA system. We found that the Golgi apparatus of the NIR-DA-treated cells were reorganized between the nucleus and plasma membrane toward the NIR laser-irradiated spot within 30 minutes (Figures 2d). The quantification analysis indicated that the significant polarization of the cells (ca. 58%) were induced to the direction toward NIR light, but random polarity in control cells without NIR light or NIR-DA (Figure S15). We next evaluated NIR light-guided directional cell migration, as determined by a time-lapse cell-tracking assay using a μ-slide device (Figure 2e). We found that most of the cells equipped with the DNA-MET chimeric receptors persistently and rapidly moved toward the NIR light irradiation spot in a time-dependent manner (Figure 2f), as opposed to a random, slow movement pattern seen in most control cells (Figure S16). Moreover, a randomly selected single cell could be guided by NIR light to move toward the irradiated spot (Figure 2g). The average velocity of NIR-DA-treated cells was ~39.4 ± 3.7 μm/h, which was significantly faster than the control cells (11.2 ± 2.2 μm/h) without irradiation (Figure 2h). Taken together, these findings demonstrate that the NIR-DA system not just triggered MET signaling, but also allowed precisely controlled NIR light-activated directional migration via such pathway activation.
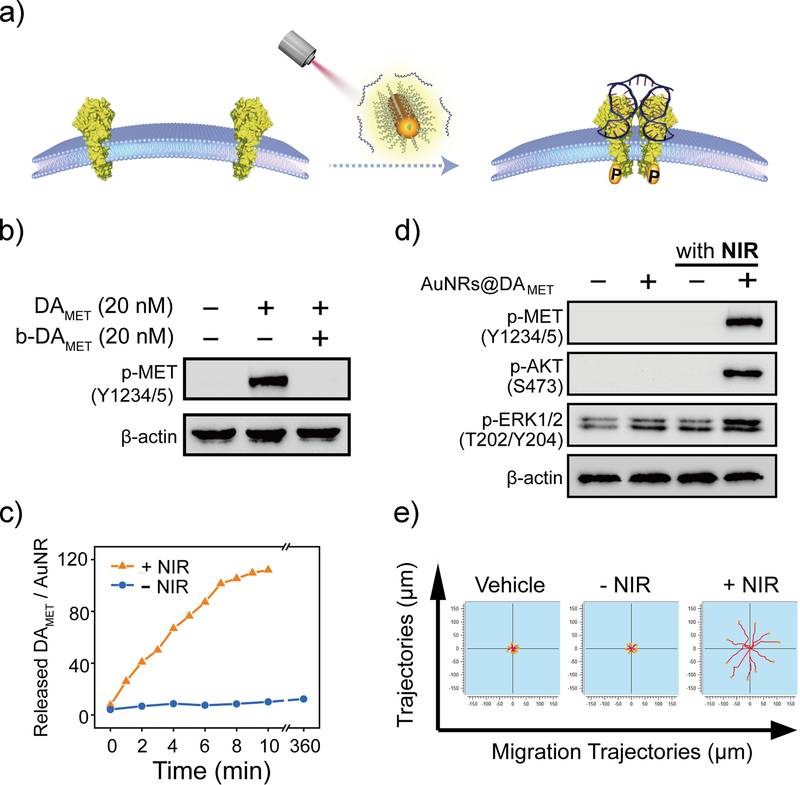
Next, we asked if the NIR-DA-controlled cell signaling and migration that we developed and validated in vitro using cultured cells could be exploited for modulating cellular behavior in deep tissues in an in vivo setting. We recognize that although the chimeric receptors allow for the specificity, selectivity and simplified design of DNA agonist in cultured cells, the requirement of selective and efficient pre-modification of the receptor on the target cells within multi-cellular tissues is a major challenge in the way of adopting the NIR-DA system to in vivo applications in living whole animals. To address this issue, we sought to further develop a NIR-DA system for directly regulating native MET (Figure 3a). The DNA agonist for native MET (DAMET, Table S3) contains two identical MET-specific aptamers that selectively bind to the extracellular domain of MET (Figure S17). Unlike the short length (28 nt) and the simple secondary structure of the DNA agonist for the DNA-MET chimeric receptor, DAMET has a long oligonucleotide chain (100 nt) with four predicted G-quadruplex structures (Figure S18), which can make it difficult to design the latent agonist complex for NIR light activation. Given that two guanine sites (G14, G32) of the MET aptamer are crucial for the formation of the G-quadruplex and for binding MET, 37 we deactivated DAMET using the blocking DNA strands hybridizing to the flanking regions of four key guanine sites (G14, G32, G64, and G82) for MET recognition (Table S2). DAMET-induced MET phosphorylation was completely diminished when all guanine sites were blocked simultaneously, but only partially inhibited when a single guanine site was blocked (Figure S19). All guanine-site-blocking DNA strands were conjugated as a long blocking strand (bDAMET), which completely abolished the DAMET-induced MET phosphorylation (Figure 3b). Therefore, we successfully optimized a blocking DNA module to prepare the latent DNA agonist for natural MET.
Figure 3. NIR-DA system for native receptor-mediated cellular signaling and behaviors.
(a) Scheme illustration of NIR-DA system for native MET. (b) The effect of DAMET-inactivating bDAMET on the MET phosphorylation. Western blotting analysis of the phosphorylation level of MET in A549 cell lysates. The A549 cells were cultured in the presence or absence of DAMET (20 nM) and bDAMET (20 nM) for 10 min. (c) Dynamic profile of NIR light-triggered release of DAMET from AuNRs (laser irradiation: 808 nm, 1.00 W/cm2). (d) NIR light-activated MET signaling using AuNRs@DAMET. Western blotting analysis of the phosphorylation level of MET (Y1234/5), AKT (S473), and ERK1/2 (T202/Y204) in A549 cell lysates. The A549 cells were cultured in the presence or absence of AuNRs@DAMET (0.5 nM each), followed by the NIR laser irradiation. (e). The migration trajectories of NIR light directed cell migration. The DU145 cells were treated with or without AuNRs@DAMET and NIR light. The time-lapse images were taken in the Cytation 5 imaging microplate reader for 6 h. The migration trajectories of cells were illustrated using ImageJ software (n = 10).
Next, we assembled the NIR-DA system for native MET using the same approach outlined earlier (Figure 3a), which involved conjugation of the DAMET/bDAMET DNA duplex on AuNRs (AuNRs@DAMET) by a thiol-Au bond. Successful conjugation of AuNRs was confirmed based on the observed change in the surface charge from positive (+26.4 mV) to negative (−8.17 mV) (Figure S20) and the increase in the hydrodynamic size by 47 nm (Figure S21). Conjugation of DAMET on AuNRs did not affect the longitudinal plasmon resonance, which remained ~810 nm (Figure S22). As seen previously, NIR light irradiation induced temperature increase, leading to subsequent de-hybridization and release of the DAMET (Figure S23). The number of DAMET per AuNR was ~124 ± 22 (Figure S24), and upon NIR light treatment the released DAMET from 0.5 nM AuNRs gradually accumulated to a concentration of ~30 nM (Figure 3c). In the presence of AuNRs@DAMET, the NIR light strongly enhanced phosphorylation of native MET and the downstream effectors, including AKT and ERK1/2 in A549 cells (Figure 3d). Finally, we confirmed that the NIR-DA system facilitated the NIR light-enhanced migration of DU145 cells (Figure 3e) and that cells migrated persistently toward the NIR light irradiated spot (Figure S25); the moving distance was increased 3-fold over control cells (Figure S26). The tissue penetration of NIR-light was investigated using the cell scattering assay, and the results indicated that NIR-light efficiently penetrated maximum 6-mm tissue to enhance the cell motility, however, the NIR light-promoted motility was totally eliminated when the tissue was thicker than 8-mm (Figure S27). Therefore, we extended the usage of the NIR-DA system to the manipulation of the native receptor-mediated signal transduction for directional migration, providing a feasible and potent strategy for the exogenous modulation of endogenous receptors in vivo.
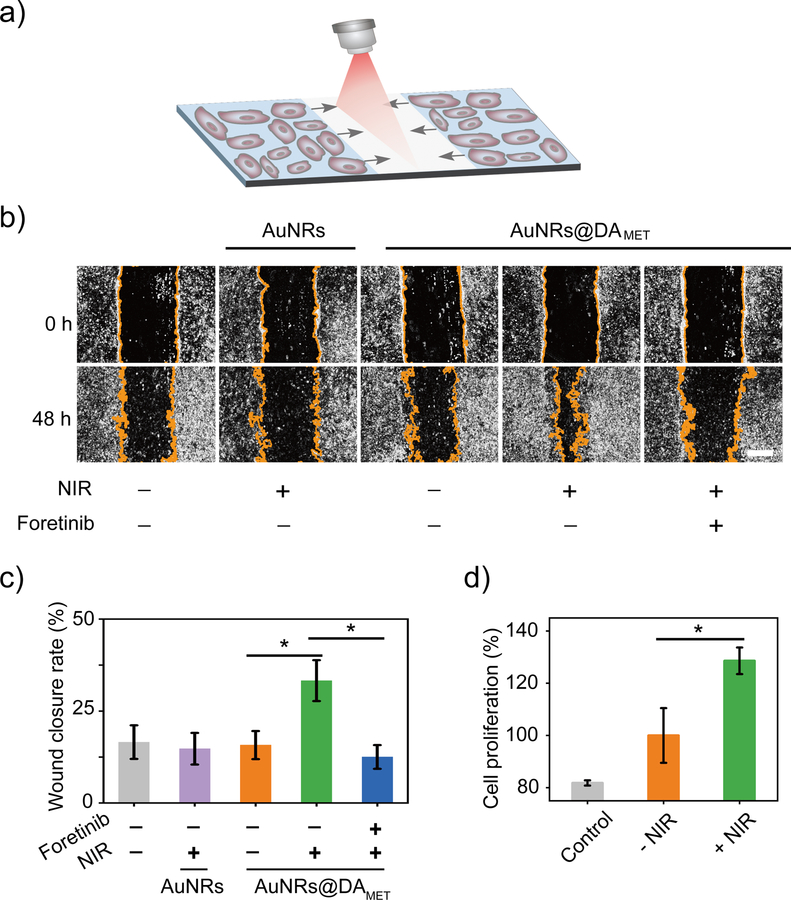
Next, we assessed the potential applications of the NIR-DA system in the optical control of MET signaling and cellular behaviors in animals. We chose to assess this in the setting of skeletal muscle regeneration in mice38–40, which requires penetrating capacities of NIR light-activation to act on muscle tissues that are under the thick skin. During injury-initiated skeletal muscle regeneration, the muscle precursor myogenic cells migrate to and differentiate at the site of injury to regenerate into functional myocytes within skeletal muscles.39, 41 Cell behaviors including proliferation and migration are essential for the wound healing process. We first tested the feasibility of the NIR-DA system in wound healing in vitro using the C2C12 cells, a skeletal muscle myogenic cell line overexpressing MET at the cell surface (Figure S28).42 The viability of C2C12 cells was not affected by the treatment of AuNRs@DAMET, and the phototoxicity of NIR light was negligible (Figure S29). In addition, the AuNRs@DAMET was stable for long time incubation in the presence of serum (Figure S30). These results demonstrated the biocompatibility and the excellent stability of the NIR-DA system for cellular and in vivo applications. Using the scratch wound assay to mimic wound healing in vitro, we investigated the effect of NIR-DA activation on wound-closure events after making an artificial wound in the monolayer of C2C12 cells (Figure 4a). In the presence of AuNRs@DAMET, NIR light significantly increased the wound closure rates (33%) of C2C12 cells (Figures 4b and 4c). The NIR light-promoted wound closure rates significantly dropped to 13% in the presence of Foretinib, a potent small molecule inhibitor for the kinase activity of MET, which indicated that MET mediated the NIR light-enhanced wound healing process (Figures 4b and 4c). Moreover, in the presence of AuNRs@DAMET, NIR light promoted cell proliferation by 130% compared to the cells without NIR light treatment (Figure 4d). These data demonstrated that the NIR-DA system could modulate the cell migration and proliferation of skeletal muscle myogenic cells, thus indicating the potential applications for the control of cellular behaviors in the muscle regeneration in vivo.
Figure 4. NIR-DA-activated cell responses of skeletal muscle myogenic cells.
(a) Scheme illustration of NIR light-activated wound healing in a scratch wound assay. (b) NIR light-enhanced wound healing. The C2C12 cells were treated with or without MET inhibitor (Foretinib, 0.5 μM). In the presence or the absence of AuNRs@DAMET or AuNRs, cells were irradiated with NIR light and the wound-closure events captured by a light microscope. The images were taken at 0 and 48 h. The yellow lines indicate boundaries between cells in the monolayer and the scratched areas uncovered by cells. Scale bar: 500 μm. (c) Quantification of relative wound closure rate. Data are presented as means ± SD (n = 5) (*P<0.05). (d) NIR light-promoted cell proliferation. The C2C12 cells were cultured in the presence of the AuNRs@DAMET (0.5 nM) and treated with NIR light for 4 minutes, then were continuously cultured for 2 days. The cell proliferation in each well (n=3) was evaluated by using Cell Counting Kit-8. Data are presented as means ± SD (*P<0.05).
To test the efficacy of NIR-DA-activation in vivo, we conducted animal experiments to investigate whether NIR-DA system could be applicable in the thick tissue to mobilize the skeletal muscle satellite cells at the injured region, promoting proliferation and myogenesis for the in-situ regeneration of muscle tissue (Figure 5a). Briefly, we used an animal model with an acute muscle injury by liquid nitrogen (Figure 5b),43 which caused the fragmentation of myofibers and massive cell death at 3 days post-injury (Figure 5d). After the injury, AuNRs@DAMET (1 nM) were injected using a long needle (inserted ~0.5 cm under the skin) to reach the injured muscle region. Because of the excellent tissue penetration, the NIR 808 nm laser caused an increase in the local temperature at the exposure region via the LSPR-mediated photothermal effect of AuNRs@DAMET (Figure 5c). We confirmed that NIR light-activated DAMET indeed triggered MET signaling in vivo, as determined by immunofluorescence staining of the muscle for phosphorylated-MET (Y1234/Y1235); NIR light dramatically increased the fluorescent signal in the exposed areas but only neglectable signal in the samples without irradiation (Figures 5d and S31). The fluorescence levels indicated by phosphorylated-MET were quantified to be 300% of that without NIR light treatment (Figure 5e), suggesting the MET signaling was significantly activated by NIR light. Three days after the injury with NIR light treatments, increased nuclei at the injury sites were observed by hematoxylin and eosin (H&E) stain, indicating NIR light-induced cell aggregation and/or proliferation at the injury site (Figures 5d).
Figure 5. NIR-DA-modulated cellular behaviors of muscle satellite cells in mice.
(a) Scheme illustration of NIR light-modulated cellular behaviors of muscle satellite cells in vivo using NIR-DA system. (b) The procedure for acute muscle injury animal model. (c) The photothermal effect of NIR light in living mice. Infrared thermal imaging of the mouse with or without the injection of AuNRs@DAMET (1.0 nM) were irradiated under 808 nm laser (1.00 W/cm2) for different periods. (d) The injured mice were injected with AuNRs@DAMET (1.0 nM) and irradiated with or without NIR light. Left: immunofluorescent analysis of p-MET in the sections from injured mice at 0.5 h post the injury, scale bar indicates 50 μm; Middle: immunofluorescence analysis of Pax7, Ki67 and MHC protein in the sections from the injured mice at 3 days post the injury; Right: histology by H&E staining of the sections from the injured mice at 3 days post the injury, scale bar indicates 500 μm. Quantification of average fluorescence intensity per cell of p-MET (e), Pax7 (f), Ki67(g), and MHC (h) at randomly selected high-power field (HPF) in the section from injured mice. Data are presented as mean ± SD (n = 5), and the statistical significance was determined using Student’s non-paired t-test, *p<0.05.
Next we asked if the NIR light-activated accumulation of muscle satellite cells at the site of injury was indeed followed by proliferation and subsequent differentiation into skeletal muscle cells. To this end, we tracked the levels of expression of Pax7, a transcription factor for myogenesis.44 It has been reported that the Pax7 positive muscle satellite cells expressed MET and respond to natural HGF for promoted proliferation and differentiation.45 During skeletal muscle regeneration, the Pax7 positive muscle satellite cells migrate to the injured area and give rise to the majority of mature skeletal muscle.38,46,47 The effect of NIR-DA activation on the cell migration, cell proliferation, and differentiation of Pax7 positive muscle satellite cells in vivo was evaluated using immunofluorescence. Three days post-injury, the Pax7 positive cells significantly increased in the injured region upon NIR-DA activation (Figures 5d, 5f and S32), suggesting that the muscle satellite cells were mobilized by NIR light. By contrast, the Pax7-positive cells were not observed at the injured region without NIR light treatment, although they were in the presence of AuNRs@DAMET. Next, the proliferative cells were labeled using a Ki67 antibody, and a significant rise in the percentage of Ki67+/Pax7+ cells was observed, indicating the NIR light promoted the proliferation of the muscle satellite cells (Figures 5d, 5g and S32). Interestingly, a population of Pax7-positive cells showed a significantly increased myosin heavy chain (MHC) expression level (Figures 5d, 5h and S33), suggesting that the migrated muscle satellite cells had undergone differentiation, an early hallmark for muscle regeneration.46 At 7 days post-injury, NIR light significantly increased cross-striated myofibers, while a large, unrepaired area at the center of the injury remained in the control mice without NIR light treatment (Figure S34). Notably, the subpopulation of Pax7+/Ki67+ satellite cells was maintained at high level in the injured regions of the NIR light-treated mice (Figure S35), implying that these cells with regenerative potential were actively participating in the process of skeletal muscle regeneration. Moreover, MHC-positive myofibers dramatically increased in the region with NIR-DA activation, while it was barely observed without NIR light treatment (Figure S36), suggesting that NIR light had activated a progressive regrowth and myogenesis program at the site of injury. These results validate the NIR-DA system in vivo as an approach to promote the muscle satellite cell to migrate and differentiate, generating new myofibers in skeletal muscles.
In summary, we have presented a novel plasmonic AuNRs-based NIR-DA system for optical activation of RTK signaling without the need for genetic manipulation to express sensitive membrane proteins, offering an easy and practical method for remote-controlled cellular behaviors. Because RTK signaling governs a very wide category of cellular functions, including motility, proliferation, and differentiation2, selective NIR-DA systems against diverse RTKs may provide a powerful tool to improve the efficiency of controlled cellular events for use in both in vitro and in vivo applications. Our strategy takes advantage of the plasmonic property of AuNRs to convert NIR light to the local heat, which activate DNA agonist for RTK signaling. With the excellent biocompatibility and tissue penetration of the NIR light, it is possible to noninvasively manipulate the cellular processes in deep tissue with the high temporal and spatial precisions, while minimally impacting the physiologic properties of normal cells. Compared with the optogenetic tools for RTK activation8,12, the NIR-DA system has the advantage in natural receptor-mediated activation of the cells under deep tissue without the need of implanted optogenetic engineered cells and optical fiber for visible or ultraviolet light illumination in vivo. This NIR-DA system can be extended to numerous desired cellular behaviors by designing a unique receptor-activating DNA agonist, for instance, the VEGFR-mediated angiogenesis. By appropriate selection of the different orthogonal DNA agonist/receptor pairs, the NIR-DA system could be customized to achieve the selective and combinatorial modulation on different target cells with various unique surface receptors. Moreover, the NIR light-triggered differential release profile and activation of DNA agonists can be customized by varying the melting temperature of the hybridized blocking DNA strands and DNA agonist or by tuning the NIR wavelength, power and irradiation periods.36 This flexibility enables selective and repetitive activation of cellular behaviors without harming the cells. Moreover, the NIR light-responsive module can be replaced by other novel nanomaterials with distinct performances in the NIR absorption range and photothermal transforming efficacy, such as MoS2,48 MXene,49, 50 or black phosphorus.51 Along with the exciting promises of NIR-DA system, the significant challenge come with the technique difficulties including how to quantitative control the release and to evaluate the biological activity of DNA agonist for long-term in vivo applications, both need to be resolved through future research. We expect that this NIR-DA system will open a new avenue for the remote regulation of cell behavior in tissue regeneration and cell-based therapy.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the National Natural Science Foundation of China (21725503, 21575038, 81730064, 81571985 and 31571368), National Science and Technology Major Project (2017ZX10202201), the Young Top-notch Talent for Ten Thousand Talent Program, and the Keypoint Research and Invention Program of Hunan Province (2017DK2011) and NIH grant (CA238042).
ABBREVIATIONS
- MET
mesenchymal-epithelial transition factor
- Akt(PKB)
protein kinase B
- ERK
extracellular regulated protein kinases
- SOS
son of sevenless
- HGF
hepatocyte growth factor
- PI3K
phosphoinositide-3 kinase
- MAPK
mitogen-activated protein kinase
- Grb2
growth factor receptor-bound protein 2
- GAB1
GRB2-associated binding protein 1
- Raf
RAF proto-oncogene serine/threonine-protein kinase
- MAPK
Mitogen-activated protein kinase
- Pax7
Paired box 7
- Ki67
Antigen KI-67
- MHC
Myosin heavy chain
- RTKs
Receptor tyrosine kinases
- PEG
polyethylene glycol
- VEGFR
vascular endothelial growth factor receptor
- DLS
dynamic light scattering
- TEM
transmission electron microscope
- LSPR
localized surface plasmon resonance
- NIR
near-infrared
- AuNRs
Au nanorods
- DNA
deoxyribonucleic acid
- FAM
6-carboxy-fluorescein
- BHQ1
Black hole quencher 1
- FITC
fluorescein isothiocyanate
Footnotes
ASSOCIATED CONTENT
Supporting Information
Please see Supporting Information (SI) for more experimental details and supplemental tables (Table S1–S3) or figures (Figure S1–S36).
The following files are available free of charge.
Supporting Information.PDF
The authors declare no competing financial interest.
REFERENCES
- (1).Arber C; Young M; Barth P Reprogramming Cellular Functions with Engineered Membrane Proteins. Curr. Opin. Biotechnol 2017, 47, 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Zinkle A; Mohammadi M A Threshold Model for Receptor Tyrosine Kinase Signaling Specificity and Cell Fate Determination. F1000 Research 2018, 7, 872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Antognazza MR; Martino N; Ghezzi D; Feyen P; Colombo E; Endeman D; Benfenati F; Lanzani G Shedding Light on Living Cells. Adv. Mater 2015, 27 (46), 7662–7669. [DOI] [PubMed] [Google Scholar]
- (4).Leopold AV; Chernov KG; Verkhusha VV Optogenetically Controlled Protein Kinases for Regulation of Cellular Signaling. Chem. Soc. Rev 2018, 47 (7), 2454–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Chen X; Wu YW Tunable and Photoswitchable Chemically Induced Dimerization for Chemo-Optogenetic Control of Protein and Organelle Positioning. Angew. Chem. Int. Ed. Engl 2018, 57 (23), 6796–6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Gorostiza P; Isacoff EY Optical Switches for Remote and Noninvasive Control of Cell Signaling. Science 2008, 322 (5900), 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Kim JM; Lee M; Kim N; Heo WD Optogenetic Toolkit Reveals the Role of Ca2+ Sparklets in Coordinated Cell Migration. Proc. Natl. Acad. Sci. U. S. A 2016, 113 (21), 5952–5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Kainrath S; Stadler M; Reichhart E; Distel M; Janovjak H Green-Light-Induced Inactivation of Receptor Signaling Using Cobalamin-Binding Domains. Angew. Chem. Int. Ed. Engl 2017, 56 (16), 4608–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Liu Q; Tucker CL Engineering Genetically-Encoded Tools for Optogenetic Control of Protein Activity. Curr. Opin. Chem. Biol 2017, 40, 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Bugaj LJ; Spelke DP; Mesuda CK; Varedi M; Kane RS; Schaffer DV Regulation of Endogenous Transmembrane Receptors through Optogenetic Cry2 Clustering. Nat. Commun 2015, 6, 6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Xu Y; Hyun YM; Lim K; Lee H; Cummings RJ; Gerber SA; Bae S; Cho TY; Lord EM; Kim M Optogenetic Control of Chemokine Receptor Signal and T-Cell Migration. Proc. Natl. Acad. Sci. U. S. A 2014, 111 (17), 6371–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Zhang K; Cui B Optogenetic Control of Intracellular Signaling Pathways. Trends Biotechnol 2015, 33 (2), 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Johnson HE; Toettcher JE Illuminating Developmental Biology with Cellular Optogenetics. Curr. Opin. Biotechnol 2018, 52, 42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Di Maria F; Lodola F; Zucchetti E; Benfenati F; Lanzani G The Evolution of Artificial Light Actuators in Living Systems: From Planar to Nanostructured Interfaces. Chem. Soc. Rev 2018, 47 (13), 4757–4780. [DOI] [PubMed] [Google Scholar]
- (15).Zhang K; Gao H; Deng R; Li J Emerging Applications of Nanotechnology for Controlling Cell-Surface Receptor Clustering. Angew. Chem. Int. Ed. Engl 2019, Article in Press. [DOI] [PubMed]
- (16).Seeman NC; Sleiman HF DNA Nanotechnology. Nat. Rev. Mater 2017, 3 (1), 17068. [Google Scholar]
- (17).Chen YJ; Groves B; Muscat RA; Seelig G DNA Nanotechnology from the Test Tube to the Cell. Nat. Nanotechnol 2015, 10 (9), 748–760. [DOI] [PubMed] [Google Scholar]
- (18).Li H; Wang M; Shi T; Yang S; Zhang J; Wang HH; Nie Z A DNA-Mediated Chemically Induced Dimerization (D-CID) Nanodevice for Nongenetic Receptor Engineering to Control Cell Behavior. Angew. Chem. Int. Ed. Engl 2018, 57 (32), 10226–10230. [DOI] [PubMed] [Google Scholar]
- (19).Ueki R; Atsuta S; Ueki A; Sando S Nongenetic Reprogramming of the Ligand Specificity of Growth Factor Receptors by Bispecific DNA Aptamers. J. Am. Chem. Soc 2017, 139 (19), 6554–6557. [DOI] [PubMed] [Google Scholar]
- (20).Ueki R; Ueki A; Kanda N; Sando S Oligonucleotide-Based Mimetics of Hepatocyte Growth Factor. Angew. Chem. Int. Ed. Engl 2016, 55 (2), 579–582. [DOI] [PubMed] [Google Scholar]
- (21).Ankenbruck N; Courtney T; Naro Y; Deiters A Optochemical Control of Biological Processes in Cells and Animals. Angew. Chem. Int. Ed. Engl 2018, 57 (11), 2768–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Wang L; Li Q Photochromism into Nanosystems: Towards Lighting up the Future Nanoworld. Chem. Soc. Rev 2018, 47 (3), 1044–1097. [DOI] [PubMed] [Google Scholar]
- (23).Chen S; Li J; Liang H; Lin XH; Li J; Yang HH Light-Induced Activation of c-Met Signalling by Photocontrolled DNA Assembly. Chemistry 2018, 24 (60), 15988–15992. [DOI] [PubMed] [Google Scholar]
- (24).Zhang Y; Huang L; Li Z; Ma G; Zhou Y; Han G Illuminating Cell Signaling with near-Infrared Light-Responsive Nanomaterials. ACS Nano 2016, 10 (4), 3881–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Hilderbrand SA; Weissleder R Near-Infrared Fluorescence: Application to in Vivo Molecular Imaging. Curr. Opin. Chem. Biol 2010, 14 (1), 71–79. [DOI] [PubMed] [Google Scholar]
- (26).Jalani G; Tam V; Vetrone F; Cerruti M Seeing, Targeting and Delivering with Upconverting Nanoparticles. J. Am. Chem. Soc 2018, 140 (35), 10923–10931. [DOI] [PubMed] [Google Scholar]
- (27).Chen S; Weitemier AZ; Zeng X; He L; Wang X; Tao Y; Huang AJY; Hashimotodani Y; Kano M; Iwasaki H; Parajuli LK; Okabe S; Teh DBL; All AH; Tsutsui-Kimura I; Tanaka KF; Liu X; McHugh TJ Near-Infrared Deep Brain Stimulation Via Upconversion Nanoparticle-Mediated Optogenetics. Science 2018, 359 (6376), 679–684. [DOI] [PubMed] [Google Scholar]
- (28).Xin H; Namgung B; Lee LP Nanoplasmonic Optical Antennas for Life Sciences and Medicine. Nat. Rev. Mater 2018, 3 (8), 228–243. [Google Scholar]
- (29).Lino MM; Simoes S; Vilaca A; Antunes H; Zonari A; Ferreira L Modulation of Angiogenic Activity by Light-Activatable Mirna-Loaded Nanocarriers. ACS Nano 2018, 12 (6), 5207–5220. [DOI] [PubMed] [Google Scholar]
- (30).Zhang P; Wang C; Zhao J; Xiao A; Shen Q; Li L; Li J; Zhang J; Min Q; Chen J; Chen HY; Zhu JJ Near Infrared-Guided Smart Nanocarriers for Microrna-Controlled Release of Doxorubicin/Sirna with Intracellular Atp as Fuel. ACS Nano 2016, 10 (3), 3637–3647. [DOI] [PubMed] [Google Scholar]
- (31).Xiao Z; Ji C; Shi J; Pridgen EM; Frieder J; Wu J; Farokhzad OC DNA Self-Assembly of Targeted near-Infrared-Responsive Gold Nanoparticles for Cancer Thermo-Chemotherapy. Angew. Chem. Int. Ed. Engl 2012, 51 (47), 11853–11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Dai W; Dong H; Guo K; Zhang X Near-Infrared Triggered Strand Displacement Amplification for Microrna Quantitative Detection in Single Living Cells. Chem. Sci 2018, 9 (7), 1753–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Lemmon MA; Schlessinger J Cell Signaling by Receptor Tyrosine Kinases. Cell 2010, 141 (7), 1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Trusolino L; Bertotti A; Comoglio PM Met Signalling: Principles and Functions in Development, Organ Regeneration and Cancer. Nat. Rev. Mol. Cell Biol 2010, 11 (12), 834–848. [DOI] [PubMed] [Google Scholar]
- (35).Huschka R; Zuloaga J; Knight MW; Brown LV; Nordlander P; Halas NJ Light-Induced Release of DNA from Gold Nanoparticles: Nanoshells and Nanorods. J. Am. Chem. Soc 2011, 133 (31), 12247–12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Goodman AM; Hogan NJ; Gottheim S; Li C; Clare SE; Halas NJ Understanding Resonant Light-Triggered DNA Release from Plasmonic Nanoparticles. ACS Nano 2017, 11 (1), 171–179. [DOI] [PubMed] [Google Scholar]
- (37).Ueki R; Sando S A DNA Aptamer to c-Met Inhibits Cancer Cell Migration. Chem. Commun. (Camb.) 2014, 50 (86), 13131–13134. [DOI] [PubMed] [Google Scholar]
- (38).Kwee BJ; Mooney DJ Biomaterials for Skeletal Muscle Tissue Engineering. Curr. Opin. Biotechnol 2017, 47, 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Wosczyna MN; Rando TA A Muscle Stem Cell Support Group: Coordinated Cellular Responses in Muscle Regeneration. Dev. Cell 2018, 46 (2), 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Loebel C; Burdick JA Engineering Stem and Stromal Cell Therapies for Musculoskeletal Tissue Repair. Cell Stem Cell 2018, 22 (3), 325–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Juhas M; Engelmayr GC Jr.; Fontanella AN; Palmer GM; Bursac N Biomimetic Engineered Muscle with Capacity for Vascular Integration and Functional Maturation in Vivo. Proc. Natl. Acad. Sci. U. S. A 2014, 111 (15), 5508–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Walker N; Kahamba T; Woudberg N; Goetsch K; Niesler C Dose-Dependent Modulation of Myogenesis by HGF: Implications for c-Met Expression and Downstream Signalling Pathways. Growth Factors 2015, 33 (3), 229–241. [DOI] [PubMed] [Google Scholar]
- (43).Hardy D; Besnard A; Latil M; Jouvion G; Briand D; Thepenier C; Pascal Q; Guguin A; Gayraud-Morel B; Cavaillon JM; Tajbakhsh S; Rocheteau P; Chretien F Comparative Study of Injury Models for Studying Muscle Regeneration in Mice. PLoS ONE 2016, 11 (1), e0147198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Chang NC; Sincennes MC; Chevalier FP; Brun CE; Lacaria M; Segales J; Munoz-Canoves P; Ming H; Rudnicki MA The Dystrophin Glycoprotein Complex Regulates the Epigenetic Activation of Muscle Stem Cell Commitment. Cell Stem Cell 2018, 22 (5), 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Morena D; Maestro N; Bersani F; Forni PE; Lingua MF; Foglizzo V; Scepanovic P; Miretti S; Morotti A; Shern JF; Khan J; Ala U; Provero P; Sala V; Crepaldi T; Gasparini P; Casanova M; Ferrari A; Sozzi G; Chiarle R; Ponzetto C; Taulli R Hepatocyte Growth Factor-Mediated Satellite Cells Niche Perturbation Promotes Development of Distinct Sarcoma Subtypes. Elife 2016, 5, e12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Tedesco FS; Dellavalle A; Diaz-Manera J; Messina G; Cossu G Repairing Skeletal Muscle: Regenerative Potential of Skeletal Muscle Stem Cells. J. Clin. Invest 2010, 120 (1), 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Machado L; Esteves de Lima J; Fabre O; Proux C; Legendre R; Szegedi A; Varet H; Ingerslev LR; Barres R; Relaix F; Mourikis P In Situ Fixation Redefines Quiescence and Early Activation of Skeletal Muscle Stem Cells. Cell Rep 2017, 21 (7), 1982–1993. [DOI] [PubMed] [Google Scholar]
- (48).Xu C; Teng Z; Zhang Y; Yuwen L; Zhang Q; Su X; Dang M; Tian Y; Tao J; Bao L; Yang B; Lu G; Zhu J Flexible MoS2-Embedded Human Serum Albumin Hollow Nanocapsules with Long Circulation Times and High Targeting Ability for Efficient Tumor Ablation. Adv. Funct. Mater 2018, 28 (45), 1804081. [Google Scholar]
- (49).Tang W; Dong Z; Zhang R; Yi X; Yang K; Jin M; Yuan C; Xiao Z; Liu Z; Cheng L Multifunctional Two-Dimensional Core-Shell MXene@Gold Nanocomposites for Enhanced Photo-Radio Combined Therapy in the Second Biological Window. ACS Nano 2019,13 (1), 284–294. [DOI] [PubMed] [Google Scholar]
- (50).Lin H; Gao S; Dai C; Chen Y; Shi J A Two-Dimensional Biodegradable Niobium Carbide (MXene) for Photothermal Tumor Eradication in NIR-I and NIR-II Biowindows. J. Am. Chem. Soc 2017, 139 (45), 16235–16247. [DOI] [PubMed] [Google Scholar]
- (51).Qiu M; Wang D; Liang W; Liu L; Zhang Y; Chen X; Sang DK; Xing C; Li Z; Dong B; Xing F; Fan D; Bao S; Zhang H; Cao Y Novel Concept of the Smart NIR-Light-Controlled Drug Release of Black Phosphorus Nanostructure for Cancer Therapy. Proc. Natl. Acad. Sci. U. S. A 2018, 115 (3), 501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.