Abstract
Purpose
Although high glucocorticoid receptor (GR) expression in early-stage estrogen receptor (ER)- negative breast cancer (BC) is associated with shortened relapse-free survival (RFS), how associated GR transcriptional activity contributes to aggressive BC behavior is not well understood. Using potent GR antagonists and primary tumor gene expression data, we sought to identify a tumor-relevant gene signature based on GR activity that would be more predictive than GR expression alone.
Design
Global gene expression and GR ChIP-sequencing were performed to identify GR-regulated genes inhibited by two chemically distinct GR antagonists, mifepristone and CORT108297. Differentially expressed genes from MDA-MB-231 cells were cross-evaluated with significantly expressed genes in GR-high versus GR-low ER-negative primary BCs. The resulting subset of GR targeted genes was analyzed in two independent ER-negative BC cohorts to derive and then validate the GR activity signature (GRsig).
Results
Gene expression pathway analysis of glucocorticoid-regulated genes (inhibited by GR antagonism) revealed cell survival and invasion functions. GR ChIP-seq analysis demonstrated that GR antagonists decreased GR chromatin association for a subset of genes. A GRsig comprised of n=74 GR activation-associated genes (also reversed by GR antagonists) was derived from an adjuvant chemotherapy-treated Discovery cohort and found to predicted probability of relapse in a separate Validation cohort (HR=1.9; p= 0.012).
Conclusions
The GRsig discovered herein identifies high-risk ER-negative/GR-positive BCs most likely to relapse despite administration of adjuvant chemotherapy. Because GR antagonism can reverse expression of these genes, we propose that addition of a GR antagonist to chemotherapy may improve outcome of these high-risk patients.
Introduction
Breast cancers (BCs) lacking expression of estrogen receptor (ER), progesterone receptor (PR), and HER2 are termed triple-negative breast cancers (TNBCs). The absence of these receptors poses a challenge in part because of the lack of druggable targets for TNBC. Resistance (de-novo or acquired) of disseminated tumor cells despite adjuvant treatment is also thought to contribute to increased relapse rates in early-stage TNBC patients. Recent efforts to distinguish the variable natural history of TNBC have used tumor gene expression profiling to divide cancers into four subtypes: basal-like-1, basal-like-2, mesenchymal, and luminal androgen receptor (LAR) (1). Additionally, genomic, epigenetic, and proteomic analyses of TNBC have revealed several potential therapeutic targets, including androgen receptor (AR), EGFR, JAK2, mTOR, PI3K, and BET family proteins (2–7). Despite these advances, outside of clinical trials, patients with early-stage TNBC still receive generic adjuvant cytotoxic chemotherapy. Therefore, the identification of targetable regulators and molecular signatures of TNBC chemoresistance remains a critical need (8).
Glucocorticoid receptor (GR) is a corticosteroid receptor with both transcription factor and chromatin remodeling functions (9). The role of GR in endocrine physiology and metabolism is cell-type specific; its role in cell survival appears to be cancer subtype-specific as well. For example, GR activation is pro-apoptotic in lymphoid malignancies (10), whereas GR activation is anti-apoptotic and its activity is associated with relapse in other cancers (11–20), including ER-negative BC (21–29). Interestingly, in ER- positive BC, GR/ER crosstalk appears to contribute to an improved patient outcome in high GR- expressing tumors (30–32), thus highlighting GR’s context-dependent function. Our laboratory and others have reported that higher tumor GR transcript (30) and protein (33) expression in early-stage ER-negative tumors is associated with shorter relapse-free survival (RFS). In a retrospective meta-analysis of tumor gene expression from n=354 ER-negative early-stage BC patients, high GR transcript expression (NR3C1, top quartile) was associated with poor long-term RFS regardless of whether patients received adjuvant chemotherapy (30). Furthermore, GR antagonism has been found to sensitize cells to chemotherapy-induced cytotoxicity in ovarian (15,18), prostate (16,17,20), and TNBC (24,25). A Phase I clinical trial of the GR/PR antagonist mifepristone (300mg/day) administered to BC patients before weekly nab- paclitaxel treatment established the safety and tolerability of this combination (34). Together, these data suggest that GR transcriptional activity plays a role in BC aggressiveness and chemoresistance, and that GR antagonism is a potential therapeutic strategy.
In this report, we derived a GR signature using GR-activated gene networks and then identified a subset of GR-activated genes whose expression changes was also reversed by GR antagonism. GR transcriptional activity was antagonized with the steroidal GR/PR antagonist mifepristone (Mif) or the highly-selective non-steroidal GR modulator CORT108297 (C297) (35). We performed studies using these GR antagonists in the context of glucocorticoid (GC)-activated GR to mimic cortisol-activated GR in vascularized patient tumors. This experimental design allowed us to identify antagonist-sensitive GC- mediated GR pathways for both mechanistic insight and to identify a GR activity signature (GRsig) for patient stratification.
GR is a widely active transcription factor with tissue-specific activities, and in the context of TNBC, GR is likely to regulate many genes that coordinately contribute to tumor viability, aggressiveness, and eventual recurrence. Therefore, we hypothesized that a network of GR target genes would be a better indicator of GR activity in TNBC than GR expression alone. We combined our analyses of antagonist-modulated GR-mediated gene expression in TNBC cells with gene expression data from primary ER-negative BCs to identify a GRsig of n=74 genes associated with poor prognosis in early-stage BC despite adjuvant chemotherapy. We then validated the GRsig in an independent patient dataset. Our results suggest that 1) the GRsig can be used to identify individual early-stage ER-negative patients with a relatively increased risk of relapse and 2) adding GR antagonism to adjuvant chemotherapy could reduce tumor GR activity, thereby increasing chemotherapy efficacy, and improving clinical outcome in poor-prognosis ER-negative BC patients.
Materials and Methods
Patients and Samples
The REMARK (‘REporting recommendations for tumor MARKer prognostic studies’) guidelines were used for the retrospective meta-analysis of tissue microarrays (TMA) in this report (36). Details regarding primary tumor microarray datasets, standard prognostic variables, adjuvant chemotherapy, and a study design flowchart is in Supplementary File 1.
Cell lines and reagents
The mesenchymal GR-high TNBC cell lines, MDA-MB-231 and SUM-159-PT, were validated and tested negative for mycoplasma throughout the course of the experiments. MDA-MB-231 cells were cultured in Dulbelcco’s Modified Eagle’s Media (DMEM, Lonza), supplemented with 10% fetal bovine serum (FBS, Gemini Bio-Products) and 1% penicillin/streptomycin (Lonza). SUM-159-PT cells were cultured in Ham’s F12 Media (Corning), supplemented with 10% FBS and 1% penicillin/streptomycin. Cells were cultured at 37°C and 5% CO2. Compounds for cell culture studies were acquired and dissolved as follows: Dexamethasone (Sigma) and Mifepristone (Enza) were dissolved into 1 mM stock solutions in ethanol (EtOH, Sigma). CORT108297 (C297, Corcept Therapeutics, Menlo Park, CA), and was dissolved at 1 mM in EtOH. Pharmaceutical-grade paclitaxel (APP Pharmaceuticals) was diluted to 1 mM in EtOH. Compounds for the fluorescent polarization assay were dissolved in dimethyl sulfoxide (DMSO) at 50 mM concentrations, including dexamethasone, mifepristone, CORT108297, CORT118335, dihydrotestosterone (DHT), and Compound A (Enzo Life Sciences). Fluorescein-dexamethasone (Invitrogen) was dissolved in DMSO in black microcentrifuge tubes at a concentration of 20 mM and further diluted as needed in water. For murine xenograft studies, pharmaceutical-grade paclitaxel (APP Pharmaceuticals) was suspended in saline and castor oil so that a 50 μL i.p. injection into a 20-gram mouse would be a 10 mg/kg dose. CORT108297 was dissolved in EtOH and suspended in sesame oil so that a 50 μL i.p. injection into a 20-gram mouse would be a 20 mg/kg dose.
In vitro GR LBD expression and purification
Details regarding the cloning of wild-type GR-LBD (amino acid residues 522–777) into a plasmid, obtaining bacmid, and SF9 transfection is described in the Supplementary Materials and Methods. After expression of GR LBD, the SF9 cells were pelleted at 3,000 RPM at 4°C, and lysed by sonication in a buffer containing 20mM TRIS pH 8.0, 500mM NaCl, 5% glycerol, 0.1% CHAPS, 0.25mM TCEP that was supplemented with protease inhibitors and 1.0μM Dex. GR-LBD was purified first using Ni-affinity chromatography followed by overnight dialysis with TEV protease to remove the His tag. Extensive dialysis into 20mM TRIS pH 8.0, 500mM NaCl, 5% glycerol, 0.1% CHAPS, 0.25mM TCEP was performed to obtain non-ligand-bound GR LBD. The final protein was obtained using size exclusion chromatography where the protein consistently eluted as a dimer. The purified proteins were concentrated to 5mg/ml, flash frozen in 50μl aliquots in N2(l), and stored at −80°C.
Ligand titration assay via fluorescence polarimetry (FP)
GR LBD was diluted in assay buffer (20mM TRIS pH 7.5, 50mM NaCl, 0.25mM TCEP) to a concentration of 50 nM. After pre-equilibration of GR LBD with both ligand (ranging from concentrations ranging from 0 to 4000 nM) and 10nM fluorescein-labeled dexamethasone (F-Dex, Life Technologies) for 30 minutes, FP signal was measured using the Beacon 2000 Fluorescence Polarization System (Invitrogen). Triplicate FP measurements were scaled to maximal FP and averaged for each ligand concentration. Dose response curves for each ligand were generated using GraphPad Prism using the log(inhibitor) vs. normalized response curve equation: (Y=100/(1+10^((LogIC50-X)*HillSlope)))
Western blot analysis of GR protein levels
MDA-MB-231 cells were grown in 10 cm dishes to 75% confluence in DMEM with 10% FBS. Next, the cells were cultured for 48h in phenol red-free DMEM with 2.5% charcoal-stripped FBS media. Cells were treated for t=30 minutes with Vehicle (EtOH) or 100nM Dex/Veh, 100 nM Dex/100 nM Mif, 100nM Dex/100nM C297, or 100nM Dex/100nM C335. Cells were washed with cold PBS, harvested and pelleted, and lysed on ice in a buffer containing 20 mM Tris, pH 7.5 containing protease/phosphatase inhibitors. Cell lysate was clarified at 15,000 rpm for 10 min and resulting lysate was quantitated using the BCA analysis. After denaturing the samples in Laemmli buffer with SDS at 95°C, the samples were resolved on an 8% SDS-page gel, which was then transferred to PVDF membrane. The membrane was blocked overnight at 4°C with 5% w/v BSA in TBST, then probed for GR (GR-XP D8H2 antibody, Cell Signaling) and beta actin (β-Actin 8226, Sigma-Aldrich). After addition of Alexafluor secondary antibodies (Invitrogen and LI-COR, respectively), the blot was imaged on the Odyssey infrared imaging system (LI-COR). Additional details of immunoblotting protocol can be found the Supplementary Materials and Methods.
Cell viability assay
MDA-MB-231 and SUM-159-PT cells (n=3 × 104) were seeded in 96-well plates. After culturing in DMEM with 10% FBS (for MDA-MB-231) or Ham’s F12 with 5% FBS (for SUM-159-PT), the cells were cultured for 48h in charcoal-stripped FBS media (2.5% for MDA-MB-231 or 5% for SUM-159-PT). Cells were treated for 72h and 96h with varying concentrations of paclitaxel (0 – 100 nM) in the presence of Vehicle/Vehicle, 100 nM Dex/Vehicle, Vehicle/1μM C297, or 100 nM Dex/1μM C297. The sulforhodamine B (SRB) assay was performed as described in the Supplementary Materials and Methods.
The experiment was performed in three biological replicates per cell line. P-values comparing the means of cell death percentages were obtained using the unpaired Student’s t-test (GraphPad).
Murine TNBC xenograft study
MDA-MB-231 tumors were established in the right pectoral mammary gland of n=23 five- or six- week old female SCID mice (Taconic). Tumor volume was measured by caliper and then calculated using the elliptical volume equation (24). When tumors reached a volume of 100 – 300 mm3, the mice were treated for five days with either 20 mg/kg/day C297 or vehicle one hour prior to 10 mg/kg/day paclitaxel or vehicle. Tumor volume was measured by caliper until reaching a volume of approximately 2000 mm3 or 40 days post-treatment initiation. Tumor data were analyzed using the repeated measures ANOVA using SigmaPlot 11.2 (Systat Software), and p-values between treatment groups over time were obtained using the Holm–Sidak post-hoc test.
Gene expression microarray and analysis
MDA-MB-231 cells were grown to 80% confluence in 15-cm dishes in DMEM with 10% FBS. After culturing cells for 48h in DMEM with 2.5% charcoal-stripped FBS, 2 × 107 cells (per condition) were treated with either Vehicle, 100 nM Dex +/− 100 nM C297 or 100 nM Mif for 4, 8, and 12h. Following compound exposure, cells were washed in PBS, and lysed in RNA lysis buffer (Qiagen) overnight at −80°C. RNA extraction, with accompanying DNase treatment, was performed using the RNeasy kit (Qiagen) following the manufacturer’s protocol. A small sample of each condition was reverse transcribed to perform Q-RT-PCR as a quality control to test GR-induction of SGK1 by Dex over vehicle, and inhibition of that induction by Mif. Duplicate microarray experiments (n=2) for Vehicle, Dex, and Dex/Mif conditions were performed along with a single experiment for the Dex/C297 treatment condition. The University of Chicago Genomics Core facility carried out the reverse transcription on the samples, followed by microarray using the Affymetrix Human U133 Plus 2.0 platform. A detailed description of the analysis of gene expression data can be found the Supplementary Materials and Methods. After RMA normalization and application of a cut-off of at least +/− 1.3-fold change difference in expression for each treatment group versus vehicle, the genes became candidates for further analysis when their expression was altered significantly by Dex, and inhibited commonly by Mif and C297 (within the same time point as Dex) by at least 25%, in any treatment group. Dex-regulated genes were overlapped with a list of n=5,170 differentially-expressed tumor-derived genes (in the same direction) from GR-high versus GR-low BCs (30). Ingenuity Pathway Analysis (Qiagen) was performed to determine relevant gene expression pathways using the Diseases and Biofunctions setting with a p-value cut-off of 0.05 (-log10=1.3).
GR ChIP-sequencing and analysis
MDA-MB-231 cells were grown to 80% confluence in 15-cm dishes in DMEM with 10% FBS, followed by 48h in DMEM supplemented with 2.5% charcoal-stripped FBS. Cells (n=4 × 107 per treatment condition) were treated with Vehicle, or 100 nM Dex +/− 100 nM C297 or 100 nM Mif for 60 minutes. Cells were cross-linked with 1% formaldehyde, quenched with glycine (final concentration of 1.25 mM), and harvested. After cell lysis with ChIP lysis buffer (Magna ChIP A Chromatin Immunoprecipitation Kit, EMD Millipore), cells were sonicated to achieve the majority of DNA fragments between 200–400 bp. GR was chromatin immunoprecipitated using 3 μg of ChIP-grade XP (D8H2) rabbit anti-GR antibody (Cell Signaling); 3 μg of rabbit IgG (Cell Signaling) was used for IgG control sample. Chromatin was eluted from GR ChIP and input samples following the manufacturer’s protocol. ChIP-sequencing and data analysis is described in depth in the Supplemental Materials and Methods. Briefly, sequences were aligned to the human genome (hg19) and peaks were called using MACS2. The peaks were then normalized to the vehicle control using deepTools2 (http://deeptools.ie-freiburg.mpg.de). ChIPseeker (37) was used to annotate peaks to nearest transcriptional start sites (TSSs) of genes.
Quantitative real-time PCR
MDA-MB-231 cells were grown to 80% confluence in 6-cm dishes in DMEM (10% FBS, 1% penicillin/streptomycin), followed by a 72h serum starvation period in charcoal-stripped DMEM (2.5% charcoal-stripped FBS, 1% penicillin/streptomycin). SUM-159PT cells were grown to 80% confluence in 6-cm dishes in Ham’s F12 (5% FBS, 1 μg/ml hydrocortisone, 5 μg/ml insulin, 1% penicillin/streptomycin), followed by a 72h serum starvation period in charcoal-stripped Ham’s F12 (5% charcoal-stripped FBS, 1% penicillin/streptomycin). Cells were treated with the following for 4, 8, and 12-hr: Vehicle (EtOH, 0.2% final volume), Veh/100 nM Dex, Dex/Mif (100 nM each), or Dex/C297 (100 nM each). Following treatment, cells were washed once with PBS and lysed in 500 μl of RLT buffer (Qiagen) supplemented with 1% 2-mercaptoethanol overnight at −80°C. Three biological replicates were performed for each compound treatment per cell line. Total RNA extraction, with accompanying DNase treatment, was performed using the Qiagen RNeasy kit (Qiagen) following manufacturer’s protocol. After reverse transcription, PerfeCTa SYBR Green FastMix (Quanta Biosciences) was used to perform quantitative real-time PCR on the resulting cDNA. Primers and controls can be found in the Supplementary Materials and Methods. Propagated error (standard deviation) in fold change was calculated and p-values were generated with the two-sample Student’s t-test with Welch’s correction for unequal variances (GraphPad).
siRNA knockdown
MDA-MB-231 cells were cultured to 80% confluence in 10-cm dishes. siRNA knockdown was carried out using the Smartpool (Dharmacon) of four siRNAs against either MCL1 or NNMT. Scrambled control pool was used as well (Dharmacon). siRNAs were introduced into cells using the RNAimax forward transfection protocol (Invitrogen). Knockdown efficiency was analyzed by Q-RT-PCR (see Methods above) normalizing NNMT siRNA pool and MCL1 siRNA pool to the Control siRNA pool. After efficient knockdown (48h), cells were treated with Veh/Veh, a range of concentrations (10 – 100nM) of paclitaxel +/− 100 nM Dex for 48h. Cell death was assessed using the sulforhodamine B assay (see Methods above). Percentage of cell death was averaged over three experiments and significance of mean cell death was analyzed using the Student’s t-test (GraphPad).
Retrospective analysis of early-stage ER-negative BC tumor NR3C1 gene expression association with RFS in TNBC subtypes
A gene expression database of TNBC gene arrays was downloaded from GEO and is summarized in Supplementary File 1. Gene expression levels were normalized using MAS5 as described previously (38). TNBC molecular subtypes were defined by Pietenpol et al (1). TNBC patients were classified according to NR3C1 gene expression (Affymetrix probeID 216321_s_at) being in the top quartile of expression versus all others. The cutoff values were determined based on all patients in a given group. RFS was estimated using the method of Kaplan-Meier and compared between patients in the top quartile of NR3C1 expression vs. all others using the logrank test. Hazard ratios (HRs) were estimated using Cox proportional hazards regression models.
Retrospective analysis of early-stage ER-negative BC tumor gene expression association with RFS in Discovery and Validation cohorts
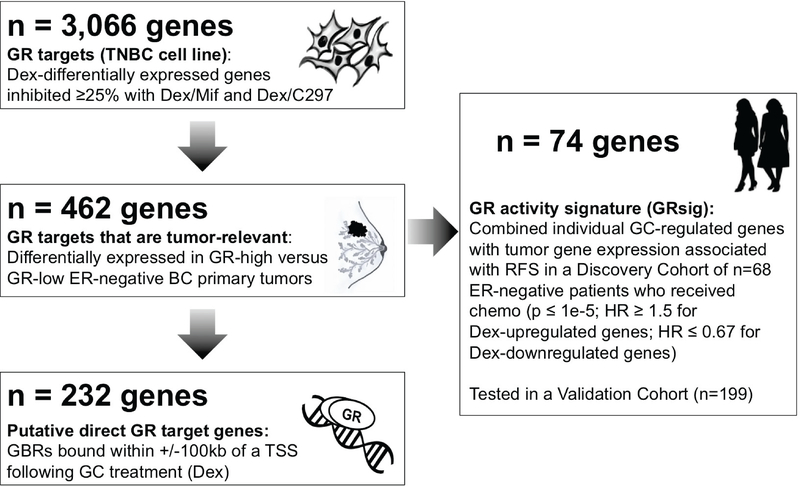
The Discovery cohort was assembled using a subset of n=68 ER-negative BC patients who received adjuvant chemotherapy (See Supplementary Table 1). Duplicate patients were removed, ESR1 status was validated, and data were normalized as previously reported (30). The independent Validation set of n=199 ER-negative BC patients who received adjuvant chemotherapy was obtained from ten studies (Supplementary Table 1); duplicate patients were removed, ESR1 status was validated, and data were normalized as previously reported (38). Per the REMARK guidelines, a description of the tumor characteristics and a study design flowchart for the Discovery and Validation cohorts can be found in Supplementary File 1. The GRsig was determined as follows: Differentially-expressed genes from the cell line microarray experiment that were induced or repressed by Dex and modulated by GR antagonists were cross-evaluated against a list of differentially-expressed GR-high vs. GR-low ER-negative primary BCs (30), to obtain a subset of n=420 genes. Next, these n=420 genes were filtered by best available Affymetrix probeID as defined by the Jetset method (39) to obtain a list of n=320 genes. Individual gene association with RFS using the Cox proportional hazards regression model with continuous expression as a predictor was determined for the n=320 genes, and a GRsig was defined as those with RFS-associated p ≤ 1 × 10−5, and a HR ≥ 1.5 for GC-induced genes or HR ≤ 0.67 (1/1.5) for Dex-repressed genes. To test the GRsig in both the Discovery and Validation cohorts, normalized expression levels of the 74 genes were added (Dex-upregulated genes) or subtracted (Dex-repressed genes) to obtain GRsig expressions. Patients were classified as having high GRsig expression if their GRsig expression was above the median GRsig expression among all n=354 ER-negative patients. RFS in each adjuvant chemotherapy group (Discovery and Validation cohorts) was estimated using the method of Kaplan-Meier, and was compared using the logrank test. Hazard ratios (HRs) were estimated using Cox regression models.
Microarray data analysis of patient-derived xenografts (PDXs) from the Mayo Clinic neoadjuvant breast cancer study (BEAUTY)
The BEAUTY trial patient-derived xenografts (PDX) were pathologically confirmed to be of human breast carcinoma origins and assayed by the Affymetrix HTA 2.0 microarray platform (40). Xenografts derived from basal TNBCs were selected for the analysis. There were a total of n=62 xenografts derived from n=13 baseline (V1) TNBC patient tumors; the biological replicates (multiple xenografts from same tumor source) were aggregated by their probeset expression means. Replicated xenografts all shared a median correlation (Spearman) above 0.85. The probe features of the array dataset were reduced to the genes provided in the GRsig. Three of these genes were assayed by two probesets each and were included if they shared a correlation (Spearman) greater than 0.65 (which excluded the MUC5AC probe). The NOX5 gene was excluded from the analysis since the probeset did not exist in the Affymetrix HTA microarray platform. The GRsig expression level was derived by summing the expression profiles for the genes designated as Dex-induced and subtracting the summarized profiles for genes designated as Dex-repressed. Samples greater than or equal to the median were classified as GRsig- high; the remainder (less than median) was considered GRsig-low. A violin plot of the expression data was generated in R using the package beanplot.
Results
High GR transcript associates with poor RFS across TNBC subtypes
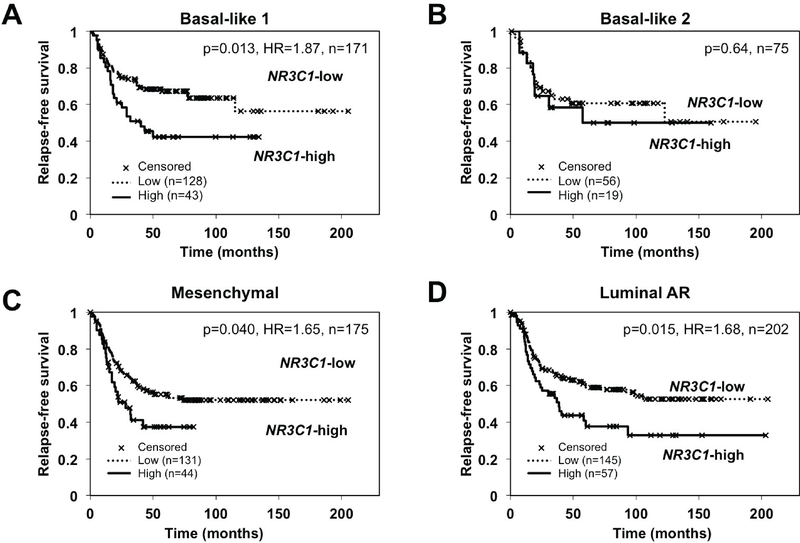
Unique gene expression signatures, discovered and refined by Pietenpol and co-workers (1), have allowed TNBCs to be classified into basal-like 1, basal-like 2, mesenchymal, and LAR subtypes, collectively named the TNBCtype-4. In light of our previous finding that high GR/NR3C1 tumor gene expression from early-stage ER-negative BC patients associated with poor RFS (30), we asked whether high tumor GR/NR3C1 transcript expression retained an association with poor outcome in these TNBC subtypes. A retrospective meta-analysis of gene expression was performed using n=623 TNBC tumors (38). Kaplan-Meier estimates of RFS in TNBC patients in the highest quartile of tumor GR/NR3C1 mRNA expression (versus all others) are shown in Figure 1 for each of TNBC subtypes: basal-like-1 (n=171), basal-like-2 (n=75), mesenchymal (n=175), and LAR (n=202). We found that high tumor GR/NR3C1 mRNA expression was significantly associated with poor RFS in the basal-like 1 (hazard ratio [HR]=1.87, p=0.013), mesenchymal (HR=1.65, p=0.040), and LAR (HR=1.68, p=0.015) subtypes. High GR/NR3C1 association with poor RFS was not significant in the basal-like 2 subtype. Together, these data suggest that GR expression levels, and by extrapolation, GR activity, can stratify most ER-negative BC patients.
Figure 1. High NR3C1 (GR) expression is associated with worse outcome in TNBC subtypes.
Kaplan-Meier estimates of relapse-free survival in patients in the top quartile (versus all others) of tumor NR3C1 expression. NR3C1 expression association with RFS was analyzed in TNBCs classified (1) as (A) basal-like 1 (NR3C1-high n=43, NR3C1-low n=128), (B) basal-like 2 (NR3C1-high n=19, NR3C1-low n=56), (C) mesenchymal (NR3C1-high n=44, NR3C1-low n=131), (D) luminal AR (NR3C1-high n=57, NR3C1-low n=145).
The selective non-steroidal GR modulator C297 is comparable to the steroidal mifepristone in GR LBD affinity and chemosensitization of TNBC cells
We next sought to understand how relatively high GR transcriptional activity (reflected by increased GR expression levels) might lead to chemoresistance and a more aggressive tumor phenotype. We used the agonist dexamethasone (Dex, 100 nM) to mimic a patient’s endogenous circulating GC and activated tumor GR. We first performed an in vitro GR ligand competition assay to choose effective antagonists for this study. Selective non-steroidal GR modulators aryl pyrazole azadecalin C297 (35), pyrimidinedione CORT118335 (C335) (41), as well as the GR/PR steroidal antagonist mifepristone (Mif), all potently displaced fluorescently-labeled Dex (F-Dex) from the GR ligand binding domain (LBD) with low nanomolar affinities (Supplementary Figures 1A and 1B). As expected, we did not observe GC competition using dihydrotestosterone (DHT as a negative control). The published GR modulator Compound A (CpdA), previously shown to displace H3-Dex in cell lysates (42), did not displace F-Dex from the GR LBD in our competition assay (Supplementary Figures 1A and 1B). This implies that regions outside the GR LBD are required for CpdA action on GR (43). We also immunoblotted for GR after thirty minutes or four hours of treatment with Dex, Dex/Mif, Dex/C297, or Dex/C335 and found that GR steady-state protein levels were not affected by the antagonists (Supplementary Figure 1C). The Western blot analysis also demonstrated the expected GC-induced degradation of GR (44). Because C335 has been reported to also bind the mineralocorticoid receptor (41), which can be expressed in TNBC, C297 and Mif were selected to further characterize GR transcriptional and functional activity.
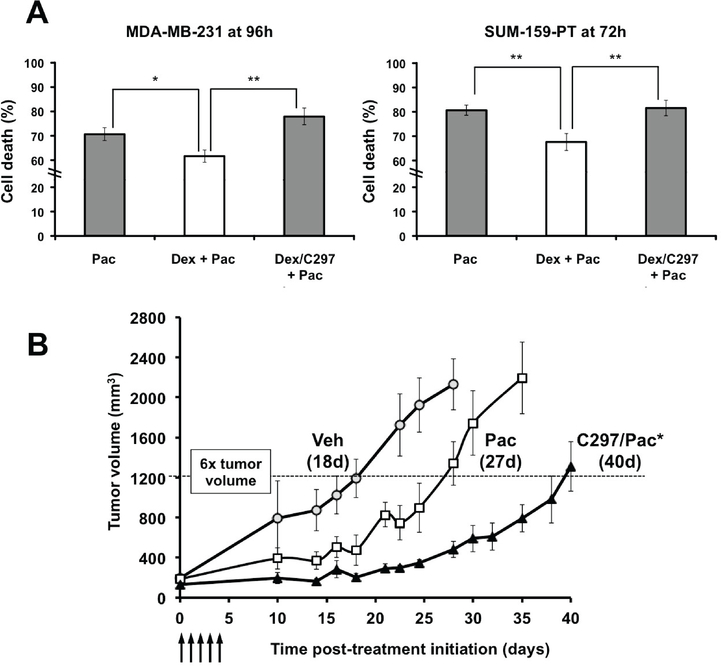
We previously found that treatment with physiological concentrations of GCs decrease TNBC sensitivity to chemotherapy in vitro and in vivo (22,23). This suggests that GR activation in TNBC may contribute to chemotherapy resistance in tumor cells following GR activation by endogenous cortisol. Indeed, we have also found that GR antagonism by Mif could counteract the effects of GC activation on tumor cell survival and thus increase paclitaxel cytotoxicity both in vitro and in vivo (24). To determine if non-steroidal C297 could likewise increase chemosensitivity in GR-positive TNBC, we first tested C297- altered paclitaxel cytotoxicity in two cell lines, MDA-MB-231 and SUM-159-PT. We observed that treatment with GC (Dex, 100nM) dampened paclitaxel (10 nM) cytotoxicity, while the addition of the GR antagonist C297 (1 μM) caused a modest, but significant, relative increase in paclitaxel cytotoxicity in vitro (Figure 2A). As was seen previously with Mif in ER-negative cell lines (21,24), C297 treatment alone did not reduce cell viability (Supplementary Figure 2A). This suggests that C297 antagonism of GR activity increases cell susceptibility to paclitaxel-induced cytotoxicity rather than causing cytotoxicity itself.
Figure 2. GR activation inhibits chemotherapy-induced cytotoxicity of cultured TNBC cells and selective GR antagonism increases sensitivity to chemotherapy in vivo.
(A) MDA-MB-231 and SUM- 159-PT cells were treated with paclitaxel alone (Pac, 10nM), with Vehicle, Dex (100nM), Dex/Pac, or C297(1μM)/Dex/Pac. C297 restored cytotoxic sensitivity at 96 hours (MDA-MB-231) and at 72 hours (SUM-159-PT) following Pac. The bars represent the average percentage cell death of n=3 independent experiments and error bars represent standard error of the mean (S.E.M). * = p value <0.05 and ** = p <0.01 (unpaired Student’s t-test) when compared to Dex/Pac. (B) MDA-MB-231 tumor xenograft re- growth is significantly inhibited by C297 (20 mg/kg/day) pre-treatment one hour prior to Pac (10 mg/kg/day) compared to Pac alone. Arrows refer to administration of Pac/Veh +/− C297/Veh. Pac-treated tumor re-growth was significantly smaller than vehicle, p<0.05, while C297/paclitaxel versus paclitaxel alone delayed post-treatment tumor re-growth significantly. The dotted line represents a 6x increase in re- growth of tumor volume; time to tumor re-growth to this size was 18d (Veh), 27d (Pac), and 40d (Pac/C297). The asterisk (*) represents p < 0.05, comparing C297/Pac to Pac alone. Both C/297 vs Pac and Pac alone vs Veh were significantly different based on a repeated measures ANOVA and the Holm- Sidak post-hoc significance test (Veh/Veh n=3, Veh/Pac n=6 and C297/Pac n=9).
Next, we studied the in vivo effect of GR activity in paclitaxel-treated GR+ TNBC-bearing female SCID mice (n=23). MDA-MB-231 xenograft tumors were established subcutaneously in the mammary fat pad of 6-week old female mice. When tumors reached a volume of 100 – 300 mm3, mice were randomly assigned to treatment groups such that each group had an approximately equal average tumor volume. Mice were then treated daily for five days with intraperitoneal C297 (or vehicle), administered one hour prior to paclitaxel. The one-hour pre-treatment with GR antagonist was intended to compete with endogenous GC (murine corticosterone and cortisol) bound to the tumor cell GR LBD and inhibit tumor GR-mediated transcription. The 5-day (5d) sequential dosing was selected to mimic the most effective weekly paclitaxel neoadjuvant and adjuvant chemotherapy schedule used in early-stage TNBC. However, extending daily treatments beyond 5d resulted in toxicity. Following cessation of the 5d treatment, time to tumor xenograft re-growth was measured to reflect time to patient relapse post- treatment (24,26). Consistent with previous in vivo results with Mif pre-treatment followed by paclitaxel (24), we observed a significantly increased time to tumor re-growth following C297/paclitaxel treatment compared to vehicle/paclitaxel (Figure 2B). Similar to the observations in vitro, C297 monotherapy did not cause a significant delay in tumor re-growth (Supplementary Figure 2B), suggesting that GR antagonism alone is neither cytotoxic nor sufficient to delay tumor progression in a TNBC model. These data are also consistent with C297 GR antagonism increasing chemotherapy sensitivity by reversing endogenous glucocorticoid-mediated expression of genes encoding anti-apoptotic proteins. Thus, as with the non-selective GR antagonist Mif, selective GR antagonism can inhibit GR-mediated chemotherapy resistance both in vitro and in vivo, thereby delaying the time to tumor re-growth.
GR antagonism identifies GR-regulated transcriptional pathways related to chemoresistance and tumor aggressiveness
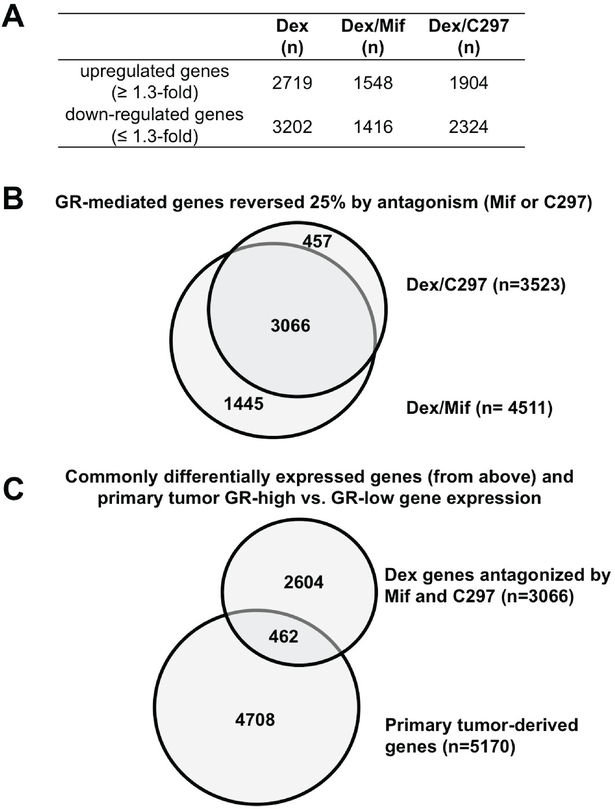
Having established that both C297 and Mif displace GC at the GR LBD, increase chemotherapy sensitivity in the context of GC-activated GR, and also delay in vivo TNBC growth in comparison to chemotherapy alone, we next sought to define which GR-regulated genes were relevant to tumor cell survival. We first used genome-wide gene expression profiling to identify GC-altered gene expression. We then used signatures of antagonist-altered GC-regulated gene expression to determine the subset of GR-regulated genes commonly antagonized by Mif or C297. Using a GR-induced or repressed transcript expression cut-off of at least +/−1.3 fold-change over vehicle treatment, GC treatment (Dex 100 nM) resulted in n=2,719 upregulated genes and n=3,202 downregulated genes at 4, 8 and 12 hours combined (Figure 3A). Markedly fewer genes were altered (in comparison to vehicle) upon co-treatment with either GR modulator (n=1,548 upregulated/1,416 downregulated for Dex/Mif, and n=1,904 upregulated/2,324 downregulated for Dex/C297, Figure 3A). Interestingly, about half of the GC-mediated genes (upregulated: 50%, n=1363; or downregulated: 41%, n=1321) were unique to Dex treatment (Supplementary Figure 3A). A principal components analysis of the differentially altered gene signatures for the three treatments revealed that the Dex/Mif signatures were more closely correlated with the Dex/C297 signatures than to the Dex signatures at their respective timepoints (Supplementary Figure 3B). These data imply that Dex/Mif and Dex/C297 antagonize the GC-induced GR transcriptional profile and modulate a common subset of genes.
Figure 3. Differentially expressed GR target genes following GR antagonism.
Genome-wide gene expression profiling was performed on MDA-MB-231 cells treated with GC (Dex) or GC/antagonist. (A) Total number of up- and downregulated genes by Dex or Dex/GR inhibitor treatment (relative to vehicle); (B) Subset of Dex-regulated genes (≥1.3-fold Dex vs. vehicle) reversed by C297 and/or Mif at 4, 8, and 12h by ≥25 percent change. (C) GR antagonist-identified genes ((B), n=3,066) overlapped with genes (n=5,170) that were differentially expressed between GR-high versus GR-low primary tumors (30). N=462 genes were overlapped.
We next sought to identify a core subset of GR-regulated genes whose activation or repression was commonly antagonized by C297 and Mif treatment. We found n=3,066 genes for which both GR modulators antagonized GR induction or repression by at least 25% (Figure 3B, Supplementary Figure 3C). Interestingly, 87% of the C297-antagonized GR-regulated genes were also regulated in the same direction by Mif, whereas about two-thirds (68%) of the Mif-antagonized GR-regulated genes were shared with C297. These data suggest that Mif is less selective for GR than C297 and/or that Mif is the more potent GR modulator at 100 nM. Because both Mif and C297 displaced GC at the GR LBD and enhanced GR+ TNBC chemosensitivity in vivo, these n=3,066 commonly GR-regulated genes were further considered as candidate GR activity genes relevant to poor prognosis ER-negative BC.
To identify the subset of the commonly antagonized GR-regulated genes (n=3,066 from Figure 3B) that might contribute to higher relapse of ER-negative BC, we next used a meta-analysis dataset of primary early-stage ER-negative tumor gene expression signatures. We previously identified n=5,170 tumor-derived genes that were differentially expressed in GR-high versus GR-low tumors from n=354 ER-negative BCs (30). We found n=462 genes were shared between the n=3,066 genes that were commonly antagonized by C297/Mif and the n=5,170 tumor-derived genes from GR-high versus GR-low primary BCs (Figure 3C). These n=462 genes were expressed in the same direction, i.e., a Dex- upregulated gene was overexpressed in the GR-high versus GR-low gene list. To better characterize the GR gene expression networks, we performed pathway analysis on the n=462 antagonist- modulated/tumor-relevant genes from Figure 3C. Exploring known pathway functions in cancer cells using Ingenuity Pathway Analysis (IPA), we found that these GR-regulated genes were significantly associated with cancer cell survival (inhibition of apoptosis), tumor cell invasion, and epithelial-to- mesenchymal transition pathways. Shown in Table 1, the combination of a positive or negative pathway activation Z-score in the GC (Dex) treatment, and a relative dampening of Z-score magnitude upon the addition of either Mif or C297, confirmed antagonism of these GR-activated and -inactivated signaling pathways. This finding suggests that antagonized GR network genes contribute to tumor relapse and chemotherapy resistance through recognized cell viability pathways. Moreover, these GR-regulated gene expression networks appeared reversible using GR antagonists.
Table 1.
GR antagonists diminish cell survival and tumor metastasis functions while promoting cell death and differentiation.
| Cell function | Activation Z-score |
Treatment time (hr) | Number of genes (n) | |||
|---|---|---|---|---|---|---|
| Dex | DexMif | Dex297 | ||||
| Dex-activated pathways | synthesis of lipid | 1.91 | 0.68 | 0.83 | 4 | 46 |
| invasion of tumor cell lines | 0.63 | −0.03 | 0.35 | 4 | 53 | |
| colony formation of tumor cell lines (invasion) | 0.14 | 0.02 | −0.52 | 4 | 24 | |
| epithelial-mesenchymal transition of breast cell lines | 0.11 | −0.34 | −0.22 | 4 | 9 | |
| metastasis of tumor cell lines | 2.05 | 1.03 | 1.22 | 8 | 16 | |
| transactivation | 1.62 | −0.44 | −0.28 | 8 | 39 | |
| cell survival | 1.53 | −1.28 | 0.69 | 8 | 99 | |
| cell proliferation of colorectal cancer cell lines | 0.72 | −1.23 | −0.71 | 8 | 27 | |
| cell transformation | 0.22 | −1.55 | −1.29 | 8 | 42 | |
| invasion of tumor cells | 1.43 | 0.26 | 0.29 | 12 | 15 | |
| growth of tumor | 0.77 | 0.47 | 0.24 | 12 | 63 | |
| growth of blood vessel | 0.44 | −0.44 | 0.22 | 12 | 10 | |
| Dex-inactivated pathways | cell death of tumor cells | −1.57 | −0.55 | −0.3 | 4 | 35 |
| cytostasis of tumor cell lines | −1.6 | 0.77 | 1.03 | 4 | 15 | |
| contact growth inhibition of tumor cell lines | −1.77 | 0.09 | 0.52 | 4 | 13 | |
| benign neoplasia | −2.35 | −0.03 | −1.49 | 4 | 63 | |
| inflammatory response | −0.14 | 2.12 | 1.94 | 8 | 54 | |
| cytostasis of tumor cell lines | −1.01 | −0.45 | 0.35 | 8 | 15 | |
| contact growth inhibition of tumor cell lines | −1.02 | −0.39 | 0.51 | 8 | 13 | |
| apoptosis | −1.06 | 0.41 | 1.11 | 8 | 162 | |
| cell death | −2.27 | 0.45 | 0.26 | 8 | 193 | |
| development of epithelial tissue (differentiation) | −1.69 | 0.19 | −1.22 | 12 | 45 | |
GR antagonism reduces GR promoter association
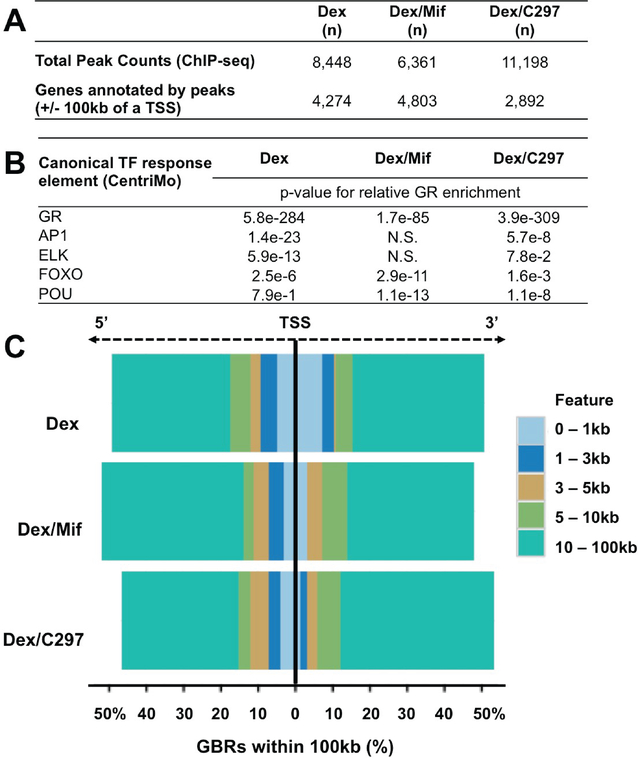
The subset of putative direct GR target genes among the n=462 GR-altered/patient-relevant genes from Figure 3C was next identified using GC-activated GR chromatin association data from MDA-MB- 231 cells. To achieve this, we performed GR ChIP-sequencing in cells treated with vehicle, GC (Dex), Dex/Mif, or Dex/C297. After normalizing GR peaks from treated conditions with vehicle, we found n=8,448 Dex genome-wide GR peaks, n=6,361 Dex/Mif GR peaks, and n=11,198 Dex/C297 GR peaks (Figure 4A, top). When examining Dex genome-wide GR peaks, we observed that only 7% (n=652) Dex GR peaks were conserved in the Dex/Mif treatment whereas 17% (n=1,434) Dex GR peaks were conserved in the Dex/C297 treatment (Supplementary Figure 4A). Motif analysis of these peaks was performed and transcription factor (TF) response elements (REs) were identified. Shown in Figure 4B, the most significant ligand-bound GR binding regions (GBRs) were found at GR response elements (GREs), regardless of treatment condition. Furthermore, we found some common GR enrichment at FOXO and POU REs in all three treatments, however these REs were less significantly represented with both Dex and Dex/C297 treatments compared to the Dex/Mif treatment. AP1 and ELK REs were only shared between Dex and Dex/C297 treatments, and were lost with Dex/Mif treatment. These data suggest that although Mif and C297 have many shared effects on GR-mediated gene expression, they appear to have distinct effects on global GR chromatin association.
Figure 4. GR chromatin association is altered by concomitant treatment with a GR antagonist.
(A) Genome-wide GR peaks and associated genes annotated to TSSs +/− 100kB of these peaks; (B) GR chromatin association with transcription factor response elements (REs) following Dex and GR antagonist treatment reveals significant changes in GR enrichment at GREs, AP1 and ELK REs compared with Dex alone (CentriMO); (C) GR chromatin association in proximal promoter regions (0–3kb from the TSS) is diminished following Dex/Mif or Dex/C297 treatment while more distal GR peak association is proportionally increased.
While we observed genome-wide relative enrichment of activated GR (upon treatment with GC) with promoter regions, there was a decrease in relative GR promoter enrichment (+/− 3kb) following co- treatment with either Mif or C297. This suggests that these antagonists preferentially decrease GR association near the transcriptional start sites (TSSs, Supplementary Figure 4B), while relatively increasing GR chromatin association at more distal (putative enhancer) regions. We next annotated GR peaks to the nearest TSSs using a maximum allowable distance of 100kB from peak to TSS (Figure 4A, bottom). When we limited the GR peak analysis to +/− 100kb of the TSS, GC treatment induced a robust genome-wide GR enrichment within 1kb of annotated TSSs, while GR association in this region was relatively decreased following the addition either GR antagonist (Figure 4C). This suggests that GR antagonists may function, at least in part, through preferentially displacing GR from proximal promoter regions. Interestingly, there was an overall increase in GR chromatin association (peak numbers) with C297 treatment; however, again the lack of peak overlap between Dex and Dex/C297 treatments represents a redirection to new chromatin regions more distal to TSSs (Figure 4A, Figure 4C, and Supplementary Figure 4B).
To identify putative direct GR target genes whose expression was antagonized by either Mif or C297, we next determined the subset of n=462 tumor-relevant genes (from Figure 3C) with Dex-GR peaks within +/− 100kb of their TSS (Figure 4A). We found n=232 putative direct GR target genes with significant Dex GBRs within 100kb, suggesting either promoter or enhancer interaction by GC-activated GR. Indeed, several previously characterized GR target genes were identified within this list, such as SGK1, DUSP1/MKP1, and GILZ/TSC22D3. Additionally, the n=232 putative direct GR target genes also include those with known involvement in cancer cell chemoresistance and evasion of apoptosis (MCL1, MUC1, GADD45B, DNAJC15/MCJ), epigenetic modification and metabolism (NMMT, SLC2A3/GLUT3, ACSL1, SP110), metastasis and invasion (CYR61, TGFB2, EIF4E, F2R/PAR1), angiogenesis (KDR, EIF4E, CALD1), and inflammation (IL15, IL1R1, IL7R, IRAK3). We selected five of these GR target genes with well-established cancer cell growth regulatory functions (SGK1, DUSP1/MKP1, TSC22D3, MCL1, NNMT) and validated antagonist-modulated gene expression by Q-RT-PCR in MDA-MB-231 or SUM-159-PT cells (Supplementary Figures 5A and 5B). Furthermore, the transient knockdown of two individual GR target genes of interest in TNBC (MCL1 and NNMT) increased paclitaxel cytotoxicity (Supplementary Figures 5C and 5D). We also selected two putative direct GR targets related to cell growth (CDKN2D) and transcriptional regulation (ZNF189) to validate by ChIP-qPCR. Dex-induced GR enrichment for ZNF189 which was inhibited by Mif and C297 (Supplementary Figure 4C). Finally, an examination of GR chromatin association within 100kb +/− of the TSS for these n=232 putative direct GR target genes revealed that the majority of Dex GBRs were lost upon Mif or C297 co-treatment (Supplementary File 2). These n=232 genes make up gene expression pathways for which GR appears to be a common upstream TF and for which GR antagonists reverse GC-mediated gene expression.
A GR activity signature (GRsig) has a stronger association with RFS than GR expression alone
We next identified a GC-mediated gene set reflective of tumor-relevant GR activity and clinical outcome. To do this, we analyzed the association between RFS and tumor expression using the n=462 (from Figure 3C) putative indirect and direct GR target genes with optimal Jetset Affymetrix probes. Next, using a Discovery cohort of n=68 ER-negative BC patients who received adjuvant chemotherapy (a dataset we previously reported (30), Supplementary Table 1), we determined individual gene association with RFS using a Cox proportional hazards regression model with continuous expression as a predictor.
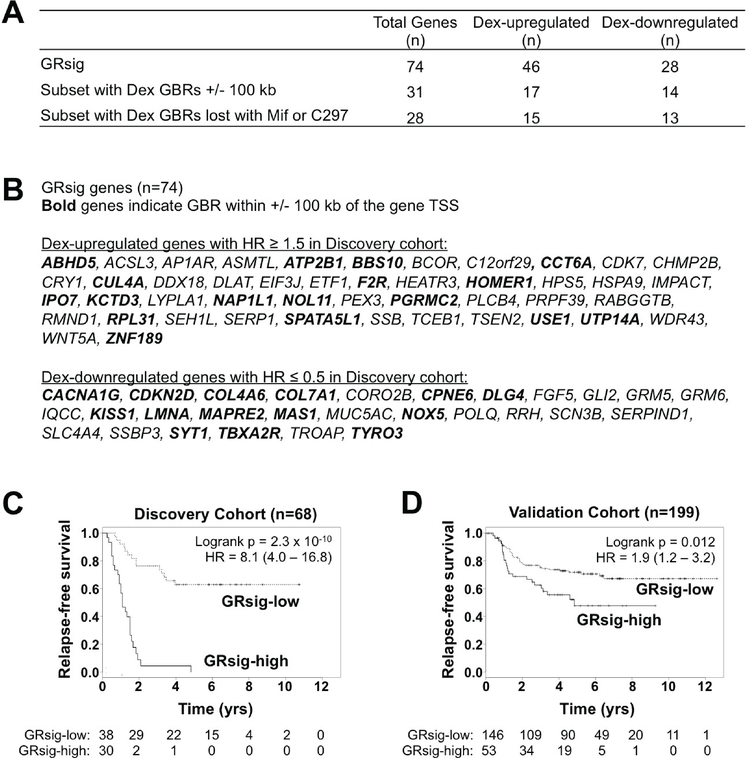
We formed a putative GR activity signature (GRsig) by selecting the most significantly RFS-associated genes using a stringent cut-off criteria including: RFS-associated p ≤ 1 × 10−5, and a HR ≥ 1.5 for GC-induced genes or HR ≤ 0.67 (1/1.5) for Dex-repressed genes (Figure 5 and Figure 6A). From this, we obtained an n=74 gene GRsig (Figure 6B and Supplementary Table 2). Of the genes in the GRsig, about 42% (n=31, were putative GR direct target genes (Figure 6A middle, and Supplementary Files 2 and 3). For these direct GRsig target genes, nearly all of the Dex GBRs (within +/− 100kb of each GRsig gene TSS) were lost upon addition of either Mif or C297 (Figure 6A bottom, Supplementary Files 2 and 3).
Figure 5. Identification schema for the GR activity signature (GRsig).
Genes that were Dex-regulated and inhibited at least 25% by Mif and C297 were identified in MDA-MB-231 cells (n=3,066). Next, the subset of genes also differentially expressed in the same direction in GR-high versus GR-low ER- negative BCs was identified (n=462). GR ChIP-seq determined putative GR direct target genes as having GR associated within 100kb of the gene TSS (n=232). A GR “activity signature” (GRsig) was identified based on their univariate association with RFS (HR ≥1.5 or HR ≤ 0.67; and p ≤ 1e-5) in the Discovery cohort of early-stage ER-negative BC patients with adjuvant chemotherapy. The GRsig comprised of n=74 genes which 1) included genes that were associated with poor RFS (HR ≥ 1.5) and were Dex- upregulated and 2) genes that were associated with improved RFS (HR ≤ 0.67) and were Dex- downregulated. This GRsig was applied to the Discovery and an independent Validation cohort of early- stage patients treated with adjuvant chemotherapy.
Figure 6. Patients with above-median expression of the 74-gene GR activity signature (GRsig) have significantly decreased relapse-free survival.
Genes in the GRsig were selected from among the n=462 tumor-relevant and antagonist-reversed Dex-regulated genes based on their univariate association with RFS in the Discovery cohort (HR ≥1.5 or HR ≤0.67; and p < 1e-5). (A) Summary of GRsig genes (top line) and their Dex-mediated up- and downregulation, and the subset of GRsig genes that are putative direct GR target genes (middle line) with loss of GR peak with Dex/antagonist treatment (bottom line). (B) List of individual GRsig genes, separated by their Dex-mediated up- or down-regulation with bolded gene names indicating putative direct GR target genes. (C, D) Kaplan-Meier estimates showing that the above-median GRsig expression (versus all others) is associated with RFS in both the Discovery and Validation cohorts.
We then compared RFS between ER-negative patients with high (above-median of all ER- negative BC patients) and low (below-median of all ER-negative BC patients) tumor GRsig expression in the same Discovery Cohort (n=68) from which the signature was derived. As expected, patients with high tumor GRsig expression had worse RFS (HR = 8.1; p = 2.3 × 10−10, Figure 6C). To validate this signature in another group of patients, we examined the GRsig in an external (non-overlapping) Validation Cohort of n=199 ER-negative BC early-stage and chemotherapy-treated patients (Supplementary Table 1). A Cox regression model revealed that patients with high tumor GRsig expression had significantly shorter time to relapse compared to those with low GRsig expression (HR= 1.9; p= 0.012, Figure 6D). Interestingly, the GRsig associated more significantly with poor RFS in the Validation Cohort compared to NR3C1 (GR) expression alone (Supplementary Figure 6). To determine if the GRsig was specific to ER-negative BCs, we split the GRsig expression at the median for n=1,024 ER-positive patients in our dataset published previously (30) and compared RFS of below and above the median tumor GRsig expression. We found no significant difference in RFS between the low- and high-GRsig expression groups, (p-value = 0.33, logrank test, Supplementary Figure 7). Taken together, these data imply that a GR signature derived from GR antagonist-reversed genes is a better indicator of pro-tumorigenic GR activity than GR expression alone. Secondly, the GRsig may stratify high-risk patients with ER-negative BC who would likely benefit from the addition of GR antagonists to standard chemotherapy.
Discussion
The identification of molecular targets that play a critical role in TNBC chemoresistance and recurrence is important for the development of more effective BC therapies. Given the diverse subtypes of TNBC, it seems unlikely that only one molecule will be a master regulator of poor prognosis. Recently, GR has been identified as an upstream regulator of important pro-oncogenic pathways through its ability to affect transcription and remodel chromatin. By immunohistochemistry, 40% of ER-negative BCs were reported to be GR-positive (at least 10% GR staining) (33). Previous reports from our laboratory (30) and others (25,33) have found a significant association between high tumor GR expression and shortened RFS in early-stage ER-negative BC patients, suggesting that GR-mediated regulation of gene expression contributes to chemotherapy resistance and shortened RFS. Because endogenous cortisol-activated GR is a transcriptional regulator of thousands of direct and indirect target genes that vary in individual cell types (45,46), identifying those GR-regulated genes that are most relevant to TNBC prognosis and treatment is challenging.
Both the GR/PR antagonist Mif (24) and the highly-selective GR antagonist C297 increase chemotherapy sensitivity in TNBC models. Here we asked whether improving chemotherapy efficacy occurs in association with antagonism of a specific subset of GR regulated genes. To define those GR targets that represent this tumor-relevant subset of GR activity, we identified GR target genes that were commonly inhibited by both C297 and Mif and further selected a subset that were also associated with high- versus low-GR expression in primary ER-negative BCs (filtering criteria shown in Figure 5). GR antagonists were powerful tools for identifying this important subset of genes because of their functional activity in increasing tumor chemosensitivity. We used primary tumor gene expression datasets to derive a 74-gene GRsig associated with shortened RFS in ER-negative BC patients who received adjuvant chemotherapy. This GRsig is hypothesized to select high-risk TNBC patients most likely to benefit from the addition of a GR antagonist to adjuvant chemotherapy.
While GR/NR3C1 cellular expression levels are predicted to correlate with GR activity (as has been shown for ER (47)), many factors contribute to an individual tumor’s GR activity level. The relative expression of nuclear receptor coregulators and cooperating transcription factors influence cell-type specific nuclear receptor activity (48,49). Other modifiers of GR activity include varying expression and activity of GR isoforms (50), post-translational GR modification (51), and the allosteric effects of chromatin landscape (52). These variables result in highly specific networks of GR target genes depending upon cellular context. For example, we previously reported that GR activation in ER+ BC increases the expression of pro-differentiating genes (31). However, as expected in this study of ER- negative BC, these pro-differentiating genes are not among the n=462 tumor-derived and GC-regulated genes shown in Figure 3C. The ER-negative GRsig derived here likely reflects gene expression specifically observed in early-stage ER-negative BCs.
Efforts to develop highly-selective, pharmacologically active GR antagonists have led to the discovery of several steroidal and non-steroidal chemical compounds. To determine clinical relevance, GR antagonists must be studied in the presence of endogenous GCs. Mechanisms of GR antagonists include both the displacement of cortisol from the GR LBD as well as active antagonism of GR-mediated transcription. Additionally, discovery of context-specific GR activity signatures can be used in the future to characterize novel GR modulators as “agonists” or “antagonists” in a cancer subtype-specific manner. Previously, GR modulators have been typically classified using in vitro binding assays and GR reporter gene assays. The resulting agonist/antagonist designation is therefore somewhat artificial, because GR modulation is entirely dependent on the context of a particular cell type.
A recently completed Phase-I clinical trial of Mif administered the day before and morning of administration of nab-paclitaxel to decrease anti-apoptotic tumor cell gene expression found that combining GR antagonism with chemotherapy appears to be safe and tolerable (34). A Phase-I clinical trial of the highly-selective GR antagonist CORT125134 [an azadecalin structurally related to C297(53)] in combination with nab-paclitaxel in solid tumors is currently underway (NCT02762981). Also, a Phase- II randomized clinical trial of Mif (versus placebo) with nab-paclitaxel in patients with advanced-stage TNBC has been recently activated (NCT02788981). While there is some concern that a potent GR antagonist might increase chemotherapy-induced side effects (Dex is used to reduce chemotherapy- associated nausea), thus far, the Phase-I studies only suggest a potential for increased cytopenias (34). This will be further investigated in an ongoing randomized Phase-II trial of nab-paclitaxel +/− Mif.
Similar to previous discovery methods for clinically useful gene expression panels [e.g. the 21- gene recurrence score for ER-positive BC (47)], the GRsig identified here includes retrospective tumor gene expression data. However, we also used potent GR antagonists (with known efficacy in TNBC models) to identify a subset of GR target genes also reversed by the antagonists. To begin to determine the predictive value of the GRsig in neoadjuvant TNBC and TNBC models, we evaluated the relative GR activity (via the GRsig) in n=64 individual patient-derived xenograft (PDX) tumors from n=13 TNBC patients enrolled in the Mayo Clinic BEAUTY trial (40,54). We found that tumors from the n=9 patients with pathological complete response (pCR) had a lower median GRsig expression than PDX tumors from n=4 non-pCR patients (Supplementary Figure 8). These preliminary data suggest that pCR TNBC has relatively low GR activity, consistent with decreased chemotherapy resistance and associated lower risk of relapse (55). In future studies, we will examine the high versus low GRsig-expressing PDX tumors for relative chemotherapy response +/− a GR antagonist, with the underlying hypothesis that GR antagonism will be effective in improving chemotherapy sensitivity in GRsig-high (i.e. relatively chemotherapy- resistant) PDX models. Ultimately, a randomized prospective clinical trial of neoadjuvant paclitaxel +/− a GR antagonist can allow testing the GRsig as a biomarker for improved outcome and RFS through addition of a GR antagonist to standard chemotherapy. The strong association of the GRsig identified here with recurrence risk in adjuvant chemotherapy-treated ER-negative BC suggests a path forward for identifying those patients at highest risk of relapse who are also more likely to benefit from selective GR antagonism.
Supplementary Material
Translational Relevance.
Glucocorticoid receptor (GR) expression is associated with poor prognosis in estrogen receptor (ER)- negative breast cancer (BC) patients. GR activation induces gene expression associated with therapy resistance and relapse, while GR antagonism improves chemotherapy sensitivity in models of triple- negative breast cancer (TNBC). After identifying antagonist-modulated GR target genes from cell line models, we uncovered a GR signature (GRsig) associated with risk of early recurrence despite adjuvant chemotherapy in ER-negative breast cancer. Derived from genes reversed by co-treatment with a GR antagonist, we predict the GRsig may be useful to identify high risk patients likely to benefit from adding GR antagonism to chemotherapy.
Acknowledgments
We thank Dr. Hazel Hunt and colleagues at Corcept Therapeutics for providing CORT108297, CORT118335, and CORT125134. We thank Dr. Pieter Faber and Dr. Jaejung Kim at The University of Chicago Genomics Core Facility, Ms. Terri Li at the University of Chicago Human Tissue Center, Dr. Doug Turnbull and Ms. Margaret Weitzman at the University of Oregon Genomics Core Facility. We also thank Dr. Kyle Hernandez and Dr. Hari Singhal for helpful bioinformatics input.
Funding: The study was supported by NIH R01 CA089208; NIH R01 CA196648; The University of Chicago Comprehensive Cancer Center NIH P30 CA014599; Susan G. Komen for the Cure IIR12223772; Prostate Cancer Foundation Movember Challenge Award; Virginia and D.K. Ludwig Fund for Cancer Research; NVKP_16–1-2016–0037 grant of the National Research, Development and Innovation Office, Hungary; Mayo Clinic Center for Individualized Medicine; Nadia’s Gift Foundation; John P. Guider; The Eveleigh Family; George M. Eisenberg Foundation for Charities; Mayo Clinic Cancer Center (CA15083–40A2); and Mayo Clinic Breast Specialized Program of Research Excellence (SPORE P50CA116201).
Footnotes
Competing interests: Drs. Kocherginsky and Conzen are listed as inventors on a patent issued “Methods and compositions related to glucocorticoid receptor (GR) antagonists and breast cancer,” which has been licensed to Corcept Therapeutics by The University of Chicago. There are no other conflicts of interest to report.
Data and materials availability: Affymetrix gene expression data and GR ChIP-sequencing data are available on NCBI GEO (GSE113571). We also received the compound CORT108297, CORT118335, and CORT125134 through an MTA with Corcept Therapeutics.
References and Notes:
- 1.Lehmann BD, Jovanovic B, Chen X, Estrada MV, Johnson KN, Shyr Y, et al. Refinement of Triple- Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS One 2016;11(6):e0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham TR, Yacoub R, Taliaferro-Smith L, Osunkoya AO, Odero-Marah VA, Liu T, et al. Reciprocal regulation of ZEB1 and AR in triple negative breast cancer cells. Breast Cancer Res Treat 2010;123(1):139–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature 2012;490(7418):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balko JM, Giltnane JM, Wang K, Schwarz LJ, Young CD, Cook RS, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov 2014;4(2):232–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barton VN, D’Amato NC, Gordon MA, Lind HT, Spoelstra NS, Babbs BL, et al. Multiple molecular subtypes of triple-negative breast cancer critically rely on androgen receptor and respond to enzalutamide in vivo. Mol Cancer Ther 2015;14(3):769–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stirzaker C, Zotenko E, Clark SJ. Genome-wide DNA methylation profiling in triple-negative breast cancer reveals epigenetic signatures with important clinical value. Mol Cell Oncol 2016;3(1):e1038424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 2016;529(7586):413–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner NC, Reis-Filho JS. Tackling the diversity of triple-negative breast cancer. Clin Cancer Res 2013;19(23):6380–8. [DOI] [PubMed] [Google Scholar]
- 9.Miranda TB, Morris SA, Hager GL. Complex genomic interactions in the dynamic regulation of transcription by the glucocorticoid receptor. Mol Cell Endocrinol 2013;380(1–2):16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt S, Rainer J, Ploner C, Presul E, Riml S, Kofler R. Glucocorticoid-induced apoptosis and glucocorticoid resistance: molecular mechanisms and clinical relevance. Cell Death Differ 2004;11 Suppl 1:S45–55. [DOI] [PubMed] [Google Scholar]
- 11.Wolff JE, Denecke J, Jurgens H. Dexamethasone induces partial resistance to cisplatinum in C6 glioma cells. Anticancer Res 1996;16(2):805–9. [PubMed] [Google Scholar]
- 12.Herr I, Ucur E, Herzer K, Okouoyo S, Ridder R, Krammer PH, et al. Glucocorticoid cotreatment induces apoptosis resistance toward cancer therapy in carcinomas. Cancer Res 2003;63(12):3112–20. [PubMed] [Google Scholar]
- 13.Zhang C, Kolb A, Buchler P, Cato AC, Mattern J, Rittgen W, et al. Corticosteroid co-treatment induces resistance to chemotherapy in surgical resections, xenografts and established cell lines of pancreatic cancer. BMC Cancer 2006;6:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C, Marme A, Wenger T, Gutwein P, Edler L, Rittgen W, et al. Glucocorticoid-mediated inhibition of chemotherapy in ovarian carcinomas. Int J Oncol 2006;28(2):551–8. [PubMed] [Google Scholar]
- 15.Goyeneche AA, Caron RW, Telleria CM. Mifepristone inhibits ovarian cancer cell growth in vitro and in vivo. Clin Cancer Res 2007;13(11):3370–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arora VK, Schenkein E, Murali R, Subudhi SK, Wongvipat J, Balbas MD, et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell 2013;155(6):1309–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isikbay M, Otto K, Kregel S, Kach J, Cai Y, Vander Griend DJ, et al. Glucocorticoid receptor activity contributes to resistance to androgen-targeted therapy in prostate cancer. Horm Cancer 2014;5(2):72–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stringer-Reasor EM, Baker GM, Skor MN, Kocherginsky M, Lengyel E, Fleming GF, et al. Glucocorticoid receptor activation inhibits chemotherapy-induced cell death in high-grade serous ovarian carcinoma. Gynecol Oncol 2015;138(3):656–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kroon J, Puhr M, Buijs JT, van der Horst G, Hemmer DM, Marijt KA, et al. Glucocorticoid receptor antagonism reverts docetaxel resistance in human prostate cancer. Endocr Relat Cancer 2016;23(1):35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kach J, Long TM, Selman P, Tonsing-Carter EY, Bacalao MA, Lastra RR, et al. Selective glucocorticoid receptor modulators (SGRMs) delay castrate-resistant prostate cancer growth. Mol Cancer Ther 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moran TJ, Gray S, Mikosz CA, Conzen SD. The glucocorticoid receptor mediates a survival signal in human mammary epithelial cells. Cancer Res 2000;60(4):867–72. [PubMed] [Google Scholar]
- 22.Wu W, Chaudhuri S, Brickley DR, Pang D, Karrison T, Conzen SD. Microarray analysis reveals glucocorticoid-regulated survival genes that are associated with inhibition of apoptosis in breast epithelial cells. Cancer Res 2004;64(5):1757–64. [DOI] [PubMed] [Google Scholar]
- 23.Pang D, Kocherginsky M, Krausz T, Kim SY, Conzen SD. Dexamethasone decreases xenograft response to Paclitaxel through inhibition of tumor cell apoptosis. Cancer Biol Ther 2006;5(8):933–40. [DOI] [PubMed] [Google Scholar]
- 24.Skor MN, Wonder EL, Kocherginsky M, Goyal A, Hall BA, Cai Y, et al. Glucocorticoid receptor antagonism as a novel therapy for triple-negative breast cancer. Clin Cancer Res 2013;19(22):6163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z, Lan X, Wu D, Sunkel B, Ye Z, Huang J, et al. Ligand-dependent genomic function of glucocorticoid receptor in triple-negative breast cancer. Nat Commun 2015;6:8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agyeman AS, Jun WJ, Proia DA, Kim CR, Skor MN, Kocherginsky M, et al. Hsp90 Inhibition Results in Glucocorticoid Receptor Degradation in Association with Increased Sensitivity to Paclitaxel in Triple- Negative Breast Cancer. Horm Cancer 2016;7(2):114–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z, Dong J, Zou T, Du C, Li S, Chen C, et al. Dexamethasone induces docetaxel and cisplatin resistance partially through up-regulating Kruppel-like factor 5 in triple-negative breast cancer. Oncotarget 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorrentino G, Ruggeri N, Zannini A, Ingallina E, Bertolio R, Marotta C, et al. Glucocorticoid receptor signalling activates YAP in breast cancer. Nat Commun 2017;8:14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Regan Anderson TM, Ma SH, Raj GV, Cidlowski JA, Helle TM, Knutson TP, et al. Breast Tumor Kinase (Brk/PTK6) Is Induced by HIF, Glucocorticoid Receptor, and PELP1-Mediated Stress Signaling in Triple- Negative Breast Cancer. Cancer Res 2016;76(6):1653–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan D, Kocherginsky M, Conzen SD. Activation of the glucocorticoid receptor is associated with poor prognosis in estrogen receptor-negative breast cancer. Cancer Res 2011;71(20):6360–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.West DC, Pan D, Tonsing-Carter EY, Hernandez KM, Pierce CF, Styke SC, et al. GR and ER Coactivation Alters the Expression of Differentiation Genes and Associates with Improved ER+ Breast Cancer Outcome. Mol Cancer Res 2016;14(8):707–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang F, Ma Q, Liu Z, Li W, Tan Y, Jin C, et al. Glucocorticoid Receptor:MegaTrans Switching Mediates the Repression of an ERalpha-Regulated Transcriptional Program. Mol Cell 2017;66(3):321–31 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abduljabbar R, Negm OH, Lai CF, Jerjees DA, Al-Kaabi M, Hamed MR, et al. Clinical and biological significance of glucocorticoid receptor (GR) expression in breast cancer. Breast Cancer Res Treat 2015;150(2):335–46. [DOI] [PubMed] [Google Scholar]
- 34.Nanda R, Stringer-Reasor EM, Saha P, Kocherginsky M, Gibson J, Libao B, et al. A randomized phase I trial of nanoparticle albumin-bound paclitaxel with or without mifepristone for advanced breast cancer. Springerplus 2016;5(1):947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clark RD, Ray NC, Williams K, Blaney P, Ward S, Crackett PH, et al. 1H-Pyrazolo[3,4-g]hexahydro- isoquinolines as selective glucocorticoid receptor antagonists with high functional activity. Bioorg Med Chem Lett 2008;18(4):1312–7. [DOI] [PubMed] [Google Scholar]
- 36.Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): explanation and elaboration. PLoS Med 2012;9(5):e1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015;31(14):2382–3. [DOI] [PubMed] [Google Scholar]
- 38.Mihaly Z, Kormos M, Lanczky A, Dank M, Budczies J, Szasz MA, et al. A meta-analysis of gene expression-based biomarkers predicting outcome after tamoxifen treatment in breast cancer. Breast Cancer Res Treat 2013;140(2):219–32. [DOI] [PubMed] [Google Scholar]
- 39.Li Q, Birkbak NJ, Gyorffy B, Szallasi Z, Eklund AC. Jetset: selecting the optimal microarray probe set to represent a gene. BMC Bioinformatics 2011;12:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu J, Qin B, Moyer AM, Sinnwell JP, Thompson KJ, Copland JA 3rd, et al. Establishing and characterizing patient-derived xenografts using pre-chemotherapy percutaneous biopsy and post- chemotherapy surgical samples from a prospective neoadjuvant breast cancer study. Breast Cancer Res 2017;19(1):130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hunt HJ, Ray NC, Hynd G, Sutton J, Sajad M, O’Connor E, et al. Discovery of a novel non-steroidal GR antagonist with in vivo efficacy in the olanzapine-induced weight gain model in the rat. Bioorg Med Chem Lett 2012;22(24):7376–80. [DOI] [PubMed] [Google Scholar]
- 42.De Bosscher K, Vanden Berghe W, Beck IM, Van Molle W, Hennuyer N, Hapgood J, et al. A fully dissociated compound of plant origin for inflammatory gene repression. Proc Natl Acad Sci U S A 2005;102(44):15827–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ronacher K, Hadley K, Avenant C, Stubsrud E, Simons SS Jr., Louw A, et al. Ligand-selective transactivation and transrepression via the glucocorticoid receptor: role of cofactor interaction. Mol Cell Endocrinol 2009;299(2):219–31. [DOI] [PubMed] [Google Scholar]
- 44.Nawaz Z, Lonard DM, Dennis AP, Smith CL, O’Malley BW. Proteasome-dependent degradation of the human estrogen receptor. Proc Natl Acad Sci U S A 1999;96(5):1858–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Haqq C, Yamamoto KR. Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc Natl Acad Sci U S A 2004;101(44):15603–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J, Kinyamu HK, Archer TK. Changes in attitude, changes in latitude: nuclear receptors remodeling chromatin to regulate transcription. Mol Endocrinol 2006;20(1):1–13. [DOI] [PubMed] [Google Scholar]
- 47.Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, et al. Prospective Validation of a 21-Gene Expression Assay in Breast Cancer. N Engl J Med 2015;373(21):2005–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Desmet SJ, Dejager L, Clarisse D, Thommis J, Melchers D, Bastiaensen N, et al. Cofactor profiling of the glucocorticoid receptor from a cellular environment. Methods Mol Biol 2014;1204:83–94. [DOI] [PubMed] [Google Scholar]
- 49.Chodankar R, Wu DY, Schiller BJ, Yamamoto KR, Stallcup MR. Hic-5 is a transcription coregulator that acts before and/or after glucocorticoid receptor genome occupancy in a gene-selective manner. Proc Natl Acad Sci U S A 2014;111(11):4007–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duma D, Jewell CM, Cidlowski JA. Multiple glucocorticoid receptor isoforms and mechanisms of post- translational modification. J Steroid Biochem Mol Biol 2006;102(1–5):11–21. [DOI] [PubMed] [Google Scholar]
- 51.Galliher-Beckley AJ, Williams JG, Cidlowski JA. Ligand-independent phosphorylation of the glucocorticoid receptor integrates cellular stress pathways with nuclear receptor signaling. Mol Cell Biol 2011;31(23):4663–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Love MI, Huska MR, Jurk M, Schopflin R, Starick SR, Schwahn K, et al. Role of the chromatin landscape and sequence in determining cell type-specific genomic glucocorticoid receptor binding and gene regulation. Nucleic Acids Res 2017;45(4):1805–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hunt HJ, Belanoff JK, Walters I, Gourdet B, Thomas J, Barton N, et al. Identification of the Clinical Candidate (R)-(1-(4-Fluorophenyl)-6-((1-methyl-1H-pyrazol-4-yl)sulfonyl)-4,4a,5,6,7,8-hexah ydro-1H- pyrazolo[3,4-g]isoquinolin-4a-yl)(4-(trifluoromethyl)pyridin-2-yl)methano ne (CORT125134): A Selective Glucocorticoid Receptor (GR) Antagonist. J Med Chem 2017;60(8):3405–21. [DOI] [PubMed] [Google Scholar]
- 54.Goetz MP, Kalari KR, Suman VJ, Moyer AM, Yu J, Visscher DW, et al. Tumor Sequencing and Patient- Derived Xenografts in the Neoadjuvant Treatment of Breast Cancer. J Natl Cancer Inst 2017;109(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Symmans WF, Wei C, Gould R, Yu X, Zhang Y, Liu M, et al. Long-Term Prognostic Risk After Neoadjuvant Chemotherapy Associated With Residual Cancer Burden and Breast Cancer Subtype. J Clin Oncol 2017;35(10):1049–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.