Abstract
Purpose
To perform the first meta-analysis of the performance of the genomic classifier test, Decipher, in men with prostate cancer postprostatectomy.
Methods
MEDLINE, EMBASE, and the Decipher genomic resource information database were searched for published reports between 2011 and 2016 of men treated by prostatectomy that assessed the benefit of the Decipher test. Multivariable Cox proportional hazards models fit to individual patient data were performed; meta-analyses were conducted by pooling the study-specific hazard ratios (HRs) using random-effects modeling. Extent of heterogeneity between studies was determined with the I2 test.
Results
Five studies (975 total patients, and 855 patients with individual patient-level data) were eligible for analysis, with a median follow-up of 8 years. Of the total cohort, 60.9%, 22.6%, and 16.5% of patients were classified by Decipher as low, intermediate, and high risk, respectively. The 10-year cumulative incidence metastases rates were 5.5%, 15.0%, and 26.7% (P < .001), respectively, for the three risk classifications. Pooling the study-specific Decipher HRs across the five studies resulted in an HR of 1.52 (95% CI, 1.39 to 1.67; I2 = 0%) per 0.1 unit. In multivariable analysis of individual patient data, adjusting for clinicopathologic variables, Decipher remained a statistically significant predictor of metastasis (HR, 1.30; 95% CI, 1.14 to 1.47; P < .001) per 0.1 unit. The C-index for 10-year distant metastasis of the clinical model alone was 0.76; this increased to 0.81 with inclusion of Decipher.
Conclusion
The genomic classifier test, Decipher, can independently improve prognostication of patients postprostatectomy, as well as within nearly all clinicopathologic, demographic, and treatment subgroups. Future study of how to best incorporate genomic testing in clinical decision-making and subsequent treatment recommendations is warranted.
INTRODUCTION
Stratifying patients’ risk of localized prostate cancer (PCa) continues to be clinically challenging.1,2 Fundamentally, National Comprehensive Cancer Network risk stratification is based on the serum prostate-specific antigen (PSA) level, staging primarily from a digital rectal examination, and the Gleason score (originally described approximately 40 years ago).3 Growing molecular evidence has raised discussion around the solitary use of routine clinicopathologic risk factors and nomograms for predicting disease progression.4,5
Decipher (GenomeDx Biosciences, Vancouver, British Columbia, Canada) is a 22-gene genomic classifier that has been developed to aid in prognostication of patients who have undergone radical prostatectomy (RP).6 It has been shown that the Decipher post-RP test discriminates risk of metastasis and PCa-specific mortality, improves accuracy of risk stratification above and beyond clinicopathologic risk factors or commonly used models, and impacts physician and patient post-RP treatment decisions.6-15 However, the cohort size and event rate in the previous validation studies did not allow for a thorough investigation into the performance of Decipher, especially within individual clinicopathologic, demographic, or treatment subgroups.
Therefore, the present meta-analysis with individual patient-level genomic and clinicopathologic data was conducted to better understand the performance of Decipher to prognosticate risk of metastases. Additionally, by pooling studies, we aimed to explore the correlation and performance of Decipher in multiple clinically relevant subgroups within a multi-institutional and multiethnic cohort of patients with adverse pathology at time of RP.
MATERIALS AND METHODS
Study Selection
Systematic literature searches were performed (July 1, 2016) using two databases (MEDLINE [via PubMed] and EMBASE) for all studies published from January 1, 2011, through July 1, 2016. Controlled vocabulary was used in the formation of the search strategy.
The search strategy contained two major components linked together with the AND operator: (1) genomic classifier AND (2) prostate. All search results were imported into a bibliographic management tool and duplicates were removed. Then, leveraging the Decipher genomic resource information database, 1,692 patients from seven independent studies of the Decipher test who underwent RP were identified.6-12 These studies were all present in the previous bibliographic search strategy and duplicates were removed electronically.
Data extraction.
Two investigators (K.Y. and D.E.S) independently reviewed each manuscript. Studies eligible for inclusion were required to (1) have used the Decipher genomic test, (2) been performed in men postprostatectomy, (3) have assessed rates of development of metastatic disease, and (4) include only patients who reached an undetectable PSA level after surgery (Fig 1). Of the seven eligible studies, patients from Erho et al6 were excluded because this study was used to discover the Decipher genomic classifier. In addition, the Klein et al9 study was excluded because of its case-control design, which is not compatible with survival analysis (Appendix Table A1, online only). After application of these requirements, five studies remained, comprising 975 patients. Patients who were not randomly selected from the case-cohort studies (n = 120) were excluded from the individual patient-level analysis to avoid bias in estimation of the hazard ratios [HRs].7,10,16 Individual patient genomic and clinicopathologic data were gathered from each study group (n = 855) after institutional review boards at the participating institutions approved the research protocol under which the data were collected. Finally, the genomic data from all prior published studies were deposited into the genomic resource information database.7,8,10-12 Approval to obtain long-term clinical outcomes data were obtained from each study’s authors, and these data were then linked to the genomic and clinical information.7,8,10-12 Data extraction and study selection followed the PRISMA-IPD (Preferred Reporting Items for a Systematic Review and Meta-analysis of Individual Participant Data) statement.17
Fig 1.
Preferred Reporting Items for Systematic Reviews and Meta-Analyses diagram of the study selection process. IPD, individual patient data.
End points.
The primary end point of the study was to determine the performance of Decipher using individual patient data (n = 855) to predict time to regional or distant metastases on multivariable analyses while adjusting for pre-RP PSA level, RP Gleason score, margin status, extracapsular extension (ECE), seminal vesical invasion (SVI), and lymph node invasion (LNI). Metastasis end point was defined as either bone, viscera, or LN metastasis documented radiographically by computed tomography or bone scan. The secondary end point was to assess the performance of Decipher to predict time to metastases by pooling study-specific HRs across all five studies (N = 975).
Preplanned analyses using the individual patient level data (n = 855) included (1) determining the correlation of Decipher with pre-RP PSA, RP Gleason score, margin status, ECE, SVI, and LNI; and (2) determining the ability of Decipher to prognosticate time to regional or distant metastases in subgroups by race, pre-RP PSA, RP Gleason score, margin status, ECE, SVI, LNI, and adjuvant/salvage treatment modality (androgen deprivation therapy [ADT] or radiation therapy [RT]).
Patients received either no postoperative treatment before metastasis onset or were treated with adjuvant RT, salvage RT, adjuvant ADT, or salvage ADT. Adjuvant RT and salvage RT were defined by initiation of therapy at PSA levels of < 0.2 and ≥ 0.2 ng/mL, respectively. Adjuvant RT or salvage RT was performed using three-dimensional conformal RT or intensity modulated radiation therapy. Adjuvant ADT and salvage ADT were defined by initiation of therapy within 12 months and after 12 months post-RP, respectively.
Calculation of Decipher Score
The expression values for the 22 prespecified biomarkers that constitute Decipher were extracted from the normalized data matrix and entered into the random forest algorithm that was locked with defined tuning and weighting parameters as previously described.6 The Decipher score read-out is a continuous score between 0 and 1, with higher scores indicating greater risk of metastasis. Previously established and locked cut points of 0.45 and 0.60 were used to categorize patients into low-, intermediate-, and high-risk groups.10,12
Statistical Analysis
Association of Decipher with individual clinicopathologic variables was assessed using the Spearman rank correlation coefficient. In time-to-event analyses, event times were defined as the time from RP to metastasis. Cumulative incidence curves were constructed using Fine-Gray competing risks analysis to estimate the risk of metastasis over time, with deaths from other causes as a competing risk.18 Time-dependent C indices were constructed using the approach described by Heagerty et al.19 The C index of the combined model was estimated by subjecting the model to bootstrapping with 100 resamples. Analysis of individual patient data used stratified Cox univariable analysis (UVA) and multivariable analysis (MVA) proportional hazards models to evaluate the association of Decipher with time to metastasis, where deaths from other causes was considered as a competing risk.20 In these analyses, institution was modeled as a stratification variable to allow for variation of underlying hazard functions by stratification levels due to the varying patient populations and referral patterns.21 In a sensitivity analysis, we fitted an MVA Cox proportional hazards model adjusting for RT and ADT as time-dependent covariates. Study-specific HRs were pooled using a random effects model according to the inverse variance method described by DerSimonian and Laird.22 Heterogeneity between studies was assessed using the I2 statistic. Egger’s regression test was used to evaluate publication bias. All statistical tests were two-sided and analyses were performed in R version 3.1 (R Foundation, Vienna, Austria).
RESULTS
Study and Patient Characteristics
Patient and study selections following the Preferred Reporting Items for a Systematic Review and Meta-analysis of Individual Participant Data statement are shown in Fig 1. Of the 855 patients with individual patient data, the median follow-up time for censored patients was 8 years (interquartile range [IQR], 5 to 11 years). Patients were treated with RP between 1990 and 2010. The median age of patients at the time of RP was 60 years (IQR, 55 to 65 years); 41.9% had ECE, 27.8% had SVI, and 26% had an RP Gleason score of 8 to 10 (Table 1). Overall, 51.3% of the patients received only prostatectomy with no additional second-line therapy.
Table 1.
Demographic and Clinical Characteristics of Included Patients and Studies
The median Decipher score for the cohort (n = 855) was 0.37 (IQR, 0.24 to 0.54). Of these patients, 60.9% (n = 520), 22.6% (n = 193), and 16.5% (n = 141) were categorized as being at, respectively, low, intermediate, and high risk by Decipher.
Correlation of Decipher With Clinicopathologic Features
Decipher was significantly, albeit only low to moderately, correlated with RP Gleason score (r = 0.27), EPE (r = 0.20), SVI (r = 0.19), and LNI (r = 0.13; all P < .001). Decipher was not significantly correlated with preoperative PSA or surgical margin status (Fig 2).
Fig 2.
Correlation of the genomic-risk score (Decipher Score) to the following clinicopathologic variables: (A) preoperative prostate-specfic antigen (PSA), (B) radical prostatectomy (RP) Gleason score, (C) surgical margins, (D) extracapsular extension, (E) seminal vesicle invasion, and (F) lymph node invasion.
Decipher As a Predictor of Metastasis
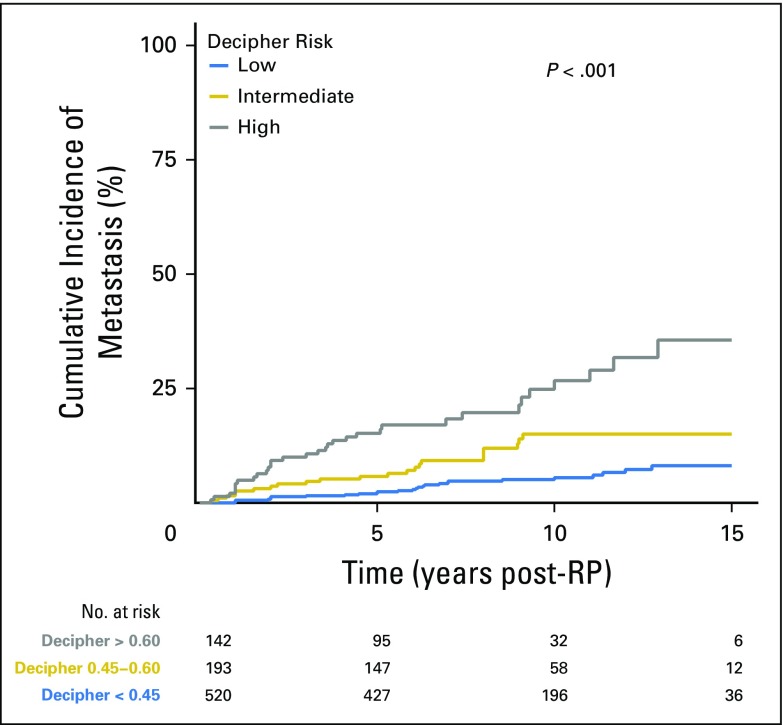
During the study, 82 patients experienced metastasis. Cumulative incidence curves demonstrated that Decipher categories significantly stratified risk of metastasis (P < .001; Fig 3). Patients categorized as low, intermediate, and high risk by Decipher had a 5-year cumulative incidence of metastasis of 2.4%, 5.8%, and 15.2%, respectively; and a 10-year rate of 5.5%, 15.0% and 26.7%, respectively.
Fig 3.
Individual patient-level analysis for the cumulative incidence of metastasis over time stratified by genomic-risk groups (also referred to as the Decipher risk categories): low, intermediate, and high. RP, radical prostatectomy.
On UVA, all variables except pre-RP PSA showed significant association with time to metastasis (Table 2). On MVA, Decipher remained a significant predictor of metastasis (P < .001). Decipher had a HR of 1.30 (95% CI, 1.14 to 1.47) per 0.1 unit increase in score. Analyzing Decipher as a categorical variable (low risk as reference), Decipher high-risk patients had the greatest hazard for metastases (HR, 3.31; 95% CI, 1.86 to 5.88; P < .001), even compared with Gleason score 8-10 (Gleason ≤ 3+4 as reference; HR 3.23; 95% CI, 1.75 to 5.93; P < .001). In a sensitivity analysis, adjusting for RT and ADT as time-dependent covariates, Decipher remained a significant predictor of metastasis (Appendix Table A2, online only). The C-index for 10-year distant metastases of the clinical model alone was 0.76 and increased to 0.81 with inclusion of Decipher.
Table 2.
Univariable and Multivariable Analysis of Metastasis Including Decipher and Clinicopathologic Risk Factors
Pooling study-specific HRs across the five studies (N = 975) demonstrated that the Decipher score was significantly associated with time to metastasis (HR, 1.52; 95% CI, 1.39 to 1.67) per 0.1-unit increase; I2 = 0%; Fig 4A).
Fig 4.
Forest plot of the Decipher score’s hazard ratio for metastasis for (A) the study-specific hazard ratio pooled across all five studies, and (B) individual patient data in preplanned demographic, treatment, and clinicopathologic subgroups. Hazard ratios are reported per 0.1 unit increase in Decipher score. Weights in (A) were calculated based on the inverse variance formulation by DerSimonian and Laird.22 Higher weights were assigned to studies with larger sample size and event rate. ADT, androgen deprivation therapy; PSA, prostate-specific antigen; RP, radical prostatectomy; RT, radiation therapy.
Performance of Decipher Within Individual Subgroups
Decipher significantly predicted risk of metastasis irrespective of pre-RP PSA levels, RP Gleason score, surgical margin status, and ECE, SVI, and LNI status (Fig 4B; Appendix Figs A1A-A1C, online only). Additionally, Decipher was associated with risk of metastasis in subgroups of white men, men treated with RP alone, those treated with RP and salvage RT, and those treated with RP and ADT (Fig 4B). For black patients (n = 106) and those of any race treated with adjuvant RT (n = 140) or ADT (n = 44), Decipher approached, but did not reach, statistical significance for predicting risk of metastasis (HR, 1.43 [95% CI, 0.95 to 2.15]; HR, 1.86 [95% CI, 0.92 to 5.62]; and HR 1.52 [95% CI, 0.97 to 2.39], respectively).
Assessment of Publication Bias
A funnel plot of the five included studies demonstrated excellent symmetry (Appendix Fig A2, online only). In this analysis, Egger regression test resulted in a P value of .60, indicating a low probability of publication bias (Appendix Fig 2).
DISCUSSION
Accurately understanding the risk of recurrence after initial therapy for cancer is critical to determine goals of care, therapeutic recommendations, and the design of future clinical trials. Men with PCa often have a long natural history after RP or definitive RT, even for disease categorized as high risk according to the National Comprehensive Cancer Network, with an estimated median time to metastatic disease of 5 to 8 years after biochemical recurrence.23,24 However, the subset of men who will eventually develop metastatic disease is poorly understood and not fully captured by clinicopathologic variables. For this reason, there have been efforts to develop companion genomic tests to provide clarity to classic phenotypic risk factors (ie, Gleason score, PSA level, and T stage) and add independent prognostic value.
Over the past several years, one of the commercially available genomic tests (Decipher) has been externally validated as a prognostic tool to predict time to metastasis in multiple independent studies.6-12 However, the sample size of each individual study limited the ability to robustly assess the correlation of Decipher with clinicopathologic risk factors, to assess the overall performance of Decipher above standard clinicopathologic risk factors, and to determine the performance of Decipher within relevant individual clinicopathologic risk groups. Our analysis provides numerous findings into the association of genomic and clinicopathologic risk and demonstrates how they are complementary and impart unique biologic information.
First, Decipher has a low-to-moderate correlation with RP Gleason score, ECE, SVI, and LNI. Additionally, there was no correlation of pre-RP PSA nor surgical margin status with Decipher score. These results demonstrate that select clinicopathologic variables trend with Decipher scores; however, there is a wide overlapping distribution of Decipher scores within each subset (ie, Gleason ≤ 3+4 v ≥ 8; Fig 2). Furthermore, it is rational that surgical margin status, a function partially dependent on the surgical procedure rather than the intrinsic biology of the disease, would not correlate with the genomic classifier results.
Second, in the present cohort of men with adverse pathology after RP, 61% were classified as low risk by Decipher, whereas only 17% were classified as high risk. Metastases rates could readily be discriminated by the Decipher score (5 year: 2.4% v 15.2% for low and high Decipher score, respectively; P < .001). Importantly, this prognostication was maintained on MVA adjusting for pre-RP PSA, RP Gleason score, margin status, ECE, SVI, and LNI (HR, 1.30 for each 0.1 increase in Decipher score). Although clinicopathologic variables perform reasonably well to predict who is at very low or very high risk of recurrence, Decipher independently improves upon this to further discriminate metastatic risk within these clinical risk groups. This observation has important implications for designing clinical trials for men with high-risk disease; use of Decipher as an entry criterion or for stratification would enrich the study population for meaningful clinical events (metastasis), thereby increasing the event rate, decreasing the needed sample size, and perhaps shortening trial length.
Last, given the large sample size of the present meta-analysis, we were able to test the prognostic benefit of Decipher in numerous preplanned analyses of distinct clinicopathologic subgroups. Decipher improved the ability to predict the cumulative incidence of metastases in nearly all subgroups based on clinicopathologic factors, treatment factors, and demographic factors. All subgroups reached statistical significance except the adjuvant RT subgroup (HR, 2.25; 95% CI, 0.90 to 5.62; P = .082) and the black race subgroup (HR, 1.43; 95% CI, 0.95 to 2.15; P = .087). Both exhibited elevated HRs associated with Decipher, and the marginally nonsignificant tests may reflect the small sample size of the subgroups. Given the recently demonstrated disparities that exist for minorities undergoing genomic sequencing, it will be important to have dedicated efforts to increase tumor profiling for racial minorities.25 Despite this, it appears that the added benefit of using the Decipher genomic classifier does not appear to be limited to a particular subgroup; rather, it appears to add consistent benefit across all subgroups (HR of all subgroups, 1.24 to 2.25).
The strength of our study is driven by the meta-analysis methodology with individual patient-level genomic and clinicopathologic data from large multi-institutional cohorts, use of a meaningful survival end point (ie, metastasis), and long-term follow-up. However, this study is not without limitations. First, the analysis is subject to the limitations associated with the retrospective design of the individual studies (Appendix Table A3, online only). Second, adjuvant and salvage treatments were not randomized and thus differed based on institutional and patient preferences. Third, our selection criteria included only patients with adverse clinicopathologic features, thus utility of Decipher in lower-risk PCa remains to be investigated. Fourth, use of imaging for restaging post-treatment was not standardized and, therefore, ascertainment bias is a potential limitation of the current analyses. Last, there are further potential social (eg, insurance and socioeconomic) factors, patient-level factors (eg, comorbidities), treatment factors (eg, duration of ADT and RT dose/field size), and tumor level factors (eg, PSA kinetics) that were not accounted for but could impact our results.
In conclusion, this study was performed to evaluate the performance of Decipher to predict metastases in men with PCa after RP. The results suggest that though Decipher only moderately correlates with clinicopathologic variables, it independently adds prognostic benefit over routine clinicopathologic variables to predict metastases and appears to add benefit across a number of clinically relevant subgroups. Decipher should be considered an additive validated test to improve prognostication in high-risk men after RP and to aid clinical decision-making and future clinical trial design.
Appendix
Fig A1.
Individual patient level analyses for the cumulative incidence of metastasis over time stratified by Decipher risk categories (low, intermediate, and high) among patients with (A) radical prostatectomy (RP) Gleason score ≤ 3+4, (B) RP Gleason score 4+3, and (C) RP Gleason score 8 to 10.
Fig A2.
Funnel plot for publication bias.
Table A1.
Demographic and Clinical Characteristics of Excluded Patients and Studies
Table A2.
Multivariable Analysis of Decipher and Clinicopathologic Risk Factors Adjusting for Radiation Treatment and Androgen Deprivation Therapy as Time-Dependent Covariates
Table A3.
National Institutes of Health Level of Evidence Scale*

Footnotes
Funded in part by the Prostate Cancer Foundation Young Investigator Award (to D.E.S).
See accompanying Editorial on page 1976
AUTHOR CONTRIBUTIONS
Conception and design: Daniel E. Spratt, Kasra Yousefi, Elai Davicioni, Eric A. Klein, R. Jeffrey Karnes, Felix Y. Feng
Administrative support: Elai Davicioni, Andrew G. Glass, Adam P. Dicker,
Provision of study materials or patients: Elai Davicioni
Collection and assembly of data: Daniel E. Spratt, Kasra Yousefi, Ashley E. Ross, Robert B. Den, Edward M. Schaeffer, Bruce J. Trock, Lucia L.C. Lam, Marguerite du Plessis, Zaid Haddad, Christine Buerki, Elai Davicioni, Sheila Weinmann, Eric A. Klein, R. Jeffrey Karnes, Felix Y. Feng
Data analysis and interpretation: Daniel E. Spratt, Kasra Yousefi, Samineh Deheshi, Bruce J. Trock, Jingbin Zhang, Firas Abdollah, Shuang G. Zhao, Lucia L.C. Lam, Voleak Choeurng, Elai Davicioni, Sheila Weinmann, Stephen J. Freedland, Felix Y. Feng
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Individual Patient-Level Meta-Analysis of the Performance of the Decipher Genomic Classifier in High-Risk Men After Prostatectomy to Predict Development of Metastatic Disease
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Daniel E. Spratt
No relationship to disclose
Kasra Yousefi
Employment: GenomeDx Biosciences
Travel, Accommodations, Expenses: GenomeDx Biosciences
Samineh Deheshi
Employment: GenomeDx Biosciences
Ashley E. Ross
Stock or Other Ownership: GenomeDx Biosciences
Robert B. Den
Consulting or Advisory Role: GenomeDx Biosciences
Speakers' Bureau: Bayer
Research Funding: Medivation/Astellas, GenomeDx Biosciences
Travel, Accommodations, Expenses: GenomeDx Biosciences
Edward M. Schaeffer
Consulting or Advisory Role: GenomeDx Biosciences
Bruce J. Trock
Consulting or Advisory Role: Champions Oncology, GenomeDx Biosciences
Research Funding: Myriad Genetics
Travel, Accommodations, Expenses: GenomeDx Biosciences
Jingbin Zhang
Employment: GenomeDx Biosciences
Andrew G. Glass
No relationship to disclose
Adam P. Dicker
Consulting or Advisory Role: Merck, Glenview Consulting, GenomeDx Biosciences, Johnson & Johnson
Travel, Accommodations, Expenses: Bayer, Merck
Other Relationship: NRG Oncology, Department of Defense-Prostate Cancer Research Program
Firas Abdollah
Consulting or Advisory Role: GenomeDx Biosciences
Speakers' Bureau: GenomeDx Biosciences
Travel, Accommodations, Expenses: GenomeDx Biosciences
Shuang G. Zhao
Employment: PFS Genomics (I)
Patents, Royalties, Other Intellectual Property: Compositions and methods for the analysis of radiosensitivity (Inst), Treatment of cancer with DNAPK inhibitors (Inst), 24-gene predictor of response to radiation therapy (Inst)
Travel, Accommodations, Expenses: GenomeDx Biosciences
Lucia L.C. Lam
Employment: GenomeDx Biosciences
Marguerite du Plessis
Employment: GenomeDx Biosciences
Voleak Choeurng
Employment: GenomeDx Biosciences
Zaid Haddad
Employment: GenomeDx Biosciences
Christine Buerki
Employment: GenomeDx Biosciences
Stock or Other Ownership: GenomeDx Biosciences
Elai Davicioni
Employment: GenomeDx Biosciences
Leadership: GenomeDx Biosciences
Stock or Other Ownership: GenomeDx Biosciences
Patents, Royalties, Other Intellectual Property: Cancer diagnostics using biomarkers 20140066323
Travel, Accommodations, Expenses: GenomeDx Biosciences
Sheila Weinmann
No relationship to disclose
Stephen J. Freedland
Stock or Other Ownership: Armune Bioscience, Parallel6
Consulting or Advisory Role: Astellas Pharma, Medivation, Janssen Biotech, Dendreon, Sanofi, Bayer, GenomeDx Biosciences, Armune Bioscience, Parallel6
Research Funding: Dendreon (Inst), Bayer (Inst), Amgen (Inst), Janssen Biotech (Inst), Myriad Genetics (Inst), GenomeDx Biosciences (Inst), Progenika (Inst)
Travel, Accommodations, Expenses: Sanofi, GenomeDx Biosciences
Eric A. Klein
Consulting or Advisory Role: Berg, GenomeDx Biosciences
Speakers' Bureau: Genomic Health, GenomeDx Biosciences
R. Jeffrey Karnes
Research Funding: GenomeDx Biosciences
Travel, Accommodations, Expenses: GenomeDx Biosciences
Felix Y. Feng
Leadership: PFS Genomics
Consulting or Advisory Role: Medivation/Astellas, GenomeDx Biosciences, Dendreon, Sanofi, EMD Serono
Research Funding: Varian Medical Systems, Celgene
REFERENCES
- 1.Eggener SE, Scardino PT, Walsh PC, et al. Predicting 15-year prostate cancer specific mortality after radical prostatectomy. J Urol. 2011;185:869–875. doi: 10.1016/j.juro.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. doi: 10.1016/j.eururo.2015.04.016. Ross AE, Yousefi K, Davicioni E, et al: Utility of risk models in decision making after radical prostatectomy: Lessons from a natural history cohort of intermediate- and high-risk men. Eur Urol 69:496-504, 2016. [DOI] [PubMed] [Google Scholar]
- 3.Carroll PR, Parsons JK, Andriole G, et al. NCCN guidelines insights: Prostate Cancer Early Detection, Version 2.2016. J Natl Compr Canc Netw. 2016;14:509–519. doi: 10.6004/jnccn.2016.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross AE, D’Amico A V, Freedland SJ. Which, when and why? Rational use of tissue-based molecular testing in localized prostate cancer. Prostate Cancer Prostatic Dis. 2016;19:1–6. doi: 10.1038/pcan.2015.31. [DOI] [PubMed] [Google Scholar]
- 5.Moschini M, Spahn M, Mattei A, et al. Incorporation of tissue-based genomic biomarkers into localized prostate cancer clinics. BMC Med. 2016;14:67. doi: 10.1186/s12916-016-0613-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erho N, Crisan A, Vergara IA, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One. 2013;8:e66855. doi: 10.1371/journal.pone.0066855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karnes RJ, Bergstralh EJ, Davicioni E, et al. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol. 2013;190:2047–2053. doi: 10.1016/j.juro.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Den RB, Feng FY, Showalter TN, et al. Genomic prostate cancer classifier predicts biochemical failure and metastases in patients after postoperative radiation therapy. Int J Radiat Oncol Biol Phys. 2014;89:1038–1046. doi: 10.1016/j.ijrobp.2014.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein EA, Yousefi K, Haddad Z, et al. A genomic classifier improves prediction of metastatic disease within 5 years after surgery in node-negative high-risk prostate cancer patients managed by radical prostatectomy without adjuvant therapy. Eur Urol. 2015;67:778–786. doi: 10.1016/j.eururo.2014.10.036. [DOI] [PubMed] [Google Scholar]
- 10.Ross AE, Johnson MH, Yousefi K, et al. Tissue-based genomics augments post-prostatectomy risk stratification in a natural history cohort of intermediate- and high-risk men. Eur Urol. 2016;69:157–165. doi: 10.1016/j.eururo.2015.05.042. [DOI] [PubMed] [Google Scholar]
- 11. doi: 10.1016/j.juro.2015.11.044. Glass AG, Leo MC, Haddad Z, et al: Validation of a genomic classifier for predicting post-prostatectomy recurrence in a community based health care setting. J Urol 195:1748-1753, 2016. [DOI] [PubMed]
- 12. doi: 10.1016/j.eururo.2016.01.008. Freedland SJ, Choeurng V, Howard L, et al: Utilization of a genomic classifier for prediction of metastasis following salvage radiation therapy after radical prostatectomy. Eur Urol 70:588-596, 2016. [DOI] [PubMed] [Google Scholar]
- 13. doi: 10.1185/03007995.2014.919908. Michalopoulos SN, Kella N, Payne R, et al: Influence of a genomic classifier on post-operative treatment decisions in high-risk prostate cancer patients: results from the PRO-ACT study. Curr Med Res Opin 30:1546-1556, 2014. [DOI] [PubMed]
- 14.Nguyen PL, Shin H, Yousefi K, et al. Impact of a genomic classifier of metastatic risk on postprostatectomy treatment recommendations by radiation oncologists and urologists. Urology. 2015;86:35–40. doi: 10.1016/j.urology.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Ross AE, Den RB, Yousefi K, et al. Efficacy of post-operative radiation in a prostatectomy cohort adjusted for clinical and genomic risk. Prostate Cancer Prostatic Dis. 2016;19:277–282. doi: 10.1038/pcan.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barlow WE, Ichikawa L, Rosner D, et al. Analysis of case-cohort designs. J Clin Epidemiol. 1999;52:1165–1172. doi: 10.1016/s0895-4356(99)00102-x. [DOI] [PubMed] [Google Scholar]
- 17.Stewart LA, Clarke M, Rovers M, et al. Preferred Reporting Items for Systematic Review and Meta-Analyses of individual participant data: The PRISMA-IPD Statement. JAMA. 2015;313:1657–1665. doi: 10.1001/jama.2015.3656. [DOI] [PubMed] [Google Scholar]
- 18.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94:496–509. [Google Scholar]
- 19.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–344. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 20.Gerds TA, Scheike TH, Andersen PK. Absolute risk regression for competing risks: Interpretation, link functions, and prediction. Stat Med. 2012;31:3921–3930. doi: 10.1002/sim.5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Therneau TM, Grambsch PM: Modeling Survival Data: Extending the Cox Model. New York, NY, Springer, 2000: p 44. [Google Scholar]
- 22.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 23.Pound CR, Partin AW, Eisenberger MA, et al. Natural history of progression after PSA elevation following radical prostatectomy. JAMA. 1999;281:1591–1597. doi: 10.1001/jama.281.17.1591. [DOI] [PubMed] [Google Scholar]
- 24.Zumsteg ZS, Spratt DE, Romesser PB, et al. The natural history and predictors of outcome following biochemical relapse in the dose escalation era for prostate cancer patients undergoing definitive external beam radiotherapy. Eur Urol. 2015;67:1009–1016. doi: 10.1016/j.eururo.2014.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spratt DE, Chan T, Waldron L, et al. Racial/ethnic disparities in genomic sequencing. JAMA Oncol. 2016;2:1070–1074. doi: 10.1001/jamaoncol.2016.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]