Abstract
The nucleolus contains multiple copies of ribosomal (r)DNA, which indicate sites of frequent replication stress and suggest the existence of a mechanism to prevent replication stress–related rDNA instability and the possibility that such a mechanism contributes to the whole genomic stability against replication stress. We have previously reported that nucleolin, a major nucleolar protein, is involved in ionizing radiation–induced DNA damage responses (DDRs) such as ataxia telangiectasia mutated (ATM)-dependent cell cycle checkpoints and homologous recombination (HR) repair. Here, we investigated the role of nucleolin in DDR due to replication stress. The results indicate that following replication stress, nucleolin interacted with the histone γH2AX, proliferating cell nuclear antigen (PCNA), and replication protein A (RPA)32, suggesting that it may be recruited to DNA damage sites on the replication fork. Furthermore, the knockdown of nucleolin by siRNA reduced the activation of ATM and RAD3-related (ATR) kinase and the formation of RAD51 and RPA32 foci after replication stress due to UV or camptothecin exposure, whereas nucleolin overexpression augmented ATR-dependent phosphorylation and RAD51 and RPA accumulation on chromatin. Moreover, these overexpressing cells seemed to increase repair activity and resistance to replication stress. Our results indicate that nucleolin plays an important role in replication stress–induced DDRs such as ATR activation and HR repair. Given that nucleolin overexpression is often observed in many types of cancer cells, our findings suggest that nucleolin is involved in the regulation of resistance to replication stress that may otherwise lead to tumorigenesis and it could be a possible target for chemotherapy and radiotherapy.
Keywords: nucleolin, nucleolus, replication stress, DNA damage, ATR
INTRODUCTION
DNA double-strand breaks (DSBs) produced by ionizing radiation (IR) or endogenous stress cause the most serious damage to genomic DNA, which can lead to cell death or genetic instability and tumorigenesis. Therefore, eukaryotic cells have developed a complex system for DSB detection called the DNA damage response (DDR), which includes cell cycle arrest at specific checkpoints, DSB repair, and restoration of cell cycle progression [1]. Cell cycle checkpoints are regulated by ataxia telangiectasia mutated (ATM) serine/threonine kinase, which is recruited to DSBs where it is activated in an MRE11/RAD50/NBS1 (MRN) complex-dependent manner and phosphorylates various DDR factors, thus controlling cell cycle progression through checkpoints [2]. DSBs are repaired by two major pathways: non-homologous end-joining (NHEJ) and homologous recombination (HR). HR needs homologous sequences generated by DNA replication and can be activated in the late S and G2 phases, whereas NHEJ is preferably employed for repair of DSB-associated DNA damages occurring in all cell cycle phases of higher eukaryotes [3].
Replication stress caused by the stall or collapse of the replication fork progression during DNA synthesis in the S phase induces DDRs, often leading to the generation of DSB damages, which are mostly repaired through the HR pathway [3]. Replication stress generates single-stranded (ss) DNA regions on the replication fork, which are then coated with replication protein A (RPA) to prevent re-annealing/degradation of ssDNA until restart of the replication fork. The recruited RPA also interacts with ATM and RAD3-related (ATR) kinase through ATR interacting protein (ATRIP) and activates the protein kinase activity of ATR, thus triggering phoshorylation of multiple effectors involved in DNA repair and cell cycle regulation [4].
It is known that multiple repeated sequences in genomic DNA are potential sites of replication stress in the S phase [5], because they often generate secondary DNA structures, which could disturb the progression of DNA replication, causing chromosomal breaks and gaps. Multiple copies of ribosomal (r)DNA existing in the nucleolus of eukaryotic nuclei [6] represent such regions vulnerable to replication stress. Thus, deletion of some rDNA copies in yeast inhibited excessive accumulation of stalled forks and prevented replication stress, which can lead to genomic instability and cell death [7, 8]. The human genome contains ~400 copies of rDNA tandemly arranged on five chromosomes in the nucleolar organizer region of the nucleolus. Nucleolin (also named NCL or C23) is a ubiquitously expressed nucleolar protein constituting 10% of the total protein in the nucleolus [9], where it functions in ribosome biogenesis, transcriptional regulation, and chromatin remodeling, and plays an essential role in the nucleolus organization. In addition, nucleolin is also present in the nucleoplasm, where it may participate in DNA replication and DDRs, including DNA repair and cell cycle checkpoints, and other stress responses [9–11]. Our previous findings indicate that nucleolin plays an important role in IR-induced DSB damage responses [10]. We showed that nucleolin was recruited to DSB damage sites created by laser micro-irradiation in the nucleoplasm and that its knockdown by siRNA arrested ATM-dependent cell cycle checkpoint progression and HR repair through the mediator of DNA damage checkpoint protein 1 (MDC1)-dependent pathway. Furthermore, our results revealed that this functional activity of nucleolin was likely associated with histone eviction from chromatin, which is important for transcriptional regulation and is increased at DSB sites [12].
Considering the multifaceted functions of nucleolin, a major nucleolar protein, we speculate that it may be closely involved in the mechanism existing in the nucleolus to counterbalance frequent-replication stress and also hypothesize that it may maintain whole genomic stability against replication stress. Therefore, we here investigated the role of nucleolin in DDRs to replication stress induced with camptothecin (CPT) or UV, especially in ATR activation and HR repair.
MATERIALS AND METHODS
Cells
HeLa, U2OS, and hTERT-immortalized human fibroblasts (48BR) were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS; Invitrogen) and antibiotics [13].
Generation of GFP-FBL–expressing cells and FLAG-NCL–expressing cells
Human fibrillarin (FBL, NM_001436) and nucleolin (NCL, NM_005381) cDNAs were amplified from a human fetal cDNA library (Clontech) by PCR with Pyrobest DNA polymerase (TAKARA), and inserted into pEGFP-C1 (Clontech) and pCMV-Tag2B-FLAG (Promega) vectors, respectively; the insertions were confirmed by DNA sequencing. U2OS cells were transfected with the generated pEGFP-FBL and pCMV-TAG2B-NCL plasmids using FugeneHD (Promega), and GFP-FBL– and FLAG-NCL–expressing cells were selected with G418.
SiRNA knockdown experiments
Sub-confluent cells were plated on culture dishes for 24 h and then transfected with NCL siRNA or negative control siRNA (Be-Bridge International Inc.) using Lipofectamine RNAiMax (Invitrogen Life Technology) [10]. Cells were re-plated after 2 days and used for western blotting or immunofluorescence the next day.
Antibodies
The following antibodies were used in this study: mouse monoclonal anti-phospho-ATM (S1981) and anti-γH2A histone family X (H2AX), and rabbit polyclonal anti-H2B (Millipore Co.); rabbit polyclonal anti-NCL, anti-phospho-RPA32, and anti-phospho-KAP1 (Bethyl Laboratories Inc.); rabbit polyclonal anti-phospho-Chk1 (S317), anti-phospho-RAD17 (S645), and anti-phospho-ATR (T1989), and mouse monoclonal anti-p53-pS15 (Cell Signaling Technology); rabbit polyclonal anti-human NBS1 (Novus Biologicals); mouse monoclonal anti-RPA32 and anti-RPA70 and rabbit polyclonal anti-ATM (Merck Millipore); mouse monoclonal anti-KAP1, anti-NBS1, anti-MRE11, and anti-RAD50 (GeneTex); rabbit polyclonal anti-RAD51 (Bioacademia); mouse monoclonal anti-beta-actin (Sigma-Aldrich); mouse monoclonal anti-p53, anti-KU70, anti-RAD17, and anti-PCNA (Santa Cruz); mouse polyclonal anti-RAD51 (Abnova); and rabbit polyclonal anti-53BP1 (Calbiochem).
Immunoprecipitation, western blotting, and immunofluorescense
Immunoprecipitation was performed as reported previously [13] by incubating the samples with monoclonal antibodies against γ-H2AX, RPA32, PCNA or RAD17. Co-immunoprecipitated proteins were detected by western blotting analysis.
Western blotting analysis of whole cell extract or chromatin fraction isolation was performed as described previously [10, 13]. Target proteins were detected with the primary antibodies listed above and secondary horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG (GE Healthcare) and visualized using the ECL™ plus chemiluminescence system (GE Healthcare). Quantification of visualized bands was carried out by ImageJ software, and the ratios to unirradiated samples of negative knockdown were calculated [13].
Immunofluorescence staining was carried out as described previously [10] using Alexa-488–conjugated anti-rabbit or alexa-594–conjugated anti-mouse IgG (Molecular Probes) to visualize the localization of the target proteins.
RESULTS
Nucleolin interacts with DDR factors on stress-damaged replication forks
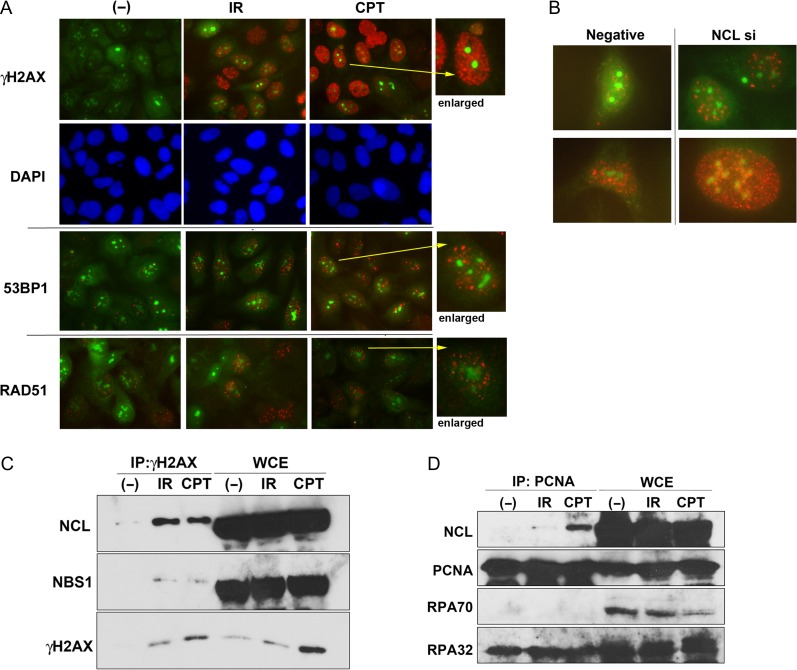
To address the mechanisms underlying nucleolus-specific DDRs preventing rDNA instability, particularly a high rate of DNA repair, we examined the formation of DDR protein foci in the nucleoli of GFP-FBL–expressing U2OS cells. As shown in Fig. 1A, several large green dots were observed in the nuclei of most cells, suggesting that GFP-FBL stained the nucleolus. When the generation of DSB-related damage caused by γ-ray irradiation or CPT exposure was visualized with anti-phosphorylated histone H2AX (γH2AX) antibodies, the results showed that both treatments increased γH2AX staining (red), which, however, did not seem to colocalize with GFP-FBL staining of the nucleolus (Fig. 1A, top panel). Furthermore, the 53BP1 foci (red) known to have formation kinetics, which were similar to that of the γH2AX foci, did not colocalize with GFP-FBL either (Fig. 1A, middle panel and Fig. S1A). RAD51 is an important HR factor and their foci (red) were detected in response to DNA damage in the nucleoplasm, but not in the nucleoli (Fig. 1A, low panel and Fig. S1B). As nucleolin is an essential nucleolar protein, we attempted the knockdown of nucleolin with siRNA in GFP-FBL–expressing U2OS cells. As a result, we found that γH2AX foci partially colocalize with GFP-FBL without DNA damage treatment (Fig. 1B), suggesting a relationship between nucleolin and the maintenance of rDNA stability in the nucleoli against replication stress.
Fig. 1.
Nucleolin (NCL) participates in replication stress–induced responses. (A) GFP-FBL–expressing U2OS cells were irradiated by γ-rays (5 Gy: γH2AX and 53BP1; 10 Gy: RAD51) following incubation for 30 min (γH2AX and 53BP1) or 4 h (RAD51), or treated with CPT (0.5 μM, 1.5 h: γH2AX and 53BP1; 2 μM, 2 h: RAD51). Then, their cells were fixed after the incubation time for IR or completion of CPT treatment, and immunostaining was performed using the indicated antibodies and visualized to the red color with the secondary antibody. The green color is derived from GFP-FBL. (B) GFP-FBL–expressing U2OS cells were transfected by nucleolin siRNA or negative control siRNA, and after 2 days these cells were fixed and immunostaining was performed using anti-γ-H2AX antibody. (C and D) Nucleolin interacts with γH2AX and PCNA following replication stress. Extracts from HeLa cells, treated with 10 Gy of γ-rays or 2 μM of CPT, were immunoprecipitated with anti-γH2AX antibody (C), anti-PCNA antibody (D), and then the immunocomplexes were detected by western blot analysis using the indicated antibodies.
As we have already reported the importance of nucleolin for general DSB damage responses such as ATM-dependent cell cycle checkpoints and DNA repair [10], we next investigated whether it is involved in general replication stress responses. It is known that the phosphorylation of H2AX, also called as γH2AX, is induced by replication stress caused by DNA damaging agents such as CPT [3, 4]. Consistent with our previous finding that nucleolin interacted with γH2AX following IR [10], the induction of replication stress by CPT also increased the interaction between γH2AX and nucleolin (Fig. 1C). Immunoprecipitation with the antibody against proliferating cell nuclear antigen (PCNA) showed that nucleolin also interacted with PCNA but not with RPA following CPT treatment, whereas IR did not induce binding between nucleolin and PCNA (Fig. 1D). As PCNA is an important regulatory factor in DNA replication, which acts on the replication fork, these results indicate that nucleolin might be recruited to the damaged forks through interaction with γ-H2AX and PCNA to play a role in cellular responses to replication stress.
Nucleolin participates in ATR-related responses
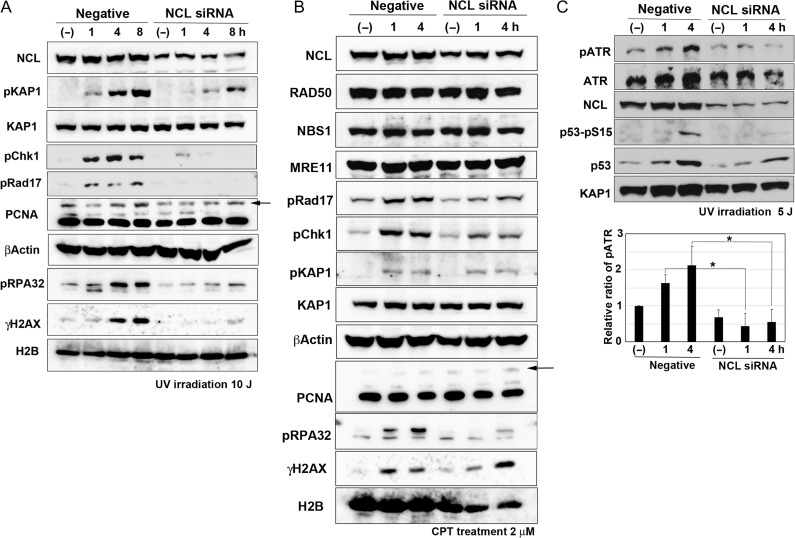
As nucleolin may be involved in replication stress–induced cellular responses, we investigated its relationship with translesion DNA synthesis (TLS). TLS allows overriding of DNA damages (i.e. UV-induced lesions) on the replication fork and the proceeding of DNA replication by switching to specific translesion DNA polymerases such as Polη, which is activated after PCNA mono-ubiquitination by RAD18 E3 ligase [14]. Therefore, we examined the mono-ubiquitination of PCNA in U2OS and 48BR cells with knockdown of nucleolin with the siRNA. However, PCNA mono-ubiquitination was either not disturbed (U2OS cells, Fig. 2A) or even increased (48BR cells, Fig. S2A) in the cells in which knockdown had occurred. As Polη/RAD18-dependent TLS dysfunction can promote mutagenesis following UV exposure, we next examined the mutation spectrum in UV-treated nucleolin-depleted cells by the SupF mutation assay [14, 15]: we observed no differences from the control (Fig. S3A), suggesting that nucleolin may not be important for Polη/RAD18-dependent TLS.
Fig. 2.
Nucleolin (NCL) contributes to ATR-dependent phosphorylation following replication stress. U2OS cells were transfected by nucelolin siRNA. After 2 days, these cells were irradiated by UV (A: 10 J; C: 5 J) or treated with 2 μM of CPT (B). Then, their cells were harvested at the indicated times and analyzed by western blot using the indicated antibodies. Arrows indicate the bands of mono-ubiquitinated PCNA in (A) and (B). Quantification of pATR of Fig. 2C and the other two experiments were carried out by ImageJ software and are shown in the lower panel of Fig. 2C (*P < 0.05).
As replication stress activates ATR through ssDNA generation [4], we examined ATR-dependent phosphorylation of its major substrates RAD17, checkpoint kinase 1 (CHK1), and RPA32 in nucleolin-reduced U2OS and 48BR cells and found that ATR activity was significantly reduced in these cells after UV exposure (Fig. 2A, Table S1A and S2A). Induction of replication stress by CPT (Fig. 2B, Table S1B and S2B) or hydroxyurea (Fig. S2C) also reduced ATR-dependent phosphorylation. We also confirmed that reduction of nucleolin with siRNA did not disturb the distribution of cells throughout the stages of the cell cycle (Fig. S3B), suggesting that such reduction of ATR-dependent phosphorylation is not due to decrease in S phase cells. Furthermore, auto-phosphorylation of ATR at Thr1989, an indicator of ATR activation [4], was reduced in nucleolin-reduced U2OS cells following treatment with CPT (Fig. 2C). This reduction of ATR auto-phosphorylation is significant, as shown in the lower panel of Fig. 2C. These results suggest that nucleolin might be an important factor in ATR activation.
Replication stress often generates secondary DSB damages and subsequently activates ATM [4], but nucleolin knockdown reduced ATM-dependent phosphorylation of KAP1 as well as ATM auto-phosphorylation (Fig. 2A and Fig. S2A–C), which is consistent with the importance of nucleolin for ATM activation after IR [10]. Although the MRN complex is essential for the IR-induced activation of ATM, the expression of MRE11, RAD50 and NBS1 did not decrease with nucleolin knockdown (Fig. 2B), suggesting that the reduction of KAP1 phosphorylation is not due to repression of the MRN complex.
Nucleolin is indispensable for replication stress–dependent HR repair
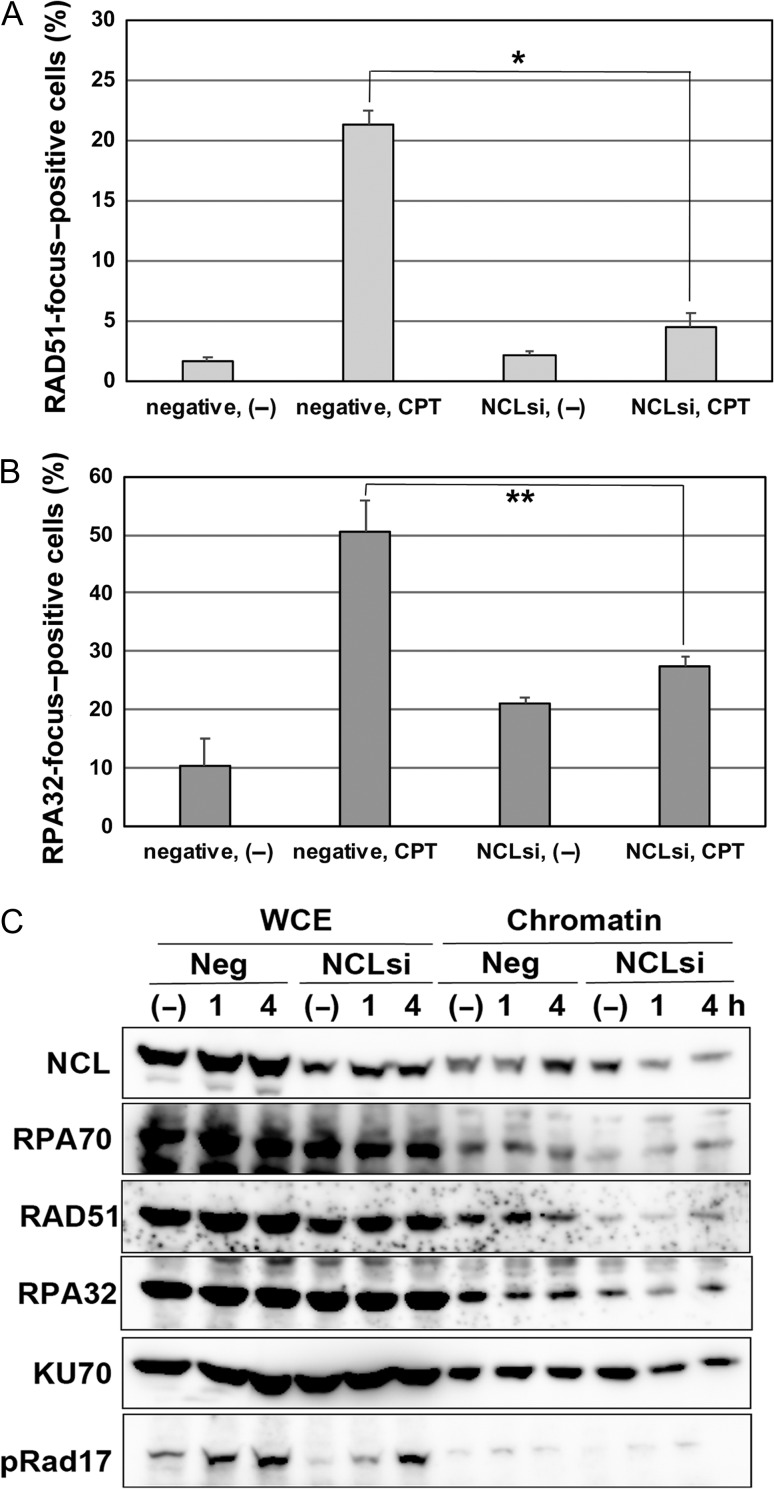
DNA damage due to replication stress is often repaired through the HR pathway [3, 16]; therefore, we next investigated the formation of RAD51 and RPA32 foci, which are indicative of HR activation. CPT treatment induced RAD51 focus formation (by >20%) in control cells but not in nucleolin-reduced cells (Fig. 3A and Fig. S4A). A similar tendency was observed for the formation of RPA32 foci, although the frequency of RPA32-positive cells was increased by nucleolin knockdown without CPT treatment (Fig. 3B and Fig. S4B).
Fig. 3.
Nucleolin (NCL) participates in the HR pathway following replication stress. (A and B) U2OS cells were transfected by nucleolin siRNA or negative control siRNA, and after 2 days these cells were treated with 2 μM of CPT. After 4 h, their cells were fixed and immunostaining was performed using the anti-RAD51 antibody (A) or the anti-RPA32 antibody (B). Then, percentages of foci-positive cells were counted under the fluorescence microscope (*P < 0.01; **P < 0.02). (C) Nucleolin is indispensable for the chromatin accumulation of HR factors. U2OS cells were transfected by nucleolin siRNA or negative control siRNA, and after 2 days these cells were treated with 2 μM of CPT. After the indicated times, their cells were harvested and their chromatin fractions were prepared. Chromatin accumulation of DDR proteins were detected by western blot analysis using the indicated antibodies.
As HR factors such as RAD51 and RPA function with their accumulation to damaged chromatin, we next examined their chromatin accumulation in nucleolin-repressed cells (Fig. 3C). Although the accumulation of RAD51, RPA 32 and RPA70 was not increased in CPT-treated control cells, it was almost abrogated in nucleolin-deficient cells. Furthermore, that of NHEJ factor KU70 was not affected by the knockdown of nucleolin. Moreover, the normal focus formation of 53BP1 (another NHEJ factor) was observed in nucleolin-deficient cells following CPT treatment (Fig. S5). Taken together, these results indicate that nucleolin might function in the activation of the HR but not the NHEJ pathway.
Nucleolin overexpression amplified DNA damage responses to replication stress
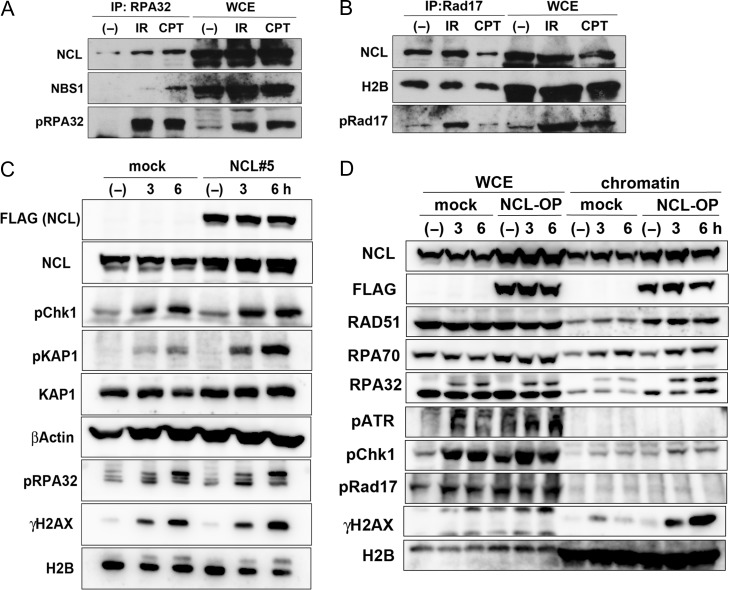
The results shown in Fig. 2 suggest that nucleolin might be important for ATR activation; we hypothesized that it was included in the active ATR complex, but we were not able to detect direct interaction of nucleolin with ATR by immunoprecipitation (data not shown). We then investigated the interaction of nucleolin with ATR regulatory factors such as RPA. Our results indicate that nucleolin associated with RPA32 in response to DNA damage caused by replication stress (CPT) or IR (Fig. 4A). Furthermore, RAD17, a factor involved in loading the RAD9-RAD1-HUS1 (9-1-1) complex to the damaged fork [4], also showed an interaction with nucleolin in untreated cells, which was decreased by CPT but not by IR (Fig. 4B). As Fig. 4B additionally shows that RAD17 also interacts with histone H2B, the binding between RAD17 and nucleolin is likely to occur on chromatin. These results suggest that nucleolin might control ATR activation through interaction with ATR regulatory factors such as RPA or RAD17.
Fig. 4.
Overexpression of nucleolin (NCL) amplified DNA damage responses with replication stress. (A and B) Nucleolin interacts with RPA and Rad17. Extracts from HeLa cells, treated with 10 Gy of γ-rays or 2 μM of CPT, were immunoprecipitated with anti-RPA32 antibody (A), or anti-Rad17 antibody (B), and then the immunocomplexes were detected by western blot analysis using the indicated antibodies. (C) Nucleolin-overexpressing or mock cells were irradiated by 5J of UV. Then, their cells were harvested at the indicated times and analyzed by western blot using the indicated antibodies. (D) Nucleolin-overexpressing (NCL#5) or mock cells were treated with 2 μM of CPT. After the indicated times, their cells were harvested and their chromatin fractions were prepared. Chromatin accumulation of DDR proteins were detected by western blot analysis using the indicated antibodies.
The results presented in Figs 2 and 3 indicate that nucleolin could be indispensable for DDRs to replication stress. Many cancer cells have increased expression of nucleolin [9], suggesting that it may be a part of the mechanism protecting tumor cells against DNA damage due to replication stress. To verify our hypothesis, we generated nucleolin (FLAG-NCL)-overexpressing U2OS cells (Fig. S6A). Clone NCL#5, which showed higher nucleolin expression than clone NCL#4, had increased ATR-dependent phosphorylation of RAD17 and RPA32 after CPT treatment (Fig. S6A, 4 hr). A similar tendency was observed after UV irradiation, which increased the phosphorylation of Chk1 and RPA32 in FLAG-NCL–overexpressing cells compared with control (Fig. 4C). Furthermore, ATR autophosphorylation was also increased by nucleolin overexpression (Fig. 4D). As ATM-dependent phosphorylation of KAP1 was also increased following replication stress (Fig. 4C and Fig. S6A), we next examined ATM activation in nucleolin-overexpressing cells and found that the phosphorylation of KAP1, H2AX and ATM was increased, whereas that of Chk1 and RPA32 was not affected in these cells after IR exposure (Fig. S6B). These results suggest that the contribution of nucleolin to ATR activation following DSB damage is less than in the replication stress response, or even dispensible. Furthermore, we found that nucleolin overexpression increased the accumulation of RAD51 and RPA32 into chromatin (Fig. 4D, NCL-OP), suggesting reinforcement of the HR pathway. Moreover, nucleolin-overexpressing cells had increased viability at 2 days after CPT treatment (Fig. S6C) and DNA repair activity, leading to reduced γH2AX at 24 h after the treatment (Fig. S6D).
Cumulatively, these results indicate that the functional activity of nucleolin could be in amplifying DDRs elicited by replication stress, which may account for enhanced resistance to replication stress in nucleolin-overexpressing cancer cells.
DISCUSSION
As the nucleolus contains multiple rDNA copies, which could generate replication stress and jeopardize rDNA stability and integrity, there should be a mechanism to prevent rDNA instability. Nucleolin is the most abundant non-ribosomal nucleolar protein functioning in ATM-dependent cell cycle checkpoints and HR repair (through histone eviction and the MDC1 pathway activated in response to IR-induced DNA damage) for the maintenance of whole-genomic stability [10]. As shown in Fig. 1A, the focus of DDR factors such as γH2AX and 53BP1 was not localized in the nucleolus; this suggests the possibility that in most of the cells the nucleoli were not hit by radiation, because the fraction of rDNA in relation to the whole genome was small. However, nucleolin knockdown allowed their localization in the nucleolus, indicating that the rDNA in the nucleolus also suffer DNA damage from irradiation or replication stress. These results also indicate that there is nucleolus-specific repair activity for rapid correction of DNA damages through nucleolin, which explains why foci formation by DDR proteins could not be observed there. These nucleolus-specific DDRs might countervail replication stress in order to maintain rDNA stability, and nucleolin may be important for this maintenance. Furthermore, we discovered a much more important role of nucleolin in the maintenance of whole-genomic stability to countervail replication stress. After replication stress, nucleolin was shown to interact with γH2AX, RPA32 and PCNA (Fig. 1C and D and 4A and B), suggesting its recruitment to the damaged replication fork. Although ATR and HR repair were activated by replication stress, nucleolin knockdown markedly reduced ATR-dependent phosphorylation (Fig. 2, Table S1 and Fig. S2) and the accumulation of HR factors RAD51 and RPA32 on chromatin (Fig. 3A and B and Fig. S4A and B), whereas nucleolin overexpression increased ATR- and HR-related responses (Fig. 4C and D). Considering that nucleolin interacts with RPA32 and that the RPA complex is critical for ATR as well as HR activation [1, 2, 4], nucleolin is suggested to be an initial regulatory factor for ATR-dependent cell cycle checkpoints as well as HR repair essential for the maintenance of whole-genomic DNA stability, including the protection of rDNA during replication stress.
It has been reported that ATR activity needs several factors [4] such as ATRIP (which interacts both with ATR and RPA coating ssDNA at the stalled replication fork, thus recruiting ATR to DNA damage sites) and TopBP1 (which binds with both ATR and the 9-1-1 clamp-like complex loaded onto the stalled fork by the RAD17–RFC complex), which contribute to stable recruitment of ATR. These events result in the catalytic activation of ATR. Recently, the importance of other RPA-binding proteins for ATR activation has been reported. Thus, it was found that NBS1 possessed an RPA-binding domain, which could enable the persistent recruitment and activation of ATR during long resection of damaged DNA ends [17]. Another RPA-binding protein, ETAA1, was shown to have a similar role in continuous ATR activation independent of the 9-1-1 complex, although ETAA1 expression is limited in several type of tumor cell lines [18, 19]. Even though the interaction of nucleolin with the RPA complex during the normal cell cycle has been reported before [20], here we show that nucleolin–RPA association is induced following replication stress, probably at damaged DNA sites of the stalled fork (Figs 1C and D and 4A). Furthermore, we found that nucleolin deficiency reduced (whereas its overexpression induced) RPA chromatin accumulation and ATR activity (Fig. 4), suggesting that nucleolin might facilitate ATR activation by the stabilization of interaction between RPA and ssDNA in chromatin containing the damaged replication fork. The regulation of damaged DNA resection to extend ssDNA is essential to RPA accumulation on chromatin following DNA damage; however, it is unknown whether nucleolin plays a role in the process, and this should be investigated to clarify the details of ATR regulation.
Although nucleolin overexpression has been shown in many cancer cells [9], its relationship with tumorigenesis is unknown. Nucleolin has histone eviction activity, important for gene transcription around the RNA polymerase II–dependent promoter region of several oncogenes [12, 21], and could promote cell proliferation and other tumorigenic processes in cancer development. Furthermore, we and others have reported that the interaction of nucleolin with DDR proteins such as RAD51, γH2AX and RAD50 could contribute to the DDR [10, 11, 22]. Currently, it is unknown whether nucleolin has the ability to reinforce resistance to DSB-mediated damage; however, our results indicate that the overexpression of nucleolin promoted DDRs elicited by replication stress, such as activation of the ATR and HR pathways. Although tumor cells suffer from increased replication stress due to a higher proliferation rate compared with normal cells, the reinforcement provided by nucleolin could contribute to replication stress resistance and promote tumorigenesis. In normal cells, nucleolin almost exclusively localizes in the nucleolus, contributing to rDNA stability, whereas its overexpression in tumor cells increases its localization in the nucleoplasm [9], where it could protect the genomic DNA of tumor cells against replication stress. We have already reported the importance of nucleolin in DSB damage responses to irradiation [10], suggesting that repression of nucleolin in cancer could enhance the effect of radiotherapy. We also clarified the importance of nucleolin in the replication stress response and its possible role in tumorigenesis in this paper. These clues might prove important for cancer chemotherapy. Further studies to clarify the detailed relationship between nucleolin overexpression and carcinogenesis might be necessary, and the future development of anti-cancer therapeutics targeting nucleolin might then be possible.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Kenshi Komatsu for his critical appraisal of the manuscript. We also thank Yukiko Hayuka and Kae Yanagida for their technical support, and Michi Tanizaki for preparing the manuscript.
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
FUNDING
This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (grant numbers JP25550026, JP15H02819, and JP18H04978) and in part by the National Institute for Fusion Science (NIFS) Collaborative Research Program (NIFS17KOCA002).
REFERENCES
- 1. Saito Y, Zhou H, Kobayashi J. Chromatin modification and NBS1: their relationship in DNA double-strand break repair. Genes Genet Syst 2016;90:195–208. [DOI] [PubMed] [Google Scholar]
- 2. Paull TT. Mechanisms of ATM Activation. Annu Rev Biochem 2015;84:711–38. [DOI] [PubMed] [Google Scholar]
- 3. Shibata A. Regulation of repair pathway choice at two-ended DNA double-strand breaks. Mutat Res 2017;803–805:51–5. [DOI] [PubMed] [Google Scholar]
- 4. Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell 2017;66:801–17. [DOI] [PubMed] [Google Scholar]
- 5. Irony-Tur Sinai M, Kerem B. DNA replication stress drives fragile site instability. Mutat Res 2018;808:56–61. [DOI] [PubMed] [Google Scholar]
- 6. McStay B. Nucleolar organizer regions: genomic ‘dark matter’ requiring illumination. Genes Dev 2016;30:1598–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Salim D, Bradford WD, Freeland A et al. . DNA replication stress restricts ribosomal DNA copy number. PLoS Genet 2017;13:e1007006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zheng DQ, Zhang K, Wu XC et al. . Global analysis of genomic instability caused by DNA replication stress in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 2016;113:E8114–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Berger CM, Gaume X, Bouvet P. The roles of nucleolin subcellular localization in cancer. Biochimie 2015;113:78–85. [DOI] [PubMed] [Google Scholar]
- 10. Kobayashi J, Fujimoto H, Sato J et al. . Nucleolin participates in DNA double-strand break–induced damage response through MDC1-dependent pathway. PLoS One 2012;7:e49245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bakkenist CJ, Kastan MB. Chromatin perturbations during the DNA damage response in higher eukaryotes. DNA Repair 2015;36:8–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Angelov D, Bondarenko VA, Almagro S et al. . Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J 2006;25:1669–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou H, Kawamura K, Yanagihara H et al. . NBS1 is regulated by two kind of mechanisms: ATM-dependent complex formation with MRE11 and RAD50, and cell cycle–dependent degradation of protein. J Radiat Res 2017;58:487–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yanagihara H, Kobayashi J, Tateishi S et al. . NBS1 recruits RAD18 via a RAD6-like domain and regulates Pol η-dependent translesion DNA synthesis. Mol Cell 2011;43:788–97. [DOI] [PubMed] [Google Scholar]
- 15. Kobayashi J, Okui M, Asaithamby A et al. . WRN participates in translesion synthesis pathway through interaction with NBS1. Mech Ageing Dev 2010;131:436–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carr AM, Lambert S. Replication stress-induced genome instability: the dark side of replication maintenance by homologous recombination. J Mol Biol 2013;425:4733–44. [DOI] [PubMed] [Google Scholar]
- 17. Shiotani B, Nguyen HD, Håkansson P et al. . Two distinct modes of ATR activation orchestrated by Rad17 and Nbs1. Cell Rep 2013;3:1651–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haahr P, Hoffmann S, Tollenaere MA et al. . Activation of the ATR kinase by the RPA-binding protein ETAA1. Nat Cell Biol 2016;18:1196–207. [DOI] [PubMed] [Google Scholar]
- 19. Bass TE, Luzwick JW, Kavanaugh G et al. . ETAA1 acts at stalled replication forks to maintain genome integrity. Nat Cell Biol 2016;18:1185–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim K, Dimitrova DD, Carta KM et al. . Novel checkpoint response to genotoxic stress mediated by nucleolin-replication protein A complex formation. Mol Cell Biol 2005;25:2463–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gaume X, Monier K, Argoul F et al. . In vivo study of the histone chaperone activity of nucleolin by FRAP. Biochem Res Int 2011;2011:187624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. De A, Donahue SL, Tabah A et al. . A novel interaction of nucleolin with Rad51. Biochem Biophys Res Commun 2006;344:206–13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.