Abstract
Exosomes and other extracellular vesicles are key players in cell-to-cell communication, and it has been proposed that they are involved in different aspects of the response to ionizing radiation, including transmitting the radiation-induced bystander effect and mediating radioresistance. The functional role of exosomes depends on their molecular cargo, including proteome content. Here we aimed to establish the proteome profile of exosomes released in vitro by irradiated UM-SCC6 cells derived from human head-and-neck cancer and to identify processes associated with radiation-affected proteins. Exosomes and other small extracellular vesicles were purified by size-exclusion chromatography from cell culture media collected 24 h after irradiation of cells with a single 2, 4 or 8 Gy dose, and then proteins were identified using a shotgun LC-MS/MS approach. Exosome-specific proteins encoded by 1217 unique genes were identified. There were 472 proteins whose abundance in exosomes was significantly affected by radiation (at any dose), including 425 upregulated and 47 downregulated species. The largest group of proteins affected by radiation (369 species) included those with increased abundance at all radiation doses (≥2 Gy). Several gene ontology terms were associated with radiation-affected exosome proteins. Among overrepresented processes were those involved in the response to radiation, the metabolism of radical oxygen species, DNA repair, chromatin packaging, and protein folding. Hence, the protein content of exosomes released by irradiated cells indicates their actual role in mediating the response to ionizing radiation.
Keywords: exosomes, head-and-neck cancer, proteomics, radiation response
INTRODUCTION
In either physiologic conditions or in response to stress factors, cells release different types of extracellular vesicles (EVs) into the extracellular microenvironment. Of these EVs, the largest attention is paid to exosomes because of their role in cell-to-cell communication and involvement in disease-related processes. Exosomes are nanometer-sized (30–120 nm) membrane-enclosed structures actively secreted by various types of normal and cancer cells, both in vivo and in vitro. They may transmit a complex network of signals driving cell death, survival, and differentiation between a secreting cell and multiple types of neighboring or distant recipient cells [1]. Among the stress factors known to affect EV-based intercellular communication is exposure to ionizing radiation (IR). Radiation can enhance the release of exosomes, and this has been observed in different types of normal and cancerous cell lines [2–4]. This phenomenon could result from activation of stress-inducible pathways of exosome secretion, including increased expression of the TSAP6 protein that is activated by the damage-induced p53 transcription factor [5]. Exosomes (and possibly other types of EVs) released from irradiated cells are putatively involved in different aspects of the systemic response to IR, including the radiation-induced bystander effect (RIBE). This phenomenon, which results in the appearance of a radiation-induced phenotype (e.g. radiation-induced cell death) in cells that were not exposed to radiation themselves, could be transmitted in a paracrine manner by different types of mediators, including cytokines, calcium fluxes, nitric oxide (NO) and radical oxygen species (ROS) [3, 6, 7]. More recently, the work of Al-Mayah and coworkers revealed the synergistic effect of both protein and RNA exosome content in inducing the RIBE and suggested that a delayed RIBE-related inflammatory response was caused not only by exosomes released by directly irradiated cells but also by exosomes secreted from bystander cells and their progeny [4]. On the other hand, it has been reported that exosomes released by irradiated cells could increase survival and modulate DNA repair in recipient cells exposed to radiation [8].
The molecular content of EVs and their influence on recipient cells primarily depends on the type and physiological status of secreting cells [9]. Therefore, any factor affecting the phenotype of a cell likely affects the molecular composition of EVs released by this cell. However, current knowledge regarding radiation-induced changes in exosome composition is rather limited and refers mainly to their proteomes. A few reports concerning exosomes released by irradiated cancer cell lines revealed an increased level of CD276 in the case of prostate cancer cells [10] and an increased level of IGFBP2 in the case of glioblastoma cells [2]. Moreover, increased levels of HSP72 was reported in exosomes purified from the serum of prostate cancer patients undergoing radiotherapy [11]. A few proteomics studies aimed at profiling the proteome of exosomes released from cells exposed to radiation in vitro were published recently. One study revealed increased levels of proteins involved in transcription and translation, chaperones, ubiquitination-related factors and proteasome components in exosomes released from FaDu cells, derived from a hypopharynx carcinoma, irradiated with a 2 Gy dose [12]. A similar analysis examined exosomes released by BHY cells, derived from a highly invasive lower alveolar carcinoma, irradiated with a 6 Gy dose. IR-modulated proteins (39 IR-upregulated and 36 IR-downregulated) were associated not only with response to stress and immunity but also to cellular adhesion and motility [13]. Here, we aimed to use a comprehensive proteomics approach to characterize the proteome of EVs released by UM-SCC6 cells, derived from a human head-and-neck squamous cell cancer located in a tongue, irradiated with different doses, and to identify proteins and their associated biological functions upregulated by IR. Head-and-neck cancer cells were selected as a relevant experimental model because radiotherapy remains the primary treatment option in this malignancy.
METHODS
Cell culture
The UM-SCC6 human head-and-neck cancer cell line (authenticated by the American Type Culture Collection service; ATCC, Manassas, USA) was used as an experimental model because these cells are characterized by the wt p53 and a negative HPV status. Cells were cultured in Dulbecco’s Minimum Essential Medium (DMEM) supplemented with 10% (v/v) fetal bovine serum. Cells were seeded and incubated for 48 h prior to irradiation with a Clinac 600 (Varian Medical Systems, Palo Alto, USA; nominal energy of photon beam 6 MV) of up to 8 Gy at a dose rate 1 Gy per min. Immediately after irradiation (or mock irradiation in the case of control samples) standard cell culture medium was replaced with fresh medium supplemented with 5% (v/v) Gibco Exosome-Depleted FBS (Thermo Fisher Scientific, Waltham, USA, A2720801).
Cell phenotyping
For the clonogenic assay, cells (plated in triplicate at 4 × 103 cells per well) were irradiated with 0, 2, 4, 6 and 8 Gy, then incubated for 10 days (every 3 days a small portion of fresh media was added). Cell colonies were stained with crystal violet solution (0.2 % (m/v) with ethanol 2 % (v/v)) and counted. For cell cycle analysis, cells (plated in triplicate at 5 × 105 cells per well) were irradiated with 0, 2, 4 and 8 Gy, then incubated for 6 or 24 h. Cells were then harvested (by trypsin treatment) and fixed overnight at –20°C with 70% ethanol, then washed and treated with RNase (100 μg/μl) for 30 min at room temperature. Finally, propidium iodide (PI) solution (50 μg/μl) was added at a ratio of 1:4 (v/v), and the content of DNA was determined with a BD FACSCanto (BD Biosciences, San Jose, USA) flow cytometer. Alternatively, freshly harvested cells were washed with PBS and suspended in PI solution (1 μg/ml) for 10 min, then analyzed with a BD FACSCanto (BD Biosciences, San Jose, USA) flow cytometer. PI-positive cells were considered dead.
Isolation of extracellular vesicles
EVs were isolated by size exclusion chromatography (SEC) from culture media 24 h after irradiation. Forty milliliters of medium (corresponding up to ~1 × 107 cells) was centrifuged sequentially at 200g (10 min), 2000g (10 min) and 10 000g (30 min) to remove contaminations like cellular debris, and then filtered with a 0.22 μm filter to remove larger EVs (e.g. putative apoptotic bodies). The filtered medium was concentrated to 1 ml using a Vivacell100 ultrafiltration unit (Sartorius, Göttingen, Germany; VC1042) then loaded onto a qEVoriginal SEC column (Izon Science LTD, Christchurch, New Zealand). Subsequent fractions of 1 ml each were eluted using PBS without divalent cations. The presence of EVs in the collected fractions was detected by Western blot using exosome markers CD9, CD63 and CD81. A fraction enriched in EVs was eluted at 5 ml after the void volume.
Western blot analysis
The concentration of proteins in the analyzed samples was assessed using the Pierce BCA Protein Assay kit (Thermo Fisher Scientific, Waltham, USA; 23 225) according to the manufacturer’s instructions. Twenty microliters of SEC fractions (corresponding to ~5–6 μg of proteins) were mixed with loading buffer to a final concentration of 2% (v/v) SDS, 0.1% (v/v) bromophenol blue, 10% (v/v) glycerol, and optionally 100 mM DTT, denatured for 5 min at 95°C and separated by 12% SDS-polyacrylamide gel electrophoresis followed by wet transfer onto nitrocellulose membranes (Thermo Fisher Scientific, Waltham, USA; 88 018). Membranes were blocked for 1 h in 5% non-fatty milk and 0.1% Tween in PBS, and then primary antibody (anti-CD63: Invitrogen, 10 628D, 1:1500; anti-CD9: Santa Cruz Biotechnology, sc-13 118, 1:800; anti-CD81: Invitrogen, 10 630D, 1:500) was added for overnight incubation at 4°C. After triplicate washes, secondary antibody conjugated with HRP was added for 1 h at room temperature. Chemiluminescence detection of bands was performed with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific, Waltham, USA; 34 095) diluted 1:10 with washing buffer. CD63 and CD81 were detected under non-reducing conditions.
Characterization of extracellular vesicles
For size and morphology assessment, exosomal samples were concentrated from 1 ml to 50 μl by using Vivaspin500 ultrafiltration units (Sartorius, Göttingen, Germany; VS0102) according to the manufacturer’s instruction. For transmission electron microscopy (TEM), equal volumes of EV sample and 4% paraformaldehyde were mixed and incubated at 4°C until analysis. Five microliters of the mixture was placed on the Formvar-carbon coated EM grids for 10 min. The grid with adsorbed EVs was transferred to a drop of uranyl-oxalate solution for 2 s and immediately washed three times in a drop of pure water. The excess of solution was removed using filter paper, and the grid was air-dried and analyzed using a transmission electron microscopy TESLA BS500 with Frame-Transfer-CCD Camera (TRS, Moorenweis, Germany). The size distribution profile of EVs was estimated by the dynamic light scattering (DLS) measurement using a Zetasizer Nano-ZS90 instrument (Malvern Instruments, Malvern, UK). Fifty microliters of EV sample was analyzed at 20°C immediately after isolation in disposable low-volume cuvettes (ZEN0118, Malvern Instruments, Malvern, UK). The dispersant refractive index was 1.330 (ICN PBS Tablets), and the equilibration time was set for 30 s. The results were the average of six measurements each consisting of 10 runs (Malvern Zetasizer Software 7.12. was used for data analysis).
Protein identification by LC-MS/MS
A modified Filter-Aided Sample Preparation (FASP) protocol [14] was used for sample processing. Four hundred microliters of EV samples (corresponding to 100–120 μg of proteins) were mixed with lysis buffer (4% SDS, 100 mM Tris/HCl pH 7.6, 0.1 M DTT) in a 1:1 ratio and incubated at 95°C for 15 min. Denatured samples were mixed (1:1; v/v) with an 8 M urea solution in 0.1 M Tris/HCl, pH 8.5 in the Microcon-30kDa centrifugal filter unit (Millipore, MRCF0R030) and centrifuged for 15 min at 14 000g. Samples were then washed with 50 mM NH4HCO3 and incubated for 30 min with 5 mM iodoacetamide at room temperature. Filter-attached proteins were washed with 50 mM NH4HCO3 and digested for 18 h with trypsin (Promega, Madison, USA; V5111), using an enzyme to protein ratio of 1:100, w/w, 37°C, 18 h. Peptides obtained after digestion were released from an ultrafiltration unit using deionized water. Peptides from each sample (100 ng) were separated on reverse phase Acclaim PepMap RSLC nanoViper C18 column (75 μm × 25 cm, 2 μm) (Thermo Fisher Scientific, Waltham, USA) using an acetonitrile gradient (from 4 to 60%, in 0.1% formic acid) and a flow rate of 300 nl/min for 230 min. The analysis was performed with the use of a Dionex Ultimate 3000 RSLC nanoLC system connected to a Q Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, USA). The spectrometer was operated in a data-dependent MS/MS mode with survey scans acquired at a resolution of 70 000 at 200 m/z in MS mode, and 17 500 at 200 m/z in MS2 mode. Spectra were recorded in the scanning range of 300–2000 m/z in the positive mode. Protein identification was performed using the Swiss-Prot human database (release 2017_03 containing 137 404 sequence entries) with a precision tolerance of 10 ppm for peptide masses and 0.08 Da for fragment ion masses.
All raw data obtained for each dataset were imported into Protein Discoverer v.1.4 (Thermo Fisher Scientific) < Thermo raw files >. Mascot engine (Matrix Science, Boston, USA) was used for database searches for protein identification and quantification. Protein was considered as positively identified if at least two peptides per protein were found by both search engines, and a peptide score reached the significance threshold FDR = 0.01 (assessed by the Percolator algorithm). The abundance of identified proteins were estimated in the Proteome Discoverer using the Precursor Ions Area detector node (Thermo Fisher Scientific, Waltham, USA), which calculates the abundance of a given protein based on the average intensity of the three most intensive distinct peptides for this protein, with further normalization to the total ion current. Data was exported to ASCII format for further analysis.
Bioinformatics analyses
Protein analysis was performed in triplicate for each experimental condition (0, 2, 4 and 8 Gy), and nine pairwise ratios for the protein level at each experimental condition were established. The distribution of such ratios for all analyzed proteins was modeled by a Gaussian mixture allowing for the quantification of the differences between compared samples. The major component located around a ratio of 1.0 was considered as the model of ‘unchanged’ proteins, and the thresholds for ‘upregulation’ and ‘downregulation’ of protein abundance were set at 4.56 and 0.21, respectively (which corresponded to cross-points between the major component and the next Gaussian); a protein was considered ‘significantly changed’ when the median fold-change ratio for all combinations of replicas exceeded the thresholds mentioned above [15]. The model-based approach was used as an alternative to the classical P-value–based approach due to sample size–limited power of statistical testing and the ‘semi-quantitative’ character of label-free proteomics implemented in the current study. A list of genes corresponding to IR-affected proteins was annotated at the Gene Ontology database (using GO Database version 05.2016, Gene Ontology Consortium). A list of genes corresponding to all identified proteins was used as the reference set, and then statistical overrepresentation of gene ontology (GO) terms associated with proteins present in a given subset was estimated based on a hypergeometrical distribution.
RESULTS
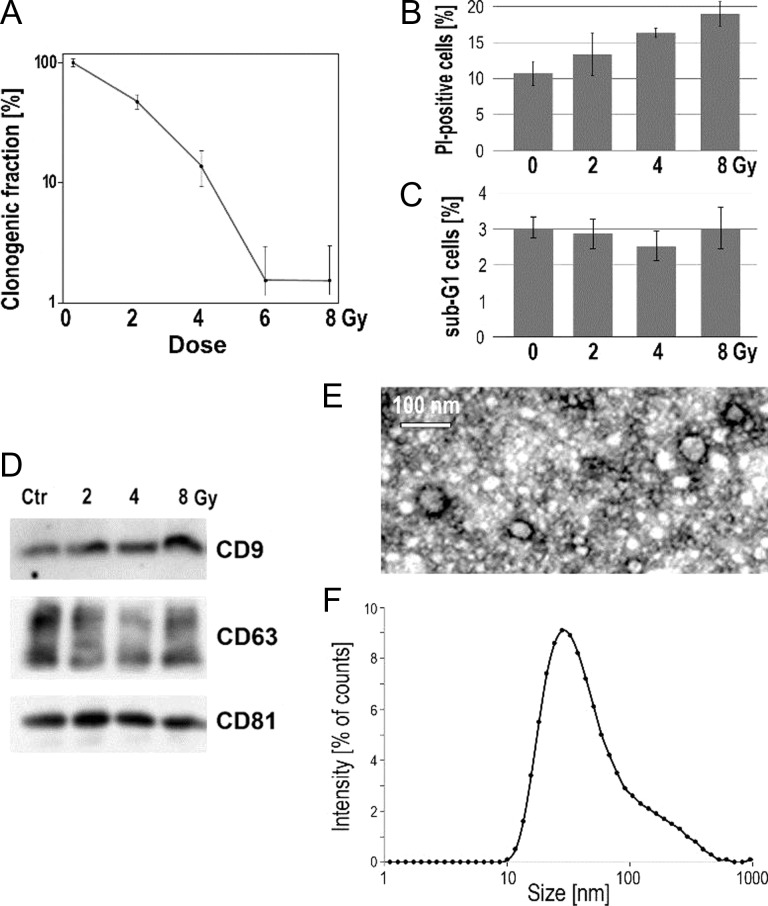
First, we analyzed the effects of IR on survival of UM-SCC6 head-and-neck cancer cells. The clonogenic assay revealed an expected dose-dependence of long-term effects of radiation: a fraction of viable clonogenic cells was approximately 50%, 10% and <1% at doses of 2, 4 and ≥6 Gy, respectively (Fig. 1A). However, the toxic effects of applied doses of radiation were comparable when analyzed within 24 h after irradiation. The fraction of cells with damaged membranes (i.e. cells permeable to propidium iodide) increased moderately 24 h after irradiation (~11% in control vs ~19% after 8 Gy; Fig. 1B). Moreover, the distribution of the cell cycle phases was examined to analyze the possible influence of radiation doses on the cell cycle arrest (looking at the G1/G2 ratio) and the induction of apoptosis (looking at the sub-G1 content). Some increase in the contribution of the G2 phase was detected 24 h after irradiation with the highest dose (G1/G2 ratio 1.2 and 0.7 for control and 8 Gy, respectively), which could reflect activation of the G2/M checkpoint (data not shown). However, when the contribution of sub-G1 cells (i.e. putatively apoptotic cells) was analyzed, a low level (<3%) of such cells was also observed after irradiation (Fig. 1C). Hence, we concluded that although the high doses of radiation used in our experimental model were generally cytotoxic, the acute effect of irradiation was moderate, and the majority of cells were viable at the time when analyzed EVs were collected (i.e. 24 h after irradiation).
Fig. 1.
The response of irradiated cells and characteristics of analyzed EVs. (A) Fraction of clonogenic cells 10 days after irradiation with different doses. (B) Fraction of PI-permeable cells 24 h after irradiation with 2, 4 and 8 Gy. (C) Fraction of sub-G1 cells 24 h after irradiation with 2, 4 and 8 Gy. (D) Level of exosome markers CD9, CD63 and CD81 in EVs released by cells within 24 h after irradiation with 2, 4 and 8 Gy. Size of EVs used for proteomics study evaluated by TEM (E) and DLS (F); EVs released by cells irradiated with 2 Gy are shown as an example.
EVs were purified from culture media contained material released by cells within 24 h after irradiation with 0, 2, 4 and 8 Gy. To confirm the presence of exosomes in the analyzed material, proteins characteristic for these vesicles were assessed by western blotting. Moreover, we verified the size of the vesicles present in the analyzed fractions using two methods considered standard in the field of EV research—imaging by TEM and physical size estimation by the dynamic light scattering (DLS) measurement. The fraction selected for analyses contained vesicles with exosome markers CD9, CD63 and CD81 (Fig. 1D), and was depleted of serum-derived components (e.g. serum albumin). In the same fraction, TEM micrographs revealed membrane-enclosed vesicles that were 40–60 nm in diameter (Fig. 1E). Moreover, DLS analysis indicated that the majority of particles present in this fraction had a diameter in the 30–40 nm range. We concluded that the analyzed fraction contained vesicles with a diameter corresponding to the diameter of (small) exosomes and positive for known exosome markers. Though a specific pathway of exosome biogenesis (i.e. via multivesicular bodies [16]) is a definitive feature of these EVs, and was not addressed in the current study, the EVs analyzed were called ‘exosomes’ afterward for simplicity (yet the term ‘small EVs’ could be also used). We observed that the size of exosomes released by control and irradiated cells was comparable when either TEM or DLS analysis was performed (not shown). Moreover, a similar level of total proteins was found in exosome-containing fractions irrespective of the cell treatment. It should be noted that a small (~10–20%) increase in the total protein level in the exosome fraction was observed after irradiation with the highest dose, yet this observation was not confirmed by any quantitative method allowing measurement of the actual number of EVs. Nevertheless, a similar amount of material released by control and irradiated cells was used for proteomic analyses.
There were 1329 proteins encoded by unique genes identified in the exosome-containing fraction of the cell culture medium collected in either experimental condition, which included 112 proteins also identified in a corresponding fraction of a unconditioned medium (supplemented with exosome-free calf serum) processed in parallel as a negative control. Therefore, 1217 non-redundant proteins were considered as exosome-specific and used in further quantitative analyses. There were 472 proteins (39% of identified proteins) whose abundances in exosomes were significantly affected by radiation (compared with an unirradiated control), including 425 IR-upregulated and 47 IR-downregulated species; all IR-affected proteins are listed in Table 1. The comparison of differences between exosomes released by cells irradiated at different doses revealed different dose-related patterns of change. These patterns, together with the numbers of corresponding proteins, are depicted in Fig. 2. The most numerous group of exosome proteins affected by radiation consisted of 369 species (30% of all detected exosome proteins) whose abundance in secreted vesicles increased at all radiation doses; this subset of proteins belong to the Mode [0 < 2,4,8] [the subset of proteins upregulated by 2, 4 and 8 Gy (2,4,8) compared with the non-irradiated control group (0)]. The second most abundant group (32 proteins) included exosome components downregulated at all radiation doses; this subset of proteins belong to the Mode [0 > 2,4,8] [the subset of proteins downregulated by 2, 4 and 8 Gy (2,4,8) compared with the non-irradiated control group (0)]. All other groups (e.g. proteins upregulated only by two higher doses; mode [0,2 < 4,8]) were less numerous, and no protein component was characterized by a clear dose-dependent mode of changes.
Table 1.
Exosome proteins upregulated and downregulated by radiation
| IR-upregulated proteins |
| Mode [0 < 2,4,8]: 100A11; ABP1; ACKR3; ACLY; ACTA1; ACTG1; ADAM30; ADRM1; AEN; AHNAK; AKAP8L; ANKFY1; ANXA1; ANXA11; APEH; APMAP; APOA4; APOM; ARF3; ARF4; ARHGEF6; ARMCX4; ARPIN; ARR3; ARRDC1; ASPN; ASS1; ATIC; ATP2B1; ATXN2; B4GALNT4; BCHE; BHLHE22; BNIP3L; BRD9; BSG; C1QTNF3; C1R; C4BPA; C8A; CALY; CAP1; CAPNS1; CASP12; CASP7; CCDC129; CCDC158; CCK; CCT2; CCT4; CCT6A; CCT7; CCT8; CD151; CD63; CD81; CD82; CDC42BPB; CENPF; CFAP65; CFHL5; CFI; CFL1; CHD4; CLIC1; CMTM2; CNOT9; COG2; COG7; COL12A1; COL17A1; COL5A2; COL6A2; COL6A3; COL6A5; COLEC10; COLEC11; COPB1; COPS2; CPB2; CSF2RB; CSRP1; CTR9; CUL5; CUX1; CYP21A2; DARS; DDIAS; DDX21; DGKZ; DHX9; DMXL1; DNAH9; DPYS; EED; EEF1A1P5; EFCAB5; EIF3C; EIF3F; EIF3L; ENO4; EPHB2; ERC2; ERCC5; ERLIN2; ESPN; ESRP2; ETNPPL; EXOC1; EXT1; F10; F13A1; FAM120A; FAM177A1; FAM3C; FAT4; FCRL6; FHAD1; FLG; FLG2; FLNA; FLNB; FMO4; FST; FUT6; FZD10; FZD2; GDPD5; GFPT1; GFRA1; GNAI2; GOLGA6L6; GPATCH11; GPC1; GPX3; GRIK4; H2AFV; HERC3; HGF; HGFAC; HIC2; HIST1H2AC; HIST1H3A; HIST1H4A; HIST2H2BF; HIST2H3PS2; HSP90AA1; HSP90AB1; HSPA8; HSPB1; HSPD1; HUWE1; IARS; IKBKE; IL1R1; ILDR2; IPO11; IST1; ITGA6; ITGB1; ITGBL1; JAK3; JMJD1C; KRT74; KRT77; KTN1; LAMA1; LAMA2; LAMA5; LAMB2; LAMB3; LAMC2; LAMP1; LIMK1; LRP2; LRP4; LRRC6; LRRC70; LTBP4; MACF1; MAPK6; MAPK8IP2; MATN2; MEGF6; MEGF8; MFGE8; MMP13; MORC4; MRC2; MROH2A; NAA16; NAP1L1; NAPA; NBR1; NCOR1; NCSTN; NDC1; NEB; NELL2; NEXN; NIPBL; NOL5A; NOS1; NOTCH1; NPTX1; NT5E; NUMA1; OMD; OR10J4; OR2A25; PAK7; PARG; PARK7; PAX6; PCSK2; PDCD6IP; PDZD2; PERP; PGM1; PHF3; PHGDH; PHKA1; PI4KB; PID1; PIGG; PIWIL1; PKM; PLCD1; PLEKHB1; PLSCR5; PLXNB2; POLI; PPM1G; PRDX1; PRDX2; PRG4; PROS1; PRSS23; PSAP; PSAT1; PSEN1; PSEN2; PSMA6; PSMC3; PSMC5; PSMC6; PSMD1; PTTG1IP; PYGB; RAB11A; RAB1A; RAB1B; RAB7A; RAC1; RAC2; RACK1; RAN; RECQL5; RGN; RGPD4; RIC8A; RNF19A; RNF6; RPL18A; RPL28; RPL30; RPL5; RPL7A; RPLP0; RPLP1; RPS18; RPS26P11; RPS27A; RPS3; RPS6; RPSA; RPTN; RRAS2; RRBP1; RTN4; RUVBL1; RYR2; S100A14; S100A2; SAP30BP; SBDS; SDC1; SERPINA5; SGCD; SHROOM2; SHROOM4; SLC1A5; SLC22A8; SLC28A2; SLC38A1; SLC38A5; SLC4A9; SLC7A1; SLC7A5; SMC5; SMCO3; SNCAIP; SORBS2; SOWAHB; SOX30; SPARCL1; SPIDR; SPOCK1; SPOCK2; SPON1; SPTBN5; SSH2; STAM; SULT2B1; SUSD5; SYCP1; SYMPK; TACSTD2; TBCD; TBCK; TBKBP1; TBR1; TCHH; TEX14; TFAP2D; TFRC; TGM6; THBD; THBS3; TIMP3; TINAGL1; TM4SF1; TM7SF3; TMPRSS6; TNFAIP2; TNFRSF19; TNKS; TOLLIP; TRIAD3; TRIML1; TROPH; TSPAN14; TSPAN9; TTC30B; TTI2; TTN; TUBA1A; U2AF1; U2SURP; UNC5CL; USP45; VIPR2; VPS4A; WBP2NL; WDSUB1; WNT7A; XPO6; XRCC5; XRCC6; ZC3H14; ZDHHC5; ZFHX3; ZFP64; ZNF25; ZNF563; ZNF671; ZNF770; ZNF804B; ZUFSP |
| Mode [0,2 < 4,8]: ABO; ANXA4; C8G; CCM2; CNGB1; COL18A1; DEFA3; F5; FBLL1; GNG12; LMNA; LRPPRC; MMRN1; NECAB2; OXCT; PRR13; QSOX1; RAB5B; RPL24; SEC14L2; SPRR2C; SRGAP3; SRI; SUPT4H1; TRPM7; TSPAN3; TTC9B; VPS37B; WWC2 |
| Mode [0,2,4 < 8]: CLDN1; CNTN4; CSMD3; DYNLL1; ETFRF1; GNG5; GUSB; KCNK1; KRT39; MAP3K7CL; MOB1B; ORAOV1; PDE4C; PGBD1; PPT1; PSMD2; RAB14; RAD51D; RBM26; RPL17; RPL36; RPS25; SET; TP53; TRIM22; UNQ3118; ZNF195 |
| IR-downregulated proteins |
| Mode [0 > 2,4,8]: ACOX3; APOA2; C1S; CASK; CIB3; COX10; CUBN; DDX58; FAM107A; FANCM; GCA; GTPBP1; IPO5; MME; PANK1; PLAT; RNF149; RPL26; RPL8; RPS14; RUVBL2; SAMD8; SETMAR; SLC34A1; SMCO2; SRL; STOX2; STRCP1; TG; THRA; TTN; ZC3H4 |
| Mode [0,2 > 4,8]: ANKRD62; BABAM1; COL9A1; HHLA1; NXF1; OAS3; SERPINB5; T |
| Mode [0,2,4 > 8]: ABI3BP; DDX41; LMCD1; LSR; PLCB4; SZT2; ZFP62 |
Presented are subsets of proteins that belong to different modes described in the text.
Fig. 2.
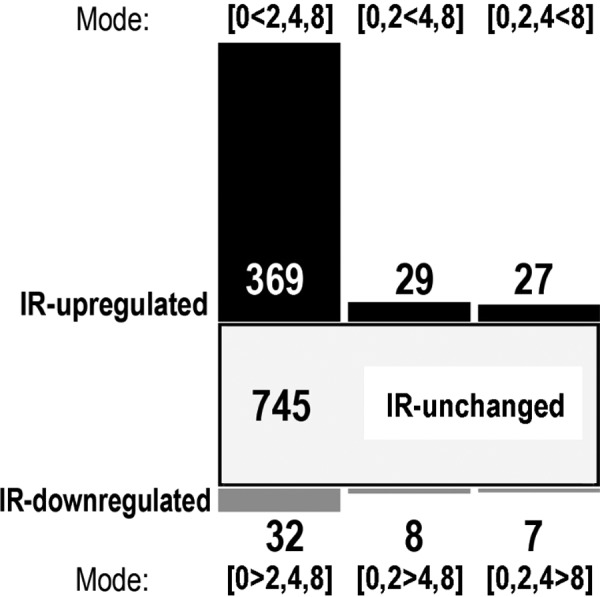
Numbers of exosome proteins whose abundances were affected by radiation; various dose-related modes of changes are shown (description in the text).
Subsequently, biological roles were attributed to exosome proteins affected by radiation using Gene Ontology tools. There was overrepresentation of IR-modulated proteins in several GO terms associated with exosome proteins. When the overrepresentation threshold was set at 1.3, five GO terms in the Biological Process (BP) domain with the largest number of IR-modulated proteins were as follows: DNA metabolic process (30 IR-affected vs 29 unaffected proteins), organelle fission (28 vs 27), regulation of vesicle-mediated transport (24 vs 21), mitotic nuclear division (23 vs 19) and DNA repair (21 vs 15). When the most significantly overrepresented GO terms were searched, a few groups of Biological Processes associated with IR-modulated proteins could be identified, including the response to IR and metabolism of ROS, DNA repair, DNA packaging, and de novo protein folding (Table 2), which were apparently associated functionally with key elements of the cellular response to radiation. A similar analysis was performed in two other GO domains—Cellular Component (CC) and Molecular Functions (MF)—with coherent results. Among GO terms showing overrepresentation of IR-modulated proteins were chromatin and proteasome localization and ATP-dependent helicase activity (Table 2).
Table 2.
Overrepresented GO terms associated with IR-modulated exosome proteins
| GO Term ID | IR-upregulated | IR-downregulated | IR-unchanged | Enrichment | P-value |
|---|---|---|---|---|---|
| GO:2000377 [BP]: regulation of ROS metabolic process | ARF4; ASS1; HSP90AA1; HSP90AB1; PARK7; PID1; RAC1; RAC2; RGN; TP53 | SZT2 | AGT; APS; CAV1; CLU; EGFR; GSTP1; INSR; PTK2B | 1.48 | 0.04 |
| GO:0006281 [BP]: DNA repair | COPS2; ERCC5; HIST1H4A; HUWE1; POLI; RAD51D; RECQL5; RPS27A; RPS3; RUVBL1; SMC5; SPIDR; SYCP1; TP53; USP45; XRCC5; XRCC6 | BABAM1; FANCM; RUVBL2; SETMAR | ATXN3; BAF53A; BRCA2; CSPG3; EP300; HERC2; MCM9; NPM1; RECQL; RFC1; SMC1A; TAOK3; TICRR; USP7; VIL2 | 1.50 | 0.01 |
| GO:0006323 [BP]: DNA packaging | AKAP8L; HIST1H3A; HIST1H4A; HIST2H2BF; HIST2H3PS2; NAP1L1; RUVBL1; SET; SYCP1; TP53 | TTN | ASH1L; CENPV; CHMP1A; HIST1H1B; HIST1H1D; NPM1 | 1.65 | 0.03 |
| GO:0010212 [BP]: response to ionizing radiation | AEN; ANXA1; NIPBL; RAD51D; SPIDR; THBD; TP53; XRCC5; XRCC6 | BABAM1 | BRCA2; DNMT3A; INTS7; MYC; TICRR | 1.70 | 0.03 |
| GO:0006458 [BP]: ‘de novo’ protein folding | CCT2; CCT4; CCT6A; CCT7; CCT8; HSPA8; HSPD1 | FYCO1; TCP1 | 2.00 | 0.02 | |
| GO:0000785 [CC]: chromatin | AKAP8L; CENPF; CHD4; CTR9; EED; H2AFV; HIST1H2AC; HIST1H3A; HIST1H4A; HIST2H2BF; HIST2H3PS2; JMJD1C; NCOR1; NIPBL; PARK7; PAX6; RAN; RUVBL1; TP53 | RUVBL2; T | AHCTF1; ANKRD17; CSNK2B; CTNNB1; DNMT3A; EIF3E; HIST1H1B; HIST1H1D; HNRNPC; MBD1; MUC1; POLR3G; RB1; TCP1; UBA1; VIL2; ZNF385A | 1.43 | 0.02 |
| GO:0031597 [CC]: cytosolic proteasome complex | PSMC3; PSMC5; PSMC6; PSMD1; PSMD2 | – | PSMC2 | 2.16 | 0.03 |
| GO:0008026 [MF]: ATP-dependent helicase activity | ABP1; CHD4; DDX21; DHX9; RECQL5; RUVBL1; XRCC5; XRCC6 | DDX41; RUVBL2 | CHD3; DDX59; EIF4A1; RECQL; YTHDC2 | 1.73 | 0.03 |
Presented are Gene Ontology (GO) terms corresponding to Biological Processes (BPs), Molecular Functions (MFs), and Cellular Components (CCs); IR = ionizing radiation.
Furthermore, considering the putative role of exosomes in transmitting the RIBE, we looked for a GO term hypothetically related to RIBE mediators, including stress-response regulators and apoptotic factors. IR-modulated proteins were not overrepresented in these processes. Interestingly, however, proteins associated with these processes (exemplified by stress-activated MAPK cascade, transcription in response to stress, apoptotic signaling, and regulation of autophagy) were detected only among IR-upregulated but not among IR-downregulated species (Table 3). Moreover, we looked for two groups of proteins naturally attributed to exosomes: tetraspanins (and other ‘exosome markers’) and heat shock proteins (Table 3). We found that the abundance of several proteins characteristic of exosomes, including CD9 and TSG101, were not altered in exosomes secreted by irradiated cells. However, an abundance of several other ‘exosome markers’, including CD63 and ALIX, increased in materials released after irradiation (no ‘exosome markers’ were IR-downregulated). This observation was coherent with data obtained by Western blot for CD9 and CD63 (Fig. 1D). Moreover, 5 out of 8 heat shock proteins (HSP90AA1, HSP90AB1, HSPA8, HSPB1, HSPD1) were markedly upregulated in exosomes released by irradiated cells (a similar effect was observed in the case of HSPA2 protein detected by western blot; not shown). Several proteins detected in exosomes released by UM-SCC6 cells were associated with immunity-related processes. This was exemplified by the GO term ‘Regulation of immune response’ (GO:0050776), consisting of 91 exosome proteins, including 34 IR-upregulated and 3 IR-downregulated species. However, none of these immunity-related processes were statistically overrepresented.
Table 3.
Selected proteins upregulated in exosomes from irradiated cells
| GO Term ID | IR-upregulated | Not affected |
|---|---|---|
| GO:0032872 [BP]: regulation of stress-activated MAPK cascade | FZD10, HGF, MAPK8IP2, NBR1, NCOR1, PRDX1, RPS3, TNFRSF19, UNC5CL, WNT7A | AMBP, CDC42, GSTP1, HRAS, MAP3K5, MAPK1, MYC, PTK2B, RIG, SDCBP, TAOK3, TRAF2, ZMYND11 |
| GO:0043618 [BP]: regulation of transcription from RNA polymerase II promoter in response to stress | NOTCH1, PSMA6, PSMC3, PSMC5, PSMC6, PSMD1, PSMD2, RPS27A, TP53 | EGLN3, EP300, HSPA5, MUC1, PSMA1, PSMA3, PSMA4, PSMA7, PSMB4, PSMB5, PSMB6, PSMC2, PSMD11, PSMD12 |
| GO:0097190 [BP]: apoptotic signaling pathway | AEN, BNIP3L, CASP12, CCK, CUL5, IKBKE, LAMP1, PERP, TP53 | BID, BRCA2, CAV1, CLU, CSPG3, EP300, EPHA2, FZD9, HRAS, KRT18, MAP3K5, PDCD6, SRGN, TIMM50, TRAF2 |
| GO:0010506 [BP]: regulation of autophagy | BNIP3L, CAPNS1, EXOC1, HGF, HSPB1, PARK7, PSAP, QSOX1, RAB1B, RAB7A, RPL28, RUVBL1, TRIM22, U2AF1 | ATP6V0A2, EIF4G1, EP300, FYCO1, HERC1, LRRK2, MET, MFN2, MHC2TA, SOGA3, VIL2 |
| Tetraspanins and other ‘exosome markers’ | CD63, CD81, PDCD6IP (ALIX), TSPAN14, TSPAN3, TSPAN9 | CD9, FLOT2, TSG101, TSPAN1, TSPAN11, TSPAN13, TSPAN4, TSPAN6 |
| heat shock proteins | HSP90AA1, HSP90AB1, HSPA8, HSPB1, HSPD1 | DNAJA2, HSP90B1, HSPA5 |
Presented are Gene Ontology (GO) terms corresponding to Biological Processes (BPs); IR = ionizing radiation.
DISCUSSION
The involvement of EVs in the cellular response to stress is not in doubt. Numerous reports have identified a role for EVs in the context of damage induced by genotoxic agents, that could be crucial for the effectiveness of some patient therapies [17–19]. The response of a tissue and a whole organism to treatment with IR could also be influenced by cell-to-cell communication mediated by EVs, however, the mechanism(s) of this phenomenon is (are) not well understood [20]. The current data regarding effects of irradiation on EV cargo, summarized by Jelonek et al. [21], shows that there is a big gap in understanding how radiation-induced changes in the composition of EVs translates into their functional importance. In the current study, an in-depth characterization of radiation-induced changes in the proteome composition of EVs was reported, which will help a hypothesis-driven exploration of the processes mediated by such vesicles.
In general, two possibly opposite roles were proposed for EVs released by irradiated cells: mediation of RIBEs (i.e. a potentially cytotoxic role) [3, 4, 7] and increased DNA damage repair and cell survival (i.e. a potentially cytoprotective role) [8]. Here, we reported that proteins involved in different aspects of DNA repair were the major group of proteins overrepresented in EVs released by irradiated cells, which supports the potential cytoprotective role of such vesicles. On the other hand, some proteins possibly involved in RIBE (e.g. apoptotic factors, factors involved in stress signaling), though not statistically overrepresented as a group, were also detected in EVs released by irradiated cells. Hence, the specific role of EVs released by irradiated UM-SCC6 cells should be addressed in functional studies, which was beyond the scope of this proteomics screen. Nevertheless, proteins involved in different aspects of the response to radiation and ROS, like the DNA damage response (e.g. p53 transcription factor–driven) and the unfolded protein response (e.g. HSPs) were represented in the protein cargo of EVs released by irradiated cells. Moreover, several lines of evidence demonstrated a functional association between radiation response and immunity [22], leaving space for a putative role of exosomes in a crosstalk between irradiated cells and immune system. However, our data did not confirm a significant overrepresentation of immunity-related species in the subset of exosome proteins upregulated by radiation in UM-SCC6 cells.
Unexpectedly, our study revealed histones and chromatin-associated factors among proteins overrepresented in EVs released from irradiated cells. The presence of nuclear proteins in exosomes and small EVs is a matter of discussion, and the International Society for EVs guide classified them in the category ‘Absent or under-represented in EVs/exosomes, but present in other types of EVs’ [23]. In general, histones are usually assigned to apoptotic bodies, possibly contaminating other fractions of vesicles. However, the presence of histones has also been reported in small EVs in various studies [24, 25]. It is important to note that the exosome purification protocol implemented in the current study should allow removing of apoptotic bodies, and the presence of these large vesicles was detected by neither TEM imaging nor DLS measurement. Moreover, a small fraction of cells in the sub-G1 phase suggested that there was a low level of radiation-induced apoptosis in UM-SCC6 cells (at a given time point), which also reduced the probability of ‘contamination’ of analyzed EVs by apoptotic bodies. Nevertheless, it was recently reported that double-stranded DNA and chromatin fragments are a natural component of exosomes that might be secreted from a cell to preserve homeostasis by removing harmful cytoplasmic DNA [26, 27]. Hence, our observation could indicate the actual presence of chromatin fragments in small EVs, which could have a functional significance for signaling between irradiated and non-irradiated cells.
The current study also revealed that the exposure of cells to stress factors affected the pattern of so-called ‘exosome markers’. Exosomes released by irradiated cells showed increased levels of certain common exosome proteins (e.g. CD63 or CD81), whereas others remained unchanged (e.g. CD9 or TSG101). Furthermore, heat shock proteins, another group of common exosome proteins, saw certain family members upregulated in EVs released by irradiated cells. Similar upregulation of HSPs in exosomes released by irradiated cells (exemplified by HSP90AA1 and HSP90AB1) was reported previously by Mutschelknaus et al. [13]. However, certain HSPs were not affected. It is worthwhile to note that a stimulus could either affect the packaging of certain proteins in a homogenous population of vesicles or production of a certain fraction of a heterogeneous population of vesicles is affected [28], and both mechanisms could be involved in specific radiation-mediated changes in the composition of exosomes.
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
FUNDING
This work was supported by Narodowe Centrum Nauki (National Science Centre, Poland): grant numbers: 2013/11/B/NZ7/01512 to MP and 2015/19/B/ST6/01736 to JP. AA was supported by ETIUDA doctoral scholarship from Narodowe Centrum Nauki (National Science Centre, Poland), grant number 2018/28/T/NZ5/00188.
REFERENCES
- 1. Bang C, Thum T. Exosomes: new players in cell–cell communication. Int J Biochem Cell Biol 2012;44:2060–4. [DOI] [PubMed] [Google Scholar]
- 2. Arscott WT, Tandle AT, Zhao S et al. . Ionizing radiation and glioblastoma exosomes: implications in tumor biology and cell migration. Transl Oncol 2013;6:638–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jella KK, Rani S, O’Driscoll L et al. . Exosomes are involved in mediating radiation induced bystander signaling in human keratinocyte cells. Radiat Res 2014;181:138–45. [DOI] [PubMed] [Google Scholar]
- 4. Al-Mayah A, Bright S, Chapman K et al. . The non-targeted effects of radiation are perpetuated by exosomes. Mutat Res 2015;772:38–45. [DOI] [PubMed] [Google Scholar]
- 5. Yu X, Harris SL, Levine AJ. The regulation of exosome secretion: a novel function of the p53 protein. Cancer Res 2006;66:4795–4801. [DOI] [PubMed] [Google Scholar]
- 6. Prise KM, O’Sullivan JM. Radiation-induced bystander signalling in cancer therapy. Nat Rev Cancer 2009;9:351–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Al-Mayah AH, Irons SL, Pink RC et al. . Possible role of exosomes containing RNA in mediating nontargeted effect of ionizing radiation. Radiat Res 2012;177:539–45. [DOI] [PubMed] [Google Scholar]
- 8. Mutschelknaus L, Peters C, Winkler K et al. . Exosomes derived from squamous head and neck cancer promote cell survival after ionizing radiation. PLoS One 2016;11:e0152213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Villarroya-Beltri C, Baixauli F, Gutiérrez-Vázquez C et al. . Sorting it out: regulation of exosome loading. Semin Cancer Biol 2014;28:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lehmann BD, Paine MS, Brooks AM et al. . Senescence-associated exosome release from human prostate cancer cells. Cancer Res 2008;68:7864–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hurwitz MD, Kaur P, Nagaraja GM et al. . Radiation therapy induces circulating serum Hsp72 in patients with prostate cancer. Radiother Oncol 2010;95:350–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jelonek K, Wojakowska A, Marczak L et al. . Ionizing radiation affects protein composition of exosomes secreted in vitro from head and neck squamous cell carcinoma. Acta Biochim Pol 2015;62:265–72. [DOI] [PubMed] [Google Scholar]
- 13. Mutschelknaus L, Azimzadeh O, Heider T et al. . Radiation alters the cargo of exosomes released from squamous head and neck cancer cells to promote migration of recipient cells. Sci Rep 2017;7:12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wiśniewski JR, Zougman A, Nagaraj N et al. . Universal sample preparation method for proteome analysis. Nat Methods 2009;6:359–62. [DOI] [PubMed] [Google Scholar]
- 15. Marczyk M, Jaksik R, Polanski A et al. . Adaptive filtering of microarray gene expression data based on Gaussian mixture decomposition. BMC Bioinformatics 2013;14:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013;200:373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Safaei R, Larson BJ, Cheng TC et al. . Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol Cancer Ther 2005;4:1595–1604. [DOI] [PubMed] [Google Scholar]
- 18. Xiao X, Yu S, Li S et al. . Exosomes: decreased sensitivity of lung cancer A549 cells to cisplatin. PLoS One 2014;9:e89534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vulpis E, Cecere F, Molfetta R et al. . Genotoxic stress modulates the release of exosomes from multiple myeloma cells capable of activating NK cell cytokine production: role of HSP70/TLR2/NF-kB axis. Oncoimmunology 2017;6:e1279372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Suchorska WM, Lach MS. The role of exosomes in tumor progression and metastasis (Review). Oncol Rep 2016;35:1237–44. [DOI] [PubMed] [Google Scholar]
- 21. Jelonek K, Widlak P, Pietrowska M. The influence of ionizing radiation on exosome composition, secretion and intercellular communication. Protein Pept Lett 2016;23:656–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lumniczky K, Candéias SM, Gaipl US et al. . Radiation and the immune system: current knowledge and future perspectives. Front Immunol 2018;8:1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lötvall J, Hill AF, Hochberg F et al. . Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles 2014;3:26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics 2008;8:4083–99. [DOI] [PubMed] [Google Scholar]
- 25. Meckes DG Jr, Gunawardena HP, Dekroon RM et al. . Modulation of B-cell exosome proteins by gamma herpesvirus infection. Proc Natl Acad Sci U S A 2013;110:E2925–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kahlert C, Melo SA, Protopopov A et al. . Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem 2014;289:3869–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Takahashi A, Okada R, Nagao K et al. . Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat Commun 2017;8:15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kowal J, Arras G, Colombo M et al. . Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A 2016;113:E968–77. [DOI] [PMC free article] [PubMed] [Google Scholar]