Abstract
The brain is the most common metastatic site of lung adenocarcinoma; however, the mechanism of this selective metastasis remains unclear. We aimed to verify the hypothesis that exposure of tumor cells to the brain microenvironment leads to changes in their gene expression, which promotes their oriented transfer to the brain. A549 and H1299 lung adenocarcinoma cells were exposed to human astrocyte-conditioned medium to simulate the brain microenvironment. Microarray analysis was used to identify differentially expressed genes, which were confirmed by quantitative real-time PCR and western blotting. Knockdown experiments using microRNAs and the overexpression of genes by cell transfection were performed in addition to migration and invasion assays. In vitro findings were confirmed in clinical specimens using immunohistochemistry. We found and confirmed a significant increase in insulin-like growth factor binding protein-3 (IGFBP3) levels. Our results also showed that the up-regulation of IGFBP3 promoted A549 cell epithelial-mesenchymal transition, migration, and invasion, while the knockdown of IGFBP3 resulted in decreased cell motility. We also found that Transforming growth factor-β (TGF-β)/Mothers against decapentaplegic homolog 4 (Smad4)-induced epithelial-mesenchymal transition was likely IGFBP3-dependent in A549 cells. Finally, expression of IGFBP3 was significantly elevated in pulmonary cancer tissues and intracranial metastatic tissues. Our data indicate that up-regulation of IGFBP3 might mediate brain metastasis in lung adenocarcinoma, which makes it a potential therapeutic target.
Keywords: brain metastasis, EMT, IGFBP3, lung adenocarcinoma
INTRODUCTION
Lung cancer is the leading cause of cancer-related deaths (Siegel et al., 2016), of which most are caused by distant metastases (Dragoj et al., 2017). The brain is the most common metastatic site of lung cancer. Among the cell types of non-small cell lung cancer (NSCLC), non-squamous cell tumors are especially linked to metastatic disease in the brain (Mujoomdar et al., 2007; Sul and Posner, 2007). Conversely, more than half of all brain metastasis originate from lung cancer (Budczies et al., 2015). In spite of recent progress in clinical diagnosis and treatment of NSCLC, effective therapeutic strategies targeting metastasis remain highly inadequate (Yousefi et al., 2017). Therefore, an in-depth understanding of the mechanism underlying NSCLC brain metastasis is crucial to identify new therapeutic targets and improve the prognosis of lung cancer.
Insulin-like growth factor binding protein-3 (IGFBP3) is the primary carrier of insulin growth factors (IGFs) in the circulation (Jogie-Brahim et al., 2009). Abnormal expression of IGFBP3 has been linked to the pathogenesis of cancers. Numerous previous studies focused on tumor suppressor activities of IGFBP3 (Jogie-Brahim et al., 2009; Li et al., 2012), but pro-tumor effects of IGFBP3 have been reported in esophageal carcinoma (Takaoka et al., 2004), breast cancer (Vestey et al., 2005), and oral squamous cell carcinoma (Yen et al., 2015). In addition, liver metastasis of pancreatic endocrine cancer (Hansel et al., 2004) and the distant metastasis of nasopharyngeal carcinoma (Bao et al., 2016) are associated with increased IGFBP3. However, the effects of IGFBP3 on brain metastasis of NSCLC have been rarely reported.
In the current study, we investigated the functional correlations between IGBBP3 and brain metastasis of lung adenocarcinoma using human tissue specimens and cell culture experiments. Our data indicated that IGFBP3 might mediate brain metastasis of lung adenocarcinoma.
MATERIALS AND METHODS
Cell culture and treatment
The human lung adenocarcinoma (LUAD) cell lines A549 and H1299 were obtained from the experimental Medicine Center, at the affiliated hospital of Southwest Medical University (Luzhou, China). The human astrocyte cell line HA1800 was obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were maintained at 37°C in 5% CO2. Unless otherwise noted, cells were cultured in complete medium containing RPMI 1640 (Hyclone, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, USA) and penicillin/streptomycin (Beyotime Biotechnology, China). Depending on the cell line, the cultures were passaged every 3–4 days using Trypsin-EDTA Solution (Beyotime Biotechnology) for cell detachment. Confluent cultures of A549 cells were maintained in RPMI containing 0.1% FBS for 24 h prior to stimulation with recombinant human TGF-β1 (PeproTech, USA) and then stimulated with 5 ng/ml of TGF-β1 for 48 h.
Preparation of conditioned medium
A549 cells, H1299 cells, and HA1800 cells were cultured to 50–60% confluency, the medium containing 10% FBS was then removed and replaced with medium containing 1% FBS, and the cells cultured for 48 h at 37°C. The A549 culture medium (A549-CM), H1299 culture medium (H1299-CM) and HA1800 culture medium (HA1800-CM) were then collected. The culture medium was filtered through 0.22 μm filters (JET BIOFIL, China) and stored at −80°C until further use.
Wound healing assay
A549 cells or H1299 cells were plated onto six-well plates. When the cell confluency reached 90%, the cell monolayers were incubated in serum-free medium for 12 h. Confluent cells were wounded using a 200 μL sterile pipette tip to ensure similar scratch widths for each well. The cells were then rinsed with phosphate-buffered saline (PBS; Maxim, China) to remove free-floating cells and debris. Finally, conditioned medium was added to each well and the cells incubated at 37°C. Wound healing was monitored, and representative scrape-lines were photographed at various time points.
Microarray data analysis
Total RNA was isolated using TRIzol reagent (Tiangen, China). The RNA quality testing, IVT target preparation (GeneChip 3’ IVT PLUS Kit), and GeneChip hybridization and scanning were commercially performed by Genechem (China). Micro RNAs (miRNAs) displaying more than a 2-fold change in expression were considered significant. The major instruments included the Thremoβ Nanodrop 2000 spectrophotometer, Agilentβ 2100 Bioanalyzer, Affymetrixβ GeneChip Hybridization Oven 645, Affymetrixβ Fluidics Station 450, and Affymetrixβ GeneChip Scanner 3000.
Quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was extracted using an RNA extraction kit (Tiangen), and the samples were stored at −80°C. The concentration of RNA was measured using a NanoDrop ND-1000 spectrophotometer. Reverse transcription was performed using a cDNA Reverse Transcription Kit (Toyobo, Japan) under the following conditions: pre-denaturation at 65°C for 5 min, 37°C for 15 min, 50°C for 5 min, and 98°C for 5 min. The reaction components were added using a SYBR Green Real-time PCR Master Mix (TOYOBO) according to the manufacturer’s instructions in a reaction volume of 20 μL. All primers were designed and synthesized by Sangon Biotech (China). The sequences of the qRT-PCR primers are shown in Table 1. Reaction conditions included 95°C for 30 s followed by 40 cycles of 95°C for 5 s, 60°C for 10 s, 72°C for 15 s. The relative expression levels of mRNA were calculated using the 2−ΔΔCt method, and β-actin was used as a reference gene. The experiments were repeated three times.
Table 1.
List of primer sequences used for qRT-PCR analysis
| Gene name | Primer sequences (5′ to 3′)a |
|---|---|
| IGFBP3 | IGFBP3 F: ATGGTCCCTGCCGTAGA |
| IGFBP3 R: CCCGCTTCCTGCCTTT | |
| E-cadherin | E-cadherin F: GAACGCATTGCCACATAC |
| E-cadherin R: ACCTTCCATGACAGACCC | |
| N-cadherin | N-cadherin F: TCCTGCTTATCCTTGTGC |
| N-cadherin R: GTCCTGGTCTTCTTCTCCT | |
| Smad1 | Smad1 F: ATGCCACTCAACGCCACTTT |
| Smad1 R: CCGCCTGAACATCTCCTCTG | |
| Smad2 | Smad2 F: AATGCCACGGTAGAAATGAC |
| Smad2 R: ACAGACTGAGCCAGAAGAGC | |
| Smad3 | Smad3 F: GGAGGAGAAATGGTGCGAGAA |
| Smad3 R: CCACAGGCGGCAGTAGAT | |
| Smad4 | Smad4 F: GGATACGTGGACCCTTCT |
| Smad4 R: CGATGACACTGACGCAAA | |
| Smad5 | Smad5 F: TAGCAGCACATATCCCAACT |
| Smad5 R: AGGCTCTTCATAGGCAACA | |
| Smad6 | Smad6 F: CCCCATCTTCGTCAACTCC |
| Smad6 R: GCTGATGCGGACGCTGTT | |
| Smad7 | Smad7 F: AACCGCAGCAGTTACCC |
| Smad7 R: CGAAAGCCTTGATGGAGA | |
| Smad8 | Smad8 F: ACATTCCAGGCTTCCTCCCG |
| Smad8 R: TGCTGTCACTCACGCACTCG | |
| β-actin | β-actin-F: GTGGACATCCGCAAAGA |
| β-actin-R: CTCGTCATACTCCTGCTTG |
F, forward; R, reverse
Lentivirus transfection
The lentivirus vector expressing human IGFBP3 (IGFBP3) and control empty vector (Control) were commercially obtained (Genechem). The primers used to amplify the IGFBP3 gene for insertion into the lentivirus vector were IGFBP3-F: 5′-ATGGTCCCTGCCGTAGA-3′ and IGFBP3-R: 5′-CCCGCTTCCT GCCTTT-3′. The lentivirus vectors (IGFBP3 or Control) were transfected into the cell culture cells according to the manufacturer’s protocol. For the establishment of the cell line stably expressing IGFBP3, 12 h after transfection the medium was replaced with medium containing 10% FBS and the cells were cultured for 72 h. The transfected cells were screened with puromycin (Clontech, USA) and the selected cells were harvested for further use. The efficiency of IGFBP3 overexpression was assessed using qRT-PCR and western blot analysis.
Transient RNA interference
Short interfering RNA (siRNA) designed against IGFBP3 (si-BP3) and nonspecific si-RNA (Con) used as negative control were purchased from RiboBio (China). A total of 3 × 105 A549 cells/well were cultured in 6-well plates and transfected with 5 μL siRNA or negative control, diluted with 120 μL 1× riboFECTδ CP Buffer (RiboBio). Then, 12 μL of riboFECTδ CP Reagent (RiboBio) was added. Finally, the volume was adjusted to 2.0 mL with serum-free medium. At 48 h post-treatment, the cells were harvested and evaluated by qRT-PCR and used in subsequent experiments. The siRNA designed against Smad4 (si-Smad4) and nonspecific siRNA (con) used as negative control were also purchased from RiboBio and transfected into cells using the same transfection process as above.
Transwell assays
Invasion and migration assays were performed using 6.5-mm diameter polycarbonate membranes with 8.0-μm pores (USA) with 30 μL Matrigel (BD Biosciences, USA) for the invasion assays and without Matrigel for the migration assays. In brief, cell suspensions (4 × 104) prepared in 200 μL serum-free medium were added into the upper chambers of the 24-well transwell culture plates for 3 h with 600 μL of complete medium containing 10% FBS being placed in the lower chambers. The cells were incubated for 24 h at 37°C to allow the cells to migrate or invade through the membrane filter. Next, the non-migratory and non-invasive cells on the upper surface were gently removed using a cotton swab. The cells that had invaded or migrated to the lower surface of the membrane were stained with 0.5% crystal violet solution (Beyotime Biotechnology) for 15 min and the cells counted using a light microscope (Nikon, Japan). Finally, cells in four random microscopic fields at 100× magnification were counted and photographed. All experiments were independently repeated in triplicate.
Western blot analysis
Proteins were extracted from the cells using RIPA buffer (Beyotime Biotechnology, China) complemented with 1% phenylmethylsulfonyl fluoride (PMSF; Beyotime Biotechnology) as a protease inhibitor. For western blot analysis, the protein extracts were subjected to electrophoresis using 10% SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes (Beyotime Biotechnology). Immunoblots were blocked with 5% non-fat milk in TBS/Tween 20 (TBST; Beyotime Biotechnology) and incubated with primary antibodies overnight at 4°C. The primary antibodies included anti-IGFBP3 (1:500, Santa Cruz Biotechnology, USA), anti-E-cadherin (1:1000, Cell Signaling Technology, USA), anti-N-cadherin (1:1000, Cell Signaling Technology), anti-Smad4 (1:1000, Abcam, USA), and anti-α-tubulin (Beyotime Biotechnology)( 1:1000, Beyotime Biotechnology). The membranes were washed with 0.1% TBST and then incubated with the appropriate secondary antibodies (Beyotime Biotechnology) for 1 h at room temperature. The bands were visualized using the Chemilmanger (TM) 5500 System (Alpha Innotech, USA). The gray value was calculated using Image J software. The experiments were repeated three times.
Tissue specimens
Patient specimens used in the study were retrieved from the archival files of the Department of Pathology, Affiliated Hospital of Southwest Medical University. A total of 15 normal lung tissue specimens and 48 specimens from patients with LUAD who had undergone no preoperative treatment and included 15 non-metastatic pulmonary cancer tissue specimens, 10 brain-metastatic pulmonary cancer tissue specimens, 15 other metastatic pulmonary cancer tissue specimens, and 8 brain metastatic lesion specimens. All human tissues were collected using protocols approved by the Ethics Committee of Affiliated Hospital of Southwest Medical University.
Immunohistochemistry (IHC)
Paraffin-embedded, 4-μm-thick tissue sections from 63 specimens were stained for IGFBP3. All of the sections were de-paraffinized in a series of xylene baths and rehydrated using a graded alcohol series. The sections were then immersed in methanol containing 0.3% hydrogen peroxide for 20 min to block endogenous peroxidase activity and incubated in 2.5% blocking serum to reduce nonspecific binding. Sections were incubated overnight at 4°C with the primary anti-IGFBP-3 antibody (1:50, Abcam). Next, the sections were incubated with horseradish peroxidase (HRP)-labeled secondary antibodies (Beyotime Biotechnology) at 37°C for 30 min and visualized with a 3,3′-diaminobenzidine (DAB) detection kit to yield a dark brown color. The sections were observed by light microscopy (Leica TE2000-S Microscope, Japan). Representative areas of each tissue section were selected, and the cells were counted in at least four randomly selected fields (at 100× and 200×). The IGFBP3 labeling index was defined as the percentage of tumor cells displaying cytoplasmic immunoreactivity and was calculated by counting the number of IGFBP3-positive stained tumor cells per total number of cells from the representative areas of each tissue section. In this study, a 25% labeling index as a cutoff point was used. On the basis of the results of the immunohistochemical staining, tissue sections showing more than 25% of positive staining were considered overexpressing of IGFBP3. All of the slides of tissues were independently evaluated and scored by two pathologists who were blinded to the clinical information of the subjects.
Statistical analysis
All statistical analyses were performed using SPSS 17.0 software. Unless otherwise noted, all data were expressed as mean ± standard error of the mean (SEM). Statistical significance between two groups was ascertained using two-tailed Student’s t-test. Two-sided p-values < 0.05 were considered as statistically significant. Comparison of immunolabeling between populations was performed using two-sided Fisher’s exact test or the Chi-Square (χ2) test.
RESULTS
HA1800 conditioned medium up-regulated IGFBP3 expression in Lung adenocarcinoma cells
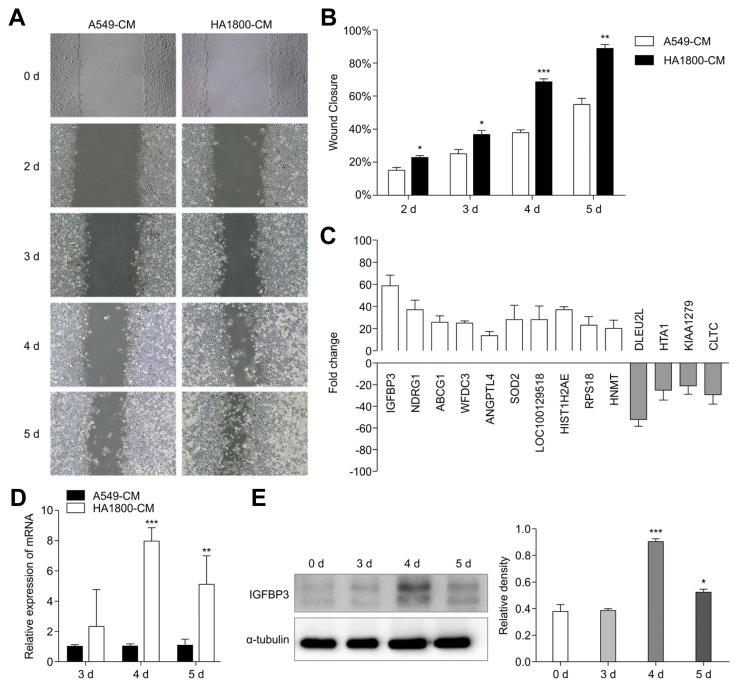
In the metastatic process, the microenvironment of the metastatic sites plays an important role for tumor cells to invade the target tissues (Fidler et al., 2002). We evaluated changes in the migration capacity of tumor cells cultured in a simulated brain microenvironment by using A549 cells exposed to HA1800-CM (experimental group) and A549-CM (control group). We then performed the wound-healing experiments by scratching the cell monolayers on the second day and monitoring the scratch healing for four consecutive days. The results demonstrated that the healing ability of the cells treated with HA1800-CM in the experimental group was significantly improved compared to that in the control group with the most significant difference being observed on the 4th and 5th day of healing. The relative wound closure rates, expressed as the percent closure, were 68.4 ± 1.6 for the experimental group and 37.75 ± 1.8 for the control group on the 4th day, and 88.17 ± 2.7 for the experimental group and 54.67 ± 3.6 for the control group on the 5th day (Figs. 1A and 1B). Since HA1800-CM promoted the migration of A549 cells, we then explored the effect of the simulated brain microenvironment on the gene expression in lung cancer cells by microarray analysis. Both the experimental and control groups were evaluated by microarray gene analysis. According to the microarray analysis, the number of differentially expressed genes between the two groups was 4,145. 1,946 genes were up-regulated (ratio > 2.0) and 2,199 genes were down-regulated (ratio < 0.5). The results showed that levels of IGFBP3, NDRG1, and other related transcripts in the experimental group were significantly higher than those in the control group (Fig. 1C). IGFBP3 increased 58.3-fold, which was the most increased gene. The role of IGFBP3 in the brain metastasis of lung cancer has been rarely reported, hence in the current study, we chose to consider IGFBP3 to confirm further that IGFBP3 expression levels were significantly up-regulated in the experimental groups compared with those in the control groups, as demonstrated by qRT-PCR (Fig. 1D) and western blot analyses (Fig. 1E). Intriguingly, in line with the results from the wound healing experiment, IGFBP3 was significantly upregulated on 4 d and 5 d post-treatment. To investigate whether the simulated brain microenvironment has the same effect on IGFBP3 in other lung adenocarcinoma cells also, we exposed H1299 lung adenocarcinoma cells to the brain microenvironment, and measured the migration ability changes by wound-healing experiments and the expression of IGFBP3 by qRT-PCR and western blot analyses. The relative wound closure rates, expressed as the percent closure, were 65.18 ± 2.165 for the experimental group (HA1800-CM) and 39.43 ± 1.543 for the control group (H1299-CM) on the 4th day, and 74.93 ± 1.715 for the experimental group and 60.42 ± 2.465 for the control group on the 5th day (Supplementary Figs. S1A and S1B). Next, we observed that, in the experimental groups, IGFBP3 expression levels were significantly up-regulated as compared to that in the control groups, as confirmed by qRT-PCR (Supplementary Fig. S1C) and western blot analyses (Supplementary Fig. S1D). These data suggested that the brain microenvironment might up-regulate the expression of IGFBP3 in A549 and H1299 lung adenocarcinoma cells, which may be involved in the progression of migration and invasion by LUAD cells. In the following experiments, we selected the commonly used A549 as the representative cell line to further explore the role of IGFBP3 in brain metastasis of lung adenocarcinoma and its potential mechanism.
Fig. 1. HA1800-CM up-regulated IGFBP3 expression in A549 cells.
(A) The wound healing of A549 cells treated with A549-CM and HA1800-CM at different time points. (B) Wound closure of A549 cells treated with A549-CM and HA1800-CM at different time points. (C) Microarray analysis showed that IGFBP3 and other related molecules were up-regulated in the experimental group (HA1800-CM). (D) IGFBP3 mRNA expression was evaluated by qRT-PCR analysis. (E) IGFBP3 protein expression was detected by western blot, α-tubulin was used as the reference protein (left). The histograms represent the protein expression levels quantified by densitometry and normalized to α-tubulin (right). Data are presented as the mean ± S.E.M. (*p < 0.05, **p < 0.01, ***p < 0.001)
Overexpression of IGFBP3 promoted epithelial-mesenchymal transition (EMT) of A549 cells
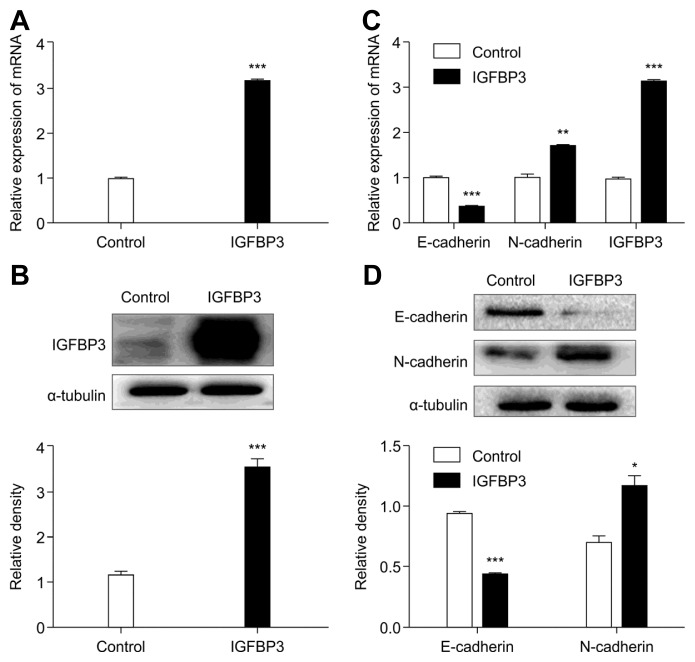
Previous studies have shown that EMT plays a vital role in IGFBP3 promotion of tumor metastasis (Natsuizaka et al., 2010). To explore whether overexpression of IGFBP3 would more clearly change cell migration, we verified the expression levels of some EMT-associated genes by qRT-PCR and western blot analyses. First, overexpression of IGFBP3 in A549 cells was induced by transfection of the lentivirus vector expressing IGFBP3. A549 cells transfected with an empty vector served as controls. Second, we performed qRT-PCR and western blot analyses to confirm the transfection efficiency (Figs. 2A and 2B). Finally, we evaluated the expression level of some EMT-associated genes. The results showed that the up-regulation of IGFBP3 was accompanied with the down-regulation of epithelial marker E-cadherin and up-regulation of mesenchymal marker N-cadherin (Figs. 2C and 2D).
Fig. 2. Role of IGFBP3 in EMT of A549 cells.
(A) IGFBP3 mRNA and (B) protein expression were detected by qRT-PCR and western blotting, respectively, after transduction with IGFBP3. The histograms represent the protein expression levels quantified by densitometry and normalized to α-tubulin. (C) Detection of expression of EMT markers by qRT-PCR and (D) western blotting after overexpression of IGFBP3 in A549 cells. The histograms represent the protein expression levels quantified by densitometry and normalized to α-tubulin. Data are presented as the mean ± S.E.M. (*p < 0.05, **p < 0.01, ***p < 0.001)
IGFBP3 accumulation was required for TGF-β-induced EMT
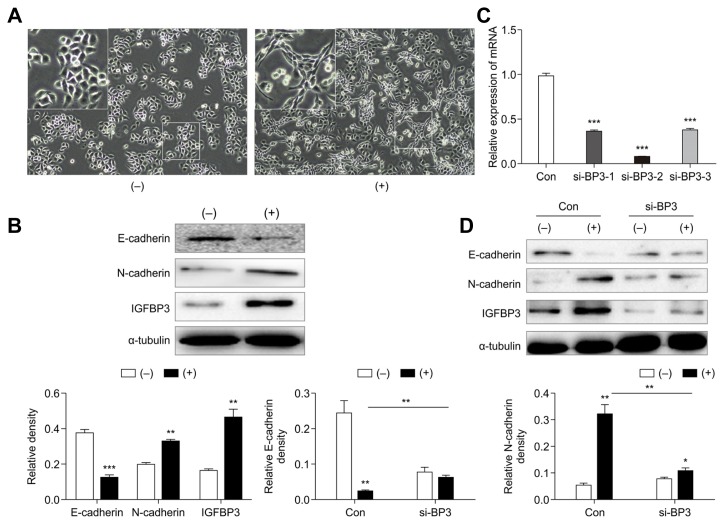
There is an increasing amount of evidence showing that TGF-β signaling is an essential inducer of EMT in various cancers, including NSCLC (David et al., 2016; Gregory et al., 2011; Li et al., 2018). Therefore, we investigated the role of IGFBP3 in TGF-β-induced EMT. The presence of TGF-β triggered the transition of A549 cells from a cobblestone-like morphology to an elongated shape that is associated with increased cell scattering (Fig. 3A). In line with the morphological changes, we also found by western blotting that the expression levels of E-cadherin were significantly decreased and those of N-cadherin were increased following TGF-β1 treatment of A549 cells at 48 h post-treatment. In addition, IGFBP3 expression levels were increased following TGF-β1 treatment (Fig. 3B).
Fig. 3. IGFBP3 accumulation was required for the induction of EMT and cell motility in response to TGF-β.
(A) A549 cells were treated with TGF-β (5 ng/mL) for 48 h, and cell morphology changes were analyzed by bright-field microscopy (magnification, 100×). (−) indicates that TGF-β1 was not added. (+) indicates the addition of 5 ng/mL TGF-β1. The magnified area in (A) corresponds to the area highlighted by the white square. (B) Western blot analysis of epithelial-mesenchymal transition marker and IGFBP3 expression in response to TGF-β induction in A549 cells (above). The histograms represent the protein expression levels quantified by densitometry and normalized to α-tubulin (below). (C) The interference efficiencies of three distinct IGFBP3-targeting siRNAs (si-BP3-1, si-BP3-2, and si-BP3-3) were evaluated by qRT-PCR after transfection into A549 cells. β-actin was used to normalize for equal loading. (D) Western blotting of E-cadherin and N-cadherin in A549 cells transfected with the IGFBP3-specific si-RNA (2 d ± TGF-β treatment; above). The histograms represent the protein expression levels quantified by densitometry and normalized to α-tubulin (below). Data are presented as the mean ± S.E.M. (**p < 0.01, ***p < 0.001)
Next, we transfected A549 cells with si-IGFBP3-RNA (si-BP3-1, si-BP3-2, or si-BP3-3) or a negative control si-RNA (Con) and then performed qRT-PCR to determine the interference efficiency of the IGFBP3-siRNA. The results showed that si-BP3-2 effectively attenuated the expression of IGFBP3 (Fig. 3C). Based on these findings, we chose si-BP3-2 for use in the subsequent experiment. Results demonstrated that the si-BP3-2-induced down-regulation of IGFBP3 remarkably decreased TGF-β-induced EMT and that si-BP3 significantly restored E-cadherin expression and impaired N-cadherin expression in the presence of TGF-β1 (Fig. 3D). These results suggested that IGFBP3 accumulation was required for the induction of EMT and cell motility in response to TGF-β.
Overexpression of IGFBP3 promoted invasion and migration by A549 cells
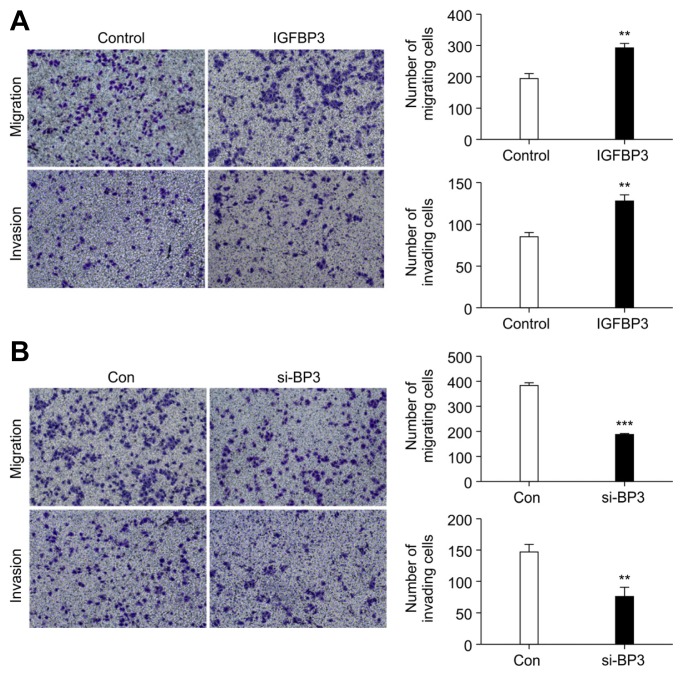
To investigate whether IGFBP3 promoted LUAD metastasis, we performed in vitro migration and invasion assays using A549 cells. As shown in Fig. 4A, migration (291.0 ± 13.65) and invasion (126.7 ± 7.796) of A549 cells were remarkably promoted by the transfection of the cells with an IGFBP3-expressing vector. In contrast, transfection with control vector had no significant effect on the migration (194.7 ± 15.62) or invasion (84.00 ± 3.606) of the cells (p < 0.01) by A549 cells.
Fig. 4. Up-regulation of IGFBP3 enhances A549 athletic ability.
(A) The migration and invasion of A549 cells overexpressing IGFBP3 were analyzed using transwell assays. (B) The migration and invasion of A549 cells transfected with si-BP3 were evaluated using transwell assays. Error bars represent the mean ± S.E.M. of three independent experiments. (**p < 0.01, ***p < 0.001)
Next, we used si-BP3-2 to interfere with the expression of IGFBP3 in the A549 cells and observe changes in both migration and invasion phenotypes. We found that migration (183.3 ± 6.936) and invasion (74.33 ± 8.570) of A549 cells were significantly inhibited by transfection with si-BP3-2 compared with the migration (380.7 ± 12.03) and invasion (145.3 ± 7.265) of the control group (Fig. 4B). These data confirmed that the up-regulation of IGFBP3 probably promoted invasion and migration by A549 cells.
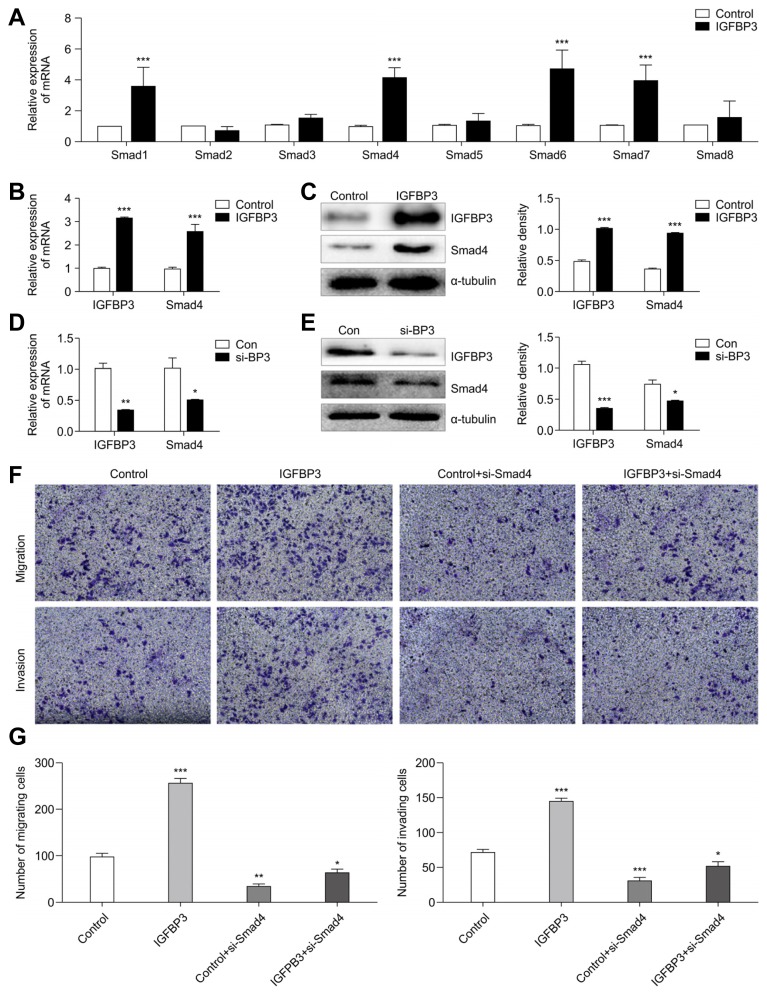
Smad4 and IGFBP3 were synergistic in the induction of A549 cell motility
Our results indicated that IGFBP3 might promote TGF-β-induced EMT of A549 cells. It has been previously shown that TGF-β-mediated EMT is primarily dependent on canonical TGF-β/Smad signaling (Bardeesy et al., 2006; Pardali and Moustakas, 2007). The aim of this series of experiments in the current study was to determine the relationship between IGFBP3 and the Smad family. We first investigated changes in the genetic expression of Smad family members when IGFBP3 expression was altered. Results from qRT-PCR showed that expression from Smad1, Smad4, Smad6, and Smad7 were higher in the IGFBP3 overexpression group compared with that in the control group (Fig. 5A). Previous studies have revealed that Smad4 is the key molecule for TGF-β-induced EMT (Cheng et al., 2015; Hesling et al., 2011) and that it promotes breast cancer bone metastasis through TGF-β-induced EMT (Deckers et al., 2006). Therefore, we next investigated whether Smad4 acted as a downstream target gene to mediate the pro-metastatic role of IGFBP3 in A549 cells. First, we detected the expression levels of Smad4 in IGFBP3-overexpressing A549 cells. The results indicated that Smad4 expression was increased when IGFBP3 was up-regulated (Figs. 5B and 5C). Inversely, the down-regulation of IGFBP3 was accompanied by the down-regulation of Smad4 (Figs. 5D and 5E). Second, we evaluated the effect of Smad4-siRNA (si-Smad4) being transiently transfected into A549 cells that had been stably transfected with IGFBP3 vector or empty control vector. As expected, following transfection with si-Smad4, even if IGFBP3 was overexpressed, migration and invasion of A549 cells were significantly inhibited (Figs. 5F and 5G). These findings confirmed that IGFBP3 might promote cell migration and invasion by targeting Smad4 in A549 cells.
Fig. 5. IGFBP3 regulates Smad4 expression.
(A) Alteration of the Smad gene family in A549 cells overexpressing IGFBP3 were evaluated by qRT-PCR analysis. β-actin was used to normalize for equal loading. (B) Smad4 mRNA and (C) protein expression levels were detected by qRT-PCR and western blotting, respectively, after transduction with IGFBP3. (D) Smad4 mRNA and (E) protein expression levels were detected by qRT-PCR and western blotting, respectively, after transfection of si-BP3. (F, G) The motility of A549 cells transfected with si-Smad4 overexpressing IGFBP3 was evaluated using transwell assays. Error bars represent the mean ± S.E.M. of three independent experiments. (**p < 0.01, ***p < 0.001)
Overexpression of IGFBP3 in Metastatic LUAD
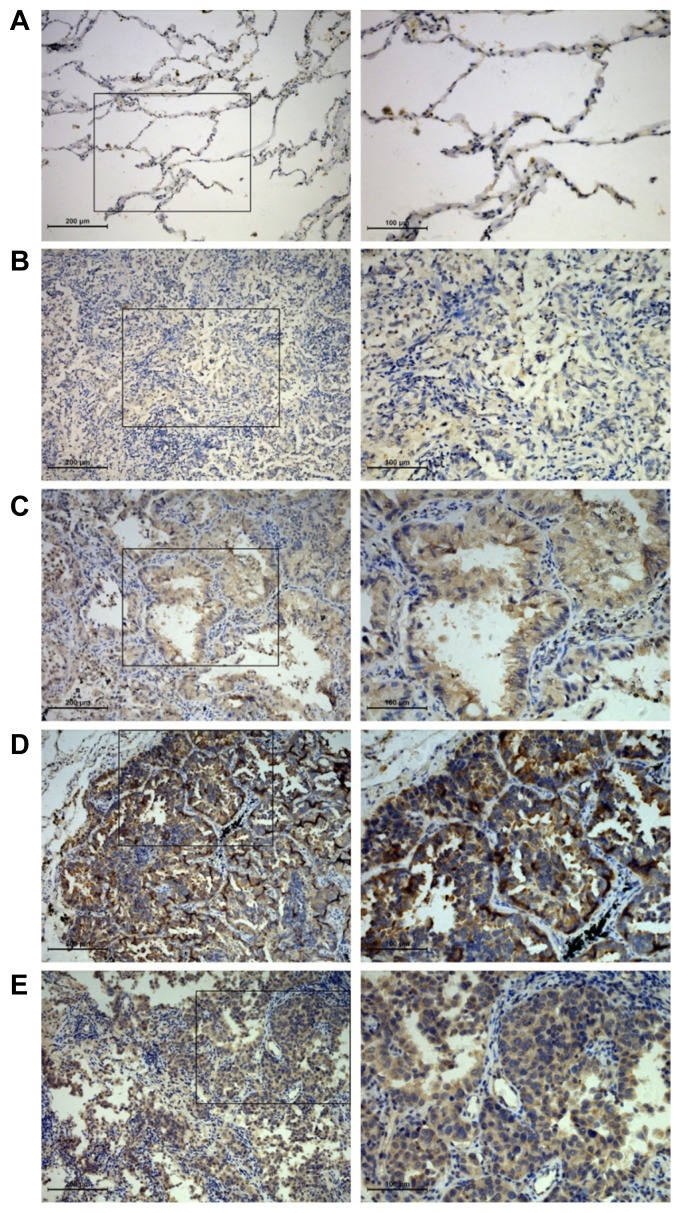
To explore the clinical significance of IGFBP3 in LUAD progression, we examined IGFBP3 expression using IHC. Examination of LUAD primary lung tissue specimens revealed that 55% (22/40) of the lesions demonstrated IGFBP3 expression. In contrast, only 13.3% (2/15) of the control group specimens expressed detectable levels of IGFBP3 (Fig. 6), which was a significant difference (Pearson Chi-Square, p = 0.006). Meanwhile, we found that lung cancer tissue specimens with distant metastasis (17/25, 68%) had higher expression levels of IGFBP3 compared to those without metastasis (5/15, 33.3%; Pearson Chi-Square, p = 0.033). This was especially the case in lung cancer tissue specimens with brain metastasis (8/10, 80%) in which IGFBP3 expression was higher than that compared with non-metastatic lung cancer tissue specimens (5/15, 33.3%; Fisher’s exact test, p = 0.041). Greater expression of IGFBP3 was also confirmed in brain metastatic lesions (7/8, 87.5%; Fisher’s exact test, p = 0.027) as compared with that of the expression in non-metastatic lung tissues (Fig. 6 and Table 2).
Fig. 6. Differential expression of IGFBP3 in lung and brain tissues of LUAD.
The representative fields of IGFBP3 expression in different groups by IHC were observed under light microscopy with 100× (left) and 200× (right) magnifications. (A) Normal lung tissues; (B) non-metastatic primary lesions; (C) other-metastatic primary lesions; (D) brain-metastatic primary lesions; (E) brain metastatic lesions. On the right column, magnifications of the areas demarcated by black squares on the left column are shown.
Table 2.
Expression of IGFBP3 in different tissues
| Control | Non-metastatic primary | Other-metastatic primary | Brain-metastatic primary | Brain metastases |
|---|---|---|---|---|
| 2/15 (13.3%) | 5/15 (33.3%) | 9/15 (60%) | 8/10 (80%) | 7/8 (83.33%) |
DISCUSSION
NSCLC represents about 85% of all pulmonary carcinomas and is the leading cause of cancer-related death worldwide (Siegel et al., 2016). Despite recent progress in the clinical treatment of NSCLC, the mortality rate has not been noticeably reduced, mainly due to the potential of tumor cells to metastasize (Dragoj et al., 2017), with the brain being the most common metastatic site of lung cancer (Sul and Posner, 2007). Early prevention and control of lung cancer is a vital node for lung cancer treatment. Thus, it is necessary to explore further the regulation of signaling molecules involved in brain metastasis of LUAD.
In the metastatic process, the microenvironment of the metastatic site plays an important role in tumor cells invasion into the target tissues (Fidler et al., 2002). In the brain, astrocytes are the most abundant cell population and contribute to the colonization, invasion, and movement of tumor cells (Balkwill and Mantovani, 2001; Fitzgerald et al., 2008). Astrocytes can produce many chemokines (Noda et al., 2012), some of which can promote the orientation of tumor cells to brain metastasis (Seike et al., 2011). Thus, in the current study, we used the astrocytes-conditioned medium to simulate the brain microenvironment. Previous studies have shown that IGFBP3 expression is increased in patients with bone metastasis of lung cancer (Sun et al., 2015), liver and lymph node metastasis of pancreatic endocrine carcinoma (Hansel et al., 2004), distant metastasis of nasopharyngeal carcinoma (Bao et al., 2016) and melanoma (Xi et al., 2006), and lymph node metastasis of oral squamous cell carcinoma (Yen et al., 2015; Zhong et al., 2008). However, the role of IGFBP3 in brain metastasis of lung cancer remains unclear. Therefore, we first evaluated the changes in the migration capacity of LUAD cells (A549 cells or H1299 cells) cultured in the astrocytes-conditioned medium (HA1800-CM) using wound healing assays. The results revealed that the healing potential of the cells treated with HA1800-CM was significantly higher compared to that in the control group treated with A549-CM or H1299-CM (Figs. 1A and 1B; Supplementary Figs. S1A and S1B). This was especially the case on the 4th and 5th days post-treatment. Hence, we hypothesized that A549 cells cultured in HA1800-CM underwent alterations in genetic expression, which promoted tumor cell migration and metastasis. Therefore, we used microarray analysis to explore the differential gene-expression changes for tumor cells in the microenvironment with astrocytes, which increased the motility of tumor cells (Fig. 1C). We found that IGFBP3 was significantly up-regulated and that it was the gene with the most significant level of increased expression. We verified the microarray data by qRT-PCR and western blot analyses (Figs. 1D and 1E). Consistent with the results of the wound healing assays, IGFBP3 was significantly elevated on days 4 and 5 post-treatment.
EMT is a key step in early tumor invasion and metastasis (Kim et al., 2018; Thiery et al., 2009) and is marked by the repression of epithelial markers and induction of mesenchymal markers (Thiery, 2002). Previous studies have shown that EMT plays an important role in IGFBP3 promotion of tumor metastasis (Natsuizaka et al., 2010). Our current study showed that the up-regulation of IGFBP3 was accompanied with the down-regulation of epithelial marker E-cadherin and the up-regulation of mesenchymal marker N-cadherin (Figs. 2C and 2D). TGF-β signaling is an essential inducer of EMT in various cancers, including NSCLC (David et al., 2016; Gregory et al., 2011). Thus, we investigated the role of IGFBP3 in TGF-β-induced EMT and found that TGF-β triggered the transition of A549 cells from a cobblestone-like morphology to an elongated shape associated with increased cell scattering (Fig. 3A). In line with the morphological changes, the expression of E-cadherin was significantly decreased, and N-cadherin was increased in A549 cells following TGF-β1 treatment (Fig. 3B), while the knockdown of IGFBP3 by si-RNA (Fig. 3C) remarkably restored E-cadherin expression and impaired N-cadherin in the presence of TGF-β1 (Fig. 3D). These results suggested that IGFBP3 accumulation was required for the induction of EMT and cell motility in response to TGF-β.
To investigate whether IGFBP3 promoted LUAD metastasis in vitro, we performed migration and invasion assays in A549 cells and found that the overexpression of IGFBP3 increased the migration and invasion of A549 cells (Fig. 4A). In contrast, knockdown of IGFBP3 resulted in the significant inhibition of A549 cell migration and invasion (Fig. 4B). Next, we started to identify potential downstream genes. Previous studies have demonstrated that Smad4 is the key molecule to TGF-β-induced EMT (Cheng et al., 2015; Hesling et al., 2011). Colon cancer lymph node metastasis and breast cancer bone metastasis are associated with up-regulation of Smad4 (Deckers et al., 2006; Ioannou et al., 2018). It has been reported that in lung metastasis of liver cancer, IGFBP3 and Smad4 are up-regulated (Yumoto et al., 2005). Therefore, we investigated whether Smad4 might be a downstream gene of IGFBP3. The results showed that Smad4 expression was increased when IGFBP3 was up-regulated (Figs. 5B and 5C) while, the down-regulation of IGFBP3 was accompanied by the down-regulation of Smad4 (Figs. 5D and 5E). After the knockdown of Smad4, the migration and invasion of A549 cells were significantly inhibited, even though IGFBP3 was overexpressed (Figs. 5F and 5G).
The in vivo significance of IGFBP-3 overexpression was further confirmed using both human lung and human brain specimens. We examined the expression of IGFBP3 in normal lung tissues, lung cancer tissues, and brain metastatic lesions by IHC. Our results indicated that compared with that of normal lung tissues, IGFBP3 was highly expressed in the tumor specimens. Moreover, lung tissues with distant metastasis had higher expression levels of IGFBP3 than did those without metastasis. This was especially the case in lung tissues with intracranial metastasis. Prominent expression of IGFBP3 was also confirmed in brain metastatic lesions.
Although the up-regulation of IGFBP3 may mediate brain metastasis in LUAD, more detailed in vivo studies on IGFBP3 are still needed. In addition, more lung adenocarcinoma cell lines will be used in our next set of studies to verify the pro-tumor brain-metastatic effects of IGFBP3 and its mechanism.
In brief, our findings demonstrated that IGFBP3 was able to promote A549 cells migration, invasion, and EMT and may play an important role in the TGF-β/Smad4 pathway. In vivo, human tissues demonstrated that IGFBP3 was highly expressed in pulmonary cancerous tissues with brain metastasis and in intracranial metastatic lesions. Therefore, IGFBP3 might serve as a potential therapeutic target for the treatment of lung adenocarcinoma brain metastasis.
Supplementary Information
ACKNOWLEDGMENTS
This work was supported by the grants from National Nature Science Foundation of China (No. 81201682), the Scientific Research Foundation of the Luzhou Science and Technology Bureau (No. 2016LZXNYD-J05), and Southwest Medical University Applied Basic Research Program Foundation (No. 201617).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Balkwill F., Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- Bao L., Liu H., You B., Gu M., Shi S., Shan Y., Li L., Chen J., You Y. Overexpression of IGFBP3 is associated with poor prognosis and tumor metastasis in nasopharyngeal carcinoma. Tumour Biol. 2016;37:15043–15052. doi: 10.1007/s13277-016-5400-8. [DOI] [PubMed] [Google Scholar]
- Bardeesy N., Cheng K.H., Berger J.H., Chu G.C., Pahler J., Olson P., Hezel A.F., Horner J., Lauwers G.Y., Hanahan D., et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budczies J., von Winterfeld M., Klauschen F., Bockmayr M., Lennerz J.K., Denkert C., Wolf T., Warth A., Dietel M., Anagnostopoulos I., et al. The landscape of metastatic progression patterns across major human cancers. Oncotarget. 2015;6:570–583. doi: 10.18632/oncotarget.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Fertig E.J., Ozawa H., Hatakeyama H., Howard J.D., Perez J., Considine M., Thakar M., Ranaweera R., Krigsfeld G., et al. Decreased SMAD4 expression is associated with induction of epithelial-to-mesenchymal transition and cetuximab resistance in head and neck squamous cell carcinoma. Cancer Biol Ther. 2015;16:1252–1258. doi: 10.1080/15384047.2015.1056418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C.J., Huang Y.H., Chen M., Su J., Zou Y., Bardeesy N., Iacobuzio-Donahue C.A., Massague J. TGF-beta tumor suppression through a lethal EMT. Cell. 2016;164:1015–1030. doi: 10.1016/j.cell.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckers M., van Dinther M., Buijs J., Que I., Lowik C., van der Pluijm G., ten Dijke P. The tumor suppressor Smad4 is required for transforming growth factor beta-induced epithelial to mesenchymal transition and bone metastasis of breast cancer cells. Cancer Res. 2006;66:2202–2209. doi: 10.1158/0008-5472.CAN-05-3560. [DOI] [PubMed] [Google Scholar]
- Dragoj M., Milosevic Z., Bankovic J., Tanic N., Pesic M., Stankovic T. Targeting CXCR4 and FAK reverses doxorubicin resistance and suppresses invasion in non-small cell lung carcinoma. Cell Oncol (Dordr) 2017;40:47–62. doi: 10.1007/s13402-016-0304-6. [DOI] [PubMed] [Google Scholar]
- Fidler I.J., Yano S., Zhang R.D., Fujimaki T., Bucana C.D. The seed and soil hypothesis: vascularisation and brain metastases. Lancet Oncol. 2002;3:53–57. doi: 10.1016/S1470-2045(01)00622-2. [DOI] [PubMed] [Google Scholar]
- Fitzgerald D.P., Palmieri D., Hua E., Hargrave E., Herring J.M., Qian Y., Vega-Valle E., Weil R.J., Stark A.M., Vortmeyer A.O., et al. Reactive glia are recruited by highly proliferative brain metastases of breast cancer and promote tumor cell colonization. Clin Exp Metastasis. 2008;25:799–810. doi: 10.1007/s10585-008-9193-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory P.A., Bracken C.P., Smith E., Bert A.G., Wright J.A., Roslan S., Morris M., Wyatt L., Farshid G., Lim Y.Y., et al. An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol Biol Cell. 2011;22:1686–1698. doi: 10.1091/mbc.e11-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansel D.E., Rahman A., House M., Ashfaq R., Berg K., Yeo C.J., Maitra A. Met proto-oncogene and insulin-like growth factor binding protein 3 overexpression correlates with metastatic ability in well-differentiated pancreatic endocrine neoplasms. Clin Cancer Res. 2004;10:6152–6158. doi: 10.1158/1078-0432.CCR-04-0285. [DOI] [PubMed] [Google Scholar]
- Hesling C., Fattet L., Teyre G., Jury D., Gonzalo P., Lopez J., Vanbelle C., Morel A.P., Gillet G., Mikaelian I., et al. Antagonistic regulation of EMT by TIF1gamma and Smad4 in mammary epithelial cells. EMBO Rep. 2011;12:665–672. doi: 10.1038/embor.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou M., Kouvaras E., Papamichali R., Samara M., Chiotoglou I., Koukoulis G. Smad4 and epithelial-mesenchymal transition proteins in colorectal carcinoma: an immunohistochemical study. J Mol Histol. 2018;49:235–244. doi: 10.1007/s10735-018-9763-6. [DOI] [PubMed] [Google Scholar]
- Jogie-Brahim S., Feldman D., Oh Y. Unraveling insulin-like growth factor binding protein-3 actions in human disease. Endocr Rev. 2009;30:417–437. doi: 10.1210/er.2008-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.M., Jung C.H., Song J.Y., Park J.K., Um H.D. Pro-apoptotic Bax promotes mesenchymal-epithelial transition by binding to respiratory complex-I and antagonizing the malignant actions of pro-survival Bcl-2 proteins. Cancer Lett. 2018;424:127–135. doi: 10.1016/j.canlet.2018.03.033. [DOI] [PubMed] [Google Scholar]
- Li C., Harada A., Oh Y. IGFBP-3 sensitizes antiestrogen-resistant breast cancer cells through interaction with GRP78. Cancer Lett. 2012;325:200–206. doi: 10.1016/j.canlet.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Li C., Wan L., Liu Z., Xu G., Wang S., Su Z., Zhang Y., Zhang C., Liu X., Lei Z., et al. Long non-coding RNA XIST promotes TGF-beta-induced epithelial-mesenchymal transition by regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer. Cancer Lett. 2018;418:185–195. doi: 10.1016/j.canlet.2018.01.036. [DOI] [PubMed] [Google Scholar]
- Mujoomdar A., Austin J.H., Malhotra R., Powell C.A., Pearson G.D., Shiau M.C., Raftopoulos H. Clinical predictors of metastatic disease to the brain from non-small cell lung carcinoma: primary tumor size, cell type, and lymph node metastases. Radiology. 2007;242:882–888. doi: 10.1148/radiol.2423051707. [DOI] [PubMed] [Google Scholar]
- Natsuizaka M., Ohashi S., Wong G.S., Ahmadi A., Kalman R.A., Budo D., Klein-Szanto A.J., Herlyn M., Diehl J.A., Nakagawa H. Insulin-like growth factor-binding protein-3 promotes transforming growth factor-{beta}1-mediated epithelial-to-mesenchymal transition and motility in transformed human esophageal cells. Carcinogenesis. 2010;31:1344–1353. doi: 10.1093/carcin/bgq108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda M., Yamakawa Y., Matsunaga N., Naoe S., Jodoi T., Yamafuji M., Akimoto N., Teramoto N., Fujita K., Ohdo S., et al. IL-6 receptor is a possible target against growth of metastasized lung tumor cells in the brain. Int J Mol Sci. 2012;14:515–526. doi: 10.3390/ijms14010515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardali K., Moustakas A. Actions of TGF-beta as tumor suppressor and pro-metastatic factor in human cancer. Biochim Biophys Acta. 2007;1775:21–62. doi: 10.1016/j.bbcan.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Seike T., Fujita K., Yamakawa Y., Kido M.A., Takiguchi S., Teramoto N., Iguchi H., Noda M. Interaction between lung cancer cells and astrocytes via specific inflammatory cytokines in the microenvironment of brain metastasis. Clin Exp Metastasis. 2011;28:13–25. doi: 10.1007/s10585-010-9354-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- Sul J., Posner J.B. Brain metastases: epidemiology and pathophysiology. Cancer Treat Res. 2007;136:1–21. doi: 10.1007/978-0-387-69222-7_1. [DOI] [PubMed] [Google Scholar]
- Sun Y., Ai X., Shen S., Gu L., Lu S. Detection and correlation analysis of serum cytokines in non-small-cell lung cancer patients with bone and non-bone metastases. Patient Prefer Adherence. 2015;9:1165–1169. doi: 10.2147/PPA.S86605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka M., Harada H., Andl C.D., Oyama K., Naomoto Y., Dempsey K.L., Klein-Szanto A.J., El-Deiry W.S., Grimberg A., Nakagawa H. Epidermal growth factor receptor regulates aberrant expression of insulin-like growth factor-binding protein 3. Cancer Res. 2004;64:7711–7723. doi: 10.1158/0008-5472.CAN-04-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery J.P. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Vestey S.B., Perks C.M., Sen C., Calder C.J., Holly J.M., Winters Z.E. Immunohistochemical expression of insulin-like growth factor binding protein-3 in invasive breast cancers and ductal carcinoma in situ: implications for clinicopathology and patient outcome. Breast Cancer Res. 2005;7:R119–129. doi: 10.1186/bcr963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y., Nakajima G., Hamil T., Fodstad O., Riker A., Ju J. Association of insulin-like growth factor binding protein-3 expression with melanoma progression. Mol Cancer Ther. 2006;5:3078–3084. doi: 10.1158/1535-7163.MCT-06-0424. [DOI] [PubMed] [Google Scholar]
- Yen Y.C., Hsiao J.R., Jiang S.S., Chang J.S., Wang S.H., Shen Y.Y., Chen C.H., Chang I.S., Chang J.Y., Chen Y.W. Insulin-like growth factor-independent insulin-like growth factor binding protein 3 promotes cell migration and lymph node metastasis of oral squamous cell carcinoma cells by requirement of integrin β1. Oncotarget. 2015;6:41837–41855. doi: 10.18632/oncotarget.5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi M., Bahrami T., Salmaninejad A., Nosrati R., Ghaffari P., Ghaffari S.H. Lung cancer-associated brain metastasis: Molecular mechanisms and therapeutic options. Cell Oncol (Dordr) 2017;40:419–441. doi: 10.1007/s13402-017-0345-5. [DOI] [PubMed] [Google Scholar]
- Yumoto E., Nakatsukasa H., Hanafusa T., Yumoto Y., Nouso K., Matsumoto E., Onishi T., Takuma Y., Tanaka H., Fujikawa T. IGFBP-3 expression in hepatocellular carcinoma involves abnormalities in TGF-B and/or Rb signaling pathways. Int J Oncol. 2005;27:1223–1230. [PubMed] [Google Scholar]
- Zhong L.P., Yang X., Zhang L., Wei K.J., Pan H.Y., Zhou X.J., Li J., Chen W.T., Zhang Z.Y. Overexpression of insulin-like growth factor binding protein 3 in oral squamous cell carcinoma. Oncol Rep. 2008;20:1441–1447. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.