Abstract
Membrane vesicles (MVs) are lumen-containing spheres of lipid bilayers secreted by all prokaryotes into the extracellular milieu. They have multifunctional roles in stress response, virulence transfer, biofilm formation, and microbial interactions. Remarkably, MVs contain various components, including lytic enzymes, genetic materials, and hydrophobic signals, at high concentrations and transfer them effectively to the target microbial cells. Therefore, MVs act as carriers for bactericidal effects, horizontal gene transfer, and quorum sensing. Although the purpose of secreted MVs remains unclear, recent reports have provided evidence that MVs selectively interact with microbial cells in order to transfer their content to the target species. Herein, we review microbial interactions using MVs and discuss MV-mediated selective delivery of their content to target microbial cells.
Keywords: bacterial interaction, DLVO theory, quorum sensing, horizontal gene transfer, membrane vesicles
Significance.
Membrane vesicles (MVs) secreted from microbes play a crucial role in membrane trafficking; however, it has not been fully elucidated how MVs transfer their content to the target microbial cells in the microbial community. We firstly applied the Derjaguin-Landau-Verwey-Overbeek theory, based on physicochemical energies, to elucidate the interaction between MVs and bacterial cells. Additionally, a ligand-receptor relationship has also been observed in MVs and the bacterial surface, and it may effectively deliver the cargo to the target microbial cells. In this mini-review, we highlight the recent progress in microbial interactions using MVs and discuss the mechanism of donor-recipient specific interactions.
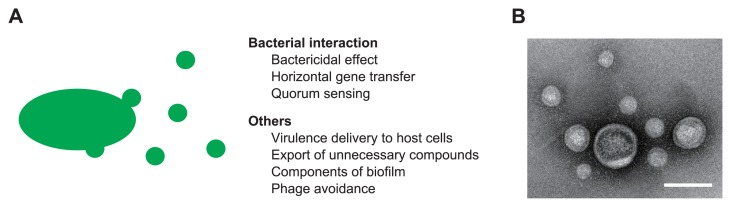
Eukaryotic and prokaryotic organisms secrete extracellular follicles with structures similar to that of the cellular membrane. These extracellular vesicles, secreted by prokaryotes, are ordinarily termed membrane vesicles (MVs) or outer membrane vesicles (OMVs), particularly by gram-negative bacteria, and their diameter ranges from 10 to 400 nm (Fig. 1). They are mainly composed of an outer membrane of proteins and phospholipids and other components such as an inner membrane and periplasmic and cytoplasmic proteins, nucleic acids, and polysaccharides [1,2]. Their composition varies, even among the same species, and depends on the growth phase and the environmental conditions [3,4]. MVs are considered to play a crucial role in stress response, virulence transfer, and biofilm formation. MVs contain DNA, RNA, and in some cases, quorum sensing signals that they transfer to other bacterial cells. MVs also mediate the cell-to-cell interaction, resulting in bacterial membrane trafficking [5]. Interior substances are maintained at high concentrations in MVs, protected from degradation by exterior stressors and enzymes; therefore, the encapsulation provides an effective means of microbial interaction in bactericidal effects, horizontal gene transfer, and quorum-sensing, in contrast to diffusion-based interaction [5] (Fig. 1). Furthermore, the characteristics of the MV surface vary among species, MV-derived bacteria, and growth conditions, and this diversity of MVs is key to the selective interaction with bacterial cells. Herein, we review the current progress on bacterial interactions using MVs and we propose a new model for selective or limited interaction in microbial communities.
Figure 1.
Bacterial vesicles. (A) Functions of membrane vesicles in prokaryotes. (B) Transmitted electron microscope image of vesicles derived from B. agrestis. The bar indicates 100 nm.
MVs kills other bacteria in different pathways
Pathogenic bacteria have emerged by implementing MVs as tools to cause infection and diseases to host cells because MVs have the ability to transmit virulence factors, mediate biofilm formation, and modulate the immune response [6]. MVs act as aggressive tools not only by disseminating virulence factors to host cells, but also by invading other species to establish the niche in multi-species coexisting environments. The first evidence of the predatory nature of MVs is a report that showed that MVs derived from Pseudomonas aeruginosa kill other bacteria [7]. Several gram-negative bacteria, and P. aeruginosa possess MV lyse properties; P. aeruginosa MVs have the ability to lyse the broadest spectrum of bacteria among all other bacteria tested [8]. The high lytic activity of P. aeruginosa MVs is mainly due to murein hydrolase, which functions as an autolysin [9], and antimicrobial quinolones [10]. Gentamicin, which is an aminoglycoside antibiotic, induces MV secretion [11] and gentamicin associated MVs also contribute to the killing of other MV recipient bacteria [12]. Lytic effects fluctuate between gram-negative and gram-positive bacteria under different proposed mechanisms [7]. Virulence factors and peptidoglycan hydrolytic enzymes are concentrated in P. aeruginosa MVs; the MV membrane fuses with the outer membrane of gram-negative bacteria, resulting in a transfer of lytic material to other bacterial cells. In contrast to gram-negative bacteria, the surface of gram-positive bacteria is covered by a peptidoglycan layer. P. aeruginosa MVs break open at the peptidoglycan layer when they are attached to gram-positive bacteria. The interior content released from MVs digest the gram-positive bacterial wall [13]. Lately, it has been shown that Bacillus subtilis is more susceptible to P. aeruginosa MVs than other gram-positive bacteria because its surface is more hydrophilic than the other tested bacteria [14]. Therefore, the physicochemical properties of the bacterial surface determine the mechanism and selectivity of the killing effect of MVs on bacterial cells.
MV-bacteria interaction leads to horizontal gene transfer
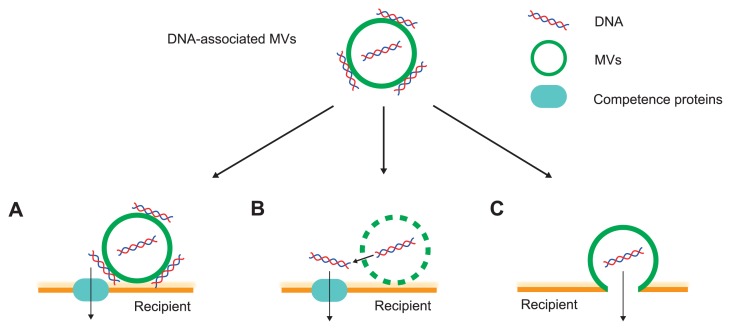
The existence of genetic material, DNA or RNA, in MVs has been previously reported by studies of MVs isolated from a variety of microbes [15]. The first study that demonstrated horizontal gene transfer (HGT) of plasmid DNA in MVs was in Neisseria gonorrhoeae [16]. Since then, HGT has been observed in gram-negative bacteria [17–22], gram-positive bacteria [23], and archaea [24,25]. MV-mediated transfer of plasmid DNA was not limited in the same species but also occurred beyond the genus [21,23,26]. A recent study showed that internal DNA has the potential for gene transfer; although most of the MV-associated DNA is located on the external surface of MVs, in contrast to its location on the internal membrane in P. aeruginosa [27]. Another study using Porphyromonas ginigivalis also showed that DNase treatment decreases the HGT efficiency via MVs by only 30% [22]. These results suggest that both the external and internal MV associated DNA are important for the MV-mediated HGT. It has been considered that there are several pathways for MV-mediated gene transfer (Fig. 2). Fulsundar et al. suggested that HGT by DNA-containing MVs is categorized into two independent pathways: (i) the DNA-containing MVs, close to the recipient cells, are lysed, and released DNA is incorporated into the recipient cells by natural competence (ii) the MV membrane is fused with the recipient cells and DNA is transferred to the recipient cells [21]. In the former case, the possibility of the occurrence of HGT depends on the natural competence of the recipients, while in the latter case, it depends on the properties of the surface. Tran and Boedicker suggested that MVs constitute a general mechanism of transfer of genetic cargo in non-specialized bacterial species [26]. They added MVs packed with plasmid from donor strains (Aeromonas veronii, Enterobacter cloacae and E. coli), to recipient strains (A. veronii, Chromobacterium violaceum, E. cloacae, E. coli, and P. aeruginosa). Plasmid transfer was observed in all combinations tested. MVs packed with plasmid DNA derived from P. aeruginosa did not lead to any DNA transfer in P. aeruginosa [28]; the efficiency of DNA transfer via MVs probably depends on the combination of MV donor and recipients, as well as on natural competency. Further work is needed to elucidate the mechanisms of HGT via MVs.
Figure 2.
Proposed routes of gene transfer using membrane vesicles (MVs). Extracellular DNA is localized at both the surface of MVs and the interior. (A) MVs are close to the recipient bacterial cells and DNA on the surface of MVs is delivered to the recipient that has competency. (B) MVs are associated with the surface of target cells, lysed and the interior DNA is leaked and transported into the recipient with natural competence. (C) Adhesion of MVs to the surface of recipient cells leads to membrane fusion and DNA delivery.
MV-mediated signal transfer to specific microbial species
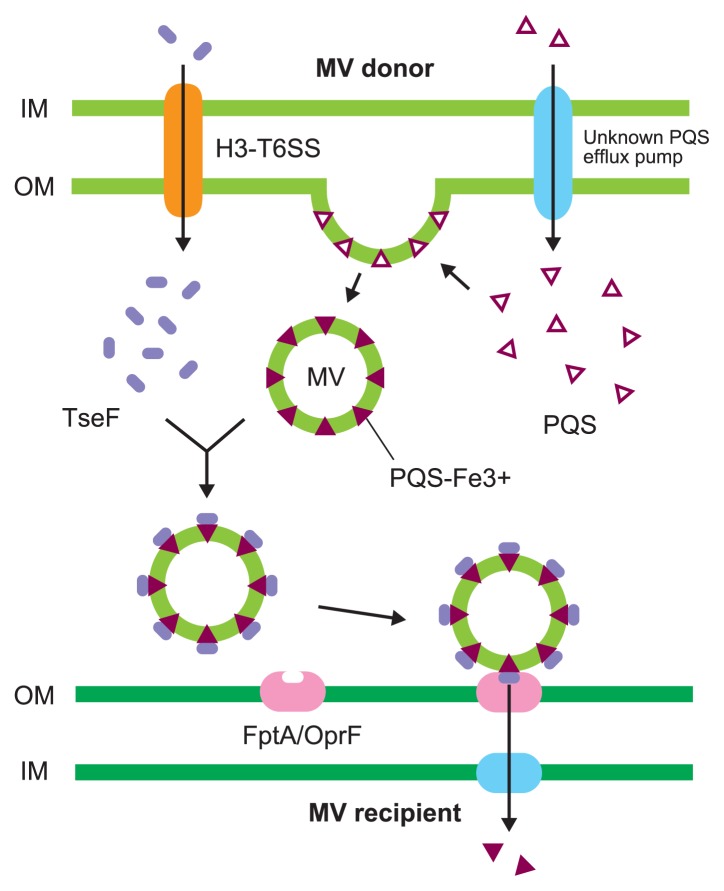
Many bacteria use extracellular signals to communicate in the microbial communities through a system called quorum sensing. The signaling molecules constitute a complex regulatory network that controls the expressions of a variety of genes including virulence-related genes. Some signal molecules are hydrophobic and associated with MVs in the extracellular environment. The first discovery of signal transfer among microbes was reported by Mashburn et al. and stated that the Pseudomonas quinolone signal (PQS), a quorum sensing signal in Pseudomonas aeruginosa, was encapsulated into MVs and thus, trafficked within the microbial population [10]. PQS is not only encapsulated in MVs but also plays a role in the increase in the rate of MV formation by being incorporated into the outer membrane and causing membrane curvature [29]. PQS strongly interacts with 4′-phosphates and acyl chains of lipid A in lipopolysaccharides (LPS), and likely increases anionic charge repulsion between neighboring LPS molecules, resulting in membrane curvature [29,30]. For this reason, exogenous PQS enhances MV production not only in P. aeruginosa but also in other bacteria [31,32]. The MV generation and the PQS export are closely related; however, the PQS export mechanism is not fully understood [33]. While strong MV producers effectively export PQS to the extracellular milieu, a large portion of PQS is localized at the inner membrane in poor MV producers. Both types of MV producers synthesize similar amounts of PQS, even though the distribution of PQS varies between them. MVs that are secreted in the stationary phase, contain more PQS and have more interactions with P. aeruginosa cells compared to those secreted in the exponential phase [4]. P. aeruginosa secretes the extracellular protein TseF through the type VI secretion system H3 (H3-T6SS), and TseF interacts with iron-binding PQS localized in MVs. The TseF-PQS-Fe3+ complex associates with the Fe (III)-pyochelin receptors FptA and OprF localized at the outer membrane [34,35]. Therefore, TseF facilitates the delivery of iron and PQS associated with MVs to recipient bacterial cells. These functions may contribute to MV-mediated specific interactions where PQS is effectively transferred to target bacteria that possess pyochelin receptors (Fig. 3).
Figure 3.
Membrane vesicle-mediated signal transfer. Pseudomonas quinolone signal (PQS) is synthesized in P. aeruginosa cytoplasm and moved out of the cell via a currently unknown export mechanism. PQS is integrated to the outer membrane of P. aeruginosa, resulting in membrane curvature and MVs secretion. PQS chelates ferric iron and extracellular protein TseF, secreted by the Type VI Secretion System H3 (H3-T6SS) associated with PQS-Fe3+. The complex of TseF and PQS-Fe3+, localized within MVs, recognizes the pyochelin receptor FptA and the porin OprF in MV recipients, and this facilitates the uptake of iron into the complex.
Another example of cell-to-cell interaction using MVs is observed in Paracoccus denitrificans. This bacterium synthesizes N-hexadecanoyl-l-homoserine lactone (C16-HSL), a long chain N-acyl-l-homoserine lactone (AHL), as a QS signal, and this hydrophobic signal, associated with MVs, may be solubilized in the aqueous environment [36]. Interestingly, MVs secreted by P. denitrificans interacted mainly with P. denitrificans cells than with the other bacterial species tested, although the mechanism responsible for this selective interaction remains unknown. Various long-chain AHLs, synthesized by this and other bacteria, are able to associate with P. denitrificans-derived MVs that can sequester various signals of the environment by using the MVs for their own gene regulation [37]. Signal transfer using MVs would be effective for intraspecies communication considering that MVs contain a high concentration of signals and are able to transfer them to the same bacterial species.
Physicochemical potential based on selective interactions between MVs and bacterial cells
The interaction energy between two particles is explained by the Derjaguin-Landau-Verwey-Overbeek (DLVO) theory, and this theory has been applied for both microbial adhesion and aggregation of lipid vesicles [38,39]. Similarly, this theory can be applied for MV interactions with bacterial cells [40]. In this theory, the interaction energy Vtotal between microbial cells and vesicles was calculated as the sum of an attractive London-van der Waals interaction energy (VA) and an electric repulsive interaction energy (VR):
| (1) |
VA was defined based on the following equation as per a previous report [39]:
| (2) |
R is the separation distance between the particles of cells and vesicles (from center to center), and expressed by the sum of their separation distance (r), the radii of the cells (a) and vesicles (a′):
| (3) |
Hamaker constant A is described by the following equation:
| (4) |
where q is the volume density and λ is London-van der Waals constant. A of the phospholipid bilayer was expressed as 4×10−19 N [41].
According to a previous report [39], VR was defined by the following equation:
| (5) |
where 𝜑 and 𝜑′ are the Stern potentials of cells and vesicles, respectively, and κ is the Debye constant. The values of the Stern potentials were used as zeta potentials (ζ) obtained experimentally. When the absolute values of 𝜑 and 𝜑′ were less than 60 mV (κr>1), VR was approximated by the following equation:
| (6) |
where ɛr is the relative permittivity of the medium, and ɛ0 is the permittivity of the vacuum. The Debye constant κ was expressed by the following equation:
| (7) |
where e is the elementary charge, NA, is Avogadro’s number, c is the ion concentration, z is the charge of ions, and k is the Boltzmann constant.
MVs derived from an enterobacterium Buttiauxella agrestis interact with limited bacterial species and this specific interaction can be explained by the DLVO theory [40]. The interaction of MVs and bacterial cells was examined using 11 MV-producing strains. It was found that MVs derived from B. agrestis interact exclusively with the same genus bacteria (Fig. 4A), whereas MVs from other bacteria do not display similar characteristics. The electron microscopic observation analysis suggests that MVs were not only attached to B. agrestis cells but also fused with them (Fig. 4B). One of the mechanisms of such a unique interaction is based on the physicochemical characteristics of B. agrestis. The cellular surface of Buttiauxella spp. possesses significantly lower zeta potential compared to that of other gram-negative bacteria. Thereby, according to the DLVO theory, the primary maximum energy between B. agrestis MVs and cells is at an extremely low level, unlike the energy of the interaction between MVs and the other bacterial cells tested in this study (Fig. 4C). These results suggest that the low primary maximum energy is one of the reasons for the specific interaction between B. agrestis MVs and cells (Fig. 4D, E). Interestingly, such specific interaction using MVs is conserved in the Buttiauxella genus [40]. The relationship between MV-cell interaction and primary maximum energy, according to the DLVO theory, showed that the low interaction energy affects the specific interaction between B. agrestis MVs and cells of Buttiauxella spp.; nevertheless, the possibility that other specific proteins may also facilitate the interaction of MVs is not excluded.
Figure 4.
The specific interaction of MVs based on DLVO theory. (A) Association of MVs derived from B. agrestis CUETM77-167 with various bacterial species: Corynebacterium glutamicum AJ2247 (C. g.), Micrococcus luteus JCM 1464 (M. l.), Bacillus subtilis C1, Flavobacterium johnsoniae JCM 8514 (F. j.), Rhizobium halotolerans JCM 17536 (R. h.), R. soli DS-42 (R. s.), Hydrogenophaga pseudoflava GA3 (H. p.), Buttiauxella agrestis CUETM77-167 (B. a.), Escherichia coli MG1655 (E. c.), Erwinia persicina HK204 (E. p.), Pseudomonas aeruginosa PAO1 (P. ae.), and P. alcaligenes JCM 20561 (P. al.) were used as recipient strains. (B) Interaction of fluorescein-4-isothiocyanate (FITC)-labelled MVs with B. agrestis cells. Bacteria-associated MVs were detected by small gold particles (black arrows) through the FITC antibody. The bar indicates 100 nm. (C) Primary maximum energy between each bacterial cell and B. agrestis MVs based on the DLVO theory. (D, E) Model for the free energy profile of the interaction between cells and MVs according to a generalized DLVO theory. (F) Relationship between MV association with cells and primary maximum energy. Blue plots show Buttiauxella strains, and red plots show other bacterial genera. Figure is reprint of Tashiro et al. [40] with modification.
The zeta potentials of bacterial cells are generally influenced by the components of LPS and capsular polysaccharides located on the cellular surfaces. The chemical characteristics of LPS and capsular polysaccharides vary among bacterial strains, including strains within the same species. It is believed that MVs are considered to be formed from the specific sites at bacterial surfaces, and therefore the surface charge of MVs is not always similar to that of origin bacterial cells. For example, the negatively charged B-band LPS is contained in MVs naturally released from P. aeruginosa, but A-band LPS is absent in those MVs [11]. Another study reported that zeta potentials of MVs released from P. aeruginosa vary according to growth phase, while those from bacterial cells are unchanged [4]. It is thought that the observed variance in zeta potentials results in decrease in electric repulsive interaction energy controlling MV-bacterium interaction in a certain bacterial species. Our results revealed that the classical form of DLVO theory was applied to the interaction between B. agrestis MVs and various bacterial cells, suggesting that hydrophobic and thermodynamic forces were not the driving force under our experimental conditions. It remains unknown, however, if this model is applicable to all MV-bacterium interactions. Bacteria possess various appendages such as flagella and pili, and it is these motility-related structures, and not Brownian motion, that organize bacterial movement and promote bacterial attachment. Given this, it is difficult to predict MV-bacterium movements using the present physico-chemical model. Further improvement of this model will promote increased understanding of MV-bacterium interactions across a broad range of species.
Conclusion and perspectives
Based on the findings of numerous laboratories, it is now clear that secreted MVs interact with limited microbial species. One of the underlying mechanisms is the physicochemical interaction energy based on the surface potentials; the DLVO theory is a method to estimate the interaction of MVs with the bacterial cells [40]. Conversely, a ligand-receptor relationship has also been observed in MVs and the bacterial surface [34], and it may contribute to specific interactions using MVs in microbial communities. Indeed, specific proteins localized on the surface of MVs derived from pathogens, increase the association with epithelial cells in the host [42–44]. The MV surface that carries specific proteins enables MVs to deliver their content to target cells; therefore, such genetic engineering could contribute to the development of MVs for drug delivery purposes and vaccines [45–47]. Understanding targeted delivery using MVs will open a new avenue for controlling specific bacterial species in the microbial community.
Acknowledgments
We gratefully thank all collaborators of the work described in this mini-review. This work was supported in part by KAKENHI (JP15K21043 and JP15H01315).
Footnotes
Conflict of Interest
The authors declare no conflict of interests.
Author Contributions
Y. T., K. T. and H. F. drafted the manuscript and prepared figures.
References
- 1.Tashiro Y, Uchiyama H, Nomura N. Multifunctional membrane vesicles in Pseudomonas aeruginosa. Environ Microbiol. 2012;14:1349–1362. doi: 10.1111/j.1462-2920.2011.02632.x. [DOI] [PubMed] [Google Scholar]
- 2.Toyofuku M, Tashiro Y, Hasegawa Y, Kurosawa M, Nomura N. Bacterial membrane vesicles, an overlooked environmental colloid: Biology, environmental perspectives and applications. Adv Colloid Interface Sci. 2015;226(Part A):65–77. doi: 10.1016/j.cis.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 3.Schooling S, Beveridge TJ. Membrane vesicles: an overlooked component of the matrices of biofilms. J Bacteriol. 2006;188:5945–5957. doi: 10.1128/JB.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tashiro Y, Ichikawa S, Shimizu M, Toyofuku M, Takaya N, Nakajima-Kambe T, et al. Variation of physiochemical properties and cell association activity of membrane vesicles with growth phase in Pseudomonas aeruginosa. Appl Environ Microbiol. 2010;76:3732–3739. doi: 10.1128/AEM.02794-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hasegawa Y, Futamata H, Tashiro Y. Complexities of cell-to-cell communication through membrane vesicles: implications for selective interaction of membrane vesicles with microbial cells. Front Microbiol. 2015;6:633. doi: 10.3389/fmicb.2015.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacDonald IA, Kuehn MJ. Offense and defense: microbial membrane vesicles play both ways. Res Microbiol. 2012;163:607–618. doi: 10.1016/j.resmic.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kadurugamuwa JL, Beveridge TJ. Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: conceptually new antibiotics. J Bacteriol. 1996;178:2767–2774. doi: 10.1128/jb.178.10.2767-2774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z, Clarke AJ, Beveridge TJ. Gram-negative bacteria produce membrane vesicles which are capable of killing other bacteria. J Bacteriol. 1998;180:5478–5483. doi: 10.1128/jb.180.20.5478-5483.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Clarke AJ, Beveridge TJ. A major autolysin of Pseudomonas aeruginosa: subcellular distribution, potential role in cell growth and division and secretion in surface membrane vesicles. J Bacteriol. 1996;178:2479–2488. doi: 10.1128/jb.178.9.2479-2488.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437:422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 11.Kadurugamuwa JL, Beveridge TJ. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J Bacteriol. 1995;177:3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadurugamuwa JL, Beveridge TJ. Natural release of virulence factors in membrane vesicles by Pseudomonas aeruginosa and the effect of aminoglycoside antibiotics on their release. J Antimicrob Chemother. 1997;40:615–621. doi: 10.1093/jac/40.5.615. [DOI] [PubMed] [Google Scholar]
- 13.Kadurugamuwa JL, Mayer A, Messner P, Sára M, Sleytr UB, Beveridge TJ. S-layered Aneurinibacillus and Bacillus spp. are susceptible to the lytic action of Pseudomonas aeruginosa membrane vesicles. J Bacteriol. 1998;180:2306–2311. doi: 10.1128/jb.180.9.2306-2311.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacDonald KL, Beveridge TJ. Bactericidal effect of gentamicin-induced membrane vesicles derived from Pseudomonas aeruginosa PAO1 on gram-positive bacteria. Can J Microbiol. 2002;48:810–820. doi: 10.1139/w02-077. [DOI] [PubMed] [Google Scholar]
- 15.Domingues S, Nielsen KM. Membrane vesicles and horizontal gene transfer in prokaryotes. Curr Opin Microbiol. 2017;38:16–21. doi: 10.1016/j.mib.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 16.Dorward DW, Garon CF, Judd RC. Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae. J Bacteriol. 1989;171:2499–2505. doi: 10.1128/jb.171.5.2499-2505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolling GL, Matthews KR. Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl Environ Microbiol. 1999;65:1843–1848. doi: 10.1128/aem.65.5.1843-1848.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yaron S, Kolling G, Simon L, Matthews K. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl Environ Microbiol. 2000;66:4414–4420. doi: 10.1128/aem.66.10.4414-4420.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiura HX, Kogure K, Hagemann S, Ellinger A, Velimirov B. Evidence for particle-induced horizontal gene transfer and serial transduction between bacteria. FEMS Microbial Ecol. 2011;76:576–591. doi: 10.1111/j.1574-6941.2011.01077.x. [DOI] [PubMed] [Google Scholar]
- 20.Rumbo C, Fernández-Moreira E, Merino M, Poza M, Mendez JA, Soares NC, et al. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob Agents Chemother. 2011;55:3084–3090. doi: 10.1128/AAC.00929-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fulsundar S, Harms K, Flaten GE, Johnsen PJ, Chopade BA, Nielsen KM. Gene transfer potential of outer membrane vesicles of Acinetobacter baylyi and effects of stress on vesiculation. Appl Environ Microbiol. 2014;80:3469–3483. doi: 10.1128/AEM.04248-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho M-H, Chen C-H, Goodwin JS, Wang B-Y, Xie H. Functional advantages of Porphyromonas gingivalis vesicles. PLoS ONE. 2015;10(e0123448) doi: 10.1371/journal.pone.0123448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klieve AV, Yokoyama MT, Forster RJ, Ouwerkerk D, Bain PA, Mawhinney EL. Naturally occurring DNA transfer system associated with membrane vesicles in cellulolytic Ruminococcus spp. of ruminal origin. Appl Environ Microbiol. 2005;71:4248–4253. doi: 10.1128/AEM.71.8.4248-4253.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaudin M, Gauliard E, Schouten S, Houel-Renault L, Lenormand P, Marguet E, et al. Hyperthermophilic archaea produce membrane vesicles that can transfer DNA. Environ Microbiol Rep. 2013;5:109–116. doi: 10.1111/j.1758-2229.2012.00348.x. [DOI] [PubMed] [Google Scholar]
- 25.Erdmann S, Tschitschko B, Zhong L, Raftery MJ, Cavicchioli R. A plasmid from an Antarctic haloarchaeon uses specialized membrane vesicles to disseminate and infect plasmid-free cells. Nat Microbiol. 2017;2:1446–1455. doi: 10.1038/s41564-017-0009-2. [DOI] [PubMed] [Google Scholar]
- 26.Tran F, Boedicker JQ. Genetic cargo and bacterial species set the rate of vesicle-mediated horizontal gene transfer. Sci Rep. 2017;7:8813. doi: 10.1038/s41598-017-07447-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bitto NJ, Chapman R, Pidot S, Costin A, Lo C, Choi J, et al. Bacterial membrane vesicles transport their DNA cargo into host cells. Sci Rep. 2017;7:7072. doi: 10.1038/s41598-017-07288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renelli M, Matias V, Lo RY, Beveridge TJ. DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology. 2004;150:2161–2169. doi: 10.1099/mic.0.26841-0. [DOI] [PubMed] [Google Scholar]
- 29.Mashburn-Warren L, Howe J, Garidel P, Richter W, Steiniger F, Roessle M, et al. Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol Microbiol. 2008;69:491–502. doi: 10.1111/j.1365-2958.2008.06302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schertzer JW, Whiteley M. A bilayer-couple model of bacterial outer membrane vesicle biogenesis. MBio. 2012;3:e00297–11. doi: 10.1128/mBio.00297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mashburn-Warren L, McLean RJ, Whiteley M. Gram-negative outer membrane vesicles: beyond the cell surface. Geobiology. 2008;6:214–219. doi: 10.1111/j.1472-4669.2008.00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tashiro Y, Ichikawa S, Nakajima-Kambe T, Uchiyama H, Nomura N. Pseudomonas quinolone signal affects membrane vesicle production in not only Gram-negative but also Gram-positive bacteria. Microbes Environ. 2010;25:120–125. doi: 10.1264/jsme2.me09182. [DOI] [PubMed] [Google Scholar]
- 33.Florez C, Raab JE, Cooke AC, Schertzer JW. Membrane distribution of the Pseudomonas quinolone signal modulates outer membrane vesicle production in Pseudomonas aeruginosa. MBio. 2017;8:e01034–17. doi: 10.1128/mBio.01034-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin J, Zhang W, Cheng J, Yang X, Zhu K, Wang Y, et al. A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat Commun. 2017;8:14888. doi: 10.1038/ncomms14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin J, Cheng J, Wang Y, Shen X. The Pseudomonas quinolone signal (PQS): not just for quorum sensing anymore. Front Cell Infect Microbiol. 2018;8:230. doi: 10.3389/fcimb.2018.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toyofuku M, Morinaga K, Hashimoto Y, Uhl J, Shimamura H, Inaba H, et al. Membrane vesicle-mediated bacterial communication. ISME J. 2017;11:1504–1509. doi: 10.1038/ismej.2017.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morinaga K, Yamamoto T, Nomura N, Toyofuku M. Paracoccus denitrificans can utilize various long-chain N-acyl homoserine lactones and sequester them in membrane vesicles. Environ Microbiol Rep. 2018;10:651–654. doi: 10.1111/1758-2229.12674. [DOI] [PubMed] [Google Scholar]
- 38.Hermansson M. The DLVO theory in microbial adhesion. Colloids Surf B: Biointerfaces. 1999;14:105–119. [Google Scholar]
- 39.Ohki S, Ohshima H. Interaction and aggregation of lipid vesicles (DLVO theory versus modified DLVO theory) Colloids Surf B Biointerfaces. 1999;14:27–45. [Google Scholar]
- 40.Tashiro Y, Hasegawa Y, Shintani M, Takaki K, Ohkuma M, Kimbara K, et al. Interaction of bacterial membrane vesicles with specific species and their potential for delivery to target cells. Front Microbiol. 2017;8:571. doi: 10.3389/fmicb.2017.00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohki S, Duzgunes N, Leonards K. Phospholipid vesicle aggregation: effect of monovalent and divalent ions. Biochemistry. 1982;21:2127–2133. doi: 10.1021/bi00538a022. [DOI] [PubMed] [Google Scholar]
- 42.Kesty N, Mason KM, Reedy M, Miller S, Kuehn M. Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J. 2004;23:4538–4549. doi: 10.1038/sj.emboj.7600471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bauman SJ, Kuehn MJ. Pseudomonas aeruginosa vesicles associate with and are internalized by human lung epithelial cells. BMC Microbiol. 2009;9:26. doi: 10.1186/1471-2180-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deo P, Chow SH, Hay ID, Kleifeld O, Costin A, Elgass KD, et al. Outer membrane vesicles from Neisseria gonorrhoeae target PorB to mitochondria and induce apoptosis. PLoS Pathog. 2018;14:e1006945. doi: 10.1371/journal.ppat.1006945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daleke-Schermerhorn MH, Felix T, Soprova Z, Ten Hagen-Jongman CM, Vikström D, Majlessi L, et al. Decoration of outer membrane vesicles with multiple antigens by using an autotransporter approach. Appl Environ Microbiol. 2014;80:5854–5865. doi: 10.1128/AEM.01941-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gujrati V, Kim S, Kim S-H, Min JJ, Choy HE, Kim SC, et al. Bioengineered bacterial outer membrane vesicles as cell-specific drug-delivery vehicles for cancer therapy. ACS Nano. 2014;8:1525–1537. doi: 10.1021/nn405724x. [DOI] [PubMed] [Google Scholar]
- 47.Baker JL, Chen L, Rosenthal JA, Putnam D, DeLisa MP. Microbial biosynthesis of designer outer membrane vesicles. Curr Opin Biotechnol. 2014;29:76–84. doi: 10.1016/j.copbio.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]