Abstract
Immunotherapy relies on the reinvigoration of immune system to combat diseases and has transformed the landscape of cancer treatments. Clinical trials using immune checkpoint inhibitors (ICI), and adoptive transfer of genetically modified T cells have demonstrated durable remissions in subsets of cancer patients. A comprehensive understanding of the polyfunctionality of T lymphocytes in ICI or adoptive cell transfer (ACT), at single-cell resolution, will quantify T-cell properties that are essential for therapeutic benefit. We briefly highlight several emerging integrated single-cell technologies focusing on the profiling of multiple properties/functionalities of T cells. We envision that these tools have the potential to provide valuable experimental and clinical insights on T-cell biology, and eventually pave the road for the discovery of surrogate T-cell biomarkers for immunotherapy.
Graphical Abstract

Introduction
Immunotherapy has revolutionized the treatment of cancer and relies on utilizing the patients’ immune system and its anti-cancer properties for therapeutic benefit [1,2]. This approach is fundamentally different from chemotherapy and even targeted therapy, both of which depend on the ability of the drug to kill the tumor cell directly [3]. Immunotherapeutic treatment is based on the recognition that there is a failure of the host immune system to control the tumor adequately, and that the goal of treatment is to facilitate resetting the dysregulated balance to enable eradication of the tumors via the host immune system [4–6]. In other words, the treatment does not work to directly kill the tumor cells but instead tries to reinvigorate the immune system to get rid of the tumors. One of the primary objectives of this approach, akin to vaccination, is the ability to establish immunological memory of the tumor, thereby enabling the immune system to seek and destroy metastases anywhere in the body and enable long-term control [7].
Although utilizing the immune system for therapeutic benefit has been around for quite some time, and proteins such as cytokines (e.g. interleukin-2) [8,9] and a suite of monoclonal antibodies (anti-CD20, anti-EGFR, etc.) [10–12] have been used clinically over the last two decades, two newer approaches to treatment — the inhibitors of checkpoint molecules [13], and the adoptive transfer of genetically modified T cells [14], have made substantial advances in the clinic. After decades of frustration with the 5-year survival rates of chemotherapy, these newer forms of immunotherapeutic treatment have altered the treatment landscape and have facilitated durable and lasting remissions in subsets of patients [15]. Both classes of treatment, immune checkpoint inhibitors (ICI) and adoptive cell transfer (ACT), critically rely on the functionality of a particular subset of lymphocytes within the immune system — the T cells. ICI aims to reinvigorate T cells and activate them to attack tumor cells and has shown clinical efficacy in various tumors, albeit in only ~20% of patients [16,17]. ACT, on the other hand, delivers ex vivo expanded (and/or genetically modified) T cells as the therapeutic and has shown complete responses in leukemias (response rate 70%) [18–22].
The introduction of immunotherapeutic molecules as drugs has facilitated new challenges and opportunities for engineers. While the potency of small-molecule-based therapies can be mapped to their mechanism of action (binding/inhibiting appropriate proteins) facilitating tumor cell killing [23,24], understanding the efficacy of ICI or ACT is a significant challenge since the mechanism of action is neither simple nor wholly defined [13,25–27]. The origin of this challenge can be mapped to our inability, to define in a comprehensive manner, all of the different T-cell functionalities that can contribute to their efficacy. T cells are essential players in the adaptive immune systems and can recognize cognate antigen through their T cell receptor (TCR) [28]. T cells bearing TCR specific for foreign or non-native peptides displayed in the context of human leukocyte antigens (HLA) get activated and can undergo a process of programmed differentiation depending on the availability of other accessory molecules including cytokines within the activating environment. Unlike antibodies, the TCR itself does not undergo somatic hypermutation subsequently, and hence can be considered a barcode to identify populations of clonally related T cells [29–31]. T cells are capable of many different functions including cytotoxicity, cytokine secretion, proliferation, and migration, which are determined by multiple cues from intrinsic properties of T cells and its environmental factors. The relative importance of these functions in defining clinical benefit is only partially understood and confounded by the differentiation status of the T cell (naïve, stem-cell-like central memory, central memory, effector memory and effector) [32,33], or by their functional status (polyfunctional, anergic, or exhausted). It is thus apparent that the availability of methods that can map all of these properties onto the same T cell will advance our understanding of the efficacy of immunotherapeutic treatments. From the perspective of the ACT, the availability of precise definitions on the properties that need to be engineered into the T-cell infusion product will facilitate consistent biomanufacturing of therapeutic products [34]. It is thus clear that immunotherapeutic treatments stand to benefit from single-cell technologies that can map the complexity of T cells. While the vast majority of advances in immunotherapeutic treatment have focused on oncology, the principles of modulating the immune system are likely to find broad applicability in other infectious diseases and autoimmunity, as well.
Single-cell technologies have attracted researchers’ attention for several decades, and there is an increasing trend for scientists to develop more accurate and sensitive, higher-throughput and automated single-cell characterization tools. These approaches allow the detection of details that cannot be revealed using traditional population-level assays [35]. Generally, these single-cell technologies are designed to capture cellular information from either the genome, transcriptome or more recently the proteome level [36]. While some assays like flow cytometry (FC) have been standardized and used even in clinical settings [37], some of the more recent single-cell technologies like mass cytometry (MC) [38], and single-cell RNA sequencing (scRNA-seq) [39] have been recently commercialized. Despite this, however, the vast majority of tools are designed in the research setting, and recent advances have enabled the integration of approaches from different omic dimensions to be able to quantify cell features simultaneously [40].
In this review, we briefly highlight several types of emerging single-cell technologies, mainly focusing on technologies that monitor multiple features (function, transcripts, phenotype, etc.) in the context of T-cell characterization. We believe that analyzing T cells at single-cell resolution will provide valuable insights on both experimental and clinical investigations, and has the potential to improve the clinical outcomes of T-cell based therapy. Furthermore, the development of multiplexed single-cell interrogations tools to explore the phenotypical and functional correlations within heterogeneous T cells populations can reveal the underlying biological networks, eventually paving the way for both a better understanding of T cells and delivering surrogate T-cell biomarkers for immunotherapy.
Protein detection from single-cell
Single cell western blotting (scWB)
Similar to the standard western blotting methodology, this approach includes protein separation based on both the affinity between the antibody and the target protein, and the relative size of protein thus minimizing concerns about antibody-cross reactivity (Figure 1A). By the application of open microwells on a polyacrylamide gel coated glass slide, single cells were deposited into individual wells, subsequently lysed, subjected to gel electrophoresis, immobilized by UV-light, and the protein detected by immune-probing. By repetition of antibody-stripping and re-probing, it could detect up to eleven different proteins across thousands of single cells in the same experiment [41–43]. By utilizing a combination of lab-on-a-disc cell device and the scWB analysis, it was possible to quantify protein from less than one hundred cells [44]. The same group further developed an approach termed single cell isoelectric focusing (scIEF) using isoelectric point (pI) difference to separate protein isoforms [45]. In this work, they reported ten cells were analyzed in the same chip as a proof-of-concept; however, the throughput can theoretically be scaled up. ScWB can be combined with flow sorting [46] or on-chip cell phenotyping [42]. This approach can be beneficial for direct measurement of proteins in a single cell, especially when the number of cells available is limited, allowing the validation of known biomarkers in a multiplexed fashion while maintaining single-cell resolution.
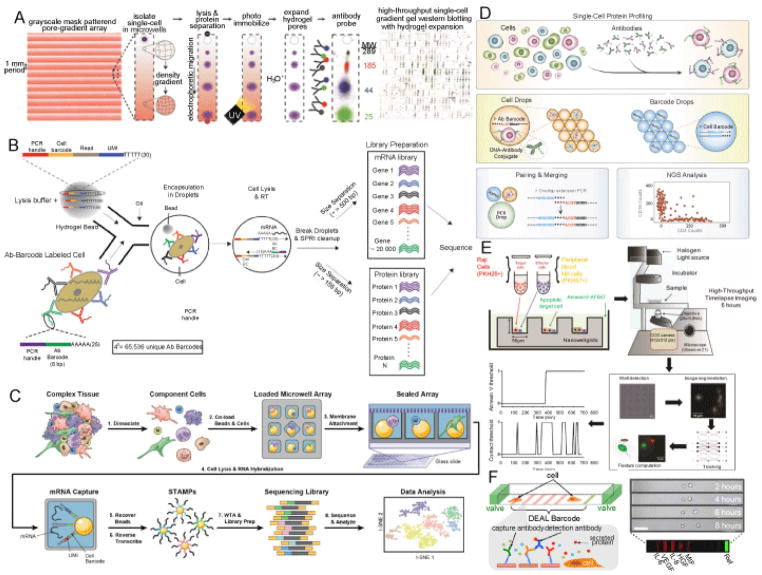
Figure 1. Single-cell technologies for multi-dimensional characterization.
(A) Workflow of scWB adapted from ref [41].
Pore-gradient gel arrays allow thousands of protein electrophoresis separations at single-cell resolution within 1 mm separation distance on a microscope glass slide. Single cells are loaded to individual microwell by gravity and are chemically lysed in situ on the chip. Once scWB is performed, the gel gets brief UV exposure for protein immobilization. Acid is used for pore size expansion, which facilitates the enhancement of local antibody concentration for immunoprobing. Fluorescence images of scWB for proteins spanning from 25–289 kDa. Closed-up false color fluorescent images represent part of arrays (over 400 lanes).
(B) Workflow of REAP-seq adapted from ref [69].
A droplet containing Ab-Barcodes (AbBCs) coated cells fuse to another discreet droplet which contains cell-barcode beads with primers. The cell is lysed once two droplets fuse, and polyadenylated mRNA and AbBC hybridize with poly(dT) primer and the extension of AbBC and complementary synthesis of transcripts can be achieved by reverse transcriptase in the same reaction. AbBC sequences (~ 155 bp) and cDNA from mRNA (~> 500 bp) are separated based on the size difference, and protein and transcript libraries are constructed and sequenced.
(C) Workflow of Seq-Well adapted from ref [72].
The complex tissue is dissociated to single-cell suspension first, and then barcoded mRNA capture beads and cells are loaded onto microwell array by gravity. The device is sealed by a semipermeable membrane to allow lysis buffer change but confine mRNA within the well. Once the beads (contained poly(dT) primers, which including cell-specific barcodes and unique molecular identifiers for each transcript) capture liberated transcripts from an individual cell, the beads are recovered from the array. Reverse transcription of bead-bound transcripts is performed in bulk, followed by library preparation, sequencing and in silico analysis.
(D) Abseq workflow adapted from ref [71].
Cells stained with DNA-conjugated antibodies are isolated in a droplet with unique cell barcoding information, and the linkage of antibody barcode and cell barcode is achieved via overextension PCR. The chimeric DNA products from over 10,000 single cells can be pooled and sequenced in parallel. The single cell protein information will be sorted by the cell barcoding. Unique molecular identifiers are utilized for PCR-bias correction.
(E) Workflow of TIMING (Time-lapse imaging in nanowell grids) adapted from ref [77].
Raji tumor cells and NK cells stained with PKH26 and PKH67 fluorescent membrane dyes respectively are loaded onto a nanowell array, which is immersed in cell culture media containing fluorescent Annexin V as cell apoptosis indicator and imaged for 6 hours by high-throughput time-lapse imaging. Imaging analysis is performed as previously described [79].
(F) Schematic of a single cell barcode chip (SCBC) and representative time-lapse images of an SCBC microchamber containing two cells. Adapted from ref [84].
Top left: schematic of a SCBC microchamber with valve and DEAL (DNA-encoded antibody library) barcodes; bottom left: immune sandwich formation indicates protein detection; top right: representative time-lapse images of an SCBC microchamber containing two cells over 8 hours; bottom right: fluorescent images of patterned barcodes of 5 detected proteins, scale bar = 100 μm.
Integration of protein detection and transcriptional profiling of single cell
Flow cytometry
FC has been widely adopted for several decades to characterize the phenotype of cells and the intracellular molecules across millions of cells. It can detect up to around 17 parameters simultaneously, which is determined by the availability of fluorescent dyes [47]. Recently, Nicolet et al were able to simultaneously profile the expression of primary human T-cell cytokines (IFN-γ, IL-2, and TNF-α) at both the protein and mRNA transcript level via integration of fluorescence in situ hybridization (FISH) and a flow cytometry-based platform [48]. This work paved a road for finding the correlation between cytokine secretion and mRNA transcripts within the same single cell.
Mass cytometry
To improve the multiplexing capacity of cytometry, heavy-metal tagged antibodies are used in mass cytometry (MC). This strategy enables the quantification of more targets on single cell simultaneously, including surface phenotypic marker characterization, intracellular protein detection, cytokine secretion, transcription factor expression, and mRNA transcripts expression [26,49–55]. Frei et al developed a method called PLAYR (proximity ligation assay for RNA) and demonstrated that this approach was able to quantify multiplexed mRNA transcripts and protein via flow cytometry or mass cytometry simultaneously [51]. The oligonucleotide labeled-fluorescence or metal tags were used to detect target transcripts. The authors validated this method by detection of 8 different mRNA transcripts and 18 proteins (cytokine + surface molecules) in LPS-stimulated PBMC for various stimulation times, and the results suggested the most LPS-responding cells were likely to be a CD14+ phenotype. Frei and colleagues expected the theoretical upper limit in the number of detected targets could be as high as 40 if combined with MC. The disadvantage of MC is that unlike FC, it is sample destructive, and thus, it is not possible to sort single cells for downstream analyses like RNA-seq.
Both FC and MC are well-developed technologies and can directly detect proteins from millions of single cells but are restricted to providing snapshots since it is not possible to track the same cell longitudinally using these methods. Despite these disadvantages, however, FC and MC are robust knowledge-based methods to identify subsets of T cells directly from tumors and hence will play an essential role in tracking the efficacy of immunotherapies.
Single-cell PCR
Unlike the PLAYR method that utilized the mass tag or fluorescent tag to capture transcript or protein abundances, other studies relied on the usage of DNA as a label to detect proteins. Although initially the profiling of mRNA and protein was achieved by splitting the cell lysate to two parts and characterizing each of them separately [56,57], Genshaft et al. presented an approach that combined the detection of protein and mRNA from same mammalian cells in a single reaction chamber in a parallel manner [58]. Modified proximity extension assays (PEA) method was used in this technology for protein detection. For each protein of interest, there were two different single-stranded oligonucleotides-labeled antibodies to detect the target protein. The 3′ end of DNA labels of this antibody pair were complementary to each other, as a result, DNA labels would hybridize once both antibodies co-localized on the target protein. The extension of DNA label complex and reverse transcription of RNA from the same cell happened simultaneously by utilizing reverse transcriptase also as DNA polymerase, followed by qPCR (FluidigmTM C1 system) to quantify protein expression and RNA abundance. By applying this approach to study protein and mRNA abundances in the PMA-stimulated MCF7 cells, they found that the correlation of mRNA and protein was variable among genes or time points: highly-expressed genes were more correlated with the corresponding protein expression in untreated cells but after simulation the lowly-expressed genes with high cell-cell variance showed the largest correlation.
ScRNA-seq
ScRNA-seq, a rapidly-growing technology can provide unbiased, high-dimensional genome-wide transcriptomic profiling of individual cells, and has emerged as a robust method to facilitate the discovery of novel cellular status [59], and provide biological insights [31,60,61]. ScRNA-seq has been extensively reviewed elsewhere [39,62–64], and we will only highlight combinations of scRNA-seq with other kinds of single-cell assays [39,62–64].
Researchers have developed several algorithms to utilize scRNA-seq data to reconstitute T cell receptor information. One advantage of obtaining TCR information at single-cell level is that the possibility to acquire the pairing detail of TCR chains (αβ, γδ). Computation approaches, such as TraCeR [65], scTCRseq [66], VDJPuzzle [67], work quite well with transcriptomic profiling results obtained from full-length mRNA transcripts. More recently, the TRAPeS pipeline was reported to enable TCR information extraction from short-read (25–30 bp) sequencing data [68]. Combining transcriptomic profiling and TCR profiling at single-cell resolution, the clonal expansion of exhausted or dysfunctional T cells was found in tumor sites, indicating the reinvigoration of T cell function may recover its anti-cancer functionality [31,61]. Owing to these emerging computational pipelines, developmental trajectories of diverse T cell population can be deciphered, holding the promise of investigating the antigen-specific T cells functions in response to diseases, and also to identify the diversity of T-cell responses within the tumor microenvironment.
Stoeckius et al and Peterson et al recently reported two closely related methods (CITE-seq and REAP-seq) for simultaneous detection of mRNA and protein [69,70] (Figure 1B). Both methods utilized a combination of unique oligonucleotide barcodes and poly (dA) sequence for indexing antibody (but using different linkers) thus enabling the detection of multiple proteins along with transcripts. Extension of DNA labels of antibodies and reverse transcription of mRNA transcripts could be achieved in the same reaction by taking advantage of the DNA polymerase function of reverse transcriptase. These two methods can be readily adapted to different high-throughput scRNA-seq platforms. Another similar technique that can be expanded to demonstrate the same capability is called Abseq (Figure 1D), which utilizes a combination of DNA-labeled antibody and droplet microfluidics [71]. One disadvantage of all three of these approaches is that the information about the spatial distribution of proteins is lost. An orthogonal method, Seq-Well (Figure 1C), takes advantage of arrays of microwells instead of droplets. The cell lysis and reverse transcriptions of mRNA are accomplished on-chip by sealing single cells and individual barcoded capture beads [72]. This assay is compatible with on-array imaging cytometry for resolving the phenotype of cells from complex samples and has the potential to obtain more information from a limited amount of samples using a single platform.
Unquestionably, the integration of transcriptomic and proteomic profiling mentioned above on the same single cell can characterize cellular response to a perturbation in a more accurate, unbiased way. However, these approaches require cell fixation or cell lysates, which exclude the possibility for tracking the dynamic transcriptomic and proteomic changes in the same cell. Although it has the advantage of being able to profile the complete transcriptome, the abundance of lowly expressed transcripts like transcription factors remains a challenge and requires pooling of data when the magnitude of change is also small. Recent reports have aimed to improve the analysis algorithms and to extract more information out of the data [73–76]. Since the cells are lysed to retrieve the mRNA, scRNA-seq ideally provides a snapshot of the cell state, inferred by the transcript profile. Immune gene signatures could be obtained from sequencing results without prior knowledge, making it ideal for the discovery of candidate biomarkers in an unbiased manner. There are disadvantages of this approach including the lack of correlation between mRNA and protein for some genes [58], the inability to directly detect post-translational modification of proteins, and a complete lack of protein localization information. Thus, an ideal implementation of scRNA-seq would be in combination with another method that directly profiles biological function.
Integrated platforms to monitor dynamic T-cell behavior and polyfunctionality
Immune cells, specifically T cells, demonstrate a variety of dynamic behaviors. From the standpoint of studying the therapeutic potential of T cells for adoptive transfer, or for identifying biomarkers of ICI, quantifying the functional status of the T cells will be essential.
Time-lapse imaging microscopy in nanowell grids (TIMING)
The characterization of the interaction between pairs of cells would benefit the understanding of how cells interact or cooperate with other cells, and help the discovery of underlying mechanisms of dynamic cell behavior. Microfluidic devices have the potential to dynamically monitor cell-cell interaction in a high-throughput manner in combination with live cell microscopy. TIMING (Figure 1E), a microwell-based platform, was reported to able to dynamically monitor cell-cell interaction, cytotoxicity, cell motility and cell survival simultaneously [77–79]. Additionally, it can integrate real-time cytokine profiling by bead-based cytokine sensors [80] or gene expression profiling by single cell retrieval via micromanipulator [81] due to the non-destructive feature of this assay. Similarly, it has also been reported that droplets can be used to co-encapsulate the two types of cells before docking to the microwells [82]. This microwell-based device was compatible with live cell imaging analysis, allowing the dynamic monitoring of cell morphology, behavior, and fate. Another recent technology, DropMap, utilized droplets to dynamically profile antibody secretion from antibody secreting cells (ASCs) [83]. Although the rate of secretion of cytokines is much lower than immunoglobulin secretion from ASCs, in principle the DropMap technology can be adapted to monitoring cytokine secretion from T cells.
Single cell barcoding chip (SCBC)
Single cell barcoding chip, pioneered by the Heath and Fan groups, is able to quantify multiple proteins from the same cell, based on the fluorescence readout and on-chip calibration (Figure 1F). SCBC consists of a collection of microchambers on the microfluidic chip (from several hundred to several thousand) to confine single cell or two cells, and one of the surfaces of the microchamber contains barcode-like patterned antibody arrays for protein capture and further detection [84–86]. Apart from protein detection, this approach entitled the monitoring cell movement of the single-cell pair along with the protein secretion [84]. Built on a similar concept, beads-on-barcode antibody microarray (BOBarray) was developed to quantify released proteins from a single cell confined in the individual well, but with modification of protein detection strategy: color-coded and sized-coded functionalized microbeads were coated on the glass slide instead of patterned antibody arrays to minimize the size of the microfluidic device [87]. SCBC technology is amenable of up to around 40-plex protein detection from a single cell and only need a small sample amount as an input; however, due to its intrinsic design, it was not designed to study dynamic or real-time protein secretion.
The advantage of these function-based single-cell assays like TIMING and SCBC is that they have the potential to reveal heterogeneity of complex biologies like motility (reflection of the homing ability of T-cell to find tumor), cytotoxicity (representative of tumor-killing functionality) or cytokine secretion (communication with other immune cells), which are potential predictive biomarker candidates of T cells-based immunotherapy, especially combined biomarkers that require simultaneous measurement of different perspectives of T-cell functionalities. One of the disadvantages of these approaches is that unlike FC/MC that are available as part of core facilities, microfluidics often requires unique expertise and infrastructure to be able to execute these assays. As mentioned above, since the ability to retrieve cells of interest has been demonstrated for at least the TIMING assay, the ability to integrate functional and transcriptional profiling at single-cell resolution might provide the in-depth insight required for defining the efficacy of immunotherapies.
Challenges
Identification and validation of biomarkers
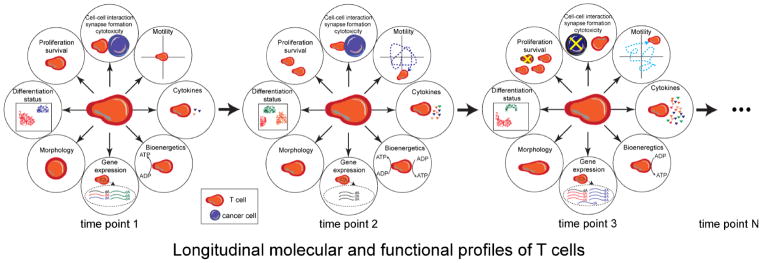
A lot of effort has been devoted to obtaining the predictive biomarkers for ICI and ACT, and candidate biomarkers are showing promising results in multiple clinical trials. Biomarkers could be in various and different formats: tumor infiltrating lymphocyte density at tumor sites, immune checkpoint expression on tumor cells, neoantigen/mutation load of tumor cells, serum protein/antibodies, circulating tumor cells, lymphocyte counts, T cell clonality, etc [88–93]. However, validated clinically actionable biomarkers for predicting the clinical outcomes of ICI/ACT are still lacking. The ligand of immune checkpoint expression of certain tumor samples is considered as one of the promising biomarkers for ICI; for instance, PD-L1 qualitative immunohistochemical assays were FDA-approved but with suboptimal sensitivity and specificity [93]. Many factors may contribute to the inconsistent performance of candidate biomarkers including the complexity of the anti-tumor response, the heterogeneity of tumor or/and patients, the inducible and non-static property of biomarkers, the suboptimal specificity and accuracy of assays. It is evident that the discovery of biomarkers with high accuracy and sensitivity will enable better patient selection into the 3000+ immunotherapy clinical trials currently underway. Since T cells are considered the major effector cells in ICI and ACT, it is not surprising that some T-cell based predictive biomarkers have been discovered [52,94]. Although profiling T cells directly from the tumor site is more indicative of T-cell functional status within the tumor, peripheral blood is a more accessible compartment with limited inconvenience to the patients. For this reason, parallel efforts are devoted to studying T cell within the blood to determine if the changes within these T cells can be used to infer changes in T-cell functionality as a result of treatment [52,94]. Unfortunately, however, T cells demonstrate a variety of dynamic behaviors, and thus mapping these to the same T cells, at single-cell resolution is a challenging problem (Figure 2).
Figure 2. Integrated and dynamic profiling of T cells.
T cells are capable of many different functions and integrate cues from both the cells and soluble factors from the microenvironment to facilitate decision-making. A complete understanding of T cells can be only accomplished by tracking dynamically cell-cell interactions (e.g. synapse formation, cytotoxicity), intrinsic and chemokine guided motility, cytokine secretion, bioenergetics, transcriptome, morphology, differentiation status and proliferation or survival. Assays that are able to provide insights into one or more of these features on the same cells, at single-cell resolution, can provide a deeper understanding of the underlying biology.
Choice of assay
The choice of the appropriate assay to define T-cell functionality is defined by a combination of multiple factors including the number of T cells available, the depth of information being sought, ease of assay standardization, and cost. ScRNA-seq is an excellent choice for the discovery of biomarkers but is still rather expensive for routine implementation. At the other end of the spectrum, FC is considered standard but it requires that the biomarkers have already been discovered using another assay. Presently, at least for monitoring ICI trials, there is no unique way to converge to a single-assay and the choice is often determined by one or more of factors listed above. ACT, on the other hand, should not be limited by cell numbers for at least the infusion product, and not surprisingly more complex assays and dynamic assays have been used to evaluate T-cell functionality [95,96].
Spatial information
All the techniques we have described work with homogenized single cells or single cells in suspension. These methods are ideal and relevant in tumor immunotherapy when profiling single T cells in peripheral blood. Thus, while the comprehensive documentation of the molecular profiles revealed by scRNA-seq is useful for identifying compositional frequencies of immune cell subsets, they cannot, however, reveal the link between the molecular profile and functional capacity, and how this is impacted by space and time. The tumor microenvironment is a three-dimensional structure composed of different kinds of cells, and it is important to document the spatial localization of immune cells within the tumor microenvironment (TME). A few in situ sequencing, proof-of-concept technologies have been demonstrated that can directly map spatial information and transcript profiles [97–99], but it remains to be seen if they can match the depth of transcript profiling afforded by even scRNA-seq. Similarly, FISH based methods that can preserve the spatial information and directly count RNA molecules down to the single-molecule level, have been reported [100–102]. The drawbacks, however, are that even with repetitive cycles of probing different mRNA molecules, the total number of unique mRNA molecules that can be detected is smaller than scRNA-seq and that researchers have to pre-determine the transcripts that are being studied.
Bioinformatics
One of the major and central challenges for realizing the potential and benefit of the next generation of single-cell technologies is matching advances in bioinformatics. As outlined above, the low number of reads per single cell, the ability to differentiate technical and biological variation, amplification biases, batch-effect, all present significant challenges to systematic data analyses. In addition, the ability to integrate single-cell data acquired across different platforms analyzing different kinds of biomolecules and functions is a complex problem, which requires adequate normalization methods and the capability to investigate the correlation among different dimensions of single-cell data [73,103–105]. The identification of conserved signatures of genes, and dimension-reduction-based visualization are the most common methods to extract information from single-cell datasets with high-dimensionality [105,106]. However, the algorithms for prediction of single-cell responses, within heterogeneous cell populations, to perturbation is not well-defined [107]. In other words, while the currently available analytic approaches are mainly focusing on descriptive analyses at single-cell resolution in vitro, it remains unclear how to utilize and integrate these single-cell data to accurately predict the behavior and fate of diverse cell populations in vivo, especially within TME, eventually serving as biomarkers to predict the clinical outcomes of cell-based therapeutics.
Table 1.
Selected examples of single-cell technologies that enable characterization of multiple features
| Technology | Throughput | Highlight | Reference | |
|---|---|---|---|---|
| FC/microscopy & protein | scWB | Up to thousands of single cells |
|
[41–46] |
| Protein&mRNA | FC | Millions |
|
[48] |
| MC | Millions |
|
[51] | |
| Single-cell PCR | 96 (using Fluidigm™ C1) |
|
[58] | |
| scRNA-seq&TCR-seq | TraCeR, scTCRseq, VDJPuzzle, TRAPeS | Depends on throughput of scRNA- seq |
|
[65–68] |
| scRNA-seq | scRNA-seq | Thousands of single cells |
|
[69,70] |
| Abseq | >10,000 cells |
|
[71] | |
| Seq-Well | ~15,000 cells |
|
[72] | |
| Integration: cell-cell interaction, protein, etc | TIMING | 20,000 cells |
|
[77–81] |
| Droplet | ~1,000 events |
|
[82] | |
| SCBC | Up to several thousands of cells |
|
[84–86] | |
| BOBarray | Several thousands of cells |
|
[87] |
Highlights.
Quantitative characterization of dynamic T cell behavior can facilitate identification of biomarkers of responses to immunotherapeutic treatment
Snapshot single-cell omics has made significant advances but lacks temporal resolution
Multi-dimensional techniques that integrate dynamic T-cell function with omics at the single-cell level are starting to emerge
Acknowledgments
We have only focused on the papers have been published in past two years, and due to space constraints have included fewer papers than would be possible in a comprehensive review. We apologize to the authors whose work has been omitted from our review. We thank Jay R Adolacion, Dr. Melisa Martinez-Paniagua, and Conrad Hom for proof reading. This work was supported by National Institutes of Health (R01CA174385), Cancer Prevention & Research Institute of Texas (RP130570), Melanoma Research Alliance Award (509800), Welch Foundation (E1774), Owens Foundation, Congressionally Directed Medical Research Programs (CA160591), and National Science Foundation (1705464).
Footnotes
Conflict of interest statement.
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
• of special interest
•• of outstanding interest
- 1.Khalil DN, Smith EL, Brentjens RJ, Wolchok JD. The future of cancer treatment: immunomodulation, CARs and combination immunotherapy. Nat Rev Clin Oncol. 2016;13:273–290. doi: 10.1038/nrclinonc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim WA, June CH. The Principles of Engineering Immune Cells to Treat Cancer. Cell. 2017;168:724–740. doi: 10.1016/j.cell.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zitvogel L, Galluzzi L, Smyth MJ, Kroemer G. Mechanism of action of conventional and targeted anticancer therapies: reinstating immunosurveillance. Immunity. 2013;39:74–88. doi: 10.1016/j.immuni.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 4.Shore ND. Advances in the understanding of cancer immunotherapy. BJU Int. 2015;116:321–329. doi: 10.1111/bju.12692. [DOI] [PubMed] [Google Scholar]
- 5.Tsai HF, Hsu PN. Cancer immunotherapy by targeting immune checkpoints: mechanism of T cell dysfunction in cancer immunity and new therapeutic targets. J Biomed Sci. 2017;24:35. doi: 10.1186/s12929-017-0341-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zarour HM. Reversing T-cell Dysfunction and Exhaustion in Cancer. Clin Cancer Res. 2016;22:1856–1864. doi: 10.1158/1078-0432.CCR-15-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell. 2017;168:707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang T, Zhou C, Ren S. Role of IL-2 in cancer immunotherapy. Oncoimmunology. 2016;5:e1163462. doi: 10.1080/2162402X.2016.1163462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenberg SA. IL-2: the first effective immunotherapy for human cancer. J Immunol. 2014;192:5451–5458. doi: 10.4049/jimmunol.1490019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim SH, Beers SA, French RR, Johnson PW, Glennie MJ, Cragg MS. Anti-CD20 monoclonal antibodies: historical and future perspectives. Haematologica. 2010;95:135–143. doi: 10.3324/haematol.2008.001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakai K, Hung MC, Yamaguchi H. A perspective on anti-EGFR therapies targeting triple-negative breast cancer. Am J Cancer Res. 2016;6:1609–1623. [PMC free article] [PubMed] [Google Scholar]
- 12.Martinelli E, De Palma R, Orditura M, De Vita F, Ciardiello F. Anti-epidermal growth factor receptor monoclonal antibodies in cancer therapy. Clin Exp Immunol. 2009;158:1–9. doi: 10.1111/j.1365-2249.2009.03992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Restifo NP, Dudley ME, Rosenberg SA. Adoptive immunotherapy for cancer: harnessing the T cell response. Nat Rev Immunol. 2012;12:269–281. doi: 10.1038/nri3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161:205–214. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schadendorf D, Hodi FS, Robert C, Weber JS, Margolin K, Hamid O, Patt D, Chen TT, Berman DM, Wolchok JD. Pooled Analysis of Long-Term Survival Data From Phase II and Phase III Trials of Ipilimumab in Unresectable or Metastatic Melanoma. J Clin Oncol. 2015;33:1889–1894. doi: 10.1200/JCO.2014.56.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, Chew A, Gonzalez VE, Zheng Z, Lacey SF, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalos M, Levine BL, Porter DL, Katz S, Grupp SA, Bagg A, June CH. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med. 2011;3:95ra73. doi: 10.1126/scitranslmed.3002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turtle CJ, Hanafi LA, Berger C, Gooley TA, Cherian S, Hudecek M, Sommermeyer D, Melville K, Pender B, Budiarto TM, et al. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J Clin Invest. 2016;126:2123–2138. doi: 10.1172/JCI85309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brentjens RJ, Davila ML, Riviere I, Park J, Wang X, Cowell LG, Bartido S, Stefanski J, Taylor C, Olszewska M, et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med. 2013;5:177ra138. doi: 10.1126/scitranslmed.3005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348:62–68. doi: 10.1126/science.aaa4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu P, Nielsen TE, Clausen MH. FDA-approved small-molecule kinase inhibitors. Trends Pharmacol Sci. 2015;36:422–439. doi: 10.1016/j.tips.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 24.Bollag G, Tsai J, Zhang J, Zhang C, Ibrahim P, Nolop K, Hirth P. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat Rev Drug Discov. 2012;11:873–886. doi: 10.1038/nrd3847. [DOI] [PubMed] [Google Scholar]
- 25.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 26.Wei SC, Levine JH, Cogdill AP, Zhao Y, Anang NAS, Andrews MC, Sharma P, Wang J, Wargo JA, Pe’er D, et al. Distinct Cellular Mechanisms Underlie Anti-CTLA-4 and Anti-PD-1 Checkpoint Blockade. Cell. 2017;170:1120–1133. e1117. doi: 10.1016/j.cell.2017.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu S, Li A, Liu Q, Li T, Yuan X, Han X, Wu K. Chimeric antigen receptor T cells: a novel therapy for solid tumors. Journal of Hematology & Oncology. 2017;10:78. doi: 10.1186/s13045-017-0444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuang M, Cheng J, Zhang C, Feng L, Xu X, Zhang Y, Zu M, Cui J, Yu H, Zhang K, et al. A novel signature for stratifying the molecular heterogeneity of the tissue-infiltrating T-cell receptor repertoire reflects gastric cancer prognosis. Sci Rep. 2017;7:7762. doi: 10.1038/s41598-017-08289-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi Q, Liu Y, Cheng Y, Glanville J, Zhang D, Lee JY, Olshen RA, Weyand CM, Boyd SD, Goronzy JJ. Diversity and clonal selection in the human T-cell repertoire. Proc Natl Acad Sci U S A. 2014;111:13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong Q, Wang C, Zhang W, Iqbal J, Hu Y, Greiner TC, Cornish A, Kim JH, Rabadan R, Abate F, et al. Assessment of T-cell receptor repertoire and clonal expansion in peripheral T-cell lymphoma using RNA-seq data. Sci Rep. 2017;7:11301. doi: 10.1038/s41598-017-11310-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tirosh I, Izar B, Prakadan SM, Wadsworth MH, 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Restifo NP, Gattinoni L. Lineage relationship of effector and memory T cells. Curr Opin Immunol. 2013;25:556–563. doi: 10.1016/j.coi.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmed R, Roger L, Costa Del Amo P, Miners KL, Jones RE, Boelen L, Fali T, Elemans M, Zhang Y, Appay V, et al. Human Stem Cell-like Memory T Cells Are Maintained in a State of Dynamic Flux. Cell Rep. 2016;17:2811–2818. doi: 10.1016/j.celrep.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maus MV, June CH. Making Better Chimeric Antigen Receptors for Adoptive T-cell Therapy. Clin Cancer Res. 2016;22:1875–1884. doi: 10.1158/1078-0432.CCR-15-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Proserpio V, Mahata B. Single-cell technologies to study the immune system. Immunology. 2016;147:133–140. doi: 10.1111/imm.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •36.Heath JR, Ribas A, Mischel PS. Single-cell analysis tools for drug discovery and development. Nat Rev Drug Discov. 2016;15:204–216. doi: 10.1038/nrd.2015.16. A comprehensive review of single-cell technologies and applications in the context of drug discovery and development, especially discusses how these technologies have applied in the field of oncology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barlogie B, Raber MN, Schumann J, Johnson TS, Drewinko B, Swartzendruber DE, Göhde W, Andreeff M, Freireich EJ. Flow cytometry in clinical cancer research. Cancer research. 1983;43:3982–3997. [PubMed] [Google Scholar]
- •38.Spitzer MH, Nolan GP. Mass cytometry: single cells, many features. Cell. 2016;165:780–791. doi: 10.1016/j.cell.2016.04.019. Provides a comprehensive review of mass cytometry, including an overview of the instrument, data analysis approaches, and the future outlook for this technology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haque A, Engel J, Teichmann SA, Lonnberg T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017;9:75. doi: 10.1186/s13073-017-0467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bock C, Farlik M, Sheffield NC. Multi-Omics of Single Cells: Strategies and Applications. Trends Biotechnol. 2016;34:605–608. doi: 10.1016/j.tibtech.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duncombe TA, Kang CC, Maity S, Ward TM, Pegram MD, Murthy N, Herr AE. Hydrogel Pore-Size Modulation for Enhanced Single-Cell Western Blotting. Adv Mater. 2016;28:327–334. doi: 10.1002/adma.201503939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang CC, Lin JM, Xu Z, Kumar S, Herr AE. Single-cell Western blotting after whole-cell imaging to assess cancer chemotherapeutic response. Anal Chem. 2014;86:10429–10436. doi: 10.1021/ac502932t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••43.Kang CC, Yamauchi KA, Vlassakis J, Sinkala E, Duncombe TA, Herr AE. Single cell-resolution western blotting. Nat Protoc. 2016;11:1508–1530. doi: 10.1038/nprot.2016.089. This paper provides detailed protocol for single-cell western blotting, a technology allows quantification of multiple proteins (especially isoforms of protein) from the same single cell and can integrate with flow sorting or on-chip cell imaging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim JJ, Sinkala E, Herr AE. High-selectivity cytology via lab-on-a-disc western blotting of individual cells. Lab Chip. 2017;17:855–863. doi: 10.1039/c6lc01333c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tentori AM, Yamauchi KA, Herr AE. Detection of Isoforms Differing by a Single Charge Unit in Individual Cells. Angew Chem Int Ed Engl. 2016;55:12431–12435. doi: 10.1002/anie.201606039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hughes AJ, Spelke DP, Xu Z, Kang CC, Schaffer DV, Herr AE. Single-cell western blotting. Nat Methods. 2014;11:749–755. doi: 10.1038/nmeth.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat Rev Immunol. 2004;4:648–655. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- ••48.Nicolet BP, Guislain A, Wolkers MC. Combined Single-Cell Measurement of Cytokine mRNA and Protein Identifies T Cells with Persistent Effector Function. J Immunol. 2017;198:962–970. doi: 10.4049/jimmunol.1601531. This paper illustrated the integration quantification of transcripts and protein of several cytokines from single T cells via a combination of fluorescence in situ hybridization (FISH) approach and flow cytometry. [DOI] [PubMed] [Google Scholar]
- 49.Bengsch B, Ohtani T, Herati RS, Bovenschen N, Chang KM, Wherry EJ. Deep immune profiling by mass cytometry links human T and NK cell differentiation and cytotoxic molecule expression patterns. J Immunol Methods. 2017 doi: 10.1016/j.jim.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chevrier S, Levine JH, Zanotelli VRT, Silina K, Schulz D, Bacac M, Ries CH, Ailles L, Jewett MAS, Moch H, et al. An Immune Atlas of Clear Cell Renal Cell Carcinoma. Cell. 2017;169:736–749. e718. doi: 10.1016/j.cell.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••51.Frei AP, Bava FA, Zunder ER, Hsieh EW, Chen SY, Nolan GP, Gherardini PF. Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nat Methods. 2016;13:269–275. doi: 10.1038/nmeth.3742. This paper demonstrates the utilization of PLAYR (proximity ligation assay for RNA) method for detection of both mRNA and protein from the same cell using either flow cytometry or mass cytometry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang AC, Postow MA, Orlowski RJ, Mick R, Bengsch B, Manne S, Xu W, Harmon S, Giles JR, Wenz B, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 2017;545:60–65. doi: 10.1038/nature22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lavin Y, Kobayashi S, Leader A, Amir ED, Elefant N, Bigenwald C, Remark R, Sweeney R, Becker CD, Levine JH, et al. Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell. 2017;169:750–765. e717. doi: 10.1016/j.cell.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matos TR, Liu H, Ritz J. Research Techniques Made Simple: Experimental Methodology for Single-Cell Mass Cytometry. J Invest Dermatol. 2017;137:e31–e38. doi: 10.1016/j.jid.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 55.Spitzer MH, Carmi Y, Reticker-Flynn NE, Kwek SS, Madhireddy D, Martins MM, Gherardini PF, Prestwood TR, Chabon J, Bendall SC, et al. Systemic Immunity Is Required for Effective Cancer Immunotherapy. Cell. 2017;168:487–502. e415. doi: 10.1016/j.cell.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Albayrak C, Jordi CA, Zechner C, Lin J, Bichsel CA, Khammash M, Tay S. Digital Quantification of Proteins and mRNA in Single Mammalian Cells. Mol Cell. 2016;61:914–924. doi: 10.1016/j.molcel.2016.02.030. [DOI] [PubMed] [Google Scholar]
- 57.Darmanis S, Gallant CJ, Marinescu VD, Niklasson M, Segerman A, Flamourakis G, Fredriksson S, Assarsson E, Lundberg M, Nelander S, et al. Simultaneous Multiplexed Measurement of RNA and Proteins in Single Cells. Cell Rep. 2016;14:380–389. doi: 10.1016/j.celrep.2015.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••58.Genshaft AS, Li S, Gallant CJ, Darmanis S, Prakadan SM, Ziegler CG, Lundberg M, Fredriksson S, Hong J, Regev A, et al. Multiplexed, targeted profiling of single-cell proteomes and transcriptomes in a single reaction. Genome Biol. 2016;17:188. doi: 10.1186/s13059-016-1045-6. This paper describes a method using DNA oligonucleotide as antibody label for simultaneous quantification of transcripts and protein from the same mammalian single cell in the same reaction chamber. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mahata B, Zhang X, Kolodziejczyk AA, Proserpio V, Haim-Vilmovsky L, Taylor AE, Hebenstreit D, Dingler FA, Moignard V, Gottgens B, et al. Single-cell RNA sequencing reveals T helper cells synthesizing steroids de novo to contribute to immune homeostasis. Cell Rep. 2014;7:1130–1142. doi: 10.1016/j.celrep.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kakaradov B, Arsenio J, Widjaja CE, He Z, Aigner S, Metz PJ, Yu B, Wehrens EJ, Lopez J, Kim SH, et al. Early transcriptional and epigenetic regulation of CD8+ T cell differentiation revealed by single-cell RNA sequencing. Nat Immunol. 2017;18:422–432. doi: 10.1038/ni.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, Kang B, Hu R, Huang JY, Zhang Q, et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell. 2017;169:1342–1356. e1316. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- ••62.Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2017 doi: 10.1038/nri.2017.76. A detailed review briefly describes several available single-cell RNA sequencing technologies and discusses how it can decipher the heterogeneity of the immune system. [DOI] [PubMed] [Google Scholar]
- 63.Picelli S. Single-cell RNA-sequencing: The future of genome biology is now. RNA Biol. 2017;14:637–650. doi: 10.1080/15476286.2016.1201618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I, Enard W. Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol Cell. 2017;65:631–643. e634. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 65.Stubbington MJT, Lonnberg T, Proserpio V, Clare S, Speak AO, Dougan G, Teichmann SA. T cell fate and clonality inference from single-cell transcriptomes. Nat Methods. 2016;13:329–332. doi: 10.1038/nmeth.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Redmond D, Poran A, Elemento O. Single-cell TCRseq: paired recovery of entire T-cell alpha and beta chain transcripts in T-cell receptors from single-cell RNAseq. Genome Med. 2016;8:80. doi: 10.1186/s13073-016-0335-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eltahla AA, Rizzetto S, Pirozyan MR, Betz-Stablein BD, Venturi V, Kedzierska K, Lloyd AR, Bull RA, Luciani F. Linking the T cell receptor to the single cell transcriptome in antigen-specific human T cells. Immunol Cell Biol. 2016;94:604–611. doi: 10.1038/icb.2016.16. [DOI] [PubMed] [Google Scholar]
- 68.Afik S, Yates KB, Bi K, Darko S, Godec J, Gerdemann U, Swadling L, Douek DC, Klenerman P, Barnes EJ, et al. Targeted reconstruction of T cell receptor sequence from single cell RNA-seq links CDR3 length to T cell differentiation state. Nucleic Acids Res. 2017;45:e148. doi: 10.1093/nar/gkx615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••69.Peterson VM, Zhang KX, Kumar N, Wong J, Li L, Wilson DC, Moore R, McClanahan TK, Sadekova S, Klappenbach JA. Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol. 2017;35:936–939. doi: 10.1038/nbt.3973. By utilizing a combination of unique oligonucleotide barcodes and poly (dA) sequence for indexing antibody, REAP-seq enables the detection of multiple proteins along with transcripts and is compatible with scRNA-seq platforms. [DOI] [PubMed] [Google Scholar]
- ••70.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. CITE-seq uses the similar concept for antibody indexing as REAP-seq to facilitate simultaneous detection of mRNA and protein from single cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •71.Shahi P, Kim SC, Haliburton JR, Gartner ZJ, Abate AR. Abseq: Ultrahigh-throughput single cell protein profiling with droplet microfluidic barcoding. Sci Rep. 2017;7:44447. doi: 10.1038/srep44447. Abseq is based on the droplet microfluidics for single cell isolation and allows high-throughput protein quantification by using DNA oligonucleotide as antibody labels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •72.Gierahn TM, Wadsworth MH, 2nd, Hughes TK, Bryson BD, Butler A, Satija R, Fortune S, Love JC, Shalek AK. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods. 2017;14:395–398. doi: 10.1038/nmeth.4179. As an orthogonal method to Abseq, Seq-well isolates single cell by patterned microwell and serves as a portable and low-cost scRNA-seq platform. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bacher R, Chu LF, Leng N, Gasch AP, Thomson JA, Stewart RM, Newton M, Kendziorski C. SCnorm: robust normalization of single-cell RNA-seq data. Nat Methods. 2017;14:584–586. doi: 10.1038/nmeth.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bacher R, Kendziorski C. Design and computational analysis of single-cell RNA-sequencing experiments. Genome Biol. 2016;17:63. doi: 10.1186/s13059-016-0927-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Poirion OB, Zhu X, Ching T, Garmire L. Single-Cell Transcriptomics Bioinformatics and Computational Challenges. Front Genet. 2016;7:163. doi: 10.3389/fgene.2016.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu AR, Wang J, Streets AM, Huang Y. Single-Cell Transcriptional Analysis. Annu Rev Anal Chem (Palo Alto Calif) 2017;10:439–462. doi: 10.1146/annurev-anchem-061516-045228. [DOI] [PubMed] [Google Scholar]
- 77.Lee CH, Romain G, Yan W, Watanabe M, Charab W, Todorova B, Lee J, Triplett K, Donkor M, Lungu OI, et al. IgG Fc domains that bind C1q but not effector Fcgamma receptors delineate the importance of complement-mediated effector functions. Nat Immunol. 2017;18:889–898. doi: 10.1038/ni.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ritthipichai K, Haymaker CL, Martinez M, Aschenbrenner A, Yi X, Zhang M, Kale C, Vence LM, Roszik J, Hailemichael Y, et al. Multifaceted Role of BTLA in the Control of CD8+ T-cell Fate after Antigen Encounter. Clin Cancer Res. 2017;23:6151–6164. doi: 10.1158/1078-0432.CCR-16-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Merouane A, Rey-Villamizar N, Lu Y, Liadi I, Romain G, Lu J, Singh H, Cooper LJ, Varadarajan N, Roysam B. Automated profiling of individual cell-cell interactions from high-throughput time-lapse imaging microscopy in nanowell grids (TIMING) Bioinformatics. 2015;31:3189–3197. doi: 10.1093/bioinformatics/btv355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••80.An X, Sendra VG, Liadi I, Ramesh B, Romain G, Haymaker C, Martinez-Paniagua M, Lu Y, Radvanyi LG, Roysam B, et al. Single-cell profiling of dynamic cytokine secretion and the phenotype of immune cells. PLoS One. 2017;12:e0181904. doi: 10.1371/journal.pone.0181904. It is the first paper that illustrating TIMING platform is compatible with cell phenotyping and dynamic monitoring of secreted protein from the same immune cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sendra VG, Lie A, Romain G, Agarwal SK, Varadarajan N. Detection and isolation of auto-reactive human antibodies from primary B cells. Methods. 2013;64:153–159. doi: 10.1016/j.ymeth.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sarkar S, Sabhachandani P, Stroopinsky D, Palmer K, Cohen N, Rosenblatt J, Avigan D, Konry T. Dynamic analysis of immune and cancer cell interactions at single cell level in microfluidic droplets. Biomicrofluidics. 2016;10:054115. doi: 10.1063/1.4964716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eyer K, Doineau RCL, Castrillon CE, Briseno-Roa L, Menrath V, Mottet G, England P, Godina A, Brient-Litzler E, Nizak C, et al. Single-cell deep phenotyping of IgG-secreting cells for high-resolution immune monitoring. Nat Biotechnol. 2017;35:977–982. doi: 10.1038/nbt.3964. [DOI] [PubMed] [Google Scholar]
- ••84.Kravchenko-Balasha N, Shin YS, Sutherland A, Levine RD, Heath JR. Intercellular signaling through secreted proteins induces free-energy gradient-directed cell movement. Proc Natl Acad Sci U S A. 2016;113:5520–5525. doi: 10.1073/pnas.1602171113. SCBC is used to detect secreted proteins from brain cancer cell pairs along with cell-cell movement monitoring. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wei W, Shin YS, Xue M, Matsutani T, Masui K, Yang H, Ikegami S, Gu Y, Herrmann K, Johnson D, et al. Single-Cell Phosphoproteomics Resolves Adaptive Signaling Dynamics and Informs Targeted Combination Therapy in Glioblastoma. Cancer Cell. 2016;29:563–573. doi: 10.1016/j.ccell.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou J, Kaiser A, Ng C, Karcher R, McConnell T, Paczkowski P, Fernandez C, Zhang M, Mackay S, Tsuji M. CD8+ T-cell mediated anti-malaria protection induced by malaria vaccines; assessment of hepatic CD8+ T cells by SCBC assay. Hum Vaccin Immunother. 2017;13:1625–1629. doi: 10.1080/21645515.2017.1304333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang L, Wang Z, Deng Y, Li Y, Wei W, Shi Q. Single-Cell, Multiplexed Protein Detection of Rare Tumor Cells Based on a Beads-on-Barcode Antibody Microarray. Anal Chem. 2016;88:11077–11083. doi: 10.1021/acs.analchem.6b03086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maleki Vareki S, Garrigos C, Duran I. Biomarkers of response to PD-1/PD-L1 inhibition. Crit Rev Oncol Hematol. 2017;116:116–124. doi: 10.1016/j.critrevonc.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 89.Xi L, Pham TH, Payabyab EC, Sherry RM, Rosenberg SA, Raffeld M. Circulating Tumor DNA as an Early Indicator of Response to T-cell Transfer Immunotherapy in Metastatic Melanoma. Clin Cancer Res. 2016;22:5480–5486. doi: 10.1158/1078-0432.CCR-16-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vasaturo A, Halilovic A, Bol KF, Verweij DI, Blokx WA, Punt CJ, Groenen PJ, van Krieken JH, Textor J, de Vries IJ, et al. T-cell Landscape in a Primary Melanoma Predicts the Survival of Patients with Metastatic Disease after Their Treatment with Dendritic Cell Vaccines. Cancer Res. 2016;76:3496–3506. doi: 10.1158/0008-5472.CAN-15-3211. [DOI] [PubMed] [Google Scholar]
- 91.Butterfield LH. The Society for Immunotherapy of Cancer Biomarkers Task Force recommendations review. Semin Cancer Biol. 2017 doi: 10.1016/j.semcancer.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Axelrod ML, Johnson DB, Balko JM. Emerging biomarkers for cancer immunotherapy in melanoma. Semin Cancer Biol. 2017 doi: 10.1016/j.semcancer.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Diggs LP, Hsueh EC. Utility of PD-L1 immunohistochemistry assays for predicting PD-1/PD-L1 inhibitor response. Biomark Res. 2017;5:12. doi: 10.1186/s40364-017-0093-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kamphorst AO, Pillai RN, Yang S, Nasti TH, Akondy RS, Wieland A, Sica GL, Yu K, Koenig L, Patel NT, et al. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1–targeted therapy in lung cancer patients. Proceedings of the National Academy of Sciences. 2017;114:4993–4998. doi: 10.1073/pnas.1705327114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ma C, Cheung AF, Chodon T, Koya RC, Wu Z, Ng C, Avramis E, Cochran AJ, Witte ON, Baltimore D, et al. Multifunctional T-cell analyses to study response and progression in adoptive cell transfer immunotherapy. Cancer Discov. 2013;3:418–429. doi: 10.1158/2159-8290.CD-12-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liadi I, Singh H, Romain G, Rey-Villamizar N, Merouane A, Adolacion JRT, Kebriaei P, Huls H, Qiu P, Roysam B, et al. Individual Motile CD4(+) T Cells Can Participate in Efficient Multikilling through Conjugation to Multiple Tumor Cells. Cancer Immunol Res. 2015;3:473–482. doi: 10.1158/2326-6066.CIR-14-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Larsson C, Grundberg I, Soderberg O, Nilsson M. In situ detection and genotyping of individual mRNA molecules. Nat Methods. 2010;7:395–397. doi: 10.1038/nmeth.1448. [DOI] [PubMed] [Google Scholar]
- 98.Ke R, Mignardi M, Pacureanu A, Svedlund J, Botling J, Wahlby C, Nilsson M. In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods. 2013;10:857–860. doi: 10.1038/nmeth.2563. [DOI] [PubMed] [Google Scholar]
- 99.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, Ferrante TC, Terry R, Jeanty SS, Li C, Amamoto R, et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mellis IA, Gupte R, Raj A, Rouhanifard SH. Visualizing adenosine-to-inosine RNA editing in single mammalian cells. Nat Methods. 2017;14:801–804. doi: 10.1038/nmeth.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shah S, Lubeck E, Zhou W, Cai L. In Situ Transcription Profiling of Single Cells Reveals Spatial Organization of Cells in the Mouse Hippocampus. Neuron. 2016;92:342–357. doi: 10.1016/j.neuron.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Moffitt JR, Hao J, Wang G, Chen KH, Babcock HP, Zhuang X. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc Natl Acad Sci U S A. 2016;113:11046–11051. doi: 10.1073/pnas.1612826113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tricot S, Meyrand M, Sammicheli C, Elhmouzi-Younes J, Corneau A, Bertholet S, Malissen M, Le Grand R, Nuti S, Luche H, et al. Evaluating the efficiency of isotope transmission for improved panel design and a comparison of the detection sensitivities of mass cytometer instruments. Cytometry A. 2015;87:357–368. doi: 10.1002/cyto.a.22648. [DOI] [PubMed] [Google Scholar]
- 104.Vallejos CA, Risso D, Scialdone A, Dudoit S, Marioni JC. Normalizing single-cell RNA sequencing data: challenges and opportunities. Nat Methods. 2017;14:565–571. doi: 10.1038/nmeth.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang B, Zhu J, Pierson E, Ramazzotti D, Batzoglou S. Visualization and analysis of single-cell RNA-seq data by kernel-based similarity learning. Nat Methods. 2017;14:414–416. doi: 10.1038/nmeth.4207. [DOI] [PubMed] [Google Scholar]
- 106.Kiselev VY, Kirschner K, Schaub MT, Andrews T, Yiu A, Chandra T, Natarajan KN, Reik W, Barahona M, Green AR, et al. SC3: consensus clustering of single-cell RNA-seq data. Nat Methods. 2017;14:483–486. doi: 10.1038/nmeth.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Su Y, Shi Q, Wei W. Single cell proteomics in biomedicine: High-dimensional data acquisition, visualization, and analysis. Proteomics. 2017:17. doi: 10.1002/pmic.201600267. [DOI] [PMC free article] [PubMed] [Google Scholar]