Abstract
Colorectal cancer (CRC) is one of the most difficult cancers to cure. An important prognostic factor is metastasis, which precludes curative surgical resection. Recent evidences show that Evodiamine (EVO) exerts an inhibitory effect on cancer cell apoptosis, migration, and invasion. In this study, we investigated the effects of EVO on the metastasis of CRC cells in vitro and in vivo. In vitro, wound-healing and transwell assay showed that migration and invasion of HT-29 and HCT-116 CRC cells were inhibited significantly by EVO. Western blot and RT-PCR showed that EVO reduced the expression of matrix metalloproteinase-9 in a dose-dependent manner. In EVO-induced cells, the intracellular NAD+/NADH ratio was increased, the level of Sirt1 was increased, and acetyl-NF-κB P65 was decreased. This process was inhibited by nicotinamide, an inhibitor of Sirt1. In vivo, EVO reduced tumor metastasis markedly. These findings provide evidences that EVO suppresses the migration and invasion of CRC cells by inhibiting the acetyl-NF-κB p65 by Sirt1, resulting in suppression of metalloproteinase-9 expression in vitro and in vivo.
Keywords: Evodiamine, invasion, migration, NF-kB, Sirt1
Introduction
Evodiamine (EVO) is a major constituent of the plant Evodia rutaecarpa. EVO exerts various biological effects, such as vasodilatory, anti-inflammatory, antitumor, and antinociception 1–3. Although some studies have shown that EVO induces apoptosis, inhibits drug resistance, and proliferation 4–6, the in-vitro and in-vivo mechanisms of EVO on migration and invasion are still unclear.
Sirtuins are the mammalian orthologues of yeast silent information regulator 2 (SIR2); they have been found to extend the life span of yeast 7. There are seven isoforms in the sirtuin family: Sirt1 to Sirt7. As a NAD-dependent histone deacetylase, Sirt1 belongs to the family of class III histone deacetylases 8. Because of cytoplasmic and nuclear localizations, Sirt1 can deacetylate various histone and nonhistone, such as p53, p300, c-MYC, and nuclear factor-κB (NF-κB) 9. NF-κB is a nuclear transcription factor. In recent years, a large number of studies have shown that NF-κB was related to the occurrence, proliferation, differentiation, apoptosis, migration, and invasion of tumors 10. Sirt1 could deacetylate the NF-κB p65 subunit under the assist of NAD+ and repress the transcription activity of NF-κB p65. Then, the transcription of specific genes associated with proliferation, migration, invasion, and metastasis is inhibited, such as IL-6 and TNF-α, VEGF, IL-1β, TF, and MMPs, respectively 11,12.
Our study focused on the interrelation between Sirt1 and the transcription activity of NF-κB p65 to explore the underlying mechanism of EVO in CRC cells’ migration and invasion in vitro and in vivo.
Materials and methods
Cell lines and culture conditions
The human colon cancer cell lines HT-29 cells and HCT-116 cells were kind gifts from Army Medical University. Both cell lines were maintained in DMEM medium containing 10% fetal bovine serum (Gibco, Carlsbad, California, USA) with 100 μg/ml penicillin and streptomycin (Gibco) and incubated at 37°C under 5% CO2.
Antibodies and chemicals
EVO (S2382, purity 99.76%; Selleck Chemicals, Houston, Texas, USA) was dissolved in dimethyl sulfoxide (Me2SO) and PBS, respectively. The concentration of Me2SO was maintained below 0.1% to avoid cytotoxicity [Nicotinamide (NAM, 47865-U, purity 99.9%; Sigma company); Cell Counting Kit-8 (Takara Bio Inc., Shiga, Japan)]. The following antibodies were used for western blot and immunofluorescence: Sirt1 and β-actin (Santa Cruz Biotechnology, Inc., Dallas, Texas, USA); NF-κB p65 and acetyl-NF-κB p65 (Cell Signaling Technology, Danvers, Massachusetts, USA); and MMP-9 (Boster Biological Technology, Wuhan, Hubei, China).
Wound-healing assay
HT-29 cells and HCT-116 cells were cultured in a six-well culture plate for 24 h. Subsequently, wounds were created in the confluent cells using a 10-µl pipette tip. The debris was removed by washing with serum-free medium, and cells were treated with EVO (control or 1.5 μmol/l). After 24 h of incubation, cells that migrated into the wounded area or protruded from the border of the wound were photographed under an inverted microscope. Each experiment was conducted independently three times.
Transwell assay
HT-29 cells and HCT-116 cells were seeded onto the upper part of a Transwell chamber (Corning Inc., Corning, New York, USA) containing a gelatin-coated polycarbonate membrane filter (pore size: 8 μm) for the invasion assays, respectively. Chambers were preconditioned using Matrigel (1 : 8; BD Biosciences, San Jose, California, USA). In brief, HT-29 cells and HCT-116 cells (2×105/ml) were seeded onto the upper chamber in 200 μl of serum-free medium containing EVO at concentrations of 0 or 1.5 μmol/l, respectively, and medium containing 10% fetal bovine serum was supplemented in the lower wells. After 24 h incubation at 37°C with 5% CO2, the noninvading cells were wiped out carefully. Then, the filters were fixed in 95% alcohol and stained with crystal violet (Sangon, Shanghai, China). Three random fields were counted per chamber using an inverted microscope (Olympus, Tokyo, Japan), and each experiment was repeated three times.
Real-time PCR
Total RNA was extracted from colorectal cancer (CRC) cell lines using Trizol reagent (Takara; Takara Bio., Shiga, Japan). Following DNaseI treatment, 2 μg of total RNA was reverse transcribed using the cDNA synthesis kit (Gene Copoeia, Rockville, Maryland, USA) to synthesize cDNA specimens. Quantitative real-time PCR (qRT-PCR) analysis of gene expression was carried out using 2 μl of cDNA and SYBR Green Supermix (Bio-Rad Laboratories Inc., Berkeley, California, USA) as recommended by the manufacturer. Primer sequences were synthesized as follows: MMP-9 primer: forward, 5′-CTCCTACTCTGCCTGCACCA-3′; reverse, 5′-CATCACCGTCGAGTCAGCTC-3′; β-actin primer: forward, 5′-CATCAAGAAGGTGGTGAAGCA-3′; reverse, 5′-CGTCAAAGGTGGAGGAGTGG-3′.
Western blot and immunofluorescence
Cells or homogenized tumor samples were washed twice with cold 1× PBS and lysed on ice for 30 min in a buffer containing 50 mmol/l Tris–HCL (pH: 7.5), 150 mmol/l NaCl, 1% Triton-X, 0.5% NaDOC, 0.1% SDS, 5 mmol/l EDTA, 50 mmol/l NaF, 1 mmol/l NaVO3, 10% glycerol, and protease inhibitors: 10 μg/ml leupeptin, 10 μg/ml phenylmethylsulfonylfluoride, and 1 μg/ml aprotinin. The lysates were cleared by centrifugation at 12 000 rpm for 30 min at 4°C. Protein concentrations were determined using a BCA Protein Assay Kit (Beyotime Biotechnology, Shanghai, China). The boiled samples were loaded on SDS-PAGE gels. Western blots were performed as described previously. Briefly, antibodies were used according to the manufacturer’s recommended conditions. Molecular weight markers were utilized to confirm the expected size of the target proteins. Immunoblots were developed using the ChemiDoc Touch Imaging System (Bio-Rad Laboratories Inc., Berkeley, California, USA). Blots were stripped with stripping solution (Bio-Rad Laboratories Inc.) at 37°C for 15 min and then reprobed with selected antibodies. Immunoblotting with antibody to β-actin provided an internal control for equal protein loading. Expression of acetylated proteins was defined as the ratio of acetylated : total protein. Immunoblotting with antibody to β-actin provided an internal control for equal protein loading. Expression of acetylated proteins was defined as a ratio of acetylated : total protein.
NAD+/NADH assay
NAD+/NADH assay was performed using the NAD+/NADH Fluorescence Assay kit (KeyGEN BioTECH, Nanjing, Jiangsu, China) according to the manufacturer’s instruction.
Animals
Female Balb/c nude mice (4 weeks) were purchased from the Animal experiment center of Chongqing Medical University and bred in the Animal experiment center. Nude mice were administered tail-vein injections of HCT-116 CRC cells (1×106 cells/injection). Ambient temperature was maintained at 25°C, and the mice were maintained on an artificial 12-h light–dark cycle, with the light period beginning at 7:00 am. The mice were divided into two groups randomly: EVO group and control group. The EVO group was treated with EVO (10 mg/kg) by filling the stomach (once a day, a total of 27 days); the control group was treated with normal saline. All protocols were reviewed and approved by the Animal experiment center of Chongqing Medical University (Chongqing, China), and the study was carried out in accordance with the principles of laboratory animal care.
Statistical analysis
Results were analyzed using SPSS 22.0 software (IBM, Armonk, New York, USA) and expressed as mean±SD. Single-factor variance analysis was used for multiple groups, and the least significant difference test was used for pairwise comparison. A P value of less than 0.05 was set as the threshold of significance.
Results
Evodiamine inhibits proliferation in CRC cells
The chemical structure of EVO (C19H17N3O) is shown in Fig. 1a. To observe the proliferation-inhibitory effects of EVO, HT-29 cells and HCT-116 cells were treated with different concentrations of EVO for 24, 48, and 72 h. The cell viability was measured using the CCK-8 assay (Beyotime, Shanghai, China). EVO could significantly inhibit the proliferation of HT-29 cells and HCT-116 cells in a dose-dependent and time-dependent manner (Fig. 1b). The half-maximal inhibitory concentration (IC50) values of EVO on HT-29 cells and HCT-116 cells were about 6 μmol/l. When cells were treated with 1.5 μmol/l EVO for 24 h, more than 90% of the cells survived. Taken together, these results indicate that EVO inhibits the proliferation of CRC cells.
Fig. 1.
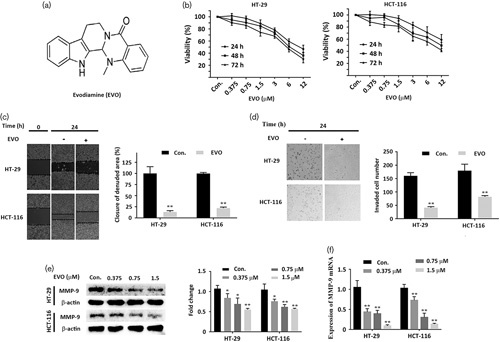
Evodiamine (EVO) inhibits cell viability, migration, and invasion in colorectal cancer cells. (a) Chemical structure of Evodiamine. (b) HT-29 cells and HCT-116 cells were treated with different concentrations (0, 0.375, 0.75, 1.5, 3, 6, and 12 μmol/l) of Evodiamine for 24, 48, and 72 h, and cell viability was detected using the CCK-8 assay. (c) Wound-healing assay. After treatment with different concentrations of Evodiamine for 24 h, the migration rates of HT-29 cells and HCT-116 cells were detected. (d) Transwell assay and the invasion rates were analyzed; the numbers of HT-29 cells and HCT-116 cells were counted. (e) Western bolt analysis of MMP-9 in Evodiamine-induced cells. Cells were treated with the indicated concentration of Evodiamine, and cell fractions were subjected to western blot by using the anti-MMP-9 antibody. (f) Real-time PCR analyses of the MMP-9 gene. Cells were treated with the indicated concentration of Evodiamine for 24 h. Values are represented as the mean±SD (n=3). *P<0.05 versus the control; **P<0.01 versus. control, a significant difference compared with the control by one‐way analysis of variance.
Evodiamine inhibits migration and invasion in CRC cells in vitro
Wound-healing assay showed that treatment with EVO (1.5 μmol/l) could significantly repress the migration of HT-29 cells and HCT-116 cells (Fig. 1c). Figure 1d shows that the invasion of HT-29 cells and HCT-116 cells was inhibited markedly by EVO (1.5 μmol/l). Compared with the control group, the numbers of HT-29 cells and HCT-116 cells adherent to the outer surface of the membrane were obviously decreased in the group treated with EVO. In addition, MMP-9 protein and mRNA levels were downregulated significantly by EVO in a concentration-dependent manner (Fig. 1e and f). Taken together, EVO could inhibit migration and invasion by downregulating MMP-9 in CRC cells in vitro.
NF-κB p65 is responsible for the inhibitory effect of Evodiamine on MMP-9 production by activating Sirt1
Western blot showed that the acetyl-p65 was decreased by EVO, but the total NF-κB p65 did not change (Fig. 2a). We further found that the intracellular NAD+/NADH ratio was increased after treatment with the indicated time and dose of EVO (Fig. 2b and c). Recent studies reported that EVO could lead to oxidative stress 13,14 and Sirt1 exerts a regulatory effect on oxidative stress by deacetylating various histone and nonhistone 15. Hence, we examined the Sirt1 by western blot and immunofluorescence staining. EVO could significantly increase the level of Sirt1 in a concentration-dependent manner (Fig. 2a) and Sirt1 showed remarkable nuclear localization (Fig. 2d). To investigate whether the downregulation of acetyl-p65 was induced by upregulating Sirt1, HT-29 cells and HCT-116 cells were treated with NAM (5 mmol/l), an inhibitor of Sirt1. The NAM inhibited the effect of EVO on Sirt1/p65 signaling (Fig. 2e). Taken together, these results indicate that EVO downregulates MMP-9 by reducing the acetyl-NF-κB p65 through activating Sirt1.
Fig. 2.
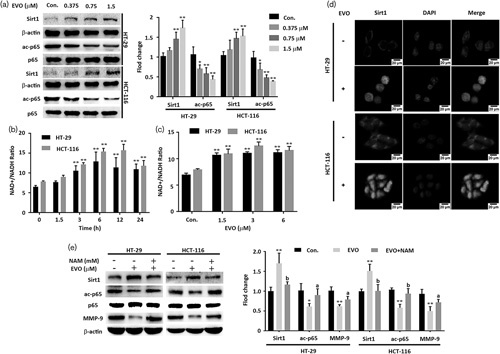
Evodiamine (EVO) increased the rate of NAD+/NADH, and activated Sirt1 signaling. (a) The rate of NAD+/NADH. HT-29 cells and HCT-116 cells were treated with Evodiamine (1.5 μmol/l) for the indicated number of hours. (b) The rate of NAD+/NADH. Cells were treated with the indicated concentration of Evodiamine. (c) Western blot analysis of Sirt1, NF-κB p65, acetyl-NF-κB p65, and MMP-9. Cells were treated with Evodiamine (0, 0.375, 0.75, and 1.5 μmol/l) for 24 h. (d) Immunofluorescence staining of Sirt1. HT-29 cells and HCT-116 cells were treated with Evodiamine (1.5 μmol/l) for 24 h. (e) Western blot analysis of Sirt1, NF-κB p65, acetyl-NF-κB p65, and MMP-9. Cells were treated with nicotinamide (5 mmol/l), followed by 1.5 μmol/l. Evodiamine for 24 h. Cell fractions were subjected to western blot using the anti-Sirt1 antibody, the anti-NF-κB p65 antibody, the anti-acetyl-NF-κB p65 antibody, and the anti-MMP-9 antibody. Data are presented as mean±SD (n=3) for each group. *P<0.01 versus the control. **P<0.05 vs. control, a significant difference compared with the control by one‐way analysis of variance. aP <0.05, bP <0.01.
Evodiamine suppresses metastasis in vivo
To explore whether EVO could suppress tumor migration and invasion in vivo, HCT-116 cells were inoculated into nude mice to construct mice models. Mice models were divided randomly into two groups: EVO group and control group. After treatment for 27 days, the nude mice were killed. The metastasis nodules in the lung and intestine were counted. In the EVO group, the mean weight of nude mice showed no significant change compared with the control group. Metastasis nodules of the lung and intestine were significantly decreased by more than 42.83 and 69.23% compared with the control group (Fig. 3c), respectively. These results indicate that EVO could suppress metastasis.
Fig. 3.
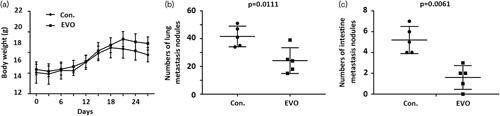
Inhibitory effects of Evodiamine (EVO) on the metastasis in nude mice. (a) The dynamic changes in body weight. (b) Metastasis nodules in the lung. (c) Metastasis nodules in the intestine. Data are presented as mean±SD (n=5) for each group.
Evodiamine activates Sirt1 signaling in vivo
In western blot analysis, Sirt1 was upregulated, and acetyl-NF-κB p65, and MMP-9 were downregulated, but the total NF-κB p65 was not affected by EVO (Fig. 4a). These results together suggest that EVO activates Sirt1 signaling in vivo.
Fig. 4.
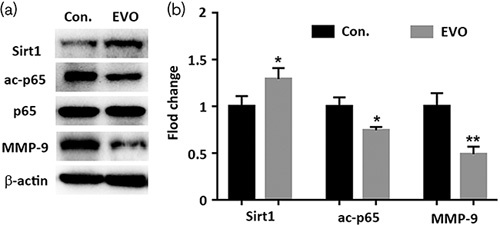
Effects of Evodiamine (EVO) on the protein expression of lung tumors nodules in nude mice. (a, b) Western blot analysis of Sirt1, NF-κB p65, acetyl-p65, and MMP-9 in the control and Evodiamine group. Tissue fractions were subjected to western blot using the anti-Sirt1 antibody, the anti-acetyl-NF-κB p65 antibody, the anti-NF-κB p65 antibody, and the anti-MMP-9 antibody. Data are presented as mean±SD (n=3) for each group. *P<0.01 versus control. **P<0.05 versus control, a significant difference compared with the control by one‐way analysis of variance.
Discussion
As the most common cancer in the world, the treatment methods of CRC include surgical treatment, radiotherapy, chemotherapy, immunotherapy, gene therapy, and traditional Chinese medicine treatment, and the inhibition of metastasis is important for the treatment of CRC 16. However, considering the drug resistance and high toxicity resulting in adverse side effects, development of novel therapeutic strategies and discovery of new drugs are essential to defeat CRC.
Recent evidences suggest that EVO exerts efficient inhibitory effect on cancer cells, for instance, lung cancer, liver cancer, gastric cancer, ovarian cancer, and CRC 3,17–20. Yang had shown that the p53 signaling pathway was activated in COLO-205 cells, and EVO could repress the proliferation of human CRC cells 21. EVO could block cell cycle and cause oxidative stress through inducing ROS 13,22. Migration and invasion are crucial for CRC, and our study investigated the effect of EVO on the migration and invasion of CRC, and the underlying mechanism.
In our study, on treatment with 1.5 μmol/l EVO for 24 h, more than 90% of the cells survived. However, at this concentration, the wound-healing experiment and transwell experiment showed that EVO could inhibit the migration and invasion of HT-29 cells and HCT-116 cells. Matrix metalloproteinases are often associated with the migration and invasion of cancer cells 23. The MMP-9 protein and mRNA were decreased. Hence, we hypothesized that the downregulation of MMP-9 transcription contributes toward the suppression effect of EVO on migration and invasion.
The transcription factor family of NF-κB consists of p65 (RelaA), RelB, c-Rel, p105/p50 (NF-κB1), and p100/52 (NF-κB2) 24. MMP-9 transcription is regulated by binding of the transcription factors NF-κB p65 to MMP-9 promoter sequences 25. The transcriptional activity of NF-κB p65 is related to acetylation and deacetylation. Our western blot showed that EVO inhibits the transcription activity of NF-κB p65 by decreasing the acetyl-NF-κB p65, and then the transcription of MMP-9 was suppressed. Interestingly, we observed that the nuclear localization of NF-κB p65 was inhibited in immunohistochemistry. It seemed that EVO could inhibit the nuclear localization of NF-κB p65, and this was involved in the acetylation of NF-κB p65. These results suggested that EVO could reduce MMP-9 protein by decreasing the transcription activity of NF-κB p65.
Sirt1, as a NAD-dependent histone deacetylase, can deacetylate various histone and nonhistone, including NF-κB 9. Hence, Sirt1 is involved in many biological processes, including metabolism, aging, oncogenesis, and cancer progression 26. Sirt1 shows a marked change in many human cancer tissues, such as ovary, liver, breast, pancreas, and colon 27–31. Also, the role of Sirt1 in carcinogenic processes is still controversial. In our study, EVO increased the expression of Sirt1 and upregulated deacetylase activity by increasing the ratio of NAD+/NADH. EVO may lead to oxidative stress, and ultimately, an increase in the ratio of NAD+/NADH. Also, the activation of Sirt1 was responsible for the protein changes of acetyl-NF-κB p65, and ultimately decreased the expression of MMP-9. In addition, NAM inhibited the effect of EVO on the acetyl-NF-κB p65. In this biological process, EVO represses the nucleus location of p65 and may involve more biomolecules, such as ΙκΒα, HDAC3, p300, and CBP 32.
A key finding in this study is that we observed a strong nuclear localization of Sirt1 in vitro and in vivo. Sirt1, with a strong nuclear localization, deacetylates NF-κB p65 at lysine 310 and p53 at lysine 382 in the nucleus 33,34. The phosphorylation may be responsible for the change in localization of Sirt1. The phosphorylation of Sirt1 (at ser27 and ser47) increases stability and nuclear localization of Sirt1, and phosphorylated Sirt1 selectively deacetylates histone H3, but not p53 33,35. EVO has the potential to affect other proteins and not only p65.
We also found that EVO markedly suppressed metastasis in the tail-vein injection model. Compared with the control group, the weight of nude mice showed no obvious change, but EVO markedly reduced the metastasis nodules in the lung and intestine. In addition, western blot analysis of nodule samples showed that EVO may inhibit MMP-9 by activating Sirt1/p65 signaling.
Conclusion
Our results indicate that EVO can repress the migration and invasion of CRC in vitro and in vivo. The underlying mechanism may involve a decrease in the acetyl-NF-κB p65, which is caused by an increase in Sirt1, which itself inhibits the expression of MMP-9. Our findings provide not only a novel target for inhibition migration and invasion of CRC but also a potential medicine for CRC.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (Grant no. 31271368). The authors would like to acknowledge the College of Life Science, Chongqing Medical University (Chongqing, China), for technical support.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Heo SK, Yun HJ, Yi HS, Noh EK, Park SD. Evodiamine and rutaecarpine inhibit migration by LIGHT via suppression of NADPH oxidase activation. J Cell Biochem 2009; 107:123–133. [DOI] [PubMed] [Google Scholar]
- 2.Chiou WF, Ko HC, Wei BL. Evodia rutaecarpa and three major alkaloids abrogate influenza A virus (H1N1)-induced chemokines production and cell migration. Evid Based Complement Alternat Med 2011; 2011:750513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kan SF, Yu CH, Pu HF, Hsu JM, Chen MJ, Wang PS. Anti-proliferative effects of evodiamine on human prostate cancer cell lines DU145 and PC3. J Cell Biochem 2007; 101:44–56. [DOI] [PubMed] [Google Scholar]
- 4.Yang ZG, Chen AQ, Liu B. Antiproliferation and apoptosis induced by evodiamine in human colorectal carcinoma cells (COLO-205). Chem Biodivers 2009; 6:924–933. [DOI] [PubMed] [Google Scholar]
- 5.Sui H, Zhou LH, Zhang YL, Huang JP, Liu X, Ji Q, et al. Evodiamine suppresses ABCG2 mediated drug resistance by inhibiting p50/p65 NF-kappaB pathway in colorectal cancer. J Cell Biochem 2016; 117:1471–1481. [DOI] [PubMed] [Google Scholar]
- 6.Chien CC, Wu MS, Shen SC, Ko CH, Chen CH, Yang LL, et al. Activation of JNK contributes to evodiamine-induced apoptosis and G2/M arrest in human colorectal carcinoma cells: a structure-activity study of evodiamine. PLoS One 2014; 9:e99729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guarente L, Franklin H. Epstein Lecture: sirtuins, aging, and medicine. N Engl J Med 2011; 364:2235–2244. [DOI] [PubMed] [Google Scholar]
- 8.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 2000; 403:795–800. [DOI] [PubMed] [Google Scholar]
- 9.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell 2003; 11:437–444. [DOI] [PubMed] [Google Scholar]
- 10.Pandey A, Goru SK, Kadakol A, Malek V, Gaikwad AB. Differential regulation of angiotensin converting enzyme 2 and nuclear factor-kappaB by angiotensin II receptor subtypes in type 2 diabetic kidney. Biochimie 2015; 118:71–81. [DOI] [PubMed] [Google Scholar]
- 11.Tilborghs S, Corthouts J, Verhoeven Y, Arias D, Rolfo C, Trinh XB, et al. The role of Nuclear Factor-kappa B signaling in human cervical cancer. Crit Rev Oncol Hematol 2017; 120:141–150. [DOI] [PubMed] [Google Scholar]
- 12.Raish M, Ahmad A, Ansari MA, Alkharfy KM, Aljenoobi FI, Jan BL, et al. Momordica charantia polysaccharides ameliorate oxidative stress, inflammation, and apoptosis in ethanol-induced gastritis in mucosa through NF-kB signaling pathway inhibition. Int J Biol Macromol 2018; 111:193–199. [DOI] [PubMed] [Google Scholar]
- 13.Yang J, Wu LJ, Tashino S, Onodera S, Ikejima T. Protein tyrosine kinase pathway-derived ROS/NO productions contribute to G2/M cell cycle arrest in evodiamine-treated human cervix carcinoma HeLa cells. Free Radic Res 2010; 44:792–802. [DOI] [PubMed] [Google Scholar]
- 14.Yang J, Wu LJ, Tashino S, Onodera S, Ikejima T. Reactive oxygen species and nitric oxide regulate mitochondria-dependent apoptosis and autophagy in evodiamine-treated human cervix carcinoma HeLa cells. Free Radic Res 2008; 42:492–504. [DOI] [PubMed] [Google Scholar]
- 15.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 2004; 303:2011–2015. [DOI] [PubMed] [Google Scholar]
- 16.Farshidfar F, Kopciuk KA, Hilsden R, McGregor SE, Mazurak VC, Buie WD, et al. A quantitative multimodal metabolomic assay for colorectal cancer. BMC Cancer 2018; 18:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen TC, Chien CC, Wu MS, Chen YC. Evodiamine from Evodia rutaecarpa induces apoptosis via activation of JNK and PERK in human ovarian cancer cells. Phytomedicine 2016; 23:68–78. [DOI] [PubMed] [Google Scholar]
- 18.Hu C, Gao X, Han Y, Guo Q, Zhang K, Liu M, et al. Evodiamine sensitizes BGC-823 gastric cancer cells to radiotherapy in vitro and in vivo. Mol Med Rep 2016; 14:413–419. [DOI] [PubMed] [Google Scholar]
- 19.Zhao N, Tian KT, Cheng KG, Han T, Hu X, Li DH, et al. Antiproliferative activity and apoptosis inducing effects of nitric oxide donating derivatives of evodiamine. Bioorg Med Chem 2016; 24:2971–2978. [DOI] [PubMed] [Google Scholar]
- 20.Lin L, Ren L, Wen L, Wang Y, Qi J. Effect of evodiamine on the proliferation and apoptosis of A549 human lung cancer cells. Mol Med Rep 2016; 14:2832–2838. [DOI] [PubMed] [Google Scholar]
- 21.Yang ZG, Chen AQ, Liu B. Antiproliferation and apoptosis induced by evodiamine in human colorectal carcinoma cells (COLO-205). Chem Biodivers 2009;6:924–933. [DOI] [PubMed] [Google Scholar]
- 22.Yang W, Ma L, Li S, Cui K, Lei L, Ye Z. Evaluation of the cardiotoxicity of evodiamine in vitro and in vivo. Molecules 2017; 22:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takada Y, Kobayashi Y, Aggarwal BB. Evodiamine abolishes constitutive and inducible NF-kappaB activation by inhibiting IkappaBalpha kinase activation, thereby suppressing NF-kappaB-regulated antiapoptotic and metastatic gene expression, up-regulating apoptosis, and inhibiting invasion. J Biol Chem 2005; 280:17203–17212. [DOI] [PubMed] [Google Scholar]
- 24.Shih RH, Wang CY, Yang CM. NF-kappaB Signaling pathways in neurological inflammation: a mini review. Front Mol Neurosci 2015; 8:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Rashed F, Kochumon S, Usmani S, Sindhu S, Ahmad R. Pam3CSK4 induces MMP-9 expression in human monocytic THP-1 cells. Cell Physiol Biochem 2017; 41:1993–2003. [DOI] [PubMed] [Google Scholar]
- 26.Moore RL, Dai Y, Faller DV. Sirtuin 1 (SIRT1) and steroid hormone receptor activity in cancer. J Endocrinol 2012; 213:37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGlynn LM, McCluney S, Jamieson NB, Thomson J, MacDonald AI, Oien K, et al. SIRT3 & SIRT7: potential novel biomarkers for determining outcome in pancreatic cancer patients. PLoS One 2015; 10:e0131344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu DF, Jiang SJ, Pan ZP, Cheng WD, Zhang WJ, Yao XK, et al. Expression and clinical significance of Sirt1 in colorectal cancer. Oncol Lett 2016; 11:1167–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung YR, Kim H, Park SY, Park IA, Jang JJ, Choe JY, et al. Distinctive role of SIRT1 expression on tumor invasion and metastasis in breast cancer by molecular subtype. Hum Pathol 2015; 46:1027–1035. [DOI] [PubMed] [Google Scholar]
- 30.Ling S, Li J, Shan Q, Dai H, Lu D, Wen X, et al. USP22 mediates the multidrug resistance of hepatocellular carcinoma via the SIRT1/AKT/MRP1 signaling pathway. Mol Oncol 2017; 11:682–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shackelford RE, Bui MM, Coppola D, Hakam A. Over-expression of nicotinamide phosphoribosyltransferase in ovarian cancers. Int J Clin Exp Pathol 2010; 3:522–527. [PMC free article] [PubMed] [Google Scholar]
- 32.Chen LF, Mu Y, Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J 2002; 21:6539–6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell 2007; 28:277–290. [DOI] [PubMed] [Google Scholar]
- 34.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J 2004; 23:2369–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ford J, Ahmed S, Allison S, Jiang M, Milner J. JNK2-dependent regulation of SIRT1 protein stability. Cell Cycle 2008; 7:3091–3097. [DOI] [PubMed] [Google Scholar]


