Abstract
Purpose:
To examine the association between expression of mutant p53 (mtp53), full length MDM2 (MDM2), and MDM2 isoform C (MDM2-C) and survival in multiethnic breast cancer patients.
Method:
A total of 787 invasive breast tumors included in a clinically-annotated multiethnic population-based tissue microarray (TMA) were screened utilizing commercially available antibodies to p53 and MDM2, and a newly developed monoclonal antibody recognizing MDM2-C.
Results:
Mutant p53 (mtp53) was more common in younger (<50 yrs) breast cancer patients. Among the 787 cases included in the study, mtp53, MDM2, and MDM2-C expression was not significantly associated with risk of overall or breast cancer-specific mortality. However when associations within individual racial/ethnic groups (White, Japanese, and Native Hawaiian) were examined, expression of MDM2-C was found to be associated with lower risk of breast cancer-specific mortality exclusively for White patients (HR=0.32, 95% CI: 0.15-0.69) and mtp53 expression was associated with higher overall mortality in Japanese patients (HR=1.63, 95% CI: 1.02-2.59). Also, Japanese patients positive for the joint expression of MDM2-C and mtp53 had a greater than 2-fold risk of overall mortality (HR=2.15, 95% CI 1.04-4.48); and White patients with positive MDM2-C and wild-type p53 expression (HR=0.28, 95% CI 0.08-0.96) were at lower risk of mortality when compared to patients negative for MDM2-C and wild-type p53 expression in their respective racial/ethnic group.
Conclusion:
Racial/ethnic differences in expression profiles of mtp53, MDM2, and MDM2-C and associations with breast cancer-specific and overall mortality. MDM2-C may have a positive or negative role in breast tumorigenesis depending on mtp53 expression.
Keywords: breast cancer, MDM2, MDM2-C, p53, multiethnic, survival
Introduction
Breast cancer is the most common cancer among women in the United States [1]. Despite improvements in survival, the disease remains the second leading cause of cancer death for women. Breast cancer incidence and survival vary dramatically by race and ethnicity [2]. White women have the highest incidence and African American women have the highest mortality rates for breast cancer in the nation. The national rates among the aggregate Asian/Pacific Islander group are generally lower than among Whites for both incidence and mortality. However, this group is comprised of very heterogeneous populations that have disparate incidence and mortality rates when evaluated as disaggregated populations. For example, Asian women have lower, and Pacific Islanders (including Native Hawaiians, Samoans, Guamanian/Chamorros) have higher incidence and mortality compared to White women [3–5]. Native Hawaiian women have amongst the highest breast cancer incidence in the nation and have a 50% higher risk of dying from breast cancer compared to White women [4,6,7].
Inactivation of the tumor suppressing p53 pathway is a common event in breast tumors. Depending on the molecular subtype, p53 mutation is observed in 12-84% of breast cancers and is correlated with poor clinical outcome [8]. In addition to p53 mutation events, p53 pathway inactivation can also result from increased levels of MDM2, an E3 ubiquitin ligase that functions as a negative regulator of p53 [9–11]. MDM2 is also capable of inhibiting the transcriptional activity of p53 [12,13].
The mdm2 gene produces at least 72 differentially spliced transcripts [14]. Expression of MDM2 isoforms, specifically isoforms A, B, and C, have been detected in multiple tumor types and associated with advanced disease and poor prognosis [15–18,14,19,20]. Exogenous expression of isoforms MDM2-A, B, and C confirms that these isoforms can promote tumorigenesis; however, the mechanism by which these isoforms do so remains poorly understood [21,19]. Recent reports indicate that expression of MDM2-B, the most common isoform in multiple tumors, is associated with accumulation of mutant p53 (mtp53) [22] and the gain of function mtp53 promotes tumorigenesis by binding to and inhibiting p53 family members (e.g., p63 and p73) [22]. MDM2-B has also been shown to function in a p53-independent manner by promoting the phosphorylation of MDM2 at Ser394/395, inhibiting its oligomerization and E3 ligase activity [23]. We have demonstrated that the MDM2-C isoform can also promote tumorigenesis in a p53-independent context, possibly through the E2F/RB pathway [24,25]. In addition to promoting cellular proliferation, MDM2 isoforms A and B have also been shown to mediate p53-dependent growth inhibition functions [26–28]. The postulated tumor suppressor capabilities for isoforms A, B, and C that lack the p53 binding domain but possess the RING finger domain necessary for MDM2 dimerization, are believed to function by sequestering MDM2 in the cytoplasm and preventing the ubiquitination and degradation of wild-type p53 [15,26,27]. Importantly, in cancers mtp53 is not degraded by MDM2 and consequently the levels of mtp53 are very high and immunohistochemistry screens detect these as mtp53 positive (often called simply p53 positive) [29–31]. The contradictory reports on the tumor promoting or inhibiting role of the MDM2 isoforms indicate the importance of the cellular mtp53 context when evaluating their function.
We examined the protein expression patterns of mtp53, MDM2, and MDM2-C isoform in breast tumor tissue. Using a population-based tissue microarray, we investigate associations with clinicopathological features, demographics (age and race/ethnicity), and breast cancer specific and overall mortality for these three protein biomarkers.
Methods:
Monoclonal Antibody Generation.
The monoclonal antibody to MDM2-C was produced at the Memorial Sloan Kettering Cancer Center Antibody and Bioresource Core Facility (Details in Supplemental Data 1). Briefly, Balb/c mice were inoculated subcutaneously with 35 μg KLH MDM2-C peptide [24], which is comprised of the 15 amino acid sequence spanning the exon 4/exon 10 junction (GCTYTMKEDLDAGVS). The best candidates were then cloned to establish a stable stock of monoclonal antibody MDM2-C clone 7C7 (mAb7C7). To confirm the specificity of the anti-MDM2-C antibody (mAb 7C7), MDM2 and MDM2-C proteins were in vitro transcribed-translated (IVT) in HeLa lysates (1-Step Human Coupled IVT Kit, Thermo Fisher) and used in a Western Blot analysis. In addition, two commercially available antibodies, mAb SMP14 (monoclonal; aa 154-167; Santa Cruz) and mAb 4B11 (monoclonal; aa 383-491; Calbiochem) were included in the Western Blot analysis.
Immunoflourescence.
Immunofluorescence was used to demonstrate cellular localization of endogenous MDM2 and MDM2-C proteins in T47D and MDA-MB-231 breast cancer cell lines. Cell were genetically engineered to express Green Fluorescence Protein (GFP). Briefly, T47D and MDA-MB-231 cells were allowed to grow on 12 well glass bottom plates (MatTek) at 50% confluency overnight. The next day cells were washed with 1× PBS and fixed with 4% paraformaldehyde (Sigma) for 15 minutes at room temperature. The plates were permeabilized with 0.5% Triton-X-100 in PBS/1% FBS for 10 minutes and incubated with anti-MDM2 (mAb SMP14), MDM2-C (mAb 7C7) and control (mouse IgG antibody) for 1 hour at room temperature. Alexa-conjugated goat anti-mouse (Invitrogen) antibody was used as secondary antibody. Cells were mounted with Vectashield mounting medium (Fisher Scientific) containing 4′,6-diamidino-2-phenylindole (DAPI). Images were collected by Nikon A1 Confocal microscope at 600× magnification.
Breast cancer tissue microarray.
The study was approved by the University of Hawaii Committee on Human Studies. The population-based TMAs were comprised of de-identified, archived formalin-fixed, paraffin-embedded (FFPE) tumor and adjacent normal tissue available through the Hawaii Tumor Registry’s Residual Tissue Repository (RTR) of the National Cancer Institute’s (NCI) Surveillance, Epidemiology, and End-Results (SEER) program. A total of 787 invasive breast cancer cases were evaluated for this study. Two sets of population-based tissue microarrays (TMA) were used to achieve this large sample size, one set included 355 invasive breast cancer cases all diagnosed in 1995 [32] and the other set included 432 invasive breast cancer cases diagnosed 2000-2004 in Hawaii. Each breast cancer case included on the TMA is represented by up to four 0.6 mm cores of tumor tissue. All cases included on the TMA are linked to the Hawaii Tumor Registry and annotated with de-identified, high-quality clinicopathological and outcome data including demographic data. Both TMAs were constructed by the University of Hawaii Cancer Center’s Pathology Core.
Immunohistochemistry.
For immunohistochemical (IHC) staining of TMA sections, 2.5 μm sections were mounted on glass slides then incubated with specific antibodies at optimized dilutions, followed by secondary antibodies, and diaminobenzidine for colorimetric detection. The antibodies recognizing full-length MDM2 (monocloncal SMP14; aa 154-167; Abeam) and p53 (monoclonal PAM801; aa 46-55; Vector Laboratories) were commercially available and IHC procedures were performed according to the manufacturers’ recommendations. The MDM2-C specific monoclonal antibody (mAb 7C7) was titered and optimized for IHC. IHC staining results were evaluated by a board certified pathologist who was blinded to clinicopathological characteristics and outcome status. Each core was scored based on the intensity of nuclear (p53) and/or cytoplasmic (MDM2 and MDM2-C) staining and the percentage of cells that stained. Cores were scored as negative (no staining) or positive. Cores comprised of <25% tumor tissue were not scored. Cases were considered positive for biomarker expression if at least one of the triplicate cores stained positive for each case.
Statistical analysis.
All analyses were performed for all study participants among the three largest racial/ethnic groups in the study (Whites, Japanese, Native Hawaiians). Tumor tissue samples with insufficient (<25%) tumor tissue or equivocal IHC results were excluded. For each of the three markers, tumor samples were graded as positive or negative for expression of the protein marker. Correlation between protein expression of MDM2, MDM2-C and p53 was assessed by pairwise Pearson’s correlation coefficients. The proportions of tumors with positive staining for MDM2, MDM2-C and p53 were compared by patient and tumor characteristics using Pearson’s chi-square test. Differences in 10-year overall survival by expression of the three markers were assessed using Kaplan-Meier estimates and log rank test. The association of protein expression of MDM2, MDM2-C and p53 with the risk of overall and breast-cancer mortality was examined using Cox proportional hazards regression. Survival period was computed from the date of cancer diagnosis to the date of death from any cause or from breast cancer to provide estimates of overall and breast cancer-specific survival, respectively. Patients alive at the date of last contact were considered censored. The proportional hazard assumption for Cox models was verified by plotting scaled Schoenfeld residuals against time to event [33]. Hazard ratios (HRs) and 95% confidence intervals (CIs) were calculated for protein expression, with negative expression as a reference category. Analyses were adjusted for patients’ age at time of diagnosis (by 5-year age groups), tumor stage (localized; regional/distant), tumor ER status (positive; negative) and PR status (positive; negative). Analyses of the associations of expression MDM2, MDM2-C and p53 with survival were also conducted for combinations of expression of MDM2 and p53, and of MDM2-C and p53, and stratified by race/ethnicity and tumor ER status. Heterogeneity across races/ethnicities was assessed using Wald tests of the cross-product interaction terms between race/ethnicity and the three markers, and subgroup analyses by race/ethnicity were conducted based on the statistically significant interaction effects. All tests were two-sided; p<0.05 was considered statistically significant. All data analyses were performed using SAS 9.4 statistical software (SAS Institute Inc.).
Results:
MDM2-C Specific Antibody Characterization
We generated a monoclonal antibody specific to the MDM2-C isoform. The immunogen peptide consists of the amino acid sequence produced from the splice junction between exon 4 and 10, which is unique to the MDM2-C isoform [24]. Specificity of the anti-MDM2-C monoclonal antibody (clone mAb 7C7) was confirmed in a Western blot using in vitro transcribed/translated (IVT) MDM2-C and MDM2 proteins (Figure 1A). Full length MDM2 (referred to as MDM2 throughout) and/or MDM2-C was detected using two commercially available antibodies, mAb SMP14 (MDM2) and mAb 4B11 (MDM2 and MDM2-C), which recognize two different epitopes of the MDM2 protein. The anti-MDM2 clone mAb SMP14 recognizes MDM2, but not MDM2-C because the epitope is encoded by exon 8, an exon that is not included in the MDM2-C isoform. The anti-MDM2 clone ab4B11 recognizes the carboxy-terminus of MDM2, included in both MDM2 and MDM2-C.
Figure 1. MDM2-C specific monoclonal antibody.
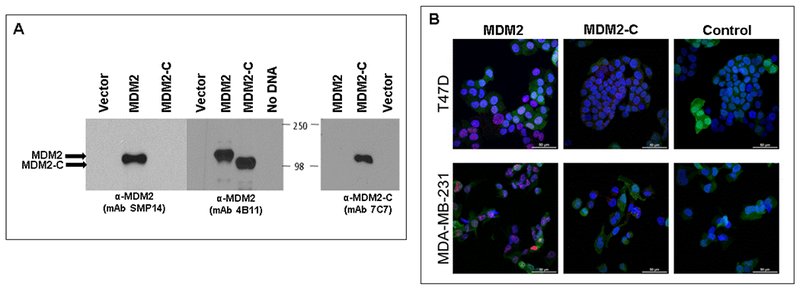
A. Western blot for MDM2 and MDM2-C. Specificity for the monoclonal antibody mAb 7C7 to recognize MDM2-C isoform and not full-length MDM2 was tested on in vitro transcribed/translated (IVT) MDM2-C and full-length MDM2. Antibody mAb SMP14 specifically recognizes full-length MDM2 and not MDM2-C and antibody mAb 4B11 recognizes both MDM2-FL and MDM2-C. B. Immunoflouresence was used to demonstrate cellular localization of endogenous MDM2 (red; nucleus) and MDM2-C (red; cytoplasm) proteins in T47D and MDA-MB-231 breast cancer cell lines. Cell lines were genetically engineered to express GFP. Alexa-conjugated goat anti-mouse was used as secondary antibody. DAPI stains the nuclei of the cells. Images were taken at 60× magnification.
We previously reported MDM2-C localization in distinct foci in the cytoplasm and nucleus in ER-positive breast cancer cells (T47D) using a different polyclonal antibody clone to MDM2-C [24]. We conducted an immunofluorescence analysis on two breast cancer cell lines, T47D (ER-positive) and MDA-MB-231 (triple negative subtype), using clone mAb 7C7 to compare the cellular localization of endogenous MDM2-C and MDM2. MDM2 was detected strongly in the nucleus of both cell lines (Figure 1B, red signal). MDM2-C was observed as punctate staining primarily in the cytoplasm of both the ER-positive and triple negative breast cancer cell lines (Figure 1B, red signal). Our results indicate that we have generated a monoclonal antibody that specifically recognizes the MDM2-C isoform, confirming the presence of cytoplasmic localization of MDM2-C in both ER-positive and triple negative breast cancer cells.
Study Population and Tumor Characteristics
The study included a total of 787 invasive breast cancer cases diagnosed in Hawaii. The majority of cases were 50 years and older (71%) and were White (26%), Japanese (31%), or Native Hawaiian (19%), reflecting three major racial/ethnic populations in Hawaii. The predominant histologic subtype of the tumors was infiltrating ductal carcinoma (83%), of localized stage (66%), well-to-moderately differentiated grade (55%), and positive for ER and PR expression (73% and 68%, respectively) (Table 1).
Table 1.
Characteristics of invasive breast cancer cases included on the tissue microarray
| No. | Percent (%)1 | |
|---|---|---|
| Age group | ||
| <50 yrs | 29 | 29.1 |
| ≥50 yrs | 558 | 70.9 |
| Race/ethnicity | ||
| White | 202 | 25.7 |
| Native Hawaiian | 150 | 19.1 |
| Japanese | 240 | 30.5 |
| Other2 | 195 | 24.8 |
| Stage3 | ||
| Localized | 518 | 65.8 |
| Regional | 246 | 31.3 |
| Distant | 22 | 2.8 |
| Unstaged | 1 | 0.1 |
| Grade | ||
| Well-differentiated | 103 | 13.1 |
| Moderately differentiated | 327 | 41.6 |
| Poorly/undifferentiated | 287 | 36.5 |
| Unknown | 70 | 8.9 |
| Histology | ||
| Infiltrating ductal carcinoma | 655 | 83.2 |
| Lobular carcinoma | 32 | 4.1 |
| Mucinous adenocarcinoma | 22 | 2.8 |
| Other | 78 | 9.9 |
| Estrogen Receptor | ||
| Positive | 572 | 72.7 |
| Negative | 171 | 21.7 |
| Unknown | 44 | 5.6 |
| Progesterone Receptor | ||
| Positive | 536 | 68.1 |
| Negative | 206 | 26.2 |
| Unknown | 45 | 5.7 |
Total percent may be higher or lower than 100 due to rounding.
Includes Chinese, Filipina, other Asian, other Pacific Islander and other racial/ethnic groups.
Based on SEER staging
Expression of MDM2, MDM2-C, and mutant p53 in breast tumor tissue
We examined expression of MDM2, MDM2-C and p53 by IHC staining of the 787 breast cancer cases included in our TMA (Figure 2). When cancer cells are positive for p53 protein expression it is generally indicative of the presence of mtp53 protein [29–31], Therefore, we denote these as p53 positive cancers as mtp53. We observed predominantly nuclear staining for MDM2 and mtp53, and cytoplasmic staining for MDM2-C in breast tumor cells (Figure 2). MDM2 was positive for expression in 80% of the tumors, MDM2-C stained positive in 82% of the tumors, and mtp53 was detected in 64% of the breast tumors (Table 2).
Figure 2. Immunohistochemical staining of MDM2, MDM2-C, and p53 proteins in breast cancer tissue.
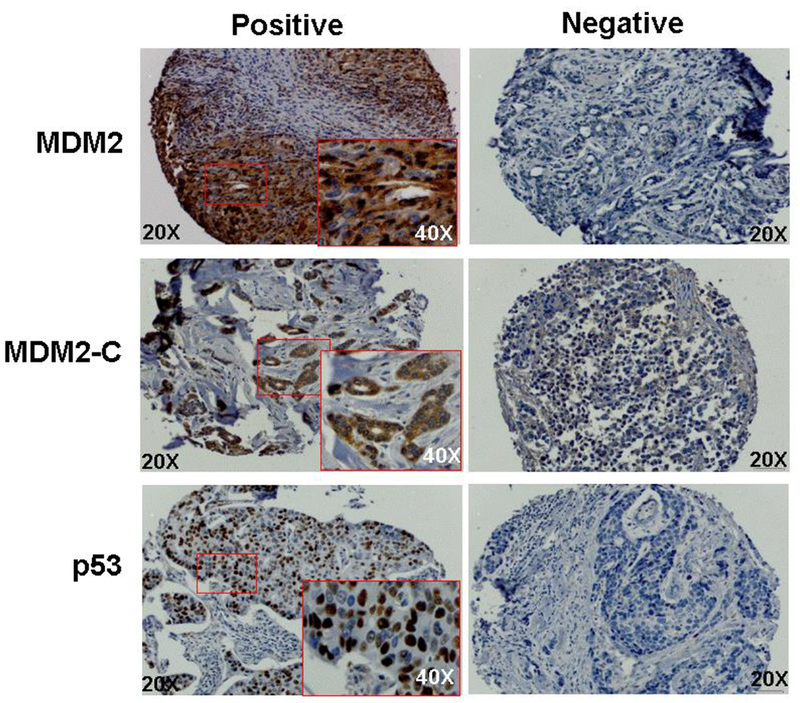
Representations of positive and negative immunohistochemical (IHC) stains of MDM2, MDM2-C, and p53 proteins in breast tumor tissue cores included in the tissue microarray. The antibodies recognizing MDM2 (mAb SMP14) and mtp53 (pAb1801). Positive MDM2 and MDM2-C protein staining was predominantly cytoplasmic and mtp53 was nuclear. Individual tissue cores at 20× magnification and for positive cores and a 40× magnification (inset) of the region highlighted in the red boxed area.
Table 2.
Distribution of MDM2, MDM2-C, p53 expression by patient characteristics
| MDM21 | MDM2-C1 | P531 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Characteristic | Positive n (%) | Negative n (%) | p-value2 | Positive n (%) | Negative n (%) | p-value2 | Positive n (%) | Negative n (%) | p-value2 |
| All cases (n=787) | 588 (80) | 152 (20) | 585 (82) | 133 (19) | 492 (64) | 273 (36) | |||
| Age | |||||||||
| <50 yrs | 179 (81) | 41 (19) | 0.40 | 180 (83) | 38 (17) | 0.62 | 159 (70) | 68 (30) | 0.03 |
| ≥50 yrs | 409 (79) | 111 (21) | 405 (81) | 95 (19) | 333 (62) | 205 (38) | |||
| Race/ethnicity3 | |||||||||
| White | 138 (76) | 44 (24) | 0.27 | 135(80) | 33 (20) | 0.95 | 122 (64) | 69 (36) | 0.90 |
| Japanese | 186 (82) | 40 (18) | 183 (81) | 42 (19) | 153 (65) | 81 (35) | |||
| Native Hawaiian | 115 (78) | 32 (22) | 116 (82) | 26 (18) | 98 (66) | 50 (34) | |||
| Stage | |||||||||
| Localized | 381 (79) | 101 (21) | 0.72 | 375 (81) | 90 (19) | 0.45 | 316 (63) | 185 (37) | 0.34 |
| Regional-Distant | 206 (80) | 51 (80) | 209 (83) | 43 (17) | 175 (67) | 88 (33) | |||
| Grade (Differentiation) | |||||||||
| Well-Moderate | 315 (78) | 88 (22) | 0.34 | 322 (83) | 64 (17) | 0.15 | 271 (65) | 146 (35) | 0.67 |
| Poor | 273 (81) | 64 (19) | 263 (79) | 69 (21) | 221 (64) | 127 (36) | |||
| Estrogen Receptor | |||||||||
| Positive | 422 (79) | 112 (21) | 0.11 | 428 (83) | 91 (17) | 0.57 | 358 (64) | 198 (36) | 0.76 |
| Negative | 139 (85) | 25 (15) | 128 (81) | 31 (19) | 109 (66) | 57 (34) | |||
Excludes cases with insufficient tissue or equivocal IHC results
P-values based on Pearson’s chi-square test
Excludes Chinese, Filipina, other Asian, other Pacific Islander, and other racial/ethnic groups
MDM2, MDM2-C and mtp53 did not have significant differences in expression patterns in breast tumors by race/ethnicity, stage, grade, or ER expression, with the exception of a significant association observed between mtp53 expression and younger age of diagnosis (p=0.03) (Table 2). When expression patterns were analyzed for the individual racial/ethnic groups, we observed significant associations for mtp53 with younger age (<50 yrs; p=0.03) and MDM2 with advanced stage (p=0.01) in tumors from Native Hawaiian patients only (Table 3).
Table 3.
Racial/Ethnic profiles of MDM2, MDM2-C, and p53 expression in breast tumors by patient and clinical characteristics
| MDM21 | MDM2-C1 | P531 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Race/Ethnicity3 | Characteristic | Positive n (%) | Negative n (%) | p-value2 | Positive n (%) | Negative n (%) | p-value2 | Positive n (%) | Negative n (%) | p-value2 |
| Native Hawaiians | Age | |||||||||
| ≥50 yrs | 45 (86) | 7 (14) | 0.07 | 43 (83) | 29 (17) | 0.81 | 41 (77) | 12 (22) | 0.03 | |
| ≥50 yrs | 70 (74) | 25 (26) | 73 (81) | 17 (19) | 57 (60) | 38 (40) | ||||
| Stage | ||||||||||
| Localized | 65 (71) | 26 (29) | 0.01 | 74 (83) | 15 (17) | 0.56 | 57 (63) | 34 (37) | 0.24 | |
| Regional/Distant | 50 (89) | 6 (11) | 42 (79) | 11 (21) | 41 (72) | 16 (28) | ||||
| Grade (Differentiation) | ||||||||||
| Well-Moderate | 50 (75) | 17 (25) | 0.33 | 54 (83) | 11 (17) | 0.69 | 48 (69) | 22 (31) | 0.57 | |
| Poor | 65 (81) | 15 (19) | 62 (80) | 15 (20) | 50 (64) | 28 (35) | ||||
| ER-status | ||||||||||
| Positive | 85 (77) | 26 (23) | 0.21 | 92 (87) | 14 (13) | 0.18 | 78 (69) | 35 (31) | 0.38 | |
| Negative | 22 (88) | 3 (12) | 19 (76) | 6 (24) | 15 (60) | 10 (40) | ||||
| Japanese | Age | |||||||||
| <50 yrs | 41 (82) | 9 (18) | 0.95 | 45 (85) | 8 (15) | 0.45 | 36 (68) | 17 (32) | 0.66 | |
| ≥50 yrs | 145 (82) | 31 (18) | 138 (80) | 34 (20) | 117 (65) | 64 (35) | ||||
| Stage | ||||||||||
| Localized | 124 (83) | 25 (17) | 0.61 | 117 (78) | 32 (22) | 0.13 | 100 (63) | 59 (37) | 0.24 | |
| Regional/Distant | 62 (80) | 15 (20) | 66 (89) | 10 (13) | 53 (71) | 22 (29) | ||||
| Grade (Differentiation) | ||||||||||
| Well-Moderate | 115 (81) | 27 (19) | 0.50 | 116 (83) | 24 (17) | 0.45 | 96 (64) | 53 (36) | 0.68 | |
| Poor | 71 (85) | 13 (16) | 67 (79) | 18 (21) | 57 (67) | 28 (33) | ||||
| ER-status | ||||||||||
| Positive | 138 (84) | 27 (16) | 0.52 | 135 (82) | 29 (18) | 0.18 | 105 (61) | 67 (39) | 0.13 | |
| Negative | 35 (79) | 9 (21) | 33 (73) | 12 (27) | 33 (73) | 12 (27) | ||||
| Whites | Age | |||||||||
| <50 yrs | 44 (76) | 14 (24) | 0.99 | 44 (83) | 9 (17) | 0.56 | 41 (68) | 19 (32) | 0.39 | |
| ≥50 yrs | 94 (76) | 30 (24) | 91 (79) | 24 (21) | 81 (62) | 50 (38) | ||||
| Stage | ||||||||||
| Localized | 96 (77) | 28 (23) | 0.46 | 90 (80) | 22 (20) | 1.00 | 84 (65) | 46 (35) | 0.76 | |
| Regional/Distant | 42 (72) | 16 (28) | 45 (80) | 11 (20) | 38 (62) | 23 (38) | ||||
| Grade (Differentiation) | ||||||||||
| Well-Moderate | 76 (73) | 28 (27) | 0.32 | 75 (80) | 18 (19) | 0.92 | 66 (63) | 39 (37) | 0.75 | |
| Poor | 62 (79) | 16 (21) | 60 (80) | 15 (20) | 56 (65) | 30 (35) | ||||
| ER-status | ||||||||||
| Positive | 96 (74) | 33 (26) | 0.11 | 95 (79) | 26 (21) | 0.26 | 88 (64) | 49 (36) | 0.57 | |
| Negative | 37 (86) | 6 (14) | 33 (87) | 5 (13) | 31 (69) | 14 (31) | ||||
Excludes tumor samples with insufficient tissue or equivocal IHC results
P-values based on Pearson’s chi-square test
Excludes Chinese, Filipina, other Asian, other Pacific Islander, and other racial/ethnic groups
We analyzed pairwise co-expression of MDM2-C, MDM2, and mtp53. These were positively correlated (p<0.001) for all cases analyzed, with a relatively stronger correlation between MDM2-C and MDM2 expression (r2=0.38; p=<0.0001). The correlation between expression of MDM2 and mtp53, and MDM2-C and mtp53 were similar (r2=0.29; p=<0.0001 and r2=0,28; p=<0.0001, respectively). Within individual racial/ethnic groups, we observed similar correlation trends for the pair-wise expression of MDM2 and MDM2-C, as well as MDM2 and mtp53 for Whites, Japanese, and Native Hawaiian women. However, among the three racial/ethnic groups we observed a weaker correlation between pair-wise expression of MDM2-C and mtp53 expression in the breast tumors in Whites (r2=0,19, p=0.01) compared to Japanese and Native Hawaiians, both with a correlation of r2=0.32 (p≤0.0001) (Supplemental Data 2).
MDM2, MDM2-C, and mutant p53 Expression and overall survival
When we examined association with overall 10-year survival for all cases, there were no significant differences in survival by expression of MDM2, MDM2-C, or mtp53 (Supplemental Data 3). Significant survival differences were also not observed by age (<50 vs. ≥50 years) or by ER-status (Supplemental Data 4). For all cases and the individual racial/ethnic populations, there was consistent trend for poorer overall survival associated with mtp53, however this association was statistically significant only among Japanese (Log rank p-value=0.03) (Supplemental Data 3).
Association of MDM2, MDM2-C, and mtp53 protein expression with mortality was also examined after adjusting for age, stage, first course treatment, ER, and PR (Table 4; Supplemental Data 5). Statistically significant heterogeneity by race/ethnicity was observed in the effect of MDM2-C. In stratified analyses, among Japanese women, mtp53 expression was still significantly associated with higher risk of overall mortality after adjusting for age, stage, first course treatment, ER and PR status (adjusted HR=1.63, 95% CI 1.02-2.59). Following adjustment, MDM2-C expression was significantly associated with lower risk of breast cancer specific mortality for Whites (adjusted HR=0.32, 95% CI 0.15-0.69). No significant associations with the three markers were observed for Native Hawaiians. In addition, we observed no significant association with the expression of the three proteins with overall or breast cancer-specific mortality stratified by ER-status (Supplemental Data 6).
Table 4.
MDM2, MDM2-C, and p53 expression in breast tumor tissue and risk of mortality by race/ethnicity
| Mortality3 | Protein Marker1 | Protein Expression | All4 | Native Hawaiian | Japanese | Whites | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR2 | 95% CI | p-value | HR2 | 95% CI | p-value | HR2 | 95% CI | p-value | HR2 | 95% CI | p-value | |||
| Overall | MDM2 | negative | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | ||||||||
| positive | 1.01 | 0.76 - 1.33 | 0.97 | 0.99 | 0.56 - 1.79 | 0.99 | 1.60 | 0.89 - 2.88 | 0.12 | 0.79 | 0.47 - 1.33 | 0.38 | ||
| MDM2-C5 | negative | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | |||||||||
| positive | 1.07 | 0.79 - 1.45 | 0.65 | 1.04 | 0.52 - 2.09 | 0.90 | 1.69 | 0.94 - 3.01 | 0.08 | 0.60 | 0.33 - 1.09 | 0.09 | ||
| P53 | negative | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | |||||||||
| positive | 1.23 | 0.97 - 1.56 | 0.08 | 1.36 | 0.80 - 2.31 | 0.25 | 1.63 | 1.02 - 2.59 | 0.04 | 1.31 | 0.83 - 2.09 | 0.25 | ||
| Breast Cancer | MDM2 | negative | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | ||||||||
| positive | 1.00 | 0.68 - 1.47 | 0.99 | 1.12 | 0.49 - 2.54 | 0.79 | 1.50 | 0.69 - 3.26 | 0.31 | 0.69 | 0.32 - 1.48 | 0.34 | ||
| MDM2-C5 | negative | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | |||||||||
| positive | 0.90 | 0.61 - 1.33 | 0.61 | 0.87 | 0.37 - 2.08 | 0.76 | 1.53 | 0.69 - 3.39 | 0.30 | 0.32 | 0.15 - 0.69 | 0.003 | ||
| P53 | negative | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | |||||||||
| positive | 1.11 | 0.81 - 1.52 | 0.53 | 1.71 | 0.824 - 3.56 | 0.15 | 1.22 | 0.65 - 2.30 | 0.53 | 1.09 | 0.56 - 2.13 | 0.80 | ||
Excludes cases with insufficient tissue or equivocal IHC results
Hazard ratio (HR); reference includes cases negative for protein(s) of interest
Models adjusted for age, stage, first course treatment, ER and PR status
Includes Chinese, Filipina, Other Asian, Other Pacific Islander and other racial/ethnic groups
P-value for interaction with race/ethnicity < 0.05, based on the Wald test of the cross-product term between the marker and race/ethnicity.
We also examined the association with the risk of mortality with combinations of joint expression of MDM2 and p53, as well as MDM2-C and p53 (Table 5). There was no association among all cases with overall or breast cancer specific mortality. However, for Whites we observed a statistically significant difference in the risk of breast cancer mortality by joint expression of MDM2-C and p53 (mtp53). A significantly decreased risk of breast cancer specific mortality for tumors that were positive for MDM2-C and negative for p53 compared to tumors that were negative for both MCM2-C and p53. Also among Whites, we observed a increased risk of overall mortality associated with tumors that were negative for MDM2 and positive for p53 (mtp53) expression compared to tumors that were negative for both MDM2 and p53 expression. A positive association was observed for Japanese, where positive expression of MDM2-C and p53 (mtp53) were associated with an increased risk of overall mortality compared to tumors that were negative for both MDM2-C and p53. No significant association with the risk of mortality was observed for any of the joint combinations of MDM2 and p53 or MDM2-C and p53 for Native Hawaiians.
Table 5.
Joint expression of MDM2 & p53 and MDM2-C & p53 and risk of mortality in breast cancer cases by race/ethnicity
| Joint Expression | Mortality3 | MDM2, MDM2C & p53 status1 | All4 | Native Hawaiians | Japanese | Whites | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR2 | 95% CI | p-value5 | HR2 | 95% CI | p-value5 | HR2 | 95% CI | p-value5 | HR2 | 95% CI | p-value5 | |||
| MDM2 & p53 | Overall | MDM2(−) & p53(−) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | ||||||||
| MDM2(−) & p53(+) | 1.53 | 0.93-2.52 | 1.19 | 0.42-3.40 | 0.95 | 0.30-2.96 | 2.70 | 1.13-6.46 | ||||||
| MDM2(+) & p53(+) | 1.27 | 0.88-1.84 | 1.10 | 0.55-2.22 | 1.96 | 0.92-4.18 | 1.23 | 0.59-2.57 | ||||||
| MDM2(+) & p53(−) | 1.04 | 0.69-1.58 | 0.21 | 0.78 | 0.32-1.88 | 0.78 | 1.16 | 0.49-2.70 | 0.08 | 1.09 | 0.47-2.53 | 0.06 | ||
| Breast Cancer | MDM2(−) & p53(−) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | |||||||||
| MDM2(−) & p53(+) | 1.64 | 0.82-3.30 | 2.15 | 0.50-9.27 | 0.52 | 0.10-2.63 | 3.31 | 0.92-11.98 | ||||||
| MDM2(+) & p53(+) | 1.26 | 0.75-2.13 | 1.58 | 0.53-4.73 | 1.37 | 0.55-3.39 | 1.15 | 0.38-3.51 | ||||||
| MDM2(+) & p53(−) | 1.23 | 0.69-2.18 | 0.58 | 1.04 | 0.28-3.84 | 0.60 | 0.98 | 0.35-2.72 | 0.51 | 1.19 | 0.34-4.11 | 0.15 | ||
| MDM2-C & p53 | Overall | MDM2-C(−) & p53(−) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | ||||||||
| MDM2-C(−) & p53(+) | 1.59 | 0.93-2.73 | 1.25 | 0.36-4.31 | 1.62 | 0.57-4.63 | 1.35 | 0.48-3.80 | ||||||
| MDM2-C(+) & p53(+) | 1.34 | 0.90-1.99 | 1.20 | 0.49-2.93 | 2.16 | 1.04-4.48 | 0.78 | 0.31-1.91 | ||||||
| MDM2-C(+) & p53(−) | 1.20 | 0.77-1.86 | 0.32 | 1.01 | 0.38-2.74 | 0.94 | 1.65 | 0.72-3.80 | 0.20 | 0.61 | 0.23-1.60 | 0.29 | ||
| Breast Cancer | MDM2-C(−) & p53(−) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | |||||||||
| MDM2-C(−) & p53(+) | 1.66 | 0.82-3.34 | 1.59 | 0.34-7.49 | 1.53 | 0.35-6.55 | 1.24 | 0.35-4.40 | ||||||
| MDM2-C(+) & p53(+) | 1.12 | 0.66-1.89 | 1.22 | 0.36-4.19 | 1.89 | 0.70-5.07 | 0.41 | 0.14-1.18 | ||||||
| MDM2-C(+) & p53(−) | 1.09 | 0.61-1.94 | 0.48 | 0.79 | 0.19-3.27 | 0.72 | 1.80 | 0.59-5.46 | 0.65 | 0.28 | 0.08-0.96 | 0.03 | ||
cases with insufficient tissue or equivocal IHC results
Hazard ratio (HR); reference includes cases negative for protein(s) of interest
Models adjusted for age, stage, first course treatment, ER and PR
Includes Chinese, Filipina, other Asian, other Pacific Islander and other racial/ethnic groups
Overall type-3 P-value based on the Wald test
Discussion
MDM2 is amplified and overexpressed in multiple tumor types, including breast cancer [34,35]. The role of MDM2 in prognosis has been inconsistent, with MDM2 expression associated with both poor and favorable prognosis [36–38,35]. It is possible that this is due to our limited understanding of the role of the multiple mdm2 splice variants. To date, at least 72 mdm2 splice variants have been identified [14]. Three MDM2 isoforms (A, B, and C) that are commonly found in multiple cancers types [14]. MDM2 isoforms have been shown to be capable of both promoting, as well as, inhibiting tumorigenesis [21,28,24,22]. The biological mechanisms underlying these two contrasting outcomes remains an area of active research. In this study we investigated the association with overall and breast cancer-specific mortality in a multiethnic population of MDM2 and MDM2-C protein in breast tumors with and without mtp53 expression.
The comparison of MDM2 and MDM2-C isoform specific antibodies enabled us to examine protein expression patterns for a multiethnic population of breast cancer cases to gain a better understanding of the role of these isoforms in relation to the expression of mtp53. When we evaluated the protein expression profiles of MDM2, MDM2-C, and mtp53 among all breast cancer cases, we found an association for p53 expression and age at diagnosis, with a higher frequency of mtp53 in younger breast cancer patients (<50 yrs). No significant differences for MDM2 and MDM2-C expression by age, stage, grade, ER or PR status, or race/ethnicity (White, Japanese, Native Hawaiian) were observed. The mdm2 gene is a transcriptional target of the ER [39] and in a comprehensive analysis of intrinsic molecular breast cancer subtypes, the mdm2 gene is found to be most commonly amplified in Luminal B and HER2-expressing subtypes, and less frequently in the Luminal A and Basal-like subtypes [8]. We did not detect a significant difference in MDM2 or MDM2-C protein levels by ER status, however we do not have intrinsic molecular subtype characterization for the breast cancer cases included on the TMA, so it is possible that there are differences in MDM2 and MDM2-C expression across molecular subtypes, which at this time we are unable to assess.
When we examined expression patterns among individual racial/ethnic groups, we found a significantly higher frequency of positive MDM2 expression in advanced stage tumors and mtp53 in breast tumors of younger Native Hawaiian patients (<50 yrs). Native Hawaiians with breast cancer have amongst the highest mortality rate in nation. Although we did not observe an association with expression of the three proteins and risk of mortality for Native Hawaiian patients in this study, it is possible that the high frequency of MDM2 expression in advanced stage and mtp53 in younger individuals for this group indicate that the disruption of the p53-mediated tumor suppressor pathway is an important component contributing to poorer survival trends for Native Hawaiian breast cancer patients.
We did not observe significant associations with expression of the three proteins and risk of overall or breast cancer-specific mortality when all cases were included. However we did observed significant associations within individual racial/ethnic groups. Surprisingly, our results indicate that MDM2-C expression is associated with better breast cancer prognosis in Whites and worse prognosis in Japanese. We observed a highly significant negative association with risk of breast cancer specific mortality for White patients with breast tumors expressing MDM2-C, and, a marginally significant positive association with overall mortality for Japanese patients (Table 4). Previous in vitro and mouse studies indicate that MDM2 isoforms A, B, and C are capable of both promoting and inhibiting tumorigenesis, in a p53-dependent and independent manner [26,27,40,28,22]. Although there are limitations to the functional interpretation of our results, perhaps the role of MDM2-C in breast tumorigenesis may depend on the status of p53 (wild type or mutant), as was previously demonstrated with MDM2 isoforms A and B. We examined association between the joint positive signal for MDM2-C and p53 as well as MDM2 and p53 expression profiles and risk of mortality for all patients and for individual racial/ethnic groups. Japanese patients with tumors expressing MDM2-C and mtp53 were at greater than 2-fold higher risk of overall mortality compared to tumors lacking MDM2-C in the presence of wild-type p53. However, this association was not observed for Whites or Native Hawaiians. In fact, for White patients, expression of MDM2-C and wild-type p53 was associated with a lower risk for breast cancer-specific mortality (Table 5).
These results support the complexity and cellular context dependence of the functional roles of MDM2, and MDM2 isoforms, to promote or inhibit tumorigenesis. Perhaps, the outcome differences in the racial/ethnic groups could be the result of differences in p53 mutation status and mutation sites, differentially impacting interaction with MDM2 and potential gain of function mtp53 events as proposed by Zheng et al. [22]. It is also possible that MDM2, and perhaps MDM2-C, are functioning in a p53 independent manner to regulate tumorigenesis through the MDM2-Rb-E2F1 axis. We recently showed that MDM2 can provoke phosphorylation of Rb and upregulate E2F1 activity in mammary cells resulting in cellular proliferation and mammary tissue architecture [25]. The role of MDM2-mediated cellular activity is further complicated because the different racial/ethnic populations have different allele frequencies for the single nucleotide polymorphism (SNP) 309 (rs2279744) in the promoter region of the mdm2 gene. This SNP impacts recruitment of the Sp1 transcription factor, resulting in an increase in MDM2 and MDM2-C expression [41,42]. The G allele at SNP309 binds SP1 with a higher affinity than the T allele and individuals of Asian ancestry have a higher frequency (54%) of the G allele compare to individuals of European (35%) or African (7%) decent [43]. We observed a higher frequency of MDM2, but not MDM2-C, tumors in Japanese patients, compared to White and Native Hawaiian patients (Table 2). Interestingly, several studies have found interactions between the MDM2 SNP309 genotype and p53-pathway status and breast cancer survival [44–46]. Similar interactions may contribute to the association differences that we are observing for the different racial/ethnic groups in our study. However, we cannot completely discount the possibility of a Type I error in our subgroup analyses. Our novel findings of the potential role of MDM2-C in breast tumors could have translational importance, because the inhibition of MDM2 function as a cancer therapeutic is an ongoing consideration [47–50]. Our findings suggest that consideration of the potential functional roles of the MDM2 isoforms need to be considered when assessing p53 status. Additional studies will need to be conducted to better understand the racial/ethnic differences associated with MDM2, MDM2-C and mtp53 and mortality for breast cancer patients.
Supplementary Material
Acknowledgements:
The authors would like to acknowledge the Memorial Sloan Kettering Cancer Center Antibody and Bioresource Core Facility for excellent assistance with generation of hybridomas, the Hawaii Tumor Registry, Residual Tissue Repository of the National Cancer Institute’s (NCI) Surveillance, Epidemiology, and End-Results (SEER) program, and the University of Hawaii Cancer Center’s Biostatistics Shared Resource and Pathology Shared Resource for their contribution to this project.
Funding:
This study was funded by the National Institute on Minority Health and Health Disparities (U54MD008149-SGP14-183; MD007599), NCI (R21CA176555-01A1; 3P30CA071789-12S7), Hawaii Community Foundation (16ADVC-78885), and the Breast Cancer Research Foundation.
Footnotes
Conflict of Interest:
The authors declare that they have no conflict of interest to disclose.
Compliance with Ethical Standards:
Ethical Approval:
This study has received ethical approval by the University of Hawaii Committee on Human Studies. This study was a retrospective study, with all subjects having been de-identified, and no formal consent was required. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. For the generation of the monoclonal antibodies, all applicable international, national, and institutional guidelines for the care and use of animals were followed.
References
- 1.American Cancer Society (2018) Cancer Facts & Figures 2018. [Google Scholar]
- 2.DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A (2017) Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin 67 (6):439–448. doi: 10.3322/caac.21412 [DOI] [PubMed] [Google Scholar]
- 3.Gomez SL, Noone AM, Lichtensztajn DY, Scoppa S, Gibson JT, Liu L, Morris C, Kwong S, Fish K, Wilkens LR, Goodman MT, Deapen D, Miller BA (2013) Cancer incidence trends among Asian American populations in the United States, 1990-2008. J Natl Cancer Inst 105 (15):1096–1110. doi: 10.1093/jnci/djt157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu L, Noone AM, Gomez SL, Scoppa S, Gibson JT, Lichtensztajn D, Fish K, Wilkens LR, Goodman MT, Morris C, Kwong S, Deapen D, Miller BA (2013) Cancer incidence trends among Native Hawaiians and other Pacific Islanders in the United States, 1990-2008. J Natl Cancer Inst 105 (15):1086–1095. doi: 10.1093/jnci/djt156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu L, Zhang J, Wu AH, Pike MC, Deapen D (2011) Invasive breast cancer incidence trends by detailed race/ethnicity and age. Int J Cancer. doi: 10.1002/ijc.26004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller B, Chu K, Hankey B, Ries L (2008) Cancer incidence and mortality patterns among specific Asian and Pacific Islander populations in the U.S. Cancer Causes Control 19 (3):227–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ooi SL, Martinez ME, Li CI (2011) Disparities in breast cancer characteristics and outcomes by race/ethnicity. Breast Cancer Res Treat 127 (3):729–738. doi: 10.1007/s10549-010-1191-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas N (2012) Comprehensive molecular portraits of human breast tumours. Nature 490 (7418):61–70. doi: 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubbutat MH, Jones SN, Vousden KH (1997) Regulation of p53 stability by Mdm2. Nature 387 (6630):299–303. doi: 10.1038/387299a0 [DOI] [PubMed] [Google Scholar]
- 10.Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W (2003) Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 302 (5652):1972–1975. doi: 10.1126/science.1091362 [DOI] [PubMed] [Google Scholar]
- 11.Pant V, Lozano G (2014) Limiting the power of p53 through the ubiquitin proteasome pathway. Genes Dev 28 (16):1739–1751. doi: 10.1101/gad.247452.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP (1996) Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274 (5289):948–953 [DOI] [PubMed] [Google Scholar]
- 13.Momand J, Zambetti GP, Olson DC, George D, Levine AJ (1992) The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69 (7):1237–1245 [DOI] [PubMed] [Google Scholar]
- 14.Rosso M, Okoro DE, Bargonetti J (2014) Splice variants of MDM2 in oncogenesis. Subcell Biochem 85:247–261. doi: 10.1007/978-94-017-9211-0_14 [DOI] [PubMed] [Google Scholar]
- 15.Bartel F, Taubert H, Harris LC (2002) Alternative and aberrant splicing of MDM2 mRNA in human cancer. Cancer Cell 2 (1):9–15 [DOI] [PubMed] [Google Scholar]
- 16.Bartel F, Taylor AC, Taubert H, Harris LC (2001) Novel mdm2 splice variants identified in pediatric rhabdomyosarcoma tumors and cell lines. Oncol Res 12 (11-12):451–457 [DOI] [PubMed] [Google Scholar]
- 17.Lukas J, Gao DQ, Keshmeshian M, Wen WH, Tsao-Wei D, Rosenberg S, Press MF (2001) Alternative and aberrant messenger RNA splicing of the mdm2 oncogene in invasive breast cancer. Cancer Res 61 (7):3212–3219 [PubMed] [Google Scholar]
- 18.Matsumoto R, Tada M, Nozaki M, Zhang CL, Sawamura Y, Abe H (1998) Short alternative splice transcripts of the mdm2 oncogene correlate to malignancy in human astrocytic neoplasms. Cancer Res 58 (4):609–613 [PubMed] [Google Scholar]
- 19.Sigalas I, Calvert AH, Anderson JJ, Neal DE, Lunec J (1996) Alternatively spliced mdm2 transcripts with loss of p53 binding domain sequences: transforming ability and frequent detection in human cancer. Nat Med 2 (8):912–917 [DOI] [PubMed] [Google Scholar]
- 20.Tamborini E, Della Torre G, Lavarino C, Azzarelli A, Carpinelli P, Pierotti MA, Pilotti S (2001) Analysis of the molecular species generated by MDM2 gene amplification in liposarcomas. Int J Cancer 92 (6):790–796. doi: 10.1002/ijc.1271 [DOI] [PubMed] [Google Scholar]
- 21.Fridman JS, Hernando E, Hemann MT, de Stanchina E, Cordon-Cardo C, Lowe SW (2003) Tumor promotion by Mdm2 splice variants unable to bind p53. Cancer Res 63 (18):5703–5706 [PubMed] [Google Scholar]
- 22.Zheng T, Wang J, Zhao Y, Zhang C, Lin M, Wang X, Yu H, Liu L, Feng Z, Hu W (2013) Spliced MDM2 isoforms promote mutant p53 accumulation and gain-of-function in tumorigenesis. Nat Commun 4:2996. doi: 10.1038/ncomms3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Comiskey DF Jr., Jacob AG, Sanford BL, Montes M, Goodwin AK, Steiner H, Matsa E, Tapia-Santos AS, Bebee TW, Grieves J, La Perle K, Boyaka P, Chandler DS (2018) A novel mouse model of rhabdomyosarcoma underscores the dichotomy of MDM2-ALT1 function in vivo. Oncogene 37 (1):95–106. doi: 10.1038/onc.2017.282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okoro DR, Arva N, Gao C, Polotskaia A, Puente C, Rosso M, Bargonetti J (2013) Endogenous human MDM2-C is highly expressed in human cancers and functions as a p53-independent growth activator. PLoS One 8 (10):e77643. doi: 10.1371/journal.pone.0077643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kundu N, Brekman A, Kim JY, Xiao G, Gao C, Bargonetti J (2017) Estrogen-activated MDM2 disrupts mammary tissue architecture through a p53-independent pathway. Oncotarget 8 (29) :47916–47930. doi: 10.18632/oncotarget.18147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dang J, Kuo ML, Eischen CM, Stepanova L, Sherr CJ, Roussel MF (2002) The RING domain of Mdm2 can inhibit cell proliferation. Cancer Res 62 (4):1222–1230 [PubMed] [Google Scholar]
- 27.Evans SC, Viswanathan M, Grier JD, Narayana M, El-Naggar AK, Lozano G (2001) An alternatively spliced HDM2 product increases p53 activity by inhibiting HDM2. Oncogene 20 (30) :4041–4049. doi: 10.1038/sj.onc.1204533 [DOI] [PubMed] [Google Scholar]
- 28.Manfredi JJ (2010) The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev 24 (15):1580–1589. doi: 10.1101/gad.1941710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davidoff AM, Herndon JE, 2nd, Glover NS, Kerns BJ, Pence JC, Iglehart JD, Marks JR (1991) Relation between p53 overexpression and established prognostic factors in breast cancer. Surgery 110 (2):259–264 [PubMed] [Google Scholar]
- 30.Davidoff AM, Kerns BJ, Iglehart JD, Marks JR (1991) Maintenance of p53 alterations throughout breast cancer progression. Cancer Research 51 (10):2605–2610 [PubMed] [Google Scholar]
- 31.Thor AD, Moore DH II, Edgerton SM, Kawasaki ES, Reihsaus E, Lynch HT, Marcus JN, Schwartz L, Chen LC, Mayall BH, et al. (1992) Accumulation of p53 tumor suppressor gene protein: an independent marker of prognosis in breast cancers. J Natl Cancer Inst 84 (11):845–855 [DOI] [PubMed] [Google Scholar]
- 32.Anderson WF, Luo S, Chatterjee N, Rosenberg PS, Matsuno RK, Goodman MT, Hernandez BY, Reichman M, Dolled-Filhart MP, O’Regan RM, Garcia-Closas M, Perou CM, Jatoi I, Cartun RW, Sherman ME (2008) Human epidermal growth factor receptor-2 and estrogen receptor expression, a demonstration project using the residual tissue repository of the Surveillance, Epidemiology, and End Results (SEER) program. Breast Cancer Res Treat 113 (1):189–196. doi: 10.1007/s10549-008-9918-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grambsch PM, Therneau TM (1994) Proportional hazards tests and diagnostics based on weighted residuals. Biometrika 81 (3):515–526 [Google Scholar]
- 34.Oliner JD, Saiki AY, Caenepeel S (2016) The Role of MDM2 Amplification and Overexpression in Tumorigenesis. Cold Spring Harb Perspect Med 6 (6). doi: 10.1101/cshperspect.a026336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turbin DA, Cheang MC, Bajdik CD, Gelmon KA, Yorida E, De Luca A, Nielsen TO, Huntsman DG, Gilks CB (2006) MDM2 protein expression is a negative prognostic marker in breast carcinoma. Mod Pathol 19 (1):69–74. doi: 10.1038/modpathol.3800484 [DOI] [PubMed] [Google Scholar]
- 36.Choschzick M, Heilenkotter U, Lebeau A, Jaenicke F, Terracciano L, Bokemeyer C, Sauter G, Simon R (2010) MDM2 amplification is an independent prognostic feature of node-negative, estrogen receptor-positive early-stage breast cancer. Cancer Biomark 8 (2):53–60. doi: 10.3233/DMA-2011-0806 [DOI] [PubMed] [Google Scholar]
- 37.Hori M, Shimazaki J, Inagawa S, Itabashi M, Hori M (2002) Overexpression of MDM2 oncoprotein correlates with possession of estrogen receptor alpha and lack of MDM2 mRNA splice variants in human breast cancer. Breast Cancer Res Treat 71 (1):77–83 [DOI] [PubMed] [Google Scholar]
- 38.Park HS, Park JM, Park S, Cho J, Kim SI, Park BW (2014) Subcellular localization of Mdm2 expression and prognosis of breast cancer. Int J Clin Oncol 19 (5):842–851. doi: 10.1007/s10147-013-0639-1 [DOI] [PubMed] [Google Scholar]
- 39.Brekman A, Singh KE, Polotskaia A, Kundu N, Bargonetti J (2011) A p53-independent role of Mdm2 in estrogen-mediated activation of breast cancer cell proliferation. Breast Cancer Res 13 (1):R3. doi: 10.1186/bcr2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huun J, Gansmo LB, Mannsaker B, Iversen GT, Ovrebo JI, Lonning PE, Knappskog S (2017) Impact of the MDM2 splice-variants MDM2-A, MDM2-B and MDM2-C on cytotoxic stress response in breast cancer cells. BMC Cell Biol 18 (1):17. doi: 10.1186/s12860-017-0134-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bond GL, Hirshfield KM, Kirchhoff T, Alexe G, Bond EE, Robins H, Bartel F, Taubert H, Wuerl P, Hait W, Toppmeyer D, Offit K, Levine AJ (2006) MDM2 SNP309 accelerates tumor formation in a gender-specific and hormone-dependent manner. Cancer Res 66 (10):5104–5110. doi: 10.1158/0008-5472.CAN-06-0180 [DOI] [PubMed] [Google Scholar]
- 42.Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, Onel K, Yip L, Hwang SJ, Strong LC, Lozano G, Levine AJ (2004) A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 119 (5):591–602. doi: 10.1016/j.cell.2004.11.022 [DOI] [PubMed] [Google Scholar]
- 43.Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR (2015) A global reference for human genetic variation. Nature 526 (7571):68–74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boersma BJ, Howe TM, Goodman JE, Yfantis HG, Lee DH, Chanock SJ, Ambs S (2006) Association of breast cancer outcome with status of p53 and MDM2 SNP309. J Natl Cancer Inst 98 (13):911–919. doi: 10.1093/jnci/djj245 [DOI] [PubMed] [Google Scholar]
- 45.Faur N, Araud L, Laroche-Clary A, Kanno J, Toutain J, Yamori T, Robert J, Le Morvan V (2009) The association between the T309G polymorphism of the MDM2 gene and sensitivity to anticancer drug is dependent on the p53 mutational status in cellular models. Br J Cancer 101 (2):350–356. doi: 10.1038/sj.bjc.6605096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van den Broek AJ, Broeks A, Horlings HM, Canisius SV, Braaf LM, Langerod A, Van’t Veer LJ, Schmidt MK (2011) Association of the germline TP53 R72P and MDM2 SNP309 variants with breast cancer survival in specific breast tumor subgroups. Breast Cancer Res Treat 130 (2):599–608. doi: 10.1007/s10549-011-1615-y [DOI] [PubMed] [Google Scholar]
- 47.Harris LC (2005) MDM2 splice variants and their therapeutic implications. Curr Cancer Drug Targets 5 (1):21–26 [DOI] [PubMed] [Google Scholar]
- 48.Shaikh MF, Morano WF, Lee J, Gleeson E, Babcock BD, Michl J, Sarafraz-Yazdi E, Pincus MR, Bowne WB (2016) Emerging Role of MDM2 as Target for Anti-Cancer Therapy: A Review. Ann Clin Lab Sci 46 (6):627–634 [PubMed] [Google Scholar]
- 49.Wade M, Li YC, Wahl GM (2013) MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 13 (2):83–96. doi: 10.1038/nrc3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao Y, Aguilar A, Bernard D, Wang S (2015) Small-molecule inhibitors of the MDM2-p53 protein-protein interaction (MDM2 Inhibitors) in clinical trials for cancer treatment. J Med Chem 58 (3):1038–1052. doi: 10.1021/jm501092z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


