Abstract
Background:
The single nucleotide polymorphism (SNP) rs2476601 of the protein tyrosine phosphatase, nonreceptor type 22 (PTPN22) gene has been presented to implicate in the pathogenesis of alopecia areata (AA) in a few association investigations with limited sample size and inconsistent conclusions.
Methods:
The aim of the current meta-analysis was to assess and synthesize the presently available data on the connection between rs2476601 and AA vulnerability. Six electronic databases, including EMBASE, PubMed, Web of Science, the Cochrane Library, Wanfang data, and the China National Knowledge Infrastructure database (CNKI), were systematically retrieved for relevant observational studies published previous to November 2018. Total odds ratios (ORs) and corresponding 95% confidence intervals (95% CIs) were analyzed to evaluate the correlation between PTPN22 polymorphism and AA. Risk of bias was estimated according to the Newcastle–Ottawa Scale (NOS). Sensitivity analyses were carried out using the RevMan 5.3 software.
Results:
In general, 5 case–control studies including 1129 AA patients and 1702 healthy control individuals were obtained for this meta-analysis. The pooled results suggested that rs2476601 SNP was significantly associated with AA susceptibility under allelic model (C vs T, OR = 0.77, 95% CI, 0.64–0.92, P = .003) and recessive model (CC vs CT + TT, OR = 0.73, 95% CI, 0.60–0.88, P = .001).
Conclusion:
On the basis of the results of the current research, the rs2476601 polymorphism of PTPN22 gene is significantly correlated with AA susceptibility. The C-allele and CC-genotype carriers at this locus have a lower risk of AA.
Keywords: alopecia areata, meta-analysis, polymorphism, PTPN22, rs2476601
1. Introduction
Alopecia areata (AA) is a nonscarring inflammatory chronic recurrent alopecia disease characterized by localized circular or oval hair loss, which can progress to severe forms with total hair loss of the scalp (alopecia totalis) or the entire body (alopecia universalis). Generally, AA affects the scalp or beard area. With a lifetime risk of approximately 1.7%,[1] AA affects both sexes equally, patients of all ages, and all ethnic groups.[2,3] The prognosis of AA is unpredictable and highly variable.
Although neither fatal or disabling nor accompanied by physical pain, AA may have a great impact on health-related quality of life (HRQOL) and mental health of patients.[4] The effect of psychological stress has been emphasized in the onset and aggravation of AA by lots of studies.[5–7] In addition, AA patients seem to be more prone to psychiatric comorbidities, including depression, anxiety, personality disorders, and social phobia than the general population.
And yet, the aetiopathogenesis of AA is not fully elucidated, which has been considered as a complex immune-mediated disorder, characterized by the presence of lymphocytes directed against the hair follicle.[8] Skin resident T cells play a vital role in the immune surveillance,[9] and the autoimmune origin hypothesis of AA is approved by the presence of CD4+ and CD8+ lymphocytes infiltration in the intrafollicular and perifollicular areas of the hair follicle.[10]
The protein tyrosine phosphatase, nonreceptor type 22 (PTPN22) gene is one of the most convincing risk factors out of the major histocompatability complex relating to autoimmune diseases. PTPN22 gene, located on chromosome 1p13.3–13.1, encodes lymphoid protein tyrosine phosphatase (LYP) exclusively expressed in immune cells[11] and negatively regulates T lymphocyte development and activation through binding with the Src homology 3 domain of C-terminal Src kinase (CSK) forming a complex.[12–15]
Up to now, the impact of PTPN22 gene polymorphism, which is also known as C1858T, rs2476601, and R620W on AA vulnerability, has been investigated by several studies; however, it still remains ambiguous on account of inconsistent and inconclusive outcomes from different studies. Due to the small sample size, the statistical capacity of a single study was insufficient. Meta-analysis of pooled data from different studies has been confirmed as an effective approach to evaluate a general risk of a polymorphism with the disease susceptibility. Hence, we conducted a meta-analysis of all available published articles on this topic to elucidate and provide comprehensive evaluation of the correlation between the PTPN22 gene polymorphisms and AA susceptibility.
2. Materials and methods
The current systematic review and meta-analysis was carried out in agreement with the Preferred Reporting Items for Systematic Review and Meta-Analyses (PRISMA) guidelines.[16] Ethical approval was not needed for this systematic review and meta-analysis owing to unnecessary data connected with individual patient information.
2.1. Literature search
The keywords alopecia areata, PTPN22, protein tyrosine phosphatase nonreceptor 22, rs2476601, and gene polymorphism were used as searching terms for electronic databases consisted of EMBASE, PubMed, the Cochrane Library, Web of Science, Wanfang data, and the China National Knowledge Infrastructure database (CNKI). Studies published up to November 2018 were included. Selected articles were not confined to any language publications, from which cited references were additionally inspected for relevant studies.
2.2. Inclusion and exclusion criteria
Studies included in the meta-analysis should satisfy the following criteria: case–control study that addressed patients with AA and healthy controls; all patients had clinically diagnosed of AA; study estimated the relationship of PTPN22 polymorphism and susceptibility to AA; study available for full text including adequate genotype distribution; the literature must be a peer-reviewed academic paper; and the latest study was selected as a result of duplicate publications in the literature.
Accordingly, studies excluded were required to meet the following criteria: no control population; reviews, repeated literature, and animal studies; and duplicate data contained in the studies.
2.3. Data extraction
In accordance with preestablished inclusion and exclusion criteria, the data were extracted by 2 authors independently from all eligible studies, and disagreements were resolved by discussion with consensus. If the 2 authors failed to reach an agreement, contradictions were determined by a third author, and the conclusive judgment was made by majority vote. The extracted data from every included articles covered the following items: first author, publication year, study design, country where the research was conducted, sample size, source of controls, total number of cases and controls, AA diagnostic criteria, and allele frequency of PTPN22 single nucleotide polymorphisms (SNPs), together with evidence of Hardy–Weinberg equilibrium (HWE) in controls.
2.4. Evaluation of study quality
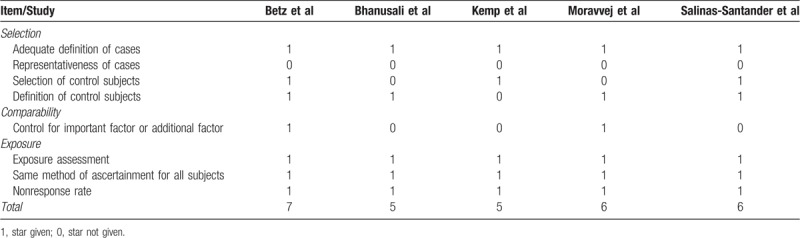
According to the Newcastle–Ottawa Scale (NOS) criteria, the methodological quality of the involved studies was estimated systematically by 2 independent investigators.[17] The NOS criteria describe 3 aspects of quality: the selection of the study groups: 0 to 4 stars; the comparability of the groups: 0 to 2 stars; and the ascertainment of either the exposure or outcome of interest for case–control studies: 0 to 3 stars. The scores range from 0 (worst) to 9 (best) stars. In addition, observational studies with fractions ≥6 were regarded as of high quality.
2.5. Statistical analysis
To estimate the between-study heterogeneity, Q-statistical test and I2 test were applied.[18]P < .10 and I2 > 50% indicated considerable heterogeneity across studies, and the random-effect model was used, otherwise the fixed-effect model was adopted.[19]
To evaluate correlation with AA susceptibility, diverse genotypic models were selected, including allelic model (C vs T), dominant model (CC + CT vs TT), homozygous model (CC vs TT), heterozygous model (CT vs TT), and recessive model (CC vs CT + TT). For each model, odds ratios (ORs) alongside corresponding 95% confidence intervals (95% CIs) and P value were measured to appraisal the association strength between rs2476601 and AA vulnerability. Forest plots were derived from Review Manager 5.33 software (Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration, 2014). Sensitivity analysis of the robustness of this meta-analysis was detected by removing the included studies one by one. Funnel plots were examined for evidence of publication bias if the number of included studies were ≥10.[20]
3. Results
3.1. Literature search
The search strategy of eligible studies was illustrated in Figure 1. In the aggregate, 40 potentially related studies were detected by the initial literature search, including 17 from ISI Web of Science, 7 from PubMed, 11 from EMBASE, 5 from Wanfang, and 0 from Cochrane Library. After the first screening, 18 duplicated records were erased. Of the remaining 22 items, a further 15 citations were excluded after reading titles and abstracts. For the rest 7 articles, another 2 ineligible articles were deleted after assessment by the predefined inclusion and exclusion criteria. Ultimately, 5 studies[21–25] were incorporated into the qualitative synthesis and meta-analysis (Fig. 1).
Figure 1.
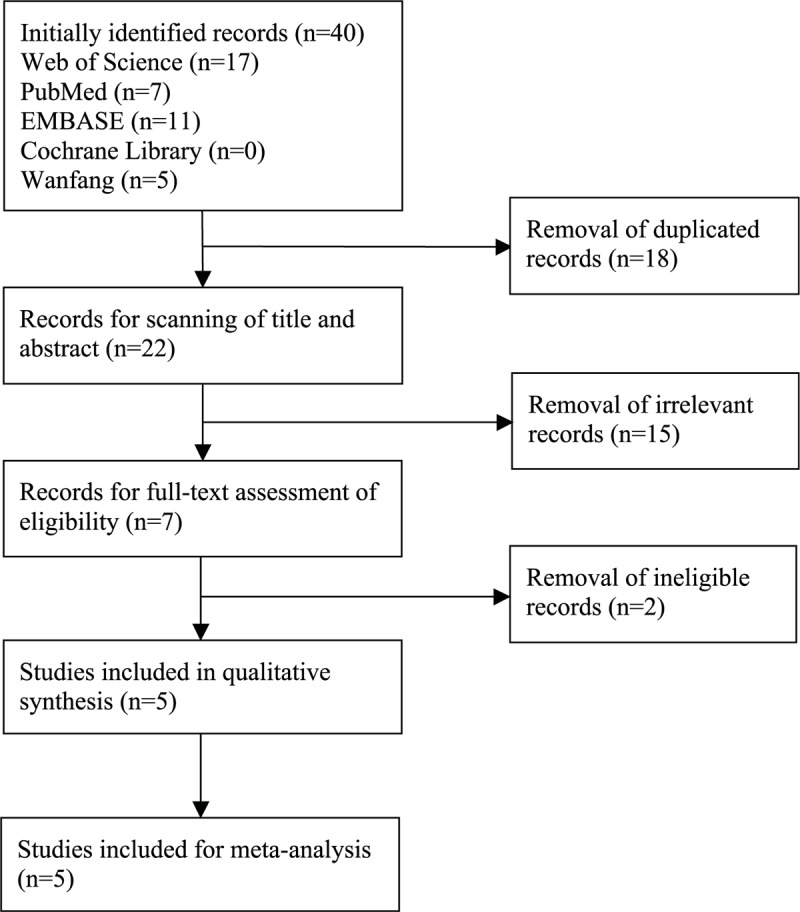
Flow chart of literature search and screen.
3.2. Main characteristics of included studies
Five case–control studies with 1129 AA cases and 1702 healthy controls were included in this meta-analysis. These researches were published between 2006 and 2018, and they were conducted in the Belgian and German,[21] the United Kingdom,[22] the United States,[23] Iran,[24] and Mexico,[25] respectively. The genotype frequencies of case and control groups were summarized in Table 1. The control subjects of each included study were in HWE. In accordance with the NOS, the involved studies gained an average of 5.8 stars for methodological quality assessment (Table 2).
Table 1.
Main characteristics of included studies and genotype frequencies in case and control groups.
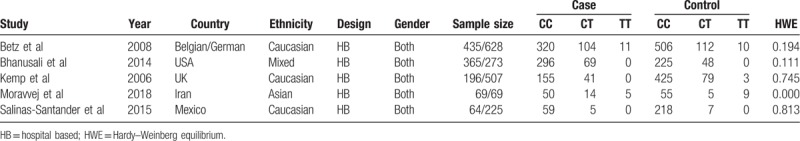
Table 2.
Quality assessment of included studies according to the Newcastle–Ottawa Scale.
3.3. Association of rs2476601 and AA
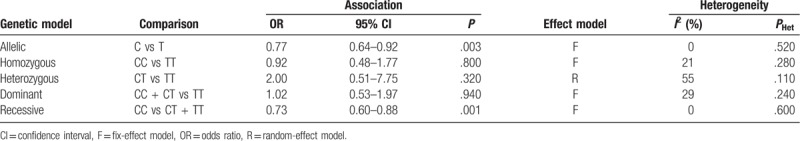
Because of significant heterogeneity was observed for heterozygous model (I2 = 55%, P = .11), random-effect model was adopted. And fixed-effect models were used for other genetic models for insignificant heterogeneity. The present study indicated significant associations between the rs2476601 SNP and AA for the allelic model (C vs T, OR = 0.77, 95% CI, 0.64–0.92, P = .003; Fig. 2) and recessive model (CC vs CT + TT, OR = 0.73, 95% CI, 0.60–0.88, P = .001; Fig. 3). However, the correlation was not observed under homozygous model (CC vs TT, OR = 0.92, 95% CI, 0.47–1.77, P = .800), heterozygous model (CT vs TT, OR = 2.00, 95% CI, 0.51–7.75, P = .320), or dominant model (CC + CT vs TT, OR = 1.02, 95% CI, 0.53–1.97, P = .940). The detailed outcomes of the meta-analysis were presented in Table 3.
Figure 2.
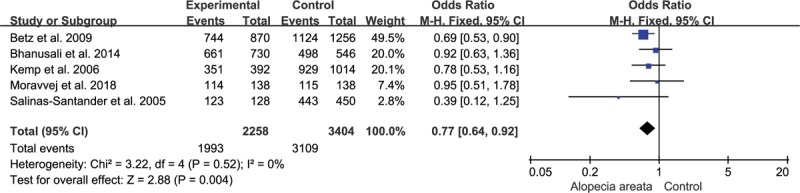
Association of rs2476601 and alopecia areata under allelic model.
Figure 3.
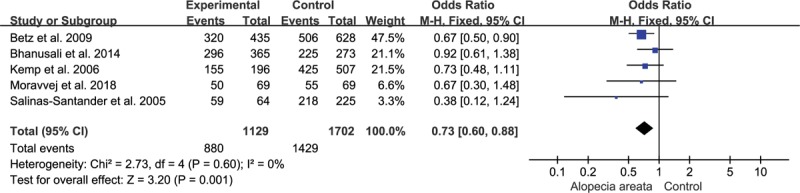
Association of rs2476601 and alopecia areata under recessive model.
Table 3.
Association between PTPN22 gene polymorphisms and alopecia areata.
3.4. Sensitivity analysis and publication bias
The leave-one-out sensitivity analysis was conducted to evaluate the influence of each individual study on the pooled OR. The analysis results reinforced the certainty and dependability of the drawn conclusion. The correlation between rs2476601 polymorphism and AA vulnerability remained irrelevant in the case of any omission of included study.
4. Discussion
Even though it is obvious that the understanding of the etiology and pathogenesis of AA can better guide its early prevention and individualized clinical treatment, researchers have not yet found the precise pathogenesis of AA.
With the development of genome-wide association studies (GWAS), researchers are paying more and more attention to the role of genetic factors in the developing of AA. Several genes associated with the human leukocyte antigen (HLA) class II with the genetic susceptibility, such as HLA-DQ3, HLA-DQ4, HLA-DQ11 AQ6, and HLA-DQ7, were found to be implicated in AA and the disease severity.[26]
However, other studies reported susceptibility loci covering the following genes, most of which could act as immune regulators controlling the activation and proliferation of regulatory T cells: PTPN22,[27] CTLA4 (cytotoxic T lymphocyte-associated antigen 4)[28]; IL2RA (IL-2 receptor A)[29]; and IKZF4 (Ikaros family zinc finger 4).[30]
With time to develop into the stage for functional studies aimed at defining molecular pathways of disease pathogenesis, PTPN22 is such a gene out of high-resolution mapping of disease risk loci. PTPN22 belongs to class I protein tyrosine phosphatases (PTPs), and its catalytic domain is highly homologous to the catalytic domains of other classical tyrosine-specific PTPs with known immune system functions, including CD45, SHP-1, and TC-PTP.[31,32]
Rs2476601 polymorphism in PTPN22 has been described as an autoimmune susceptibility locus, which codes for an arginine (R) to tryptophan (W) amino acid substitution in codon 620. The change perturbs the interaction between lymphoid tyrosine phosphate and CSK which decrease thresholds for T-cell antigen receptor signaling. Hence, T-cell activation arises in a way out of control, triggering a hyperimmune response,[33] then inducing the production of autoantibodies by the hyperresponsive B cells.[34] Consequently, this process may lead to the eventual progression to multiple human autoimmune diseases.[35] Owing to the association with many other autoimmune diseases as mentioned before, it is another support for the hypothesis of an autoimmune origin of AA.
The relationship between rs2476601 polymorphism and disease predisposition is well defined for type 1 diabetes mellitus (T1D),[35] rheumatoid arthritis (RA),[36] systemic lupus erythematosus (SLE),[37] vitiligo,[38] Graves’ disease,[39] Hashimoto's thyroiditis,[40] and AA.[22] Quantity of associations has been established in numerous populations. Notably, PTPN22 polymorphism explicates a T1D locus confirmed by genetic linkage analysis.[41] The carrying of the PTPN22 polymorphism variant increases the risk of disease 1.3 to 2 times each allele, owing to the study and the disease, with the strongest effects being observed in T1D, RA, and vitiligo. Even so, a weaker effect has been identified in systemic sclerosis.[42] PTPN22 polymorphism also predisposes individual to a cooccurrence of major autoimmune diseases, including T1D and thyroid disease[43] or RA and T1D.[44] In spite of this, a significant protective effect has been observed in Behcet's disease.[45] Especially, a small but forceful protective effect for PTPN22 polymorphism is detected in Crohn's disease (CD) as well.[46]
Regarding a common theme among the variety of disorders related with the PTPN22 gene mutation, the associations between PTPN22 polymorphisms and AA risk have been intensively studied. Previous researches have investigated the impact of rs2476601 polymorphism on AA in different populations and disease severity, the findings of which are debatable. Salinas-Santander et al[25] and El-Zawahry et al[47] have proposed a significantly association between rs2476601 and patchy AA risk in Latin American and African populations, respectively. However, studies carried out by Betz et al[21] and Kemp et al[22] in European cohort, as with study conducted by Bhanusali et al[23] across North American Caucasian and non-Caucasian cohort, failed to confirm these data in a mild or patchy form of AA, whereas significant association was found in patients with severe disease presentation or with familial history. Besides, an investigation conducted recently by Moravvej et al[24] in an Iranian population validated no significant difference between patchy AA patients and control groups, supporting the results presented by European and North American cohorts. The conflicting results may be caused by factors such as heredity, ethnicity, region, environment, and so on.
Based on these data, a comprehensive evaluation is proposed by this meta-analysis aiming to quantitatively analyze the association between rs2476601polymorphism and AA susceptibility, hence enlarging the overall sample size and improving statistical power. According to inclusion and exclusion criteria, a total of 1129 cases and 1702 controls from 5 hospital-based case–control studies[21–25] were selected and analyzed to provide a wide-ranging assessment of the association. The present study presented that there was a significant statistical correlation between PTPN22 gene polymorphism and AA susceptibility, and that the C-allele and CC-genotype carriers had a lower risk of AA.
Despite the present study has combined the existing evidence of the association of rs2476601 polymorphism and AA, several drawbacks should be acknowledged. First, the number of investigated studies and participants was limited, which might cause insufficient statistical power to guarantee the robustness of our findings. Second, the publication bias, which could lead to the overestimation of effects in a meta-analysis, was also an unavoidable problem. Third, the majority of the included studies were carried out in Caucasian populations, and only one was performed in Asian population. Thus, the current outcome might be more applicable to Europeans. Last, AA is a multifactorial disease that caused by both environmental and genetic factors, whereas the effect of environmental factor could not be analyzed because of insufficient data.
5. Conclusion
The findings of the current investigation suggested a significant association between the rs2476601 polymorphism of PTPN22 gene and AA. The C-allele and CC-genotype carriers at this locus have a lower risk of AA. To substantiate the specific gene-disease association between PTPN22 polymorphisms and susceptibility of AA, multi-center, large-sample studies in different ethnicities are strongly recommended.
Author contributions
Z-XL and W-JC formed the idea to this study, retrieved the eligible literature, conducted statistical analyses, and drafted the article. Y-JW, LJ, and CX participated in the study design and estimated the data from involved articles. X-JK and J-QL seriously revised this manuscript. X-JK had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. All of authors carefully read and approved the final version of this article.
Conceptualization: Zi-Xian Lei, Wen-Jing Chen, Xiao-Jing Kang.
Data curation: Zi-Xian Lei, Wen-Jing Chen, Yan-Jun Wang, Lan Jin, Xiao-Jing Kang.
Formal analysis: Zi-Xian Lei, Wen-Jing Chen, Jun-Qin Liang.
Funding acquisition: Xiao-Jing Kang.
Investigation: Wen-Jing Chen, Yan-Jun Wang, Lan Jin, Xiao-Jing Kang.
Methodology: Zi-Xian Lei, Wen-Jing Chen, Yan-Jun Wang, Lan Jin, Chen Xu.
Project administration: Wen-Jing Chen, Jun-Qin Liang, Xiao-Jing Kang.
Software: Zi-Xian Lei, Wen-Jing Chen, Chen Xu.
Supervision: Jun-Qin Liang, Xiao-Jing Kang.
Validation: Jun-Qin Liang, Yan-Jun Wang, Lan Jin, Chen Xu, Xiao-Jing Kang.
Visualization: Zi-Xian Lei, Wen-Jing Chen, Yan-Jun Wang, Lan Jin, Chen Xu.
Writing – original draft: Zi-Xian Lei, Wen-Jing Chen, Xiao-Jing Kang.
Writing – review and editing: Zi-Xian Lei, Wen-Jing Chen, Jun-Qin Liang, Xiao-Jing Kang.
Footnotes
Abbreviations: AA = alopecia areata, CI = confidence interval, CNKI = China National Knowledge Infrastructure, CSK = C-terminal Src kinase, GWAS = genome-wide association studies, HLA = human leukocyte antigen, HWE = Hardy–Weinberg equilibrium, LYP = lymphoid protein tyrosine phosphatase, NOS = Newcastle–Ottawa Scale, OR = odds ratio, PRISMA = Preferred Reporting Items for Systematic Review and Meta-Analyses, PTPN22 = protein tyrosine phosphatase, nonreceptor type 22, PTPs = protein tyrosine phosphatases, RA = rheumatoid arthritis, SLE = systemic lupus erythematosus, SNP = single nucleotide polymorphism, T1D = type 1 diabetes mellitus.
Z-XL and W-JC equally contributed to this work.
The study was supported by the National Natural Science Foundation of China (Grant no. 81760563).
The authors have no conflicts of interest to disclose.
References
- [1].Betz RC, Petukhova L, Ripke S, et al. Genome-wide meta-analysis in alopecia areata resolves HLA associations and reveals two new susceptibility loci. Nat Commun 2015;6:5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Alzolibani AA. Epidemiologic and genetic characteristics of alopecia areata (part 1). Acta Dermatovenerol Alp Pannonica Adriat 2011;20:191–8. [PubMed] [Google Scholar]
- [3].Strazzulla LC, Wang EHC, Avila L, et al. Alopecia areata: Disease characteristics, clinical evaluation, and new perspectives on pathogenesis. J Am Acad Dermatol 2018;78:1–2. [DOI] [PubMed] [Google Scholar]
- [4].Rencz F, Gulacsi L, Pentek M, et al. Alopecia areata and health-related quality of life: a systematic review and meta-analysis. Br J Dermatol 2016;175:561–71. [DOI] [PubMed] [Google Scholar]
- [5].Garcia-Hernandez MJ, Ruiz-Doblado S, Rodriguez-Pichardo A, et al. Alopecia areata, stress and psychiatric disorders: a review. J Dermatol 1999;26:625–32. [DOI] [PubMed] [Google Scholar]
- [6].Gupta MA, Gupta AK, Watteel GN. Stress and alopecia areata: a psychodermatologic study. Acta Derm Venereol 1997;77:296–8. [DOI] [PubMed] [Google Scholar]
- [7].Manolache L, Benea V. Stress in patients with alopecia areata and vitiligo. J Eur Acad Dermatol Venereol 2007;21:921–8. [DOI] [PubMed] [Google Scholar]
- [8].Galan-Gutierrez M, Rodriguez-Bujaldon A, Moreno-Gimenez JC. [Update on the treatment of alopecia areata]. Actas Dermosifiliogr 2009;100:266–76. [PubMed] [Google Scholar]
- [9].Smith SE, Neier SC, Davis TR, et al. Signalling protein complexes isolated from primary human skin-resident T cells can be analysed by Multiplex IP-FCM. Exp Dermatol 2014;23:272–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rodriguez TA, Duvic M. Onset of alopecia areata after Epstein-Barr virus infectious mononucleosis. J Am Acad Dermatol 2008;59:137–9. [DOI] [PubMed] [Google Scholar]
- [11].Burn GL, Svensson L, Sanchez-Blanco C, et al. Why is PTPN22 a good candidate susceptibility gene for autoimmune disease? FEBS Lett 2011;585:3689–98. [DOI] [PubMed] [Google Scholar]
- [12].Hill RJ, Zozulya S, Lu YL, et al. The lymphoid protein tyrosine phosphatase Lyp interacts with the adaptor molecule Grb2 and functions as a negative regulator of T-cell activation. Exp Hematol 2002;30:237–44. [DOI] [PubMed] [Google Scholar]
- [13].Hasegawa K, Martin F, Huang G, et al. PEST domain-enriched tyrosine phosphatase (PEP) regulation of effector/memory T cells. Science (New York, NY) 2004;303:685–9. [DOI] [PubMed] [Google Scholar]
- [14].Begovich AB, Carlton VE, Honigberg LA, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet 2004;75:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bottini N, Musumeci L, Alonso A, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet 2004;36:337–8. [DOI] [PubMed] [Google Scholar]
- [16].Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol 2009;62:1006–12. [DOI] [PubMed] [Google Scholar]
- [17].Wells GA, Shea B, O’Connell D, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomized studies in meta-analyses. Available at: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. [Google Scholar]
- [18].Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539–58. [DOI] [PubMed] [Google Scholar]
- [20].Ioannidis JP, Trikalinos TA. The appropriateness of asymmetry tests for publication bias in meta-analyses: a large survey. CMAJ 2007;176:1091–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Betz RC, Konig K, Flaquer A, et al. The R620W polymorphism in PTPN22 confers general susceptibility for the development of alopecia areata. Br J Dermatol 2008;158:389–91. [DOI] [PubMed] [Google Scholar]
- [22].Kemp EH, McDonagh AJ, Wengraf DA, et al. The non-synonymous C1858T substitution in the PTPN22 gene is associated with susceptibility to the severe forms of alopecia areata. Hum Immunol 2006;67:535–9. [DOI] [PubMed] [Google Scholar]
- [23].Bhanusali DG, Sachdev A, Olson MA, et al. PTPN22 profile indicates a novel risk group in Alopecia areata. Hum Immunol 2014;75:81–7. [DOI] [PubMed] [Google Scholar]
- [24].Moravvej H, Tabatabaei-Panah PS, Abgoon R, et al. Genetic variant association of PTPN22, CTLA4, IL2RA, as well as HLA frequencies in susceptibility to alopecia areata. Immunol Invest 2018;47:666–79. [DOI] [PubMed] [Google Scholar]
- [25].Salinas-Santander M, Sanchez-Dominguez C, Cantu-Salinas C, et al. Association between PTPN22 C1858T polymorphism and alopecia areata risk. Exp Ther Med 2015;10:1953–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Price VH, Colombe BW. Heritable factors distinguish two types of alopecia areata. Dermatol Clin 1996;14:679–89. [DOI] [PubMed] [Google Scholar]
- [27].Maine CJ, Hamilton-Williams EE, Cheung J, et al. PTPN22 alters the development of regulatory T cells in the thymus. J Immunol 2012;188:5267–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].John KK, Brockschmidt FF, Redler S, et al. Genetic variants in CTLA4 are strongly associated with alopecia areata. J Invest Dermatol 2011;131:1169–72. [DOI] [PubMed] [Google Scholar]
- [29].Redler S, Albert F, Brockschmidt FF, et al. Investigation of selected cytokine genes suggests that IL2RA and the TNF/LTA locus are risk factors for severe alopecia areata. Br J Dermatol 2012;167:1360–5. [DOI] [PubMed] [Google Scholar]
- [30].Petukhova L, Duvic M, Hordinsky M, et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature 2010;466:113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Alonso A, Sasin J, Bottini N, et al. Protein tyrosine phosphatases in the human genome. Cell 2004;117:699–711. [DOI] [PubMed] [Google Scholar]
- [32].Tonks NK. Protein tyrosine phosphatases—from housekeeping enzymes to master regulators of signal transduction. FEBS J 2013;280:346–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rieck M, Arechiga A, Onengut-Gumuscu S, et al. Genetic variation in PTPN22 corresponds to altered function of T and B lymphocytes. J Immunol 2007;179:4704–10. [DOI] [PubMed] [Google Scholar]
- [34].Zikherman J, Hermiston M, Steiner D, et al. PTPN22 deficiency cooperates with the CD45 E613R allele to break tolerance on a non-autoimmune background. J Immunol 2009;182:4093–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Onengut-Gumuscu S, Ewens KG, Spielman RS, et al. A functional polymorphism (1858C/T) in the PTPN22 gene is linked and associated with type I diabetes in multiplex families. Genes Immun 2004;5:678–80. [DOI] [PubMed] [Google Scholar]
- [36].Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Harley JB, Alarcon-Riquelme ME, Criswell LA, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet 2008;40:204–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jin Y, Birlea SA, Fain PR, et al. Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. N Engl J Med 2010;362:1686–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Velaga MR, Wilson V, Jennings CE, et al. The codon 620 tryptophan allele of the lymphoid tyrosine phosphatase (LYP) gene is a major determinant of Graves’ disease. J Clin Endocrinol Metab 2004;89:5862–5. [DOI] [PubMed] [Google Scholar]
- [40].Criswell LA, Pfeiffer KA, Lum RF, et al. Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am J Hum Genet 2005;76:561–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Smyth DJ, Cooper JD, Howson JM, et al. PTPN22 Trp620 explains the association of chromosome 1p13 with type 1 diabetes and shows a statistical interaction with HLA class II genotypes. Diabetes 2008;57:1730–7. [DOI] [PubMed] [Google Scholar]
- [42].Zheng J, Ibrahim S, Petersen F, et al. Meta-analysis reveals an association of PTPN22 C1858T with autoimmune diseases, which depends on the localization of the affected tissue. Genes Immun 2012;13:641–52. [DOI] [PubMed] [Google Scholar]
- [43].Dultz G, Matheis N, Dittmar M, et al. The protein tyrosine phosphatase non-receptor type 22 C1858T polymorphism is a joint susceptibility locus for immunthyroiditis and autoimmune diabetes. Thyroid 2009;19:143–8. [DOI] [PubMed] [Google Scholar]
- [44].Liao KP, Gunnarsson M, Kallberg H, et al. Specific association of type 1 diabetes mellitus with anti-cyclic citrullinated peptide-positive rheumatoid arthritis. Arthritis Rheum 2009;60:653–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Baranathan V, Stanford MR, Vaughan RW, et al. The association of the PTPN22 620W polymorphism with Behcet's disease. Ann Rheum Dis 2007;66:1531–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet 2008;40:955–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].El-Zawahry BM, Azzam OA, Zaki NS, et al. PTPN22 gene polymorphism in Egyptian alopecia areata patients and its impact on response to diphencyprone immunotherapy. Gene 2013;523:147–51. [DOI] [PubMed] [Google Scholar]


