Supplemental Digital Content is available in the text
Keywords: acetylcholine receptors, CHRNA7, non-small cell lung cancer, survival analysis
Abstract
Acetylcholine receptors (AChRs), including nicotinic acetylcholine receptors (nAChRs) and muscarinic acetylcholine receptors (mAChRs), are highly expressed in bronchial epithelial cells.
We used The Cancer Genome Atlas (TCGA) data set to evaluate the expression pattern and prognostic value of the AChR gene family in non-small cell lung cancer (NSCLC). The mined data was validated by quantitative real-time polymerase chain reaction (qRT-PCR) and immunohistochemistry (IHC).
The survival analysis of TCGA data set showed that only CHRNA7 in the AChR gene family affected prognosis in both lung adenocarcinoma and lung squamous cell carcinoma. Furthermore, qRT-PCR proved that CHRNA7 was significantly upregulated in tumor tissues compared with matched normal tissues at mRNA level (P = .001). The expression level of α7 nAChR (encoded by CHRNA7) in 141 patients was measured by IHC and a high expression of α7 nAChR was associated with unfavorable prognosis (P = .008). Multivariate analysis showed that α7 nAChR was an independent prognostic factor (HR = 2.041; 95% CI 1.188-3.506; P = .007).
α7 nAChR was upregulated in NSCLC and was associated with unfavorable prognosis. This gene may be a potential target for lung cancer treatment.
1. Introduction
It is estimated that lung cancer accounts for more than one-quarter of all cancer deaths, which is higher than breast, prostate, and colon cancer combined.[1] Non-small cell lung cancer (NSCLC) is a major part of all lung cancer cases and is highly associated with cigarette smoking.[2] Although sophisticated combinations of surgery, radiation, and targeted chemotherapies have been developed for lung cancer treatment, mortality remains high.[3] Given the heterogeneity of lung cancer, new targets are urgently needed. Recently, growing knowledge of cholinergic signaling provides potential new therapies.
Acetylcholine receptors (AChRs), including nicotinic acetylcholine receptors (nAChR) and muscarinic acetylcholine receptors (mAChR), which are highly expressed in bronchial epithelial cells can be activated by acetylcholine (ACh).[4] Nicotine, the major addictive element in cigarettes, can also activate nAChRs with much higher affinity than ACh.[5] Although there is still no evidence that nicotine is a carcinogen, nicotine-specific metabolites named 4-(methyInitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and N-nitrosonornicotine (NNN) can definitely induce lung cancer owing to their binding to nAChRs.[6] Nicotine can also promote cancer cell survival, proliferation, angiogenesis, invasion, and epithelial to mesenchymal transition (EMT) through nAChRs.[7,8] In addition, the activation of the M2 muscarinic receptor (M2R) by non-neuronal ACh induces EMT in NSCLC cells and blocking the M2R signaling suppresses lung cancer cell migration and invasion.[9,10]
Genome-wide association studies (GWAS) proved for the first time that variations in genomic regions located on chromosome 15q24–25 are associated with nicotine dependence and lung cancer risk.[11–13] Although there have been several genomic variation studies investigating the relationships between the single-nucleotide polymorphisms (SNPs) of AChR and lung cancer risk, the functions of the AChR gene family are still unclear. Nicotine can bind to α7 nAChR and activate several tumor proliferation-related signal pathways.[14] In addition, the mitogenic effects of nicotine were previously shown to be mediated via the α7 nAChR subunit and result in the enhanced recruitment of E2F1 and Raf-1 on proliferative promoters in NSCLC cell lines and human lung tumors.[15]
Since the AChR gene family is closely related to tumor progression, this family may be good targets for lung cancer treatment. However, until now, we cannot find any studies that have comprehensively evaluated the expression and prognostic value of this gene family. In the present study, we used The Cancer Genome Atlas (TCGA) dataset to conduct differentially expressed gene (DEG) and survival analysis of the AChR gene family in NSCLC. We found that only α7 nAChR was associated with survival in both adenocarcinoma and squamous carcinoma. Furthermore, we tried to use quantitative real-time polymerase chain reaction (qRT-PCR) and immunohistochemistry (IHC) to validate the expression difference and prognostic value of α7 nAChR in NSCLC.
2. Materials and methods
2.1. Alterations of the AChR gene family in lung cancer
We studied the genomic alterations (amplification, deep deletion, missense mutation, mRNA upregulation, mRNA downregulation) that occurred in the AChR gene family in Lung Adenocarcinoma (TCGA, Nature 2014) and Lung Squamous Cell Carcinoma (TCGA, Nature 2012) case sets using cBioPortal (http://www.cbioportal.org/).[16,17] Advanced cancer genomic data visualization was obtained with the help of the Onco Query Language (OQL). OncoPrints show distinctive genomic alterations, including somatic mutations, copy number variations (CNV), and mRNA expression changes. OncoPrints (different levels of zoom) were generated using cBioPortal.
2.2. Survival analysis and differentially expressed genes
We obtained mRNA expression data and related survival data of lung cancer from the Genomic Data Commons (GDC) data portal
(https://cancergenome.nih.gov/newsevents/newsannouncements/genomic-data-commons-launch).[18,19] DEG analysis was performed using the R DEseq package. The heatmap was generated by HemI (Heatmap Illustrator, version 1.0).[20] The survival analysis was performed with normalized data using the DESeq method. Receiver operating characteristic (ROC) curves were used to identify the optimal cutoff points.[21,22] To avoid the emergence of bias, a running log-rank test was used at intervals between the 5th percentile and the 95th percentile of the normalized expression of each gene. The cutoff value of each gene was defined when the log-rank statistical value was maximum. The analyses were performed using the Statistical Package for Social Sciences (SPSS) program, version 20.0, in English.
2.3. Frozen tissue samples
Twenty-nine pairs of primary NSCLC and matched normal bronchiolar epitheliums were obtained from patients in Shandong Provincial Hospital Affiliated to Shandong University from 2012 to 2013 with informed consent. All tissue samples were from untreated patients undergoing surgery and all of the clinicopathologic information (age, gender, pathology, differentiation, invasion depth, and lymph node metastasis) was available. The study was approved by the Hospital's Ethical Review Committee. All samples were snap frozen in liquid nitrogen and stored at −80°C until the extraction of ribonucleic acid (RNA).
2.4. RNA isolation and quantitative reverse transcriptase-PCR
Quantitative real-time PCR was used to quantify cholinergic gene expression in the tumor samples and matched distant normal lung tissues as previously described.[23] The total RNA in the tumor samples and normal lung tissues were prepared with RNA Lyzol (Shanghai ExCell Biology, Inc.). RNA quality was confirmed in an Agilent 2100 Bioanalyzer (Agilent Technologies). Quantitative real-time PCR assay kits were purchased from Takara Bio (Dalian, China). The primers for real-time PCR were as follows: CHRNA7 (forward: 5’-ACATGCGCTGCTCGCCGGGA-3’; reverse: 5’-GATTGTAGTTCTTGACCAGCT-3’); GAPDH (forward: 5’-CATGAGAAGTATGACAACAGCCT-3’; reverse: 5’-AGTCCTTCCACGATACCAAAGT-3’). All reactions were run in triplicate and RNA levels were normalized to GAPDH. In addition, no-reverse transcriptase controls were run with all RNAs to check for genomic DNA contamination.
2.5. Patients
A total of 141 patients with NSCLC underwent R0 resection at Shandong Provincial Hospital Affiliated to Shandong University from January 2009 to December 2011. No patients had received preoperative adjuvant therapy. Our research was approved by the Ethical Committee of Shandong Provincial Hospital affiliated to Shandong University, and informed written consent was obtained from each patient. Altogether, 141 primary NSCLC specimens were examined for IHC. They were fixed in 10% phosphate-buffered formalin and embedded in paraffin. We made serial sections of 4 μm thickness for IHC. All patients were pathologically staged according to the tumor-node-metastasis (TNM) classification system of the American Joint Committee for Cancer.
2.6. Immunohistochemistry
An anti-α7-nAChR antibody, which was a rabbit polyclonal antibody specific for human α7 nAChR, was purchased from Abcam Biotechnology (ab10096, USA). The procedure for IHC has been described previously.[24] Briefly, all sections firstly underwent deparaffinization and rehydration, and were then heated in a 1 mmol/L ethylenediaminetetraacetic acid (EDTA) buffer (water bath, 96–98°C) for 15 minutes in order to retrieve the antigens. A 3% hydrogen peroxide was used to quench the endogenous peroxidase activity, and nonspecific binding was blocked by 10% normal goat serum. We used the primary anti-α7-nAChR antibody at a dilution of 1:250. The primary antibody was replaced by normal serum or phosphate-buffered saline (PBS) as a negative control.
2.7. Immunostaining evaluation
The criterion for a positive reaction of α7 nAChR was clear cytoplasm and nucleus staining. The samples with more than 10% of the tumor cells stained were considered to be α7 nAChR positive carcinomas. The criteria used for quantitating immunohistochemical staining was described previously, which included the staining intensity and the percentage of positive cells stained.[24] A range of 0 to 3 was defined for classifying the intensity of the staining: 0-absence of staining; 1-weak staining; 2-moderate staining; and 3-intense staining. Furthermore, the extent of the staining was scored as 0 (<10%), 1 (11–25%), 2 (26–50%), 3 (51–75%), and 4 (76–100%) for evaluation. The final scores were calculated by multiplying the staining intensity by the extension. In this study, final scores of 0 to 7 were stratified as low expression and scores of 8 to 12 as high expression. The results were assessed by 2 pathologists (X. Qu and GY. Ma), who were blinded to the patients’ background. If there were disagreements, both pathologists performed another review for these samples in order to obtain a conclusive judgment.
2.8. Statistical analysis
Pearson chi-square and Fisher exact tests were used to evaluate the clinicopathologic significance of enrolled patients’ characteristics in NSCLC. Univariable Cox regression tests were used to evaluate correlations between single variable and overall survival (OS). The correlations between multiple variables and OS were measured by multivariate Cox regression analysis. Survival analysis and curves were established using the Kaplan-Meier method, and the comparison of differences between groups was made using the log-rank test. All of the statistical analyses were performed using SPSS software (version 20.0). Two-sided P values were calculated, and P values less than .05 were considered a symbol of significant difference.
3. Results
3.1. Alterations of the AChR gene family in lung cancer
The cBioPortal for Cancer Genomics (http://cbioportal.org) provides a Web resource for exploring, visualizing, and analyzing multidimensional cancer genomics data.[16,17] We acknowledge the TCGA Research Network for generating TCGA datasets. We tried to use the Lung Adenocarcinoma (LUAD, TCGA, Nature 2014) and Lung Squamous Cell Carcinoma (LUSC, TCGA, Nature 2012) case sets to generate OncoPrints. As shown in Supplementary Figure 1, there were more alterations in LUAD than LUSC. The most altered genes in LUAD were CHRM3 (16%) and CHRNB2 (17%). However, in LUSC, the most altered genes were CHRM3 (13%) and CHRM4 (10%). Interestingly, mutual exclusivity and co-occurrence analysis showed that that CHRM3 and CHRNB2 tended to have co-occurrent alteration (amplification) in LUAD (P < .001). Similarly, CHRM5 and CHRNA7 also had co-occurrent alteration (deep deletion) in LUAD (P < .001). In LUSC, CHRNA6, and CHRNB3 seemed to have similar alterations (P < .001). We also studied the relationship between the AChR family and smoking history. However, as shown in Supplementary Figures 2 and 3, the relationship between gene expression and smoking was not significant. However, since we cannot get the detailed data from cBioPortal, the relationship needs to be further studied.
3.2. Prognostic value of the AChR gene family in LUAD and LUSC
We first examined the prognostic value of each gene in LUAD and LUSC. The ROC curves for each gene in LUAD and LUSC are shown in Supplementary Figure 4. As shown in Supplementary Figure 5, survival analysis showed that high expression of CHRM2 (P = .007), CHRM3 (P = .005), CHRNA1 (P < .001), CHRNA2 (P = .025), CHRNA6 (P < .001), CHRNB3 (P = .015) or CHRNE (P = 0.010) was associated with favorable prognosis in LUAD. However, high expression of CHRNA5 (P < .001) or CHRNA7 (P = 0.010, Fig. 1A) was associated with unfavorable prognosis. Compared with LUAD, only CHRNA7 (P = .019, Fig. 1B) and CHRNA10 (P = .005) were associated with prognosis of LUSC (Supplementary Figure 6). Although the P value for CHRNA1 was also lower than .05, the number of patients with high expression was much lower than patients with low-expression, which may produce bias.
Figure 1.
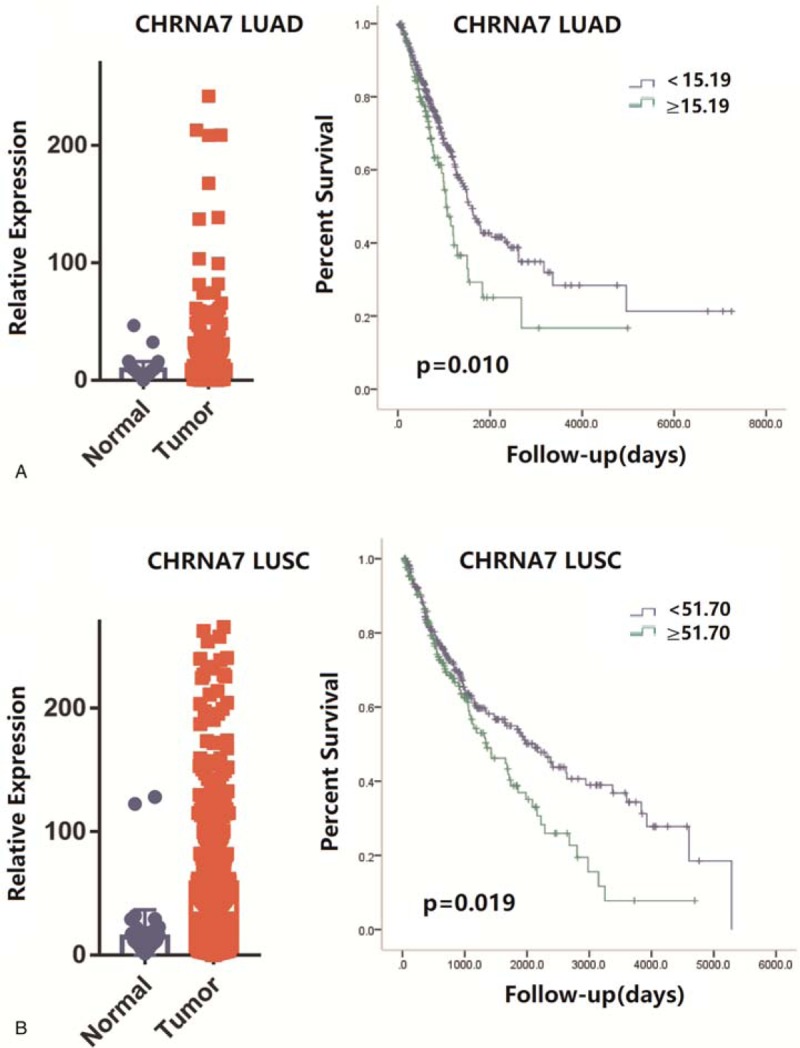
A. (Left) Scatter diagram showing the expression of CHRNA7 in lung adenocarcinoma (TCGA, Nature 2014) case set. (Right) The prognostic value of CHRNA7 in lung adenocarcinoma (TCGA, Nature 2014) case set. The P values are from stratified log-rank test. Kaplan-Meier survival curve for overall survival in LUAD patients stratified by CHRNA7. B. (Left) Scatter diagram showing the expression of CHRNA7 in lung squamous cell carcinoma (TCGA, Nature 2012). (Right) The prognostic value of CHRNA7 in lung squamous cell carcinoma (TCGA, Nature 2012). The P values are from stratified log-rank test. Kaplan-Meier survival curve for overall survival in LUSC patients stratified by CHRNA7. LUAD = lung adenocarcinoma, LUSC = lung squamous cell carcinoma, TCGA = the cancer genome atlas.
3.3. CHRNA7 was upregulated in tumor tissue compared with matched normal tissue
CHRNA7 is one of the most-studied genes among the AChR gene family. Several studies have reported that CHRNA7 can affect tumor proliferation, invasion, and is associated with chemotherapy resistance.[7,8,25] Since the expression of CHRNA7 may be affected by smoking status, we collected 29 pairs of tumor and matched normal tissue to investigate differential expression. The matched tumor and normal tissue can avoid the bias induced by smoking status. The clinicopathologic findings of 29 patients are shown in Table 1. Of the 29 pairs, CHRNA7 was up-regulated in 26 pairs (Fig. 2A). The CHRNA7 expression was significantly higher in tumor tissues than in matched normal tissues (P = .0013). We also tried to investigate the relationship between CHRNA7 and other pathological factors. However, given the limitation of patient number, the expression level of CHRNA7 was not associated with pathological stage or tumor size (data not shown).
Table 1.
Patients and tumor characteristics (total patients = 29).
Figure 2.

A. Expression level of CHRNA7 in NSCLC tissue specimens by qRT-PCR. Total RNA was isolated from normal and lung cancer tissues. CHRNA7 expression was analyzed by qRT-PCR and normalized to GAPDH expression. B. Expression status of α7 nAChR in NSCLC via IHC. (Left) Low expression of α7 nAChR; (Right) High expression of α7 nAChR. C. (Left) Survival curve of patients with low and high α7 nAChR expression; (Right) Recurrence free survival curve of patients with low and high α7 nAChR expression. IHC = immunohistochemistry , nAChR = nicotinic acetylcholine receptors, NSCLC = non-small cell lung cancer, qRT-PCR = quantitative real-time polymerase chain reaction.
3.4. Upregulation of α7 nAChR was associated with unfavorable prognosis
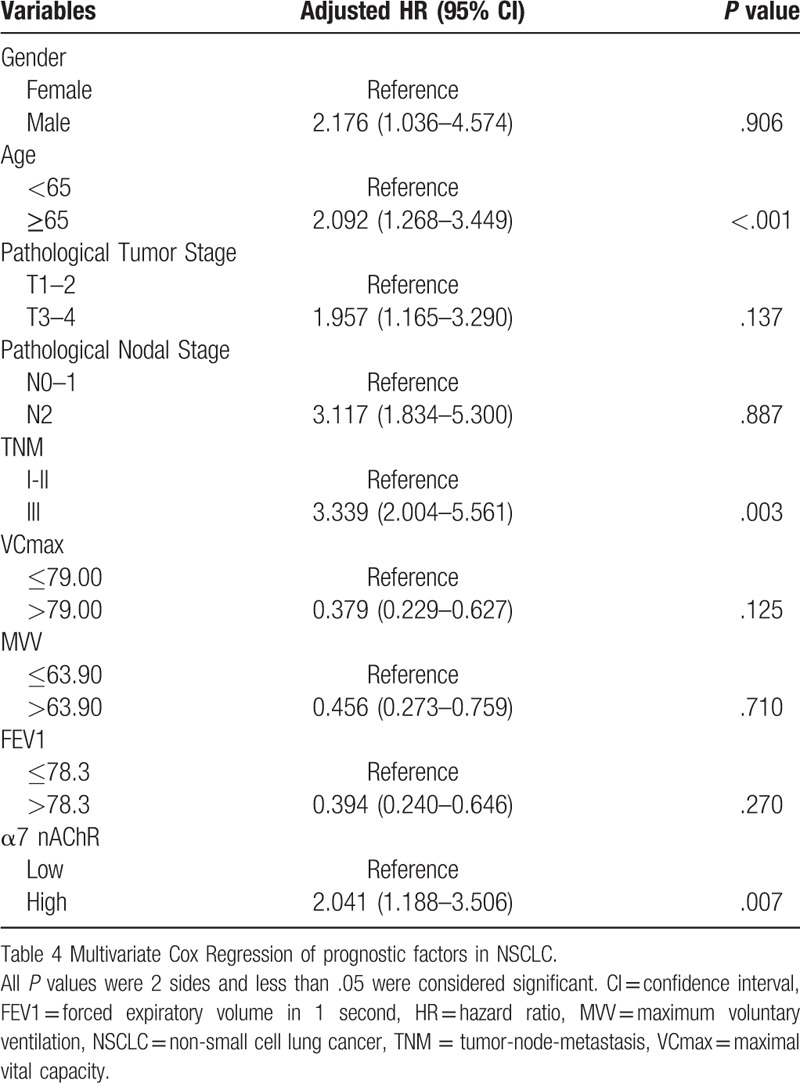
To investigate the prognostic value of this gene at the protein level, a total of 141 patients with NSCLC who underwent R0 resection were enrolled in the study. The clinicopathologic findings of the 141 patients are shown in Table 2. The expression of CHRNA7 was detected by IHC (Fig. 2B). The 83 patients showed high expression of α7 nAChR. No patients had received preoperative adjuvant therapy. As shown in Figure 2C, Kaplan-Meier survival analysis showed that high expression of α7 nAChR in NSCLC was associated with unfavorable prognosis (P = .008) and early recurrence (P = .003). Univariate Cox regression analysis of the influence of α7 nAChR staining scores’ influence and other related factors on OS in NSCLC patients shown in Table 3. We included all of the significant factors from the univariate analysis in the multivariate Cox regression analysis and the result showed that TNM stage, age at diagnosis, and α7 nAChR expression status were independent risk factors for prognosis in NSCLC patients (P = .003, P < .001, and P = .007, respectively, Table 4). We also analyzed the association between pulmonary function indexes and the expression of α7 nAChR. However, the expression of α7 nAChR had no effects on pulmonary function. Although the prognostic value of FEV1, VCmax, and MVV were significant in the univariate analysis, they were not independent prognostic factors in the multivariate analysis.
Table 2.
Patients and tumor characteristics (total patients = 141).
Table 3.
Univariate Cox Regression of prognostic factors in NSCLC (total patients = 141).

Table 4.
Multivariate Cox Regression of prognostic factors in NSCLC (total patients = 141).
3.5. Differentially expressed genes in TCGA data set
To further analyze the differential expression of the AChR gene family, we obtained separate mRNA expression data of LUAD and LUSC from the GDC data portal.[18,19] The raw count data was normalized using the DESeq method. DEG analysis was performed using the R DEseq package. An absolute value of log2 fold change >1 and adjusted P value < .05 were considered significant. As shown in Supplementary Figure 7, in the LUAD case set, we found that CHRM1 (log2 fold change = −4.42, adjusted P value = 2.86 × 10–12), CHRM2 (log2 fold change = −3.41, adjusted P value = 9.05 × 10–5), and CHRNA2 (log2 fold change = −3.59, adjusted P value = 5.45 × 10–5) were significantly downregulated compared with normal tissues. Additionally, we also found that CHRNA9 (log2 fold change = 5.15, adjusted P value < .001), CHRNA5 (log2 fold change = 3.61, adjusted P value = 2.09 × 10–7), and CHRNB4 (log2 fold change = 3.45, adjusted P value = .027) were significantly upregulated in tumor tissues. In LUSC, as shown in Supplementary Figure 8, we found that CHRM1 (log2 fold change = −4.02, adjusted P value = 5.80 × 10–10) and CHRM2 (log2 fold change = −4.56, adjusted P value = 2.19 × 10–10) were down regulated and CHRNB4 (log2 fold change = 4.81, adjusted P value = 5.06 × 10–7), CHRNB2 (log2 fold change = 3.40, adjusted P value = .002), CHRNA5 (log2 fold change = 3.17, adjusted P value = 6.74 × 10–5), and CHRM3 (log2 fold change = 1.43, adjusted P value < .001) were up regulated. According to the DEG analysis, CHRM1 and CHRM2 were down regulated and CHRNA5 and CHRNB4 were up regulated in both LUAD and LUSC.
4. Discussion
In the current study, we investigated the prognostic value of the AChR gene family in NSCLC using TCGA data-set and found that only CHRNA7 was a prognostic factor in both LUAD and LUSC. Using qRT-PCR, we then found that CHRNA7 was upregulated in tumor tissues compared with matched normal tissues. IHC data analysis of the present patient cohort showed that α7 nAChR was an independent prognostic factor in NSCLC. Importantly, our data has provided new insights into the AChR gene family.
Gene alteration analysis showed that there were more alterations (somatic mutations, copy number variations, and mRNA expression changes) in LUAD than LUSC. Interestingly, mutual exclusivity and co-occurrence analysis showed that that CHRM3 and CHRNB2, or CHRM5 and CHRNA7, tend to have co-occurrent alterations (amplification) in LUAD (P < .001). In LUSC, CHRNA6 and CHRNB3 seemed to have similar alterations (P < .001). Although the structure and biological function of nAChR and mAChR were different, they may be regulated by each other. It should be very interesting to investigate the significance of these co-occurrent alterations.
DEG analysis of TCGA data set showed that CHRM1 and CHRM2 were down regulated in both LUAD and LUSC. In LUAD, survival analysis also showed that low expression of CHRM1 or CHRM2 was associated with unfavorable prognosis. However, the mechanisms are still unclear. Especially for CHRM1, until now we cannot find any research on its functions in tumor cells. In addition, blocking M2 AChR signaling inhibits tumor growth and reverses EMT in NSCLC.[9,10] This is controversial to the present findings. However, our data was at the mRNA level. It should be very interesting to investigate their expression via IHC or western-blot in tumor and matched normal tissues. This controversy may be induced by post-translation modification. As a result, it is important to investigate their functions in cancer cells.
GWAS have shown for the first time with strong evidence that variations in genomic regions located on chromosome 15q24–25 are associated with nicotine dependence and lung cancer risk.[11–13] There are three nAChR genes (CHRNA5, CHRNA3, and CHRNB4) at this locus. Although several studies have shown that gene variations at this cluster are associated with lung cancer risk, little research has focused on their expression and lung cancer mortality risk.[26–28] Here, for the first time, we have shown that CHRNA5 and CHRNB4 were up regulated in both LUAD and LUSC. Furthermore, the upregulation of CHRNA5 and CHRNB4 were associated with unfavorable prognosis. However, CHRNA3 is not a prognostic factor in both LUAD and LUSC. Seung et al showed that the unmethylation of the CHRNB4 gene was an unfavorable prognostic factor in NSCLC.[29] However, the detailed biological functions of CHRNB4 are still unclear. SNPs rs421629 on 5p15.33 and rs1948, rs660652, rs8040868, and rs2036527 on 15q25.1 previously identified as lung cancer risk or nicotine-addiction modifiers were associated with tumor DNA methylation levels in the promoters of TERT and CHRNB4.[30] In addition, CHRNB4 knockdown in NSCLC cell lines resulted in a reduced proliferation and propensity to form colonies.[30] Also, nicotine inhibits cisplatin-induced apoptosis via regulating α5 nAChR/AKT signaling in human gastric cancer cells.[31] Nicotine induces HIF-1α and VEGF expression in NSCLC through α5 nAChR.[32] However, the function of α5 nAChR is also controversial. Krais et al showed that CHRNA5 was a negative regulator of nicotine signaling in normal and cancer bronchial cells.[33] In non-transformed bronchial cells and in lung cancer cell lines, silencing CHRNA5 or inhibiting receptors containing α5 nAChR with a-conotoxin MII exerted a nicotine-like effect, with increased motility and invasiveness in vitro and increased calcium influx.
The α7 nAChR is the most-studied receptor in this family. Its expression and function has previously been studied and summarized.[34] David et al compared the expression of nAChR subunits between tumor and matched normal tissue and found a significant upregulation of the β4 subunit and a concomitant decrease in α4 levels.[35] Another study showed that all lung cancer tissues expressed mRNA encoding α7 nAChR and the resultant proteins in the following rank: squamous carcinoma more than > adenocarcinoma > squamous carcinoma from non-smokers > large cell carcinoma > carnification > and pulmonary chondroid hamartoma.[36] As summarized by Schuller, nicotine and NNK will upregulate the expression of α7 nAChR in tumor cells. However, except tumor cells, the expression of α7 nAChR will also be upregulated in human bronchial epithelial and endothelial cells exposed to nicotine.[37] In addition, except nicotine, the exposure of small airway epithelial cells to estrogen in vitro upregulates and sensitizes the α7 nAChR in these cells.[38] Since the expression of α7 nAChR can be regulated by several reagents in normal and tumor cells, it is important to investigate its expression in tumor and matched normal tissue. In line with this, we designed the present experiments to study the differential expression of α7 nAChR in matched tissues. Although this is not the first time the upregulation of α7 nAChR in tumor tissue has been reported, we have provided new evidence that this gene is upregulated compared with matched normal tissues. According to this and its important function, α7 nAChR may be a good target for lung cancer treatment. It has the greatest Ca2+ permeability and significantly affects cell invasion, migration, and EMT.[7,8,39] The upregulated calcium activates the PKC and subsequent MEK-ERK signaling cascade.[40,41] Studies have shown that α7 is the main nAChR subunit that mediates the proliferative effects of nicotine in cancer cells.[42] Further studies have also shown that nicotine promotes the binding of ARRB1 to α7 nAChR and that the nuclear translocation of ARRB1 induces the increased expression of proliferative and survival genesin NSCLCs.[15,25] To our surprise, our data from TCGA showed that there was no expression difference between tumor and normal tissues. However, α7 nAChR was a prognostic factor in both LUAD and LUSC.
Limitations of the present study may include:
-
1.
the data provided by TCGA is at the mRNA expression level, but not the protein expression level;
-
2.
the normal tissues in TCGA data-set are not matched by tumor tissue one by one;
-
3.
the amount of normal tissue is much less than tumor tissue.
Considering all the possibilities, we tried to use matched normal and tumor tissues to investigate the expression difference of α7 nAChR. Our data showed that α7 nAChR was significantly upregulated in tumor tissues. Although the data was different from TCGA, we think it was reasonable since this expression data came from matched tumor and normal tissues. Since α7 nAChR was upregulated in lung cancer, we tried to investigate the prognostic value of α7 nAChR via IHC. High expression of α7 nAChR was associated with unfavorable prognosis in lung cancer. This indicates that α7 nAChR is a potential drug target for lung cancer treatment.
Several studies have investigated the expression and function of α7 nAChR in different types of tumor. Because of its high calcium permeability, which modulates intracellular signaling molecules, α7 nAChR has been implicated in lung tumorigenesis.[43,44] The copy number variations (CNV-3956) of α7 nAChR was associated with an increased risk of lung cancer and the poor survival of lung cancer patients.[45] Plummer et al found that α7 nAChR was ubiquitously expressed in both normal and cancer lung cells (squamous, carcinoid, adenocarcinoma, large cell carcinoma, and small cell lung cancer) which confirmed its involvement in lung biology and lung cancer development.[46] For the first time, we found that α7 nAChR was upregulated in tumor tissues compared with matched normal tissues and proved that α7 nAChR was an independent prognostic factor.
Our study was based on TCGA datasets which included more than 10,000 cases of human cancer including over 25 different cancer types. Datasets including the RNA-Seq, miRNA-Seq, Exon-Seq, somatic mutations, methylation, and CNV for each case are publically available via TCGA data portal (https://tcgadata.nci.nih.gov/tc-ga/tcgaHome2.jsp) and the UCSC Cancer Genomics Hub (https://cghub.ucsc.edu).[47] In our study, 533 LUAD tissue samples and 502 LUSC tissue samples were enrolled in the DEG analysis. We also conducted qRT-PCR and IHC to confirm the expression and prognostic value of α7 nAChR in our dataset. This gave us strong evidence that α7 nAChR plays important roles in the progression of lung cancer. However, there were also several limitations in the present study:
-
1.
the data provided by TCGA was at the mRNA expression level, but not at the protein expression level;
-
2.
the normal tissues in TCGA data-set were not matched by tumor tissue one by one;
-
3.
the amount of normal tissue was much less than tumor tissue;
-
4.
there was a lack of experiments to prove the function of α7 nAChR in cell lines; and
-
5.
there was a lack of chemotherapy or radiotherapy after surgery. Further studies should focus on the mechanism of α7 nAChR in lung cancer.
The present study has provided strong evidence that α7 nAChR was upregulated in lung cancer and associated with worse prognosis. Importantly, this gene may be a good target for lung cancer treatment and further research should focus on developing new inhibitors of α7 nAChR.
Acknowledgment
We gratefully acknowledge the contributions from the TCGA Research Network and we really acknowledge all patients enrolled in this study. We also acknowledge Dr. Danli Sheng for her great contributions on modifying the language.
Author contributions
Conceptualization: Guoyuan Ma, Delin Ji, Xiao Qu, Qi Liu, Jiajun Du.
Data curation: Delin Ji, Xiao Qu.
Formal analysis: Delin Ji, Guanghui Wang.
Funding acquisition: Jiajun Du.
Investigation: Delin Ji, Jiajun Du.
Methodology: Guoyuan Ma, Delin Ji, Xiao Qu, Guanghui Wang.
Project administration: Delin Ji, Guanghui Wang.
Resources: Guoyuan Ma, Delin Ji, Shaorui Liu, Xudong Yang, Qi Liu, Jiajun Du.
Software: Shaorui Liu, Xudong Yang.
Supervision: Jiajun Du.
Validation: Xiao Qu, Jiajun Du.
Visualization: Xiao Qu, Jiajun Du.
Writing – original draft: Guoyuan Ma, Xiao Qu, Shaorui Liu, Xudong Yang, Guanghui Wang, Qi Liu, Jiajun Du.
Writing – review & editing: Guoyuan Ma, Xiao Qu, Shaorui Liu, Xudong Yang, Guanghui Wang, Qi Liu, Jiajun Du.
Supplementary Material
Footnotes
Abbreviation: ACh = acetylcholine, AChRs = acetylcholine receptors, CNV = copy number variations, DEG = differentially expressed genes, EDTA = ethylenediaminetetraacetic acid, EMT = epithelial to mesenchymal transition, GDC = Genomic Data Commons, GWAS = genome-wide association studies, HemI = heatmap illustrator, IHC = immunohistochemistry, LUAD = lung adenocarcinoma, LUSC = lung squamous cell carcinoma, M2R = M2 muscarinic receptor, mAChR = muscarinic acetylcholine receptors, nAChR = nicotinic acetylcholine receptors, NNK = nicotine-specific metabolites named 4-(methyInitrosamino)-1-(3-pyridyl)-1 -butanone, NNN = N-nitrosonornicotine, NSCLC = non-small cell lung cancer, OQL = The Onco Query Language, OS = overall survival, PBS = phosphate-buffered saline, qRT-PCR = quantitative real-time polymerase chain reaction, RNA = ribonucleic acid, ROC = receiver operating characteristic, SNPs = single-nucleotide polymorphisms, SPSS = statistical package for social sciences, TCGA = the cancer genome atlas, TNM = tumor-node-metastasis.
Guoyuan Ma and and Delin Ji contributed equally to this work.
This work was supported by National Natural Science Foundation of China (81672288), National Natural Science Foundation of China (81602009), and Provincial science and technology development plan of Shandong (2015GSF118063).
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7–30. [DOI] [PubMed] [Google Scholar]
- [2].Chen Z, Fillmore CM, Hammerman PS, et al. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 2014;14:535–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin 2016;66:271–89. [DOI] [PubMed] [Google Scholar]
- [4].Spindel E. Cholinergic targets in lung cancer. Curr Pharm Des 2016;22:2152–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gotti C, Zoli M. Nicotine inside neurons. Oncotarget 2016;7:81977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Grando SA. Connections of nicotine to cancer. Nat Rev Cancer 2014;14:419–29. [DOI] [PubMed] [Google Scholar]
- [7].Zhang C, Ding X-P, Zhao Q-N, et al. Role of α7-nicotinic acetylcholine receptor in nicotine-induced invasion and epithelial-to-mesenchymal transition in human non-small cell lung cancer cells. Oncotarget 2016;7:59199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Pillai S, Trevino J, Rawal B, et al. β-Arrestin-1 mediates nicotine-induced metastasis through E2F1 target genes that modulate epithelial–mesenchymal transition. Cancer Res 2015;75:1009–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhao Q, Yue J, Zhang C, et al. Inactivation of M2 AChR/NF-κB signaling axis reverses epithelial-mesenchymal transition (EMT) and suppresses migration and invasion in non-small cell lung cancer (NSCLC). Oncotarget 2015;6:29335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhao Q, Gu X, Zhang C, et al. Blocking M2 muscarinic receptor signaling inhibits tumor growth and reverses epithelial-mesenchymal transition (EMT) in non-small cell lung cancer (NSCLC). Cancer Biol Ther 2015;16:634–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 2008;452:633–7. [DOI] [PubMed] [Google Scholar]
- [12].Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 2008;452:638–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25. 1. Nature Genet 2008;40:616–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Heusch WL, Maneckjee R. Signalling pathways involved in nicotine regulation of apoptosis of human lung cancer cells. Carcinogenesis 1998;19:551–6. [DOI] [PubMed] [Google Scholar]
- [15].Dasgupta P, Rastogi S, Pillai S, et al. Nicotine induces cell proliferation by β-arrestin–mediated activation of Src and Rb–Raf-1 pathways. J Clin Invest 2006;116:2208–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. AACR 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBio Portal. Sci Signal 2013;6:l1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Network CGAR. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Network CGAR. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Deng W, Wang Y, Liu Z, et al. HemI: a toolkit for illustrating heatmaps. PloS One 2014;9:e111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Van der Schouw Y, Verbeek A, Ruijs J. ROC curves for the initial assessment of new diagnostic tests. Fam Pract 1992;9:506–11. [DOI] [PubMed] [Google Scholar]
- [22].Metz CE. Basic principles of ROC analysis. Paper presented at: Seminars in nuclear medicine 1978. [DOI] [PubMed] [Google Scholar]
- [23].Ni Y, Meng L, Wang L, et al. MicroRNA-143 functions as a tumor suppressor in human esophageal squamous cell carcinoma. Gene 2013;517:197–204. [DOI] [PubMed] [Google Scholar]
- [24].Ma H, Wang L, Zhang T, et al. Loss of β-arrestin1 expression predicts unfavorable prognosis for non-small cell lung cancer patients. Tumour Biol 2016;37:1341–7. [DOI] [PubMed] [Google Scholar]
- [25].Dasgupta P, Rizwani W, Pillai S, et al. ARRB1-mediated regulation of E2F target genes in nicotine-induced growth of lung tumors. J Natl Cancer Inst 2011;103:317–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wu C, Hu Z, Yu D, et al. Genetic variants on chromosome 15q25 associated with lung cancer risk in Chinese populations. Cancer Res 2009;69:5065–72. [DOI] [PubMed] [Google Scholar]
- [27].Flora AV, Zambrano CA, Gallego X, et al. Functional characterization of SNPs in CHRNA3/B4 intergenic region associated with drug behaviors. Brain Res 2013;1529:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Qu X, Wang K, Dong W, et al. Association between two CHRNA3 variants and susceptibility of lung cancer: a meta-analysis. Sci Rep 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yoo SS, Lee SM, Do SK, et al. Unmethylation of the CHRNB4 gene is an unfavorable prognostic factor in non-small cell lung cancer. Lung Cancer 2014;86:85–90. [DOI] [PubMed] [Google Scholar]
- [30].Scherf DB, Sarkisyan N, Jacobsson H, et al. Epigenetic screen identifies genotype-specific promoter DNA methylation and oncogenic potential of CHRNB4. Oncogene 2013;32:3329–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jia Y, Sun H, Wu H, et al. Nicotine inhibits cisplatin-induced apoptosis via regulating α5-nAChR/AKT signaling in human gastric cancer cells. PloS One 2016;11:e0149120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ma X, Jia Y, Zu S, et al. Alpha5 nicotinic acetylcholine receptor mediates nicotine-induced HIF-1α and VEGF expression in non-small cell lung cancer. Toxicol Appl Pharmacol 2014;278:172–9. [DOI] [PubMed] [Google Scholar]
- [33].Krais AM, Hautefeuille AH, Cros M-P, et al. CHRNA5 as negative regulator of nicotine signaling in normal and cancer bronchial cells: effects on motility, migration and p63 expression. Carcinogenesis 2011;32:1388–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Schuller HM. Is cancer triggered by altered signalling of nicotinic acetylcholine receptors? Nat Rev Cancer 2009;9:195–205. [DOI] [PubMed] [Google Scholar]
- [35].Lam DC-l, Girard L, Ramirez R, et al. Expression of nicotinic acetylcholine receptor subunit genes in non–small-cell lung cancer reveals differences between smokers and nonsmokers. Cancer Res 2007;67:4638–47. [DOI] [PubMed] [Google Scholar]
- [36].Paleari L, Catassi A, Ciarlo M, et al. Role of alpha7-nicotinic acetylcholine receptor in human non-small cell lung cancer proliferation. Cell Prolif 2008;41:936–59. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [37].Wang Y, Pereira E, Maus A, et al. Human bronchial epithelial and endothelial cells express α7 nicotinic acetylcholine receptors. Molr Pharmacol 2001;60:1201–9. [DOI] [PubMed] [Google Scholar]
- [38].Al-Wadei HA, Al-Wadei MH, Masi T, et al. Chronic exposure to estrogen and the tobacco carcinogen NNK cooperatively modulates nicotinic receptors in small airway epithelial cells. Lung Cancer 2010;69:33–9. [DOI] [PubMed] [Google Scholar]
- [39].Fucile S. Ca 2+ permeability of nicotinic acetylcholine receptors. Cell Calcium 2004;35:1–8. [DOI] [PubMed] [Google Scholar]
- [40].Schuller HM. Neurotransmitter receptor-mediated signaling pathways as modulators of carcinogenesis. Neuronal Activity in Tumor Tissue. Karger Publishers; 2007;39:45–63. [DOI] [PubMed] [Google Scholar]
- [41].Improgo MR, Tapper AR, Gardner PD. Nicotinic acetylcholine receptor-mediated mechanisms in lung cancer. Biochem Pharmacol 2011;82:1015–21. [DOI] [PubMed] [Google Scholar]
- [42].Egleton RD, Brown KC, Dasgupta P. Nicotinic acetylcholine receptors in cancer: multiple roles in proliferation and inhibition of apoptosis. Trends Pharmacol Sci 2008;29:151–8. [DOI] [PubMed] [Google Scholar]
- [43].Grozio A, Paleari L, Catassi A, et al. Natural agents targeting the α7-nicotinic-receptor in NSCLC: A promising prospective in anti-cancer drug development. Int J Cancer 2008;122:1911–5. [DOI] [PubMed] [Google Scholar]
- [44].Brown KC, Perry HE, Lau JK, et al. Nicotine induces the up-regulation of the α7-nicotinic receptor (α7-nAChR) in human squamous cell lung cancer cells via the Sp1/GATA protein pathway. J Biol Chem 2013;288:33049–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yang L, Lu X, Qiu F, et al. Duplicated copy of CHRNA7 increases risk and worsens prognosis of COPD and lung cancer. Eur J Hum Genet 2015;23:1019–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Plummer HK, Dhar M, Schuller HM. Expression of the α7 nicotinic acetylcholine receptor in human lung cells. Respir Res 2005;6:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Peng L, Bian XW, Xu C, et al. Large-scale RNA-seq transcriptome analysis of 4043 cancers and 548 normal tissue controls across 12 TCGA cancer types. Sci Rep 2015;5:13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


