Abstract
EAG1 channels belong to the KCNH family of voltage gated potassium channels. They are expressed in several brain regions and increased expression is linked to certain cancer types. Recent cryo-EM structure determination finally revealed the structure of these channels in atomic detail, allowing computational investigations. In this study, we performed molecular dynamics simulations to investigate the ion binding sites and the dynamical behavior of the selectivity filter. Our simulations suggest that sites S2 and S4 form stable ion binding sites, while ions placed at sites S1 and S3 rapidly switched to sites S2 and S4. Further, ions tended to dissociate away from S0 within less than 20 ns, due to increased filter flexibility. This was followed by water influx from the extracellular side, leading to a widening of the filter in this region, and likely non-conductive filter configurations. Simulations with the inactivation-enhancing mutant Y464A or Na+ ions lead to trapped water molecules behind the SF, suggesting that these simulations captured early conformational changes linked to C-type inactivation.
Keywords: Molecular dynamics simulation, KCNH1 channel, Ion binding sites, Y464A mutant, Na+ binding sites, C-type inactivation
1. Introduction
EAG channels belong to the KCNH family, which includes EAG (ether-à-go-go), ERG (EAG-related gene) and ELK (EAG-like K+) channels [1]. Mammalian EAG1 channels are strongly expressed in several brain regions, including brain stem, cerebellum, hippocampus and olfactory bulb [2], but the specific physiological roles are still not completely understood. More is known about their role in disease. Increased expression of these proteins is linked to certain cancer types including prostate, colon, ovary, melanoma, liver and thyroid cancer cells. Inhibition by blockers, antibodies, and siRNA has been shown to decrease proliferation of tumor cell lines (for recent review see Ref. [3]). Recently, de novo mutations in EAG1 have been shown to cause Temple-Baraitser syndrome, a developmental disorder characterized by intellectual disability and epilepsy [4]. Thus, these channels have great therapeutic potential.
EAG1 channels have unique biophysical properties, quite distinct from the closely related hERG1 channel [5]. Depolarization leads to relatively fast activation currents, which exhibit a prominent Cole-Moore shift, i.e., activation is delayed and slowed when currents are elicited from more negative holding potentials [6,7]. EAG1 channels show intrinsic, voltage-dependent, slow, C-type inactivation [8] in contrast to hERG1 channels, which exhibit very fast, voltage-dependent C-type inactivation [5]. The mechanistic and structural basis of C-type inactivation was extensively studied in several K+ channels using site-directed mutagenesis, X-ray crystallography, and molecular dynamics (MD) simulations. These studies suggest that C-type inactivation reduces K+ conductance via subtle voltage-dependent conformational changes of the selectivity filter, which can be preceded by a transient loss of K+ selectivity [9–15]. C-type inactivation in most K+ channels, including the closely related hERG1 channels, can be slowed by elevated extracellular [K+] and extracellular TEA [9,16]. Further, a number of K+ channels were reported to become transiently Na+ conductive, upon depletion of K+ ions, due to conformational changes in the selectivity filter, leading to C-type inactivation [17,18]. hEAG1 inactivation is unique, since it is not slowed by elevated extracellular [K+] or extracellular TEA, but can be strongly enhanced by mutation of residue Tyr464, located in the S6 helix (see Fig. 1A), or a small molecule activator [8].
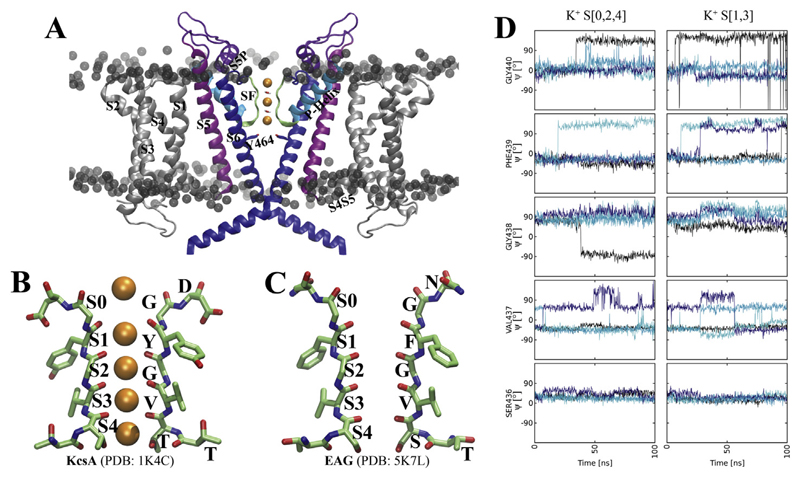
Fig. 1. Schematic representation of the transmembrane domain (TMD) of rEAG1.
A) side view of the TMD of rEAG1, with helices S1-S6 labelled. The phospholipid head groups are shown as grey spheres. The filter K+ ions are coloured orange. B) side view of two opposing selectivity filter subunits of KcsA with K+ ions shown as orange spheres. C) side view of two opposing selectivity filter subunits of rEAG1. D) Ψ angles of SF residues over simulation time plotted in different colours.
In 2016, the Mackinnon lab solved the closed state structure of the rat orthologue of EAG1 (rEAG1) at 3.78 Å resolution using single-particle cryo–electron (cryo-EM) microscopy [19]. This structure reveals that similar to other K+ channels, EAG channels are formed by four subunits surrounding a central pore. Each subunit contains six transmembrane segments S1–S6 (Fig. 1 A). The cytosolic regions contain a Per–Arnt–Sim domain at the N-terminus [20], a C-terminal C-linker domain, and a cyclic nucleotide-binding homology domain at the C-terminus [21,22]. Unlike other Kv channels, the S4-S5 linker connecting the pore module with the voltage-sensing module, is only a short, 5-residue loop, which interacts with its own subunit as shown in Fig. 1A [19].
EAG1 channels display a conventional selectivity filter (SF) structure, as can be seen in other K+ channel crystal structures (Fig. 1B and C). The high resolution structure of the bacterial K+ channel KcsA revealed that backbone carbonyl oxygen atoms from each subunit assemble to form six ion binding sites, one outside the filter (denoted S0), four in the filter (denoted S1–S4) and one in the cavity [14]. MD simulations of different K+ channels, revealed that K+ ions can either occupy sites S1 and S3 or sites S2 and S4, while transitions between these two states have been observed to occur on the nanosecond timescale (for recent review on simulations see Ref. [23]). This indicates an almost barrier-less diffusion of ions.
The highly conserved signature sequence TXGYG, where X represents any hydrophobic residue, diverges to TSVGFG in the KCNH family, with the conserved tyrosine replaced by a phenylalanine (Fig. 1C). The resolution of the EAG structure is too low to observe actual ion binding to the selectivity filter. Nevertheless, it is reasonable to assume that similar ion binding modes as in KcsA exist (shown in Fig. 1B).
Numerous MD simulations on different K+ channels have provided detailed insights into the behavior of the SF [10,12,13,23–27]. So far, no simulation studies have focused on the dynamics of the selectivity filter of the EAG1 channel.
Thus, the aim of the current study was to investigate the ion binding sites and dynamical behavior of the selectivity filter of the EAG1 channel, using atomistic MD simulations on the 100 ns time scale.
2. Methods
2.1. MD simulations
MD simulations were performed using the program Gromacs version 5.1.2 [28] with the same parameters as described previously [29]. Briefly, the transmembrane part of the EAG1 channel (pdb code: 5K7L) was embedded in a lipid bilayer consisting of 456 1-Palmitoyl-2-oleoylphosphatidylcholine lipids and solvated using the TIP3P water model [30], with a potassium chloride concentration of 150 mM. The amber99sb force field [31] was used for the protein. Berger lipid parameters were used for the membrane [32,33].
2 times 100-ns MD simulations were performed with different ion configurations (S1,S3; S0,S2,S4; S0-S4; as well as the Y464A mutant channel (ions in S0,S2,S4) and with Na+ ions in the SF (placed at sites S0,S2,S4). Before each simulation, the protein atoms were restrained by a force constant of 1000 kJ/mol/nm2 to their initial position, and 1000 conjugate gradient energy-minimization steps were performed, followed by 5 ns of equilibrium simulation.
3. Results/discussion
The conformational flexibility of the EAG1 structure in the closed state was analysed using MD simulations. Repeated simulations of the channel, embedded in a lipid-bilayer membrane, with different ion binding configurations, identified in previous simulations [25,34,35] were performed. Due to electrostatic repulsion, it was suggested that ions are located at alternate sites within the selectivity filter, separated by water molecules. However, more recently an alternative, direct knock-on mechanism without alternating water molecules in the filter was proposed [36]. Thus, we performed simulations with different filter ion configurations and monitored their time dependent behavior. Specifically, in system 1 (denoted S[0,2,4]) ions where placed at site S0, S2 and site S4, while water molecules where placed at sites S1 and S3 (for starting configurations see Fig. 2B). In system 2 (denoted S [1,3]), ions where placed at sites S1 and S3, with water molecules at sites S0, S2 and S4 (Fig. 2F). In system 3 (denoted S[0,1,2,3,4]), ions where placed at sites S0–S4, without water molecules (shown in Supplemental Fig. 1). The stability of the different systems, measured as the root mean square deviation (RMSD) as a function of time is plotted in Supplemental Fig. 3. With RMSD values of 3 Å, the different closed state systems are comparable to previous values for low resolution structures [37].; K+ Ion Coordination in the Selectivity Filter.
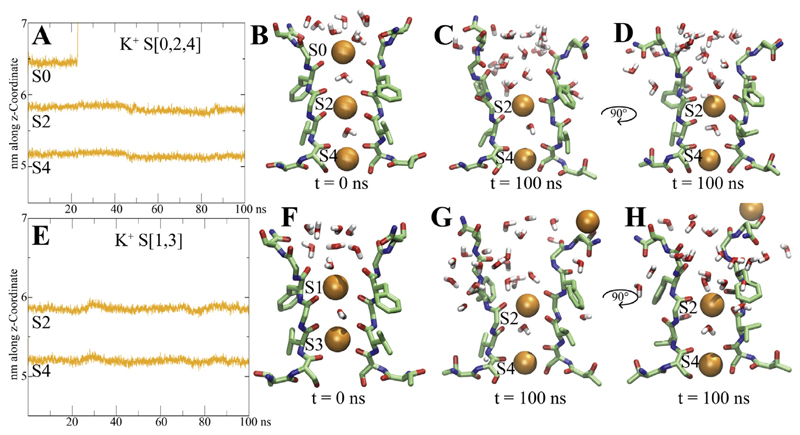
Fig. 2. Ion binding sites and dynamics over simulation time.
A, E) Positions of K+ ions along the z-coordinate are plotted over simulation time B, F) starting configurations for systems S[0,2,4] and S [1,3]. C,D, G and H) end states after 100 ns, two opposing subunits shown each. K+ ions are shown as orange spheres, water molecules within 6 Å are shown in stick representation.
To investigate the behavior of the ions in the different binding sites, we monitored the ion positions over time, as shown in Fig. 2 A, E and Supplemental Fig. 1. No ion-conduction event was observed within 100 ns, due to the closed state of the activation gate, a dewetted cavity and the lack of ion gradient or applied voltage. Nevertheless, individual ion movements were observed in the selectivity filter.
In both S[0,2,4] simulations, ions at site 2 and 4 remained stably bound to their sites, due to optimal coordination of backbone carbonyls and side-chain hydroxyl groups of S436, with coordination numbers of 6–8. In contrast, the ion from site S0 dissociated into the extracellular solvent within short time (after 23ns in run 1 and ~ 1ns in run2), due to increased flexibility of the selectivity filter, as measured via monitoring the angles of the SF backbone carbonyl atoms over time. As can be seen in Fig. 1D, two carbonyl groups of residue G440 and up to two carbonyl groups of F439 rotate away from the pore axis (‘flip’), indicated by large rotations of the Ψ angles from their starting positions. This leads to defective ion coordination at site 0, which is rapidly propagated to site S1 (Fig. 2CD). Generally, a widening of the upper part of the selectivity filter was observed in both runs, accompanied by increased water influx, and complete loss of ion coordination for sites S0 and S1.
Interestingly, a similar behavior of the SF was reported for the closely related hERG (Kv11.1) channel in MD simulations based on homology models [10,25]. This suggests that increased flexibility of the filter might be an intrinsic feature of the whole KCNH family. Based on computational investigations from Ceccarini et al., 2012 [25], focusing on the conductance mechanism of the closely related hERG1 channel, it was suggested that the filter might visit partially unfolded states during permeation, distinct from non-conductive states. Rather, these states could reflect low conductance states, explaining the low conductivity of these channels. The conductance rates for hEAG1 and hERG1 are similar. However there is a clear difference in the rate of inactivation, which is unusually fast in hERG1 [38], but very slow in EAG1 [8] channels.
To further prope if the observed structural changes in the SF are due to increased filter flexibility, or possibly represent early conformational changes during inactivation, we monitored the number of water molecules behind the SF (see Fig. 3). Water influx (three water molecules) behind the cavity has been previously shown to be linked to C-type inactivation in KcsA [12]. However, in contrast to KcsA, the filter geometry at site S1 is not “narrow” or “pinched” [11], instead the filter is dilated at the extracellular side, as has been previously suggested as alternative conformational change during C-type inactivation by Hoshi et al. [39]. However, only one water molecule was bound behind the SF, suggesting that the SF might not be in an early inactivated state. Additional simulations, particularly with an open state conformation and applied driving force will be needed to further test if the observed filter states represent low conductive states, or early inactivated states.
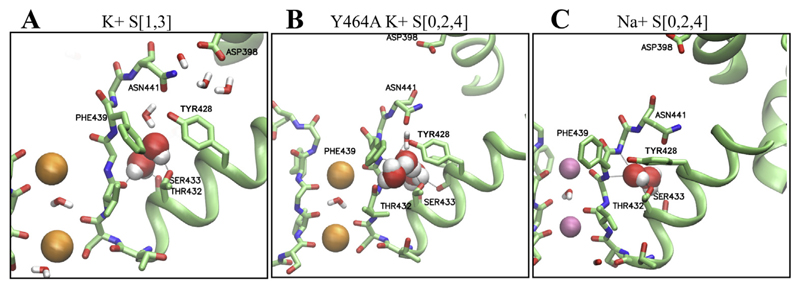
Fig. 3. Trapped water molecules in the cavity behind the SF.
A) the trapped water molecules in system S [1,3] are shown as spheres. Hydrogen bonds are indicated as black dotted lines. B,C) trapped water molecules in the Y464A inactivation enhancing mutant and C) with Na+ ions (pink spheres) bound to the SF.; Na+ binding to the EAG1 filter; Several K+ channels were reported to become transiently Na+ conductive, upon depletion of K+ ions due to conformational changes in the selectivity filter, during inactivation gating [17,18].
To discriminate protein-mechanistic from ion-induced effects possibly influencing filter flexibility, we performed additional simulations with varying ion occupancy [1,3] and [0, 1, 2, 3, 4] in the filter (see Fig. 2E and Supplemental Fig. 1 for starting configurations. Analysis of the trajectories revealed that irrespective of the starting system, coordination of ions at site 0 rapidly was lost, due to conformational changes of the carbonyl backbone oxygens starting at G440. These changes rapidly propagated to site S1. In particular, the carbonyl group of F439 revealed increased flexibility as shown in Fig. 1D (right side) and Supplemental Fig. 2. In system S [1,3] ions instantaneously (within the first 20 ps) switched to positions 2–4, where they were stably coordinated over 100 ns Similar loss of ion coordination at site S0 and S1 and filter flexibility was observed in system S[01234], as shown in Supplemental Fig. 1. This further supports that increased filter flexibility might indeed be an inherent property of the KCNH family, not dependent on the ion configurations bound to the SF. Nevertheless, the filter geometry was clearly influenced by the starting ion configurations. In contrast to the S[0,2,4] system, water influx of at least two molecules behind the SF was observed in all S [1,3] and S[0,1,2,3,4] runs, as can be seen in Fig. 3. Based on simulations on KcsA [12], it is tempting to speculate that the conformational changes observed in systems S [1,3] and S[0,1,2,3,4,] represent early inactivated states.
Hydrogen bond networks around the SF have been implicated in C-type inactivation in different K+ channels [11,13,15]. To test, if changed hydrogen bond networks behind the SF, led to conformational changes, enabling water leakage behind the SF, we monitored the hydrogen bonds between residues Y428, N441 and D398 as a function of time. As shown in Supplemental Fig. 4, there is no clear difference in the hydrogen bond network observable between the different simulation systems.
Thus to further probe, if the observed filter changes could represent early inactive conformations of the EAG1 channel, we introduced the Y464A mutation located in the S6 helix (see Fig. 1A for location), which was previously shown to enhance C-type inactivation [8].; Behavior of the Y464A inactivation-enhancing mutant.
Simulations were started with K+ ions placed at sites 0, 2 and 4 and two times 100 ns simulations were performed. Similar to WT simulations, the carbonyl oxygens of G440 were displaced outwards in the Y464A mutant, which lead to an increased diameter of the selectivity filter and destroyed ion coordination sites at position S0 (Fig. 4A, D, D). After K+ dissociation, water molecules entered the filter and led to further carbonyl group flips, at sites S1 and distortion of the upper part of the selectivity filter as shown in Fig. 4C and D and Supplemental Figs. 1 and 2. Again, water molecules occupied the space behind the filter, possibly rendering the channel non-conductive, due to early conformational changes during C-type inactivation (see Fig. 3B).
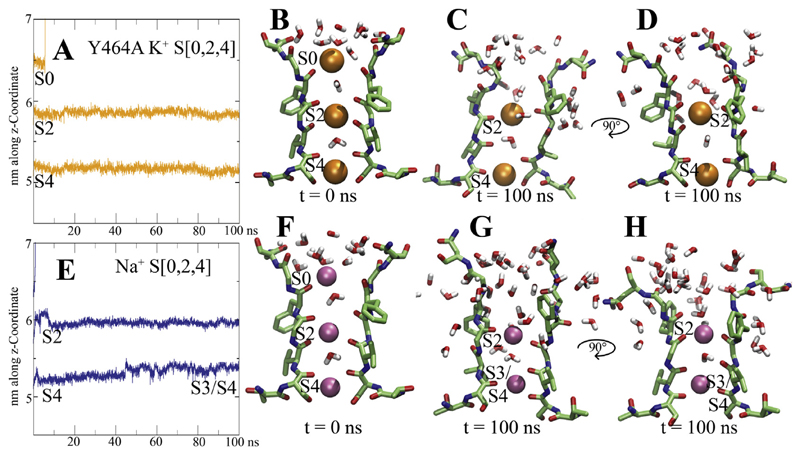
Fig. 4. Ion binding sites and dynamics of the Y464A and Na+ systems.
A, E) Position of the different K+ ions along the z-coordinate are plotted over simulation time B, F) starting configurations for systems Y464A and with Na+ ions, respectively. C,D, G, H) end states after 100 ns, two opposing subunits shown each. K+ ions are shown as orange spheres, water molecules within 6 Å are shown in stick representation, Na+ ions are shown as pink spheres.
Thus, we investigated the influence of Na+ ions on the EAG1 filter geometry. To this end, we placed Na+ ions at sites S0, S2 and S4, separated by water molecules and performed 2 times 100 ns MD simulations. The conformational changes observed in these simulations are clearly distinct from all above described systems. As can be seen in Fig. 4E, two of the three sodium ions remained bound to the filter over 100 ns, while the ion placed at site S0 rapidly dissociated. In both runs, the Na+ ion bound to site four, gradually moved upwards to site three, where it was coordinated by carbonyl groups from S634 and V635 after 100 ns The ion placed at site S2 moved upward slightly within the first 10 ns, leading to coordination of only four carbonyl oxygen groups of residue G438. This novel binding site was stable for the last 90 ns Additional ion coordination was achieved via water molecules, which passed from the extracellular site to sites S3 and S4. Despite different ion coordination behavior, the final filter geometry is not so different from systems S [1,3] and S[0,1,2,3,4]. Again, water influx behind the SF was observed (shown in Fig. 3C), suggestive of conformations representing early inactivation.
Summarizing, our studies revealed that the EAG1 selectivity filter is intrinsically highly dynamic, in agreement with previous theoretical studies on the closely related hERG1 channel [10,25]. In all six WT runs, ion coordination at sites S0 and S1 was rapidly disrupted. A similar behavior was observed in the Y464A mutant runs and even when replacing filter K+ ions with Na+ ions.
In contrast, but consistent with previous theoretical studies (for recent review see Ref. [23]), ions were favourably bound to sites S2 and S4. Further, our simulations reveal that the starting ion configurations influence, if water molecules could become trapped behind the SF. It is tempting to speculate that the end states of systems S [1,3] and S[0,1,2,3,4] represent early inactivated states of the EAG1 SF, since at least two water molecules are buried behind the SF (maintained over the whole simulation time) in all these simulations. It has been previously shown in μs long MD simulations that three water molecules bound behind the SF can lock the filter of KcsA in the inactivated state [12].
Further studies, including free energy calculations are necessary to analyse in depth, why site S0 is energetically less favourable for K+ coordination, as suggested by our unbiased free MD simulations. It has to be stressed that longer simulation times, preferably with an open state conformation will be needed to further validate our observations, based on the recently solved closed state cryo-EM structure of rEAG1.
Supplementary Material
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.bbrc.2017.01.064.
Acknowledgements
We thank Eva-Maria Plessl-Zangerl for help with scripting. This work was supported by the Austrian Science Fund (Grant No. W1232). The computational results presented have been achieved using the Vienna Scientific Cluster (VSC).
Abbreviations
- Cryo-EM
Cryo-electron microscopy
- SF
selectivity filter
- MD
molecular dynamics simulation
- EAG
ether-à-go-go
- TMD
transmembrane domain
- RMSD
root mean square deviation
Footnotes
Transparency document
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.bbrc.2017.01.064.
References
- [1].Warmke JW, Ganetzky B. A family of potassium channel genes related to eag in Drosophila and mammals. Proc Natl Acad Sci U S A. 1994;91:3438–3442. doi: 10.1073/pnas.91.8.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Martin S, Lino de Oliveira C, Mello de Queiroz F, Pardo LA, Stühmer W, Del Bel E. EAG1 potassium channel immunohistochemistry in the CNS of adult rat and selected regions of human brain. Neuroscience. 2008;155:833–844. doi: 10.1016/j.neuroscience.2008.05.019. [DOI] [PubMed] [Google Scholar]
- [3].Pardo LA, Stühmer W. The roles of K(+) channels in cancer. Nat Rev Cancer. 2014;14:39–48. doi: 10.1038/nrc3635. [DOI] [PubMed] [Google Scholar]
- [4].Simons C, Rash LD, Crawford J, Ma L, Cristofori-Armstrong B, Miller D, Ru K, Baillie GJ, Alanay Y, Jacquinet A, Debray F-G, et al. Mutations in the voltage-gated potassium channel gene KCNH1 cause Temple-Baraitser syndrome and epilepsy. Nat Genet. 2015;47:73–77. doi: 10.1038/ng.3153. [DOI] [PubMed] [Google Scholar]
- [5].Vandenberg JI, Perry MD, Perrin MJ, Mann SA, Ke Y, Hill AP. hERG K(+) channels: structure, function, and clinical significance. Physiol Rev. 2012;92:1393–1478. doi: 10.1152/physrev.00036.2011. [DOI] [PubMed] [Google Scholar]
- [6].Terlau H, Ludwig J, Steffan R, Pongs O, Stühmer W, Heinemann SH. Extracellular Mg2+ regulates activation of rat eag potassium channel. Pflugers Arch. 1996;432:301–312. doi: 10.1007/s004240050137. [DOI] [PubMed] [Google Scholar]
- [7].Lin MCA, Papazian DM. Differences between ion binding to eag and HERG voltage sensors contribute to differential regulation of activation and deactivation gating. Channels Austin Tex. 2007;1:429–437. doi: 10.4161/chan.1.6.5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Garg V, Sachse FB, Sanguinetti MC. Tuning of EAG K(+) channel inactivation: molecular determinants of amplification by mutations and a small molecule. J Gen Physiol. 2012;140:307–324. doi: 10.1085/jgp.201210826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].López-Barneo J, Hoshi T, Heinemann SH, Aldrich RW. Effects of external cations and mutations in the pore region on C-type inactivation of Shaker potassium channels. Recept Channels. 1993;1:61–71. [PubMed] [Google Scholar]
- [10].Stansfeld PJ, Grottesi A, Sands ZA, Sansom MSP, Gedeck P, Gosling M, Cox B, Stanfield PR, Mitcheson JS, Sutcliffe MJ. Insight into the mechanism of inactivation and pH sensitivity in potassium channels from molecular dynamics simulations. Biochem (Mosc.) 2008;47:7414–7422. doi: 10.1021/bi800475j. [DOI] [PubMed] [Google Scholar]
- [11].Cuello LG, Jogini V, Cortes DM, Perozo E. Structural mechanism of C-type inactivation in K+ channels. Nature. 2010;466:203–208. doi: 10.1038/nature09153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ostmeyer J, Chakrapani S, Pan AC, Perozo E, Roux B. Recovery from slow inactivation in K+ channels is controlled by water molecules. Nature. 2013;501:121–124. doi: 10.1038/nature12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Köpfer DA, Hahn U, Ohmert I, Vriend G, Pongs O, de Groot BL, Zachariae U. A molecular switch driving inactivation in the cardiac K+ channel HERG. PLoS One. 2012;7:e41023. doi: 10.1371/journal.pone.0041023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- [15].Pless SA, Galpin JD, Niciforovic AP, Kurata HT, Ahern CA. Hydrogen bonds as molecular timers for slow inactivation in voltage-gated potassium channels. eLife. 2013;2:e01289. doi: 10.7554/eLife.01289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Grissmer S, Cahalan M. TEA prevents inactivation while blocking open K+ channels in human T lymphocytes. Biophys J. 1989;55:203–206. doi: 10.1016/S0006-3495(89)82793-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Starkus JG, Kuschel L, Rayner MD, Heinemann SH. Macroscopic Na+ currents in the “Nonconducting” Shaker potassium channel mutant W434F. J Gen Physiol. 1998;112:85–93. doi: 10.1085/jgp.112.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gang H, Zhang S. Na+ permeation and block of hERG potassium channels. J Gen Physiol. 2006;128:55–71. doi: 10.1085/jgp.200609500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Whicher JR, MacKinnon R. Structure of the voltage-gated K+ channel EAG1 reveals an alternative voltage sensing mechanism. Science. 2016;353:664–669. doi: 10.1126/science.aaf8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Morais Cabral JH, Lee A, Cohen SL, Chait BT, Li M, Mackinnon R. Crystal structure and functional analysis of the HERG potassium channel N terminus: a eukaryotic PAS domain. Cell. 1998;95:649–655. doi: 10.1016/s0092-8674(00)81635-9. [DOI] [PubMed] [Google Scholar]
- [21].Brelidze TI, Gianulis EC, DiMaio F, Trudeau MC, Zagotta WN. Structure of the C-terminal region of an ERG channel and functional implications. Proc Natl Acad Sci U S A. 2013;110:11648–11653. doi: 10.1073/pnas.1306887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Marques-Carvalho MJ, Sahoo N, Muskett FW, Vieira-Pires RS, Gabant G, Cadene M, Schönherr R, Morais-Cabral JH. Structural, biochemical, and functional characterization of the cyclic nucleotide binding homology domain from the mouse EAG1 potassium channel. J Mol Biol. 2012;423:34–46. doi: 10.1016/j.jmb.2012.06.025. [DOI] [PubMed] [Google Scholar]
- [23].Furini S, Domene C. K(+) and Na(+) conduction in selective and nonselective ion channels via molecular dynamics simulations. Biophys J. 2013;105:1737–1745. doi: 10.1016/j.bpj.2013.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Oakes V, Furini S, Pryde D, Domene C. Exploring the dynamics of the TWIK-1 channel. Biophys J. 2016;111:775–784. doi: 10.1016/j.bpj.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ceccarini L, Masetti M, Cavalli A, Recanatini M. Ion conduction through the hERG potassium channel. PLoS One. 2012;7:e49017. doi: 10.1371/journal.pone.0049017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Domene C, Grottesi A, Sansom MSP. Filter flexibility and distortion in a bacterial inward rectifier K+ channel: simulation studies of KirBac1.1. Biophys J. 2004;87:256–267. doi: 10.1529/biophysj.104.039917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Domene C, Klein ML, Branduardi D, Gervasio FL, Parrinello M. Conformational changes and gating at the selectivity filter of potassium channels. J Am Chem Soc. 2008;130:9474–9480. doi: 10.1021/ja801792g. [DOI] [PubMed] [Google Scholar]
- [28].Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- [29].Linder T, Wang S, Zangerl-Plessl E-M, Nichols CG, Stary-Weinzinger A. Molecular dynamics simulations of KirBac1.1 mutants reveal global gating changes of Kir channels. J Chem Inf Model. 2015;55:814–822. doi: 10.1021/acs.jcim.5b00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- [31].Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins Struct Funct Bioinform. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Berger O, Edholm O, Jähnig F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys J. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cordomí A, Caltabiano G, Pardo L. Membrane protein simulations using amber force field and berger lipid parameters. J Chem Theory Comput. 2012;8:948–958. doi: 10.1021/ct200491c. [DOI] [PubMed] [Google Scholar]
- [34].Aqvist J, Luzhkov V. Ion permeation mechanism of the potassium channel. Nature. 2000;404:881–884. doi: 10.1038/35009114. [DOI] [PubMed] [Google Scholar]
- [35].Stansfeld PJ, Gedeck P, Gosling M, Cox B, Mitcheson JS, Sutcliffe MJ. Drug block of the hERG potassium channel: insight from modeling. Proteins Struct Funct Bioinform. 2007;68:568–580. doi: 10.1002/prot.21400. [DOI] [PubMed] [Google Scholar]
- [36].Köpfer DA, Song C, Gruene T, Sheldrick GM, Zachariae U, de Groot BL. Ion permeation in K+ channels occurs by direct Coulomb knock-on. Science. 2014;346:352–355. doi: 10.1126/science.1254840. [DOI] [PubMed] [Google Scholar]
- [37].Stary A, Wacker SJ, Boukharta L, Zachariae U, Karimi-Nejad Y, Aqvist J, Vriend G, de Groot BL. Toward a consensus model of the HERG potassium channel. ChemMedChem. 2010;5:455–467. doi: 10.1002/cmdc.200900461. [DOI] [PubMed] [Google Scholar]
- [38].Smith PL, Baukrowitz T, Yellen G. The inward rectification mechanism of the HERG cardiac potassium channel. Nature. 1996;379:833–836. doi: 10.1038/379833a0. [DOI] [PubMed] [Google Scholar]
- [39].Hoshi T, Armstrong CM. C-type inactivation of voltage-gated K+ channels: pore constriction or dilation? J Gen Physiol. 2013;141:151–160. doi: 10.1085/jgp.201210888. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.