Abstract
Epigenetic regulatory mechanisms, encompassing diverse molecular processes including DNA methylation, histone post-translational modifications, and non-coding RNAs, are essential to numerous processes such as cell differentiation, growth and development, environmental adaptation, aging, and disease states. In many cases, epigenetic changes occur in response to environmental cues and lifestyle factors, resulting in persistent changes in gene expression that affect vascular disease risk over the lifetime of the individual. Biological aging, a powerful cardiovascular risk factor, is partly genetically determined yet strongly influenced by traditional risk factors, reflecting epigenetic modulation. Quantification of specific DNA methylation patterns may serve as an accurate predictor of biological age, a concept known as the “epigenetic clock”, which could help to refine cardiovascular risk assessment. Epigenetic reprogramming of monocytes rewires cellular immune signaling and induces a metabolic shift toward aerobic glycolysis, thereby increasing innate immune responses. This form of “trained epigenetic memory” can be maladaptive, thus augmenting vascular inflammation. Somatic mutations in epigenetic regulatory enzymes leads to clonal hematopoiesis of indeterminate potential, a precursor of hematological malignancies and a recently recognized cardiovascular risk factor; moreover, epigenetic regulators are increasingly being targeted in cancer therapeutics. Thus, understanding epigenetic regulatory mechanisms lies at the intersection between cancer and cardiovascular disease and is of paramount importance to the burgeoning field of cardio-oncology.
Epigenetic regulation refers to changes in gene expression and ensuing phenotypes that occur independent of alterations in the DNA sequence per se, encompassing different levels of epigenetic modulation, including DNA methylation, post-translational histone modifications and non-coding RNAs. Epigenetic mechanisms have been shown to be associated with diverse human diseases such as cancer, cardiovascular disease, autoimmune diseases and Alzheimer’s dementia. This review is focused on key concepts pertaining to epigenetic regulatory mechanisms in vascular disease; the reader is referred to recent comprehensive review articles pertaining to the epigenetics of atherosclerosis for a more detailed overview of the topic.1,2
1. Brief review of epigenetic modifications
1.1. DNA methylation
In mammalian cells, DNA methylation is the most widely studied epigenetic modification;3 it involves the covalent binding of a methyl group to the 5’-position of cytosine residues at CpG (Cytosine preceding Guanosine) islands, which most typically suppresses transcription by impeding the binding of transcription factors to cis-DNA binding elements. Most CpG-rich genomic regions are sites of transcription initiation and are approximately 200–300 base pairs in length, with a CG content of approximately 50%, predominantly non-methylated. DNA methylation is catalyzed by three different DNA methyltransferases (DNMTs); DNMT1 is the maintenance enzyme, and its inhibition leads to passive demethylation, while DNMT3a and DNMT3b are associated with de novo methylation. Methylated CpG islands can be oxidized by ten–eleven translocation (TET) family of enzymes through the base excision repair pathway, leading to active demethylation and gene reactivation. It is also important to point out that RNA can be methylated/demethylated, thereby regulating gene expression, which represents a less well studied epigenetic regulatory mechanism.4
1.2. Histone post-translational modifications in chromatin
Histone modifications lead to changes in chromatin structure to render it “active” (euchromatin), in which DNA is accessible to transcriptional factors, or “inactive” (heterochromatin), in which DNA is inaccessible to transcriptional factors. Eight different types of modifications catalyzed by distinct enzymes have been described. The two most widely studied histone modifications are methylation and acetylation; less well studied modifications include phosphorylation, sumoylation, ubiquitination, ADP-ribosylation, deimination, and proline isomerization.5 Histone methyltransferases (HMTs) facilitate histone methylation through the transfer of one, two, or three methyl groups from S-adenosyl-L-methionine to lysine or arginine residues of histone proteins. Histone methylation can be associated with either gene activation or repression depending on the amino acid residue modified and the extent of methylation. For example, tri-methylation of histone 3 at lysine 27 (H3K27me3) is a repressive mark enriched at inactive or silent gene promoters, while di- and tri- methylation of lysine 4 at histone 3 (H3K4me2, and H3K4me3 respectively) are typically enriched at transcriptionally active gene promoters. Histone acetylation is the enzymatic addition of an acetyl group (COCH3) from acetyl coenzyme A by HATs. Generally, acetylation of gene promoters correlates with transcriptional activation such as the acetylation of histone 3 at lysine 9 (H3K9ac), which is typically enriched at transcriptionally active gene promoters, whereas its removal is associated with gene repression.
1.3. Non-coding RNAs
Accumulating evidence points to the role of non-coding RNAs (ncRNAs) in various physiological and pathological processes and in response to environmental stress. ncRNAs are classified as infrastructural (i.e., small nuclear and nucleolar RNAs, ribosomal RNAs) and regulatory RNAs [i.e., microRNAs (miRNAs), long non-coding RNAs (lncRNAs), Piwi-interacting RNAs (piRNAs), small interfering RNAs (siRNAs)]. ncRNAs are under intense investigation as biomarkers and modulators of vascular disease, as well as targets for therapeutic intervention. The reader is referred to recent comprehensive review articles pertaining to this topic for a detailed overview.6
2. Epigenetics and vascular disease risk
While there is a heritable component to vascular disease risk, studies on identical twins who share the same genome yet exhibit different vascular disease event rates and life spans suggest the importance of a non-genetic component of vascular disease risk in humans.7 Indeed, while advancement in cardiovascular disease (CVD) genomic research has progressed rapidly over the past decade, resulting in the development of commercially available genetic risk assays, the performance of these assays has been weaker than anticipated, and they have not yet been widely adopted as clinically applicable tools.8 Furthermore, lifestyle factors have been shown to be independently associated with susceptibility to CVD, regardless of an individual’s genetic risk. In several large studies, participants with an elevated genetic risk of CVD leading a favorable lifestyle had nearly a 50% lower relative risk of coronary artery disease compared to those leading an unfavorable lifestyle.9–12 Conversely, the presumably protective effects of a low genetic risk score were abrogated in participants who lead an unfavorable lifestyle. These findings underscore the importance of environmental-genetic interactions in defining vascular disease risk, with a significant component linked to epigenetic mechanisms. Interestingly, emerging evidence suggests that the effects of traditional risk factors, such as hypercholesteremia, on CVD risk differ between elderly and young patients, pointing to unidentified risk factors that contribute to CVD in elderly population.13,14 These observations attest to our limited understanding of the mechanisms by which age, genetic risk, and lifestyle factors interact to convey CVD risk. Emerging data suggest that the interaction between these factors is regulated by epigenetic mechanisms that may be quantified to predict CVD risk and discover novel therapeutic targets.
3. Chronological vs DNA methylation age: the epigenetic clock
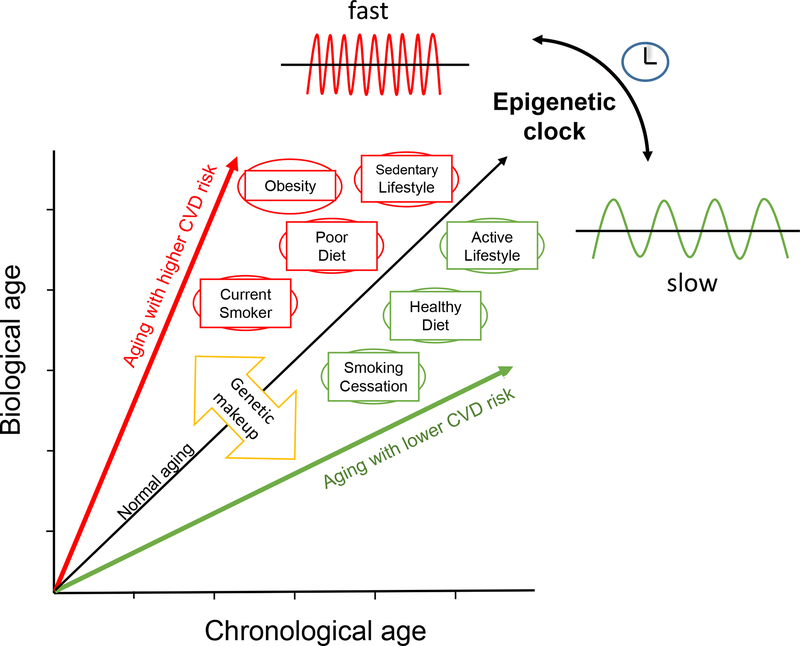
Age is a strong and independent risk factor for CVD, even after statistical adjustment of traditional CVD risk factors.15,16 Biological aging is in part genetically determined but is also strongly influenced by traditional risk factors, which can accelerate the aging process as well as CVD onset and progression. It thus stands to reason that chronological age is an imprecise determinant of biological age and thus lacks fidelity as a numerical marker of vascular disease risk. Recently, the concept of an “epigenetic clock” based on DNA methylation levels has emerged as a potentially powerful and accurate determinant of biological age, and a promising tool to understand mechanisms of age-related vascular disease risk.
During aging, global DNA hypomethylation has been observed in humans and various animal models. However, age-dependent DNA hypermethylation occurs preferentially at certain CpG islands, particularly in Polycomb target genes, which are known to regulate chromatin remodeling and gene silencing. DNA methylation is not a static modification, but rather is dynamically controlled by enzymatic deposition and removal by DNMTs and TET family members, respectively. Recent studies indicate that the methylation status of a substantial proportion of CpGs oscillates with a circadian rhythm, even in tissues comprised of largely slow or non-proliferating cells. Rulands et al. determined the oscillation rate to be 2–3/hr in stem cells exiting pluripotency, pointing to a dynamic process independent of DNA replication.17 While many age-related epigenetic changes depend on tissue type, recent studies suggest that age-dependent CpG methylation can be defined independently of sex, tissue type, disease state, and array platform. In this regard, Horvath developed an age predictor by combining publicly available DNA methylation data sets, accrued in over 7000 healthy non-cancer samples from 82 individual data sets, to define DNA methylation levels in over 30 different tissues and cell types collected from children and adults.18 He used a subset of these data to construct a biological age predictor (training data set) by regressing a calibrated version of chronological age on the methylation data from 21,369 CpG probes. This led to identification of 353 CpGs that collectively function as an “epigenetic clock”, which has subsequently been interrogated and validated in several different data subsets. For example, DNA methylation age defined by the epigenetic clock was highly accurate and performed remarkably well in most tissues and cell types tested (age correlation = 0.96, error = 3.6 years); DNA methylation age did not vary significantly across sorted blood cells with different lifespans; clock age was close to zero for embryonic and induced pluripotent stem cells and correlated with cell passage number. The epigenetic clock also gave rise to a highly heritable measure of age acceleration. Since its inception, numerous epidemiological studies have tested Horvath’s DNA methylation clock, demonstrating its accuracy to predict risk for age-related diseases and all-cause mortality.
Smoking is both a major risk factor for CVD and a source of accelerated biological aging based on DNA methylation analyses19,20 and a novel combined biochemical and hematologic clock.21 Interestingly, smoking cessation may be able to reverse some of these age-related changes, consistent with epidemiologic evidence that the CVD risk attributed to smoking declines over time in ex-smokers.22 Assessment of genomic DNA methylation in well-defined cohorts may also help to identify age-independent methylation marks associated with smoking-related CVD events23 These findings suggest that utilization of an epigenetic clock and related DNA methylation data may be helpful to refine CVD risk assessment, illuminate the pathogenic mechanisms that underlie CVD risk factors, and perhaps tailor therapies according to individual risk profiles (Figure 1).
Figure 1.
The interaction between genetic and environmental factors modulates CVD risk and concordantly alters the oscillation rate of the epigenetic clock, which functions as a timekeeper of biological age.
4. Epigenetic memory
Epigenetic inheritance is an essential mechanism that allows stable propagation of gene activity states from one generation of cells to the next. While the precise mechanisms are not yet elucidated, recent studies suggest two primary layers of epigenetic inheritance. The first is termed transgenerational memory, which represents mitotically heritable changes in gene expression and physiology in response to factors or events experienced by the previous generation. Multiple studies have reported this type of memory in animal models as well as humans, most notably the Dutch Hunger Winter Families Study in the western Netherlands.24,25 The Dutch hunger winter famine occurred near the end of World War Two, during late 1944/early 1945, when food rations fell as low as 500 kcal/day. Intrauterine exposure to the famine had differential effects on the body weight of affected newborns depending on their gestational age during exposure to the famine; for example, early gestational exposure produced slightly heavier and larger newborns, whereas exposure to the famine during mid or late gestation resulted in newborns that were lighter, shorter, thinner, and had a smaller head circumference as compared with sibling newborns that did not experience intrauterine exposure to famine. Thanks to exceedingly accurate and detailed medical records obtained during and after the war, plus the fact that many of these individuals have remained in the area throughout their lifetimes, researchers have been able to evaluate their DNA methylation patterns in relation to disease developing later in life. Interestingly, over the ensuing decades, individuals exposed to the famine during early gestation exhibited a three-fold increased risk of coronary heart disease, increased total cholesterol, triglycerides, and low-density lipoprotein (LDL) cholesterol levels, and were more obese compared to their siblings not exposed to the famine during early gestation.26 Remarkably, approximately 50 years after the famine occurred, specific differences in DNA methylation patterns in genes associated with metabolism were detected only in those individuals exposed to the famine during early gestation.27 Moreover, their second-generation offspring also exhibited higher neonatal adiposity and elevated risk of metabolic disease compared with the offspring of unexposed individuals.28 Tobacco exposure during gestation may also accelerate the epigenetic clock of newborns, suggesting a transgenerational heritable component of smoking-related CVD risk.29 These findings suggest that environmental factors may actually be able to contribute to heritable CVD risk, thus blurring the distinction between genetic and epigenetic heritability. Whether transgenerational epigenetic memory can be modulated by lifestyle changes or pharmacological agents warrants further investigation.
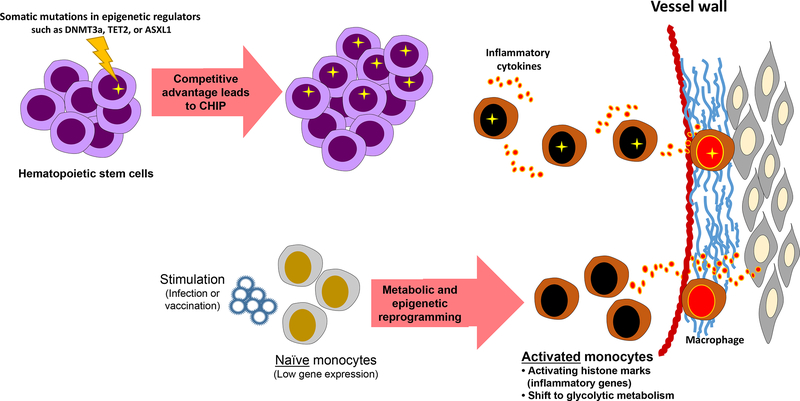
Alternatively, mitotically inherited memory is the heritable transcriptional regulation and responsiveness to developmental and environmental cues. A prime example has been elucidated recently in the epigenetic reprogramming of monocytes, which has challenged the dogma that innate immune system lacks immunological memory (Figure 2). Perhaps the best evidence of this phenomenon in humans comes from studies with bacille Calmette-Guérin (BCG) vaccination, a live attenuated vaccine against tuberculosis (TB) that has been shown to protect against childhood mortality independent of its effect against TB. BCG vaccination in healthy adults led to epigenetic reprogramming of monocytes, which exhibited an enhanced and lasting pro-inflammatory phenotype that might render non-specific protection against reinfections.30 This and other mechanistic studies have uncovered epigenetic programs that rewire cellular immune signaling via histone modifications to induce a shift of cellular metabolism from oxidative phosphorylation toward aerobic glycolysis, which increases the capacity of innate immune cells to respond to stimuli.31 When inappropriately activated, trained immunity programs can become maladaptive. Various levels of experimental evidence support the concept that exposure of the innate immune system to certain micro-organisms triggers a prolonged state of hyperactivation, with increased production of proatherogenic cytokines/chemokines and increased foam cell formation.32 Whether the metabolic shift of monocytes influences the epigenetic reprogramming of trained immunity is yet to be determined; however, recent evidence indicates an important role for metabolism in regulating chromatin structure. For example, the ratio of α-ketoglutarate and succinate controls the activity of two epigenetic regulators, the JMJ family of lysine demethylases and the TET family of methyl-cytosine hydroxylases. These enzymes require α-ketoglutarate as a cofactor, whereas succinate limits their activity.33 Additionally, macrophage stimulation causes an elevation in the level of succinate, which in turn inhibits JMJD3, leading to enhanced H3K27me3 of genes associated with the M2 phenotype, thus suppressing their expression and sustaining a proinflammatory phenotype of trained macrophages upon re-stimulation.
Figure 2.
Somatic mutations and environmental stimuli (such as vaccines, infections, hyperglycemia) lead to activation of immune cells via epigenetic mechanisms to drive inflammation in vascular disease. DNMT3a, DNA methyltransferases 3a; TET2, tet methylcytosine dioxygenase 2; ASXL1, additional sex combs like 1, CHIP, clonal hematopoiesis of indeterminate potential.
Perhaps the strongest evidence of the importance of epigenetic memory in vascular disease risk comes from diabetics with a history of poor glycemic control, who exhibit persistent vasculopathies even after subsequent intensive glycemic control. Conversely, a period of good glycemic control early during the disease course may confer vasculoprotective benefit later in life, even if glycemic control is not maintained. This phenomenon has been termed ‘metabolic memory’. The first demonstration of metabolic memory in diabetic patients came from several studies, including the United Kingdom Prospective Diabetes Study (UKPDS), that reported reduced diabetic-induced microvascular disease if adequate glycemic control was achieved early in the disease course.34 In this trial, newly diagnosed diabetics who underwent intensive treatment had fewer vascular complications and adverse clinical outcomes over time as compared to diabetics who underwent standard treatment, even when glycemic control was similar in the two groups during the long-term follow-up that ensued. Mechanistic understanding of metabolic memory in humans comes from epigenetic profiling of type 1 diabetics with history of poor glycemic control who exhibited progression of nephropathy and retinopathy despite subsequent intensive therapy. Several histone post-translational modifications were identified in white blood cells, including enrichment in H3K9ac at promoters of key inflammatory genes in monocytes from cases compared with controls.35 This may be relevant to diabetic vasculopathy, which is characterized by chronic inflammation as evidenced by increased macrophage infiltration and pro-inflammatory gene expression in blood vessels, kidneys, and other target organs. Recent studies implicate NF-κB as a major mediator of inflammatory gene expression in vascular endothelial cells and monocytes in response to elevated glucose. Mechanistically, high glucose increased inflammatory gene and NF-κB active subunit (p65) expression by promoting SET7 recruitment and H3K4me1 enrichment at their promoters.36 Further, animal models and in vitro studies have implicated metabolic memory in the persistently abnormal expression of fibrotic, pro-oxidant and inflammatory genes in smooth muscle and endothelial cells.37
5. Complex interacting epigenetic mechanisms in vascular diseases
Given the multiple types of epigenetic regulatory mechanisms that contribute to vascular diseases, it is not surprising that these mechanisms can interact in a complex manner to modulate gene expression and vascular pathology. Cardenas et al. reported such an interactive epigenetic regulatory mechanism leading to vascular smooth muscle cell (VSMC) dysfunction in genetic models of thoracic aortic aneurysm (TAA).38 The group identified upregulated expression of the histone deacetylase enzyme histone deacetylase 9 (HDAC9) in diverse VSMC models of TAA and demonstrated a fundamentally important role for HDAC9 in disease pathogenesis. Mechanistically, they identified a lncRNA, MALAT1, which forms a chromatin-remodeling complex with HDAC9 and the brahma-related gene 1 protein (BRG1), thereby regulating the promoters of key cytoskeletal and contractile genes, resulting in pathological VSMC dedifferentiation. Deletion of either MALAT1 or HDAC9 reduced aneurysm formation in conjunction with increased contractile gene expression and decreased elastin fragmentation, matrix metalloproteinase activity, TGF-β signaling and VSMC proliferation. Interesingly, HDAC9 gene polymorphisms have also been associated with risk of atherosclerosis and ischemic stroke in human population studies.39,40
MANTIS, a lncRNA controlled by the histone demethylase JARID1B, was found to be downregulated in patients with idiopathic pulmonary arterial hypertension41. Deletion of MANTIS in endothelial cells ex vivo and in vivo reduced angiogenesis, which was mechanistically linked to an interaction with BRG1.41 Neumann et al. demonstrated that the lncRNA GATA6-AS regulates endothelial gene expression in vitro and in vivo by interacting with the epigenetic regulator lysyl oxidase like 2 (LOXL2), which can induce demethylation of H3K4 marks by deaminating H3K4me3, suggesting a putative link between GATA6-AS and histone modifying mechanisms42. Taken together, these studies indicate complex interacting epigenetic regulatory mechanisms in vascular diseases.
6. Clonal hematopoiesis of indeterminate potential (CHIP)
Aging is also associated with somatic mutations in DNA that have been linked to many diseases, including cancer. Somatic mutations that confer competitive advantages to highly proliferative cells, such as hematopoietic stem cells, lead to expansion of mutant clones and consequently somatic genome mosaicism. Clonal hematopoiesis is often the result of somatic mutations in cancer-related genes; given the importance of aging in vascular disease, and the common co-occurrence of cancer and vascular disease, clonal hematopoiesis has recently been linked to atherosclerotic CVD. Indeed, whole-exome sequencing of blood cells in 7 independent cohorts of >5000 cancer-free individuals revealed that the presence of single-nucleotide variants and small insertions or deletions in hematopoiesis-related genes is associated with a substantial increase in deaths caused by myocardial infarction or stroke among subjects exhibiting >10% relative frequency of the mutant allele).43 Consistent with this finding, a significant association was found between the detection of somatic mutations in blood cells and the future incidence of atherosclerotic diseases (coronary artery disease and stroke): almost half of mutation carriers developed atherosclerotic CVD during the study, compared with ≈20% of non-carriers, which translates into >2 fold increased risk of developing atherosclerotic CVD. Thus, carrying somatic mutations in hematopoiesis-related genes has a comparable or even greater risk than that of various conventional risk factors. Interestingly, the top three most frequently mutated genes identified as candidate drivers of clonal hematopoiesis are epigenetic regulators, namely DNA methyltransferase 3α [DNMT3A], tet methylcytosine dioxygenase 2 [TET2], and additional sex combs like 1 [ASXL1] (Figure 2). A more recent study has confirmed the existence of independent associations between mutations in each of these 3 genes and coronary artery disease, with median variant allele fraction ranging from 10% to 20%.44 Moreover, studies in animal models of CHIP, in which TET2 mutant hematopoietic stem cells were introduced into hypercholesterolemic mice, have likewise demonstrated a significant increase in atherosclerotic disease burden in association with increased monocyte inflammatory activity.45
7. Impact on cardio-oncology: targeting epigenetic regulators in the cancer clinic
Epigenetic regulators are increasingly being targeted in cancer therapeutics, with five FDA approved medications at the time of this review and many others in clinical trials. These medications target the disordered pattern of DNA cytosine methylation and hydroxymethylation, as well as histone acetylation and methylation. Given the fact that the same epigenetic machinery plays a role in vascular disease, it is highly possible that off-target vascular effects of these therapies could be encountered. Conversely, it is conceivable that certain types of epigenetically-targeted cancer therapies could concomitantly diminish vascular pathology, thereby leading to the potential to “re-purpose” them to treat vascular disease. These concepts are of obvious importance to the field of cardio-oncology and should produce ample opportunities for collaborative clinical/translational research. Many challenges complicate the translation of epigenetic regulatory therapies to cancer therapeutics, such as the interacting complexity of epigenetic regulators; presence of alternative compensatory pathways; and the variability in specific somatic mutations of epigenetic genes that potentially may impact therapeutic responses. Integrating genetic risk calculators with epigenomic profiling is needed to optimize cancer therapeutics, and it likewise may help advance CVD risk assessment and discovery of novel therapeutic targets.
8. Evidence of epigenetic regulation of diverse vascular diseases
Most of the published literature focuses on epigenetic regulatory mechanisms in the context of atherosclerosis and coronary disease; however, it is important to point out that intensive research efforts have also recently been focused on understanding epigenetic regulation of many other forms of vascular disease, such as aneurysms. Aneurysms are irreversible and progressive enlargements in vascular diameter resulting from weakening of the vascular wall, potentially leading to rupture with fatal consequences. Aneurysms can occur throughout the arterial tree and, like atherosclerosis, are caused by a combination of genetic and environmental factors. In addition to the aforementioned role of HDAC9/MALAT1 in epigenetic regulation of thoracic aortic aneurysm, there is abundant evidence linking epigenetic mechanisms to aortic dissection, abdominal aortic aneurysms and cerebral aneurysms.46 Epigenetic mechanisms have also been implicated in stroke pathogenesis and recovery, with most of the emphasis thus far on DNA methylation and histone modifications.47 There is also considerable evidence linking epigenetic mechanisms to pulmonary arterial hypertension.48 These and other studies illustrate the broad importance of epigenetic regulation in numerous types of vascular disease.
9. Conclusions
Mounting evidence points to the importance of epigenetic regulation in promoting or preventing vascular diseases. Studying environment-gene interactions in the context of vascular diseases risk has revealed key regulatory mechanisms and underscored the dynamic nature of epigenetic modifications occurring over long periods of time, and the potential impact of epigenetic modifications accruing prenatally that may contribute to heritable CVD risk. These mechanisms are often complex, interacting across different tissue types, and altered by somatic mutations. Identifying and quantifying key epigenetic modifications may help to more precisely define biological age, CVD risk, and disease mechanisms. The recent therapeutic targeting of epigenetic regulators in the field of oncology has important implications for CVD risk and complications, and thus impacts the burgeoning field of cardio-oncology.
Highlights.
Epigenetic regulation is essential to diverse cellular mechanisms such as cell differentiation, growth and development, environmental adaptation, aging, and disease states.
Epigenetic changes occur in response to environmental cues and lifestyle factors, resulting in persistent changes in gene expression.
DNA methylation age may serve as a predictor of biological age, a concept known as the “epigenetic clock”, which could help to refine cardiovascular risk assessment.
Trained epigenetic memory in monocytes by epigenetic reprogramming increases innate immune responses, which if maladaptive, can augment vascular inflammation.
Somatic mutations in epigenetic regulatory enzymes leads to clonal hematopoiesis of indeterminate potential, a precursor of hematological malignancies and a recently recognized cardiovascular risk factor.
Epigenetic regulatory mechanisms lie at the intersection between cancer and cardiovascular disease and are of great importance to the field of cardio-oncology.
Acknowledgements
None.
Sources of funding: this study was funded by grants HL124097, HL126949, HL134354 and AR070029 from the National Institutes of Health (N.L.W).
Abbreviations
- BRG1
Brahma-related gene 1 protein
- CHIP
Clonal hematopoiesis of indeterminate potential
- CpG
Cytosine preceding Guanosine
- CVD
Cardiovascular disease
- DNMT
DNA methyltransferase
- H3K4me2
Di-methylation of lysine 4 at histone 3
- H3K4me3
Tri-methylation of lysine 4 at histone 3
- H3K9ac
Acetylation of histone 3 at lysine 9
- H3K27me3
Tri-methylation of histone 3 at lysine 27
- HDAC9
Histone deacetylase 9 (HDAC9)
- HMT
Histone methyltransferase
- lncRNA
Long non-coding RNA
- ncRNA
Non-coding RNA
- LDL
Low-density lipoprotein
- TET
Ten-eleven translocation
- TAA
Thoracic aortic aneurysm
- VSMC
Vascular smooth muscle cel
Footnotes
Disclosure
None.
References
- 1.Khyzha N, Alizada A, Wilson MD, Fish JE. Epigenetics of atherosclerosis: emerging mechanisms and methods. Trends Mol Med. 2017;23:332–347. [DOI] [PubMed] [Google Scholar]
- 2.Johan M Lorenzen JM, Martino F, Thum T. Epigenetic modifications in cardiovascular disease. Basic Res Cardiol. 2012;107:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen GB, Tost J A summary of the biological processes, disease-associated changes, and clinical applications of DNA methylation. Methods Mol Biol. 2018;1708:3–30. [DOI] [PubMed] [Google Scholar]
- 4.Traube FR, Carell T. The chemistries and consequences of DNA and RNA methylation and demethylation. RNA Biol. 2017;14:1099–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kouzarides T Chromatin modifications and their function. Cell 2007;128:693–705. [DOI] [PubMed] [Google Scholar]
- 6.Gangwar RS, Rajagopalan S, Natarajan R, Deiuliis JA. Noncoding RNAs in cardiovascular disease: pathological relevance and emerging role as biomarkers and therapeutics. Am J Hypertens. 2017;31:150–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zdravkovic S, Wienke A, Pedersen NL, Marenberg ME, Yashin AI, De Faire U. Heritability of death from coronary heart disease: a 36-year follow-up of 20 966 Swedish twins. J Intern Med. 2002;252:247–254. [DOI] [PubMed] [Google Scholar]
- 8.Schnabel RB, Baccarelli A, Lin H, Ellinor PT, Benjamin EJ. Next Steps in Cardiovascular Disease Genomic Research--Sequencing, Epigenetics, and Transcriptomics. Clin Chem. 2012;58:113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khera AV, Emdin CA, Drake I, et al. Genetic Risk, Adherence to a Healthy Lifestyle, and Coronary Disease N Engl J Med. 2016;375:2349–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abraham G, Havulinna AS, Bhalala OG, et al. Genomic prediction of coronary heart disease. Eur Heart J. 2016;37:3267–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sotos-Prieto M, Baylin A, Campos H, Mattei J. Lifestyle cardiovascular risk score, genetic risk score, and myocardial infarction in hispanic/latino adults living in Costa Rica. J Am Heart Assoc. 2016;5:e004067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Said MA, Verweij N, van der Harst P. Associations of combined genetic and lifestyle risks with incident cardiovascular disease and diabetes in the UK biobank study. JAMA Cardiol. 2018;3:693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prospective Studies Collaboration, Lewington S, Whitlock G, Clarke R, Sherliker, Emberson J, Halsey J, Qizilbash N, Peto R, Collins R. Blood cholesterol and vascular mortality by age, sex, and blood pressure: a meta-analysis of individual data from 61 prospective studies with 55 000 vascular deaths. Lancet. 2007;370:1829–1839. [DOI] [PubMed] [Google Scholar]
- 14.Sniderman AD, Islam S, McQueen M, Pencina M, Furberg CD, Thanassoulis G, Yusuf S. Age and cardiovascular risk attributable to apolipoprotein B, low-density lipoprotein cholesterol or non-high-density lipoprotein cholesterol. J Am Heart Assoc. 2016;5:e003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang TJ, Gona P, Larson MG, Tofler GH, Levy D, Newton-Cheh C, Jacques PF, Rifai N, Selhub J, Robins SJ, Benjamin EJ, D’Agostino RB, Vasan RS. Multiple biomarkers for the prediction of first major cardiovascular events and death. N Engl J Med. 2006;355:2631–2639. [DOI] [PubMed] [Google Scholar]
- 16.Cook NR, Buring JE, Ridker PM. The effect of including C-reactive protein in cardiovascular risk prediction models for women. Ann Intern Med. 2006;145:21–29. [DOI] [PubMed] [Google Scholar]
- 17.Rulands S, Lee HJ, Clark SJ, Angermueller C, Smallwood SA, Krueger F, Mohammed H, Dean W, Nichols J, Rugg-Gunn P, Kelsey G, Stegle O, Simons BD, Reik W. Genome-scale oscillations in DNA methylation during exit from pluripotency. Cell Syst. 2018;7:63–76.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horvath S DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siemelink MA, van der Laan SW, Haitjema S, et al. Smoking is associated to DNA methylation in atherosclerotic carotid lesions. Cir Genom Precis Med. 2018;11:e002030. [DOI] [PubMed] [Google Scholar]
- 20.Zhong J, Agha G, Baccarelli AA. The role of DNA methylation in cardiovascular risk and disease: methodological aspects, study design, and data analysis for epidemiological studies. Cir Res. 2016;118:119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su D, Wang X, Campbell MR, Porter DK, Pittman GS, Bennett BD, Wan M, Englert NA, Crowl CL, Gimple RN, Adamski KN, Huang Z, Murphy SK, Bell DA. Distinct epigenetic effects of tobacco smoking in whole blood and among leukocyte subtypes. PloS one. 2016;11:p.e0166486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mons U, Müezzinler A, Gellert C, et al. Impact of smoking and smoking cessation on cardiovascular events and mortality among older adults: meta-analysis of individual participant data from prospective cohort studies of the CHANCES consortium. BMJ. 2015;350:h1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernández-Sanlés A, Sayols-Baixeras S, Curcio S, Subirana I, Marrugat J, Elosua R. DNA methylation and age-independent cardiovascular risk, an epigenome-wide approach: the REGICOR study (REgistre GIroní del COR). Arterioscler Thromb Vasc Biol. 2018;38:645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stein ZA, Susser M, Saenger G, Marolla F. Famine and human development: The Dutch hunger winter of 1944–1945. New York, NY, US: Oxford University Press; 1975. [Google Scholar]
- 25.Lumey LH, Stein AD, Susser E. Prenatal famine and adult health. Annu Rev Public Health. 2011;32:237–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Painter RC, Roseboom TJ, Bleker OP. Prenatal exposure to the Dutch famine and disease in later life: an overview. Reprod Toxicol. 2005;20:345–352. [DOI] [PubMed] [Google Scholar]
- 27.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105:17046–17049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veenendaal MV, Painter RC, de Rooij SR, Bossuyt PM, van der Post JA, Gluckman PD, Hanson MA, Roseboom TJ. Transgenerational effects of prenatal exposure to the 1944–45 Dutch famine. BJOG. 2013;120:548–553. [DOI] [PubMed] [Google Scholar]
- 29.Javed R, Chen W, Lin F, Liang H. Infant’s DNA methylation age at birth and epigenetic aging accelerators. Biomed Res Int. 2016;2016:4515928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bekkering S, Blok BA, Joosten LAB, Riksen NP, van Crevel R, Netea MG. In vitro experimental model of trained innate immunity in human primary monocytes. Clin Vaccine Immunol. 2016;23:926–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Heijden CDCC, Noz MP, Joosten LAB, Netea MG, Riksen NP, Keating ST. Epigenetics and trained immunity. Antioxid Redox Signal. 2018;29:1023–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christ A, Bekkering S, Latz E, Riksen NP. Long-term activation of the innate immune system in atherosclerosis. Semin Immunol. 2016;28:384–393. [DOI] [PubMed] [Google Scholar]
- 33.Liu PS, Wang H, Li X, et al. α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat Immunol. 2017;18:985–994. [DOI] [PubMed] [Google Scholar]
- 34.UK Prospective Diabetes Study (UKPDS) Group. Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). Lancet. 1998;352:837–853. [PubMed] [Google Scholar]
- 35.Miao F, Chen Z, Genuth S, et al. Evaluating the role of epigenetic histone modifications in the metabolic memory of type 1 diabetes. Diabetes. 2014;63:1748–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El-Osta A, Brasacchio D, Yao D, Pocai A, Jones PL, Roeder RG, Cooper ME, Brownlee M. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med. 2008;205:2409–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddy MA, Zhang E, Natarajan R. Epigenetic mechanisms in diabetic complications and metabolic memory. Diabetologia. 2014;58:443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lino Cardenas CL, Kessinger CW, Cheng Y, et al. An HDAC9-MALAT1-BRG1 complex mediates smooth muscle dysfunction in thoracic aortic aneurysm. Nat Commun. 2018;9:1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bellenguez C, Bevan S, Gschwendtner A, et al. Genome-wide association study identifies a variant in HDAC9 associated with large vessel ischemic stroke. Nat Genet. 2012;44:328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Markus HS, Mäkelä KM, Bevan S, Raitoharju E, Oksala N, Bis JC, O’donnell C, Hainsworth A, Lehtimäki T. Evidence HDAC9 genetic variant associated with ischemic stroke increases risk via promoting carotid atherosclerosis. Stroke. 2013;44:1220–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leisegang MS, Fork C, Josipovic I, et al. Long noncoding RNA MANTIS facilitates endothelial angiogenic function. Circulation. 2017;136:65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neumann P, Jaé N, Knau A, Glaser SF, Fouani Y, Rossbach O, Krüger M, John D, Bindereif A, Grote P, Boon RA, Dimmeler S. The lncRNA GATA6-AS epigenetically regulates endothelial gene expression via interaction with LOXL2. Nat Commun. 2018;9:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuster JJ, Walsh K. Somatic mutations and clonal hematopoiesis: unexpected potential new drivers of age-related cardiovascular disease. Circ Res. 2018;122:523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jaiswal S, Natarajan P, Silver AJ, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017; 377:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fuster JJ, MacLauchlan S, Zuriaga MA, et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science. 2017;355:842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim HW, Stansfield BK. Genetic and epigenetic regulation of aortic aneurysms. Biomed Res Int. 2017;2017:7268521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hwang JY, Aromolaran KA, Zukin RS. Epigenetic mechanisms in stroke and epilepsy. Neuropsychopharmacology. 2012;38:167–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gamen E, Seeger W, Pullamsetti SS. The emerging role of epigenetics in pulmonary hypertension. Eur Respir J. 2016;48:903–917. [DOI] [PubMed] [Google Scholar]