Abstract
Intercellular communication governs complex physiologic processes ranging from growth and development to the maintenance of cellular and organ homeostasis. In nearly all Metazoa, Receptor Tyrosine Kinases (RTKs) are central players in these diverse and fundamental signaling processes. Aberrant RTK signaling is at the root of many developmental diseases and cancers and it remains a key focus of targeted therapies, several of which have achieved considerable success in patients. These therapeutic advances in targeting RTKs have been propelled by numerous genetic, biochemical, and structural studies detailing the functions, molecular mechanisms of regulation and activation of RTKs. The latter in particular have proven to be instrumental for the development of new drugs, selective targeting of mutant forms of RTKs found in disease, and counteracting ensuing drug resistance. However, to this day, such studies have not yet yielded high resolution structures of intact RTKs that encompass the extracellular and intracellular domains and the connecting membrane-spanning transmembrane domain. Technically challenging to obtain, these structures are instrumental to complete our understanding of the mechanisms by which RTKs are activated by extracellular ligands and of the effect of pathological mutations that do not directly reside in the catalytic sites of tyrosine kinase domains. In this review, we focus on the recent progress towards obtaining such structures and the insights already gained by structural studies of the subdomains of the receptors that belong to the HER/EGFR, Insulin Receptor, and PDGFR RTK families.
Introduction
The human proteome contains 58 receptor tyrosine kinases (RTKs) classified into 20 subfamilies (1). These integral membrane proteins are receptors for soluble extracellular or membrane-embedded ligands that control receptor activation through the modulation of receptor oligomerizations states. Typically, ligand binding induces receptor dimerization and/or leads to higher-order clustering which switches on the activity of the intracellular kinase domains resulting in receptor phosphorylation (Figure 1A). Enhanced receptor phosphorylation allows for the recruitment of downstream signaling pathway components and their subsequent phosphorylation. Functions of RTKs are critically important in development, with many of the receptors also playing key roles in the maintenance of organismal homeostasis through adulthood (2, 3). The abnormal activation of the homeostatic signals or reactivation of those RTKs which signal primarily during development are detrimental to humans and results in a number of diseases. Hence, RTKs have been an interest of therapeutic efforts for decades and on the front lines of structural studies for just as long (4). These studies have revealed that all RTKs feature a broadly conserved domain architecture with an N-terminal ligand-binding extracellular domain (ECD), a single-pass hydrophobic helical transmembrane domain (TMD), an intracellular juxtamembrane domain (JMD), a tyrosine kinase domain (KD), and a C-terminal tail (C-tail) typically predicted to be largely unstructured (2).
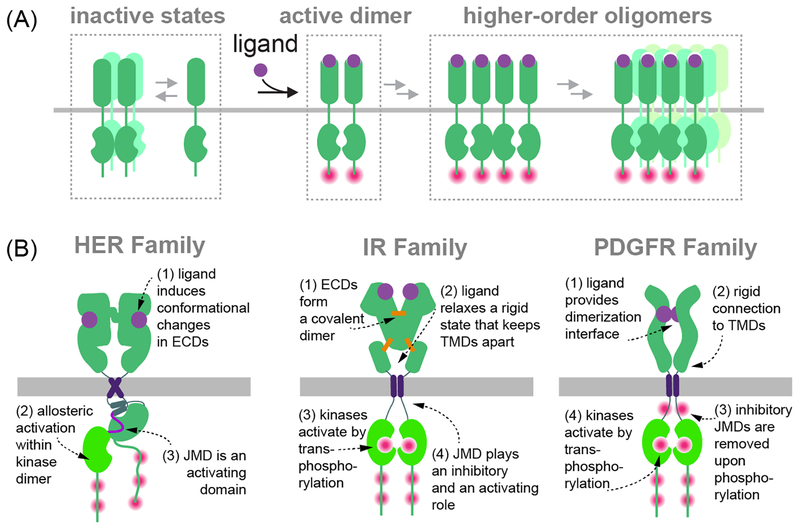
Figure 1. Ligand-induced activation of RTKs.
(A) Multiple modes of oligomerization have been proposed at different steps of RTK activation in response to ligand binding. In general, ligand binding promotes the formation of a productive oligomer in which an active conformation of the kinase is stabilized. Pink dots depict sites of autophosphorylation. (B) Summary of unique features of the activation mechanisms operative in the HER (EGFR/ErbB), IR and PDGFR receptor families.
Structures of RTKs’ subdomains have showcased a variety of mechanisms that control receptor activation and emphasize that despite conserved domain composition, individual RTKs have evolved unique modes of regulation to control the amplitude of activity and temporal precision of signaling (Figure 1B). In an effort to unify some of these principles, these modes have been classified into four primary mechanisms by which ligand binding mediates receptor oligomerization: 1) indirect ligand-mediated receptor dimerization in which a ligand does not directly engage in the dimerization interface, 2) direct receptor-mediated dimerization in which ligand forms the dimerization interface, 3) ligand and receptor mediated dimerization in which both ligand and receptor contribute to the dimerization interface, and 4) ligand and receptor mediated dimerization facilitated by accessory molecules (5). Regardless of the model, the role of a ligand often extends beyond passive dimerization and serves to disrupt an autoinhibited conformation of the receptor that prevents its activation in a productive manner. Notably, some receptors are known, or are predicted, to already form oligomers (mostly dimers) prior to ligand engagement, and in those cases the critical role of ligand binding is to promote a conformational change in addition to higher order oligomerization in some cases.
As much as different mechanisms guide activation of receptor ectodomains by their ligands, the intracellular kinase domain modules also employ different strategies for catalytic activation. Known mechanisms can be generalized into two distinct modes. The first, and more common mechanism, is phosphorylation-dependent and entails auto-phosphorylation of the catalytically important activation loop within the kinase domain, and/or other regions within the intracellular domain, as a mechanism for stabilization of kinase active conformation following ligand stimulation. The second, less common among RTKs, described for the human Epidermal Growth Factor Receptor (HER/EGFR) family, is phosphorylation-independent. In HER receptors, active kinase conformation is achieved instead through an allosteric effect of binding of one kinase domain to another, and stabilization of the catalytic elements in a manner analogous to the effect of phosphorylation in other kinases (6).
Existing high resolution structural insights into ligand binding interaction modes with the extracellular domains and into catalytic mechanisms of kinase domain activation derive solely from the studies of those domains alone. However, key to RTK activation is the communication between ligand binding events on the outside of the cell and kinase activation in the cytosol. Examples of RTKs, such as the Insulin Receptor, which constitutively exists in a dimeric form and does not alter its oligomeric state upon ligand binding, emphasize that signal transduction across the membrane relies on concerted conformational changes in all receptor domains to support receptor activation. The notable occurrence of mutations in the transmembrane domains of RTKs associated with human diseases points to important roles that TMDs play to ensure the proper communication between the ECDs and the intracellular domains (ICDs) (7). Revealing how TMDs structurally contribute to the communication between the ECDs and ICDs is essential to uncovering the mechanisms underlying the pathological effects of such mutations. Lastly, for most RTKs, the stoichiometry of inactive to active oligomeric states is still very much a matter of debate, and often single molecule cell-based measurements of the distribution of full-length receptor oligomeric states cannot be accurately modeled by existing structures. Hence, how all receptor domains simultaneously interact with each other prior to and post ligand binding, ultimately answering the fundamental question of how RTK signal transduction occurs across a membrane, can only be revealed by structures of full-length receptors.
While remarkable progress towards obtaining high resolution structures of multi-span transmembrane proteins has been made over the last decade using both X-ray crystallography and cryo-electron microscopy (cryoEM) approaches (8, 9), such structures of full-length RTKs have yet to be solved. Experimental progress to this end has proven difficult owing to challenges in expressing, solubilizing, and stabilizing full-length RTKs in homogeneous conformational and oligomeric states. In this review, we discuss the progress towards revealing structures of full-length RTKs and the novel knowledge gleaned from such efforts for three RTK families: the HER/EGFR family, the Insulin Receptor (IR) family and Platelet-Derived Growth Factor Receptor (PDGFR) family, all of which apply unique mechanistic features for activation (Figure 1B).
EGFR: Breaking symmetry for activation
Knowledge learned from structures of individual domains.
The HER/ErbB family of receptor tyrosine kinases consist of four highly related members (EGFR, HER2, HER3 and HER4), which are indispensable for embryonic development and frequently misregulated in diseases through mutations, gene amplification, and overexpression. Alterations in EGFR and HER2 are most frequent. EGFR is over-expressed in 60% of non-small cell lung cancer (NSCLC) tumors and mutated in 25% of glioblastoma, and less frequently in tumors of the GI tract (10–12). HER2 amplification accounts for over one in five cases of breast cancer, and in recent years HER2 mutations have been increasingly found across different tumor types (13–15). HER3 is a unique member of the HER family due to the lack of catalytic activity, but nevertheless plays an essential role in the activation of other HER family members during development (16). In adults, under pathological conditions, HER3 overexpression hyperactivates the HER2 and EGFR signaling pathways and contributes to resistance to treatments targeting HER2-overexpressing breast cancers, and NSCLC tumors carrying activating EGFR mutations (17, 18). Heterodimerization of HER receptors is a characteristic feature of this family and is particularly essential for signaling by the catalytically impaired HER3 or by HER2, which is an orphan receptor with no known ligand. HER receptor heterodimers generate unique signaling outputs determined by the combinatorial signaling properties of the receptor pair. For example, the HER2/HER4 heterodimer plays a role in cardiomyocyte and neural development and the EGFR/HER3 heterodimer has been implicated in driving some forms of pancreatic cancer and resistance to anti-HER2 breast cancer treatments (19–21).
HER receptors share overall domain structure, revealed over the years by X-ray crystallography and NMR solution structures of individual domains. In the ECD region, all four HER family members contain four conserved subdomains: 2 leucine-rich domains (subdomains I and III) and 2 cysteine-rich domains (subdomains II and IV). A comparison of the apo-EGFR ECD and EGF-bound EGFR ECD crystal structures revealed a significant conformational change within the ECD induced by ligand binding. In the unliganded state, subdomain II intramolecularly folds onto subdomain IV and a motif in subdomain II termed the “dimerization arm” is obscured in this “tethered” conformation (22–24) (Figure 2A). When the ligand contacts the binding site in subdomains I and III, subdomains II and IV coordinate a 130° rigid-body rearrangement to expose the dimerization arm in the “extended” conformation (Figure 2A). Exposed dimerization arms of adjacent monomers intermolecularly contact each other to form a heart-shaped back-to-back EGFR ECD homodimer (25). Interestingly, the ECD dimer interface is built entirely by the ECD, and the ligand does not directly participate in the interface. The ligand, therefore serves as an allosteric activator that stabilizes the EGFR ECD in the extended state leading to receptor dimerization. The structural analysis of ECDs of other HER receptors is less extensive, but points to the conservation of ligand-induced conformational changes during receptor dimerization, with one exception. The ECD of the orphan receptor, HER2, seems to adopt only an extended conformation (26). The extended HER2 state slightly deviates from the ligand-bound extended conformations of other HER receptors and its unique features likely contribute to the fact that HER2 extracellular domain does not constitutively dimerize. As a result, under physiologic levels of expression HER2 signaling is dependent on heterodimerization with other HER receptors.
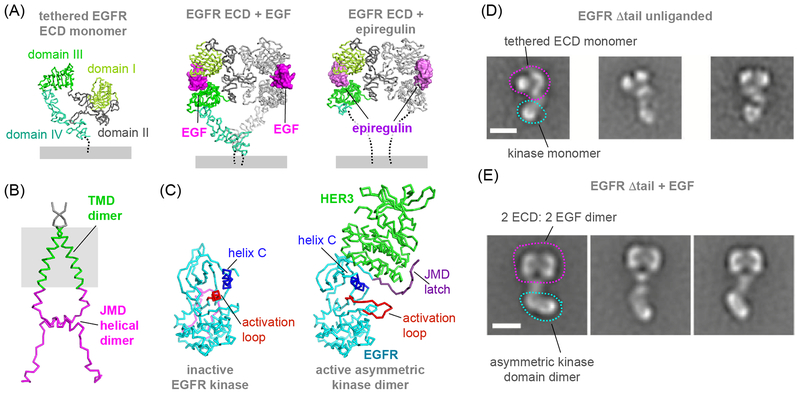
Figure 2. Towards the structure of the full-length EGFR.
(A) Ribbon representation of the crystal structures of the unliganded EGFR ectodomain (PDB ID: 1NQL), EGF ectodomain bound to EGF (PDB ID: 3NJP) or bound to epiregulin (PDB ID: 5WB7). (B) Cartoon representation of the NMR structure of the EGFR TMD/JMD dimer solved in lipid bicelles (PDB ID: 2M20). (C) Ribbon representation of the crystal structures of the inactive EGFR V924R kinase domain (PDB ID: 5CNO) and an active EGFR kinase domain shown in the asymmetric dimer form in complex with the HER3 kinase, which serves here as an allosteric activator of EGFR (PDB ID: 4RIW). (D) Representative NS-EM class averages of unliganded EGFR Δtail receptors solubilized in DDM (figure adapted with permission from Mi et al, 2011). (F) Representative NS-EM class averages of EGFR Δtail receptor dimers in the presence of EGF (figure adapted with permission from Mi et al, 2011).
Although critical links between ligand-induced ECD subdomain reorganization and intracellular KD activation remain unknown, NMR spectroscopy and cross-linking studies on peptides corresponding to the receptor transmembrane domains (TMDs) provide evidence that the single-pass helical TMD, especially in the context of a receptor dimer, may play an active role as a conduit for the extracellular to intracellular relay in signal. All HER receptors contain a putative dimerization motif, originally described in glycophorin A, the GxxxG motif where “x” is any hydrophobic amino acid. EGFR, HER2, and HER4 TMDs have two such GxxxG motifs, one located at the N-terminus of TMD and the other in the C-terminal region (27). Curiously, the catalytically inactive HER3 has only the N-terminal motif. Crosslinking of EGF-bound EGFR dimers on the cell surface showed homodimeric contacts within the N-terminal GxxxG motif thus correlating receptor activation with a specific TMD conformation (28). Molecular dynamics simulations and mutagenesis studies further support a role for the C-terminal GxxxG motif in stabilizing an inactive EGFR dimer conformation (29, 30). The selective interactions between the two defined motifs is thought to impact the relative orientation of the transmembrane domains in the plasma membrane, which in turn can affect the conformations of the JMD and KD (Figure 2B).
In the first EGFR kinase domain crystal structure, the EGFR kinase adopted an active conformation in the absence of activation loop phosphorylation (31). This distinct feature of EGFR led to a hypothesis that EGFR might be constitutively in an “on” state. However, a structure of the EGFR kinase in complex with lapatinib visualized an autoinhibited state of the kinase (32) and multiple activating mutations within EGFR were discovered in NSCLC patients that mapped to the key inhibitory interactions visualized in the EGFR-lapatinib structure (33). These findings supported the notion that under homeostatic conditions, EGFR remains in the autoinhibited state (Figure 2C). Analysis of crystal lattice interactions in EGFR structures in which the kinase domain adopted an active conformation led to identification of an asymmetric dimeric assembly of kinases that supported stabilization of an active state in one of the kinases (Figure 2C). This extensive and hydrophobic dimer interface was validated by mutagenesis as essential for ligand-induced EGFR activation (6). In the asymmetric dimer, kinase activity is induced in one partner (the receiver) that engages its N-lobe with the C-lobe of another kinase (the activator), a mechanism reminiscent of cyclin-dependent kinases (CDKs) activation by their allosteric activators, cyclins (6, 34). This mechanism is conserved in the HER family, and underlies activation of HER2 and HER4 kinases, and of HER3-containing heterodimers (35–37). In such heterodimers, the catalytically inactive HER3 adopts the position of an allosteric activator and serves to stabilize an active conformation of its dimerizing partner’s kinase domain. In fact, one of the determinants of HER3’s lack of activity is the absence of an intact receiver interface. A recent study proposed that under certain circumstances, such as the presence of lapatinib which binds HER2 with high affinity, the HER2/HER3 kinase domain heterodimer may also adopt a head-to-head orientation upon stimulation with the HER3 ligand, neuregulin (NRG) (38). This interface was first observed in the crystal lattice of the HER3 kinase domain structure supporting head-to-head HER3 homodimers (39). The relevance of this dimeric organization of HER kinases for receptor signaling remains unknown.
Subsequent studies expanded the boundaries of the EGFR asymmetric dimer to encompass the intracellular juxtamembrane domain (JMD), a segment that connects the kinase domain with the transmembrane domain. The JMD domain of the receiver kinase was found in crystal structures of EGFR and HER4 to interact with the C-lobe of the activator kinase in the asymmetric kinase dimer (40, 41). This interaction further stabilizes the dimer and significantly increases dimerization affinity. It also participates in stabilization of HER3-containing asymmetric heterodimer with EGFR (36). In this case, the JMD of EGFR which takes a receiver position in the heterodimer binds to the C-lobe of HER3. The inactive HER3 receptor, which is deficient as a receiver kinase, carries sequence alterations in the JMD region relative to other HER receptors, likely because it evolved without pressure to conserve determinants of the functional receiver. Together with the kinase domain, the JMD region establishes an asymmetry of receptor interactions on the cytosolic face resulting in one receiver interface, one activator interface, one JMD domain and corresponding C-lobe patch unoccupied. This open-ended state has long been proposed to drive multimerization of HER receptors, the evidence of which is briefly discussed in the next paragraph.
Receptor Multimerization.
To date, numerous cell imaging approaches have provided evidence that following ligand binding HER receptors form higher-order receptor assemblies on cell surface. Early data point to an EGF-induced assembly of EGFR tetramers (42, 43), whereas more recent high resolution fluorescent microscopy imaging reveals the formation of progressively larger EGFR clusters (5–20 receptors) upon ligand exposure (43–46). While our understanding of the composition, organization, and regulation of these receptor assemblies is far from complete, molecular dynamics simulations studies revealed a putative oligomerization domain within subdomain IV of the EGFR ECD, which is distinct from the canonical ECD dimerization interface (45). Mutation of this interface reduced EGFR multimerization observed through stepwise photobleaching in Xenopus oocytes (45). Higher order oligomerization has also been observed for other members of the HER family, including HER2 and HER3 (44, 47) and the RNA aptamer-based experiments have identified a multimerization site in the subdomain IV of HER3 in the context of HER2-HER3 hetero-oligomers (48).
Multiple variables have been shown to influence the extent of higher order-oligomerization of HER family of receptors, including kinase activity, the nature of a dimerization partner and of a bound ligand. For example, the addition of a kinase inhibitor or the ablation of kinase activity by mutations in the asymmetric dimer interface reduced EGFR cluster formation, suggesting an important role of the kinase domain-mediated interactions in stabilizing higher-order oligomers (44, 49). In another example, addition of neuregulin-1 (NRG1), the cognate ligand for HER3, stimulated HER3 clustering only in the presence of HER2 but not of EGFR demonstrating that oligomerization states might be unique for different receptor pairs (44). Finally, some data indicate that the local receptor milieu and ligand-specificity not only influence receptor clustering but also the mechanism of kinase activation in the clusters, as HER3 phosphorylation by EGFR proceeds through different mechanisms in complexes induced by EGF compared to the ones induced by NRG (44).
Not surprisingly, higher-order oligomers have been linked to distinct downstream signaling outcomes. For example, inhibition of EGFR multimerization through mutations in subdomain IV selectively reduces phosphorylation of a proximal C-tail tyrosine and downstream extracellular signal-regulated kinase (ERK) phosphorylation (45). In another example, an RNA aptamer that selectively inhibits formation of higher order HER2/HER3 oligomers but not their heterodimerization negatively affected only HER2 phosphorylation but preserved HER3 phosphorylation (48). EGFR, which can be selectively locked in dimers that do not progress to higher order oligomers by covalent interaction with orthogonal dimeric ligands, undergoes efficient autophosphorylation but the signal does not propagate downstream to the Ras and PI3K pathways (50). These findings indicate that indeed, dimerization may represent just one step in the assembly of a signaling-competent receptor unit. Given the important relationship between the organization of receptors at the membrane and downstream signaling, structural efforts geared towards characterization of the putative higher order oligomeric states will likely provide invaluable insights into receptors function, and will likely necessitate structural characterizations of nearly full-length receptor fragments.
Visualization of Full-Length EGFR and Coupling Across the Membrane.
The atomic-scale resolution structural studies on isolated receptor domains have relied exclusively on X-ray crystallography and NMR (in the case of the TMD and JMD). While these studies unveiled many key aspects of the EGFR activation mechanism, the fundamental question of how the domains organize and mediate activation in the intact receptor remains unanswered. For any RTK, solving a high-resolution full-length structure is a considerable challenge due to numerous technical hurdles ranging from low expression yield, challenging protein purification, poor stability of the purified protein and low crystallization probability due to substantial conformational dynamics of full-length receptors. Currently, there is considerable hope in the field that several of these hurdles can be overcome by applying single-particle cryo-electron microscopy (cryo-EM) for data collection and structure determination. As of now, four out of 58 RTKs have been successfully purified and characterized at resolution limits of ~20Å by negative-stain EM (NS-EM) (51–54). These attempts have given us invaluable glimpses into the effects of ligand binding on dimerization and the stability of active complex formation.
In a pioneering study four decades ago, Cohen, Gill, and colleagues reported the first isolation of a catalytically active, detergent-solubilized endogenous full-length EGFR (55). The sample was obtained from Triton X-100 solubilized A-431 cell membranes by affinity purification using immobilized EGF. This detergent-solubilized EGFR retained catalytic activity and the capacity to bind EGF, but the sample was not pure enough to pursue thorough biochemical and structural analysis. Almost three decades later, Springer and colleagues employed a similar detergent solubilization strategy to purify recombinantly expressed EGFR encompassing intact ECD, TMD, JMD, and KD, missing only the C-terminal tail predicted not to have any secondary structure (51), and thus denoted EGFR Δtail. The obtained quantity of relatively pure EGFR Δtail receptor was sufficient for its reconstitution and enzymatic characterization in a number of detergent and lipid conditions. Kinase activity could be measured when EGF-bound EGFR Δtail dimers were reconstituted in lipid vesicles or nanodiscs, and interestingly, the presence of the Triton X-100 detergent preserved catalytic activity better than n-Dodecyl β-D-maltoside (DDM), a common detergent used in membrane protein structural biology (56). These studies underscored the critical role of the reconstitution conditions for biophysical characterization of RTKs.
Increased yields and homogeneity of the recombinant EGFR Δtail enabled visualization of particles in DDM micelles by NS-EM. Low resolution projections of previously published EGFR ECD crystal structures correlated well with ECD masked class-averages of NS-EM particles, bolstering confidence in the approach. Using existing crystal structures of isolated domains, Springer and colleagues provided the first glimpses of a monomeric nearly full-length EGFR with clear NS-EM class-averaged projection density for ECD and KD in the same particle (Figure 2D) (51). The addition of EGF resulted in a noticeable shift to a species with a higher Stokes radius on gel-filtration and the putative ECD of resultant class-averages cross-correlated well with the back-to-back EGF:ECD 2:2 homodimer crystal structure (Figure 2E). Notably, the EGFR Δtail homodimer class-averages showed a consistent ECD homodimer coupled to three distinct intracellular domain configurations of kinase domains: (1) rod-shaped, (2) circular/heart-shaped, and (3) disjointed densities. The rod-shaped kinase domain arrangement was in agreement with the asymmetric kinase dimer, such that it no longer formed in the presence of a kinase domain dimerization interface-disrupting mutation (V924R). Although unverified, the heart-shaped kinase density was speculated to represent an “inactive” symmetric kinase dimer while the disjointed densities were suggestive of a kinase dissociated state. This significant conformational heterogeneity of the intracellular domains in the receptor dimer likely captured different receptor activation states, which was quite obvious even in the absence of atomic resolution.
Small molecule inhibitors that stabilize an active conformation of the EGFR kinase domain result in formation of predominantly rod-shaped kinase domain structures in NS-EM averages of the EGFR Δtail, indicative of the active asymmetric kinase dimers (51). This stabilizing effect was reported even in a ligand-independent manner (57). In these ligand-free EGFR dimers, ectodomains adopted multiple conformations, each time coupled to a single asymmetric kinase dimer. These findings suggest that, in the absence of the constrains of the physiological milieu, kinase domain-mediated interactions are sufficient to promote receptor dimerization in an inside-out signaling mode, without engagement of ECDs. This observation is consistent with experiments showing that the oncogenic mutations can promote kinase dimerization even in the presence of the ECD dimer blocking therapeutic antibody, cetuximab (57, 58).
The direct visualization of multiple intracellular domain configurations in the presence of one consistent extracellular domain arrangement, and vice-versa, by NS-EM supports the existence of a loose linkage between the extracellular and intracellular domains in the full-length EGFR. In the structure of the EGFR ECD homodimer bound to EGF, the seven residue-long linker that connects the ECD to TMD is disordered, arguing against a rigid connection between these two domains (28). The flexibility of the linker and the variability in linker length across the HER family further support its passive role in receptor activation. While, it is possible that in the presence of transmembrane or intracellular receptor domains, the linker region adopts a defined conformation, the biochemical studies on full-length EGFR and molecular dynamics simulations of the isolated EGFR transmembrane domains corroborate the model of loose linkage (59). Isolated transmembrane domains from HER receptors efficiently dimerize in vitro (60), but disulfide cross-linking of systematically inserted cysteine residues within the ECD domain IV and linker region and within the TMD of EGFR showed that only the first two N-terminal helical turns of the TMD come in close proximity upon EGF stimulation (28). These findings contrast with the presence of extensive transmembrane helical interactions observed in other GxxxG motif-containing single-span membrane helices present in proteins such as Glycophorin A and Integrins (61, 62). Furthermore, no single mutation can be identified within TMD or Domain IV (C2) interface that reduces ligand-dependent dimerization and signaling by EGFR (28). Altogether these findings suggest that the TMD-mediated interactions are not a primary driving force of receptor dimerization but likely play a role in concert with other receptor domains in stabilization of the active state. Their structural interrogation in the context of a full-length receptor structure will be of essence to delineate their actual contribution to dimerization.
Understanding the structural connection between the ECD, TMD and ICD domains has important ramifications for defining if and how EGFR activation is regulated by a transition between ligand-free preformed “inactive” dimers to ligand-induced active dimers. Several studies have provided evidence for the existence of such inactive dimers (40, 42, 63, 64). It is difficult to rationalize how the independence of extracellular and intracellular components would prevent aberrant kinase activation within the inactive dimer. One possibility is that inactive-active dimer transitions are regulated by electrostatic interactions with the plasma membrane, a variable that was not faithfully recapitulated in the NS-EM experiments in which membrane environment was approximated by a detergent micelle. Long time scale molecular dynamics (MD) simulations of full-length EGFR embedded in a membrane show that electrostatic interactions between the intracellular domain EGFR and the inner membrane leaflet are critical in the maintenance of the equilibrium between a ligand-free inactive dimer and ligand-bound active dimer (65). These studies suggested that in the absence of ligand the JMD interacts with the inner membrane leaflet while the ECD domains hold the N-termini of the TMD domains apart favoring the TMD interactions through the C-terminal regions. The addition of EGF causes the ectodomain to reposition the TMD from a C-terminal dimer (proposed to be inactive) to an N-terminal dimer (active), as previously observed by cysteine mutagenesis (28). These changes are associated with the loss of interactions between the JMD domain and the membrane, facilitating the formation of the juxtamembrane latch and asymmetric kinase dimer. The simulations also supported an autoinhibitory role of the apo ECD (not bound to a ligand) in preventing receptor activation. This more “rigid” coupling model of the cross-talk between the extracellular and intracellular receptors domains suggested by the MD simulations has been further supported by in vivo experiments on intracellular JMD-KD constructs and direct visualization of an “active” conformation of TM-JMD dimer in lipid bicelles by NMR (30). The complete understanding of how these domains are coupled with each other in the full-length receptor will be greatly facilitated by its structure.
EGFR has multiple activating ligands that vary in structure and nature of induced downstream signals (66). These variegated responses to ligands might be in part a result of unique conformations adopted by EGFR depending on the nature of a bound ligand. Seven growth factor ligands are known to activate EGFR: the high affinity ligands: EGF, transforming growth factor-α (TGFα), betacellulin (BTC), and heparin binding EGF-like growth factor (HB-EGF), and low affinity ligands: epiregulin (EREG), amphiregulin (AREG), and epigen (EPGN). Bipartite tetracysteine display-based experiments revealed that stimulation with EGF, but not with TGFα, supports formation of the antiparallel coiled-coil dimer by the JMD domains, indicating that activation of the receptor by ligands does not always follow the same mechanism and that these ligand-specific structural modes might be responsible for unique signaling outcomes (67, 68). Most recently, structures of the EGFR ECD with low affinity ligands EREG and EPGN revealed that they bind to the ECD in conformations distinct from those stabilized by high affinity ligands such as EGF and TGFα (69). EGF and TGFα cause a bend in subdomain II enabling extensive homodimer contacts. The result is a symmetric, heart-shaped ligand-bound ECD homodimer (Figure 2A). In contrast, EREG enforces a less extensive asymmetric homodimer interface through an unbent subdomain II closely reminiscent of the Drosophila EGFR (dEGFR) ECD dimer bound to the ligand, Spitz (70). Lacking the bend in subdomain II also, EPGN-bound ECD of EGFR does not even crystallize as a dimer, but as a monomer analogous to the ECD of the orphan receptor, HER2, which does not homodimerize (71, 72). The inability to induce stable dimer conformation, rather than lower binding affinity, seems to be the primary reason why EREG and EPGN are deficient in inducing EGFR dimerization compared to other high and low affinity ligands: EGF, TGFα, and AREG (69). The ability to promote only transient rather than stable dimerization has been proposed as a mechanism that explains the unique effects EREG and EPGN have on downstream EGFR signaling. Whether unique structural features of EREG and EPGN-induced ECD dimers are also coupled to unique conformational changes in the TMD and kinase domain regions awaits analysis that can only be brought by a structure of full-length EGFR.
Insulin Receptor: Conformational rearrangements within the Covalent Dimer
Insights from the Domain Structures.
The Insulin Receptor Family consists of three members: Insulin Receptor (IR), Insulin-like Growth Factor-1 Receptor (IGF1R), and Insulin Receptor-Related Receptor (InsRR). Insulin and IGF-1 serve as cognate ligands for IR and IGF1R, respectively. Like HER2, InsRR is considered to be an orphan receptor but, uniquely, it becomes activated by elevated pH and may play a role in the compensatory response to metabolic alkalosis by the kidney (73). One α and one β subunit comprise each IR, IFG1R, and InsRR monomer. The entire α subunit exists extracellularly whereas the β subunit contains a short ECD, TMD, JMD, KD, and C-tail (Figure 3A). An extracellular covalent disulfide bond connects the two subunits to form a mature receptor, also referred to as αβ monomer (74). Collectively, the extracellular portion of the αβ monomer contains the following subdomains: two leucine-rich domains (L1 and L2), a cysteine-rich region (CR), two fibronectin type III domains (Fn1 and Fn2) and an insert domain (IDα) on the α monomer, and ID-β and Fn3 on the β monomer (Figure 3A and B). The insert domain is located within the Fn2 domain and contains the proteolytic site which generates α and β monomers upon cleavage. Additional disulfide bridges (two for IR and four for IGF1R) connect the α subunits from adjacent αβ monomers to form the physiologic α2β2 heterotetramer (functional dimer) (75). As a result, unlike most other RTKs, receptors in the IR family exist on the membrane as covalently linked dimers.
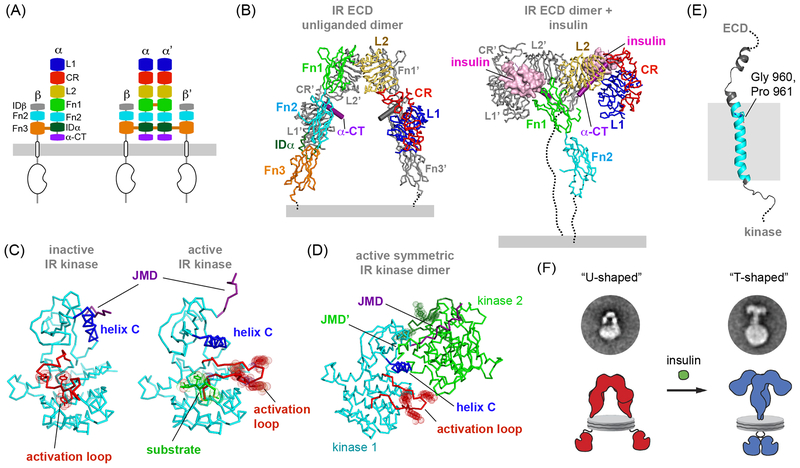
Figure 3. Towards the structure of the full-length Insulin Receptor.
(A) Left, subdomain organization within the extracellular portion of the IR αβ monomer. Right, disulfide bonds between two αβ monomers form the covalent IR dimer. (B) Ribbon representation of the crystal structures of the unliganded IR ectodomain (PDB ID: 4ZXB, Fab molecules present in this structure have been removed for clarity) and bound to Insulin (PDB ID: 6CEB), demonstrating the characteristic U-shaped (OFF) and T-shaped (ON) architecture of the ECD dimer. (C) Ribbon representation of the crystal structures of the inactive IR kinase domain (PDB ID: 1IRK) and fully active IR kinase domain (PDB ID: 1IR3) in which all activation loop tyrosines are phosphorylated. In both structures activation loop tyrosines are shown in stick and dot representation. (D) Ribbon representation of the crystallographic dimer formed by the two IR kinase domains in the active conformation via exchange of the JMD domain (PDB ID: 4XLV). The JMD domain is marked in purple in the cyan-colored IR kinase monomer. In both monomers activation loop tyrosines are shown in stick and dot representation. (E) Cartoon representation of the NMR structure of the IR TMD domain solved in micelles (PDB ID: 2MFR) reveals a kink in the N-terminal portion of the TM helix, attributed to the presence of Gly 960 and Pro 961. (F) Representative NS-EM class averages of glycosylated full-length IR receptor dimers in the absence and presence of bound insulin, reconstituted in lipid nanodiscs (figure adapted with permission from Gutmann et al, 2018).
Constitutive dimerization of the Insulin Receptor Family members poses a critical question of basal state regulation: how is kinase transphosphorylation prevented between two monomers already in close proximity in the absence of ligand? While very little is known about regulation of InsRR, the activation mechanisms of IR and IGF1R have been studied quite extensively and they are highly conserved among the two receptors. The emerging picture of IR family activation posits that in the basal state, the receptor is interlocked in the inactive conformation by a set of autoinhibitory interactions established by both the extracellular and intracellular modules. When triggered by binding of their cognate ligands, insulin or IGF1, respectively, the IR and IGF1R receptors undergo a conformational change that releases these autoinhibitory interactions activating the kinase domains. The intracellular locks located within the juxtamembrane and kinase domain have been understood quite well at the structural level. In the inactive state, the activation loop physically blocks the substrate binding site on the kinase domain and restricts ATP binding to the nucleotide pocket (Figure 3C) (76). Following autophosphorylation on three tyrosine residues, the activation loops of IR and IGF1R kinases undergo a major conformational change that opens up the substrate binding and ATP binding sites (Figure 3C) (77). These three phosphorylation events are required to fully activate the kinase (78). The N-terminally adjacent juxtamembrane domains of IR and IGF1 receptors provide additionally autoinhibitory interactions prior to ligand binding. Specifically, a highly conserved tyrosine residue located within the JMD (Y984 in IR) makes several hydrophobic interactions with the kinase N-lobe that stabilizes an unproductive conformation of helix C (79). This JMD-imposed autoinhibition is released in the fully autophosphorylated states of the IR and IGF1R kinase domains, and in the structures in which the intact JMD segment is present, this is achieved through the formation of a symmetric dimer mediated by a new conformation of JMD. In this dimer, the JMDs do not form intramolecular autoinhibitory interactions with their respective kinase domains but cross over to interact intermolecularly with the N-lobe of another kinase (Figure 3C) (80). In this context, the JMDs form distinct interactions with the αC helices than those observed in the inactive state and contribute to stabilization of the active kinase conformation. Thus, in IR and IGF1R the JMD segment serves both inhibitory and activating roles and emerges as a critical structural component coupling ligand binding to kinase activation.
Over the years, extensive structural and biochemical studies have been undertaken in an effort to understand how ligand binding to the constitutive dimeric IR and IGF1R receptors initiates the conformational changes in the ECD that are ultimately propagated to the kinase and juxtamembrane domains resulting in kinase activation (81). Mutagenesis studies revealed that there are two symmetric ligand binding sites within the IR and IGF1 receptors, and each of these sites is composed of two interfaces called Site 1 and Site 2 that encompass residues from both αβ monomers within the α2β2 dimer (82). Site 1 is composed of residues contributed by the L1 domain on one αβ monomer and by the residues from αCT domain on the other αβ monomer. Site 2 is composed of residues from the Fn1 and Fn2 domains on the opposite αβ monomer that contributed the L1 interface. When both interfaces are engaged with a ligand, the ligand binds with high affinity. The lower affinity ligand interactions reflect engagement of the ligand with only one of the interfaces. Hence, there is a number of binding modes that the ligand may adopt when engaging the receptor dimer but receptor activation is achieved through high affinity binding when both interfaces of the composite ligand binding site are engaged (83). Additionally, there is negative cooperativity between the two equivalent composite insulin binding sites implying an asymmetric state within the receptor dimer upon ligand binding (84). Thus, the current model of IR activation by insulin is that a single molecule of insulin binds to one of the two equivalent composite sites (each comprised of both Sites 1 and 2) with a 1:1 insulin:IR stoichiometry and that the binding of a second molecule of insulin to the other composite site exhibits reduced affinity as compared to the first.
Until recently, there were no structures of the full-length liganded ectodomains of the IR family of receptors, and the proposed models describing ligand-induced conformational changes were derived from the analysis of the crystal structures of the unliganded ectodomain of the IR α2β2 dimer and of fragments of the IR ectodomain, bound to insulin (85–87). These fragments encompassed first four domains of IR (L1, CR, L2 and Fn1) and the αCT domain fused at the C-terminus of Fn1. In the unliganded state the ECD adopts an inverted “U”-shaped conformation (or “V”-shaped conformation) in which reciprocal interactions between the L2 domain of one αβ monomer and Fn1 domain of another αβ monomer form the base of the U shape and are oriented most distally part from the membrane, while the Fn2 and Fn3 domains form arms of the U shape and connect to the membrane (Figure 3B). As a result, the C-termini of the ECD in the unliganded state are significantly separated (by ~120Å). This separation would structurally affect the relative orientation of the TMD, JMD and KD domains, which based on the structures of unliganded ECDs are predicted to also be separated in the unliganded receptor. The structures of the liganded IR ECD fragments, while revealing the molecular basis for ligand binding to the receptor, did not provide conclusive insight into how the overall conformation of the ECD might change upon ligand binding. Based on these structures and biochemical studies (88), it was originally proposed that only small conformational changes occur in the ECDs upon ligand binding, including a slight twist within the ECDs upon binding of the first ligand to allow for the binding of another ligand molecule resulting with little change in relative TMD separation within the α2β2 dimer (81, 89).
This model has been revised by an elegant reexamination of the insulin bound IR and unliganded ECDs structures and accompanying biochemical studies which support an alternative model for ligand-dependent receptor activation (90). Reinterpretation of the insulin-bound IR ECD structure revealed a potential hinge-like motion in the ECD upon ligand binding. Such motion would bring the TMDs into close proximity to each other in the presence of ligand. The striking ECD motion supports a model stating that physical separation of the TMDs may be a mechanism by which IR family members remain inactive without ligand. Several lines of evidence corroborate this model. Deletion of all or part of the ECD increases receptor autophosphorylation, indicating that the function of the ECD in the absence of ligand is autoinhibitory (91, 92). While the solution structure of the IR TMD in detergent micelles, solved by NMR, yielded a monomeric state of the TMD (Figure 3E), cross-linking studies are consistent with the propensity of the TMD to dimerize, in the micelle environment (93). Finally, the first structures of the full-length IR ECD in complex with insulin recently solved by cryoEM (94, 95) revealed significant conformational changes that are induced within the α2β2 dimer upon ligand binding (Figure 3B). These changes include movement of the L1, CR and L2 subdomains of one αβ monomer and the Fn3 of another αβ monomer, together with the αCT domain of one of the monomers to generate the ligand binding site. Because of the disulfide bonding between two αβ monomers, these conformational changes are predicted to significantly affect both monomers, and influence the orientation of both Fn3 domains, which are most proximal to the plasma membrane, and hence propagate to the TMD domains. Interestingly, one of the constructs used to produce a cryo-EM ectodomain structure features a C-terminal leucine zipper that holds the two Fn3 domains in close proximity and this construct recapitulated high affinity for insulin as observed with full-length IR (95). Satisfyingly, these conformational changes become rationalized through recent advancements in structural analysis of full-length IR discussed next.
Structural snapshots of full-length IR receptors.
The incomplete understanding of the conformational changes induced by ligand binding to the covalent dimers of the Insulin Receptor Family members and the critical role of conformational coupling within the constitutive dimer for the maintenance of low basal kinase activity, provides a highly motivating incentive towards obtaining high resolution full-length structures of these receptors in the presence and absence of a ligand. The EM-rooted efforts towards this goal span nearly four decades. The first EM studies of ligand-bound full-length IR revealed a T-shaped conformation of the α2β2 heterotetramer, both in detergent and upon reconstitution in liposomes (96–98). These pioneering studies hinted to a lack of substantial conformational shift within ectodomains upon ligand binding, lending more credence to the “twist” model of ligand binding in which invoked conformational changes during receptor activation are subtle.
Most recent NS-EM analysis of full-length IR reconstituted in lipid nanodiscs hints to a different mechanism and corroborates the insights gained through crystallographic and EM structural analyses conducted on isolated domains by depicting a significant conformational shift in relative positions of TMDs upon insulin binding (54). Nanodiscs are circular pieces of lipid bilayer in which lipids are encircled by helical membrane scaffold proteins (MSPs) and which spontaneously assemble around purified recombinant membrane proteins upon removal of a surfactant (99). Both the size and lipid composition of the nanodiscs may be modulated, rendering them a powerful tool for reconstitution of membrane proteins for biophysical and structural studies. The nanodiscs employed to visualize full-length IR also significantly helped to unambiguously demarcate the relative positions of the TMDs in the receptor dimer. In the absence of insulin, NS-EM class-averages of IR particles converged to a U-shaped density with the majority of these particles connected to two circular densities consistent with the presence of two nanodiscs per receptor particle (Figure 3F). The dimensions of the U-shaped projection densities are in close agreement with crystal structures of the apo-ECD dimer. The remarkable presence of two distinct nanodiscs per receptor in class averages can be interpreted as a sign of significant separation of TMDs in the absence of ligand, as originally suggested by the structure of the apo-IR receptor (85) and later reasserted by Kavran and colleagues (90). The addition of insulin dramatically shifts the conformational distribution from mostly two-nanodisc U-shaped averages to one-nanodisc T-shaped averages. The single nanodisc in insulin bound particle averages suggests the close juxtaposition of TMDs induced by ligand binding. These T-shaped averages are reminiscent of the earlier EM studies and are consistent with the recently solved cryo-EM structures of the full ectodomains of the insulin-bound IR (94).
Although still not at atomic resolution, the work by Gutmann and colleagues significantly improved our mechanistic understanding of IR activation mechanism and further illustrated the power of full-length RTK structural biology in elucidating the effects of ligand on receptor structure. It is now evident that ligand binding in the IR family precipitates a significant conformational change within the ectodomains that enables TMD dimerization and likely activation of the kinase domains by close juxtaposition. Based on these observations, the separation of kinase domains by widely splayed TMDs provides one mechanism by which the covalent dimeric receptors of the IR family maintain a low basal activity in the absence of ligand. Additional mechanisms in addition to TMD separation involving the interaction between the intracellular JMD and the KD likely further contribute to maintaining low basal trans-phosphorylation activity. A high-resolution full-length structure of an IR family receptor will be invaluable in revealing additional mechanisms that control ligand-dependent activation of this RTK class.
PDGFRβ and c-KIT: Membrane proximal regions show off in the full-length arena
Insights from the Domain Structures.
The PDGFR family is more diverse than the EGFR and IR families and consists of five broadly related receptors: PDGFRα, PDGFRβ, KIT receptor (also known as stem cell factor (SCF) receptor), colony-stimulating factor 1 receptor (CSF1R), and the Fms-like tyrosine kinase 3 receptor (FLT3, also known as FLK2) (2). The unifying feature of all five receptors is the presence of five immunoglobulin-like (Ig-like) domains (D1–D5) within the ectodomain, and the intracellular juxtamembrane domain module (JMD) followed by the kinase domain (Figure 4A). The kinase domains of the PDGFR family of receptors feature an insertion of linker domains with variable intervening sequences within the kinases’ C-lobes (2). One remarkable property of this family of receptors is their responsiveness to structurally diverse ligands, such as short chain α-helical bundle cytokines that activate KIT, CSF1R, and FLT3 or structurally unrelated β-strand cystine-knot fold growth factor ligands bind PDGFRα and PDGFRβ (100, 101).
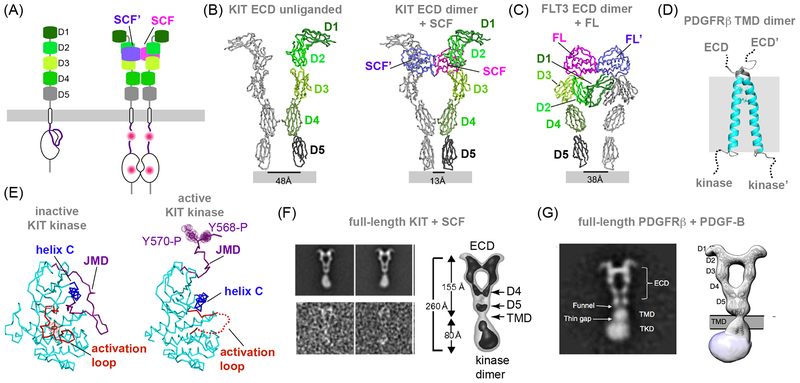
Figure 4. Towards the structures of the full-length PDGFR family of receptors.
A) Left, five Ig-like subdomains constitute the extracellular domain of receptors in the PDGFR family. Right, contacts made by the Ig-like subdomains in the liganded structure of the KIT extracellular domain. Tyrosine phosphorylation (marked by pink dots) in the JMD and kinase regions releases the autoinhibition and activates the kinase. (B) The left panel shows a ribbon representation of the crystal structure of the unliganded KIT ectodomain (PDB ID: 2EC8). This construct crystallizes as a monomer, depicted here using color code introduced in (A). A second monomer (colored grey) shown in the left panel was generated by the alignment of the 2EC8 monomer structure on the KIT ectodomain active dimer induced by the binding of the KIT ligand, SCF, shown on the right panel in ribbon representation (PDB ID: 6CEB). This alignment illustrates that ligand binding changes the relative orientation of the D4 and D5 domains promoting homotypic interactions between these domains in the liganded KIT dimer. (C) Ribbon representation of the crystal structure of the FLT3 ectodomain dimer in complex with the FL ligand (PDB ID: 3QS9). (D) Cartoon representation of the NMR structure of the PDGFRβ TMD domain solved in micelles (PDB ID: 2L6W) reveals a potential dimerization interface between two TM helices. Two alanine residues in this interface are depicted in stick and dot representation. (E) Ribbon representation of the crystal structures of the inactive KIT kinase domain (PDB ID: 1T45) showcasing the inhibitory interactions made by the JMD domain (left), and of the active KIT kinase domain (PDB ID: 1PKG) (right). Two phosphorylated tyrosine residues in the JMD segment of KIT are depicted in stick and dot representation. (F) Left panel, upper row shows representative NS-EM class averages of full-length KIT. The lower row shows representative raw particles. Right panel depicts a three-dimensional reconstruction and domain assignment of SCF- induced dimeric KIT complex. The ECD, TM, membrane proximal D4 and D5 domains and kinase dimer are marked (figure adapted with permission from Opatowsky et al, 2014). (G) The left panel shows a representative 2D NS-EM class average of the full-length PDGFRβ dimer in complex with PDGF-B. Individual receptor domains are marked. The right panel shows the corresponding 3D reconstruction with atomic models of the ECD and TMD obtained by X-ray crystallography fitted into the density (figure adapted with permission from Chen et al, 2015).
Numerous structural studies of the PDGFR family members revealed that all known PDGFR family ligands form dimers themselves and promote receptor dimerization by directly bridging two receptors together (100). This binding mode is quite distinct from that observed in the HER/EGFR and IR families. Despite conserved domain composition of the ECDs, there is a remarkably diversity and plasticity with which ectodomain/ligand complexes assemble in the PDGFR family (101). In the crystal structures of KIT and PDGFRβ ectodomains the D2 and D3 Ig-like domains make extensive interactions with the cognate dimeric ligands (Figure 4B). Although the relative orientation of the D2 and D3 Ig-like domains does not significantly change between the ligand-free and ligand-bound conformations of the ECD, ligand binding has an allosteric effect on the orientation of the distal D4 and D5 Ig-like domains. The D4 and D5 domains, which are most proximal to the plasma membrane, are brought into closer proximity in the liganded receptor dimer. This results in homotypic contacts between the D4 and D5 domains observed in the structures of the liganded KIT ectodomain dimer (102). These “zipper-like” interactions contributed by the Ig-like subdomains of the ectodomain concluding in the homotypic interactions within the membrane proximal regions are predicted to propagate across the plasma membrane resulting in kinase activation (103). Importantly, several activating KIT mutations identified in various human cancers localize in the D5 domain (104) and modulate the strength of the homotypic D5–D5 contacts as well as cooperation between the D4 and D5 domains (105), suggesting that these mutations directly compromise the receptor’s activation mechanism.
The “zipper-like” interactions are predicted to operate for all PDGFR family members (103), except for FLT3. In FLT3, the interactions between the dimeric ligand and receptor ectodomains are more compact (Figure 4C). Most importantly, the homotypic interactions between the membrane proximal domains do not form and the residues that participate in such interactions in other PDGFR family of receptors are not conserved in FLT3 (106). Interestingly, there is a functional murine isoform of FLT3 in which the D5 domain is entirely missing (107), suggesting that the D5 domain is largely dispensable for FLT3 activation. It is still possible that homotypic interactions between the membrane-proximal regions in the FLT3 receptor are promoted by other receptor domains, including the TMD and intracellular modules. These relationships will only be possible to assess in the context of full-length receptor structures. The homotypic interactions observed in the ectodomain structures of the KIT receptor are poised to promote proximity of the TMDs, underscoring the potential role of active coupling between the conformation of ectodomains and the interaction between TM helices in signal transduction across the membrane. The isolated TM module of PDGFRβ has the propensity to dimerize in the in vitro reconstituted membranes (108) and is dimeric according to molecular dynamics simulations and NMR measurements in lipid bilayers (109) (Figure 4D). An activating mutation within the TM domain module has been identified in KIT in mastocytosis (110). Interestingly, this mutation results in a replacement of a phenylalanine to a cysteine (F522C) in a region of the TMD proximal to the extracellular membrane leaflet, suggesting a potential role of this mutation in creating non-physiological disulfide bonds.
In stark contrast to the documented activating role of JMD in EGFR and IR families of receptors, this region in the PDGFR family receptors is only known to participate in kinase inhibition. Through a set of elaborate and remarkably conserved interactions spanning across N- and C-lobe of the kinase domains, the JMD stabilizes the inactive conformation of KIT, FLT3 CSFR1 and PDGFR kinases (Figure 4E) (101, 111). Upon ligand binding sequential tyrosine autophosphorylation within the JMD dislodges the JMD from the intradomain active site crevice between the kinase domain N- and C-lobes, allowing the activation loop to adopt an active conformation (112, 113). The autoinhibitory role of the JMD might be of particular importance for the maintenance of low basal activity of the PDGFR family of RTKs due to the absence of other autoinhibitory interactions, such those established by the ectodomains in the IR and HER/EGFR families. Consequently, multiple mutations in the JMD regions of the PDGFR family members have been reported in diseases, including tandem duplications within the FLT3 JMD accounting for 15–30% of acute myeloid leukemia (AML) cases (114) and point mutations in the JMD of PDGFRα in gastrointestinal stromal tumors (GISTs) (115).
Structural snapshots of full-length KIT and PDGFRβ receptors.
The demonstrated structural diversity of ligand/ectodomain interaction modes and the extent to which membrane proximal regions engage in homotypic interactions suggests that fundamental mechanisms for activation of the PDGFR family members will be unique and distinct for each receptor. How these interactions couple to the activation of the intracellular domains, which in receptors like KIT and PDGFRβ have been shown to form stable complexes (116, 117), remains unknown. Low resolution structural analyses of two full-length PDGFR family receptors provide first clues to the answers to these questions. The NS-EM projections of KIT in a 2:2 complex with stem cell factor (SCF) (52), and of PDGFRβ in a 2:2 complex with the platelet derived growth factor B (PDGF-B) (53), reveal ectodomain dimeric contacts previously underappreciated by crystal structures (Figure 4F–G). In the liganded crystal structure of KIT ectodomain dimer, the ligand mediates all dimer contacts (Figure 4B) (102). The ectodomain crystal structure closely agrees with the full-length NS-EM projection density within the region spanning the D1–D3 Ig-like domains but the superposition of the crystal structure over the projection density demonstrates a notable angular divergence in D4 and D5 relative to D1–D3 suggestive of even more extensive homotypic interactions than originally suggested by crystal structures (Figure 4F). It is likely that the presence of transmembrane and intracellular domains further constrains D4 and D5 orientation extending their homotypic interactions.
Full-length PGDFRβ projection density and corresponding 3D reconstruction further reinforce that homotypic contacts between the membrane-proximal D4 and D5 Ig-like domains within the receptor ectodomains is a shared feature within the PDGFR family (with the notable exception of FLT-3) (Figure 4G). Homotypic contacts provide a structural rationalization for puzzling PDGFRβ oncogenic mutations in the D4 and D5 domain and the biochemical observation that PDGF has a lower affinity for the PDGFR fragment that contains only D1–D3 domains fragment compared to the full ectodomains spanning the D1–D5 domains (101). In the case of PDGFRβ, D4 and D5 may act to optimally position D1–D3 for ligand-receptor interactions. Incidentally, affinity of the KIT ectodomain for its cognate ligand, SCF, is the same with and without the presence of the D4 and D5 domains, suggesting inherent variations between activation mechanisms of different members of the PDGFR family, despite overall structural similarities (53, 118).
The presence of membrane-proximal homotypic ectodomain interactions within a liganded receptor dimer support the hypothesis that activation of PDGFR family of receptors relies on the juxtaposition of transmembrane and intracellular components as proposed in the HER/EGFR and IR families. The 3D reconstruction of the NS-EM data from the analysis of full-length PDGFRβ shows that volumes corresponding to the ectodomains converge into a “funnel” before entering the putative detergent micelle indicating increase in proximity of extracellular juxtamembrane components upon ligand binding (Figure 4G) (53). 2D projections of c-KIT similarly demonstrate a constriction at the dimeric interface preceding density corresponding to the detergent micelle (Figure 4F) (52). In both NS-EM studies, the densities corresponding to the kinase domains appear larger than the dimensions of a single KIT or PDGFR kinase domain and two closely positioned kinase domains may be accommodated in the density. The relatively low resolution of these studies prevents high confidence modeling of a kinase dimer. Intriguingly, a modeled PDGFR asymmetric arrangement of kinase domains, reminiscent of the asymmetric dimerization interface of the EGFR kinase domains, fits in the density quite well. Further studies are needed to explore the possibility of such a dimer, and thus far, there is no biochemical data supporting the existence and importance of an asymmetric kinase domain arrangement for activation of PDGFR family of kinases.
Conclusions
The three RTK families discussed in this review exemplify uniqueness of ectodomain architecture, ligand binding modes and kinase-mediated interactions. The kinase domains of the HER/EGFR family do not require activation loop phosphorylation for activity unlike the other RTKs and their ligands do not participate in the extracellular dimer interface. IR is a constitutive covalent dimer, activated via structural rearrangements induced by still incompletely understood ligand binding poses. The cognate ligands of the PDGFR family of RTKs directly bridge receptor monomers in the dimer, and through close juxtaposition of the transmembrane domains, promote release of kinase autoinhibition via phosphorylation of the juxtamembrane domain. Ultimately, high-resolution full-length structures of RTKs are essential to reveal how these unique structural features act in concert to mediate signaling across membrane in each individual case. Any future high-resolution structural efforts will likely need to consider modelling the membrane environment to account for regulatory interactions at the protein/membrane interface that have been implied by a number of insightful studies (30, 65). For a number of RTKs, the presence of non-catalytic co-receptors and cytosolic cofactors might also be necessary to stabilize conformational states for high resolution determination. The technological developments of cryo-EM mark this as an exciting time to finally visualize these complex interactions, uncovering a deeper understanding of RTK activation which may reveal nuances in currently established dogma about their homeostatic and oncogenic signaling.
Acknowledgements
We sincerely apologize to all colleagues whose work was omitted in this review due to space constraints. N.J. is supported by a grant from the National Institute of General Medical Sciences (R01 GM109176) and the Susan G. Komen Foundation Training Grant (CCR14299947). T.M.T. acknowledges support from the National Cancer Institute (F32 CA216928).
References
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The Protein Kinase Complement of the Human Genome. Science. 2002;298:1912–34. [DOI] [PubMed] [Google Scholar]
- 2.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141(7):1117–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neben CL, Lo M, Jura N, Klein OD. Feedback regulation of RTK signaling in development. Dev Biol. 2019;447(1):71–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arteaga CL, Engelman JA. ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell. 2014;25(3):282–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Receptor Tyrosine Kinases: Structure, Functions and Role in Human Disease. New York: Springer. [Google Scholar]
- 6.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125(6):1137–49. [DOI] [PubMed] [Google Scholar]
- 7.Li E, Hristova K. Receptor Tyrosine Kinase Transmembrane Domains: Function, Dimer Structure and Dimerization. Cell Adhesion and Migration. 2010;4(2):249–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai XC, McMullan G, Scheres SH. How cryo-EM is revolutionizing structural biology. Trends Biochem Sci. 2015;40(1):49–57. [DOI] [PubMed] [Google Scholar]
- 9.Vinothkumar KR. Membrane protein structures without crystals, by single particle electron cryomicroscopy. Curr Opin Struct Biol. 2015;33:103–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bethune G, Bethune D, Ridgway N, Xu Z. Epidermal growth factor receptor (EGFR) in lung cancer: an overview and update. Jorunal of Thoracic Disease. 2010;2(1):48–51. [PMC free article] [PubMed] [Google Scholar]
- 11.Maire CL, Ligon KL. Molecular pathologic diagnosis of epidermal growth factor receptor. Neuro Oncol. 2014;16 Suppl 8:viii1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwak EL, Jankowski J, Thayer SP, Lauwers GY, Brannigan BW, Harris PL, et al. Epidermal growth factor receptor kinase domain mutations in esophageal and pancreatic adenocarcinomas. Clin Cancer Res. 2006;12(14 Pt 1):4283–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bose R, Kavuri SM, Searleman AC, Shen W, Shen D, Koboldt DC, et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013;3(2):224–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greulich H, Kaplan B, Mertins P, Chen TH, Tanaka KE, Yun CH, et al. Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc Natl Acad Sci U S A. 2012;109(36):14476–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arteaga CL, Sliwkowski MX, Osborne CK, Perez EA, Puglisi F, Gianni L. Treatment of HER2-positive breast cancer: current status and future perspectives. Nat Rev Clin Oncol. 2011;9(1):16–32. [DOI] [PubMed] [Google Scholar]
- 16.Erickson S, O’Shea S, Ghaboosi N, Loverro L, Frantz G, Bauer M, et al. ErbB3 is required for normal cerebellar and cardiac development: a comparison with ErbB2- and heregulin-deficient mice. Development. 1997;124:4999–5001. [DOI] [PubMed] [Google Scholar]
- 17.Sergina NV, Rausch M, Wang D, Blair J, Hann B, Shokat KM, et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445(7126):437–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Oh Park J, et al. MET Amplification Leads to Gefitinib Resistance in Lung Cancer by Activating ERBB3 Signaling. Science. 2007;316:1039–43. [DOI] [PubMed] [Google Scholar]
- 19.Frolov A, Schuller K, Tzeng C-WD, Cannon EE, Ku BC, Howard JH, et al. ErbB3 expression and dimerization with EGFR influence pancreatic cancer cell sensitivity to erlotinib. Cancer Biology & Therapy. 2014;6(4):548–54. [DOI] [PubMed] [Google Scholar]
- 20.Gassmann M, Casagranda F, Orioli D, Simon H, Lai C, Klein R, et al. Aberrant Neural and Cardiac Development in Mice Lacking the ErbB4 Neuregulin Receptor. 1995;378:390–4. [DOI] [PubMed] [Google Scholar]
- 21.Zhang N, Chang Y, Rios A, An Z. HER3/ErbB3, an emerging cancer therapeutic target. Acta Biochim Biophys Sin (Shanghai). 2016;48(1):39–48. [DOI] [PubMed] [Google Scholar]
- 22.Burgess A, Cho H-S, Eigenbrot C, Ferguson K, Garrett T, Leahy DJ, et al. An Open-and-Shut Case? Recent Insights into the Activation of EGF/ErbB Receptors. Molecular Cell. 2003;12:541–52. [DOI] [PubMed] [Google Scholar]
- 23.Ferguson K, Berger M, Mendrola J, Cho H-S, Leahy DJ, Lemmon MA. EGF Activates Its Receptor by Removing Interactions with Autoinhibit Ectodomain Dimerization. Molecular Cell. 2003;11:507–17. [DOI] [PubMed] [Google Scholar]
- 24.Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim J-H, et al. Crystal Structure of the Complex of Human Epidermal Growth Factor and Receptor Extracellular Domains. Cell. 2002;110:775–87. [DOI] [PubMed] [Google Scholar]
- 25.Kovacs E, Zorn JA, Huang Y, Barros T, Kuriyan J. A structural perspective on the regulation of the epidermal growth factor receptor. Annu Rev Biochem. 2015;84:739–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho H-S, Mason K, Ramyar K, Stanley AM, Gabelli S, Denney D Jr, et al. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421(6924):756–60. [DOI] [PubMed] [Google Scholar]
- 27.Teese MG, Langosch D. Role of GxxxG Motifs in Transmembrane Domain Interactions. Biochemistry. 2015;54(33):5125–35. [DOI] [PubMed] [Google Scholar]
- 28.Lu C, Mi LZ, Grey MJ, Zhu J, Graef E, Yokoyama S, et al. Structural evidence for loose linkage between ligand binding and kinase activation in the epidermal growth factor receptor. Mol Cell Biol. 2010;30(22):5432–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arkhipov A, Shan Y, Kim ET, Shaw DE. Membrane interaction of bound ligands contributes to the negative binding cooperativity of the EGF receptor. PLoS Comput Biol. 2014;10(7):e1003742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Endres NF, Das R, Smith AW, Arkhipov A, Kovacs E, Huang Y, et al. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell. 2013;152(3):543–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem. 2002;277(48):46265–72. [DOI] [PubMed] [Google Scholar]
- 32.Wood E, Truesdale A, McDonald O, Yuan D, Hasell A, Dickerson S, et al. A Unique Structure for Epidermal Growth Factor Receptor Bound to GW572016 (Lapatinib): Relationships among Protein Conformation, Inhibitor Off-Rate, and Receptor Activity in Tumor Cells Cancer Research. 2004;64:6652–9. [DOI] [PubMed] [Google Scholar]
- 33.Lynch T, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating Mutations in the Epidermal Growth Factor Receptor Underlying Responsiveness of Non-Small-Cell Lung Cancer to Gefitinib. The New England Journal of Medicine. 2004;350(21):2129–39. [DOI] [PubMed] [Google Scholar]
- 34.Jura N, Zhang X, Endres NF, Seeliger MA, Schindler T, Kuriyan J. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol Cell. 2011;42(1):9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu C, Tarrant MK, Choi SH, Sathyamurthy A, Bose R, Banjade S, et al. Mechanism of activation and inhibition of the HER4/ErbB4 kinase. Structure. 2008;16(3):460–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Littlefield P, Liu L, Mysore V, Shan Y, Shaw DE, Jura N. Structural analysis of the EGFR/HER3 heterodimer reveals the molecular basis for activating HER3 mutations. Science Signaling. 2015;7(354). [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 37.Monsey J, Shen W, Schlesinger P, Bose R. Her4 and Her2/neu tyrosine kinase domains dimerize and activate in a reconstituted in vitro system. J Biol Chem. 2010;285(10):7035–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Claus J, Patel G, Autore F, Colomba A, Weitsman G, Soliman TN, et al. Inhibitor-induced HER2-HER3 heterodimerisation promotes proliferation through a novel dimer interface. Elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jura N, Shan Y, Cao X, Shaw DE, Kuriyan J. Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc Natl Acad Sci U S A. 2009;106(51):21608–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jura N, Endres NF, Engel K, Deindl S, Das R, Lamers MH, et al. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell. 2009;137(7):1293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Red Brewer M, Choi SH, Alvarado D, Moravcevic K, Pozzi A, Lemmon MA, et al. The juxtamembrane region of the EGF receptor functions as an activation domain. Mol Cell. 2009;34(6):641–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sako Y, Minoguchi S, Yanagida T. Single-molecule imaging of EGFR signalling on the surface of living cells. Nature Cell Biology. 2000;2. [DOI] [PubMed] [Google Scholar]
- 43.Clayton AH, Walker F, Orchard SG, Henderson C, Fuchs D, Rothacker J, et al. Ligand-induced dimer-tetramer transition during the activation of the cell surface epidermal growth factor receptor-A multidimensional microscopy analysis. J Biol Chem. 2005;280(34):30392–9. [DOI] [PubMed] [Google Scholar]
- 44.van Lengerich B, Agnew C, Puchner EM, Huang B, Jura N. EGF and NRG induce phosphorylation of HER3/ERBB3 by EGFR using distinct oligomeric mechanisms. Proceedings of the National Academy of Sciences. 2017;114(14):E2836–E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang Y, Bharill S, Karandur D, Peterson SM, Marita M, Shi X, et al. Molecular basis for multimerization in the activation of the epidermal growth factor receptor. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Needham SR, Roberts SK, Arkhipov A, Mysore VP, Tynan CJ, Zanetti-Domingues LC, et al. EGFR oligomerization organizes kinase-active dimers into competent signalling platforms. Nat Commun. 2016;7:13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagy P, Claus J, Jovin TM, Arndt-Jovin DJ. Distribution of resting and ligand-bound ErbB1 and ErbB2 receptor tyrosine kinases in living cells using number and brightness analysis. Proc Natl Acad Sci U S A. 2010;107(38):16524–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Q, Park E, Kani K, Landgraf R. Functional isolation of activated and unilaterally phosphorylated heterodimers of ERBB2 and ERBB3 as scaffolds in ligand-dependent signaling. Proc Natl Acad Sci U S A. 2012;109(33):13237–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clayton A, Tavarnesi M, Johns T. Unligated Epidermal Growth Factor Receptor Forms Higher Order Oligomers within Microclusters on A431 Cells That Are Sensitive to Tyrosine Kinase Inhibitor Binding. Biochemistry. 2007;47:4589–97. [DOI] [PubMed] [Google Scholar]
- 50.Liang SI, van Lengerich B, Eichel K, Cha M, Patterson DM, Yoon TY, et al. Phosphorylated EGFR Dimers Are Not Sufficient to Activate Ras. Cell Rep. 2018;22(10):2593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mi LZ, Lu C, Li Z, Nishida N, Walz T, Springer TA. Simultaneous visualization of the extracellular and cytoplasmic domains of the epidermal growth factor receptor. Nat Struct Mol Biol. 2011;18(9):984–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Opatowsky Y, Lax I, Tome F, Bleichert F, Unger VM, Schlessinger J. Structure, domain organization, and different conformational states of stem cell factor-induced intact KIT dimers. Proc Natl Acad Sci U S A. 2014;111(5):1772–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen PH, Unger V, He X. Structure of Full-Length Human PDGFRbeta Bound to Its Activating Ligand PDGF-B as Determined by Negative-Stain Electron Microscopy. J Mol Biol. 2015;427(24):3921–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gutmann T, Kim KH, Grzybek M, Walz T, Coskun U. Visualization of ligand-induced transmembrane signaling in the full-length human insulin receptor. J Cell Biol. 2018;217(5):1643–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cohen S, Carpenter G, King LJ. Epidermal Growth Factor-Receptor-Protein Kinase Interactions. The Journal of Biological Chemistry. 1980;255(10):4834–42. [PubMed] [Google Scholar]
- 56.Mi LZ, Grey MJ, Nishida N, Walz T, Lu C, Springer TA. Functional and Structural Stability of the Epidermal Growth Factor Receptor in Detergent Micelles and Phospholipid Nanodiscs. Biochemistry. 2008;47:10314–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu C, Mi LZ, Schurpf T, Walz T, Springer TA. Mechanisms for kinase-mediated dimerization of the epidermal growth factor receptor. J Biol Chem. 2012;287(45):38244–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z, Longo PA, Tarrant MK, Kim K, Head S, Leahy DJ, et al. Mechanistic insights into the activation of oncogenic forms of EGF receptor. Nat Struct Mol Biol. 2011;18(12):1388–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bessman NJ, Lemmon MA. Finding the missing links in EGFR. Nat Struct Mol Biol. 2012;19(1):1–3. [DOI] [PubMed] [Google Scholar]
- 60.Mendrola JM, Berger MB, King MC, Lemmon MA. The single transmembrane domains of ErbB receptors self-associate in cell membranes. J Biol Chem. 2002;277(7):4704–12. [DOI] [PubMed] [Google Scholar]
- 61.Mineev KS, Bocharov EV, Volynsky PE, Goncharuk MV, Tkach EN, Ermolyuk YS, et al. Dimeric Structure of the Transmembrane Domain of Glycophorin A in Lipidic and Detergent Environments. 2011;3(2). [PMC free article] [PubMed] [Google Scholar]
- 62.Li R, Gorelik R, Nanda V, Law PB, Lear JD, DeGrado WF, et al. Dimerization of the transmembrane domain of Integrin alphaIIb subunit in cell membranes. J Biol Chem. 2004;279(25):26666–73. [DOI] [PubMed] [Google Scholar]
- 63.Clayton AH, Orchard SG, Nice EC, Posner RG, Burgess AW. Predominance of activated EGFR higher-order oligomers on the cell surface. Growth Factors. 2008;26(6):316–24. [DOI] [PubMed] [Google Scholar]
- 64.Chung I, Akita R, Vandlen R, Toomre D, Schlessinger J, Mellman I. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464(7289):783–7. [DOI] [PubMed] [Google Scholar]
- 65.Arkhipov A, Shan Y, Das R, Endres NF, Eastwood MP, Wemmer DE, et al. Architecture and membrane interactions of the EGF receptor. Cell. 2013;152(3):557–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harris RC, Chung E, Coffey RJ. EGF receptor ligands. Exp Cell Res. 2003;284(1):2–13. [DOI] [PubMed] [Google Scholar]
- 67.Doerner A, Scheck R, Schepartz A. Growth Factor Identity Is Encoded by Discrete Coiled-Coil Rotamers in the EGFR Juxtamembrane Region. Chem Biol. 2015;22(6):776–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scheck RA, Lowder MA, Appelbaum JS, Schepartz A. Bipartite tetracysteine display reveals allosteric control of ligand-specific EGFR activation. ACS Chem Biol. 2012;7(8):1367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Freed DM, Bessman NJ, Kiyatkin A, Salazar-Cavazos E, Byrne PO, Moore JO, et al. EGFR Ligands Differentially Stabilize Receptor Dimers to Specify Signaling Kinetics. Cell. 2017;171(3):683–95 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alvarado D, Klein DE, Lemmon MA. ErbB2 resembles an autoinhibited invertebrate epidermal growth factor receptor. Nature. 2009;461(7261):287–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ferguson KM, Darling PJ, Mohan MJ, Macatee TL, Lemmon MA. Extracellular domains drive homo- but not hetero-dimerization of erbB receptors. Embo J. 2000;19(17):4632–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, et al. The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Mol Cell. 2003;11(2):495–505. [DOI] [PubMed] [Google Scholar]
- 73.Deyev IE, Sohet F, Vassilenko KP, Serova OV, Popova NV, Zozulya SA, et al. Insulin receptor-related receptor as an extracellular alkali sensor. Cell Metab. 2011;13(6):679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sparrow LG, McKern NM, Gorman JJ, Strike PM, Robinson CP, Bentley JD, et al. The Disulfide Bonds in the C-terminal Domains of the Human Insulin Receptor Ectodomain. The Journal of Biological Chemistry. 1997;272:29460–7. [DOI] [PubMed] [Google Scholar]
- 75.Hubbard SR. The insulin receptor: both a prototypical and atypical receptor tyrosine kinase. Cold Spring Harb Perspect Biol. 2013;5(3):a008946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hubbard SR, Wei L, Ellis L, Hendrickson W. Crystal Structure of the Tyrosine Kinase Domain of the Human Insulin Receptor. Nature. 1994;372. [DOI] [PubMed] [Google Scholar]
- 77.Hubbard SR. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. The EMBO Journal. 1997;16(18):5573–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Favelyukis S, Till JH, Hubbard SR, Miller WT. Structure and autoregulation of the insulin-like growth factor 1 receptor kinase. Nat Struct Biol. 2001;8(12):1058–63. [DOI] [PubMed] [Google Scholar]
- 79.Li S, Covino ND, Stein EG, Till JH, Hubbard SR. Structural and biochemical evidence for an autoinhibitory role for tyrosine 984 in the juxtamembrane region of the insulin receptor. J Biol Chem. 2003;278(28):26007–14. [DOI] [PubMed] [Google Scholar]
- 80.Cabail MZ, Li S, Lemmon E, Bowen ME, Hubbard SR, Miller WT. The insulin and IGF1 receptor kinase domains are functional dimers in the activated state. Nat Commun. 2015;6:6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ward CW, Menting JG, Lawrence MC. The insulin receptor changes conformation in unforeseen ways on ligand binding: sharpening the picture of insulin receptor activation. Bioessays. 2013;35(11):945–54, 10.1002/bies.201370111. [DOI] [PubMed] [Google Scholar]
- 82.Whittaker L, Hao C, Fu W, Whittaker J. High-affinity insulin binding: insulin interacts with two receptor ligand binding sites. Biochemistry. 2008;47(48):12900–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.De Meyts P, Whittaker J. Structural biology of insulin and IGF1 receptors: implications for drug design. Nat Rev Drug Discov. 2002;1(10):769–83. [DOI] [PubMed] [Google Scholar]
- 84.De Meyts P. Insulin and its receptor: structure, function and evolution. Bioessays. 2004;26(12):1351–62. [DOI] [PubMed] [Google Scholar]
- 85.McKern NM, Lawrence MC, Streltsov VA, Lou MZ, Adams TE, Lovrecz GO, et al. Structure of the insulin receptor ectodomain reveals a folded-over conformation. Nature. 2006;443(7108):218–21. [DOI] [PubMed] [Google Scholar]
- 86.Smith BJ, Huang K, Kong G, Chan SJ, Nakagawa S, Menting JG, et al. Structural resolution of a tandem hormone-binding element in the insulin receptor and its implications for design of peptide agonists. Proc Natl Acad Sci U S A. 2010;107(15):6771–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Menting JG, Whittaker J, Margetts MB, Whittaker LJ, Kong GK, Smith BJ, et al. How insulin engages its primary binding site on the insulin receptor. Nature. 2013;493(7431):241–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Whitten AE, Smith BJ, Menting JG, Margetts MB, McKern NM, Lovrecz GO, et al. Solution structure of ectodomains of the insulin receptor family: the ectodomain of the type 1 insulin-like growth factor receptor displays asymmetry of ligand binding accompanied by limited conformational change. J Mol Biol. 2009;394(5):878–92. [DOI] [PubMed] [Google Scholar]
- 89.Ward CW, Lawrence MC. Similar but different: ligand-induced activation of the insulin and epidermal growth factor receptor families. Curr Opin Struct Biol. 2012;22(3):360–6. [DOI] [PubMed] [Google Scholar]
- 90.Kavran JM, McCabe JM, Byrne PO, Connacher MK, Wang Z, Ramek A, et al. How IGF-1 activates its receptor. Elife. 2014;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tamura S, Fujita-yamaguchi Y, Larner J. Insulin-like Effect of Trypsin on the Phosphorylation of Rat Adipocyte Insulin Receptor. The Journal of Biological Chemistry. 1983;258:14749–52. [PubMed] [Google Scholar]
- 92.Shoelson SE, White MF, Kahn RC. Tryptic activation of the insulin receptor. Proteolytic truncation of the alpha-subunit releases the beta-subunit from inhibitory control. The Journal of Biological Chemistry. 1988;263:4852–60. [PubMed] [Google Scholar]
- 93.Li Q, Wong YL, Kang C. Solution structure of the transmembrane domain of the insulin receptor in detergent micelles. Biochim Biophys Acta. 2014;1838(5):1313–21. [DOI] [PubMed] [Google Scholar]
- 94.Scapin G, Dandey VP, Zhang Z, Prosise W, Hruza A, Kelly T, et al. Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis. Nature. 2018;556(7699):122–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weis F, Menting JG, Margetts MB, Chan SJ, Xu Y, Tennagels N, et al. The signalling conformation of the insulin receptor ectodomain. Nat Commun. 2018;9(1):4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Christiansen K, Tranum-Jensen J, Carlsen J, Vinten J. A model for the quaternary structure of human placental insulin receptor deduced from electron microscopy. Proceedings of the National Academy of Sciences. 1991;88:249–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Woldin C, Hing F, Lee J, Pilch P, Shipley G. Structural Studies of the Detergent-solubilized and vesicle-reconstituted insulin receptor. J Biol Chem. 1999;274(49):34981–92. [DOI] [PubMed] [Google Scholar]
- 98.T-J J, C K, C J, B G, V J. Membrane topology of insulin receptors reconstituted into lipid vesicles. Journal of Membrane Biology. 1994;140(3):215–23. [DOI] [PubMed] [Google Scholar]
- 99.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. irected Self-Assembly of Monodisperse Phospholipid Bilayer Nanodiscs with Controlled Size. J AM CHEM SOC. 2003;126:3477–87. [DOI] [PubMed] [Google Scholar]
- 100.Chen PH, Chen X, He X. Platelet-derived growth factors and their receptors: structural and functional perspectives. Biochim Biophys Acta. 2013;1834(10):2176–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Verstraete K, Savvides SN. Extracellular assembly and activation principles of oncogenic class III receptor tyrosine kinases. Nat Rev Cancer. 2012;12(11):753–66. [DOI] [PubMed] [Google Scholar]
- 102.Yuzawa S, Opatowsky Y, Zhang Z, Mandiyan V, Lax I, Schlessinger J. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130(2):323–34. [DOI] [PubMed] [Google Scholar]
- 103.Yang Y, Yuzawa S, Schlessinger J. Contacts between membrane proximal regions of the PDGF receptor ectodomain are required for receptor activation but not for receptor dimerization. Proc Natl Acad Sci U S A. 2008;105(22):7681–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Forbes S, Clements J, Dawson E, Bamford S, Webb T, Dogan A, et al. Cosmic 2005. British Journal of Cancer. 2006;94(2):318–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reshetnyak AV, Opatowsky Y, Boggon TJ, Folta-Stogniew E, Tome F, Lax I, et al. The strength and cooperativity of KIT ectodomain contacts determine normal ligand-dependent stimulation or oncogenic activation in cancer. Mol Cell. 2015;57(1):191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Verstraete K, Vandriessche G, Januar M, Elegheert J, Shkumatov AV, Desfosses A, et al. Structural insights into the extracellular assembly of the hematopoietic Flt3 signaling complex. Blood. 2011;118(1):60–8. [DOI] [PubMed] [Google Scholar]
- 107.Lavagna C, Marchetto S, Birnbaum D, Rosnet O. Identification and Characterization of a Functional Murine FLT3 Isoform Produced by Exon Skipping. The Journal of Biological Chemistry. 1995;270(7):3165–71. [DOI] [PubMed] [Google Scholar]
- 108.Oates J, King G, Dixon AM. Strong oligomerization behavior of PDGFbeta receptor transmembrane domain and its regulation by the juxtamembrane regions. Biochim Biophys Acta. 2010;1798(3):605–15. [DOI] [PubMed] [Google Scholar]
- 109.Muhle-Goll C, Hoffmann S, Afonin S, Grage SL, Polyansky AA, Windisch D, et al. Hydrophobic matching controls the tilt and stability of the dimeric platelet-derived growth factor receptor (PDGFR) beta transmembrane segment. J Biol Chem. 2012;287(31):26178–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Akin C, Fumo G, Yavuz A, Lipsky P, Neckers L, Metcalfe D. A novel form of mastocytosis associated with a transmembrane c-kit mutation and response to imatinib. Blood. 2004;103(8):3222–5. [DOI] [PubMed] [Google Scholar]
- 111.Liang L, Yan XE, Yin Y, Yun CH. Structural and biochemical studies of the PDGFRA kinase domain. Biochem Biophys Res Commun. 2016;477(4):667–72. [DOI] [PubMed] [Google Scholar]
- 112.Mol CD, Lim KB, Sridhar V, Zou H, Chien EY, Sang BC, et al. Structure of a c-kit product complex reveals the basis for kinase transactivation. J Biol Chem. 2003;278(34):31461–4. [DOI] [PubMed] [Google Scholar]
- 113.Mol CD, Dougan DR, Schneider TR, Skene RJ, Kraus ML, Scheibe DN, et al. Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase. J Biol Chem. 2004;279(30):31655–63. [DOI] [PubMed] [Google Scholar]
- 114.Meshinchi S, Appelbaum FR. Structural and functional alterations of FLT3 in acute myeloid leukemia. Clin Cancer Res. 2009;15(13):4263–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: origin and molecular oncology. Nature Reviews Cancer. 2011;11(12):865–78. [DOI] [PubMed] [Google Scholar]
- 116.Zaman G, Vink P, van den Doelen A, Veeneman G, Theunissen H. Tyrosine Kinase Activity of Purified Recombinant Cytoplasmic Domain of Platelet-Derived Growth Factor Biochemical Pharmacology. 1999;57:57–64. [DOI] [PubMed] [Google Scholar]
- 117.Lam LP, Chow RY, Berger SA. A transforming mutation enhances the activity of the c-Kit soluble tyrosine kinase domain. Biochemical J. 1999:131–8. [PMC free article] [PubMed] [Google Scholar]
- 118.Lemmon MA, Pinchasi D, Zhou M, Lax I, Schlessinger J. Kit Receptor Dimerization Is Driven by Bivalent Binding of Stem Cell Factor. The Journal of Biological Chemistry. 1997;272(10):6311–7. [DOI] [PubMed] [Google Scholar]