Abstract
Cachexia represents a leading cause of morbidity and mortality in various cancers, chronic inflammation and infections. Understanding of the mechanisms that drive cachexia has remained limited, especially for infection-associated cachexia (IAC). Here we describe a model of reversible cachexia in mice with chronic viral infection and identify an essential role for CD8+ T cells in IAC. Cytokines linked to cancer-associated cachexia did not contribute to IAC. Instead, virus-specific CD8+ T cells caused morphological and molecular changes in the adipose tissue, which led to depletion of lipid stores. These changes occurred at a time point that preceded the peak of the CD8+ T cell response and required T cell–intrinsic type 1 interferon signaling and antigen-specific priming. Our results link systemic antiviral immune responses to adipose-tissue remodeling and reveal an underappreciated role of CD8+ T cells in IAC.
Keywords: Cachexia, chronic virus infection, CD8 T cells, type I interferon, lipolysis, lymphocytic choriomeningitis virus
Patients suffering from chronic illnesses such as infections and malignancies are often confronted with cachexia, a multifactorial syndrome that aggravates the underlying disease and worsens prognosis1,2. Cachexia manifests with both behavioral and metabolic symptoms, the severity of which vary between diseases and individuals, making it difficult to define and diagnose3,4. Cachectic patients exhibit anorexia, anhedonia and lethargy, as well as unintentional loss of over 5% of their body weight3,4. This is caused by a rapid depletion of fat and lean mass, which not only impacts the patient’s quality of life, but is often a leading cause of morbidity and mortality2,3,5. Conventional nutritional support does not reverse cachexia. While several treatment options such as agonists for the orexigenic hormone ghrelin are explored, standards of care to prevent or alleviate cachexia remain ill-defined1,6.
The emergence of cachexia accompanies a surge of pro-inflammatory cytokines such as TNF, IFN-γ, IL-6 and IL-17,8. Depending on the disease model, these cytokines can be secreted by either tumor cells, in the case of cancer, host immune cells and/or cells involved in metabolic regulation, such as adipocytes9,10. Murine models of cancer-associated cachexia (CAC) have greatly improved our understanding of the mechanisms contributing to weight loss5,10,11. However, appropriate models to study cachexia in the context of infection have not been developed. In this study we employed a murine benchmark model of chronic viral infection to interrogate the molecular and cellular requirements for infection-associated cachexia (IAC). We designed an integrative approach of genetic, dietary and pharmacological perturbations to uncover both shared and unique properties of CAC and IAC. This led to the characterization of morphological, molecular and metabolic changes in adipose tissue, and identified the main immune drivers of IAC, providing much-needed molecular insights into the pathophysiology of cachexia.
Results
Infection with LCMV clone 13 leads to reversible cachexia
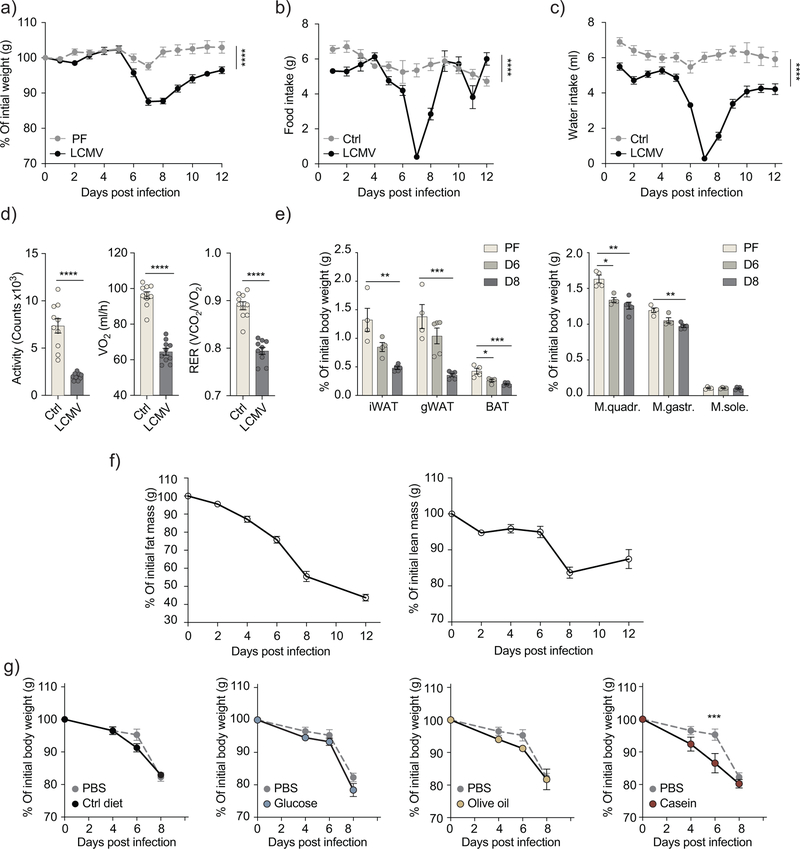
Wild-type C57BL/6J mice infected with 2×106 FFU of the chronic strain clone 13 of the lymphocytic choriomeningitis virus (LCMV) exhibited 15–20% weight loss within the first week of infection (Fig. 1a). To assess whether this is a manifestation of cachexia or a direct result of anorexia, we fed uninfected wild-type mice with the same amount of chow diet consumed by LCMV-infected mice for two weeks post-infection (pair-feeding). Between 6 and 8 days after infection the pair-fed uninfected group lost less than 3% of the initial weight compared to over 10% lost by infected mice (Fig. 1a, b), indicating that the anorectic feeding behavior could not account for the full extent of weight loss observed in the LCMV-infected mice. To characterize the infection-induced pathophysiology, we used metabolic cages to monitor LCMV-infected mice compared to age- and sex-matched uninfected mice. Between day 6 and 8 after infection, mice exhibited signs of lethargy and showed reduced food and water intake, activity and energy expenditure compared to uninfected mice (Fig. 1b–d and Supplementary Fig. 1a). The respiratory exchange ratio (RER), calculated from ratio of CO2 emission to O2 consumption, showed a significant reduction between day 6 and 8 post-infection (Fig. 1d and Supplementary 1a), suggesting that LCMV-infected mice relied on fat metabolism as the main energy substrate. These pathophysiological changes preceded the peak of the viral load and of the adaptive immune response, which happened at day 8–12 (Supplementary Fig. 1b–d). After day 8 mice slowly started to regain weight, despite the continuous presence of the virus (Fig. 1a–c and Supplementary Fig. 1a–d)12.
Figure 1: Infection with LCMV clone 13 leads to transient cachexia.
a) Body weight kinetics for C57BL6/J wild-type mice infected with 2×106 FFU of LCMV-Cl13, in comparison with pair-fed mice (n=5). b-c) Food intake (b) and water intake (c) of LCMV-infected mice in comparison with uninfected controls. (n=10) (p-values: ****<0.0001, two-way ANOVA), Data are representative of three independent experiments for a) and b), c) represents a single experiment. d) Activity, oxygen consumption and respiratory exchange ratio (RER) in LCMV-infected mice compared to uninfected controls (n=10). Data shown represent the average of day 6, 7 and 8 post-infection (p-values: ****<0.0001) from a single experiment. e) Percentage of tissue weight normalized to body weight prior to infection in inguinal, gonadal and interscapular brown adipose tissue (p-values: **0.002, ***0.0006, *0.0172, ***0.0009 unpaired two tailed Student’s t-test), as well as quadriceps, gastrocnemius and soleus muscles (p-values: *0.039, **0.0035, ***0.0001 unpaired two tailed Student’s t-test) at day 7 for pair-fed uninfected mice and day 6 and 8 post-infection for LCMV-infected mice (n=5). f) Body composition as measured in live un-anesthetized LCMV-infected mice using EchoMRI (n=5). e-f) shows representative data of two independent experiments g) Body weight kinetics in LCMV-infected mice supplemented with the equivalent of 1kcal of indicated diet through oral gavage daily between day 4 and 7 post-infection (n=4). (p-values: ***0.0003 two-way ANOVA, Bonferroni correction). Data are representative of three independent experiments. All data shows mean ± s.e.m.
By comparing the body composition, we found that both the fat and lean (muscle) mass were severely depleted in the first week following infection compared to the pair-fed uninfected mice (Fig. 1e,f). The fat tissue, however, underwent an earlier and more severe depletion compared to muscles, starting at 4 days after infection (Fig. 1f). This depletion over time affected different fat depots, including the inguinal white adipose tissue (iWAT), gonadal (gWAT) and interscapular brown adipose tissue (BAT) (Fig. 1e). To assess the loss of muscle mass we examined quadriceps (M.quadr.), gastrocnemius (M.gastr.) and soleus (M.sole.) muscles, which represent both slow- and fast-twitch muscles. The muscle mass of quadriceps and gastrocnemius was reduced in infected mice compared to pair-fed mice (Fig. 1e). This was associated with decreased expression of the myoblast differentiation marker, MyoD1 and the increased expression of Fbxo32 and Trim63, encoding Atrogin1 and Murf1 respectively, involved in proteasomal degradation in all three muscles in the infected mice compared to pair-fed mice (Supplementary Fig. 1e–g).
To evaluate the relationship between the viral inoculum and the weight loss, we titrated the infection dose of LCMV clone 13, ranging from 2×106 FFU to 2×102 FFU. There was a marked amelioration of weight loss at the lower virus inocula, doses below 2×104 FFU showed little to no weight loss (Supplementary Fig. 1h). These lower virus inocula are known to be associated with accelerated viral clearance13. Infection with the acute strain of LCMV (ARM) resulted in similar weight kinetics to low dose LCMV clone 13 infection (Supplementary Fig. 1i), suggesting a possible link between determinants for viral persistence and the development of cachexia.
To examine whether weight loss was ameliorated by nutrient supplementation, we used oral gavage to administer a daily caloric intake of 1kcal of either a chow-like control diet, glucose, olive oil, or casein to LCMV clone 13-infected mice between day 4 and 8 after infection. Oral gavage supplementation did not alleviate weight loss compared to PBS-supplemented control (Fig. 1i). On the contrary, all gavage-supplemented groups recovered at a slower rate in comparison to PBS-supplemented LCMV-infected mice (Supplementary Fig. 1j), indicating that virus-induced weight loss cannot be prevented by nutritional supplementation. These results reveal several hallmarks of cachexia in LCMV clone13-infected mice including anorexia, metabolic dysfunction as well as depleted fat and muscle mass that cannot be reversed by nutritional supplementation.
Viral infection triggers adipose tissue remodeling and lipolysis
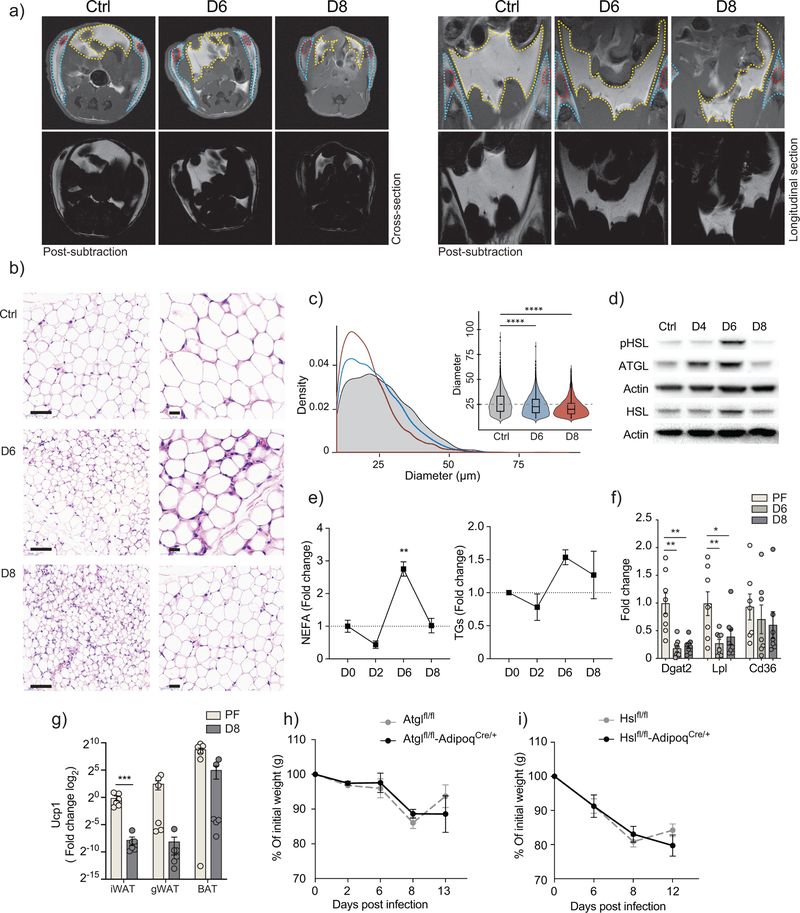
Both subcutaneous and visceral compartments of the adipose tissue in LCMV-infected mice underwent depletion compared to uninfected control (Fig. 2a). Using histological analysis to examine the morphological changes in the inguinal adipose tissue of LCMV-infected compared to uninfected mice, we observed a significant reduction in the size of adipocytes at day 6 and 8 after infection (Fig. 2b,c). Next, we determined the protein abundance and activation status of ATGL (adipose triglyceride lipase) and HSL (hormone sensitive lipase), two enzymes central to lipolysis,14 in inguinal adipose tissue at day 0, 4, 6, and 8 after LCMV infection. Expression of ATGL and HSL, as well as that of p-HSL phosphorylated at residue Ser660, were increased in inguinal adipose tissue upon viral infection compared to uninfected mice (Fig. 2d), suggesting increased lipolysis during LCMV infection, peaking at day 6 after infection. This was corroborated by elevated levels of non-esterified fatty acids (NEFA) in the serum of LCMV-infected mice compared to uninfected controls (Fig. 2e). In addition, reduced mRNA expression of the lipoprotein lipase (Lpl), and to a lesser extent, of the lipid scavenging receptor Cd36 in inguinal fat, together with the increased levels of triglyceride in the serum of LCMV-infected mice compared to pair-fed uninfected mice (Fig. 2e,f), indicated a reduction in the rate of lipid uptake in adipose tissue and/or lipid mobilization in the liver. Decreased mRNA expression of Dgat2, the rate limiting enzyme in triglyceride biosynthesis (Fig. 2f), also suggested a lower rate of lipid synthesis in LCMV-infected mice15. Notably, inguinal fat pads in infected mice showed increased vascularization (Supplementary Fig. 2b,c), which may be involved in inflammation-mediated tissue remodeling. Together, this data show that LCMV clone 13 infection lead to depletion of lipid stores in adipose tissue by modulating key regulators of lipid metabolism.
Figure 2: infection-associated cachexia triggers severe adipose tissue remodeling, and increased lipolysis.
a) Representative MRI cross-sections (left), and longitudinal sections (right). Displayed sections were selected to show iLNs on uninfected controls and LCMV-infected mice at 6 and 8 days post-infection. (Bottom) images shows the fat compartment, after subtracting fat-suppressed acquisitions from the matching non-suppressed images. Images were acquired from a single experiment where (n=3) b) Representative H/E staining of inguinal fat pads from uninfected controls and mice infected at 6 and 8 days post-infection. Images were acquired from a single experiment where (n=3). c) 4–5 images were collected from each inguinal fat pad, at 20X magnification (n=3). The diameter of each adipocyte was measured, and the distribution and median values were evaluation across all conditions. Violin plots represent the density of data points in each condition and the minimum and maximum values. The inner box lots are bound by the upper (75%) and lower (25%) quantile, and the median is represented by an inner horizontal line. Cut-offs were set to include all cells between 10 and 100 μm in diameter (p-values: ****< 0.0001 one-way ANOVA, Bonferroni correction). d) Protein expression of Hormone Sensitive Lipase (HSL), phosphorylated-HSL (pHSL) and Adipose Triglyceride Lipase (ATGL), within the inguinal fat pad of control and infected mice at indicated timepoints (n=3). Data are representative of three independent experiments e) Serum non-esterified fatty acids (NEFA) and triglyceride levels of control and infected mice along 8 days post-infection (n=3). Data are representative of two independent experiments (p-values: **0.002 one-way ANOVA, Bonferroni correction). f) Fold change of inguinal fat pat mRNA expression of scavenger receptor (Cd36) (n=7), Lipoprotein Lipase (Lpl) (n=7) and Diglyceride Acyltransferase 2 (Dgat2) (n=8) as measured by qPCR from infected mice in comparison with pair-fed mice. Data are pooled from two independent experiments (p-values: **0.0021, **0.0038, **0.0069, *0.0279 two-way ANOVA, Bonferroni correction). g) Fold change of Uncoupling protein 1 (Ucp1) mRNA expression of control and infected mice in indicated fat pads (n=6). Data are pooled from two independent experiments (p-values: ***0.0002 unpaired two tailed Student’s t-test). h) Body weight kinetics in adipose tissue-specific knockouts of either ATGL (n=3) or HSL (n=4) compared to flox controls. Data are representative of two and independent experiments. e-i) shows mean ± s.e.m.
To investigate whether the virus-induced adipose tissue wasting followed similar mechanisms as cancer-associated cachexia (CAC), we examined the mRNA expression of Ucp1, as an indicator of adipose tissue beiging. mRNA expression of Ucp1 was marginally lower in the interscapular, inguinal and gonadal fat of infected mice compared to pair-fed mice (Fig. 2g). Based on models of cachexia in Lewis lung carcinoma (LLC) and B16 melanoma in Atgl−/− and Hsl−/− mice16, the absence of ATGL or HSL was sufficient to ameliorate weight loss in cachectic mice, more profoundly in Atgl−/−, indicating an essential role for ATGL and HSL in CAC. To determine whether these lipases were required for infection-associated cachexia (IAC), we infected Atglflfl AdipoqCre/+ and Hslfl/flAdipoqCre/+ mice, which have a conditional ablation of ATGL and HSL in adipocytes driven by the adiponectin (Adipoq) promoter. We found that the ablation of either lipase did not reduce the extent of weight loss during LCMV infection compared to the floxed control group (Fig. 2h), suggesting that neither lipase was required for IAC.
The hormone ghrelin and adipokines such as leptin and adiponectin are considered active modulators of both metabolic and immune processes11,17,18. The amount of ghrelin in the serum and the expression of adiponectin in inguinal adipose tissue were similar in LCMV-infected and uninfected mice (Supplementary Fig. 2d,f). Notably, circulating leptin levels showed a reduction upon LCMV-infection, whereas mRNA expression of leptin was increased in inguinal adipose tissue at day 6 (Supplementary Fig. 2e,f), suggesting leptin could be involved in mediating adipose tissue wasting during IAC. However, leptin-deficient LepOb/Ob mice lost weight at a similar rate as LepOb/+ mice following LCMV infection, and displayed anorexia (Supplementary Fig. 2g). We also quantified the loss of fat and lean mass over 8 days after infection in LepOb/Ob and LepOb/+ mice and found that fat and lean compartments were equally depleted in both groups (Supplementary Fig. 2h), suggesting that although leptin expression was increased in the adipose tissue, it did not mediate the activated lipolytic state nor the disrupted feeding behavior during LCMV infection. These results indicate that IAC triggers structural and metabolic reprogramming in adipose tissue between day 6 and 8 after infection.
Infection-associated cachexia is driven by type 1 IFN and CD8+ T cells
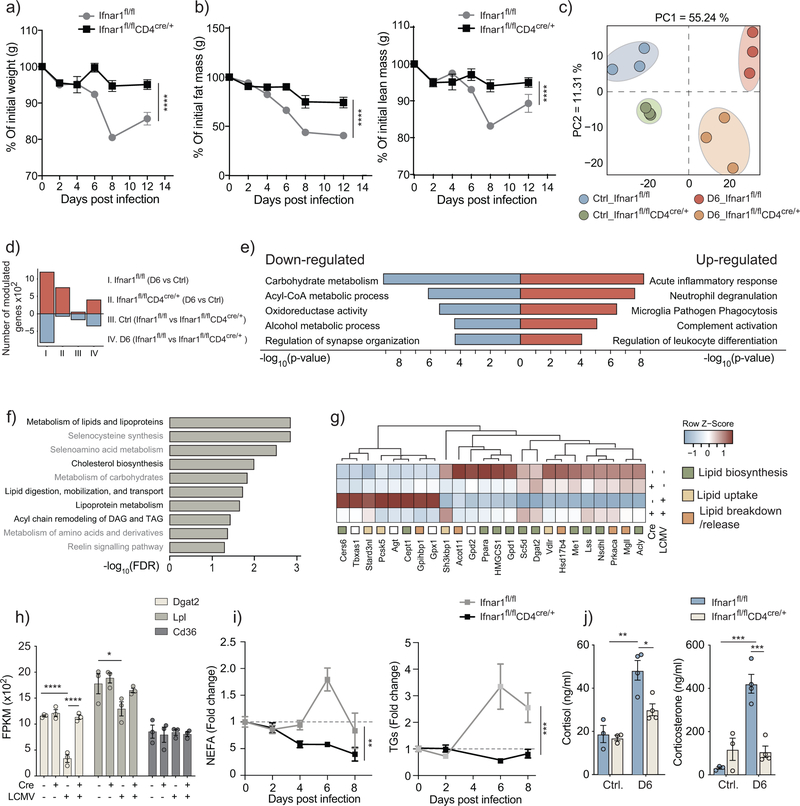
In various models of cachexia and adipose tissue remodeling, cytokines such as IFN-γ, TNF and IL-6 trigger weight loss, acting either independently or in consortium with other cytokines and immune cells8. Bead-based multiplex assays for 32 cytokines as well as ELISA assays for IFN-α and IFN-β revealed a highly dynamic pattern for serum cytokines and indicated the induction of IFN-γ, TNF, IL-6, IFN-α and IFN-β, among others within the first 96 hours after LCMV clone 13 infection of wild-type mice (Fig. 3a, Supplementary Fig. 2a and Supplementary Table 1). Cytokines implicated in CAC, such as IL-1β and LIF8,19 showed no increase in the serum of LCMV-infected mice (Supplementary Fig. 3a and Supplementary Table 1). Pathways associated to pro-inflammatory cytokines such as IFN-γ, TNF, and IL-6 have been linked to CAC8,20. Neutralization of IFN-γ, TNF and IL-6 by genetic ablation using Ifng−/−, Tnf−/− and TnfrI−/− mice or antibody-mediated blockade initiated one day prior to infection, then continued every second day, did not ameliorate weight loss in LCMV-infected mice (Fig. 3b,c). Infected mice in which we simultaneously depleted IFN-γ and TNF using neutralizing antibodies lost up to 20% of their body weight by day 8 after infection (Fig. 3b). Type 1 IFNs were detectable in the serum of LCMV-infected mice within the first two days of infection (Fig. 3a)21. Genetic ablation of IFNAR signaling using Ifnar1−/− mice resulted in ameliorated weight loss by approximately 10% at 8 days after infection compared to wild-type LCMV-infected mice (Fig. 3d and Supplementary Fig. 3b,c), implicating type 1 IFN responses in IAC.
Figure 3: Type I IFN and CD8 T cells play critical roles in inducing infection-associated cachexia.
a) Serum cytokine concentration of LCMV-infected wild-type mice (n=4). Data are representative of a single experiment. b) Body weight of LCMV-infected mice, either genetically ablated from IFNγ or TNF (n=4), or treated with anti-IFNγ or anti-TNF depleting antibodies (n=5), or a combination of both (n=9). Data are representative of a single experiment for genetic knockouts, two independent experiments for single antibody treatment, and pooled data from two independent experiments for the double depletion of IFNγ and TNF. c) Mice treated with anti-IL-6 depleting antibodies (n=5). d) Ifnar1−/− mice (n=4) (p-values: ****< 0.0001 two-way ANOVA) e) Mice treated with anti-CD4 depleting antibodies (n=4). f) Cd8−/− mice and mice treated with anti-CD8 depleting antibodies (n=4) (p-values: *0.027, *0.046 two-way ANOVA). c-f) are representative of two independent experiments. Data shows mean ± s.e.m.
Next, we determined whether a particular T cell population mediated weight loss during LCMV-infection. This was done through the intravenous injection of depleting antibodies for CD8+ T cells and CD4+ T cells two and one days respectively prior to LCMV-infection (Fig. 3e, 3f), or infection of Cd8−/− mice (Fig. 3f). The depletion of CD4+ T cells in LCMV-infected mice did not protect against weight loss (Fig. 3e, and Supplementary Fig. 3d–f), however, depletion of CD8+ T cells resulted in little to no weight loss compared to wild-type controls (Fig. 3f, and Supplementary Fig. 3d–3f). These results suggested a key role for CD8+ T cells in IAC.
To examine whether CD8+ T cells drove weight loss in other viral infections, we infected Rag2−/− mice, which lack both T and B lymphocytes, with a sublethal dose of influenza virus strain PR/8. While LCMV-infected Rag2−/− mice were protected from weight loss (Supplementary Fig. 3g), PR/8-infected Rag2−/− mice continued to lose weight after 8 days while infected wild-type mice started to regain weight. (Supplementary Fig. 3h). These results indicated that weight loss during influenza virus infection occurs via different mechanisms compared to those mediating weight loss during LCMV infection. Uninfected wild-type mice fed the same amount of food as that consumed by PR/8-infected wild-type mice had identical weight loss as the infected group (Supplementary Fig. 3i–j), suggesting weight loss in PR/8-infected mice was due to anorexia. These findings highlight the variety of mechanisms by which weight loss occurs in different infection settings.
CD8+ T cell-intrinsic IFNAR1 alters lipid metabolism during infection
To further investigate the relationship between type 1 IFN signaling, CD8+ T cells and the wasting of the adipose tissue, we used Ifnar1fl/flAdipoqCre/+ mice, which lack IFNAR1 expression in adipocytes, and Ifnar1fl/flCd4Cre/+ mice, in which IFNAR1 is specifically deleted in CD4+ and CD8+ T cells. The LCMV-infected Ifnar1fl/flAdipoqCre/+ mice showed a similar weight loss as the floxed control group (Supplementary Fig. 4a), while the LCMV-infected Ifnar1fl/flCd4Cre/+ mice were protected from weight loss, and preserved both fat and lean mass (Fig. 4a,b), indicating that type 1 IFN signaling in CD8+ T cells was required for IAC. LCMV-infected Ifnar1fl/fl and Ifnar1fl/flCd4Cre/+ mice had comparable numbers of CD8+ T cell in spleens and inguinal lymph nodes (iLNs) at day 6 after infection, the time point cachexia is initiated (Supplementary Fig.4b–c). However, Ifnar1fl/flCd4Cre/+ CD8+ T cells did not upregulate the activation marker CD44 and could not launch antigen-specific responses as indicated by the absence of GP33+ and NP396+ CD8+ T cells (Supplementary Fig. 4b,c). Expansion of CD8+ T cell was impaired at day 8 after LCMV infection in the spleens of Ifnar1fl/flCd4Cre/+ compared to Ifnar1flfl (data not shown), indicating that CD8+ T cell-intrinsic signaling through IFNAR was required for T cell activation and expansion22,23.
Figure 4: CD8 T cells modulate adipose tissue lipid metabolism in a type I IFN-dependent manner.
Analysis of Ifnar1fl/flCD4cre/+ mice compared to Ifnar1fl/fl controls. a) Body weight kinetics of LCMV-infected mice (n=4) (p-values: ****< 0.0001 two-way ANOVA). Data are representative of four independent experiments and show mean ± s.e.m. b) Body composition as measured using EchoMRI in live un-anesthetized LCMV-infected mice (n=4) (p-values: ****< 0.0001 two-way ANOVA). Data are representative of a single experiment and show mean ± s.e.m. c-h) RNA-seq analysis of inguinal fat pads of LCMV-infected and uninfected Ifnar1fl/flCD4cre/+ mice compared to Ifnar1fl/fl controls at 6 days post-infection (n=3). Data shows principal component analysis (PCA) (c), number of modulated genes (d), and significantly modulated genes (<0.05 Adjusted p-value, log fold change > 0.7 or < −0.7) in the limma implementation of 2×2 factorial interaction model. Top five enriched ClueGo pathways for up- and down-regulated genes (e). Enrichment of top 10 modulated metabolic pathways (f). Heatmap of the top modulated genes involved in lipid biosynthesis, uptake, or breakdown and release (g). RNA expression of Cd36, Lpl, and Dgat2 (h) (p-values: **** < 0.0001, *0.015 two-way ANOVA, Bonferroni correction). i) Serum triglyceride and non-esterified fatty acid levels (n=4) (p-values: **0.0027, ***0.0004 two-way ANOVA). j) Circulating levels of cortisol and corticosterone, as measured by ELISA (n=4) (p-values: **0.0010, *0.0185, ***0.0001, ***0.0004 two-way ANOVA, Bonferroni correction). h-j) data are representative of two independent experiments, and show mean ± s.e.m.
To gain further insights into the changes taking place in the adipose tissue of LCMV-infected mice, we performed RNA-seq analysis of bulk-adipose tissue from inguinal fat pads of Ifnar1fl/fl and Ifnar1fl/fl Cd4cre/+ mice 6 days after infection. Principal component analysis showed differential clustering between uninfected and LCMV-infected mice on PC1 and between Ifnar1fl/fl and Ifnar1fl/fl Cd4cre/+ on PC2 (Fig. 4c and Supplementary Fig. 4d). Pathway enrichment analysis of infection-induced gene modulation showed an increase in immune processes in Ifnar1fl/fl mice compared to Ifnar1fl/fl Cd4cre/+, and highlighted multiple changes in metabolic processes within the top 5 pathways downregulated (Fig. 4e). Metabolic pathway enrichment analysis with the Reactome Metabolic database found that 5 of the top 10 pathways enriched in Ifnar1fl/fl mice compared to Ifnar1fl/fl Cd4cre/+ during infection were involved in lipid metabolism (Fig. 4f). The expression of genes involved in the biosynthesis and uptake of lipids was predominantly down-regulated in in adipose tissue of LCMV-infected Ifnar1fl/fl compared to LCMV-infected Ifnar1fl/fl Cd4cre/+ (Fig. 4g). Notably, the expression of Dgat2, the rate limiting enzyme in TG synthesis, showed the anticipated reduction upon infection in Ifnar1fl/fl mice and this was absent in infected Ifnar1fl/fl Cd4cre/+ mice (Fig. 4g–h). Similarly, Lpl (encoding lipoprotein lipase) expression decreased during infection but not in Ifnar1fl/fl Cd4cre/+ mice. Expression of Cd36 was not affected in all conditions (Fig. 4h). Circulating levels of NEFA and TG showed no increase during infection in Ifnar1fl/fl Cd4cre/+ mice compared to Ifnar1fl/fl mice (Fig. 4i).
The lipolysis of adipose tissue is triggered by a variety of hormones and signaling molecules, such as glucocorticoids, which induce the expression of ATGL and HSL, thyroid stimulating hormones (TSH) and catecholamines, which all induce the phosphorylation of HSL by interacting with the TSH receptor (TSHR) in the case of TSH or with beta-adrenergic receptors (β-AR) in the case of catecholamines24. Quantification of circulating cortisol and corticosterone at day 6 after LCMV infection indicated increased levels of cortisol and corticosterone in Ifnar1flfl mice compared to uninfected- Ifnar1flfl mice, which was abrogated in Ifnar1fl/flCd4+/+ mice (Fig. 4j and Supplementary Fig. 4d). Circulating amounts of free thyroxine (fT4) and free triiodothyronine (fT3) remained relatively constant across all conditions (Supplementary Fig. 4d). Furthermore, serum concentrations of norepinephrine were slightly reduced in Ifnar1flfl mice upon infection, but where increased in infected Ifnar1fl/flCd4cre/+ mice compared to uninfected mice at day 6 post infection (Supplementary Fig. 4d). Norepinephrine amounts in the inguinal adipose tissue remained relatively constant throughout all conditions (Supplementary Fig. 4d). Downstream of these signaling hormones, expression of Abrd2 mRNA (encoding the β-AR) was highly increased in the inguinal fat of LCMV-infected compared to uninfected Ifnar1flfl mice, as were protein levels of p-HSL (Supplementary Fig. 4d). Both increases were abrogated in the inguinal fat of Ifnar1fl/flCd4cre/+ mice (Supplementary Fig. 4d), consistent with reduced lipolysis in these mice. Expression of ATGL was increased and expression of lipid droplet-associated protein perilipin was decreased similarly in LCMV-infected Ifnar1fl/flCd4Cre/+ and Ifnar1fl/fl mice (Supplementary Fig. 4d). The hydrolytic activity of ATGL is controlled by its coactivator, CGI-5814. Expression of Abhd5 mRNA (encoding CGI-58) increased in inguinal adipose tissue of LCMV-infected Ifnar1fl/fl, but not LCMV-infected Ifnar1fl/flCd4cre/+ mice compared to uninfected mice (Supplementary Fig. 4d). These observations indicate that the correlation between the increased amounts of glucocorticoids in the serum and the upregulation of lipolysis in LCMV-infected mice was dependent on IFNAR signaling in CD8+ T cells.
CD8+ T cells trigger cachexia through antigen-specific activation
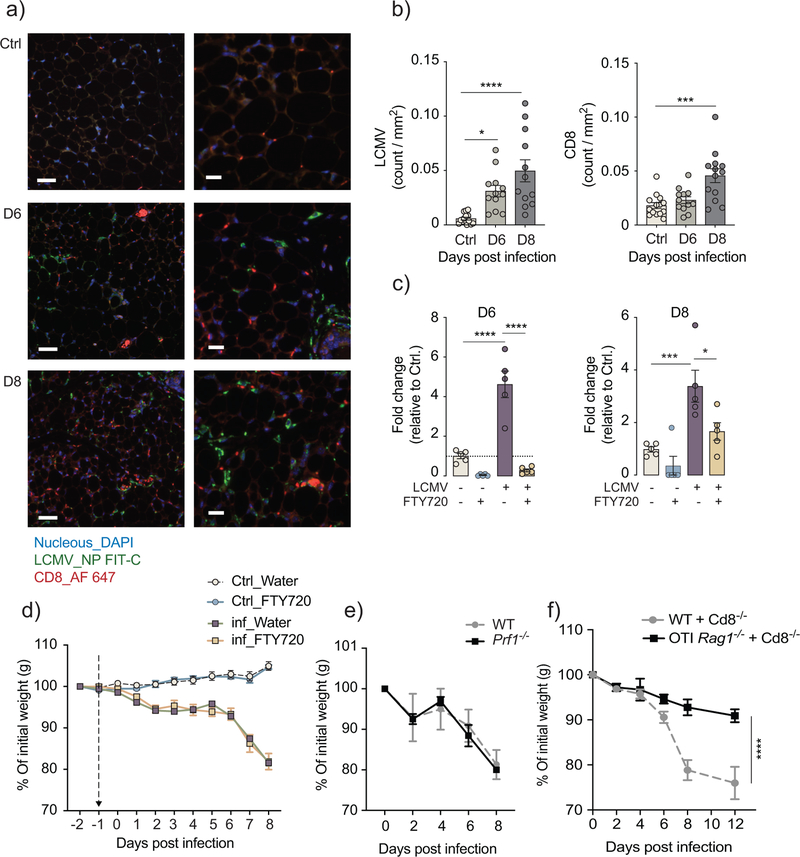
We next examined the kinetics of viral infection and T cell infiltration into the inguinal fat of LCMV-infected mice. Using immunofluorescent co-staining for the LCMV_NP+ and CD8+ T cells, we detected the presence of viral nucleoprotein and CD8+ cells at day 6 and 8 post-infection (Fig. 5a,b), consistent with detection of LCMV_NP mRNA (encoding LCMV nucleoprotein) by qPCR from inguinal, gonadal and brown adipose tissue of infected wild-type mice (Supplementary Fig. 1c). To address the role of T cell infiltration into adipose tissue, we treated infected and control mice with the immunosuppressive drug FTY720, which transiently blocks the egress of lymphocytes from the lymphoid organs without interfering with T cell priming25. Flow cytometry indicated a reduction of virus-specific GP33+ and NP396+ CD8+ T cells in the blood and spleen of FTY720-treated mice, and their accumulation in iLN (Fig. 5c and Supplementary Fig. 5a–d). Treatment with FTY720 did not affect weight loss in infected mice compared to sham-treated mice (Fig. 5d), suggesting no role or only a minor role of local T cell infiltration in triggering adipose tissue wasting. In line with this, LCMV-infected Prf1−/− mice, which lack perforin, an important mediator of cytotoxic activity in CD8+ T cells, had a similar weight loss kinetics as the infected wild-type mice (Fig. 5e), suggesting that CD8+ T cells triggered cachexia independently of cytotoxic cell-to-cell interaction.
Figure 5: CD8 T cells trigger cachexia during the early stage of T cell priming and antigen recognition.
a) Immunohistochemistry co-staining with DAPI, LCMV_NP (FITC), and CD8+ T cells (Alexa Fluor 647) in inguinal fat pads of LCMV-infected mice ay 6 and 8 days compared to uninfected mice (n=3). b) LCMV-NP+ and CD8+ staining from 4–5 images per inguinal fat pad (n=3), as quantified using cell profiler. (p-values: *0,0138, ****<0,0001, ***0,0001 one-way ANOVA, Bonferroni correction) a-b) data are representative of a single experiment. c) Fold change in circulating CD8+ T cells on 6 and 8 days post infection relative to uninfected control after daily gavage administration of either FTY720 (0.3mg/kg) or water (n=5). (p-values: **** < 0.0001, ***0.0004, *0.0143 two-way ANOVA, Bonferroni correction). d) Body weight kinetics of infected FTY720-treated and water-treated compared to uninfected controls (n=5). Data are representative of two independent experiments. e) Body weight kinetics of Prf1−/− mice compared to wild-type controls (n=4). Data are representative of a single experiment. f) Body weight kinetics in WT+Cd8−/− and OT-I Rag1−/−+Cd8−/− chimeric mice infected with LCMV (n=6), data are pooled from two independent experiments. b-f) data shows mean ± s.e.m.
To address whether CD8+ T cells trigger adipose tissue wasting as a result of direct antigen stimulation or in response to bystander activation induced by the inflammatory milieu, we generated mixed bone marrow chimeras. This was done using wild-type (CD45.2+) recipients, and congenic (CD45.1+) donors, where the mice received mixed bone marrows at 1:1 ratios of either OT-I Rag1−/− and Cd8−/− or wild-type and Cd8−/−. OT-I Rag1−/− mice carry a transgenic T cell receptor specific for the ovalbumin-derived peptide SIINFEKL (OVA257–264) and do not have a residual antigen-specific repertoire due to loss of endogenous recombination events on the Rag1−/− background. The addition of bone marrow from Cd8−/− mice compensated for the lack of the CD4 T cells and B cells in the OT-I Rag1−/− mice. After confirming the success of the bone marrow reconstitution using congenic markers (Supplementary Fig. 5e–f), chimeric mice were infected with LCMV clone 13 and monitored up to 12 days after infection. Using FACS analysis, we confirmed the lack of LCMV-specific CD8+ GP33+ T cells in the spleens of OT-I Rag1−/−+Cd8−/− compared to wild-type+Cd8−/− control (Supplementary Fig. 5g,h). OT-I Rag1−/−+Cd8−/− chimeric mice were completely protected against LCMV-induced weight loss, while the wild-type+Cd8−/− chimeric mice lost approximately 20% of their body weight (Fig. 5f,g), indicating that antigen-specific priming of CD8+ T cells was required for LCMV-induced weight loss. Together, this data suggests that CD8 T cells triggered weight loss during the early stages of activation, following antigen-specific stimulation and is likely to occur independent of direct cell-to-cell interaction.
Discussion
In this study we employed a model of chronic viral infection to elucidate the inflammatory drivers of infection-associated cachexia (IAC).
IAC manifested 6 days after infection, preceding the peak of the adaptive immune response at day 12, then showed gradual resolution thereafter, which contrasts the terminal course of cancer-associated cachexia (CAC)3,5. We reported here a role for type I IFNs and CD8+ T cells in triggering IAC, but not IL-6, TNF and IFN-γ, cytokines that often mediate CAC6,8. The manifestation of IAC required IFNAR1 signaling specifically within T cells not adipocytes, in addition to antigen-specific CD8+ T cell receptor priming. Deletion of the cytolytic effector molecule perforin or the pharmacological blockade of T cell egress from lymphatic organs did not prevent cachexia, which suggested mechanisms other than classical CD8+ T cell-mediated cytotoxicity.
During CAC adipose lipid storage is depleted through increased ATGL- and HSL-mediated lipolysis, resulting in elevated circulating NEFA, a process which we also observed during LCMV-induced cachexia14,15. In contrast to CAC, however, we did not observe increased adipose tissue beiging and thermogenesis26,27. We found a depletion of inguinal, gonadal and brown adipose tissue, accompanied by increased vascular density at 6 and 8 days after infection, mainly in the inguinal compartment. Such remodeling may facilitate nutrient shuttling from the adipose tissue and modulate its exposure to metabolic and/or hormonal regulators, which could further amplify its depletion. Increased vascularization is a feature of obesity28; other characteristics of obesity, such as the inflammatory milieu and the metabolic changes observed during IAC, can be the cause of systemic inflammation, dyslipidemia and metabolic dysfunctions across different organs29.
Expression of leptin, a key regulator of appetite, lipolysis and the immune response17, showed a marked increase in adipose tissue of infected mice. However, the genetic ablation of leptin in Lepob/ob did not prevent weight loss and anorexia during LCMV infection. We observed a correlation between increased lipolysis and the increased circulating glucocorticoids, which are central drivers of the stress-response program and can induce lipolysis by activating ATGL and HSL24. In addition, glucocorticoids have endogenous roles in regulating systemic metabolism and are well known immunomodulatory molecules30–32, which situates them as potential candidates in mediating immunometabolic crosstalk during IAC.
Previous studies examining LCMV-associated morbidity used intracranial infection with LCMV Armstrong and reported weight loss that occurred as a direct result of anorexia mediated by MHCII-restricted CD4+ T cells33–35, which contrasts the weight loss independent of anorexia observed in our model. Force-feeding the mice during LCMV infection, or the supplementation with specific nutrients did not alleviate weight loss, but instead hampered recovery, particularly upon administration of glucose and casein. This is contrary to observations during infection with the influenza virus or injection of poly(I:C), in which administration of glucose and casein improved survival36. These differences may be due to distinct metabolic demands during different infections, influenced by viral life cycle, tropism and/or virus-induced immune responses, leading to either anorexia or cachexia. In line with this, Rag2−/− mice were not protected from weight loss during influenza infection. The detrimental effects of nutrient supplementation suggest that fasting metabolism is advantageous for efficient recovery. Fasting metabolism and caloric restriction had positive effects on survival and immune function in other models of bacterial infection and cancer36,37 and may have beneficial effects for cancer patients38,39. The underlying mechanisms of nutrient redistribution during fasting in infection- and cancer-associated cachexia warrant further investigations.
CD8+ T cells are drivers of immunopathology in chronic infections such as LCMV and HIV40. Studies have shown that virus clearance or persistence is determined early on during T cell receptor activation, though the exact mechanisms are not well understood12,41,42. We speculate that the onset of cachexia during the early stages of infection with LCMV clone 13 may be linked with the establishment of viral persistence. To that point, mice infected with either the acute Armstrong strain of LCMV or with low doses of LCMV clone 13 did not show weight loss. These observations raise the question of the potential evolutionary advantages of cachexia. Because of the correlation between cachexia and worsened prognosis in cancer patients, the syndrome has been presumed detrimental7. However, this assumption lacks a thorough mechanistic foundation and remains a matter of debate43. In infection, cachexia could serve an immunomodulatory role by tempering the immune response to limit immunopathology, similar to T cell exhaustion44. Alternatively, cachexia could facilitate a rapid increase of nutrient accessibility to fuel adaptive immune responses and/or other inflammatory processes. This would explain the excessive release of NEFA at day 6 after infection, a timepoint when energy metabolism shifts towards fat utilization. As of yet, relatively little is known about the regulation of nutrient redistribution across organs in inflammatory conditions, and how that may influence the immune response, metabolism and tissue repair45.
The current study contributes to understanding the mechanisms that induce cachexia in different inflammatory and disease settings. Studies of cachexia and its context-dependent pathogenesis can better the understanding of pathophysiological processes and systemic immunometabolism46,47. Comparative studies in infection and cancer could provide invaluable insights into the pathogenesis of cachexia and possible novel therapeutic strategies.
Methods
Contact for Resource and Reagent Sharing:
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Corresponding Contact, Andreas Bergthaler (abergthaler@cemm.oeaw.ac.at). Additional experimental information can be found in the Life Sciences Reporting Summary accompanying this study.
Experimental Model and Subject Details:
Mice:
In this study we used both wild type and genetically modified mouse models all on C57BL/6 background. These mice were bred under specific pathogen-free conditions at the Institute for Molecular Biotechnology of the Austrian Academy of Sciences, Vienna, Austria, as well as the Central Facility for Animal Research and Scientific Animal Welfare (ZETT) in Dusseldorf, Germany, and the Zentrales Tierlabor at the German Cancer Research Center (DKFZ) in Heidelberg. Genetically modified mice include: Ifnγ−/− 48 (003288-JAX), Tnf−/− 49 (005540- JAX), TnfrI−/− 50 (003242-JAX), Ifnar1−/− 51 (032045- JAX), Ifnar1fl/fl 52 (028256-JAX), Ifnar1fl/fl-AdipoqCre/+, Atglfl/fl-AdipoqCre/+ 53,54 (024278-JAX, 010803-JAX), Hslfl/fl-AdipoqCre//+ 54 (010803-JAX), Ifnar1fl/fl-Cd4Cre/+ 55 (017336-JAX), Ob/+ and Ob/Ob 56 (000632-JAX), Cd45.1 57 (MGI:4819849), Cd8−/− 58 (002665-JAX), Rag2−/− 59 (008449-JAX), OT-I Rag1−/− CD45.1 60,61 (003831-JAX) (002216-JAX), Prf1−/− 62 (002407-JAX). The Ifnar1fl/fl-Cd4Cre/+ mice were kindly provided by the lab of D. Pinschewer in Zürich, Switzerland. Ob/+ and Ob/Ob mice were purchased from Jackson Laboratories. Ifnar1fl/fl-AdipoqCre/+ mice were generated in house by crossing Ifnar1fl/fl mice to Hslfl/fl-AdipoqCre/+ mice, and then crossing out the Hslfl/fl allele. Animal experiments were performed at the Department for Biomedical Research of the Medical University of Vienna, Vienna, Austria, the Central Facility for Animal Research and Scientific Animal Welfare (ZETT) in Dusseldorf, Germany, the Division of Chronic Inflammation and Cancer, German Cancer Research Center, (DKFZ), Heidelberg, Germany and the Institute of Systems Biology in Seattle, USA. Mice were housed in individually ventilated cages, and experiments were performed in compliance with the respective animal experimental licenses (BMWFW-66.009/0199-WF/V/3b/2015, BMWFW-66.009/0395-WF/V/3b/2015, BMWFW-66.009/0379-WF/V/3b/2016, BMWFW-66.009/0360-WF/V/3b/2017 BMWFW-66.009/0318-II/3b/2012, A25, A504, A479, A424 respectively 35–9185.81/G-152/16, 35–9185.81/G-152/16, 01.24A.10) approved by the institutional ethical committees and guidelines and the Austrian respective, the German animal protection law and the state of Baden-Württemberg, as well as the Institutional Animal Care and Use Committees of the Institute of Systems Biology in Seattle. Within each experiment, mice were both age- and sex-matched, however, male and female mice were used interchangeably in between experiments, and results showed no sex-dependent differences in any of the measured parameters. During pair-feeding experiments, mice were housed individually to enable precise food intake measurements. The mice were allowed to adapt to solitary housing for minimally three days before the start of the experiment. After which, infected mice received food ad-libitum, whereas the pair-fed mice were placed on a restricted diet to match the daily intake of the control group. To generate chimeric mice, bone marrow cells were isolated from donor mice by flushing the femur and tibia with 10%FCS, β-mercaptoethanol (βME), 1% Penicillin-Streptomycin (P/S), and 2mM L-Glutamine. Cells were ran through strainer, centrifuged and then frozen in 10% DMSO, 75% FCS, 15% media. Media consisted on RPMI (10%FCS, βME, 1%P/S and 2mM L-glutamine, Penicillin-Streptomycin (P/S). The cells were then stored at −80°C until transfer. Recipient mice were subjected to 10.5Gy irradiation using BIOBEAM GM 2000 (Gamma Medical Services) one day post irradiation, mice received 1×107 cell from donor mice in a 1:1 ratio from donor mice. Mice were then allowed a period of recovery, before receiving two rounds of 200μg of anti-CD90 treatment. To examine the efficiency of the procedure, we performed FACS analysis of CD45.1 to CD45.2 ratio. For the adoptive transfer experiments, splenocytes from CD45.1 WT mice were transferred in a 1:1 ratio to WT and H2Kb−/−Db−/− mice one day prior to LCMV infection. At 8 days post infection mice were sacrificed and spleens were used for FACS analysis to test the number of CD8+CD45.1+ and CD8+CD45.2+ T cells.
Virus:
C57BL/6 mice were infected intravenously (i.v.) with high dose (2×106 focal-forming-units, FFU) LCMV strain clone 13, except when indicated otherwise. For dose titration experiment the virus was titrated by a factor of 10 to get the following inoculums: 2×106 FFU, 2×105 FFU, 2×104 FFU, 2×103 FFU and 2×102 FFU. For acute LCMV infection, mice were infected with high dose 2×106 FFU of LCMV strain ARM. Viral loads were determined using focal-forming assay (FFA)12, or via qPCR of LCMV nuclear protein. Probe: [6FAM]-CTTGCCGACCTCTTCAATGCGCAA-[BHQ1], forward primer: ACTGACGAGGTCAACCCGG, reverse primer: CAAGTACTCACACGGCATGGA. Influenza infections were performed using 1010.2 EID50/ml titer of PR8 strain of influenza.
Mouse Calorimetry:
Mice were individually placed in the PhenoMaster (TSE Systems) cages two days prior to infection and allowed to acclimate to the new environment. After infection, metabolic parameters such as food and water intake, oxygen consumption (VO2), carbon dioxide production (VCO2), respiratory exchange ratio (RER) and activity were measured every 12 minutes (5 measurements every hour) for 12 consecutive days.
Feeding experiments:
During pair-feeding experiments, all mice were placed in single housing for minimally three days before the start of the experiment, and monitored daily throughout the experimental course. Each day the food intake of LCMV-infected mice was measured (ad libitum diet), and a matching amount was given to the Pair-fed control mice (restricted diet) up to the point of harvest. For gavage feeding experiments, mice were weighed prior to infection and, subsequently, infected at the same time. The gavage took place starting from 4 until 7 days post infection with continuous access to chow diet ad libitum. During the gavage period mice received 0.5ml of either PBS or respective diet twice a day, constituting a supplementation of 1kcal/day. Abbot Promote-high protein diet (1kcal/ml) was used as the control diet due to the high compatibility with the nutritional profile of chow diet (Abbot Promote: calorie composition: 25% protein, 23% fat and 52% carbohydrates, chow diet: calorie composition: 24% protein, 18% fat and 58% carbohydrates)36.
Antibodies and Pharmachological Treatments:
Rat IgG1 isotype (MOPC-21, BE0083-BioXcell), Hamster IgG1 isotype (BE0091-BioXcell), Anti-TNFα (XT3.11, Rat IgG1, BE0058-BioXcell), anti-IFNγ (XMG1.2, Rat IgG1, BE0055-BioXcell), anti-IL1α (ALF-161, Hamster IgG1, BE0243-BioXcell), anti-IL6 (MP5–20F3, Rat IgG1, BE0046-BioXcell), anti-CD4 (YTS191, Rat IgG2b, BE0119-BioXcell), anti-CD8 (YTS169.4, Rat IgG2b, BE0117-BioXcell), anti-CD90 (T24, Rat IgG2b, BE0212-BioXcell). 0.5mg of anti-IFNγ, anti-TNFα, anti-IL6 neutralizing antibodies and their Rat IgG1 isotype control were intraperitonially (i.p.) injected one day prior to LCMV infection, and where re-administered every second day up to 7 days post infection. For anti-IL1α and its Hamster IgG1 isotype control, 0.2mg were i.p. injected every 3 days, starting 2 days prior to LCMV infection. For antibody depletion of CD4 and CD8 T cells, 0.2mg of mAbs were injected i.v. 2 and 1 days prior to infection. FACS analysis were performed from blood to confirm the efficiency to CD4+ and CD8+ T cell depletion.
FTY720 administration was performed as described in25. Mice received a daily gavage of 0.3mg/kg of FTY720 (SML0700, Sigma-Aldrich, US) dissolved in sterile water. The treatment was administered one day prior to infection and carried on until 8 days post infection. Blood Samples were drawn at 6 and 8 days post infection, and mice were scarified at 8 days post infection via cervical dislocation. Spleen and inguinal lymph nodes were subsequently taken for FACS analysis.
Imaging:
We used the EchoMRI™-100H (EchoMRI LCC, Houston, TX, USA) to longitudinally monitor body composition in a non-invasive manner in awake non-anesthetized mice. The results displayed shows the average of three consecutive measurements for each mouse within an experimental group during each timepoint. For MRI imaging, mice were euthanized using cervical dislocation prior to imaging and kept at 4°C until imaged. MRI measurements were performed on a 9.4 T animal MRI (BioSpec 94/30USR Bruker Biospin, Ettlingen Germany) equipped with a B-GA12SHP gradient and an 1H volume coil with an inner diameter of 40 mm (RFRES 400 1H 075/040 QSN TR, Bruker Biospin, Ettlingen Germany). A multi spin echo (MSME) sequence (repetition time[TR]= 500 ms, echo time[TE]=7.86 ms, averages =4) was used with and without fat suppression (fat suppression bandwidth=1401.17 Hz). A set of 24 Slices was acquired in coronal direction with a resolution of 100 μm2 in plain and a slice thickness on 1 mm. Total acquisition time was 16 minutes. To evaluate the size of the fat compartments, the fat suppressed images are subtracted from corresponding images without fat suppression. Postprocessing was performed using ParaVision V6 (Bruker Biospin, Ettlingen Germany).
Histology:
Mice were anesthetized using 15% Ketamine, 5% Xylazine in PBS, and then perfused with 20ml of 4% Paraformaldehyde (PFA) in PBS. Isolated tissue was then incubated in 4% PFA in PBS, before being transferred to PBS.
For H/E staining, histological evaluation was performed on 3μm-thick sections stained with hematoxylin-eosin (H-E). 4 to 5 representative images (20× magnification) were collected from each fat pad for quantitative analysis using ImageJ-Adiposoft plugin. Cutoffs were set to include all cells larger than 10μm and smaller than 100μm in diameter. Images were run through the automated Adiposoft analysis and then manually adjusted as needed.
For immunofluorescence staining, heat-induced antigen retrieval (Citrate buffer, pH6) and unspecific binding blocking (FCS block plus goat-anti mouse Fab block) was performed on PFA-fixed sections. Sections were subsequently incubated with following primary antibodies: rabbit anti-LCMV nucleoprotein sera (1:4000, generated by prime-boost immunization against purified LCMV-NP) and rat anti-CD8a (1:1000, eBioscience, #4SM15) diluted in DAKO RealTM Antibody Diluent (#S2022). Bound antibodies were visualized with appropriate species-specific Alexa Fluor 647 anti rat and Alexa Fluor 488 anti-rabbit (1:200, Jackson Immuno Research). Nuclei were stained with DAPI (1:2000, Invitrogen). Immunostained sections were scanned using Pannoramic 250 FLASH II (3DHISTECH) Digital Slide Scanner with objective magnification of 20 x. 4–5 representatives images were captured at 20X magnification and the image analysis was done using Cell Profiler 3.0.
Serum measures:
Blood samples were collected via tail vein for longitudinal measurements. For end point measurements, animals were anesthetized using 15% ketamine, 5% xylazine in PBS, blood was immediately drawn from the vena cava, and the animals where then euthanized via cervical dislocation. Triglyceride levels in the serum were determined using a Cobas C311 Analyzer (Roche). Non-esterified fatty acid (NEFA) content was evaluated enzymatically using NEFA kit (Wako Chemicals, Reagents:434–91795, Standard 270–77000).
We used the MILLIPLEX MAP Mouse Cytokine/Chemokine Magnetic Bead Panel-Premixed 32 Plex (MCYTMAG-70K-PX32, Millipore), to quantify a wide range of circulating cytokines and chemokines. Analysis was done following manufacturer’s instructions. To detect circulating levels of IFNα, we performed ELISA using rat anti-mIFN-α capture antibody (PBL Interferon Source 22100–1), rabbit anti-mIFN-α detection antibody (PBL Interferon Source 32100–1), anti-rabbit HRP secondary antibody (Jackson ImmunoResearch 711–036–152) and TMB solution (Life Tech 002023). For IFNβ measures we used, rat anti-mIFN-β capture antibody (PBL Interferon Source 22400–1), rabbit anti-mIFN-β detection Ab (PBL Interferon Source 32400–1), anti-rabbit HRP secondary antibody (Jackson ImmunoResearch 711–036–152) and TMB solution (Life Tech 002023).
Different ELISA kits were used to measure leptin, cortisol, corticosterone, freeT3, FreeT4 and Norepinephrine, according to manufacturer’s instructions. For leptin (PK-EL-68232DM, PromoCell, Germany), cortisol (EIAHCOR, Thermo Scientific, US), corticosterone (ADI-900–097, Enzo Biochem, US), free Triiodothyronine (fT3) (KET0004, Abbkine, China), free thyroxine (fT4) (CSB-E05080m, Cusabio, US), and noradrenaline (NE) (BA E-5200, Labor Diagnostika Nord, LDN, Germany).
Tissue measures:
Following euthanasia by cervical dislocation, all harvested tissue was instantly rinsed in PBS and then snap frozen in liquid nitrogen, until further processing. Importantly, inguinal lymph nodes were always separated from the inguinal adipose tissue during the harvest. RNA was isolated from adipose tissue for real-time PCR analysis using QIAzol lysis reagent and following the manufacturer’s instructions (Qiagen). However, after homogenization, an additional centrifugation step was required to remove excess oil layer. We then reverse transcribed the RNA samples into cDNA using the First Strand cDNA synthesis Kit (Fermentas). After which, we used TaqMan Fast Universal PCR Mastermix and TaqMan Gene Assays for Rplp0 (Mm00725448_s1), Cd36 (Mm00432403_m1), Lpl (Mm00434764_m1), Dgat2 (Mm00515643_m1), Ucp1 (Mm01244861_m1), Lep (Mm00434759_m1), and Adipoq (Mm00456425_m1).
For protein isolation from adipose tissue, samples were homogenized in 1ml sucrose buffer (250mM Sucrose, 1mM DTT, 1mM EDTA, 1:1000 protease inhibitor cocktail, pH7), then centrifuged at 4°C, 1000g for 10min. The infranatant layer was collected, and protein content was determined using Coomassie Plus Bradford Assay Kit (Thermo Scientific). 20–40μg of protein were used for SDS-Page analysis using NuPAGE® Novex 4–12% Bis-Tris Gels (Life Tech), Westran® Clear signal PVDF membranes (Whatman) and the following primary antibodies: anti-ATGL (2138S, cell signaling), anti-HSL (4107S, cell signaling), anti-phospho-HSL Ser660 (4126S, cell signaling), anti-D418-Perlipin (3470S, cell signaling) and anti-β-Actin (ab8224, Abcam). As a secondary antibody, we used horseradish peroxidase-conjugated anti-rabbit antibody (P0448, Dako). Detection of the luminescence signal was performed using the ChemiDoc XRS+ (Bio-Rad Laboratories, US) following the immunoreaction using Pierce ECL-Western blotting detection reagent (Thermo Scientific) and Amersham ECL select Western blotting detection reagent (GE Healthcare Life Sciences).
To quantify noradrenaline levels in iWAT, fat tissue was first weighed, cut into smaller pieces with scissors and then homogenized in noradrenaline extraction buffer (1 mM EDTA; 10mM HCl; 4 mM Sodium metabisulfite) using a mixer mill (TissueLyser II, Qiagen, Germany). Homogenates were rested for 1 hour at 4°C and adjusted to the same weight per volume percentage by adding deionized water (final volume per sample was 1 mL). To this mixture 200 μL chloroform were added and centrifugation was performed for 15 min at 13.000 g at 4 °C. The aqueous supernatant was quantified for noradrenaline using the corresponding ELISA kit.
Flow Cytometry analysis:
Single cell suspensions of inguinal lymph nodes (iLN) and spleen were prepared through mechanical disruption against 40–70μm cell strainers. Total cell count was quantified using Casy Cell Counter (Innovatis). To access immune cells in circulation, blood samples were collected in MEM-Heparin (1000U), and treated with RBC lysis buffer (eBioscience). Samples were then treated with anti-CD16/32 FcR-Block (clone 93, eBioscience), and subsequently stained with the respective antibodies as indicated in each experiment. CD8b.2: Pacific Blue (clone: 53–5.8), CD8a: PE-Cy7 (clone: 53–6.7), PerCP-Cy5 (clone:53–6.7), FITC (clone: 53–6.7), AF700 (clone: 53–6.7). CD4: Pacific Blue (clone: RM4–4). CD3: APC (clone: 17A2), PE-Cy7 (clone: 145–2C11). CD45.1: Pacific Blue (clone: A20), PE-Cy7 (clone:A20). CD45.2: PE (clone 104), APC (clone:104). CD44: BV605 (clone: IM7). Fixable Viability Dye eFluor™ 780 (APC-Cy7, Life Technologies). In order to enumerate the number of virus specific CD8+ T cells, samples were incubated with fluorophore-labelled GP33-specific tetramer at 37°C for 15min prior to FcR-block treatment. GP33-specific tetramers were obtained through the NIH Tetramer Core Facility, US. To quantify cell/μl of blood, a precise amount of blood was either directly pipetted into MEM-Heparin, or collected in EDTA-coated tubes and then transferred to MEM-Heparin. 123count™ eBeads Counting Beads (01–1234 Invitrogen) were used to quantify cell numbers according to manufacturer’s instructions.
RNA-sequencing and data processing:
To establish the best quality of RNA for RNA-sequencing analysis, and avoid RNA degradation as a result of the heat generated from homogenization, lymph node-free inguinal adipose tissue was smashed into powered over dry ice, using pre-cooled BioMasher (Kimble and Chase). After which 1ml of QIAzol lysis reagent (Qiagen) was added, and RNA was isolated following manufacturer’s instructions, with an additional starting centrifugation step to remove oil layer. Total RNA was quantified using Qubit 2.0 Fluorometric Quantitation system (Life Technologies), whereas the RNA integrity number (RIN) was determined using Experion Automated Electrophoresis System (Bio-Rad).
The sequencing libraries for the 12 samples were pooled and sequenced on Illumina HiSeq 3000/4000 instrument using 50bp single-end chemistry. Calling of the bases by the Illumina Realtime Analysis software were converted into BAM format using Illumina2bam and demultiplexed using BamIndexDecoder (https://github.com/wtsi-npg/illumina2bam)). The Tuxedo suite was performed for the RNA-seq analysis pipeline. Reads were mapped on the mouse reference genome (Mus musculus, Ensembl e87, December 2016) using TopHat2 (v2.0.10). Cufflinks (v2.2.1) was employed to assemble transcripts from spliced read alignments, using the Ensembl e87 transcriptome as the reference as well as de novo assembly of transcript models. Further, differential analysis of gene expression was quantified with Cuffdiff (v2.2.1). Transcriptome sets of all replicates for each sample group were combined with Cuffmerge. Expression values in graphs are reported as FPKM (fragments per kilobase pf transcript per million). Genes are considered expressed when they have FPKM values superior or equal to 1.
Principal component analysis (PCA) was performed on the gene set with a minimum average expression level across conditions of 5 FPKM. Only the 10% most variable (computed on the coefficient of variance) genes were considered for the PCA analysis. Heatmaps of different gene sets FPKM expression values were performed using a pearson distance measure with an average clustering method.
An additional RNA-seq analysis was performed at the transcriptome level in order to implement an interaction model (2 × 2 factorial design). The gene expression on the mouse Ensembl e87 transcripts was quantified from the previously TopHat2 mapped reads with featureCounts (v1.5.3). Raw read counts are further normalized with the voom function of the limma package 63. Normalized expression values, reported as log2 counts per million (CPM), were farther processed through limma’s empirical Bayes models. We implemented limma’s interaction model as a two (uninfected T cell-Ifnar1+/+, uninfected T cell-Ifnar1Δ/Δ) by two (uninfected, 6 days post LCMV infected) factorial design. Genes differentially modulated in the interaction model have been selected based on a minimum log2 CPM of 0, a minimum log2 fold-change absolute value of 0.6 and a maximum adjusted p-value of 0.05.
We performed enrichment analyses on the genes differentially modulated in the interaction model. Separately, up- and down-regulated genes were analyzed with Cytoscape ClueGO (v2.3.3), based on GO (Biological Processes, Molecular Functions, Immune System Process), InterPro, KEGG, Reactome and Wiki Pathways. Terms were called enriched based on a maximum p-value of 0.05 and a minimum of 3% gene overlap. GO Term Fusion and grouping has been performed. Enriched groups where farther ranked according to the group Bonferroni step-down adjusted p-value. Group lead terms were defined inside each group as the term with the lowest adjusted p-value of enrichment. In-depth metabolic Reactome pathway enrichment was performed with Cystoscape Reactome Functional Interaction app. Pathways with a maximum of False Discovery Rate (FDR) of 0.05 are retained.
Quantification and Statistical Analysis:
Results are displayed as mean ± SEM and were statistically analyzed as detailed in the figure legends using GraphPad Prism version 7.0. Statistically significant p-values were indicated as follows: * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001.
Data and Software Availability
Accession number for the row data of the RNAseq: GSE118819.
Supplementary Material
Acknowledgements
We thank S. Niggemeyer, S. Jungwirth and J. Riede for animal husbandry. The authors would like to thank L. Bakiri, V. Baracos, Y. Belkaid, R. Medzhitov, G. Superti-Furga, E. Wagner and R. M. Zinkernagel for valuable feedback and discussions.
This project has received funding from the European Research Council (ERC) under the European Union’s Seventh Framework Programme and Horizon 2020 research and innovation program (grant agreement No 677006, “CMIL” to A.B., No 340896 and “LipoCheX” to R. Z.,) from the German Research Council (SFB974, KFO217, LA-2558/5–1, and Jürgen Manchot Graduate School MOI III to P.A.L.), from the Austrian Science Fund (FWF P26766, to T.S.) and from the US National Institutes of Health (R01AI032972, U19AI100627 to A.Ad.). A.L. and M.S. are supported by DOC fellowships of the Austrian Academy of Sciences.
Footnotes
The Authors declare no competing interests.
References
- 1.Von Haehling S & Anker SD Treatment of cachexia: An overview of recent developments. Int. J. Cardiol 184, 726–742 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Baracos VE, Martin L, Korc M, Guttridge DC & Fearon KCH Cancer-associated cachexia. Nat. Rev. Dis. Prim 4, 1–18 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Fearon K et al. Definition and classification of cancer cachexia: an international consensus. Lancet Oncol 12, 489–495 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Kotler D Challenges to Diagnosis of HIV-Associated Wasting. JAIDS J. Acquir. Immune Defic. Syndr 37, S280–S283 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Tisdale MJ Cachexia in cancer patients. Nat. Rev. Cancer 2, 862–871 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Porporato PE Understanding cachexia as a cancer metabolism syndrome. Oncogenesis 5, 200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morley JE, Thomas DR & Wilson M-MG Cachexia: pathophysiology and clinical relevance. Am J clin Nutr (2006). [DOI] [PubMed] [Google Scholar]
- 8.Fearon KCH, Glass DJ & Guttridge DC Cancer cachexia: Mediators, signaling, and metabolic pathways. Cell Metab 16, 153–166 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Vaitkus JA & Celi FS The role of adipose tissue in cancer-associated cachexia. Exp. Biol. Med 242, 473–481 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petruzzelli M & Wagner EF Mechanisms of metabolic dysfunction in cancer-associated cachexia. Genes and Development 30, 489–501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flint TR, Fearon DT & Janowitz T Connecting the Metabolic and Immune Responses to Cancer. Trends Mol. Med 23, 451–464 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Bergthaler A et al. Viral replicative capacity is the primary determinant of lymphocytic choriomeningitis virus persistence and immunosuppression. Proc. Natl. Acad. Sci. U. S. A 107, 21641–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stamm A, Valentine L, Potts R & Premenko-Lanier M An intermediate dose of LCMV clone 13 causes prolonged morbidity that is maintained by CD4+ T cells. Virology 425, 122–132 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zechner R et al. FAT SIGNALS - Lipases and lipolysis in lipid metabolism and signaling. Cell Metab 15, 279–291 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsoli M et al. Depletion of white adipose tissue in cancer cachexia syndrome is associated with inflammatory signaling and disrupted circadian regulation. PLoS One 9, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das SK et al. Adipose triglyceride lipase contributes to cancer-associated cachexia. Science (80-. ) 333, 233–238 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Ruud J & Brüning JC Metabolism: Light on leptin link to lipolysis. Nature 527, 43–44 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Baatar D, Patel K & Taub DD The effects of ghrelin on inflammation and the immune system. Mol. Cell. Endocrinol 340, 44–58 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Kandarian SC et al. Tumour-derived leukaemia inhibitory factor is a major driver of cancer cachexia and morbidity in C26 tumour-bearing mice. J. Cachexia. Sarcopenia Muscle 1–12 (2018). doi: 10.1002/jcsm.12346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel HJ & Patel BM TNF-α and cancer cachexia: Molecular insights and clinical implications. Life Sci 170, 56–63 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Bhattacharya A et al. Superoxide Dismutase 1 Protects Hepatocytes from Type I Interferon-Driven Oxidative Damage. Immunity 43, 974–986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curtsinger JM, Valenzuela JO, Agarwal P, Lins D & Mescher MF Cutting Edge: Type I IFNs Provide a Third Signal to CD8 T Cells to Stimulate Clonal Expansion and Differentiation. J. Immunol 174, 4465–4469 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Kolumam GA, Thomas S, Thompson LJ, Sprent J & Murali-Krishna K Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J. Exp. Med 202, 637–650 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu C et al. Direct Effect of Glucocorticoids on Lipolysis in Adipocytes. Mol. Endocrinol 23, 1161–1170 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinschewer DD et al. FTY720 Immunosuppression Impairs Effector T Cell Peripheral Homing Without Affecting Induction, Expansion, and Memory. J. Immunol 164, 5761–5770 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Petruzzelli M et al. A switch from white to brown fat increases energy expenditure in cancer-associated cachexia. Cell Metab 20, 433–447 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Kir S et al. PTH/PTHrP Receptor Mediates Cachexia in Models of Kidney Failure and Cancer (2016). doi: 10.1016/j.cmet.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao Y Angiogenesis and vascular functions in modulation of obesity, adipose metabolism, and insulin sensitivity. Cell Metab 18, 478–489 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Vegiopoulos A, Rohm M & Herzig S Adipose tissue: between the extremes. EMBO J 36, 1999–2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quatrini L et al. Endogenous glucocorticoids control host resistance to viral infection through the tissue-specific regulation of PD-1 expression on NK cells. Nat. Immunol 19, 954–962 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller a H. et al. Effects of viral infection on corticosterone secretion and glucocorticoid receptor binding in immune tissues. Psychoneuroendocrinology 22, 455–74 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Jamieson AM, Yu S, Annicelli CH & Medzhitov R Influenza Virus-Induced Glucocorticoids Compromise Innate Host Defense against a Secondary Bacterial Infection. Cell Host Microbe 7, 103–114 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doherty PC, Hou S & Southern PJ Lymphocytic choriomeningitis virus induces a chronic wasting disease in mice lacking class I major histocompatibility complex glycoproteins. J Neuroimmunol 46, 11–17 (1993). [DOI] [PubMed] [Google Scholar]
- 34.Hildeman D & Muller D Immunopathologic Weight Loss in Intracranial LCMV Infection Initiated by the Anorexigenic Effects of IL-1β. Viral Immunol 13, 273–285 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Kamperschroer C & Quinn DG The Role of Proinflammatory Cytokines in Wasting Disease During Lymphocytic Choriomeningitis Virus Infection. J. Immunol 169, 340–349 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Wang A et al. Opposing Effects of Fasting Metabolism on Tissue Tolerance in Bacterial and Viral Inflammation. Cell 166, 1512–1525.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pietrocola F et al. Caloric Restriction Mimetics Enhance Anticancer Immunosurveillance. Cancer Cell 30, 147–160 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nencioni A, Caffa I, Cortellino S & Longo VD Fasting and cancer: molecular mechanisms and clinical application. Nat. Rev. Cancer (2018). doi: 10.1038/s41568-018-0061-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Longo VD & Mattson MP Fasting: Molecular mechanisms and clinical applications. Cell Metabolism 19, 181–192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rouse BT & Sehrawat S Immunity and immunopathology to viruses: What decides the outcome? Nat. Rev. Immunol 10, 514–526 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Virgin HW, Wherry EJ & Ahmed R Redefining Chronic Viral Infection. Cell 138, 30–50 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Sullivan BM, Teijaro JR, De La Torre JC & Oldstone MBA Early Virus-Host Interactions Dictate the Course of a Persistent Infection. PLoS Pathog 11, 1004588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Danai LV et al. Altered exocrine function can drive adipose wasting in early pancreatic cancer. Nature 558, 600–604 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wherry EJ & Kurachi M Molecular and cellular insights into T cell exhaustion HHS Public Access. Nat Rev Immunol 15, 486–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perry RJ et al. Hepatic acetyl CoA links adipose tissue inflammation to hepatic insulin resistance and type 2 diabetes. Cell 160, 745–758 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Norata GD et al. The Cellular and Molecular Basis of Translational Immunometabolism. IMMUNI 43, 421–434 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Buck MD, Sowell RT, Kaech SM & Pearce EL Metabolic Instruction of Immunity. Cell 169, 570–586 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Accession number for the row data of the RNAseq: GSE118819.