Abstract
Regulatory T cells (Treg cells) maintain host self-tolerance but are a major barrier to effective cancer immunotherapy. Treg cells subvert beneficial anti-tumor immunity by modulating inhibitory receptor expression on tumor-infiltrating lymphocytes (TILs); however, the underlying mediators and mechanisms have remained elusive. Here we found that the cytokines IL-10 and IL-35 (Ebi3–IL-12α heterodimer) were divergently expressed by Treg cell subpopulations in the tumor microenvironment (TME) and cooperatively promoted intratumoral T cell exhaustion by modulating multiple inhibitory receptor expression and exhaustion-associated transcriptomic signature of CD8+ TILs. While expression of BLIMP1 (encoded by Prdm1) was a common target; IL-10 and IL-35 differentially affected effector T cell versus memory T cell fates, respectively, highlighting their differential, partially overlapping but non-redundant regulation of anti-tumor immunity. Our results reveal previously unappreciated cooperative roles for Treg cell-derived IL-10 and IL-35 in promoting BLIMP1-dependent exhaustion of CD8+ TILs that limits effective anti-tumor immunity.
Regulatory T cells (Treg cells) are a specialized suppressive CD4+ T cell population capable of limiting deleterious immune responses to self and foreign antigens that underlie autoimmune and chronic inflammation1,2. Conversely, Treg cells suppress beneficial anti-tumor immunity and thereby pose an impediment to effective immunotherapies3,4. Indeed, increased Treg cells frequencies and a reduced CD8+/Treg cell-ratio in tumors are linked to poor prognosis in multiple cancers3,5. Although Treg cell depletion dramatically reduces tumor burden, the ensuing autoimmune sequelae limit the utility of this approach in the clinic6,7. Hence, current trials are evaluating strategies targeting receptors preferentially enriched on intratumoral Treg cells (CCR4, GITR, OX40, CTLA4)8. Treg cells utilize a plethora of suppressive mechanisms – inhibitory cytokine secretion, metabolic disruption, modulation of antigen-presenting cell (APC) function, and cytolysis of effector cells; often in a target cell and tissue-specific manner1. While targeting suppressive mechanisms selectively or predominantly employed by intratumoral Treg cells would be efficacious, the precise mechanisms and predominant suppressive mediators that promote T cell exhaustion and dominant suppression in the tumor microenvironment (TME) remain poorly defined.
One of the major contact-independent modes of Treg cell suppression is via secretion of inhibitory cytokines (IL-10, IL-35, and TGF-β)1,9. The established suppressive cytokine duo (IL-10 and TGF-β) plays a critical role in steady-state immune homeostasis and taming exuberant responses at environmental interfaces. Perturbation of IL-10 and TGF-β signaling improved the function of exhausted T cells in chronic viral infections10–12. Co-targeting Treg cells or IL-10 along with the PD-1 immune checkpoint pathway resulted in a synergistic reversal of T cell exhaustion13,14. IL-35 is required for maximal Treg cell suppressor function and contributes to the regulatory milieu in numerous disease states15,16. Previous work demonstrated enrichment of IL-35 expression on tumor-infiltrating Treg cells in B16 tumors and a role of IL-35 in promoting inhibitory receptor expression (PD-1, TIM3, LAG3) on intratumoral T cells7. Thus, while Treg cells and suppressive cytokines have been linked to T cell exhaustion in chronic settings, the molecular mechanisms and the relative contribution of Treg cell-derived suppressive cytokines underlying Treg cell-induced exhaustion remain obscure.
In this study, we report a divergent and largely non-overlapping IL-10 and IL-35 expression pattern on Treg cells infiltrating both murine tumors as well as patients with non-small cell lung cancer (NSCLC). We further demonstrate that these IL-10+ and Ebi3+ Treg cell populations are not distinct lineages, but rather transitory states with concordant transcriptional and T cell receptor (TCR) profiles. This transition is inducible via TCR stimulation of purified Treg cell subpopulations in vitro, potentially reflecting plasticity in inhibitory cytokine expression in chronic environments. While the two cytokines target a common BLIMP1 axis to promote the exhausted intratumoral T cell state; IL-10 and IL-35 differentially impacted effector versus memory generation, respectively. Our results uncover the previously unappreciated adaptive plasticity of IL-10 and IL-35 inhibitory cytokine expression by Treg cell subpopulations in the TME and how they cooperatively impinge on T cell function to promote exhaustion and limit anti-tumor immunity.
Results
Divergent IL-10 and IL-35 expression on Treg cells in murine tumors and NSCLC patients
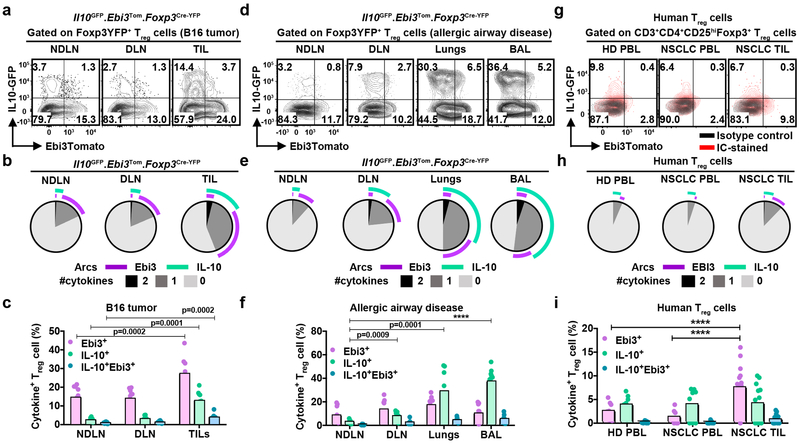
Intratumoral Treg cells are more suppressive than their peripheral counterparts in both murine models and human tumors owing to their heightened activation status3,4. Thus, we reasoned that the major suppressive mediators might be co-expressed on intratumoral Foxp3+ Treg cells to maximize their functional impact. To address this hypothesis, we generated triple reporter mice (Il10GFP.Ebi3Tom.Foxp3Cre-YFP; Il10 for IL-10, Ebi3 for IL-35, and Foxp3 for Treg cells, respectively7,17,18; Fig. 1a, Supplementary Fig. 1a). We observed largely segregated expression of IL-10 and Ebi3 by Treg cells in various lymphoid and non-lymphoid organs, with minimal co-expression (Supplementary Fig. 1b); consistent with a previous report19. The only tissues with a notable (>5%) population of Treg cells co-expressing IL-10 and Ebi3 at steady state were skin and lamina propria, likely due to elevated IL-10+ Treg cells at these environmental interfaces (Supplementary Fig. 1b). Specifically, in the B16 TME, there was an approximately 4-fold and 2-fold intratumoral enrichment of IL-10+ (GFP+YFP+) and Ebi3+ (Tom+YFP+) Treg cells, respectively, compared to the periphery (Fig. 1a-c).
Figure 1: Reciprocal expression of IL-10 and IL-35 on both mouse and human Treg cells.
a, Representative flow plots depicting the expression and distribution of IL-10+ and Ebi3+ cells within Foxp3YFP+ Treg cells, isolated from B16-tumor bearing Il10GFP.Ebi3Tom.Foxp3Cre-YFP mice 14 days post tumor inoculation. The expression of IL-10 (GFP+) and IL-35 (Ebi3-Tomato+) assessed in the non-draining axillary and brachial lymph nodes (NDLN), draining inguinal lymph nodes (DLN) and B16 tumor-infiltrating lymphocytes (TIL).
b, SPICE plots depicting co-expression pattern of IL-10 and Ebi3 on TIL Treg cells as in (a).
c, Scatter-bar plot depicting percent distribution of cytokine single- and double-positive Treg cells as in (a) (n=11 mice). Bars represent mean values. Statistical significance was determined by Two-way ANOVA with Holm-Sidak multiple comparisons (p-values as indicated).
d, Representative flow plots depicting expression and distribution of IL-10+ and Ebi3+ cells within Foxp3-YFP+ Treg cells, isolated from day 14 allergic inflammation induced Il10GFP.Ebi3Tom.Foxp3Cre-YFP mice. NDLN, DLN, Lungs, and Bronchoalveolar lavage fluid (BAL).
e, SPICE plots depicting co-expression pattern of IL-10 and Ebi3 on Treg cells as described in (d).
f, Scatter-bar plot depicting percentage of cytokine single- and double-positive Treg cells as described in (d) (n=12 mice). Bars represent mean values. Statistical significance was determined by Two-way ANOVA with Holm-Sidak multiple comparisons (****p<0.0001 and other p-values as indicated).
g, Representative flow plots depicting the expression and distribution of IL-10 and IL-35 in human Treg cells (CD4+CD25HIFoxp3+) obtained from healthy donor (HD) PBMC (PBL), NSCLC PBL, or NSCLC TILs. Cells were stimulated overnight with plate-bound anti-CD3 and anti-CD28 in the presence of hIL2, followed by four hours of stimulation with PMA-ionomycin prior to surface and intracellular (IC)-staining for IL-10 and IL-35 (EBI3) expression analysis. Isotype control (black) and IC-stained (red) are overlaid. Data representative of three independent experiments.
h, SPICE plots depicting co-expression pattern of IL-10 and EBI3 on Treg cells as described in (g).
i, Scatter-bar graph depicting percent distribution of inhibitory cytokines as in (g). Bars represent mean values HD PBL (n=9), NSCLC PBL (n=9), and NSCLC TIL (n=16). Statistical significance was determined by Two-way ANOVA with Holm-Sidak multiple comparisons (****p<0.0001).
Furthermore, this divergent inhibitory cytokine distribution in Treg cells was not a characteristic limited to the somewhat TH1-polarized TME, as we noted a similarly divergent expression pattern in a TH2-polarized fungal protease (Aspergillus oryzae) model of allergic airway disease (Fig. 1d-f)20. The enrichment of IL-10+ Treg cells was more pronounced in the allergic environment than the B16 TME, resulting in an approximately 10-fold expansion compared to the periphery, supporting the notion of adaptive plasticity in the relative abundance of IL-10+ and IL-35+ subpopulations depending on the nature of the inflammatory microenvironment. Treg cell-restricted deletion of Ebi3 in Il10GFP.Ebi3L/L-Tom.Foxp3Cre-YFP mice did not result in a compensatory change in the distribution of these Treg cell subpopulations in either TH1- or TH2-polarized inflammatory environment (Supplementary Fig. 1c-h). Importantly, human Treg cells from healthy donors and non-small cell lung cancer (NSCLC) patients also showed a similar segregated IL-10 and IL-35 expression pattern with a minimal percentage of double cytokine positive Treg cells (Fig. 1g-i). In this case, there was also an increased percentage of Ebi3+ intratumoral Treg cells, highlighting the translational importance of understanding the developmental and functional relationship between these intratumoral Treg cell-subpopulations. Collectively, these data highlight preferential enrichment and divergent inhibitory cytokine expression pattern on mouse and human intratumoral Treg cells.
Adaptive plasticity of cytokine expression and comparable transcriptome of intratumoral IL-10+ and IL-35+ Treg cell subpopulations
To dissect the divergent cytokine expression pattern and the preferential generation of single inhibitory cytokine positive Treg cell subpopulations, we performed single-cell RNAseq (scRNAseq) comparing Treg cells isolated from naive, unchallenged lymph nodes (LNs) or day 14 B16 tumors from Foxp3Cre-YFP mice (Fig. 2a). IL-10+ and Ebi3+ Treg cells exhibited comparable transcriptomic signatures (Fig. 2b-c). Although unsupervised K-means clustering identified six unique Treg cell clusters (Supplementary Fig. 2a), the distribution ratio of IL-10+ versus Ebi3+ Treg cell subpopulations among those clusters was comparable, especially for intratumoral Treg cells (Supplementary Fig. 2b). Consistent with the scRNAseq analyses, T cell receptor beta chain (TCRβ)-sequencing of IL-10+ and Ebi3+ Treg cell subpopulations isolated from non-draining LNs (NDLN) and B16 tumors from Il10GFP.Ebi3Tom.Foxp3Cre-YFP mice showed a comparable clonal enrichment (Supplementary Fig. 2c). We did not observe any bias in TCR Vβ-gene usage or CDR3-length, which can play a role in regulating T cell development and differentiation via TCR affinity and signaling strength21,22 (Fig. 2d, Supplementary Fig. 2d). Furthermore, substantial TCR clonal overlap noted among intratumoral Treg cell subpopulations is consistent with the lack of distinct transcriptomic signatures (Supplementary Fig. 2e). TCR signaling plays a crucial role in the production of both IL-10 and IL-35[23]. Importantly, we noted that TCR-stimulation in vitro was sufficient to induce Il10 and Ebi3 expression in purified Treg cell subpopulations (Fig. 2e), inferring extensive developmental plasticity. There was a preferential upregulation of Ebi3 expression observed during in vitro fate mapping, which is consistent with a progressive enrichment of Ebi3+ (either IL-10−Ebi3+ or IL-10+Ebi3+) Treg cells revealed by diffusion pseudotime analysis (Fig. 2f-g). Furthermore, while it has been reported that the development and function of Ebi3+ Treg cells are independent of BLIMP1 expression19, our results indicate that BLIMP1 may also play a key role in supporting the maintenance and functions of Ebi3+ Treg cells as approximately 90% of TIL Treg cells express BLIMP1 and 30–50% of which are Ebi3+ (Supplementary Fig. 2f, Fig. 1a-c). These observations suggest that TCR signaling, and perhaps the suppressive tumor milieu, might drive this adaptive plasticity and lead to the generation of Treg cell subpopulations marked by transitory IL-10 and IL-35 expression.
Figure 2: Adaptive plasticity of IL-10+ and IL-35+ Treg cells.
a, scRNAseq tSNE plots of bulk Treg cells from naive LNs or 14 days post inoculation B16 tumors (TIL Treg cells) from Foxp3Cre-YFP mice.
b, scRNAseq tSNE plot depicting the expression of Il10 and Ebi3 in individual Treg cells overlaid on the same tSNE plot as in (a).
c, Heat maps contrasting the top 30 genes selected based on the differential expression analysis of Treg cells utilizing the two-sided Negative Binomial Exact test, demonstrating the lack of distinct transcriptional signatures. p-values were adjusted to control the false discovery rate (FDR) set at 0.05. Treg cells were first stratified into IL-10−Ebi3−, IL-10+Ebi3−, IL-10−Ebi3+, and IL-10+Ebi3+ as described in (b). (Naive LN): IL-10−Ebi3− (n=717 cells), IL-10+Ebi3− (n=3 cells), IL-10−Ebi3+ (n=83 cells), and IL-10+Ebi3+ (n=3 cells). (TIL): IL-10−Ebi3− (n=491 cells), IL-10+Ebi3− (n=88 cells), IL-10−Ebi3+ (n=491 cells), and IL-10+Ebi3+ (n=111 cells).
d, Bar graphs with overlaid scatter-dots depicting the TCR Vβ gene usage, comparing the Treg cells subpopulations and effector T cells. Three independent B16 tumor-bearing Il10GFP.Ebi3Tom.Foxp3Cre-YFP mice were used to harvest Treg cell subpopulations for sequencing without pooling. Bars represent mean values.
e, In vitro tracing of adaptive plasticity in cytokine expression by TCR-stimulation. (top): Naive Treg cells from LN and spleens were double-sort purified and stimulated with anti-CD3/CD28-coated beads in the presence of hIL2 and CD11c+ cells for 72 hours, followed by FACS analysis. (bottom): Stacked bar graph summarizing four independent experiments. Statistical significance was determined by Two-way ANOVA with Holm-Sidak multiple comparisons (#p=0.0073, *p=0.0016, **=P=0.0017, ***p=0.0002, ****p<0.0001).
f, Diffusion pseudo-time analysis depicting the stochastic oscillation of IL-10 and IL-35 (Ebi3) expression based on the transcriptomic features of sequenced Treg cells from the scRNAseq experiment.
g, Stacked-bar graph demonstrating the distribution of indicated Treg subpopulations along the Pseudotime projection as analyzed in (f). The Pseudotime projection was evenly divided into 10 fractions and the percent distribution of each Treg cell subpopulation was calculated.
We also performed bulk RNAseq-based transcriptome analysis on purified LN and tumor (TIL) Treg cell subpopulations defined by Il10 and Ebi3 reporter expression (Supplementary Fig. 3a), to further investigate the transcriptomic relationship between these Treg cell subpopulations with greater sequencing depth. Principal component analysis (PCA) of differentially regulated genes clearly separated LN vs. TIL Treg cells, irrespective of cytokine expression pattern (Supplementary Fig. 3a). Consistent with the scRNAseq data, bulk profiling revealed no striking transcriptomic differences between intratumoral Treg cell subpopulations (Supplementary Fig. 3b). Modest differences were noted in expression patterns of genes encoding co-signaling molecules (Cd226, Tnfsf11, Cd2, Lag3, Cd27, Cd28), likely reflecting their relative suppressive capacity noted in vitro (Supplementary Fig. 3c-f). Collectively, the concordant transcriptional and TCR profiles highlight these IL-10+ and Ebi3+ Treg cell subpopulations as plastic transitory states, rather than distinct subsets.
Cooperative regulation of anti-tumor immunity by IL-10+ and IL-35+ Treg cells
It was previously shown that mice with a Treg cell restricted Ebi3 deletion, and thus loss of IL-35 production, exhibit reduced tumor burden7. To assess the functional impact of IL-10- and IL-35-producing Treg cell subpopulations on the TME, we compared the growth rate of three transplantable tumor models; B16 and BrafPten (clone 24) melanoma models and EL4 thymoma in mice with Treg cells that are unable to produce IL-10 (Il10L/L.Foxp3Cre-YFP), IL-35 (Ebi3L/L-Tom.Foxp3Cre-YFP) or either cytokine (Il10L/L.Ebi3L/L-Tom.Foxp3Cre-YFP). Treg cell-restricted deletion of Il10 or Ebi3 resulted in a comparable reduction in tumor growth (Fig. 3a-c). Dual-deletion did not exhibit a significant additive or synergistic reduction in tumor burden in both the melanoma models and only a marginal effect on EL4, suggesting that Treg cell-derived IL-10 and IL-35 might regulate anti-tumor immunity through a common, co-operative pathway.
Figure 3: Cooperative, redundant regulation of CD8+ TILs by Treg cell-derived IL-10 and IL-35.
(a-c): Tumor growth curve of Foxp3Cre-YFP, Il10L/L.Foxp3Cre-YFP, Ebi3L/L-Tom.Foxp3Cre-YFP, and Il10L/L.Ebi3L/L-Tom.Foxp3Cre-YFP mice inoculated with 1.25×105 tumor cells intradermally. (a) B16 (**p=0.0019, ***p=0.0004, ****p<0.0001), (b) BrafPten clone 24 (*p=0.0263, ****p<0.0001), (c) EL4 (#p=0.0054, *P=0.0047, **p=0.0043, ***p=0.0003, ****p<0.0001). Data averaged from 3 independent experiments with an indicated total number of mice per genotype. Each measurement time point represents mean value with s.e.m. error bar. Statistical significance was determined by Two-way ANOVA with Holm-Sidak multiple comparisons.
d, Representative flow plots depicting the expression of inhibitory receptors (PD-1 versus other inhibitory receptors: TIM3, LAG3, TIGIT, and 2B4) on CD8+ T cells infiltrating d14 B16 tumor-bearing Foxp3Cre-YFP (n=13), Il10L/L.Foxp3Cre-YFP (n=9), Ebi3L/L-Tom.Foxp3Cre-YFP (n=11), and Il10L/L.Ebi3L/L-Tom.Foxp3Cre-YFP (n=10) mice. SPICE plots depict the co-expression of multiple inhibitory receptors.
e, Scatter-bar graphs tabulating the percent distribution of inhibitory receptor-negative (0 & 1 inhibitory receptor-expressing effector-like) and multi-inhibitory receptor+ (3-5 inhibitory receptor-expressing exhausted) CD8+ TILs as in (d). Bars represent mean values. Statistical significance was determined by One-way ANOVA with Holm-Sidak multiple comparisons (****p<0.0001 and other p-values indicated).
f, Representative flow plots depicting expression of inhibitory receptors (PD-1 versus the other inhibitory receptors – TIM3, LAG3, TIGIT and 2B4) on CD4+Foxp3− T cells harvested from d14 B16 tumor-bearing Foxp3Cre-YFP (n=13), Il10L/L.Foxp3Cre-YFP (n=9), Ebi3L/L-Tom.Foxp3Cre-YFP (n=11), and Il10L/L.Ebi3L/L-Tom.Foxp3Cre-YFP (n=10) mice. SPICE plots depicting expression and co-expression of multiple inhibitory receptors on CD4+Foxp3− TILs.
g, Bar graphs representing the percent distribution of inhibitory receptor-negative (0 & 1 inhibitory receptor-expressing effector-like) and multi-inhibitory receptor+ (4 & 5 inhibitory receptor-expressing exhausted) CD4+Foxp3− T cells as in (f). Bars represent mean values. Statistical significance was determined by One-way ANOVA with Holm-Sidak multiple comparisons (****p<0.0001 and other p-values indicated).
(d-g) Data averaged from 3 independent experiments.
IL-35 can modulate inhibitory receptor expression on CD8+ and CD4+ TILs7. A substantial fraction of CD8+ and CD4+ TILs in B16 tumor-bearing control Foxp3Cre-YFP mice were PD-1hi and co-expressed multiple inhibitory receptors (TIM3, LAG3, TIGIT and 2B4, referred to as PD-1himulti-inhibitory receptor+ TILs) (Fig. 3d-g, Supplementary Fig. 4a-b). Consistent with previous observations, Ebi3L/L-Tom.Foxp3Cre-YFP exhibited a drastic reduction of CD8+ PD-1himulti-inhibitory receptor+ TILs on day 14. Interestingly, Il10L/L.Foxp3Cre-YFP mice also exhibited a reduction in the CD8+ PD-1himulti-inhibitory receptor+ TILs, albeit to a lesser extent. Mice with Treg cell-specific deletion of both II10 and Ebi3 (Il10L/L.Ebi3L/L-Tom.Foxp3Cre-YFP) demonstrated a near complete loss of the PD-1hi and multi-inhibitory receptor+ populations and significant enrichment of inhibitory receptor-negative and PD-1int CD8+ T cells (Fig. 3d-e, Supplementary Fig. 4a-b). B16 TIL analysis at a later time point (day 20) revealed a less extensive reduction of inhibitory receptor expression on CD8+ TILs from Il10L/L.Foxp3Cre-YFP mice while CD8+ TILs from Ebi3L/L-Tom.Foxp3Cre-YFP and Il10L/L.Ebi3L/L-Tom.Foxp3Cre-YFP mice still exhibited diminished inhibitory receptor-expression (Supplementary Fig. 4c-d). Although the overall extent of PD-1hi and multi-inhibitory receptor expression was not as prominent on CD4+ TILs, Treg cell-restricted deletion of Il10 and Ebi3 showed a comparable, substantial impact on PD-1himulti-inhibitory receptor+ CD4+ TILs at both time points as their CD8+ counterparts (Fig. 3f-g, Supplementary Fig. 4e-f). These data suggest that while both Treg cell-derived IL-10 and IL-35 function cooperatively in driving inhibitory receptor induction on TILs, IL-35 may play a more dominant role.
We also analyzed the impact of inhibitory cytokine-driven multi-inhibitory receptor induction on CD8+ T cell differentiation as well as other cell types in the TME. Increased cellularity and cytokine production were noted in tumors of Il10L/L.Foxp3Cre-YFP mice (Fig. 4a-b, Supplementary Fig. 5a-b), while enhanced TCM differentiation was noted in both Ebi3L/L-Tom.Foxp3Cre-YFP and Il10L/L.Ebi3L/L-Tom.Foxp3Cre-YFP TMEs (Fig. 4c). We also examined myeloid populations and observed a comparable reduction of PD-L1 expression and increase in T cell stimulatory molecules such as MHC-II and CD80 following the loss of Treg cell-derived IL-10 or IL-35 (Supplementary Fig. 5c-e). There was an increase in the M1-like tumor-associated macrophage (TAM) population in Il10L/L.Foxp3Cre-YFP mice which conversely was decreased in Ebi3L/L-Tom.Foxp3Cre-YFP and Il10L/L.Ebi3L/L-Tom.Foxp3Cre-YFP mice (Supplementary Fig. 5f)24,25. Collectively, these data point to the differential impact of Treg cell-restricted deletion of these two cytokines on the TME, with an apparent greater impact of IL-10 in limiting effector function and proliferation, whereas IL-35 seems to limit memory differentiation.
Figure 4: Non-redundant regulation of anti-tumor immunity by IL-10+ and IL-35+ Treg cells.
a, Representative flow plots depicting the expression of IFN-γ and TNF-α. Scatter-bar plots representing the percent distribution of CD8+ TILs expressing IFN-γ and/or TNF-α infiltrating d20 B16 tumor-bearing Foxp3Cre-YFP (n=10), Il10L/L.Foxp3Cre-YFP (n=12), Ebi3L/L-Tom.Foxp3Cre-YFP (n=12), and Il10L/L.Ebi3L/L-Tom.Foxp3Cre-YFP (n=9) mice.
b, Representative flow plots depicting the expression of IFN-γ and TNF-α. Scatter-bar plots representing the percent distribution of CD4+Foxp3− TILs expressing of IFN-γ and/or TNF-α as in (a). Foxp3Cre-YFP (n=10), Il10L/L.Foxp3Cre-YFP (n=12), Ebi3L/L-Tom.Foxp3Cre-YFP (n=12), and Il10L/L.Ebi3L/L-Tom.Foxp3Cre-YFP (n=9).
c, Representative flow plots depicting gating strategy to identify CD8+ TCM TILs (CD44+KLRG1−CD127+CD62L+TCF1+) (left). The percent distribution of CD8+ TCM was tabulated in scatter-bar graphs (right), analyzed 14 days (Foxp3Cre-YFP (n=8), Il10L/L.Foxp3Cre-YFP (n=9), Ebi3L/L-Tom.Foxp3Cre-YFP (n=10), and Il10L/L.Ebi3L/L-Tom.Foxp3Cre-YFP (n=12) mice) and 18 days (Foxp3Cre-YFP (n=12), Il10L/L.Foxp3Cre-YFP (n=13), Ebi3L/L-Tom.Foxp3Cre-YFP (n=16), and Il10L/L.Ebi3L/L-Tom.Foxp3Cre-YFP (n=10) mice) post-B16 tumor inoculation.
(a-c) Data averaged from 3 independent experiments. Bars represent mean values. Statistical significance was determined by One-way ANOVA with Holm-Sidak multiple comparisons (****p<0.0001 and other p-values indicated).
Treg cell-derived IL-35 and IL-10 can directly induce inhibitory receptor expression on intratumoral CD8+ T cells
We next investigated whether IL-10 and IL-35 were directly impacting CD8+ T cells in the TME or if this regulation required an intermediate or accessory cell. We created an experimental system in which only CD8+ T cells were unable to respond to IL-10 or IL-35 utilizing a Rag1−/− reconstitution model (Fig. 5a). Reconstitution with IL-35 receptor-deficient (CD4Cre.Il6stL/L.IL12rb2−/−; referred to as IL-35R−/−)26 or IL-10 receptor-deficient (Il10rb−/−; referred to as IL-10R−/−)27 CD8+ T cells recapitulated the reduced B16 tumor growth observed in mice with Treg cells that cannot make IL-35 and IL-10, respectively (Fig. 5b). Although inhibitory receptor expression was substantively reduced on IL-35R.KO CD8+ T cells, there was only a partial reduction on IL-10R.KO CD8+ T cells assessed at both d14 and d18 timepoints (Fig. 5c-d, Supplementary Fig. 5g-h). These results in the Rag1−/− reconstitution model were consistent with our analysis of intact mice, showing a ‘weaker’ effect of Treg cell-derived IL-10 relative to IL-35 on inhibitory receptor expression (Fig. 3d-e, Supplementary Fig. 4c-d), limiting the possibility that an accessory cell is involved in this process. These data further support the notion that both Treg cell-derived IL-10 and IL-35 can directly regulate multi-inhibitory receptor expression on intratumoral CD8+ cells.
Figure 5: Direct impact of IL-10 and IL-35 signaling on CD8+ TILs.
a, Experimental schematic of adoptive transfer system to generate an environment in which only CD8+ T cells lack IL-10R or IL-35R.
b, Tumor growth curve of Rag1−/− mice that were sequentially reconstituted with CD8-depleted splenocytes and CD8+ T cells from wild-type (WT) (n=11), IL-10R−/− (Il10rb−/−) (n=8), or IL-35R−/−(CD4Cre.Il6stL/L.Il12rb2 KO) (n=7) mice followed by B16 intradermal inoculation. Each measurement time point represents mean value with s.e.m. error bar. Statistical significance was determined by Two-way ANOVA with Holm-Sidak multiple comparisons (****p<0.0001).
c, Representative flow plots depicting the expression of PD-1 against the other 4 inhibitory receptors (TIM3, LAG3, TIGIT, 2B4) and SPICE plots showing the multi-inhibitory receptor expression on the donor CD8+ T cells infiltrating B16 tumor-bearing reconstituted Rag1−/− mice 18 days post-tumor inoculation as described in (a).
d, Scatter-bar graphs representing the percent distribution of donor CD8+ TILs based on the number of inhibitory receptors expressed. (WT: n=10, IL-10R−/−: n=6, IL-35R−/−: n=7). Bars represent mean values. Statistical significance was determined by One-way ANOVA with Holm-Sidak multiple comparisons (****p<0.0001 and other p-values indicated).
(a-d) Data averaged from 2 independent experiments.
Treg cell-derived IL-10 and IL-35 co-opt the BLIMP1-regulated exhaustion module to drive T cell dysfunction
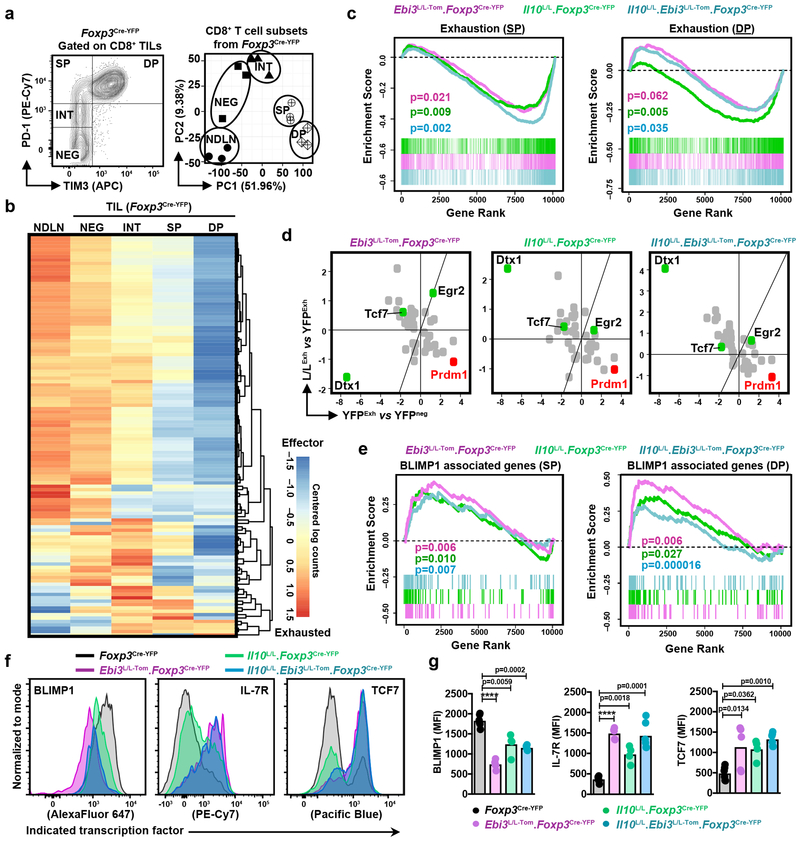
We next probed the mechanistic underpinnings of IL-10 and IL-35-driven multi-inhibitory receptor upregulation to assess if these Treg cell subpopulations employed similar or distinct downstream mechanisms to drive T cell dysfunction. We performed RNAseq with CD8+ TIL-subsets based on the expression of PD-1 and TIM3: (i) PD-1hiTIM3+ double positive (DP), (ii) PD-1hiTIM3− single positive (SP), (iii) PD-1int, and (iv) PD-1neg T cell subsets (Fig. 6a, Supplementary Fig. 6a). PCA of these subsets from control Foxp3Cre-YFP mice segregated them into defined clusters based on location (NDLN vs. TIL) and state of exhaustion (NEG, INT, SP vs. DP), highlighting the distinct transcriptional signatures specific to each subset (Fig. 6a-b). Differential gene expression analysis comparing the CD8+ TIL subsets from single and dual Treg cell cytokine-deficient mice to corresponding wild-type counterparts revealed that the alteration of the TME by Treg cell-restricted deletion of IL-10 and/or IL-35 influenced the transcriptome of all the CD8+ subsets (Supplementary Fig. 6b-d). In particular, gene set enrichment analysis (GSEA) confirmed significant downregulation of an exhaustion signature in SP and DP CD8+ subsets from Il10L/L.Foxp3Cre-YFP, Ebi3L/L-Tom.Foxp3Cre-YFP, and Il10L/L.Ebi3L/L-Tom.Foxp3Cre-YFP mice, relative to the control CD8+ T cells (Fig. 6c). This exhaustion gene signature of CD8+ TIL subsets displayed strong congruence to the published data from LCMV infection28, indicative of a core molecular program that drives PD-1int to PD-1hi transition in chronic settings (Supplementary Fig. 6e). These results further support the ability of both IL-10+ and IL-35+ Treg cell subpopulations to drive the PD-1int to PD-1hi transition thereby promoting the multi-inhibitory receptor+ TIL state (Fig. 6c).
Figure 6: Treg cell-derived IL-10 and IL-35 regulate CD8+ TILs through BLIMP1–inhibitory receptor axis.
(a-e) Data averaged from 5 independent RNASeq experiments: Foxp3Cre-YFP (n=3), Il10L/L.Foxp3Cre-YFP (n=2), Ebi3L/L-Tom.Foxp3Cre-YFP (n=3), and Il10 L/L.Ebi3L/L-Tom.Foxp3Cre-YFP CD8s (n=4).
a, The gating strategy for double-sorted CD8+ subsets for RNAseq. (PD-1negTIM3−, NEG; PD-1intTIM3−, INT; PD-1hiTIM3−, SP; and PD-1hiTIM3+, DP). PCA plot depicting the NDLN and CD8+ TILs subsets from Foxp3Cre-YFP. Each symbol represents an independently sequenced replicate.
b, Heat map of tumor-exhaustion gene signature mapped to the chronic LCMV exhaustion program, representing the NDLN and CD8+ TIL subsets from B16-tumor-bearing Foxp3Cre-YFP mice.
c, GSEA of tumor-exhaustion gene signature derived from SP (left) and DP (right) subsets of Foxp3Cre-YFP mice; p-values as indicated.
d, XY plots representing differentially expressed transcription factors (TFs) in SP subsets. X axis represents differentially expressed genes in the PD-1hi (SP and DP) subsets relative to NEG subsets from control Foxp3Cre-YFP mice; Y axis represents differentially expressed genes in the Treg cell cytokine deficient environments relative to control Foxp3Cre-YFP counterparts. Only significantly changed TFs are listed. We only considered TFs that were differentially expressed in exhausted cells (PD1hi, DP and SP) with a minimum log2 fold-change of 1.5, which was further filtered for ones that were differentially expressed across genotypes in the SP subset with the same cutoffs.
e, GSEA of BLIMP1 associated gene signature in SP (left) and DP (right) subsets of Foxp3Cre-YFP mice; p-values as indicated.
(c, e) Statistical significance was determined by Rank Sum Test With Correlation test in the limma R package with correction for loss in degrees of freedom due to correlation among genes. All p-values are two sided. No correction for multiple hypothesis was performed.
f, Representative histograms for validation of BLIMP1 and its associated gene (TCF7 and IL-7R) at the protein level in the CD8+ TILs from B16 tumor-bearing Foxp3Cre-YFP (n=6), Il10L/L.Foxp3Cre-YFP (n=4), Ebi3L/L-Tom.Foxp3Cre-YFP (n=5), and Il10L/L.Ebi3L/L-Tom.Foxp3Cre-YFP (n=6) mice from two independent experiments.
g, Scatter-bar plots tabulating the expression levels of BLIMP1, IL-7R, and TCF7 as in (f). Statistical significance was determined by One-way ANOVA with Holm-Sidak multiple comparisons (****p<0.0001 and other p-values as indicated).
We next interrogated the transcription factors that were upregulated during the PD-1int to PD-1hi transition in control CD8+ T cells and assessed their expression in mice lacking IL-10 and/or IL-35 in Treg cells. The transcription factor Prdm1 (encoding BLIMP1) was one of the top genes induced during this transition and was significantly reduced in SP and DP CD8+ T cells from Il10L/L.Foxp3Cre-YFP, Ebi3L/L-Tom.Foxp3Cre-YFP, and Il10L/L.Ebi3L/L-Tom.Foxp3Cre-YFP mice (Fig. 6d). Reduced Prdm1 expression was accompanied by an upregulation of its target genes (many of which are characteristic of memory T cells: Tcf7, Id3, Il7r)29 (Fig. 6d-g), which is in agreement with an increase in CD8+ central memory T cells (TCM) (Fig. 4c). We also noted upregulation of a PD-1 blockade-responsive TCF-1+CXCR5+ memory gene signature30 in CD8+ T cells from Il10L/L.Foxp3Cre-YFP, Ebi3L/L-Tom.Foxp3Cre-YFP, and Il10L/L.Ebi3L/L-Tom.Foxp3Cre-YFP mice (Supplementary Fig. 6f). Collectively, these data suggest common modulation of the BLIMP1 axis by the Treg cell-derived IL-10 and IL-35 for optimal induction of an exhaustion gene signature in intratumoral CD8+ T cells.
Direct regulation of BLIMP1 locus by Treg cell-derived IL-10 and IL-35
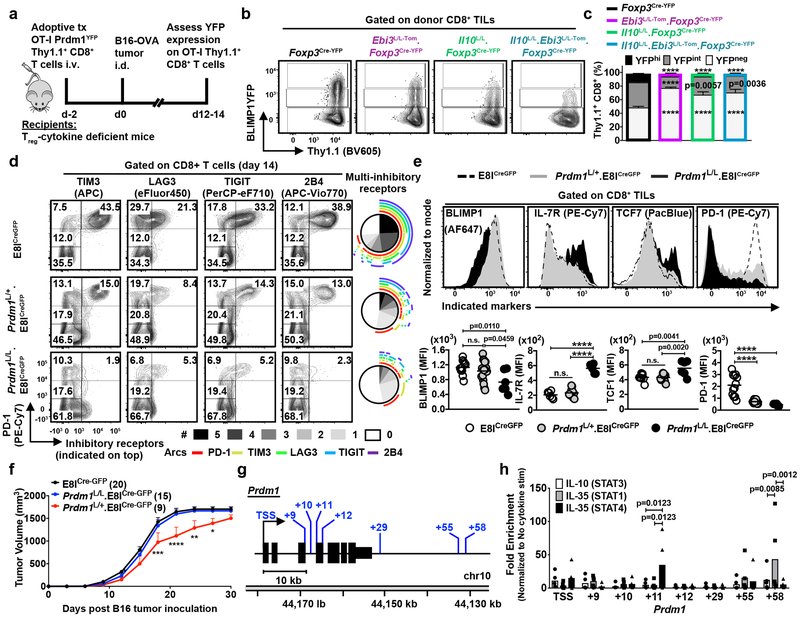
BLIMP1 is a well-characterized regulator of terminal differentiation for both T and B cell lineages, and it has been reported to drive inhibitory receptor expression and exhaustion of CD8+ T cells in chronic viral infection31. However, its role in intratumoral T cell exhaustion and whether it links Treg cell-derived cytokines with inhibitory receptor expression is unknown. Analysis of B16 tumor-bearing Prdm1YFP reporter mice revealed that BLIMP1 expression was strongly correlated with PD-1hi and multi-inhibitory receptor+ CD8+ TILs (Supplementary Fig. 7a-c) and BLIMP1 protein was significantly reduced in the TME that lacks Treg cell-derived IL-10 or IL-35 (Supplementary Fig. 7d). Adoptively transferred OT-I.Prdm1YFP transgenic CD8+ T cells failed to up-regulate high BLIMP1 expression in B16-OVA tumor-bearing Il10L/L.Foxp3Cre-YFP, Ebi3L/L-Tom.Foxp3Cre-YFP, and Il10L/L.Ebi3L/L-Tom.Foxp3Cre-YFP mice compared to Foxp3Cre-YFP counterparts, further corroborating the role of both Treg cell-derived IL-10 and IL-35 to cooperatively promote the intratumoral multi-inhibitory receptor+ TIL state via BLIMP1 induction (Fig. 7a-c).
Figure 7: Direct and cooperative regulation of BLIMP1–inhibitory receptor axis in CD8+ TILs by IL-10 and IL-35.
a, Experimental design schematic of BLIMP1YFP (Prdm1YFP)xOT1 adoptive transfer model.
b, Representative flow plots depicting expression of BLIMP1YFP on Prdm1YFP.OT-I Thy1.1+ CD8+ T cells adoptively transferred into Foxp3Cre-YFP (n=11), Il10L/L.Foxp3Cre-YFP (n=10), Ebi3L/L-Tom.Foxp3Cre-YFP (n=10), and Il10L/L.Ebi3L/L-Tom.Foxp3Cre-YFP (n=6) mice B16-OVA tumors.
c, Stacked bar graph depicting BLIMP1 expression in the donor CD8+ T cells as in (b). Statistical significance was determined by Two-way ANOVA with Holm-Sidak multiple comparisons (****p<0.0001 and other p-values as indicated).
d, Representative flow and SPICE plots depicting the expression of inhibitory receptors (PD-1, TIM3, LAG3, TIGIT, and 2B4) on CD8+ TILs from B16-tumor-bearing E8ICre-GFP (n=9), Prdm1L/+.E8ICre-GFP (n=12), and Prdm1L/L.E8ICre-GFP (n=10)mice.
e, Representative histograms depicting expression levels of BLIMP1, IL-7R and TCF7 on CD8+ TILs from B16 tumor-bearing E8ICre-GFP (n=8), Prdm1L/+.E8ICre-GFP (n=12), and Prdm1L/L.E8ICre-GFP (n=6) mice (top). For PD1, E8ICre-GFP (n=12), Prdm1L/+.E8ICre-GFP (n=12), and Prdm1L/L.E8ICre-GFP (n=6) mice. Scatter plots tabulating the percent distributions (bottom). Statistical significance was determined by One-way ANOVA with Holm-Sidak multiple comparisons (****p<0.0001 and other p-values as indicated).
f, Tumor growth curve of E8ICre-GFP (n=20), Prdm1L/+.E8ICre-GFP (n=15), and Prdm1L/L.E8ICre-GFP (n=9) mice inoculated with 1.25×105 cells of B16 tumors intradermally. Data averaged from 2 independent experiments. Statistical significance was determined by Two-way ANOVA with Holm-Sidak multiple comparisons (*p=0.0165, **p=0.0008, ***p=0.0002, ****p<0.0001).
g, Depiction of locus surrounding mouse Prdm1. The annotated STAT-binding sites were investigated here.
h ChIP analysis of IL-10-mediated STAT3 binding and IL-35-mediated STAT1/STAT4 binding to Prdm1 locus of CD8+ T cells activated for 48 hrs with anti-CD3, anti-CD28, and IL-2, followed by 4 days expansion with IL-2, resting in serum-free media for 2 hrs, then stimulation with IL-10 or IL-35 for 30 mins at 37°C. Chromatin was immunoprecipitated with anti-STAT1, anti-STAT3, anti-STAT4, or monoclonal rabbit IgG isotype control for real-time RT-PCR. Fold enrichment was calculated based on the isotype controls, which was then normalized to the no-cytokine stimulation (No-Stim) controls. Data averaged from 5 independent experiments. Bars represent mean values. Statistical significance was determined by Two-way ANOVA with Holm-Sidak multiple comparisons (p-values as indicated).
We next assessed inhibitory receptor expression in mice with a CD8+ T cell-restricted Prdm1 deletion (Prdm1L/L.E8ICreGFP) to validate whether BLIMP1 has a cell-intrinsic role in inhibitory receptor-regulation in tumors. Bi-allelic deletion of BLIMP1 in CD8+ T cells resulted in a substantial loss of multi-inhibitory receptor+ CD8+ TILs, mirroring our observations in Treg cell-mutant mice, while heterozygous Prdm1L/+.E8ICreGFP mice exhibited a substantive but intermediate inhibitory receptor reduction (Fig. 7d, Supplementary Fig. 7e). Consistent with the Treg cell cytokine-deficient mice, expression of memory-associated genes (IL-7R and TCF7) were also upregulated in Prdm1L/L.E8ICreGFP mice (Fig. 7e). It has been reported previously that bi-allelic deletion of BLIMP1 results in a loss of effector functions in CD8+ T cells in the context of chronic viral infection despite diminished inhibitory receptor expression32. Indeed, no loss of tumor growth was observed in Prdm1L/L.E8ICreGFP mice (Fig. 7f). Induced BLIMP1-haploinsufficiency32 or double-deletion of BLIMP1 and c-Maf33 in CD8+ T cells was required to reinvigorate the cytotoxic effector function and memory-potential, as c-Maf is highly upregulated in the absence of BLIMP1 and is capable of regulating a largely overlapping network of genes, maintaining the exhausted phenotype33. Consistent with these observations, heterozygous Prdm1L/+.E8ICreGFP mice exhibited improved B16 tumor control comparable to Treg cell cytokine-deficient mice (Fig. 7f).
Lastly, ChIP-qPCR revealed that IL-10-induced STAT3[34] and IL-35-induced STAT1 and STAT4[26] were differentially enriched at STAT-binding sites within the Prdm1 gene locus (Fig. 7g-h), suggesting that IL-10 and IL-35 may act directly on CD8+ T cells to modulate the BLIMP1-inhibitory receptor axis. Overall, these data confirm an intrinsic role of BLIMP1 in regulating inhibitory receptor expression of T cells in tumors and validate BLIMP1 as a direct downstream target of both Treg cell-derived cytokines, IL-10 and IL-35, in their cooperative promotion of multi-inhibitory receptor expression on TILs (Supplementary Fig. 7f).
Discussion
Collectively, our data support a model in which different subpopulations of intratumoral Treg cells produce IL-10 and IL-35 while exhibiting adaptive plasticity in their cytokine production, which seems to favor single rather than double inhibitory cytokine-secreting states. Although these two inhibitory cytokines cooperatively regulate the BLIMP1-inhibitory receptor axis in CD4+ and CD8+ TILs and exhibit largely overlapping functions such as induction of an inhibitory receptor module (including PD-1, LAG3, TIM3, TIGIT, 2B4), IL-35 appears to play a greater role in inhibitory receptor induction and limiting TCM differentiation whereas IL-10 plays a greater role in regulating cytokine production and effector function. Cooperatively, they have a substantive impact on anti-tumor immunity. Loss of Treg cell-derived IL-10 and IL-35 also results in (i) significant downregulation of the exhaustion gene signature, (ii) upregulation of a memory-associated transcriptional profile, and (iii) development of a CXCR5+ signature in CD8+ TILs that is observed following PD-1 checkpoint blockade30. These data suggest that IL-10- or IL-35-targeted immunotherapy may have a broader therapeutic impact than previously appreciated.
The physiological requirement for inducible, transitional, divergent intratumoral Treg cell subpopulations that shift between IL-10 and IL-35 single inhibitory cytokine expressing states remains unclear. Adaptive plasticity in Treg cell function has been reported in a number of inflammatory scenarios2. While expression of Foxp3 and TCR signaling drive the core suppressive module23,35,36, Treg cells retain developmental plasticity to adapt to their microenvironment leading to their acquisition of additional suppressive modules characterized by expression of transcription factors, miRNAs, chemokine receptors and suppressive mediators. The current study exemplifies this plasticity in Treg cell fates in the context of the tumor microenvironment, wherein priming in response to tumor antigens and/or the suppressive tumor milieu leads to the generation of IL-10+ and Ebi3+ Treg cells for optimal tumor-induced immune suppression. Reciprocal IL-10 and IL-35 expression on Treg cells has also been reported to play a role in the maintenance of immune tolerance19. In this context, differential TCR signal strength led to the generation of distinct effector IL-10+ and IL-35+ Treg cell subsets that function in a complimentary fashion in the control of autoimmunity. In fact, such heterogeneity in inhibitory cytokine expression expands beyond the Treg cell lineage and has also been noted for regulatory plasma cells. During Salmonella enterica Typhimurium infection, different subsets of CD138hi plasma cells, depending upon their maturation level, expressed either IL-10 or IL-35, and very few co-transcribed Il10, Ebi3, and Il12a together37. This functional segregation may represent a unifying theme of regulatory cell subsets for maximizing immunoregulation offering a “last-resort” to subvert beneficial anti-tumor or detrimental autoreactive T cell responses while limiting collateral tissue damage. While the mechanism that limits the accumulation of dual inhibitory cytokine expressing Treg cell subpopulations will require further analysis, it is possible that IL-10 and IL-35 cooperatively limit Treg cell development and expansion in a cell-intrinsic manner limiting the abundance of IL-35/IL-10-dual expressing Treg cells.
Despite the differential inhibitory cytokine expression pattern, comprehensive profiling of IL-10+ and Ebi3+ Treg cells did not reveal striking transcriptional and TCR differences between these sub-populations. These results differ from previous observations wherein differences were noted in transcription factor dependency, chemokine receptor expression, and activation status of IL-10+ and IL-35+ Treg cells isolated from secondary lymphoid organs19. In this study, BLIMP1 expression marked the IL-10+ Treg cell subset while IL-35+ Treg cells were predominantly BLIMP1-independent. In contrast, we noted that over 90% of Treg cells express BLIMP1 in B16 tumors, indicating that BLIMP1 may also play a role in regulating the development and function of IL-35+ Treg cells as approximately 30–50% of these intratumoral Treg cells express IL-35. It is conceivable that following priming in response to tumor antigens, the Treg cell repertoire that migrates to the tumor is fixed in its core transcriptional identity, while still retaining plasticity in inhibitory cytokine expression. Interestingly, Treg cell-restricted IL-35 depletion did not lead to a compensatory increase in IL-10+ Treg cells. While we were unable to assess the effect of Treg cell-restricted IL-10 loss on IL-35+ Treg cells, it has been suggested that mice with BLIMP1-deficient Treg cells (unable to produce IL-10) exhibited a 2-fold enrichment of IL-35+ Treg cells19. Thus it is possible that if Treg cell-restricted IL-10 deficiency led to a compensatory increase in IL-35+ Treg cells in our B16 tumor model, that may explain multiple aspects: (i) the less dramatic loss of multi-inhibitory receptor-positive TILs in Il10L/L.Foxp3Cre-YFP as well as Rag1−/− reconstitution studies with IL-10R−/− CD8+ T cells, (ii) consistent and more dramatic loss of multi-inhibitory receptor+ TILs in Ebi3L/L-Tom.Foxp3Cre-YFP and Il10L/L.Ebi3L/L-Tom.Foxp3Cre-YFP mice, respectively, and (iii) lack of synergy or additivity in B16 tumor control in Il10L/L.Ebi3L/L-Tom.Foxp3Cre-YFP mice. This may in part also underlie the non-overlapping regulation of effector versus memory responses by the two cytokines, with relatively low BLIMP1 protein reduction in Il10L/L.Foxp3Cre-YFP mice compared to Ebi3L/L-Tom.Foxp3Cre-YFP and Il10L/L.Ebi3L/L-Tom.Foxp3Cre-YFP mice, recapitulating BLIMP1 haploinsufficiency32 and translating to increased effector responses and decreased memory generation. Although BLIMP1 is well-established as a driver of inhibitory receptor expression, a recent report demonstrated that another transcription factor, c-MAF, sufficiently promoted inhibitory receptor expression in the absence of BLIMP1, by impacting overlapping transcriptional networks. However, our RNAseq dataset did not reveal significant modulation of Maf expression in CD8+ TILs from Treg cell cytokine-deficient mice relative to Foxp3Cre-YFP counterparts, highlighting specific modulation of the BLIMP1-axis by IL-35 and IL-10 in cooperative regulation of T cell exhaustion.
In summary, the adaptive plasticity and cooperative regulation of anti-tumor immunity by IL-10+ and IL-35+ Treg cells pose another tumor-immune evasive strategy and potential resistance mechanism to immunotherapy. Understanding this complex Treg cell-driven regulatory circuitry in the TME may inform the rational design of combinatorial modalities targeting Treg cells and their downstream mediators that promote T cell exhaustion to maximize responsiveness to checkpoint blockade and other immunotherapies while limiting adverse events and the risk of inflammatory or autoimmune complications.
Data availability
Bulk RNAseq and single-cell RNAseq data will be deposited in the Gene Expression Omnibus (GEO), and the accession codes will be provided. The RNAseq data sets reported by other studies used to cross-examine with our sequencing data in this study were obtained from GSE9650 and GSE84105. The main data supporting the findings of this study are available within the article and its supplementary figures. Data are available from the corresponding authors upon appropriate and reasonable request.
Methods
Mice
Unless otherwise specified, all experimental procedures were performed on 5–8 week old laboratory mice housed in Helicobacter/MNV free SPF facilities at the University of Pittsburgh in accordance with the current National Institutes of Health guidelines and with the approval and guideline of the Institutional Animal Care and Use Committee (IACUC) of the University of Pittsburgh. Female mice were used for tumor growth and RNA sequencing experiments, while both male and female mice were used for tumor-infiltrating lymphocyte flow cytometry analysis.
Il10GFP.Ebi3Tom.Foxp3Cre-YFP mice were generated by crossing Il10GFP (Vert-X17; Jackson Laboratory) to Ebi3Tom.Foxp3Cre-YFP (developed in our laboratory7). Ebi3L/L-Tom mice were developed in our laboratory and have been previously described7. Il10L/L (provided by Werner Muller (Germany))38 and Ebi3L/L-Tom were crossed to Foxp3Cre-YFP mice18 to generate Treg cell-specific cytokine deletion strains. Prdm1YFP and Prdm1L/L strains were provided by Amanda Poholek at University of Pittsburgh. E8ICre mice (provided by Dan R. Littman, New York University; previously described39) and OT-I transgenic mice (obtained from Jackson Laboratory) were crossed to Prdm1L/L and Prdm1YFP strains, respectively, to generate E8ICre.Prdm1L/L and Prdm1YFP.OT-I mice. Rag1−/− and IL-10R−/− (Il10rb−/−) mice were purchased from Jackson Laboratory. IL-35R−/− (CD4Cre.Il6stL/L.IL12rb2−/−) mice were generated as described previously26.
Human specimen collection and processing
All specimens were acquired under University of Pittsburgh approved Institutional Review Board (IRB) protocol, and informed consent was obtained from all patients. All lung tumors were processed to single-cell suspension by a combination of mechanical and enzymatic digestion. For enzymatic digestion, Liberase (Sigma Aldrich) was used at 50 μg/mL for 15 minutes at 37°C in RPMI medium (Lonza) to release tumor infiltrating lymphocytes (TIL). For healthy donor peripheral blood samples, Ficoll Paque Plus (GE Healthcare) was used to separate peripheral blood leukocytes (PBL) and lysis buffer (BD Biosciences) was used to remove red blood cells when necessary.
Staining for EBI3 and IL-10 in human specimens
Lung TIL and HD PBL were incubated in complete RPMI (cRPMI) medium (10% FBS (Atlanta Biologicals), non-essential amino acids (Sigma Aldrich), L-glutamine (Lonza) and penicillin/streptomycin (Corning) and sodium pyruvate (Sigma Aldrich)) with TCR stimulation: plate-bound anti-CD3 (0.5 μg/mL, Invitrogen, clone: OKT3) and soluble anti-CD28 (1 μg/mL Invitrogen, clone: CD28.2) and 200U/ml rhIL-2 overnight followed by four hours of stimulation with PMA/ionomycin and GolgiStop. The cells were then harvested and stained with viability dye and fluorochrome-conjugate antibodies. For intracellular staining, fixation/permeabilization kit (eBiosciences) was used according to manufacturer’s protocol.
Cell lines and tumor processing
B16-F10 cells (referred to as B16) and EL4 cells were purchased from ATCC (Manassas, Virginia) and cultured in cRPMI. BrafPten (clone24) and B16-OVA cells were kindly provided by Greg Delgoffe (University of Pittsburgh) and cultured in cDMEM (Lonza) and G418-supplemented cRPMI (1mg/mL), respectively. BrafPten (clone 24) was originally generated by A.V. Menk and G.M. Delgoffe by cloning a cell line from a tumor harvested from BRafCA.PtenL/L.TyrCreERT2 mice induced with 4-OHT administration.
Mice received an intradermal inoculation (i.d.) of tumor cells (1.25×105) on day 0. Tumors were measured every 3 days in two dimensions using a digital caliper and expressed as tumor volume (mm3; defined as the Square of Smaller diameter x Larger diameter/2).
Tumors were excised on day 14 or d20 for end-point experimental analyses. Excised solid tumors were minced into small pieces and digested with Collagenase type IV (200 U/mL) and Dispase (1 U/mL) in cDMEM for 30 minutes at 37°C. Digested cell suspension was then processed through 70 μm cell-strainer and washed with cDMEM. Isolated cells were then used in various assays. Red blood cells were lysed with Gey’s solution when appropriate.
Fungal protease-induced allergic airway disease
Mice received 7 intranasal challenges with a mixture comprised of a fungal protease derived from Aspergillus oryzae and Ovalbumin, every alternate day for two weeks. On day 14, mice were intubated, tracheas were cannulated and flushed with PBS (twice) for isolation of broncho-alveolar lavage (BAL) fluid, prior to isolation of lungs and the draining (mediastinal) and non-draining (inguinal) lymph nodes. Lungs were perfused with 10 mL PBS injected through the right ventricle of the heart using a 20 gauge needle immediately after BAL fluid collection. Harvested lungs were minced into small pieces and digested in 5 mL capped polystyrene tube with 1 mg/mL Collagenase D (Roche) in 2 mL PBS for 45 minutes at 37°C. Digested cell suspension was then processed through 70 μm cell-strainer and washed with cDMEM. Isolated cells were then used in flow cytometry analysis. Red blood cells were lysed with Gey’s solution when appropriate.
Flow cytometry
Single-cell suspensions from mouse or human specimens were stained with live/dead exclusion dye, followed by fluorochrome-conjugated antibodies in the presence of 5% normal mouse serum. TruNuclear transcription factor staining kit (Biolegend) was used for intracellular staining. Flow cytometry data were acquired on BD Fortessa and analyzed by FlowJo (Treestar, Inc.). Pie charts were created using the SPICE program40.
Processing and analysis of scRNAseq data
We used Cell Ranger (v2.0) (10X Genomics) to analyze sequencing data generated from Chromium Single Cell 3’ RNA-seq libraries41. We first ran “cellranger mkfastq” on the Illumina BCL output folder to generate fastq files. We next generated the UMI count matrix with “cellranger count” for each library. The data were normalized with R package cellrangerRkit and visualized via t-distributed stochastic neighbor embedding (t-SNE). The diffusion pseudo-time analysis was run with R package Destiny42,43. Briefly, we built a transition matrix for all cells based on their adjacency using a locally-scaled Gaussian kernel and determined the diffusion components according to the eigenvectors of the matrix. Then, a new matrix M was generated by removing the first eigenvalue of the original transition matrix and the diffusion pseudo-time was calculated as a distance metric between the rows of M. Lastly, we plotted the trajectory lines, which were fitted by the locally weighted scatterplot smoothing (LOESS) regression using the previously calculated values. For the differential expression analyses between the IL-10 and Ebi3 gene-expressing cells in LN and Tumor Treg cells, we used the “sseq” method from the standard R package CellrangerRkit. Additionally, top expressed genes from differential expression analysis were determined by performing the two-sided Negative Binomial Exact test44 and the p-values were adjusted to control the false discovery rate (FDR)45.
RNASeq profiling and data analysis
Treg cell fractions and NDLN Foxp3YFP CD4+ Teff control were FACS sorted from day 14 B16 tumor-bearing Il10GFP.Ebi3Tom.Foxp3Cre-YFP mice. CD8+ T cell subsets were FACS sorted from day 14 B16-bearing WT control and Treg cell cytokine-deficient mice.
Each cell fraction was double-sorted to ensure high purity (>95%) and directly lysed using the Clontech SMART-Seq v4 (Treg cells) or v3 (CD8) kit for cDNA synthesis. Libraries were prepared using Nextera XT DNA Library Preparation kit (Illumina), normalized at 2nM using Tris-HCl (10mM, pH 8.5) with 0.1% Tween20, diluted and denatured to a final concentration of 1.8nM. Cluster generation and 75bp paired-end dual-indexed sequencing were performed on Illumina NextSeq 500 system.
RNASeq data were aligned to the mm10 genome using the STAR aligner46 and quantified against the Refseq gene models47 using featureCounts48. The number of uniquely aligned reads ranged from 10 to 12 million. The raw data counts were processed for differential expression using the “voom” function49 in the limma R package50,51 with the robust model option. Gene set enrichment testing was performed using the “Rank Sum Test With Correlation” function in the limma R package, which automatically corrects enrichment statistic inflation due to correlation among genes. We used the GSEA-style enrichment score for visualization of pathway enrichment results52.
We defined the tumor-specific exhaustion signature to be the set of genes that are overexpressed in the PD-1hi state (both DP and SP) when compared to PD-1neg state in Foxp3Cre-YFP mice at fold change of 4 and q-value FDR of 0.05. Tumor exhaustion signature was aligned with the chronic LCMV exhaustion profile dataset (GEO accession number: GSE9650)28. The CXCR5+ PD-1-responsive CD8+ T cell signature was derived from GEO accession number: GSE84105[30].
Code availability
Computational and mathematical codes utilized in the RNAseq analyses supporting the findings of this study are available within the article. Additional information is available from corresponding author upon reasonable and appropriate request.
In vitro micro-suppression assay
Sorted Treg cell subpopulations from NDLN and B16 tumors were co-cultured with CellTrace Violet (Life Technologies)-labeled CD4+Foxp3− responder T cells (Tresponders) in the presence of mitomycin-C-treated TCRβ-depleted splenocytes and anti-CD3ε (1 μg/mL) for 72 hrs at 37°C as previously described7.
TCRseq and data analysis
Treg cell fractions and IL-10−Ebi3−Foxp3YFP− CD4+ Teff control were purified by FACS from day 14 B16-bearing Il10GFP.Ebi3Tom.Foxp3Cre-YFP mice. DNA was purified using QIAamp DNA Micro Kit (QIAGEN), and TCRbeta-enriched library was generated with TCRbeta immunoSeq (Adaptive Biotechnologies), following the manufacturer’s protocol. Cluster generation and sequencing were performed on Illumina high output NextSeq 500 system. The data were analyzed using immunoSeq Analyzer (Adaptive Biotechnologies).
Adoptive transfer experiments
BLIMP1YFP.OT-1 transfer experiment: Recipient mice received intravenous injections of 0.5×106 BLIMP1YFP CD8+ T cells isolated by negative selection from spleen and lymph nodes of Prdm1YFP.OT-I.Thy1.1 mice, followed by i.d. injection of 2.5×105 B16-OVA cells two days following adoptive transfer. Tumors and NDLN were harvested at day 14 post-tumor inoculation for assessment of BLIMP1YFP induction.
Rag1 KO reconstitution experiment: Rag1 KO recipient mice received intravenous injections of CD8-depleted splenocytes isolated from Foxp3Cre-YFP.Thy1.1 mice containing 1×106 Treg cells day −8, followed by injection of 6×106 WT, IL-10R.KO, or IL-35R KO.CD8+ T cells on day −1 and i.d. B16 tumor inoculation (1.25×105 cells/mouse) on day 0. Tumor size was measured every 3 days until day 18 post-tumor inoculation. Tumors were harvested on day 14 or day 18 for end-point experimental analysis.
ChIP-qPCR
Naive CD8+ T cells were purified and activated for 2 days with plate-bound anti-CD3 (3 μg/mL) and anti-CD28 (5 μg/mL) supplemented with 50 U/mL hIL2, followed by expansion for 4 days in hIL2-containing cRPMI. Following serum-deprivation for 3 hours, cells were pulsed with recombinant IL-10 or IL-35 for 30 mins prior to fixation with 1% formaldehyde. 15×106 cells per sample were sonicated with Bioruptor Pico in shearing buffer. ChIP was performed overnight for STAT1 (D1K9Y), STAT3 (124H6), STAT4 (C46B10), and IgG (Cell Signaling Technology) using Protein A Dynabeads (Thermo Fisher). EvaGreen-based qPCR was performed using primers previously described34.
Statistical analysis
Except for RNAseq and scRNAseq data analysis, GraphPad Prism software was used to determine the statistical significance. Group means were compared with two-tailed Student’s t-test when only two experimental groups were involved. Tumor growth was analyzed using Two-way ANOVA with multiple comparisons correction with sequential time point measurements. For other analysis, One-way or Two-way ANOVA with multiple comparison correction. All p-values were two-sided, and statistical significance assessed at or below 0.05.
Reporting summary
Additional experimental design detail of this study is available in the Life Sciences Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
The authors wish to thank H. Shen, D. Falkner and A. Yates from the Immunology Flow Core for cell sorting, E. Brunazzi and the staff of the Division of Laboratory Animals for animal husbandry, A. Cillo for helpful suggestions regarding scRNAseq analysis, and W. Horne, J. Kolls and University of Pittsburgh HSCRF Genomics Research Core for assistance with sequencing, and A. Menk and G. Delgoffe at the Universtiy of Pittsburgh for generation of and providing the BrafPten (clone24) cell line for tumor growth experiments. The authors also wish to thank the Department of Cardiothoracic Surgery at the University of Pittsburgh, in particular, Ms. J. Ward for her help in coordination and gathering patient consents, as well as the Department of Cardiothoracic Surgery at the University of Colorado and the University of Colorado SPORE for providing some samples. This work was supported by the National Institutes of Health (R01 CA203689 and P01 AI108545 to D.A.A.V.), NCI Comprehensive Cancer Center Support CORE grant (CA047904 to D.A.A.V.) and an SRA from Tizona Therapeutics. This work also benefitted from the Immunology Department Flow Cytometry Core SPECIAL BD LSR FORTESS ATM funded by NIH 1S10OD011925–01 (L. Borghesi, Department of Immunology). This project also used the Hillman Cancer Center Immunologic Monitoring and Cellular Products Laboratory that is supported in part by award P30 CA047904.
Footnotes
Competing financial interest statement
The authors declare competing financial interests. D.A.A.V. and C.J.W. have submitted patents covering IL-35 that are pending and are entitled to a share in net income generated from licensing of these patent rights for commercial development.
References:
- 1.Vignali DA, Collison LW & Workman CJ How regulatory T cells work. Nature reviews. Immunology 8, 523–532 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sawant DV & Vignali DA Once a Treg, always a Treg? Immunological reviews 259, 173–191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka A & Sakaguchi S Regulatory T cells in cancer immunotherapy. Cell research 27, 109–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu C, Workman CJ & Vignali DA Targeting regulatory T cells in tumors. The FEBS journal 283, 2731–2748 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Curiel TJ, Coukos G, Zou L et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nature medicine 10, 942–949 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Shimizu J, Yamazaki S & Sakaguchi S Induction of tumor immunity by removing CD25+CD4+ T cells: a common basis between tumor immunity and autoimmunity. Journal of immunology (Baltimore, Md. : 1950) 163, 5211–5218 (1999). [PubMed] [Google Scholar]
- 7.Turnis ME, Sawant DV, Szymczak-Workman AL et al. Interleukin-35 Limits Anti-Tumor Immunity. Immunity 44, 316–329 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shitara K & Nishikawa H Regulatory T cells: a potential target in cancer immunotherapy. Annals of the New York Academy of Sciences 1417, 104–115 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Sawant DV, Hamilton K & Vignali DA Interleukin-35: Expanding Its Job Profile. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research 35, 499–512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brooks DG, Trifilo MJ, Edelmann KH et al. Interleukin-10 determines viral clearance or persistence in vivo. Nature medicine 12, 1301–1309 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ejrnaes M, Filippi CM, Martinic MM et al. Resolution of a chronic viral infection after interleukin-10 receptor blockade. The Journal of experimental medicine 203, 2461–2472 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tinoco R, Alcalde V, Yang Y et al. Cell-intrinsic transforming growth factor-beta signaling mediates virus-specific CD8+ T cell deletion and viral persistence in vivo. Immunity 31, 145–157 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks DG, Ha SJ, Elsaesser H et al. IL-10 and PD-L1 operate through distinct pathways to suppress T-cell activity during persistent viral infection. Proceedings of the National Academy of Sciences of the United States of America 105, 20428–20433 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Penaloza-MacMaster P, Kamphorst AO, Wieland A et al. Interplay between regulatory T cells and PD-1 in modulating T cell exhaustion and viral control during chronic LCMV infection. The Journal of experimental medicine 211, 1905–1918 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collison LW, Chaturvedi V, Henderson AL et al. IL-35-mediated induction of a potent regulatory T cell population. Nature immunology 11, 1093–1101 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bettini M, Castellaw AH, Lennon GP et al. Prevention of autoimmune diabetes by ectopic pancreatic beta-cell expression of interleukin-35. Diabetes 61, 1519–1526 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madan R, Demircik F, Surianarayanan S et al. Nonredundant roles for B cell-derived IL-10 in immune counter-regulation. Journal of immunology (Baltimore, Md. : 1950) 183, 2312–2320 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubtsov YP, Rasmussen JP, Chi EY et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Wei X, Zhang J, Gu Q et al. Reciprocal Expression of IL-35 and IL-10 Defines Two Distinct Effector Treg Subsets that Are Required for Maintenance of Immune Tolerance. Cell reports 21, 1853–1869 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Kheradmand F, Kiss A, Xu J et al. A protease-activated pathway underlying Th cell type 2 activation and allergic lung disease. Journal of immunology (Baltimore, Md. : 1950) 169, 5904–5911 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Moran AE & Hogquist KA T-cell receptor affinity in thymic development. Immunology 135, 261–267 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen G, Yang X, Ko A et al. Sequence and Structural Analyses Reveal Distinct and Highly Diverse Human CD8(+) TCR Repertoires to Immunodominant Viral Antigens. Cell reports 19, 569–583 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine AG, Arvey A, Jin W & Rudensky AY Continuous requirement for the TCR in regulatory T cell function. Nature immunology 15, 1070–1078 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noy R & Pollard JW Tumor-associated macrophages: from mechanisms to therapy. Immunity 41, 49–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genard G, Lucas S & Michiels C Reprogramming of Tumor-Associated Macrophages with Anticancer Therapies: Radiotherapy versus Chemo- and Immunotherapies. Frontiers in immunology 8, 828 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collison LW, Delgoffe GM, Guy CS et al. The composition and signaling of the IL-35 receptor are unconventional. Nature immunology 13, 290–299 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spencer SD, Di Marco F, Hooley J et al. The orphan receptor CRF2–4 is an essential subunit of the interleukin 10 receptor. The Journal of experimental medicine 187, 571–578 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wherry EJ, Ha SJ, Kaech SM et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27, 670–684 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Rutishauser RL, Martins GA, Kalachikov S et al. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity 31, 296–308 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Im SJ, Hashimoto M, Gerner MY et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xin A, Nutt SL, Belz GT & Kallies A Blimp1: driving terminal differentiation to a T. Advances in experimental medicine and biology 780, 85–100 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Shin H, Blackburn SD, Intlekofer AM et al. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity 31, 309–320 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chihara N, Madi A, Kondo T et al. Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature 558, 454–459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poholek AC, Jankovic D, Villarino AV et al. IL-10 induces a STAT3-dependent autoregulatory loop in TH2 cells that promotes Blimp-1 restriction of cell expansion via antagonism of STAT5 target genes. Science immunology 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fontenot JD, Gavin MA & Rudensky AY Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nature immunology 4, 330–336 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Hori S, Nomura T & Sakaguchi S Control of regulatory T cell development by the transcription factor Foxp3. Science (New York, N.Y.) 299, 1057–1061 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Shen P, Roch T, Lampropoulou V et al. IL-35-producing B cells are critical regulators of immunity during autoimmune and infectious diseases. Nature 507, 366–370 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roers A, Siewe L, Strittmatter E et al. T cell-specific inactivation of the interleukin 10 gene in mice results in enhanced T cell responses but normal innate responses to lipopolysaccharide or skin irritation. The Journal of experimental medicine 200, 1289–1297 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seo W, Muroi S, Akiyama K & Taniuchi I Distinct requirement of Runx complexes for TCRbeta enhancer activation at distinct developmental stages. Scientific reports 7, 41351 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roederer M, Nozzi JL & Nason MC SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A 79, 167–174 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng GX, Terry JM, Belgrader P et al. Massively parallel digital transcriptional profiling of single cells. Nature communications 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Angerer P, Haghverdi L, Buttner M et al. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics (Oxford, England) 32, 1241–1243 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Haghverdi L, Buttner M, Wolf FA et al. Diffusion pseudotime robustly reconstructs lineage branching. Nature methods 13, 845–848 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Yu D, Huber W & Vitek O Shrinkage estimation of dispersion in Negative Binomial models for RNA-seq experiments with small sample size. Bioinformatics (Oxford, England) 29, 1275–1282 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klipper-Aurbach Y, Wasserman M, Braunspiegel-Weintrob N et al. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Medical hypotheses 45, 486–490 (1995). [DOI] [PubMed] [Google Scholar]
- 46.Dobin A, Davis CA, Schlesinger F et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pruitt KD, Tatusova T & Maglott DR NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Research 35, D61–65 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao Y, Smyth GK & Shi W featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics (Oxford, England) 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Law CW, Chen Y, Shi W & Smyth GK voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ritchie ME, Phipson B, Wu D et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu D & Smyth GK Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Research 40, e133 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subramanian A, Tamayo P, Mootha VK et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bulk RNAseq and single-cell RNAseq data will be deposited in the Gene Expression Omnibus (GEO), and the accession codes will be provided. The RNAseq data sets reported by other studies used to cross-examine with our sequencing data in this study were obtained from GSE9650 and GSE84105. The main data supporting the findings of this study are available within the article and its supplementary figures. Data are available from the corresponding authors upon appropriate and reasonable request.