Abstract
Deflecting biomineralized crystals attached to vestibular hair cells are necessary for maintaining balance. Zebrafish (Danio rerio) are useful organisms to study these biomineralized crystals called otoliths, as many required genes are homologous to human otoconial development. We sought to identify and characterize the causative gene in a trio of homozygous recessive mutants, no content (nco) and corkscrew (csr), and vanished (vns), which fail to develop otoliths during early ear development. We show that nco, csr, and vns have potentially deleterious mutations in polyketide synthase (pks1), a multi-modular protein that has been previously implicated in biomineralization events in chordates and echinoderms. We found that Otoconin-90 (Oc90) expression within the otocyst is diffuse in nco and csr; therefore, it is not sufficient for otolith biomineralization in zebrafish. Similarly, normal localization of Otogelin, a protein required for otolith tethering in the otolithic membrane, is not sufficient for Oc90 attachment. Furthermore, eNOS signaling and Endothelin-1 signaling were the most up- and down-regulated pathways during otolith agenesis in nco, respectively. Our results demonstrate distinct processes for otolith nucleation and biomineralization in vertebrates and will be a starting point for models that are independent of Oc90-mediated seeding. This study will serve as a basis for investigating the role of eNOS signaling and Endothelin-1 signaling during otolith formation.
Keywords: inner ear, otolith, biomineralization, calcium carbonate, polyketide synthase, zebrafish, endothelin-1, eNOS
1. Introduction
Otoconia and otoliths act as a mass load that increase the sensitivity of mechanosensory hair cells to the effects of gravity and linear acceleration in mammals and fish, respectively. While the morphology of otoconia (“ear particles”) and otoliths (“ear stones”) differ, the initial formation of bio-crystals rely on many homologous proteins [1].
Zebrafish otoliths are primarily composed of calcium carbonate (CaCO3), in the form of aragonite, which accounts for ~99% of the total otolithic mass with the remainder consisting of proteins called otoconins [2, 3]. Further analysis of teleost otoliths has identified more than 380 protein components [4]. Based on the level of protein expression or changes in the rate of otolith growth, the polymorph of calcium carbonate crystals can change [1, 5]. For example, knockdown of Starmaker results in otoliths made of calcite rather than aragonite [6]. There are three pairs of otoliths in zebrafish, which include the sagittae, lapilli, and asterisci. While the lapillus and sagitta nucleate early in zebrafish development, the asteriscus does not form until 11–12 days in development [7]. The center of the otoliths contains a proteinaceous core that acts as a site for otolith nucleation and biomineralization. This matrix lays the foundation for further otolith growth, which is mediated by daily deposition of additional otoconins and calcium carbonate molecules [2]. Otolith nucleation occurs when the otolith precursor particles (OPPs) bind to the tips of the immotile kinocilia of tether cells within the otic vesicle [8, 9]. Subsequent studies have demonstrated that the critical period of otolith seeding and nucleation starts at 18–18.5 hpf (hours post fertilization) and ceases by 24 hpf [1, 8, 10–12].
In mammalian inner ear development, Otoconin-90 (Oc90; the major protein component of otoconia) is necessary for otoconial seeding and nucleation [13–15]. Oc90 can bind Otolin-1 (Otol1) to establish a protein-rich matrix that serves as a scaffold for subsequent deposition of calcium carbonate [16, 17]. Additionally, in vitro studies have suggested that Oc90 and Otol1 act synergistically to modulate otoconial crystal morphology [17]. While Oc90 is not the major protein component in zebrafish otoliths, it plays an important role in otolith seeding and early development as oc90-morphants do not develop otoliths [1, 18]. While additional gene mutations have been identified that lead to otolith agenesis in zebrafish [19–24], the genes responsible for several zebrafish otolith mutants have been undetermined.
In this study, we sought to identify and characterize the causative gene in a trio of zebrafish mutants, no content (nco) corkscrew (csr), and vanished (vns), which fail to develop otoliths during early inner ear development. We provide genetic evidence that the causative gene is polyketide synthase (pks1; currently wu:fc01d11), a candidate gene that was previously identified as a key factor of biomineralization in Japanese medaka (Oryzias latipes) and sea urchin (Hemicentrotus pulcherrimus) [25]. Furthermore, we offer potential signaling pathways for pks1 function during inner ear development in the zebrafish.
2. Materials and Methods
Husbandry and maintenance
All zebrafish were maintained in a temperature-controlled (28.5°C) and light-controlled (14h on/10h off) room per standardized conditions. nco strain (jj149) was generated by an ENU screen on the AB background and obtained from ZIRC (Eugene, OR, USA)[26]. csr was a spontaneous mutant generated in a bre-KO2/ntl-GFP line (AB background). vns was a spontaneous mutant generated in a AB/TL background. All protocols were approved by Creighton University and the University of Michigan Animal Care and Use Committees.
Whole genome and RNA-sequencing
Mutant nco embryos and wild-type (WT) clutchmates were phenotyped and collected during the critical period of otolith nucleation and seeding (24 hours post fertilization, hpf) and the whole embryo lysates (n=50) were submitted for RNA sequencing. Analysis was completed using MMAPPR (Mutation Mapping Analysis Pipeline for Pooled RNA-seq) as previously described [24]. Whole genome sequencing of csr phenotypically-mutant embryos (n=150) was performed and analyzed using MegaMapper as previously described [27]. Common SNPs were removed by the Single Nucleotide Polymorphism Database (dbSNPs). Reference sequences for both experiments were mapped to Zv9. All sequencing was conducted at the University of Nebraska Medical Center Genomics Core Facility. Accession numbers for nco RNA-seq and csr genome sequencing will be provided during review.
mRNA and plasmid DNA rescue
WT mRNA and pks1L905P were synthesized using mMessage Machine from a clone provided by Dr. Hiroyuki Takeda (University of Tokyo), cleaned on an RNeasy column, and subsequently injected into single-cell csr and nco embryos. Naked plasmid of the medaka pks1 clone was injected into vns embryos. Overall penetrance of otolith formation was determined in all three mutants. Site-directed mutagenesis (Agilent) was used to generate the mutant clone containing the causative mutation in csr (pks1L905P in Japanese medaka; pks1A911P in zebrafish). Primers used for site-directed mutagenesis were:
pks1_L905P_Forward: 5′-GATATGGCGTGATGTCCGGTGACAGGTTGAAGATC-3′
pks1_L905P_Reverse: 5′-ATCTTCAACCTGTCACCGGACATCACGCCATATC-3′
Pathway analysis
Pathway analysis of nco was performed using Ingenuity Pathway Analysis (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis [28]. The Ensembl Gene IDs were assigned to each gene and uploaded to IPA. Cut-off for gene expression analysis was set at 0.75 RPKM. The calculated z-score indicates a pathway with genes exhibiting increased mRNA levels (positive) or decreased mRNA levels (negative). No change in mRNA levels results in a z-score of zero.
Genotyping
csr, nco, and vns samples were PCR-amplified and submitted for Sanger sequencing using the following primers:
nco_Forward: 5′-GGGAGGATGCTTGTTGTTGG-3′
nco_Reverse: 5′-GTGGCCCAGAATAGGATCCA-3′
csr_Forward: 5′-AAGACGGGGACATGACTCAG-3′
csr_Reverse: 5′-TTCAACAAACAGTGCTCCGG-3′
vns_Forward: 5′-GCCATCATTGGAATTGGATG-3′
vns_Reverse: 5-GGTGTTCCAGTCCCATGAGC-3′
RT-PCR
All RNA was extracted from Danio rerio wild-type embryos (A/B strain). After collecting embryos at the separate time-points, the samples were homogenised in lysis buffer from the Quick- RNA MiniPrep kit (Zymo Research-R1054) and RNA was extracted following protocol provided by the manufacturer. The RNA samples were then DNase treated using TURBO™ DNase (ThermoFisher, AM2238) as per manufacturer instructions, in order to remove any genomic contamination that may be present in the RNA. cDNA synthesis was achieved using the GoScript™ Reverse Transcription System (Promega, A5001) and followed the protocol provided by the manufacturer.
MiniPrep kit (Zymo Research-R1054) and RNA was extracted following protocol provided by the manufacturer. The RNA samples were then DNase treated using TURBO™ DNase (ThermoFisher, AM2238) as per manufacturer instructions, in order to remove any genomic contamination that may be present in the RNA. cDNA synthesis was achieved using the GoScript™ Reverse Transcription System (Promega, A5001) and followed the protocol provided by the manufacturer.
actb1_Forward: 5′-CTTCCAGCCTTCCTTCCT-3′
actb1_Reverse: 5′-CCACCGATCCAGACGGAGTA-3′
pks1_Forward: 5′-GAATTTTCTGCCGAGTAGAACAAAG-3′
pks1_Reverse: 5′-TCTGCATGTCAGGCGATCAG-3′
RT-PCR on the cDNA samples was carried out using the GoTaq® G2 Flexi DNA Polymerase (Promega, M7805) and PCR was done following the protocol provided by the manufacturer, using the primers stated above. The RT-PCR samples were then run on a 2% agarose gel.
Immunofluorescence
csr and nco embryos were collected during key stages in early inner ear development, fixed with hydrogel and washed in CHAPS-based (1% by weight) CLARITY-clearing solution [29]. Embryos were decalcified with EDTA (120 mM in 0.1% PBS-Triton) before blocking (0.1% PBS-Triton with 3.33% sheep serum and 3.33% BSA), incubating in primary and secondary antibodies diluted in blocking buffer, mounting in 50% Glycerol-PBS solution, and imaging by confocal microscopy (Leica TCS SP8). Affinity-purified rabbit polyclonal antibodies were generated to Otogelin (CGNRVDGPSASKG; 1:1000) or Oc90 (CNTQSDTVDRKPTQSKPQ; 1:1000) by conventional methods (GenScript, USA) and directly labelled before immunofluorescence. Other antibodies used were Keratan Sulfate (MZ15; 1:2000; DSHB), Hair Cell Specific-1 (HCS-1; 1:500; DSHB), and acetylated-tubulin (1:500; Sigma T6793). Phalloidin (ThermoFisher A12379) was used at a concentration of 1:500.
Mitotracker staining
Mitotracker Red (ThermoFisher #M22425) was resuspended in DMSO (0.25 mM) and diluted to 200 nM in E3 embryo medium. nco and csr embryos were then incubated in the dark for 20 minutes before removing Mitotracker solution and replacing with fresh E3 embryo medium. Samples were allowed to stabilize in the dark for 30 minutes before imaging at 21 hpf. Embryos were then phenotyped at 27 hpf.
Exogenous salt solutions
To test the effects of exogenous ions on otolith formation, embryos were kept in E3 Medium until early gastrulation (~10 hpf). Embryos were washed, dechorionated, and transferred to 1X Basic Solution (58 mM NaCl, 0.4 mM MgSO4 and 5 mM HEPES) supplemented with 0.7 mM potassium chloride, 0.6 mM calcium nitrate or 0.6 mM calcium chloride. Embryos were then transferred to fresh 1X Basic Solution with respective supplement for the remaining development. Embryos were scored by the presence or absence of otoliths at 27 hpf and genotyped using High Resolution Melt analysis.
Statistical analyses
Statistical significance was calculated using Fisher’s Exact Test, G-test for Independence, and Chi-Squared Distribution.
3. Results
3.1. csr and nco are genetically-linked
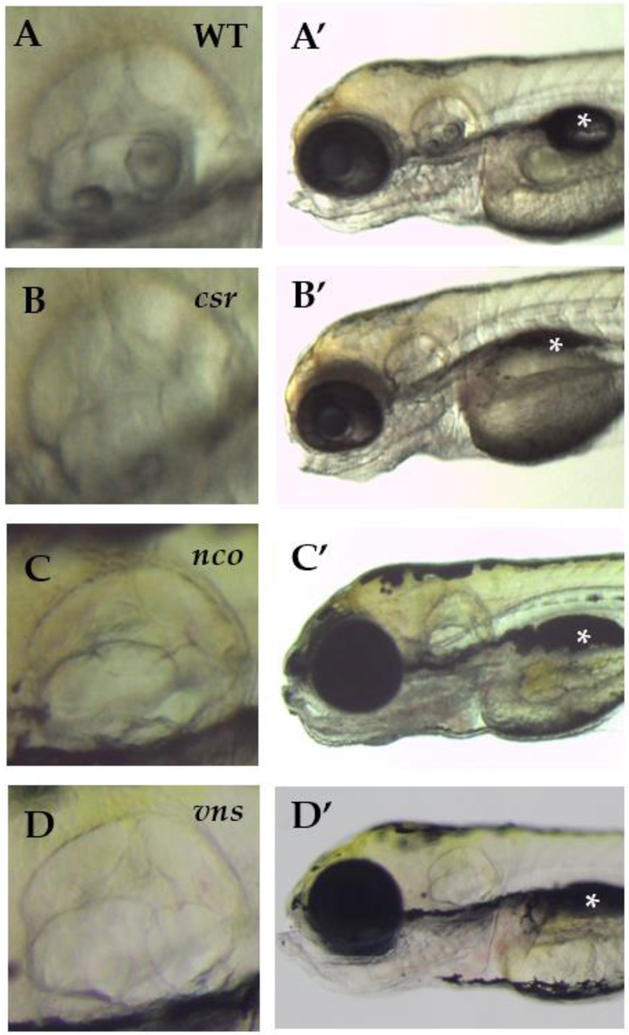
The most apparent phenotype of the homozygous recessive csr, nco, and vns mutants is that they fail to form otoliths (lapillus and sagitta) or any observable complex calcium deposits within the inner ear (Fig. 1A–D; Table S1). Furthermore, the mutant larvae are homozygous lethal by 7 days post fertilization (dpf) as the swim bladder fails to inflate (Fig. 1A′–D′) and they are unable to feed. As a result, we do not know whether asteriscus formation is affected. While it is still unknown why the swim bladder fails to inflate when otoliths are absent, it is a common phenotype in other mutants with otolith agenesis [18–24]. Due to this commonality within csr and nco, we sought to determine if these phenotypes would complement each other. The results of the complementation test showed that some offspring failed to develop otoliths (29.25%; n=106; Table S1), supporting that nco and csr likely are allelic.
Figure 1:
(A-D) The csr, nco, and vns mutant phenotypes fail to form otoliths within the inner ear. However, semicircular canal formation appears to be normal. (A′-D′) All mutants fail to inflate their swim bladders, which is lethal. Imaged at 5 days post fertilization (dpf). Magnification 6.3X. (*) indicates swim bladder.
3.2. Exogenous ions influence otolith nucleation in csr embryos; not nco or vns embryos
As an aquatic species, the environment of zebrafish can be easily controlled and adapted to assess its impact on embryonic development. Previously, small molecules have been used to block otolith development by inhibiting otolith nucleation [10]. We hypothesized that there was an error in ion homeostasis that could be affected by exogenous solutions. In water treatments supplemented with calcium chloride (n=51), we found a significant decrease in csr penetrance in homozygous embryos (χ2=19.27, df=6; p=0.0037) compared to treatments supplemented with potassium chloride (n=46) or calcium nitrate (n=54). Additionally, we observed no significant change in nco mutant phenotype penetrance for water treatments supplemented with potassium chloride (17.76%; n=107), calcium chloride (16.67%; n=120) or calcium nitrate (16.9%; n=112)(G-test; p=0.975). Similarly, the penetrance of otolith formation in vns was not affected by exogenous salts (data not shown).
Building on the hypothesis that there was an error in ion homeostasis, Mitotracker was used to mark mitochondria-rich cells (i.e. presumptive ionocytes) in csr and nco embryos. While nco embryos appear normal, we observed that csr embryos show a lack of Mitotracker localization at 21 hpf (Fig. S1). Altogether, this suggests the nature of the nco and csr mutation, while likely allelic, are inherently different.
3.3. Potentially deleterious mutations identified in polyketide synthase for csr, nco, and vns
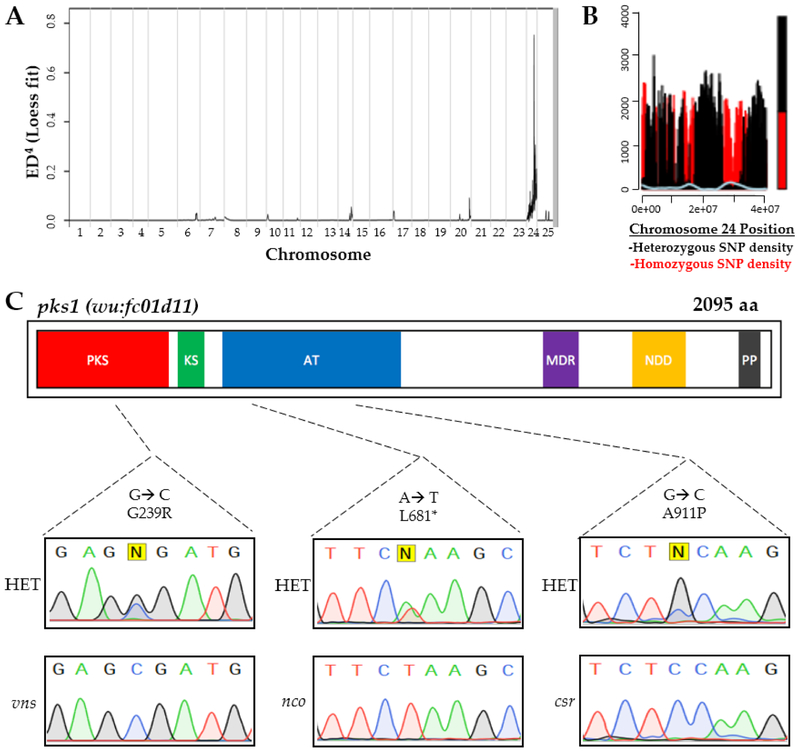
To positionally clone the gene responsible for nco and csr, we used complementary approaches for each strain. MMAPPR analysis of nco-derived RNA sequencing (Fig. 2A) [24] and MegaMapper analysis of csr-derived whole genome sequencing (Fig. 2B) [27] both identified a genomic region with high homology surrounding the pks1 locus. While several other genes were in that region, a previous study on otolith biomineralization in Japanese medaka made pks1 the likely gene candidate [25]. Potentially deleterious mutations were identified in pks1 for csr (A911P) and nco (L681*), which were both located within a conserved acyl transferase domain (Fig. 2C). Furthermore, a deleterious mutation in vns (G239R) was serendipitously found to be linked to a neighboring gene during a separate study. The deleterious point mutation was identified by Sanger sequencing of the pks1 locus and confirmed by relatively high penetrance of otolith agenesis (95%).
Figure 2:
Complementary approaches for causative gene discovery. MMAPPR analysis of RNA sequencing data for nco (A) and whole genome homology mapping for csr (B) identified regions of high homology on the 24th chormosome near the pks1 locus (~33 Gb). (C) Deleterious mutations were identified in pks1 for nco and csr within the acyl transferase (AT) domain and vns within the polyketide synthase (PKS) domain. Sanger sequencing confirmed SNPs in csr, nco, and vns mutants. Other domains include Ketoacyl Synthetase (KS), Medium Chain Reductase (MDR), NAD(P)-dependent dehydrogenase (NDD), and Phosphopanthetheine-Binding (PP).
3.4. Japanese medaka pks1 mRNA or plasmid DNA rescues otolith biomineralization in csr, nco, and vns
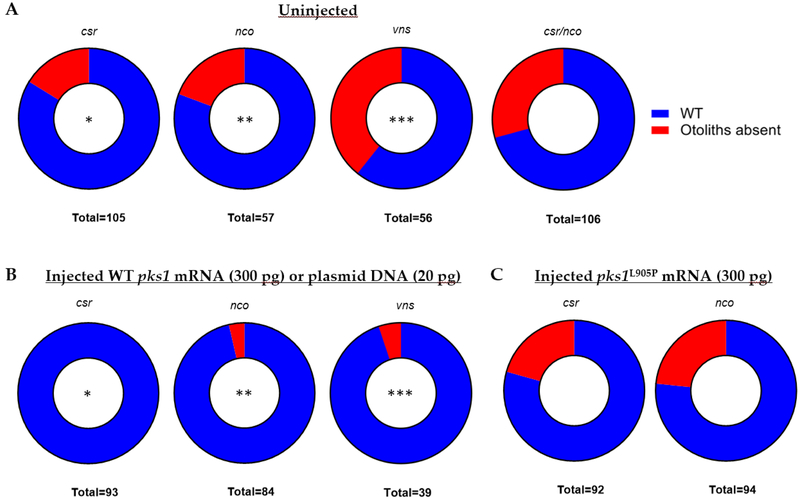
While the last common ancestor of Japanese medaka and zebrafish was estimated to be 150 million years ago [30], we sought to assess if the function of pks1 within the inner ear is conserved. We injected Japanese medaka pks1 mRNA or DNA into single-cell embryos of csr, nco, and vns heterozygous incrosses. Microinjection of Japanese medaka pks1 mRNA (300 ng/μL) rescued otolith biomineralization in both csr (p<0.0001; χ2<0.0001; n=93) and nco (p=0.0032; χ2=0.0022; n=84) mutants (Fig. 3B; Table S1). Additionally, microinjection of the Japanese medaka pks1 plasmid (20 ng/uL) provided by Dr. Takeda rescued otolith biomineralization in vns (p<0.0001; χ2=0.0004; n=39). Using site-directed mutagenesis, we introduced the non-synonymous mutation (A911P) in csr to the Japanese medaka mRNA construct (L905P). We repeated injections into single-cell embryos and failed to rescue otolith biomineralization in csr and nco. WT medaka pks1, but not pks1L905P, rescued otolith biomineralization in csr and nco embryos (Fig. 3C; Table S1).
Figure 3:
WT pks1 nucleic acid rescues otolith formation in csr, nco, and vns. (A) Normal frequencies of mutant phenotypes in each uninjected strain. All four pairings follow homozygous recessive mode of inheritance. (B) Results of injected embryos show that Japanese medaka pks1 mRNA (300 pg) rescues both csr and nco mutants and pks1 DNA (20 pg) rescues vns mutants. (*, p < 0.0001, paired t-test)(**, p < 0.0032, paired t-test)(***, p = 0.0001, paired t-test),. Site-directed mutagenesis was used to introduce a conserved mutation in csr (A911P) into the Japanese medaka construct (L905P) (C) Injection of pks1L905P (300 pg) fails to rescue csr or nco mutant phenotypes.
3.5. Ingenuity pathway analysis of nco embryos
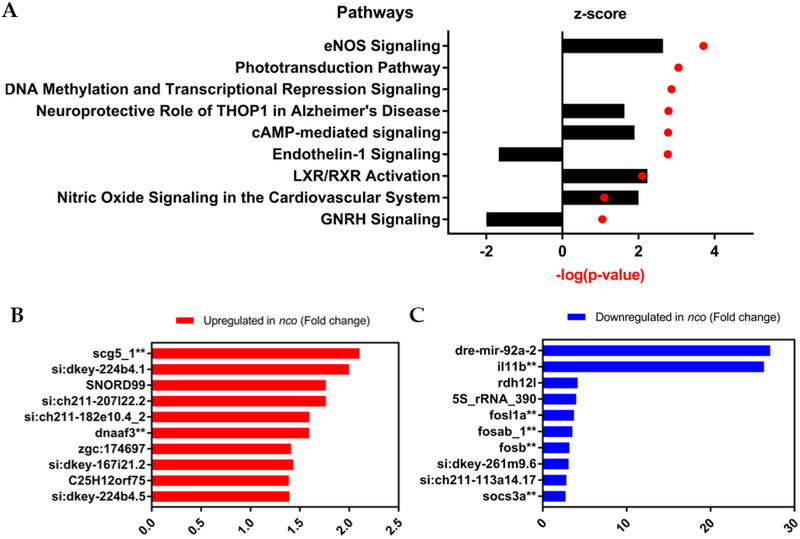
While pks1 is thought to produce an otolith nucleation factor [25], its broader role during inner ear development is unknown. Ingenuity Pathway Analysis of nco at 24 hpf identified eNOS and Endothelin-1 signaling as the top up- and down-regulated pathways, respectively (Fig. 4A). Among the down regulated genes was rdh12l, a gene adjacent to pks1, suggesting that there is local control of transcription at that locus. mir-92a, the top down-regulated gene, has a predicted binding site in the 3′UTR of rdh12l (Fig. S2) [31]. In addition, several genes listed in the top ten up- or down-regulated lists are also enriched in adult mechanosensory hair cells such as il11b, fosab, fosb, fosl1a, socs3a, scg5, and dnaaf3 (Figs. 4B–C) [32]. Of these genes, il11b is up-regulated during neuromast hair cell regeneration [33]. Notably, dnaaf3 causes primary ciliary dyskinesia and morpholino knockdown of dnaaf3 causes abnormal otolith growth [34]. While its role in inner ear development is unknown, scg5 is expressed within the anterior and posterior poles of the otic placode during the critical period of otolith nucleation [35].
Figure 4:
Gene expression and pathway analysis of nco embryos. (A) Ingenuity Pathway Analysis shows the top up-regulated and down-regulated pathways, which are eNOS Signaling and Endothelin-1 Signaling, respectively. Positive z-score indicated increased mRNA levels. Negative z-score indicates decreased mRNA levels. No change in mRNA levels results in a z-score of zero. (B) Differential gene expression in the top up-regulated genes. (C) Differential gene expression in the top down-regulated genes. (**, expressed in adult zebrafish mechanosensory hair cells) [32].
3.6. Aberrant expression of proteins involved in otolith development in csr and nco
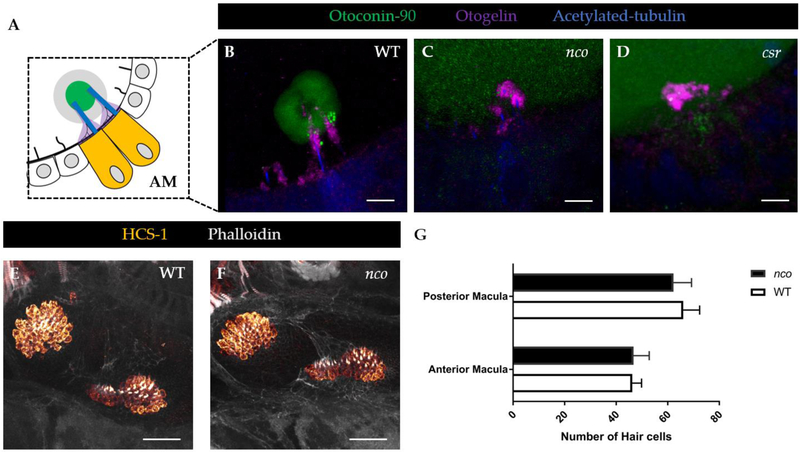
In mammalian inner ear development, Oc90 is necessary for otoconial seeding and nucleation [13, 14]. Similarly, the role of Oc90 is evolutionarily-conserved in zebrafish and has been previously thought to be necessary for otolith nucleation [18]. Using immunofluorescence (IF), we saw diffuse expression of Oc90 in csr and nco otocysts (Figs. 5B–D), which demonstrated that Oc90 expression within the otocyst is not sufficient for otolith biomineralization in zebrafish. Similarly, normal localization of Otogelin (Otog), a protein required for otolith tethering in the otolithic membrane is not sufficient for Oc90 attachment. Additionally, other otoconins that are important for calcium deposition and growth were detected with diffuse expression within the otocyst such as Starmaker and Keratan Sulfate (data not shown) [36, 37].
Figure 5:
Aberrant expression of proteins invovled in otolith development in csr and nco. (A) Schematic of anterior macula (AM) tethered to otolith at 27 hpf. (B) In WT, Otoconin-90 (Oc90) is expressed within the mineralized otolith, which is situated atop the otolithic membrane (Otogelin, or Otog), at 27 hpf. Scale bar = 5 μm. (C-D) Oc90 has diffuse expression within the otocyst of csr and nco. In csr and nco, Otog is localized near the apical surface of hair cells. (E-F) Expression showing hair cells in WT and nco larvae at 5dpf. Scale bar = 25 μm. (G) Quantification of hair cell numbers in the posterior and anterior macula of WT and nco (n = 4).
3.7. Polyketide synthase as an otolith precursor binding factor?
Otolith nucleation is thought to be mediated by a tether-cell specific otolith precursor binding factor (OPBF), which lays the foundation for the successive biomineralization of the otolith [9, 11, 38]. The presence of an OPBF was proposed almost two decades ago and its identification proves to be elusive [38]. Recent studies suggest that one or more OPBFs are expressed by tether-cells and help to mediate otolith nucleation by binding other OPPs [9, 11, 39].
We sought to assess if pks1 or its enzymatic product is a tether-cell specific nucleation factor. While medaka has diffuse pks1 mRNA expression in the otic epithelium [25], we hypothesized that the expression might be restricted to hair cells. First, using publicly available RNA-seq data, we found that pks1 mRNA is enriched (7.46-fold increase) in adult mechanosensory hair cells compared to support cells within the zebrafish inner ear (Table S2). Additionally, this data suggests pks1 mRNA to be transcriptionally regulated in support cells. Support cells predominantly express a 300bp region of the 5′UTR of the pks1 transcript while hair cells express the full open reading frame [32]. A search for transcriptional regulatory motifs in the 5′UTR of pks1 found a predicted binding site for TCF-3 [40], a transcription factor highly expressed in adult mechanosensory hair cells [32]. While the role of TCF-3 in the inner ear is unknown, it is expressed within the otic vesicle during the critical period of otolith nucleation [35].
Then, we demonstrated that the total number of hair cells remain unchanged during early development in nco, suggesting there are no differences in tether cell maturation and maintenance (Figs. 5E–G). Using RT-PCR, we detected pks1 mRNA during the critical period of otolith nucleation (Fig. S3). However, in situ data showed ubiquitous expression of pks1 in the otic vesicle of zebrafish [25]. While pks1 might be enriched in adult hair cells, early expression shows that it is ubiquitously expressed in the otic vesicle and, therefore, not the tether-cell specific OPBF.
4. Discussion
The homozygous recessive mutants csr, nco, and vns were chosen for this study because each lack the necessary factors such as an OPBF for otolith seeding and biomineralization. To determine the genes responsible for otolith agenesis in these mutants, we used two complementary approaches. The first approach was Whole Genome Sequencing of the csr mutant genome to identify regions of high homology. This indeed was difficult as the csr background strain was heavily inbred, resulting in multiple peaks of high homology. Since we demonstrated csr and nco are genetically-linked, we sought to further clarify the responsible locus using a second method (i.e. RNA-seq of the nco transcriptome) for comparison. This result pinpointed a region of high homology near the end of the 24th chromosome. While deciphering potentially deleterious mutations within that region, we focused on pks1 following evidence that it is responsible for otolith nucleation in Japanese medaka [25]. While these species are evolutionarily divergent, the shared phenotype between medaka and our mutants suggested that the role of pks1 is conserved. As a result, we chose to use medaka pks1 nucleic acid to rescue otolith formation in csr, nco and vns mutants. Similarities can also be drawn with other zebrafish mutants such as keinstein, which has diffused expression of Starmaker within the otocyst and exhibits similar circling swimming behaviors [41, 42]. Furthermore, keinstein may be another pks1 allele due to its predicted chromosomal location [43].
While WT medaka pks1 rescues otolith biomineralization in csr and nco, differences in penetrance of exogenous ions on otolith formation suggested the nature of each mutation is fundamentally different. This was confirmed by Sanger sequencing that nco has a premature stop codon while csr likely makes a defective protein that may be stabilized by exogenous ions. This defective protein may be the explanation for the differences in Mitotracker localization in csr. Due to its surface stain expression, we hypothesize that Mitotracker was localized to mitochondria-rich ionocytes [44]. Ionocytes have previously been implicated in otolith formation as mutations in gcm2, which is responsible for ionocyte maturation, leads to otolith agenesis [20, 45]. We hypothesize that the endolymph in csr and nco mutants has the necessary components for otolith nucleation [2] but lack a trigger factor produced by pks1. The absence of pks1 does not visibly appear to affect hair cell development that are required for otolith nucleation either [9]. It has been previously suggested that apolipoprotein could potentially bind polyketide synthase [4, 25]. Given our RNA-seq analysis of nco, we see no significant change in any apolipoprotein expression. Publicly-available in situ data does not support Apolipoprotein expression within the inner ear [35]. Additionally, IF of csr and nco embryos demonstrated that expression of a critical otoconial seeding protein, Oc90, within the otocyst is not sufficient for otolith biomineralization in the presence of the otolithic membrane.
One caveat is that the penetrance of otolith formation is influenced by the genetic background of zebrafish. When treated with the small molecule 31N3, WT embryos in the AB/EKW background fail to develop otoliths [10]. However, 31N3 fails to inhibit otolith formation in the TL and TU strains, suggesting that there are potential genetic modifiers that influence otolith nucleation in these backgrounds. While the csr mutation (A911P) leads to otolith agenesis in the AB background, homozygosity at the locus is compatible with proper development in the AB/TL background (data not shown). This suggests csr may be a hypomorphic allele and the AB background can overcome the loss of Pks1 function with enhanced ion flux. Ironically, the mutant phenotype was lost when csr was outcrossed to the WIK background. It was only until csr was backcrossed to the AB background that the mutants were recovered. Altogether, we suggest that the AB background heavily influences the penetrance of otolith formation.
While pks1 likely acts as an enzyme whose expression is enriched in adult mechanosensory hair cells [32], its product is required for otolith nucleation in zebrafish. However, the molecular function of pks1 remains unknown. Using nco RNA-seq data, we performed an Ingenuity Pathway Analysis, which identified eNOS and Endothelin-1 signaling as the most up- and down-regulated pathways, respectively. eNOS signaling could be impacted by pks1 metabolites such as iromycin, which has been shown to inhibit this pathway [46]. Both eNOS and Endothelin-1 have been implicated in inner ear development and function. Notably, it has been demonstrated that these pathways are inversely related in sensorineural hearing loss [47]. An example of this is Waardenburg syndrome, caused by mutations in endothelins, which cause abnormal pigmentation and sensorineural hearing loss [48]. During early development, Endothelin-1 mRNA turns on during the critical period of otolith nucleation [35, 49] and is detected in the otic vesicle at 24 hpf [50]. Endothelin-1 and its receptor (ednraa) are both enriched in adult zebrafish inner ear support cells [32]. Additionally, Endothelin-1 has been identified as a potential modifier of osteoblast function to increase bone mineralization [51]. Furthermore, Endothelin-1 has been implicated with the FOS-family of genes (fosab, fosb, and fosl1a) and socs3a, which are all differentially expressed in nco at 24 hpf. These genes are all part of a regulatory network during hypergravity-mediated bone formation [52]. Furthermore, the presence of osteoblast-associated proteins within teleost otoliths suggest a common mechanism between bone mineralization and otolith biomineralization [4]. Future studies will attempt to clarify the roles of Endothelin-1 and eNOS signaling pathways during biomineralization events.
Supplementary Material
Highlights.
Mutations in polyketide synthase lead to otolith agenesis in zebrafish
Neither Otoconin-90 or Otogelin are sufficient for otolith seeding and tethering
Endothelin-1 and eNOS signaling likely mediate otoconia biomineralization
Acknowledgments:
We recognize the University of Nebraska Medical Center Genomics Core Facility for assistance with sequencing and bioinformatics. We thank Dr. Hiroyuki Takeda from the University of Tokyo for supplying the Japanese medaka pks1 mRNA construct. We acknowledge Creighton University Integrated Biomedical Imaging Facility for assistance with confocal microscopy. Finally, we express gratitude to the members of the Kramer Lab at Creighton University, the Shavit Lab at University of Michigan, and the Wilkinson Lab at Royal Holloway University of London for their support with zebrafish husbandry.
Funding: The Kramer lab is grateful for funding through grants from the State of Nebraska (LB-692), the National Center for Research Resources (5P20RR018788–09), and the National Institute of General Medical Sciences (8 P20 GM103471–09). The Shavit lab acknowledges support from National Heart, Lung, and Blood Institute grants (R01HL124232 and HL125774).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: The authors declare no conflict of interest.
References
- 1.Lundberg YW, et al. , Mechanisms of otoconia and otolith development. Dev Dyn, 2015. 244(3): p.239–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Payan P, et al. , Endolymph chemistry and otolith growth in fish. Comptes Rendus Palevol, 20043(6–7): p. 535–547. [Google Scholar]
- 3.Borelli G, et al. , Biochemical relationships between endolymph and otolith matrix in the trout (Oncorhynchus mykiss) and turbot (Psetta maxima). Calcif Tissue Int, 2001. 69(6): p. 356–64. [DOI] [PubMed] [Google Scholar]
- 4.Thomas ORB, et al. , The inner ear proteome of fish. Febs j, 2019. 286(1): p. 66–81. [DOI] [PubMed] [Google Scholar]
- 5.Reimer T, et al. , Rapid growth causes abnormal vaterite formation in farmed fish otoliths. J Exp Biol, 2017. 220(Pt 16): p. 2965–2969. [DOI] [PubMed] [Google Scholar]
- 6.Söllner C, et al. , Control of Crystal Size and Lattice Formation by Starmaker in Otolith Biomineralization. Science, 2003. 302(5643): p. 282–286. [DOI] [PubMed] [Google Scholar]
- 7.Haddon C and Lewis J, Early ear development in the embryo of the zebrafish, Danio rerio. J Comp Neurol, 1996. 365(1): p. 113–28. [DOI] [PubMed] [Google Scholar]
- 8.Riley BB, et al. , A critical period of ear development controlled by distinct populations of ciliated cells in the zebrafish. Dev Biol, 1997. 191(2): p. 191–201. [DOI] [PubMed] [Google Scholar]
- 9.Stooke-Vaughan GA, et al. , The role of hair cells, cilia and ciliary motility in otolith formation in the zebrafish otic vesicle. Development, 2012. 139(10): p. 1777–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson RT, et al. , Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc Natl Acad Sci U S A, 2000. 97(24): p. 12965–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stooke-Vaughan GA, et al. , Otolith tethering in the zebrafish otic vesicle requires Otogelin and alpha-Tectorin. Development, 2015. 142(6): p. 1137–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riley BB, Genes Controlling the Development of the Zebrafish Inner Ear and Hair Cells, in Current Topics in Developmental Biology. 2003, Academic Press; p. 357–388. [DOI] [PubMed] [Google Scholar]
- 13.Zhao X, et al. , Otoconin-90 deletion leads to imbalance but normal hearing: a comparison with other otoconia mutants. Neuroscience, 2008. 153(1): p. 289–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao X, et al. , Gene targeting reveals the role of Oc90 as the essential organizer of the otoconial organic matrix. Dev Biol, 2007. 304(2): p. 508–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, et al. , Otoconin-90, the mammalian otoconial matrix protein, contains two domains of homology to secretory phospholipase A2. Proc Natl Acad Sci U S A, 1998. 95(26): p. 15345–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deans MR, Peterson JM, and Wong GW, Mammalian Otolin: a multimeric glycoprotein specific to the inner ear that interacts with otoconial matrix protein Otoconin-90 and Cerebellin-1. PLoS One, 2010. 5(9): p. e12765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreland KT, et al. , In vitro calcite crystal morphology is modulated by otoconial proteins otolin-1 and otoconin-90. PLoS One, 2014. 9(4): p. e95333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petko JA, et al. , Otoc1: a novel otoconin-90 ortholog required for otolith mineralization in zebrafish. Dev Neurobiol, 2008. 68(2): p. 209–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes I, et al. , Otopetrin 1 is required for otolith formation in the zebrafish Danio rerio. Dev Biol,2004. 276(2): p. 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stawicki TM, et al. , The zebrafish merovingian mutant reveals a role for pH regulation in hair cell toxicity and function. Dis Model Mech, 2014. 7(7): p. 847–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sumanas S, Larson JD, and Miller Bever M, Zebrafish chaperone protein GP96 is required for otolith formation during ear development. Dev Biol, 2003. 261(2): p. 443–55. [DOI] [PubMed] [Google Scholar]
- 22.Kiss PJ, et al. , Inactivation of NADPH oxidase organizer 1 results in severe imbalance. Curr Biol, 2006. 16(2): p. 208–13. [DOI] [PubMed] [Google Scholar]
- 23.Colantonio JR, et al. , The dynein regulatory complex is required for ciliary motility and otolith biogenesis in the inner ear. Nature, 2008. 457: p. 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill JT, et al. , MMAPPR: Mutation Mapping Analysis Pipeline for Pooled RNA-seq. Genome Research, 2013. 23(4): p. 687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hojo M, et al. , Unexpected link between polyketide synthase and calcium carbonate biomineralization. Zoological Lett, 2015. 1(1): p. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schibler A and Malicki J, A screen for genetic defects of the zebrafish ear. Mech Dev, 2007. 124(7–8): p. 592–604. [DOI] [PubMed] [Google Scholar]
- 27.Obholzer N, et al. , Rapid positional cloning of zebrafish mutations by linkage and homozygosity mapping using whole-genome sequencing. Development, 2012. 139(22): p. 4280–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kramer A, et al. , Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics, 201430(4): p. 523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung K, et al. , Structural and molecular interrogation of intact biological systems. Nature, 2013. 497(7449): p. 332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirchmaier S, et al. , The Genomic and Genetic Toolbox of the Teleost Medaka (Oryzias latipes). Genetics, 2015. 199(4): p. 905–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ulitsky I, et al. , Extensive alternative polyadenylation during zebrafish development. Genome Res, 2012. 22(10): p. 2054–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barta CL, et al. , RNA-seq transcriptomic analysis of adult zebrafish inner ear hair cells. Sci Data, 2018. 5: p. 180005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang L, et al. , Gene-expression analysis of hair cell regeneration in the zebrafish lateral line. Proceedings of the National Academy of Sciences, 2014. 111(14): p. E1383–E1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchison HM, et al. , Mutations in axonemal dynein assembly factor DNAAF3 cause primary ciliary dyskinesia. Nature Genetics, 2012. 44: p. 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thisse B, et al. , Expression of the zebrafish genome during embryogenesis. 2001: ZFIN Direct Data Submission. [Google Scholar]
- 36.Yang H, et al. , Matrix recruitment and calcium sequestration for spatial specific otoconia development. PLoS One, 2011. 6(5): p. e20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sollner C, et al. , Control of crystal size and lattice formation by starmaker in otolith biomineralization. Science, 2003. 302(5643): p. 282–6. [DOI] [PubMed] [Google Scholar]
- 38.Riley BB and Grunwald DJ, A mutation in zebrafish affecting a localized cellular function required for normal ear development. Dev Biol, 1996. 179(2): p. 427–35. [DOI] [PubMed] [Google Scholar]
- 39.Yu X, et al. , Cilia-driven fluid flow as an epigenetic cue for otolith biomineralization on sensory hair cells of the inner ear. Development, 2011. 138(3): p. 487–94. [DOI] [PubMed] [Google Scholar]
- 40.Chang TH, et al. , An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics, 2013. 14 Suppl 2: p. S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sollner C, et al. , Mutated otopetrin 1 affects the genesis of otoliths and the localization of Starmaker in zebrafish. Dev Genes Evol, 2004. 214(12): p. 582–90. [DOI] [PubMed] [Google Scholar]
- 42.Whitfield TT, et al. , Mutations affecting development of the zebrafish inner ear and lateral line. Development, 1996. 123: p. 241–54. [DOI] [PubMed] [Google Scholar]
- 43.Geisler R, et al. , Large-scale mapping of mutations affecting zebrafish development. BMC Genomics, 2007. 8: p. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Esaki M, et al. , Mechanism of development of ionocytes rich in vacuolar-type H(+)-ATPase in the skin of zebrafish larvae. Developmental biology, 2009. 329(1): p. 116–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumai Y, Kwong RWM, and Perry SF, A role for transcription factor glial cell missing 2 in Ca2+ homeostasis in zebrafish, Danio rerio. Pflügers Archiv - European Journal of Physiology, 2015. 467(4): p. 753–765. [DOI] [PubMed] [Google Scholar]
- 46.Surup F, et al. , The iromycins, a new family of pyridone metabolites from Streptomyces sp. I. Structure, NOS inhibitory activity, and biosynthesis. J Org Chem, 2007. 72(14): p. 5085–90. [DOI] [PubMed] [Google Scholar]
- 47.Liu Q, et al. , [The study on plasma ET and NO of patients with sudden hearing loss]. Lin Chuang Er Bi Yan Hou Ke Za Zhi, 2003. 17(11): p. 668–9. [PubMed] [Google Scholar]
- 48.Pingault V, et al. , Review and update of mutations causing Waardenburg syndrome. Hum Mutat, 2010. 31(4): p. 391–406. [DOI] [PubMed] [Google Scholar]
- 49.White RJ, et al. , A high-resolution mRNA expression time course of embryonic development in zebrafish. Elife, 2017. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller CT, et al. , sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch development. Development, 2000. 127(17): p. 3815–3828. [DOI] [PubMed] [Google Scholar]
- 51.Johnson MG, et al. , Big endothelin changes the cellular miRNA environment in TMOb osteoblasts and increases mineralization. Connect Tissue Res, 2014. 55 Suppl 1: p. 113–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aceto J, et al. , Zebrafish Bone and General Physiology Are Differently Affected by Hormones or Changes in Gravity. PLoS ONE, 2015. 10(6): p. e0126928. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.