Abstract
The recent outbreaks of Ebolavirus (EBOV) in West Africa underscore the urgent need to develop an effective EBOV vaccine. Here, we report the development of synthetic nanoparticles as a safe and highly immunogenic platform for vaccination against EBOV. We show that a large recombinant EBOV antigen (rGP) can be incorporated in a configurational manner into lipid-based nanoparticles, termed interbilayer-crosslinked multilamellar vesicles (ICMVs). The epitopes and quaternary structure of rGP were properly maintained on the surfaces of ICMVs formed either with or without nickel nitrilotriacetic acid (NTA)-functionalized lipids. When administered in mice, rGP-ICMVs without NTA-lipids efficiently generated germinal center B cells and polyfunctional T cells while eliciting robust neutralizing antibody responses. This study suggests the potential of vaccine nanoparticles as a delivery platform for configurational, multivalent display of large subunit antigens and induction of neutralizing antibody and T cell responses.
Graphical Abstract
Two forms of lipid-based nanoparticles (ICMVs) loaded with recombinant Ebola virus glycoprotein (rGP) were produced, characterized, and administered to mice for analysis of immune responses. rGP ICMVs promoted germinal center B cell formation, antibodies with neutralizing-capacity, and IFN-γ+ T-cell responses. Graphical abstract: Figure 1, 6C, 6H, & 7B.
Introduction
Since the emergence of Ebolavirus (EBOV) in 1976, approximately 13,000 lives have been lost to EBOV, with mortality rates of 25–90% among infected individuals.1 The 2014 Ebola virus (EBOV) outbreak in West Africa led to unprecedented cases of Ebola virus disease (EVD) resulting in ~11,000 deaths, an approximate 7-fold increase compared to all previous incidents combined.1 This outbreak also marked the first inter-continental cases of EVD, prompting a worldwide response to the endemic. Therefore, there is an urgent need to develop an effective vaccine against EBOV.
To that end, several vaccine candidates for EBOV have progressed to clinical trials, most of which are so far based on recombinant viral constructs expressing the EBOV envelope glycoprotein (GP). As the only surface-expressed EBOV antigen, GP is naturally presented as a heavily glycosylated trimer and is required for both target cell binding and viral membrane fusion, making it an ideal vaccine target.2–4 While the recombinant Vesicular Stomatitis Virus vaccine candidate (rVSV-EBOV) has been shown to induce potent immune responses after a single dose in clinical trials, numerous side effects have been reported among healthy adults, including acute arthritis and skin lesions.5–7 Additionally, the potential for toxic side effects in infants and immunocompromised individuals remain as major concerns.6 Alternatively, non-replicating recombinant adenovirus vectors co-expressing GP are a safer alternative, but issues with potency require booster vaccinations, and in many cases pre-existing or post exposure anti-vector immunity may affect vaccine efficacy.8,9
In contrast, EBOV subunit vaccines may offer a safer alternative for inducing immune responses against the antigen of interest.10–13 However, subunit vaccines can generally suffer from weak and transient immune responses. While many laboratories have focused on engineering vaccine delivery platforms that can improve immunogenicity of subunit antigens,14–18 it remains unclear how in vivo performance is dictated by antigen configuration on vaccine delivery vehicles. Here, we aimed to develop synthetic nanoparticles as a safe and highly immunogenic platform for vaccination against EBOV and to examine how antigen orientation impacts cellular and humoral immune responses. We have previously reported the development of lipid-based nanoparticles, called interbilayer-crosslinked multilamellar vesicles (ICMVs).19 ICMVs have been shown to elicit potent cellular and humoral immune responses with a range of antigens, including peptides and recombinant proteins.19–22 Yet, it remains to be seen how to preserve the epitope configuration and orientation of large recombinant proteins, such as EBOV GP trimer, while maintaining their immunogenicity in vivo.
Here, we report that a recombinant EBOV GP (rGP) with minimal recombinant alterations and no chemical cross-linking can be incorporated into ICMVs while preserving its epitope configuration and orientation. Specifically, we tested rGP encapsulated in two different ICMV variants, with or without NTA-functionalized lipids (NTA ICMVs and ICMVs, respectively). Introduction of NTA-lipid is thought to allow binding of poly-histidine tagged rGP on the surfaces of NTA ICMVs.22 We then performed detailed immunofluorescence analyses with monoclonal antibodies specific for linear or configurational rGP epitopes both on a bulk sample and a single particle level. Our results indicated that the quaternary structure of rGP was properly maintained on the surfaces of both ICMV formulations. Mice vaccinated with both forms of rGP nanoparticles carrying an immunostimulatory adjuvant, MPLA (a Toll-like receptor-4 agonist) increased humoral and cellular immune responses, compared with the soluble control. In particular, the rGP ICMV + MPLA group potently induced germinal center (GC) B cells and polyfunctional T cells while generating immune sera with neutralizing-capacity.
Methods
Synthesis of rGP-loaded ICMVs.
ICMVs were synthesized as reported before,19,20 with some modifications. Brief, 1,2-dioleoyl-snglycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[4-(p-maleimidophenyl)butyramide] sodium salt (MPB), and 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] nickel salt (DOGS NTA) (Avanti Polar Lipids), were mixed in either 50:50:0 or 48:50:2 molar ratios and dried to produce thin films. Some samples were added with 0.2 molar % of a lipophilic fluorophore 1,1’-Dioctadecyl-3,3,3’,3’-Tetramethylindodicarbocyanine, 4-Chlorobenzenesulfonate Salt (DiD, Thermo Scientific). The dried films were hydrated in the presence of recombinant Ebola virus glycoprotein (rGP) produced with a 6-histidine protein purification tag, as we have reported previously.23 The resulting multilamellar vesicles (MLVs) were probe tip sonicated (QSonica) to produce unilamellar vesicles (ULVs). We then added CaCl2 and dithiothreitol (DTT) at 33mM and 1.24mM, respectively, for inducing vesicle fusion and crosslinking. The resulting ICMVs were centrifuged at 18000 r.c.f. for 4 minutes at 4 °C to pellet the ICMVs, and the supernatant containing unloaded rGP was decanted. Finally, the ICMV pellets were washed with DNA grade water (Fisher) and suspended in 0.22 µm filtered PBS (Gibco). Monophosphoryl lipid A (MPLA) (Avanti Polar Lipids) was added to the initial phospholipid mixture for production of ICMVs, and if necessary, additional MPLA was added for an injection dose of 2.5 µg.
Characterization of ICMVs.
Antigen loading in ICMVs was assessed by poly-acrylamide gel electrophoresis under non-reducing conditions (NR PAGE). Briefly, samples were prepared following manufacturer’s instructions, loaded into Bolt 4–12% Bis-Tris Plus gels (Invitrogen), and ran for 35 minutes at 200V in MOPS running buffer (Novex). Protein content was assessed by Coomassie Brilliant Blue R-250 staining (Fisher), imaged with FluorChem M (Protein Simple), and quantified using ImageJ software. For Blue Native PAGE, ICMVs were run on NativePAGE™ Novex® Bis-Tris gel system (Life Technologies). Briefly, samples were diluted in Native PAGE sample buffer, bath sonicated to disrupt aggregates, and select samples (indicated in figures) were incubated with N-Dodecyl β-D-maltoside (DDM, Invitrogen) at a 1.11% working concentration for 30 minutes on ice. Immediately before loading onto gels (3–12% Bis-Tris), Coomassie G-250 was added to the samples following manufacturer’s instructions. Samples that were not detergent incubated were prepared in sample buffer and remained on ice until loaded. Gels were run at room temperature using dark blue cathode buffer for approximately 100 minutes. Gels were destained according to manufacturer’s instructions, and protein migration was assessed by silver staining (Thermo Fisher). Mass spectrometry analysis of rGP content was performed as described previously.23 Particle diameter and zeta potential were measured by dynamic light scattering (DLS) using a Malvern ZetaSizer Nano ZSP. Samples were diluted in 0.22 µm filtered diluent for DLS analysis, recollected, and diluted further for individual particle sizing by nanoparticle tracking analysis via a NanoSight NS300 instrument (Malvern).
Immunofluorescence stained nanoparticles were prepared by incubating equal volumes of DiD-embedded, rGP-displaying ICMVs overnight at 4 °C in 0.04 mg/mL antigen-specific primary monoclonal antibodies 6D8 or 13C6 (USAMRIID), or mouse IgG1, κ isotype (BP Pharmingen) in FACS buffer (1% BSA in PBS). After washing, the particles were incubation with F(ab’)2 α-mouse IgG-PE secondary antibody (eBioscience) for one hour at room temperature. After washing, fluorescence signal on particles was measured at 488/578 and 644/670 nm for PE and DiD, respectively. After immunostaining assay, the same samples were analyzed on a Beckman Coulter MoFlo Astrios with M1 and M2 masks. NanoFACS data was analyzed via FloJo software.
Animals.
Female 8–12-weeks-old C57BL/6 mice were obtained from Charles River. Research was conducted under an IACUC approved protocol in compliance with the Animal Welfare Act, PHS Policy, and other Federal statutes and regulations. The United States Army Medical Research Institute of Infectious Diseases (USAMRIID) IACUC committee approved this protocol, and experiments were conducted at an Association for Assessment and Accreditation of Laboratory Animal Care, International accredited facility. Mice were primeboost vaccinated at a three-week interval using bi-lateral subcutaneous injections at the tail base. Antigen and adjuvant amounts were administered at 3 µg and 2.5 µg, respectively, for both vaccinations.
Analysis of antibody titers and neutralization
Immune serum was collected and analyzed for IgM and IgG titers using ELISA plates coated with 2 μg/ml rGP. Secondary antibodies included goat anti-mouse IgG-HRP (Southern Biotech 1030–05), IgG1-HRP (Southern Biotech 1070–05), IgG2c-HRP (Southern Biotech 1079–05), and IgM-HRP (1030–05). Plates were developed using Sure Blue TMB 1-component substrate and optical density (O.D.) was read at 450 nm on a SpectraMax M5 (Molecular Devices). End-point titers were defined as the background plus 0.2. O.D.
Percent neutralization of serum antibodies was assessed by enhanced green fluorescent protein (eGFP) fluorescence of residually infected Freestyle™ 293-F cells (ThermoFisher). Briefly, recombinant vesicular stomatitis virus expressing both Ebola virus glycoprotein and eGFP (rVSV-GP) (a kind grift from Kartik Chandran, Albert Einstein College of Medicine) were incubated with serial dilutions of vaccination serum and 5% v/v guinea pig complement (Cedarlane Laboratories) for 1 hour at 37 °C. The rVSV-GP/serum mixtures were added to 100uL suspensions of Freestyle™ 293-F cells at a concentration of 1×106 cells/mL and incubated for 18–20 hours at 37°C. Percentage of infection, i.e. eGFP expression, was measured using a BD FACSCanto II, and, neutralization was calculated by normalizing the infection percentages to the ICMV + MPLA control group.
Analysis of B-cell and T-cell responses.
For GC B cell analysis, single-cell suspensions of draining lymph nodes were washed with FACS buffer (PBS, 0.5% BSA and 2 mM EDTA), Fc-blocked (Miltenyi anti- CD16/CD32), and stained with B220 (BD, #RA3–6B2), CD95 (BD, #Jo2), CD138 (BD, #281–2), T & B Cell Activation Antigen (BD, #GL-7), and viability dye (Live/Dead Aqua, Invitrogen). Samples were run on a BD FACS Canto II and analyzed using FlowJo software (Tree Star, Inc.). For B cell ELISpot assay, splenocytes were seeded at 2.5×105 cells/well into rGP-coated ELISpot plates. For T cell ELISpot assay, splenocytes were suspended in complete medium (RPMI with 20 U/mL mouse recombinant IL2 (Life Technologies), 2 μg/mL mouse CD49d (BD), and 2 μg/mL mouse CD28 (BD)), and plated at 2.5×105 cells/well pre-coated with IFNγ capture antibody (clone AN18). EBOV GP peptide (Mimotopes, WIPYFGPAAEGIYTE) was added at 4 μg/mL. After 16 h incubation at 37°C, both B-cell and T-cell ELISpot plates were developed per the manufacturer’s instructions, followed by analysis using a CTL ImmunoSpot instrument (Cellular Technology Limited).
For intracellular cytokine staining (ICS) assay, splenocytes were cultured at 106 cells/mL in complete media with 1X protein transport inhibitor cocktail (eBioscience). Cells were re-stimulated with EBOV GP peptide, WIPYFGPAAEGIYTE at 2 μg/mL. After five hours, cells were stained with Live/Dead Aqua and antibodies against CD3 (BD Clone 500A2), CD8 (BD Clone 53–6.7), and CD4 (BD Clone RM4–5). After fixation and permeabilization, cells were stained for IFNγ (BD Clone XMG1.2), IL2 (BD Clone JES6–5H4), and TNFα-PE (BD Clone MP6-XT22) and analyzed on BD FACSCanto II.
Statistical analysis.
Determination of statistical significance was performed using Prism 7.0.3. One-way or two-way ANOVA significance tests with Tukey’s post-hoc multiple comparison test was used for group-wise analysis as indicated in the figure legends. Statistical significance is indicated as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Results
Recombinant GP and nanoparticle design
EBOV GP viral spike is displayed as a trimer of GP1/GP2 heterodimers embedded in the viral surface by a transmembrane domain (TM) of GP2 (Figure 1A). EBOV rGP used for ICMV formulations was modified to generate a soluble form by removal of the transmembrane domain (TM) from the C terminus of GP2 (Figure 1B). Additionally, a polyhistidine-tag (his-tag) was inserted at the C terminus of truncated GP2 for purification. The resulting GP1 and GP2 are bridged together by a disulfide bond, forming EBOV rGP monomer (~150 kDa MW). rGP monomers self-assemble into multimeric complexes as shown by non-denaturing and non-reducing Blue Native PAGE, a widely used assay for examining multi-protein complexes24 (Figure 2B). Since rGP has 5 disulfide bonds (including the disulfide bond holding GP1 and GP2 together), rGP loaded into the “standard” ICMV formulation could be conjugated to maleimide-functionalized lipids in ICMVs,20 resulting in uncontrolled display of antigen (Figure 1C). On the other hand, we hypothesized that the addition of NTA-functionalized lipids (in addition to the structural, maleimide-functionalized lipid) in NTA ICMVs could allow more concerted display of rGP (Figure 1C).
Figure 1. Antigen and nanoparticle design.
(A) Illustration of ZEBOV envelope glycoprotein (GP) displayed on the viral envelope as a native trimer (red brackets indicate the GP1/GP2 disulfide bond). (B) Illustration recombinant modifications to produce rGP (TM, transmembrane domain). (C) Diagram of expected rGP display on an ICMV or NTA ICMV (DTT, dithiothreitol).
Figure 2. Preservation of rGP incorporated into ICMVs.
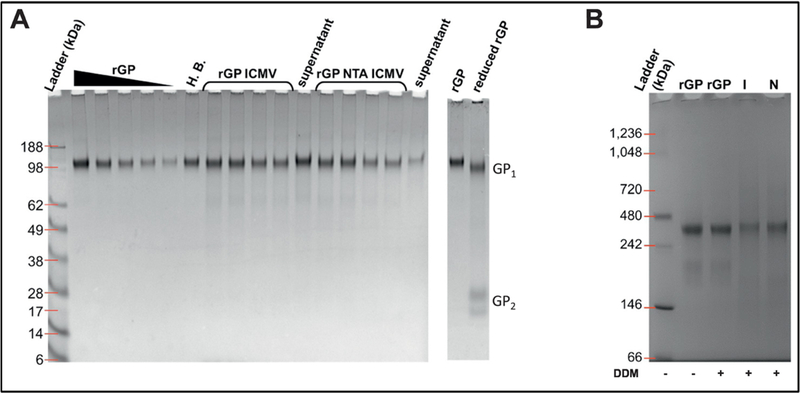
(A) Non-reducing PAGE of rGP formulations. ICMVs and NTA ICMVs were ran in two sets of duplicate volumes alongside their respective supernatants (unloaded rGP), the SOSIP containing hydration buffer (H.B.), and decreasing rGP standard. The two lanes at far right depict the migration of reduced rGP as bands of GP1 and GP2. (B) Blue Native PAGE of stock rGP, rGP ICMVs (I), and rGP NTA ICMVs (N). Samples that were incubated with DDM are indicated by a “+” sign.
Loading and preservation of rGP in ICMVs
ICMV synthesis may affect the tertiary and quaternary structure of rGP. We initially produced rGP ICMV formulations using the standard synthesis method with 2.46 mM of DTT used as a crosslinker of the opposing lipid layers within ICMVs, but this resulted in reduction of the disulfide bond holding the GP1/GP2 together (data not shown). We titrated down the concentration of DTT to 1.24 mM in order to remove excess thiols not required for interbilayer crosslinking. This led to preservation of the GP1/GP2 disulfide bond as demonstrated by NR-PAGE (Figure 2A). rGP formulated into either ICMVs or NTA ICMVs migrated at molecular weights similar to the original stock rGP without any fragmentation or reduction. In contrast, fully reduced rGP dissociated as separate GP1 and GP2 fragments (far right lane in Figure 2A).
We assessed whether the quaternary structure of rGP was maintained during the ICMV formulation by Blue Native PAGE. To analyze individual rGP complexes, we disrupted the ICMV formulations using a mild detergent, dodecyl β-D-maltoside (DDM) immediately before BN PAGE analysis. rGP from both traditional ICMVs or NTA-linked ICMVs appeared primarily around 400 kDa (Figure 2B). Interestingly, soluble rGP incubated with or without DDM had 400 kDa MW bands as well as prominent bands at lower MW, which were absent in the lanes for rGP ICMVs or rGP NTA ICMVs (Figure 2B). These results indicated that rGP was stably loaded in ICMVs and NTA ICMVs without any significant fragmentation or dissociation of the GP1/GP2 heterodimer. To determine whether rGP was displayed on the surfaces of ICMVs, we “tagged” surface-bound rGP by incubating ICMVs with membrane-impermeable PEG-NHS. NR-PAGE analysis showed significant increase in MW of rGP on ICMVs and NTA ICMVs after tagging with PEG-NHS (Supplementary Fig. 1), indicating that the majority of rGP was surface-displayed on both ICMV formulations.
We tested the effect of the initial protein loading amount and the addition of DOGS-NTA on the loading efficiency of the EBOV rGP in ICMV formulations. Batches of ICMVs and NTA ICMVs were produced using 20 µg or 40 µg rGP, and incorporation of antigen was determined by NR-PAGE (Figure 1, Table 1). While doubling the initial loading amount of rGP did not significantly alter the loading efficiencies (17.2 ± 8.4% and 15.0 ± 6.5% for ICMVs, and 26.1 ± 6.8% and 32.8 ± 5.1% for NTA ICMVs, p values > 0.05), the total amount of rGP incorporated into a batch of ICMVs and NTA ICMVs increased by ~1.8 and ~2.5 fold, respectively (Table 1). Addition of DOGS-NTA to ICMVs contributed to the loading efficiency and mass content; however, the increase was only statistically significant for particle produced with 40 µg rGP (p < 0.001). Based on the increased antigen content, ICMVs and NTA ICMVs produced with 40 µg rGP were selected for further in vitro characterization and in vivo immunogenicity studies.
Table 1. rGP loading in nanoparticles.
Loading efficiencies are reported as percent of initial rGP used for production. Measurements reported as mean ± standard deviation. Statistical analysis performed by two-way ANOVA, followed by Tukey’s multiple comparisons test. *p < 0.05, ** p < 0.01, *** p < 0.001.
| n | Loading Efficiency (%) | rGP per batch (μg) | |||
|---|---|---|---|---|---|
| 20µg rGP ICMV | 3 | 17.2 ± 8.4 | 3.4 ± 1.7 | ||
| 40µg rGP ICMV | 5 | 15.0 ± 6.5 | 6.0 ± 2.6 | ||
| 20µg rGP NTA ICMV | 4 | 26.1 ± 6.8 | 5.2 ± 1.4 | ||
| 40µg rGP NTA ICMV | 4 | 32.8 ± 5.1 | 13.1 ± 2.1 | ||
Measurements reported as sample mean ± standard deviation
Characterization of ICMVs
We utilized dynamic light scattering (DLS) to examine the size, zeta potential, and unilamellar vesicle (ULV) properties of traditional ICMVs or NTA ICMVs loaded with rGP. The average diameter of ULVs and NTA ULVs were 63.4 ± 6.9 nm and 57.9 ± 1.4 nm, respectively, which increased approximately by 55 nm after processing them into respective ICMVs (117.2 ± 10.1 nm and 117.5 ± 17.6 nm), consistent with previous reports19 (Figure 3A). Both ICMVs and NTA ICMVs exhibited homogenous particle sizes, as evidenced by average polydispersity indices of 0.17 ± 0.02 and 0.18 ± 0.01, respectively, along with negative zeta potentials (−22.3 ± 1.4 mV and −21.7 ± 1.3 mV, Figure 3A, B). Because DLS measures bulk samples and may generate data skewed towards larger particle sizes, we validated our results with individual particle-based analysis. The data indicated that the size distribution of ULVs and NTA ULVs as well as their respective ICMV formulations correlated well with the DLS results (Figure 3A, C).
Figure 3. Size distributions of rGP ULV and ICMV formulations.
(A) Table of intensity-based nanoparticle sizes, and polydispersity indices and zeta potentials from Zetasizer measurements, along with number-based nanoparticle diameters by nanoparticle tracking analysis. Measurements reported as mean ± standard deviation. (B) Intensity-based and (C) number-based size distributions of antigen-loaded ICMV formulations, compared with the respective ULVs.
Antigen conformation and display on ICMVs
Antigen-loaded ICMVs and NTA ICMVs were evaluated by a microplate-based indirect immunofluorescence staining assay20 to examine display and preservation of rGP epitopes on the surfaces of nanoparticles. First, we sought to measure the nanoparticle recovery during the immunofluorescence assay since this procedure requires multiple antibody incubation and washing steps. We added a trace amount of DiD, a lipophilic fluorophore, during the ICMV synthesis, and DiD-embedded, rGP-displaying ICMVs were then incubated with antibodies as detailed in the Methods section, followed by determining DiD fluorescence signal of the processed nanoparticles (Figure 4A). ICMVs that did not go through the antibody incubation and washing steps were used as “Unprocessed” samples to calculate the particle recovery. Processed ICMV and NTA ICMV samples exhibited an average retention of 44 ± 3% and 59 ± 2%, respectively, and similar retention rates were observed with or without the presence of antibodies.
Figure 4. Bulk analysis of rGP display on nanoparticle surfaces.
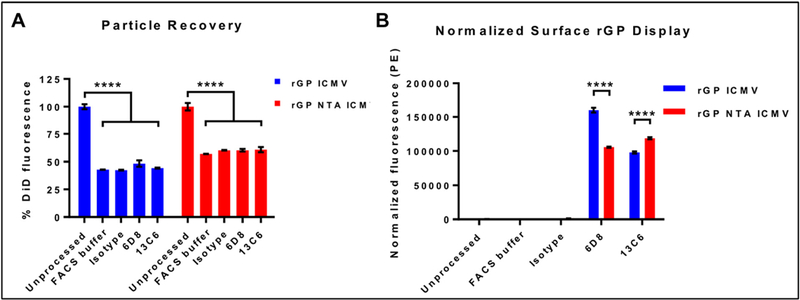
(A) Particle recovery after immunostaining process by retention of DiD signals in unprocessed and processed nanoparticles. (B) Normalized fluorescence signals of PE-labeled secondary antibodies bound to nanoparticles incubated with the indicated primary antibodies. Measurements reported as mean ± SEM. Statistical analysis performed by two-way ANOVA, followed by Tukey’s multiple comparisons test. ***p < 0.001.
We next evaluated ICMVs and NTA ICMVs for preservation and accessibility of epitopes by indirect measurement of bound EBOV GP-specific mouse monoclonal IgG antibodies (mAbs), 6D8 or 13C6. Specifically, 6D8 antibody binds to a linear epitope on GP1 393–401, while 13C6 recognizes a quaternary epitope within the GP1/GP2 glycan cap25,26. We measured the phycoerythrin (PE) fluorescence signal of secondary antibodies on ICMVs and normalized the value to the particle retention (as measured by the DiD recovery in Figure 4A). The PE signal associated with the linear antibody (6D8) was ~1.5-fold higher on ICMVs than NTA ICMVs, while that for the conformational antibody (13C6) was ~1.2-fold higher on NTA ICMVs than ICMVs (p < 0.0001, Figure 4B).
To further investigate display and preservation of rGP epitopes on a single particle level, we adapted a flow cytometry-based assay, called NanoFACS.27–31 Using this method, we were able to examine individual nanoparticles and quantify both DiD and PE fluorescence of each particle (Figure 5A). We first confirmed similar DiD content between individual particles that were processed with different antibodies (Figure 5B). Interestingly, while linear antibody binding was not significantly different between the two formulations, we observed ~30% higher binding of the conformational antibody on NTA ICMVs, compared with ICMVs (p < 0.01, Figure 5C).
Figure 5. Display of rGP on single nanoparticles via NanoFACS analysis.
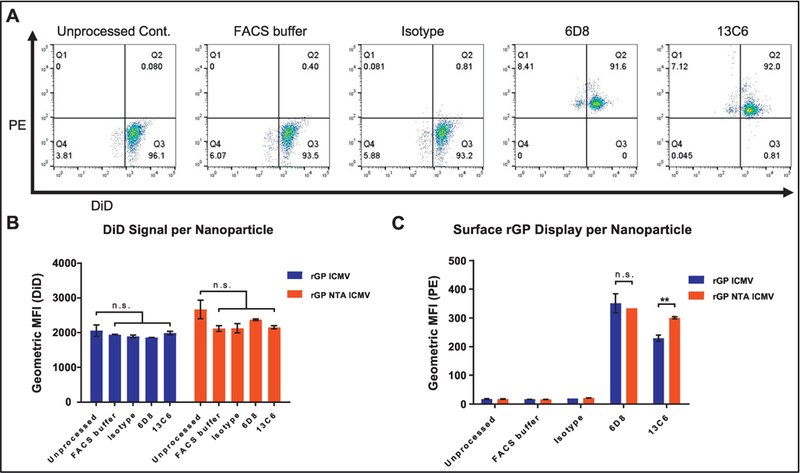
(A) Representative flow cytometry plots of PE and DiD signals for immunostained rGP ICMVs. (B) DiD fluorescence of individual particles. (C) Fluorescence signal of PE-labeled secondary antibodies bound to single particles. (B-C) Measurements reported as geometric mean ± SEM. Statistical analysis performed by two-way ANOVA, followed by Tukey’s multiple comparisons test. n.s. (not significant). **p < 0.01.
Neutralizing antibody responses elicited by ICMVs
We performed vaccination studies using EBOV rGP nanoparticle formulations incorporated with a potent adjuvant molecule, MPLA.20 The addition of MPLA improved rGP encapsulation in ICMV and NTA ICMVs (Supplemental Table 1). Mice were immunized subcutaneously on day 0 and day 21, and serum samples were collected on day 35. Analysis of immune sera with ELISA indicated that mice immunized with all vaccine formulations had low levels of anti-EBOV GP IgM on day 35 (Figure 6B). On the other hand, we observed ~5-log average anti-EBOV GP IgG end point titers for both nanoparticle groups and the soluble rGP group formulated with MPLA (rGP + MPLA) (Figure 6C). Analysis of the immune sera for anti-EBOV GP IgG1 and IgG2c subclasses demonstrated similar trends, with the addition of MPLA significantly boosting antibody responses (Figure 6D, E). In particular, mice vaccinated with rGP ICMV + MPLA had the highest anti-EBOV GP IgG1 and IgG2c titers, with 6.0-fold and 7.8-fold improvement, compared with the soluble rGP + MPLA control group (p < 0.05 and p < 0.01 for IgG1 and IgG2c titers, respectively, Figure 6D, E).
Figure 6. Analyses of immune sera after vaccination.
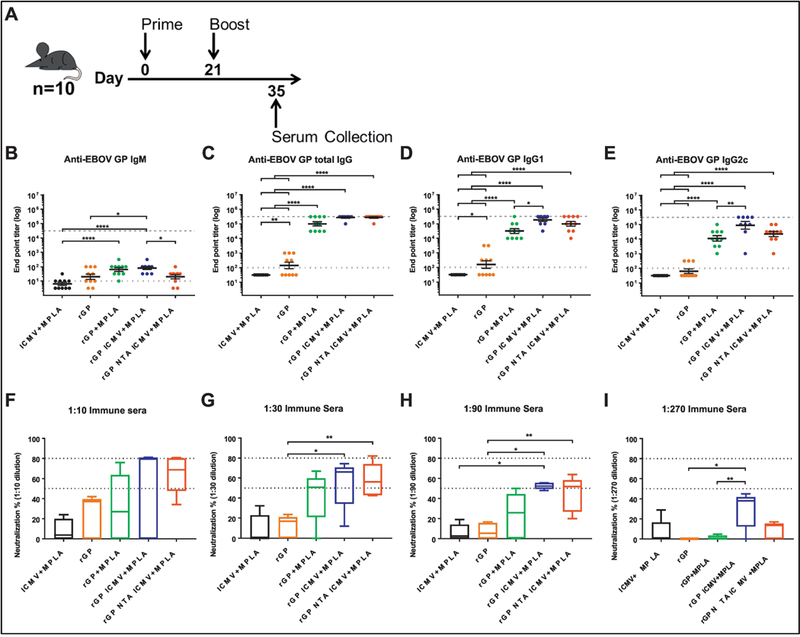
(A) Mice (n = 10/group) were vaccinated subcutaneously at the tail base with either blank ICMV + MPLA, rGP, rGP + MPLA, rGP ICMV + MPLA, or rGP NTA ICMV + MPLA on day 0 and 21. Antigen and adjuvant doses were 3 µg rGP and 2.5 µg MPLA, respectively, for both injections. Serum was collected two weeks after final vaccination. (B-E) EBOV GP-specific IgM, total IgG, IgG1, or IgG2c antibody responses were measured by ELISA. Dotted and dashed lines represent minimum and maximum dilutions tested, respectively. Measurements reported as geometric mean ± SEM. Non-seroconverted serum samples were assigned log-values of 0.5 (IgM) or 1.5 (IgG and subclasses) for graphical representation and statistical analysis. (F-I) Box and whisker plots of rVSV-GP neutralization by diluted serum from five randomly selected mice from each group. Percent neutralization was determined by residual infectivity of sample groups compared to naïve serum. (B-I) Statistical analysis performed by one-way ANOVA, followed by Tukey’s multiple comparisons test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Antibody effector function was evaluated by a neutralization assay with hybrid VSV co-expressing EBOV GP and eGFP reporter protein (rVSV-EBOV GP, as described in the Supplementary Methods). Median neutralization values ranging 50–80% were observed with 10, 30, and 90-fold diluted sera from both antigen-loaded ICMV formulations (Figure 6F–I). On the other hand, the median neutralization for the soluble rGP + MPLA group peaked at 51% at a 30-fold dilution and decreased to the basal level upon further dilution (Figure 6F–I). Notably, whereas the soluble rGP + MPLA vaccine group lost the neutralizing activity at 270-fold serum dilution, the rGP ICMV + MPLA group still exhibited 38% neutralization (p < 0.01, Figure 6I).
Activation of adaptive immune responses
To further study adaptive immune responses elicited by ICMVs, mice were vaccinated as above, and secondary lymph organs were harvested 10 days after the final vaccination (Figure 7A). The frequency of EBOV GP-specific splenic B cells was quantified by an IgG ELISpot assay (Figure 7B). Significantly higher frequencies of antigen-specific B-cells were observed for the nanoparticle groups, compared with the soluble antigen or vehicle control groups (Figure 7B). The frequency of total B cells and GC B cells in the draining lymph nodes were enumerated via flow cytometry analysis (Figure 7C). While total B cells numbers were similar between all groups (Figure 7D), the frequencies of GC B cells were significantly higher for both ICMV formulations, compared with the vehicle control or soluble antigen (p < 0.05, Figure 7E).
Figure 7. B cell activation and germinal center (GC) formation.
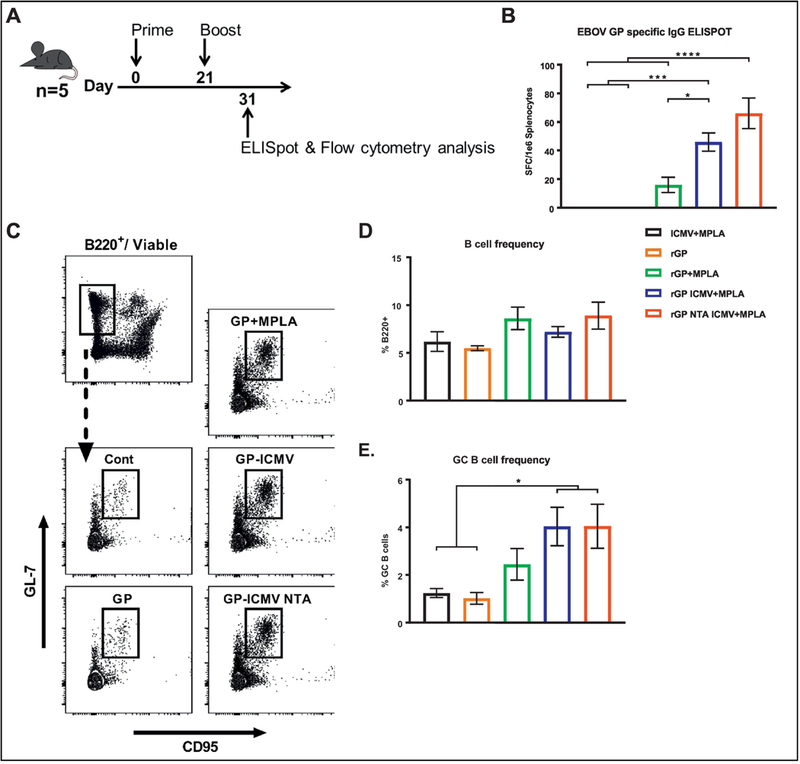
(A) Mice (n = 5/group) were vaccinated subcutaneously at the tail base with either blank ICMV + MPLA, rGP, rGP + MPLA, rGP ICMV + MPLA, or rGP NTA ICMV + MPLA on day 0 and 21. Antigen and adjuvant doses were 3 µg rGP and 2.5 µg MPLA, respectively, for both injections. Draining (inguinal) lymph nodes and spleens were harvested 10 days after the final vaccination for ELISpot, intracellular cytokine staining (ICS), and GC analyses. (B) EBOV GP antigen-specific B cell frequencies from harvested spleens were enumerated by ELISpot. (C) Representative flow cytometry gating of harvested B cells from draining lymph nodes of vaccinated mice. Total B cell frequencies were gated on viable/B220+ population, and GC B cells were additionally gated on GL-7+/CD95+ population. (D-E) Relative frequency of viable total B cells and GC B cells from draining lymph nodes. (B, D-E) Measurements reported as mean ± SEM. Statistical analysis performed by one-way ANOVA, followed by Tukey’s multiple comparisons test. *p < 0.05, ***p < 0.001, ****p < 0.0001.
We further evaluated the induction of EBOV-specific T cell responses. ELISpot performed on splenocytes re-stimulated with a GP-specific peptide revealed that the ICMV vaccine group generated a 5.8-fold higher frequency of IFN-γ producing splenic T-cells, compared with the soluble control group (p < 0.05, Figure 8A). In addition, there was a trend for an increased frequency of IFN-γ producing splenic T-cells for the NTA ICMV vaccine group, compared with the soluble control group (Figure 8A). We also evaluated production of IFN-γ, IL2, and TNF-α by splenic CD4+ and CD8+ T cells using intracellular cytokine staining (ICS) (Figure 8B). The ICMV vaccine group (without the NTA-lipid) induced the highest frequency of polyfunctional IFN-γ+/IL2+/TNF-α+ CD4+ T cells (Figure 8C) and also the highest frequency of CD4+ T cells expressing at least two of the Th1-associated IFN-γ, IL2, and TNF-α cytokines (Figure 8D). There was a trend for increased frequency of polyfunctional IFN-γ+/IL2+/TNF-α+ CD8+ T cells for both rGP nanoparticle groups (Figure 8E, F).
Figure 8. T cell responses in spleens of vaccinated mice.
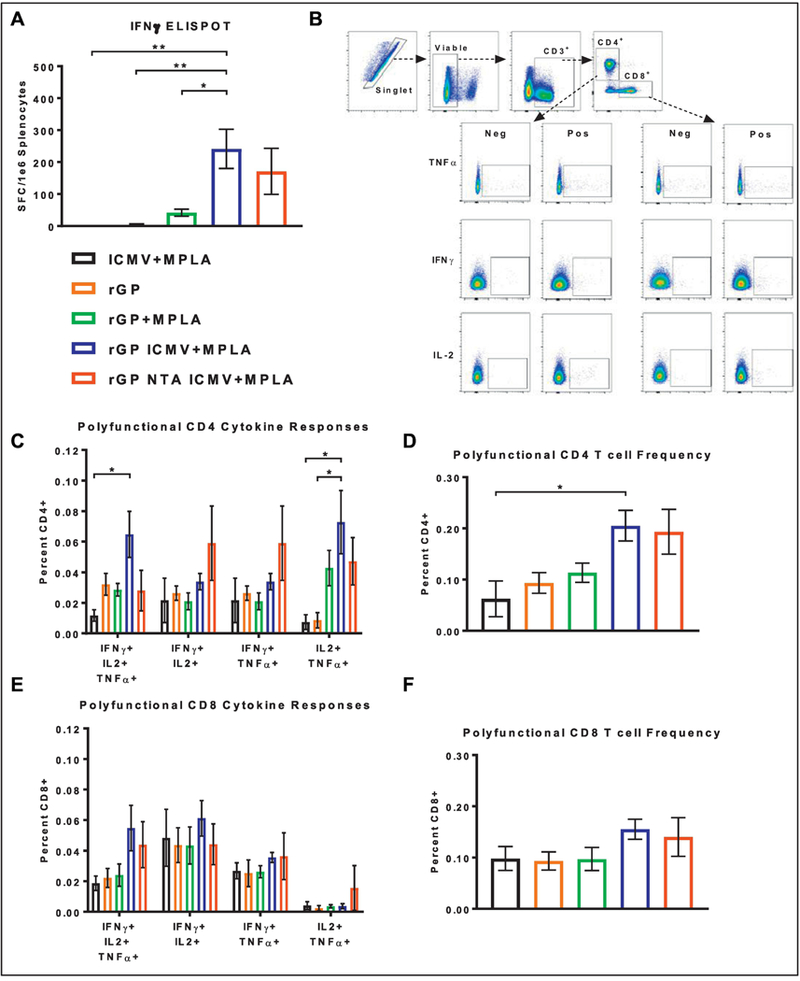
(A) IFN-γ-producing T cell frequencies from harvested spleens were enumerated by ELISpot. (B) Representative flow cytometry gating of ICS stained CD4+ and CD8+ T cells from vaccinated mice (Pos. columns) and naïve mice (Neg. columns). (C, E) Relative frequency of CD4+ and CD8+ polyfunctional T-cell subsets. (D, F) Relative frequency of combined CD4+ and CD8+ polyfunctional T-cell responses from spleens of vaccinated mice. Measurements reported as mean ± SEM. Statistical analysis performed by one-way (A, D, F) or two-way (C, E) ANOVA, followed by Tukey’s multiple comparisons test. *p < 0.05, **p < 0.01.
Discussion
Our objective in this study was to assess immunogenicity of synthetic nanoparticles displaying a recombinant EBOV GP antigen in a configurational manner. Optimization of the synthesis process preserved the inter-GP1/GP2 disulfide bond and the quaternary structure of rGP (Figure 2). Furthermore, two ICMV formulations loaded with rGP were characterized using antigen-specific antibodies and shown to present rGP in a configurational manner with slight differences in epitope presentation. When administered in mice, both ICMV formulations induced robust anti-EBOV GP-specific humoral responses. In particular, the rGP-ICMV + MPLA vaccine group generated immune sera with the highest neutralizing-capacity, robust GC B cell formation, and polyfunctional T cell responses.
Initial characterization of ICMV formulations revealed that the addition of NTA-lipid to ICMVs increased the loading efficiency of rGP via the selective interaction with the polyhistidine tag of rGP (Table 1), consistent with a previous report.32 Additionally, we evaluated rGP epitopes by immunofluorescence staining and observed reverse trends for linear and conformational EBOV GP-specific antibodies to either formulation (Figures 4, 5). Notably, while NTA ICMVs loaded ~2-fold more rGP than ICMVs, this trend did not translate to 6D8 or 13C6 binding. These observations may be explained in part by the high rGP density and concerted orientation on NTA ICMVs. Prior research has shown that 6D8 binds to the flexible mucin-like domain on the periphery of EBOV GP, while 13C6 binds to the glycan cap domain at an angle in line with the GP vertical axis,33,34 suggesting that the 6D8 epitope would be more susceptible to occlusion by neighboring rGP complexes whereas the 13C6 epitope would be less affected. However, while 13C6 epitopes are available for binding, the overall rGP density may lead to steric hindrance, limiting binding of all accessible epitopes.35 Interestingly, we observed that after a prime-boost vaccination, mice generated potent isotype-switched antibodies against rGP for both nanoparticle groups as well as the rGP + MPLA control (Figure 6B–E). However, higher levels of antigen-specific IgG1 and IgG2c responses were observed in ICMV vaccine group. When vaccination serum was tested for neutralization, immune sera from the ICMV group had the greatest neutralization capacity across the most dilutions, compared with those from the soluble control or the NTA ICMV group (Figure 6F–I).
Further interrogation of the immune responses demonstrated that only the ICMV vaccine group (without NTA-lipid) generated statistically significant increases in both B cell and polyfunctional T cell responses, compared with the control groups (Figures 7, 8). This was a surprise as we expected the increased antigen density and concerted display of rGP on NTA ICMVs would enhance B cell and T cell responses. The unexpected lower performance of the NTA ICMV group may be attributed to (1) the high surface antigen density, (2) nanoparticle aggregation, and/or (3) instability of NTA-polyhistidine tag interactions in vivo. (1) As shown in Table 1, NTA ICMVs were loaded with approximately two-fold more rGP than ICMVs; however, we did not observe a two-fold increase in antibody binding to rGP NTA ICMVs during immunostaining (Figures 4, 5), indicating potential steric hindrance due to high antigen density on NTA ICMVs or masked epitopes.35 (2) We also observed that NTA ICMVs tended to aggregate more than ICMVs in vitro. While the aggregates could be disrupted by bath sonication, this could adversely affect both particle drainage to lymph nodes and proper epitope display to B-cell receptors in vivo. (3) Lastly, it is possible that NTA-polyhistidine tag was not strong enough to withstand the in vivo condition, leading to loss of antigens, as recently demonstrated with liposome formulations.36 While our proof-of-concept results presented here are encouraging, further analysis and optimization are required to address these remaining questions.
In conclusion, we have shown that rGP can be incorporated into ICMVs without adversely affecting the quaternary structure or key conformational epitopes. Two ICMV variants (standard and NTA containing) also provided the means to control rGP loading and potentially surface orientation. Administration of rGP-loaded nanoparticles displaying rGP in a multivalent, configurational manner induced GC B cells, neutralizing antibody responses, and polyfunctional T cell responses in mice. Analysis of long-term immune responses will be a main focus of future studies. Additionally, efforts are underway to optimize nanoparticle formulations to generate Pan-filovirus (e.g. EBOV, Sudan and Marburg) ICMV vaccines.
Supplementary Material
Acknowledgements and financial conflict of interest
This work was supported in part by NIH (R01AI127070, R01EB022563, and R01HL125555) and the Defense Threat Reduction Agency (DTRACB3947, C.L.C) J.J.M. is a Young Investigator supported by the Melanoma Research Alliance (348774), DoD/CDMRP Peer Reviewed Cancer Research Program (W81XWH-16-1-0369), Emerald Foundation, and NSF CAREER Award (1553831). Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the US Department of the Army or the US Department of Defense. J.J.M. is an inventor on patents related to the ICMV technology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.WHO. Ebola virus disease, <http://www.who.int/en/news-room/fact-sheets/detail/ebola-virus-disease> (2018).
- 2.Hood CL, Abraham J, Boyington JC, Leung K, Kwong PD, Nabel GJ. Biochemical and structural characterization of cathepsin L-processed Ebola virus glycoprotein: implications for viral entry and immunogenicity. J Virol 2010;84:2972–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 2008;454:177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JE, Saphire EO. Ebolavirus glycoprotein structure and mechanism of entry. Future Virol 2009;4:621–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agnandji ST, Huttner A, Zinser ME, Njuguna P, Dahlke C, Fernandes JF et al. Phase 1 Trials of rVSV Ebola Vaccine in Africa and Europe. N Engl J Med 2016;374:1647–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coller BG, Blue J, Das R, Dubey S, Finelli L, Gupta S et al. Clinical development of a recombinant Ebola vaccine in the midst of an unprecedented epidemic. Vaccine 2017;35:4465–4469. [DOI] [PubMed] [Google Scholar]
- 7.Regules JA, Beigel JH, Paolino KM, Voell J, Castellano AR, Hu Z et al. A Recombinant Vesicular Stomatitis Virus Ebola Vaccine. N Engl J Med 2017;376:330–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stanley DA, Honko AN, Asiedu C, Trefry JC, Lau-Kilby AW, Johnson JC et al. Chimpanzee adenovirus vaccine generates acute and durable protective immunity against ebolavirus challenge. Nat Med 2014;20:1126–1129. [DOI] [PubMed] [Google Scholar]
- 9.Geisbert TW, Bailey M, Hensley L, Asiedu C, Geisbert J, Stanley D et al. Recombinant adenovirus serotype 26 (Ad26) and Ad35 vaccine vectors bypass immunity to Ad5 and protect nonhuman primates against ebolavirus challenge. J Virol 2011;85:4222–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji Y, Lu Y, Yan Y, Liu X, Su N, Zhang C et al. Design of Fusion Proteins for Efficient and Soluble Production of Immunogenic Ebola Virus Glycoprotein in Escherichia coli. Biotechnol J 2018:e1700627. [DOI] [PubMed]
- 11.Konduru K, Shurtleff AC, Bradfute SB, Nakamura S, Bavari S, Kaplan G. Ebolavirus Glycoprotein Fc Fusion Protein Protects Guinea Pigs against Lethal Challenge. PLoS One 2016;11:e0162446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehrer AT, Wong TS, Lieberman MM, Humphreys T, Clements DE, Bakken RR et al. Recombinant proteins of Zaire ebolavirus induce potent humoral and cellular immune responses and protect against live virus infection in mice. Vaccine 2018;36:3090–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rios-Huerta R, Monreal-Escalante E, Govea-Alonso DO, Angulo C, Rosales-Mendoza S. Expression of an immunogenic LTB-based chimeric protein targeting Zaire ebolavirus epitopes from GP1 in plant cells. Plant Cell Rep 2017;36:355–365. [DOI] [PubMed] [Google Scholar]
- 14.Rao M, Matyas GR, Grieder F, Anderson K, Jahrling PB, Alving CR. Cytotoxic T lymphocytes to Ebola Zaire virus are induced in mice by immunization with liposomes containing lipid A. Vaccine 1999;17:2991–2998. [DOI] [PubMed] [Google Scholar]
- 15.Rao M, Bray M, Alving CR, Jahrling P, Matyas GR. Induction of Immune Responses in Mice and Monkeys to Ebola Virus after Immunization with Liposome-Encapsulated Irradiated Ebola Virus: Protection in Mice Requires CD4+ T Cells. Journal of Virology 2002;76:9176–9185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bengtsson KL, Song H, Stertman L, Liu Y, Flyer DC, Massare MJ et al. Matrix-M adjuvant enhances antibody, cellular and protective immune responses of a Zaire Ebola/Makona virus glycoprotein (GP) nanoparticle vaccine in mice. Vaccine 2016;34:1927–1935. [DOI] [PubMed] [Google Scholar]
- 17.Fan Y, Moon JJ. Particulate delivery systems for vaccination against bioterrorism agents and emerging infectious pathogens. Wiley Interdiscip Rev Nanomed Nanobiotechnol 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chahal JS, Khan OF, Cooper CL, McPartlan JS, Tsosie JK, Tilley LD et al. Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and Toxoplasma gondii challenges with a single dose. Proc Natl Acad Sci U S A 2016;113:E4133–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moon JJ, Suh H, Bershteyn A, Stephan MT, Liu H, Huang B et al. Interbilayer-crosslinked multilamellar vesicles as synthetic vaccines for potent humoral and cellular immune responses. Nat Mater 2011;10:243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon JJ, Suh H, Li AV, Ockenhouse CF, Yadava A, Irvine DJ. Enhancing humoral responses to a malaria antigen with nanoparticle vaccines that expand Tfh cells and promote germinal center induction. Proc Natl Acad Sci U S A 2012;109:1080–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li AV, Moon JJ, Abraham W, Suh H, Elkhader J, Seidman MA et al. Generation of effector memory T cell-based mucosal and systemic immunity with pulmonary nanoparticle vaccination. Sci Transl Med 2013;5:204ra130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pejawar-Gaddy S, Kovacs JM, Barouch DH, Chen B, Irvine DJ. Design of lipid nanocapsule delivery vehicles for multivalent display of recombinant Env trimers in HIV vaccination. Bioconjug Chem 2014;25:1470–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cazares LH, Ward MD, Brueggemann EE, Kenny T, Demond P, Mahone CR et al. Development of a liquid chromatography high resolution mass spectrometry method for the quantitation of viral envelope glycoprotein in Ebola virus-like particle vaccine preparations. Clin Proteomics 2016;13:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wittig I, Braun HP, Schagger H. Blue native PAGE. Nature protocols 2006;1:418–428. [DOI] [PubMed] [Google Scholar]
- 25.Shedlock DJ, Bailey MA, Popernack PM, Cunningham JM, Burton DR, Sullivan NJ. Antibody-mediated neutralization of Ebola virus can occur by two distinct mechanisms. Virology 2010;401:228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson JA, Hevey M, Bakken R, Guest S, Bray M, Schmaljohn AL et al. Epitopes involved in antibody-mediated protection from Ebola virus. Science 2000;287:1664–1666. [DOI] [PubMed] [Google Scholar]
- 27.Nolte-’t Hoen EN, van der Vlist EJ, Aalberts M, Mertens HC, Bosch BJ, Bartelink W et al. Quantitative and qualitative flow cytometric analysis of nanosized cell-derived membrane vesicles. Nanomedicine 2012;8:712–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Vlist EJ, Nolte-’t Hoen EN, Stoorvogel W, Arkesteijn GJ, Wauben MH. Fluorescent labeling of nano-sized vesicles released by cells and subsequent quantitative and qualitative analysis by high-resolution flow cytometry. Nature protocols 2012;7:1311–1326. [DOI] [PubMed] [Google Scholar]
- 29.Danielson KM, Estanislau J, Tigges J, Toxavidis V, Camacho V, Felton EJ et al. Diurnal Variations of Circulating Extracellular Vesicles Measured by Nano Flow Cytometry. PLoS One 2016;11:e0144678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pasalic L, Williams R, Siupa A, Campbell H, Henderson MJ, Chen VMY. Enumeration of extracellular vesicles by a new improved flow cytometric method is comparable to fluorescence mode nanoparticle tracking analysis. Nanomedicine 2016;12:977–986. [DOI] [PubMed] [Google Scholar]
- 31.van der Pol E, Coumans FA, Grootemaat AE, Gardiner C, Sargent IL, Harrison P et al. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J Thromb Haemost 2014;12:1182–1192. [DOI] [PubMed] [Google Scholar]
- 32.Ingale J, Stano A, Guenaga J, Sharma SK, Nemazee D, Zwick MB et al. High-Density Array of Well-Ordered HIV-1 Spikes on Synthetic Liposomal Nanoparticles Efficiently Activate B Cells. Cell reports 2016;15:1986–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davidson E, Bryan C, Fong RH, Barnes T, Pfaff JM, Mabila M et al. Mechanism of Binding to Ebola Virus Glycoprotein by the ZMapp, ZMAb, and MB-003 Cocktail Antibodies. J Virol 2015;89:10982–10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murin CD, Fusco ML, Bornholdt ZA, Qiu X, Olinger GG, Zeitlin L et al. Structures of protective antibodies reveal sites of vulnerability on Ebola virus. Proc Natl Acad Sci U S A 2014;111:17182–17187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng W The Density Code for the Development of a Vaccine? J Pharm Sci 2016;105:3223–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watson DS, Platt VM, Cao L, Venditto VJ, Szoka FC Jr. Antibody response to polyhistidine-tagged peptide and protein antigens attached to liposomes via lipid-linked nitrilotriacetic acid in mice. Clin Vaccine Immunol 2011;18:289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


