Abstract
The body’s microbiome represents an actively regulated network of novel mechanisms that potentially underlie the etiology and pathophysiology of a wide range of diseases. For complex brain disorders such as schizophrenia, understanding the cellular and molecular pathways that intersect the bidirectional gut-brain axis is anticipated to lead to new methods of treatment. The means by which the microbiome might differ across neuropsychiatric and neurological disorders are not known. Brain disorders as diverse as schizophrenia, major depression, Parkinson’s disease and multiple sclerosis appear to share a common pathology of an imbalanced community of commensal microbiota, often measured in terms of a leaky gut phenotype accompanied by low level systemic inflammation. While environmental factors associated with these disease states might contribute to intestinal pathologies, products from a perturbed microbiome may also directly promote specific signs, symptoms and etiologies of individual disorders. We hypothesize that in schizophrenia, it is the putatively unique susceptibility related to genes that modulate the immune system and the gut-brain pleiotropy of these genes which leads to a particularly neuropathological response when challenged by a microbiome in dysbiosis. Consequences from exposure to this dysbiosis may occur during pre- or post-natal time periods and thus may interfere with normal neurodevelopment in those who are genetically predisposed. Here, we review the evidence from the literature which supports the idea that the intersection of the microbiome and immune gene susceptibility in schizophrenia is relevant etiologically and for disease progression. Figuring prominently at both ends of the gut-brain axis and at points in between are proteins encoded by genes found in the major histocompatibility complex (MHC), including select MHC as well as non-MHC complement pathway genes.
Keywords: gut-brain axis, neurodevelopment, synapses, immune system, enteric nervous system, complement C4, gene-environmental interactions
Introduction
Schizophrenia is a severe psychiatric disorder that is characterized by a series of complex and debilitating symptoms, such as delusions, hallucinations, anhedonia, social withdrawal and disorganized thought processes (APA, 2013). The neurobiological mechanisms orchestrating the causes and pathophysiology of this disorder are not known. There are many implicated pathways and causality is likely multifactorial. Schizophrenia has a very evident familial component, but twin studies demonstrate that this heritability is not fully penetrant (Cannon et al., 2000; Kato et al., 2005; Tsujita et al., 1998). Current hypotheses regarding etiology focus on genetic and environmental factors that may interact during sensitive neurodevelopmental time points (Demjaha et al., 2012; European Network of National Networks studying Gene-Environment Interactions in et al., 2014; Kavanagh et al., 2015; Modinos et al., 2013; Nimgaonkar et al., 2017; Schizophrenia Working Group of the Psychiatric Genomics, 2014; Tsuang, 2000; Ursini et al., 2018). Toward this end, the immune system has become a highly-interrogated candidate for studies of schizophrenia susceptibility. This immune focus derives from evidence of environmental immune challenges as risk factors for the disease and on associations of the disorder with the immune gene-rich 6p21–6p22 chromosomal regions (International Schizophrenia et al., 2009; Jones et al., 2005; Kirch, 1993; Knuesel et al., 2014; Meyer, 2013; Muller, 2014; Rothermundt et al., 2001; Shi et al., 2009; Stefansson et al., 2009; Torrey and Peterson, 1976; Yolken and Torrey, 2008). Supporting an immune dysfunction hypothesis for schizophrenia are findings that immune proteins encoded in the 6p21–6p22 region also work in various central nervous system (CNS) capacities including the formation and maintenance of properly functioning synapses (Boulanger, 2009).
An important regulator of immune system homeostasis is the gastrointestinal (GI) mucosa and the large population of resident microbes that help to establish and coordinate the body’s immune tolerance and defense. This process may unfold insufficiently if there is a predisposing polymorphism or mutation in a gene that modifies immunological functions (i.e. an immune gene) and/or if exposures to environmental factors such as diet, infection, stress, toxins, and medications disrupt the normal functioning and balance of the gut microbiome (Abegunde et al., 2016; Sandhya et al., 2016; Wu et al., 2011). At risk is the ability of the host to launch an appropriate immune response against pathogens and the ability to accurately recognize self vs non-self cellular and molecular entities (Chistiakov et al., 2014; Dinan and Cryan, 2015; Sandhya et al., 2016; Wekerle, 2017). The timing of the environmental trigger is a particularly important consideration given the dynamism of gene activities during neurodevelopment. An environmental exposure during pregnancy or adolescence in a genetically susceptible individual could result in an inadequately seeded microbiome that has lifelong health effects.
Experimental findings from mechanistic studies in rodent models demonstrate that direct CNS effects are tangible consequences of a gut microbiome that is perturbed during early neurodevelopment or in adulthood (Codagnone et al., 2018; Dinan et al., 2014) (Collins et al., 2012; Diaz Heijtz et al., 2011; Erny et al., 2015; Foster and McVey Neufeld, 2013; Hsiao et al., 2013; Luczynski et al., 2016; Sampson and Mazmanian, 2015; Stilling et al., 2014). Translational support from clinical populations is more difficult to obtain, especially data that might reflect endophenotypes characteristic of schizophrenia and that are not the product of medication. In this paper, we examine the evidence that a gut microbiome in dysbiosis and the dysregulation of immune genes known to be associated with schizophrenia are integrally linked. We review the neurobiology of select immune genes implicated from genetic susceptibility studies of schizophrenia and evaluate gene duplicity of function peripherally and in the CNS in the context of a gut-immune-brain interactome. To the extent possible in this still burgeoning field, we address microbial interactions with these gene products at both the gut and CNS sites.
Overview of immune gene dysregulation in schizophrenia
In individuals with schizophrenia, genome-wide association studies (GWAS) revealed a significant disease connection driven by a structurally complex region of chromosome 6 (Corvin and Morris, 2014; International Schizophrenia et al., 2009; Kodavali et al., 2014; Sekar et al., 2016; Shi et al., 2009; Stefansson et al., 2009). This large stretch contains over 400 genes that encompass the major histocompatibility complex (MHC), the proteins of which are encoded by the human leukocyte antigen (HLA) genes (Horton et al., 2004). Complement C4 loci, the most recent genes demonstrating strong biological and functional associations with schizophrenia, are also located in this region (Sekar et al., 2016). Among brain pathologies evident in schizophrenia are deficient densities of dendritic spines in the prefrontal cortex and hippocampus, and this neuropathology is thought to be a product of aberrantly accelerated synaptic pruning during adolescence (Bennett et al., 2013; Clarke et al., 2018). Although the functions of MHC and complement pathway proteins are most well understood in the peripheral immune system, both actively participate in homologous processes in the brain including synapse formation, tagging, and removal (Boulanger, 2009; Fourgeaud and Boulanger, 2007; Glynn et al., 2011; Goddard et al., 2007; Huh et al., 2000; McAllister, 2014; Presumey et al., 2017; Shatz, 2009; Stephan et al., 2013; Stevens et al., 2007).
The Major Histocompatibility Complex in schizophrenia and in the gut
The MHC proteins are found on cell surfaces where they bind and deliver self and non-self antigens to appropriate T-cells. Class I MHC molecules are recognized by cytotoxic CD8(+) T-cells, and class II MHC are recognized by CD4(+) helper T-cells. Class III MHC encode cytokines, heat shock proteins and several complement components including C4 (Murphy et al., 2012). While the primarily described function of MHCI class proteins, in particular, has been to process and present antigens in systemic circulation in support of establishing and effecting immunity, rodent studies show that these proteins are also involved in such CNS activities as neurogenesis, neuronal migration and synaptic pruning and plasticity (Glynn et al., 2011; Goddard et al., 2007; Huh et al., 2000; McAllister, 2014; Shatz, 2009). As indicated earlier, these neurobiological processes are highly relevant to schizophrenia and its neurodevelopmental origins (Balu and Coyle, 2011; Mei and Xiong, 2008; Stephan et al., 2006). In rodents, MHCI expression in the CNS is dynamic with elevated levels early during development and which generally taper off upon adulthood (Chacon and Boulanger, 2013; Needleman et al., 2010). During aging, expression levels increase again, apparently as a function of glial cell expression, whereas expression earlier in the lifespan appears to be predominantly neuronal. MHCI may also be expressed by astrocytes when the brain’s immune system is activated (Mokhtari and Lachman, 2016; Sobue et al., 2018).
If antigens are mislabeled when immunity is being primed, the host may become susceptible to infectious disease, cancer and autoimmunity. Susceptibility to these conditions may be driven or otherwise compounded by a genetic predisposition encoded by the HLA genes. A number of common infections – Toxoplasma gondii, cytomegalovirus, Herpes Simplex Virus Type I - are thought to persist in schizophrenia as evident from persistent elevations of antibody levels in individuals with the disorder compared to controls (Hornig, 2013; Yolken and Torrey, 2008). Studies that incorporate HLA typing found that certain of these polymorphisms were significantly associated with specific neurotropic infections and that this HLA-based susceptibility was altered in schizophrenia (Avramopoulos et al., 2015; Parks et al., 2018). These results support and extend findings from earlier studies that also detected associations between HLA polymorphisms and exposure to neurotropic viruses in schizophrenia (Bamne et al., 2012; Kim et al., 2007; McAllister, 2014; Shirts et al., 2008). As such, the extent to which a pathogenic agent or other condition can activate the immune response, including an imbalanced commensal microbial composition, may be governed by HLA susceptibilities.
A study in mice which evaluated possible mechanisms by which MHC genes might influence the gut microbiome found that specific genotypes modulated antibody responses against gut commensals, thus providing protection against or rendering susceptibility to enteric infection (Kubinak et al., 2015). Furthermore, it is this immune response directed toward gut bacteria that is thought to perpetrate inflammatory bowel diseases, in part, through altered MHCII activation of CD4(+) T cells (Thelemann et al., 2014). In humans, HLA haplotypes are definitively associated with the autoimmune intestinal disorder, celiac disease (Cukrowska et al., 2017; Karhus et al., 2018; Leonard et al., 2017), and with some cases of inflammatory bowel diseases such as Crohn’s disease and ulcerative colitis (Gomes et al., 2018; Jung et al., 2016; Lee et al., 2018; Palmieri et al., 2017; Saito et al., 2018). Interestingly, these bowel disorders in turn are overrepresented in individuals with schizophrenia (Fadgyas-Stanculete et al., 2014; Filipovic and Filipovic, 2014; Gupta et al., 1997; Vaknin et al., 2004; Vu et al., 2014).
The complement system in schizophrenia and in the gut
The complement system is composed of three pathways (classical, lectin and alternative) that function to amplify and direct the innate immune response in order to protect the host from potentially dangerous antigens such as infectious pathogens, antigenic foods and cellular debris. Complement components also work with the adaptive immune system to respond to foreign proteins and form immune complexes with antibody-bound antigens. The complement system antigen clearing process culminates with the activation and recruitment of phagocytes to aid removal of antigenic culprits by the membrane attack complex (Murphy et al., 2012).
As observed for MHCI proteins, components of the classical complement pathway including C1q, C3, C4, are involved with synapse activities during neurodevelopment and to a less well characterized extent during adulthood. This brain complement system encodes components that participate in an immune signaling cascade and results in the formation and directed pruning of synapses during development, in early neurodegenerative diseases and in response to inflammation and exposure to brain pathogens in adults. (Hong et al., 2016; Lui et al., 2016; Vasek et al., 2016) (Bialas and Stevens, 2013; Boulanger, 2009; Fourgeaud and Boulanger, 2007; Nimgaonkar et al., 2017; Presumey et al., 2017; Ransohoff and Stevens, 2011; Schafer et al., 2012; Stephan et al., 2013; Stevens et al., 2007). In experimental models of neurodevelopment, C1q and C3 are highly expressed in rodent cortex at postnatal day 5 with a reduction to baseline amounts at postnatal day 30 (Stevens et al., 2007). While C1q and C3 are generally not detectable in the normal adult mouse brain, there is evidence for CNS expression of complement components following injury, immune challenge and aging (Bellander et al., 2001; Mocco et al., 2006; Stephan et al., 2013; Xiao et al., 2016).
Research of the complement pathways in schizophrenia received a significant boost by the findings of Sekar et al (2016), who showed that the number of copies of complement C4 genes dictated the amount of protein that was produced, with greater levels associated with a greater risk of schizophrenia (Sekar et al., 2016). In a clinical study that incorporated brain imaging, C4 copy number was positively correlated with neuropil contraction, an indicator of increased pruning, or decreased formation of synapses, in a number of different brain regions including the prefrontal cortex (Prasad et al., 2018). Similarly, predicted C4 RNA expression based on copy number was associated with reduced cortical activation and memory deficits in psychosis patients compared to healthy controls (Donohoe et al., 2018). Other preliminary clinical findings suggest that the expression of complement genes C5 and SERPING1 was associated with cortical thickness, another indicator of possible overpruning in schizophrenia (Allswede et al., 2018). The role of the complement system in schizophrenia and its relationship to infection-based hypotheses is further reviewed in more detail elsewhere (Nimgaonkar et al., 2017; Presumey et al., 2017).
People with inflammatory bowel diseases often have psychiatric comorbidities (Bernstein et al., 2018). The complement pathway is involved in inflammatory bowel disease pathogenesis; thus, there is precedence that complement activation peripherally and perhaps in response to gut dysbiosis and inflammation could trigger gut-brain axis pathways. In pediatric inflammatory bowel disease, C4 copy number was positively correlated with inflammatory indices and decreased microbial diversity (Nissila et al., 2017). In another study of pediatric inflammatory bowel disease, the combination of both HLA and complement C4 copy number typing identified a haplotype that was strongly linked to disease and not to controls (Kolho et al., 2016). C4 copy number was further implicated as a susceptibility target for Crohn’s disease, indicating copy number elevations may amplify the response to infections and the development of autoimmune diseases in general (Cleynen et al., 2016). In individuals with Crohn’s disease, C3 mRNA was expressed in epithelial cells located at crypts of diseased tissue, whereas C4 was expressed throughout the intestinal epithelium in both normal and diseased tissue (Laufer et al., 2000). Thus, the role of complement is complex with some studies reporting both protective and pathogenic effects (Elvington et al., 2015; Schepp-Berglind et al., 2012). In a comparison of mice lacking the C1q/mannose binding lectin genes to those deficient in the C3 gene, the induction of colitis by dextran sulfate sodium was only successful in the C1q/MBL double deficient mice, although ultimately all complement deficient mice died. Interestingly, antibiotic treatment prevented this lethal effect suggesting a role for the gut microbiome in complement regulation (Schepp-Berglind et al., 2012). Yadav et al (2017) recently demonstrated in an animal model expressing a specific multiple sclerosis-associated HLA and T-cell receptor haplotype that gut dysbiosis triggered experimental autoimmune encephalomyelitis in adolescence and young adulthood but not afterwards. This dysbiosis induced the expression of complement C3 locally in the GI tract as well as systemically (Yadav et al., 2017). Finally, in gastric biopsies of children with autism, Crohn’s disease, Helicobacter pylori infection and normal controls, distinct gastritis typified each disease state compared to controls. In autism, however, 20/25 children were also uniquely distinguished by a CD8-dominated gastritis and colocalization of IgG and C1q on the subepithelial basement membrane (Torrente et al., 2004). This study is particularly intriguing given suggestions that autism and schizophrenia may be linked on a spectrum of shared genes and/or environmental interactions (Cattane et al., 2018). Determination of the disease phenotype as autism or schizophrenia may simply be variation of the presence and timing of environmental modifications on a similar genetic template.
At the crossroads of intestinal pathologies, immune genes and schizophrenia
Chronic low-grade inflammation is increasingly observed in schizophrenia and it may typify a phenotype that is associated with especially poor outcome over the course of the disorder (Bechter, 2013; Bulzacka et al., 2016; Fond et al., 2018a; Fond et al., 2018b). As described above, immune proteins that are genetically associated with schizophrenia are operative in inflammatory processes that occur systemically as well as in the gut. Demonstrating an interactive and definitive connection between GI inflammation and immune gene susceptibility specifically in schizophrenia, however, is difficult and to date can only be inferred from associative data. Reports of significant gastrointestinal inflammation associated with the disease state, and not the result of a medication effect, support a GI phenotype for schizophrenia (Severance et al., 2012a; Severance et al., 2015). The microbiome and its dysbiosis is specifically implicated in schizophrenia based on case-control analyses of markers of microbial translocation and changes in microbial taxa associated with the disorder (Castro-Nallar et al., 2015; He et al., 2018; Lv et al., 2017; Schwarz et al., 2018; Severance et al., 2016a; Severance et al., 2013; Shen et al., 2018; Yolken et al., 2015; Yuan et al., 2018). One study extends this association to the complement system where increased rates of circulating immune complexes containing complement C1q bound to food antigen antibodies, a source of GI inflammation, were found in individuals with schizophrenia compared to controls (Severance et al., 2012b).
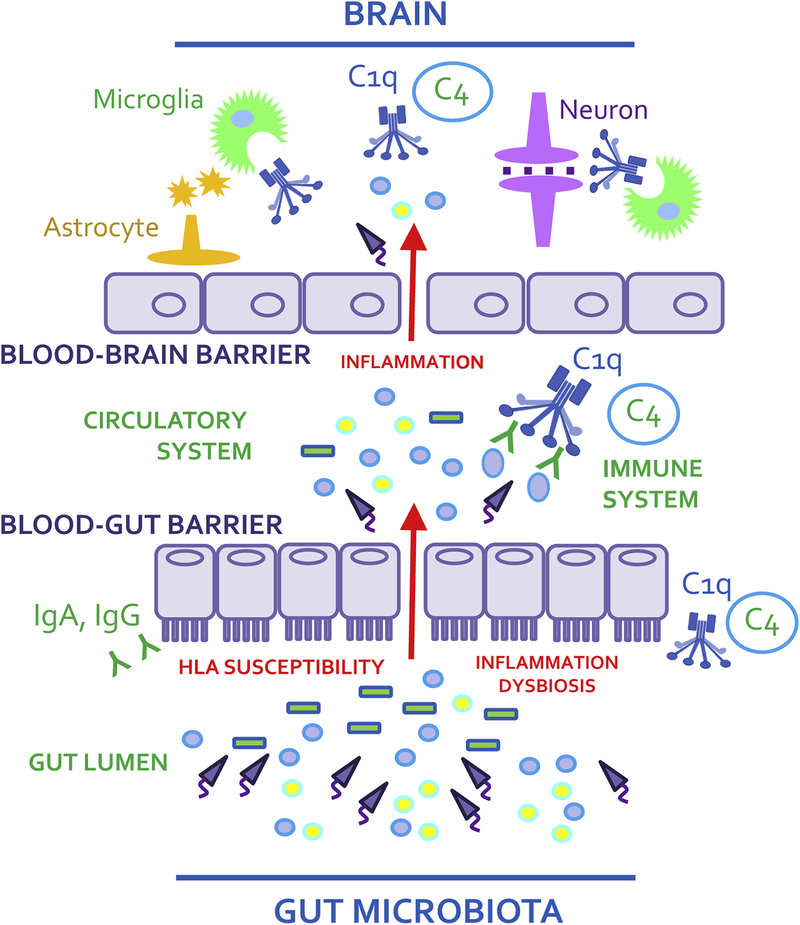
The pertinent question, therefore, is could activation of peripheral MHC and complement pathways by an imbalanced microbiome result in homologous immune molecule activation in the brain and specifically disrupt synapse function and neural circuitry? There are hypothetical scenarios by which a microbiome-gut-brain mechanism might unfold to trigger activation of brain MHC and complement. A promising candidate mechanism, as depicted in Figure 1, involves HLA-driven disease-associated endothelial barrier defects present in a cascade involving gut dysbiosis, inflammation, and the translocation of microbes and their products (metabolites, toxins, neurotransmitters) into systemic circulation where microbes or microbial products may access the brain (Pollak et al., 2018; Severance et al., 2016b). This scenario has been studied using as an opportune model, the neurotropic parasite Toxoplasma gondii, to explore how enteric infection leads to CNS infection via defects in blood-gut and blood-brain barrier integrity (Kannan et al., 2017). Coinciding with this experimental infectious process was the expression of multiple complement components systemically and in the CNS where deposition of complement C1q in the vicinity of nearby synapses was observed (Xiao et al., 2016). The translational application of these findings is supported by numerous epidemiology investigations that implicate Toxoplasma gondii infection as a risk factor for the development of schizophrenia (Torrey et al., 2012). One study showed that individuals who were seropositive for this parasite not only had higher rates of blood-gut and blood-brain barrier permeabilities, but also exhibited elevated levels of antibodies reactive against NMDA receptors (Kannan et al., 2017). Epidemiological accounts of autoimmune comorbidities in schizophrenia are extensive, can be related to complement dysfunction and microbial dysbiosis, and are reviewed in this context elsewhere (Severance et al., 2018).
Figure 1.
The gut microbiome and immune gene pleiotropy along the gut-brain axis in schizophrenia. The proposed model illustrates an overview of microbial dysbioses leading to systemic inflammation and permeabilized blood-gut and blood-brain barriers in HLA-susceptible individuals. As resident microbes translocate into circulation, the immune response, as initiated by the complement pathway, is activated. With access to the brain, gut-derived molecules can enter, signaling astrocytes (orange) and microglia (green) to respond to invaders. Subsequent complement expression in the vicinity of neurons (purple) may render susceptible synapses to inappropriate pruning.
Perinatal exposure to microbial dysbiosis
As indicated, the precise timing of the gut microbiome perturbation is a critical issue for neurodevelopmental brain disorders, with evidence that inflammation during sensitive time periods in early life can lead to such disorders as autism, cerebral palsy, epilepsy and schizophrenia (Jiang et al., 2018). Epidemiological studies have long implicated maternal immune activation, such as exposure to the infectious disease process, as a risk factor for the development of schizophrenia (Allswede et al., 2016; Blomstrom et al., 2012; Brown et al., 2004a; Brown et al., 2000; Brown et al., 2004b; Buka et al., 2008; Ellman et al., 2009; Karlsson et al., 2012; Mortensen et al., 2010; Pedersen et al., 2011; Severance et al., 2014; Xiao et al., 2009). A long literature of preclinical work directly supports brain behavioral and biochemical deficits in offspring exposed prenatally to immune activation (Brown and Derkits, 2010; Estes and McAllister, 2016; Labouesse et al., 2015; Meyer, 2014). Recent reports indicate that this exposure to prenatal immune activation results in the upregulation of complement components C1q and C4 in cortical regions of the brains of offspring (Duchatel et al., 2018; Han et al., 2017). In a study of clinical samples, maternal C1q was also elevated peripherally at time of birth in mothers whose children went on to develop serious psychoses as adults compared to mothers whose children were mentally healthy as adults (Severance et al., 2014). This inappropriate activation of complement during development has obvious detrimental implications regarding impaired construction of properly sculpted neuronal networks. In individuals with elevated C4 copy numbers, the associated increased expression could subject the brain to overpruning (Sekar et al., 2016).
The maternal microbiome is predominantly vertically transferred to offspring and as such is well-staged to severely impact offspring immune gene expression if the microbiome tips toward an inflammatory state. Such microbiome shifting sources might include exposures to stress, pathogens, altered diet, and medications (Codagnone et al., 2018; Jasarevic et al., 2018). In mice reared in a germfree environment, microglial function was particularly associated with significantly altered transcriptomics and structural densities in male embryos and in female adults; an accompanying transcriptomic survey of human fetal microglia was consistent with the mouse results (Thion et al., 2018). The sex specificity of the microglial functional response has been demonstrated previously (Desbonnet et al., 2014; Erny et al., 2015), and has interesting implications for schizophrenia which is often thought to materialize differently in males vs females (Mendrek and Mancini-Marie, 2016; Shimamoto and Rappeneau, 2017). Microglia perform immune surveillance in the CNS and when activated, release toxic or protective agents in an effort to phagocytose tissue debris and pathogens. Microglia are themselves modulators of synapse form and function including mediation of complement expression. Thus, any alterations or depletions of these brain immune cells likely influences the ensuing presence of active complement, as indicated from studies of the retinogeniculate model, which show that the removal of synapses by microglia was dependent on the complement cascade (Schafer et al., 2012).
Conclusions
Remaining issues relating to the interconnections between immune genes, the microbiome and schizophrenia include ones of causality. Important questions are those regarding whether or not immune associations in schizophrenia are truly representative of disease pathophysiology or if rather they reflect stress, medication effects, lifestyle factors and exposures common in less than ideal environments. Other brain disorders in addition to schizophrenia which exhibit significant associations with a disrupted microbiome include autism, multiple sclerosis, Alzheimer’s disease, Parkinson’s disease, amyotrophic lateral sclerosis and Huntington’s disease (Tremlett et al., 2017). Consistent findings across these brain disorders is the presence of dysbiosis, inferred based on biomarkers of a leaky gut or case-control differences in the compositions of taxa of bacteria, viruses and other microbes. Changes in the gut microbiome have also been associated with obesity, diabetes, autoimmune disease and asthma (Tremlett et al., 2017), which are all risk factors for and/or are common comorbidities found in schizophrenia and other psychiatric disorders (Annamalai et al., 2017; Benros et al., 2011; Eaton et al., 2006; Henderson et al., 2015; Wu et al., 2018). Thus, a prevailing question is which came first, the disorder or the dysbiosis? Stress is a major mediator of gut dysfunction and permeability; as such, dysbiotic comorbidities may be prevalent in many disorders because of exposure to any number of different forms of stress or other environmental factors. It is possible that microbiome involvement in schizophrenia is not causal to the disorder, but rather it contributes to or amplifies psychiatric and cognitive symptoms, perhaps in people with predisposing genotypes. A genetic template that makes the host susceptible to aberrant responses to immune challenges in a manner that alters synapse function is, however, consistent with both a pathophysiological and etiologic role. To accelerate further traction, this field of study requires prospective and longitudinal cohorts with accompanying microbial biospecimens for use in population based studies. In so doing, it may be determinable whether certain microbial measures, GI conditions or exposures convey a future risk for schizophrenia. Ultimately, it is hoped that the microbial mechanisms affecting the gut-brain axis can be harnessed in such a way as to result in novel methods to treat and manage symptoms that currently compromise the mental health of so many people around the world.
Acknowledgements
This work was supported by a NIMH P50 Silvio O. Conte Center at Johns Hopkins (grant# MH-94268) and by the Stanley Medical Research Institute. The funding sources had no involvement in study design; collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Footnotes
Declaration of interest: Dr. Yolken is a member of the Stanley Medical Research Institute Board of Directors and Scientific Advisory Board. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies. Dr. Severance does not report any potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abegunde AT, Muhammad BH, Bhatti O, & Ali T (2016). Environmental risk factors for inflammatory bowel diseases: Evidence based literature review. World Journal of Gastroenterology : WJG, 22, 6296–6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allswede DM, Buka SL, Yolken RH, Torrey EF, & Cannon TD (2016). Elevated maternal cytokine levels at birth and risk for psychosis in adult offspring. Schizophr Res, 172, 41–45. [DOI] [PubMed] [Google Scholar]
- Allswede DM, Zheutlin AB, Chung Y, Anderson K, Hultman CM, Ingvar M, et al. (2018). Complement gene expression correlates with superior frontal cortical thickness in humans. Neuropsychopharmacology, 43, 525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annamalai A, Kosir U, & Tek C (2017). Prevalence of obesity and diabetes in patients with schizophrenia. World J Diabetes, 8, 390–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APA. (2013). Diagnostic and statistical manual of mental disorders, 5th edition: Dsm-5 (5th ed.). Arlington, VA: American Psychiatric Publishing. [Google Scholar]
- Avramopoulos D, Pearce BD, McGrath J, Wolyniec P, Wang R, Eckart N, et al. (2015). Infection and inflammation in schizophrenia and bipolar disorder: A genome wide study for interactions with genetic variation. PLoS One, 10, e0116696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balu DT, & Coyle JT (2011). Neuroplasticity signaling pathways linked to the pathophysiology of schizophrenia. Neuroscience and Biobehavioral Reviews, 35, 848–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamne M, Wood J, Chowdari K, Watson AM, Celik C, Mansour H, et al. (2012). Evaluation of hla polymorphisms in relation to schizophrenia risk and infectious exposure. Schizophr Bull, 38, 1149–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechter K (2013). Updating the mild encephalitis hypothesis of schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry, 42, 71–91. [DOI] [PubMed] [Google Scholar]
- Bellander BM, Singhrao SK, Ohlsson M, Mattsson P, & Svensson M (2001). Complement activation in the human brain after traumatic head injury. Journal of Neurotrauma, 18, 1295–1311. [DOI] [PubMed] [Google Scholar]
- Bennett MR, Farnell L, & Gibson WG (2013). Fiber pathway pathology, synapse loss and decline of cortical function in schizophrenia. PLoS One, 8, e60518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benros ME, Nielsen PR, Nordentoft M, Eaton WW, Dalton SO, & Mortensen PB (2011). Autoimmune diseases and severe infections as risk factors for schizophrenia: A 30-year population-based register study. The American Journal of Psychiatry, 168, 1303–1310. [DOI] [PubMed] [Google Scholar]
- Bernstein CN, Hitchon CA, Walld R, Bolton JM, Sareen J, Walker JR, et al. (2018). Increased burden of psychiatric disorders in inflammatory bowel disease. Inflamm Bowel Dis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialas AR, & Stevens B (2013). Tgf-beta signaling regulates neuronal c1q expression and developmental synaptic refinement. Nat Neurosci, 16, 1773–1782. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Blomstrom A, Karlsson H, Wicks S, Yang S, Yolken RH, & Dalman C (2012). Maternal antibodies to infectious agents and risk for non-affective psychoses in the offspring--a matched case-control study. Schizophrenia Research, 140, 25–30. [DOI] [PubMed] [Google Scholar]
- Boulanger LM (2009). Immune proteins in brain development and synaptic plasticity. Neuron, 64, 93–109. [DOI] [PubMed] [Google Scholar]
- Brown AS, Begg MD, Gravenstein S, Schaefer CA, Wyatt RJ, Bresnahan M, et al. (2004a). Serologic evidence of prenatal influenza in the etiology of schizophrenia. Arch Gen Psychiatry, 61, 774–780. [DOI] [PubMed] [Google Scholar]
- Brown AS, Cohen P, Greenwald S, & Susser E (2000). Nonaffective psychosis after prenatal exposure to rubella. The American Journal of Psychiatry, 157, 438–443. [DOI] [PubMed] [Google Scholar]
- Brown AS, & Derkits EJ (2010). Prenatal infection and schizophrenia: A review of epidemiologic and translational studies. Am J Psychiatry, 167, 261–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AS, Hooton J, Schaefer CA, Zhang H, Petkova E, Babulas V, et al. (2004b). Elevated maternal interleukin-8 levels and risk of schizophrenia in adult offspring. The American Journal of Psychiatry, 161, 889–895. [DOI] [PubMed] [Google Scholar]
- Buka SL, Cannon TD, Torrey EF, Yolken RH, & Collaborative Study Group on the Perinatal Origins of Severe Psychiatric, D. (2008). Maternal exposure to herpes simplex virus and risk of psychosis among adult offspring. Biol Psychiatry, 63, 809–815. [DOI] [PubMed] [Google Scholar]
- Bulzacka E, Boyer L, Schurhoff F, Godin O, Berna F, Brunel L, et al. (2016). Chronic peripheral inflammation is associated with cognitive impairment in schizophrenia: Results from the multicentric face-sz dataset. Schizophr Bull, 42, 1290–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon TD, Huttunen MO, Lonnqvist J, Tuulio-Henriksson A, Pirkola T, Glahn D, et al. (2000). The inheritance of neuropsychological dysfunction in twins discordant for schizophrenia. Am J Hum Genet, 67, 369–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Nallar E, Bendall ML, Perez-Losada M, Sabuncyan S, Severance EG, Dickerson FB, et al. (2015). Composition, taxonomy and functional diversity of the oropharynx microbiome in individuals with schizophrenia and controls. PeerJ, 3, e1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattane N, Richetto J, & Cattaneo A (2018). Prenatal exposure to environmental insults and enhanced risk of developing schizophrenia and autism spectrum disorder: Focus on biological pathways and epigenetic mechanisms. Neuroscience and Biobehavioral Reviews. [DOI] [PubMed] [Google Scholar]
- Chacon MA, & Boulanger LM (2013). Mhc class i protein is expressed by neurons and neural progenitors in mid-gestation mouse brain. Mol Cell Neurosci, 52, 117–127. [DOI] [PubMed] [Google Scholar]
- Chistiakov DA, Bobryshev YV, Kozarov E, Sobenin IA, & Orekhov AN (2014). Intestinal mucosal tolerance and impact of gut microbiota to mucosal tolerance. Frontiers in Microbiology, 5, 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke DJ, Chohan TW, Kassem MS, Smith KL, Chesworth R, Karl T, et al. (2018). Neuregulin 1 deficiency modulates adolescent stress-induced dendritic spine loss in a brain region-specific manner and increases complement 4 expression in the hippocampus. Schizophr Bull. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleynen I, Konings P, Robberecht C, Laukens D, Amininejad L, Theatre E, et al. (2016). Genome-wide copy number variation scan identifies complement component c4 as novel susceptibility gene for crohn’s disease. Inflamm Bowel Dis, 22, 505–515. [DOI] [PubMed] [Google Scholar]
- Codagnone MG, Spichak S, O’Mahony SM, O’Leary OF, Clarke G, Stanton C, et al. (2018). Programming bugs: Microbiota and the developmental origins of brain health and disease. Biol Psychiatry. [DOI] [PubMed] [Google Scholar]
- Collins SM, Surette M, & Bercik P (2012). The interplay between the intestinal microbiota and the brain. Nature reviews. Microbiology, 10, 735–742. [DOI] [PubMed] [Google Scholar]
- Corvin A, & Morris DW (2014). Genome-wide association studies: Findings at the major histocompatibility complex locus in psychosis. Biol Psychiatry, 75, 276–283. [DOI] [PubMed] [Google Scholar]
- Cukrowska B, Sowinska A, Bierla JB, Czarnowska E, Rybak A, & Grzybowska-Chlebowczyk U (2017). Intestinal epithelium, intraepithelial lymphocytes and the gut microbiota - key players in the pathogenesis of celiac disease. World Journal of Gastroenterology : WJG, 23, 7505–7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demjaha A, MacCabe JH, & Murray RM (2012). How genes and environmental factors determine the different neurodevelopmental trajectories of schizophrenia and bipolar disorder. Schizophr Bull, 38, 209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbonnet L, Clarke G, Shanahan F, Dinan TG, & Cryan JF (2014). Microbiota is essential for social development in the mouse. Mol Psychiatry, 19, 146–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, et al. (2011). Normal gut microbiota modulates brain development and behavior. Proceedings of the National Academy of Sciences of the United States of America, 108, 3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinan TG, Borre YE, & Cryan JF (2014). Genomics of schizophrenia: Time to consider the gut microbiome? Molecular Psychiatry, 19, 1252–1257. [DOI] [PubMed] [Google Scholar]
- Dinan TG, & Cryan JF (2015). The impact of gut microbiota on brain and behaviour: Implications for psychiatry. Current Opinion in Clinical Nutrition and Metabolic Care, 18, 552–558. [DOI] [PubMed] [Google Scholar]
- Donohoe G, Holland J, Mothersill D, McCarthy-Jones S, Cosgrove D, Harold D, et al. (2018). Genetically predicted complement component 4a expression: Effects on memory function and middle temporal lobe activation. Psychol Med, 48, 1608–1615. [DOI] [PubMed] [Google Scholar]
- Duchatel RJ, Meehan CL, Harms LR, Michie PT, Bigland MJ, Smith DW, et al. (2018). Increased complement component 4 (c4) gene expression in the cingulate cortex of rats exposed to late gestation immune activation. Schizophr Res. [DOI] [PubMed] [Google Scholar]
- Eaton WW, Byrne M, Ewald H, Mors O, Chen CY, Agerbo E, et al. (2006). Association of schizophrenia and autoimmune diseases: Linkage of danish national registers. Am J Psychiatry, 163, 521–528. [DOI] [PubMed] [Google Scholar]
- Ellman LM, Yolken RH, Buka SL, Torrey EF, & Cannon TD (2009). Cognitive functioning prior to the onset of psychosis: The role of fetal exposure to serologically determined influenza infection. Biological Psychiatry, 65, 1040–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvington M, Schepp-Berglind J, & Tomlinson S (2015). Regulation of the alternative pathway of complement modulates injury and immunity in a chronic model of dextran sulphate sodium-induced colitis. Clin Exp Immunol, 179, 500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erny D, Hrabe de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, et al. (2015). Host microbiota constantly control maturation and function of microglia in the cns. Nature Neuroscience, 18, 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes ML, & McAllister AK (2016). Maternal immune activation: Implications for neuropsychiatric disorders. Science, 353, 772–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Network of National Networks studying Gene-Environment Interactions in, S., van Os J, Rutten BP, Myin-Germeys I, Delespaul P, Viechtbauer W, et al. (2014). Identifying gene-environment interactions in schizophrenia: Contemporary challenges for integrated, large-scale investigations. Schizophr Bull, 40, 729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadgyas-Stanculete M, Buga AM, Popa-Wagner A, & Dumitrascu DL (2014). The relationship between irritable bowel syndrome and psychiatric disorders: From molecular changes to clinical manifestations. Journal of Molecular Psychiatry, 2, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovic BR, & Filipovic BF (2014). Psychiatric comorbidity in the treatment of patients with inflammatory bowel disease. World Journal of Gastroenterology : WJG, 20, 3552–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fond G, Godin O, Boyer L, Berna F, Andrianarisoa M, Coulon N, et al. (2018a). Chronic low-grade peripheral inflammation is associated with ultra resistant schizophrenia. Results from the face-sz cohort. European Archives of Psychiatry and Clinical Neuroscience. [DOI] [PubMed] [Google Scholar]
- Fond G, Lancon C, Auquier P, & Boyer L (2018b). C-reactive protein as a peripheral biomarker in schizophrenia. An updated systematic review. Front Psychiatry, 9, 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JA, & McVey Neufeld KA (2013). Gut-brain axis: How the microbiome influences anxiety and depression. Trends in Neurosciences, 36, 305–312. [DOI] [PubMed] [Google Scholar]
- Fourgeaud L, & Boulanger LM (2007). Synapse remodeling, compliments of the complement system. Cell, 131, 1034–1036. [DOI] [PubMed] [Google Scholar]
- Glynn MW, Elmer BM, Garay PA, Liu XB, Needleman LA, El-Sabeawy F, et al. (2011). Mhci negatively regulates synapse density during the establishment of cortical connections. Nature Neuroscience, 14, 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard CA, Butts DA, & Shatz CJ (2007). Regulation of cns synapses by neuronal mhc class i. Proceedings of the National Academy of Sciences of the United States of America, 104, 6828–6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes RG, Brito CAA, Martinelli VF, Santos RND, Gomes F, Peixoto CA, et al. (2018). Hla-g is expressed in intestinal samples of ulcerative colitis and crohn’s disease patients and hla-g5 expression is differentially correlated with tnf and il-10 cytokine expression. Human Immunology, 79, 477–484. [DOI] [PubMed] [Google Scholar]
- Gupta S, Masand PS, Kaplan D, Bhandary A, & Hendricks S (1997). The relationship between schizophrenia and irritable bowel syndrome (ibs). Schizophr Res, 23, 265–268. [DOI] [PubMed] [Google Scholar]
- Han M, Zhang JC, & Hashimoto K (2017). Increased levels of c1q in the prefrontal cortex of adult offspring after maternal immune activation: Prevention by 7,8-dihydroxyflavone. Clin Psychopharmacol Neurosci, 15, 64–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Kosciolek T, Tang J, Zhou Y, Li Z, Ma X, et al. (2018). Gut microbiome and magnetic resonance spectroscopy study of subjects at ultra-high risk for psychosis may support the membrane hypothesis. European Psychiatry : the Journal of the Association of European Psychiatrists, 53, 37–45. [DOI] [PubMed] [Google Scholar]
- Henderson DC, Vincenzi B, Andrea NV, Ulloa M, & Copeland PM (2015). Pathophysiological mechanisms of increased cardiometabolic risk in people with schizophrenia and other severe mental illnesses. Lancet Psychiatry, 2, 452–464. [DOI] [PubMed] [Google Scholar]
- Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, et al. (2016). Complement and microglia mediate early synapse loss in alzheimer mouse models. Science, 352, 712–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig M (2013). The role of microbes and autoimmunity in the pathogenesis of neuropsychiatric illness. Current opinion in rheumatology, 25, 488–795. [DOI] [PubMed] [Google Scholar]
- Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, et al. (2004). Gene map of the extended human mhc. Nat Rev Genet, 5, 889–899. [DOI] [PubMed] [Google Scholar]
- Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, et al. (2013). Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell, 155, 1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh GS, Boulanger LM, Du H, Riquelme PA, Brotz TM, & Shatz CJ (2000). Functional requirement for class i mhc in cns development and plasticity. Science, 290, 2155–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia C, Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, et al. (2009). Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature, 460, 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasarevic E, Howard CD, Morrison K, Misic A, Weinkopff T, Scott P, et al. (2018). The maternal vaginal microbiome partially mediates the effects of prenatal stress on offspring gut and hypothalamus. Nature Neuroscience. [DOI] [PubMed] [Google Scholar]
- Jiang NM, Cowan M, Moonah SN, & Petri WA Jr. (2018). The impact of systemic inflammation on neurodevelopment. Trends in Molecular Medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AL, Mowry BJ, Pender MP, & Greer JM (2005). Immune dysregulation and self-reactivity in schizophrenia: Do some cases of schizophrenia have an autoimmune basis? Immunology and Cell Biology, 83, 9–17. [DOI] [PubMed] [Google Scholar]
- Jung ES, Cheon JH, Lee JH, Park SJ, Jang HW, Chung SH, et al. (2016). Hla-c*01 is a risk factor for crohn’s disease. Inflamm Bowel Dis, 22, 796–806. [DOI] [PubMed] [Google Scholar]
- Kannan G, Gressitt KL, Yang S, Stallings CR, Katsafanas E, Schweinfurth LA, et al. (2017). Pathogen-mediated nmda receptor autoimmunity and cellular barrier dysfunction in schizophrenia. Translational Psychiatry, 7, e1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karhus LL, Thuesen BH, Skaaby T, Rumessen JJ, & Linneberg A (2018). The distribution of hla dq2 and dq8 haplotypes and their association with health indicators in a general danish population. United European Gastroenterol J, 6, 866–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson H, Blomstrom A, Wicks S, Yang S, Yolken RH, & Dalman C (2012). Maternal antibodies to dietary antigens and risk for nonaffective psychosis in offspring. Am J Psychiatry, 169, 625–632. [DOI] [PubMed] [Google Scholar]
- Kato T, Iwamoto K, Kakiuchi C, Kuratomi G, & Okazaki Y (2005). Genetic or epigenetic difference causing discordance between monozygotic twins as a clue to molecular basis of mental disorders. Mol Psychiatry, 10, 622–630. [DOI] [PubMed] [Google Scholar]
- Kavanagh DH, Tansey KE, O’Donovan MC, & Owen MJ (2015). Schizophrenia genetics: Emerging themes for a complex disorder. Mol Psychiatry, 20, 72–76. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Shirts BH, Dayal M, Bacanu SA, Wood J, Xie W, et al. (2007). Are exposure to cytomegalovirus and genetic variation on chromosome 6p joint risk factors for schizophrenia? Ann Med, 39, 145–153. [DOI] [PubMed] [Google Scholar]
- Kirch DG (1993). Infection and autoimmunity as etiologic factors in schizophrenia: A review and reappraisal. Schizophr Bull, 19, 355–370. [DOI] [PubMed] [Google Scholar]
- Knuesel I, Chicha L, Britschgi M, Schobel SA, Bodmer M, Hellings JA, et al. (2014). Maternal immune activation and abnormal brain development across cns disorders. Nature reviews. Neurology, 10, 643–660. [DOI] [PubMed] [Google Scholar]
- Kodavali CV, Watson AM, Prasad KM, Celik C, Mansour H, Yolken RH, et al. (2014). Hla associations in schizophrenia: Are we re-discovering the wheel? American journal of medical genetics. Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics, 165B, 19–27. [DOI] [PubMed] [Google Scholar]
- Kolho KL, Paakkanen R, Lepisto A, Wennerstom A, Meri S, & Lokki ML (2016). Novel associations between major histocompatibility complex and pediatric-onset inflammatory bowel disease. J Pediatr Gastroenterol Nutr, 62, 567–572. [DOI] [PubMed] [Google Scholar]
- Kubinak JL, Stephens WZ, Soto R, Petersen C, Chiaro T, Gogokhia L, et al. (2015). Mhc variation sculpts individualized microbial communities that control susceptibility to enteric infection. Nat Commun, 6, 8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labouesse MA, Langhans W, & Meyer U (2015). Long-term pathological consequences of prenatal infection: Beyond brain disorders. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology, 309, R1–R12. [DOI] [PubMed] [Google Scholar]
- Laufer J, Oren R, Goldberg I, Horwitz A, Kopolovic J, Chowers Y, et al. (2000). Cellular localization of complement c3 and c4 transcripts in intestinal specimens from patients with crohn’s disease. Clin Exp Immunol, 120, 30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Yang SK, Hong M, Jung S, Kim BM, Moon JW, et al. (2018). An intergenic variant rs9268877 between hla-dra and hla-drb contributes to the clinical course and long-term outcome of ulcerative colitis. Journal of Crohn’s & Colitis. [DOI] [PubMed] [Google Scholar]
- Leonard MM, Sapone A, Catassi C, & Fasano A (2017). Celiac disease and nonceliac gluten sensitivity: A review. JAMA : the Journal of the American Medical Association, 318, 647–656. [DOI] [PubMed] [Google Scholar]
- Luczynski P, Whelan SO, O’Sullivan C, Clarke G, Shanahan F, Dinan TG, et al. (2016). Adult microbiota-deficient mice have distinct dendritic morphological changes: Differential effects in the amygdala and hippocampus. The European Journal of Neuroscience, 44, 2654–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui H, Zhang J, Makinson SR, Cahill MK, Kelley KW, Huang HY, et al. (2016). Progranulin deficiency promotes circuit-specific synaptic pruning by microglia via complement activation. Cell, 165, 921–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv F, Chen S, Wang L, Jiang R, Tian H, Li J, et al. (2017). The role of microbiota in the pathogenesis of schizophrenia and major depressive disorder and the possibility of targeting microbiota as a treatment option. Oncotarget, 8, 100899–100907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister AK (2014). Major histocompatibility complex i in brain development and schizophrenia. Biol Psychiatry, 75, 262–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei L, & Xiong WC (2008). Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci, 9, 437–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendrek A, & Mancini-Marie A (2016). Sex/gender differences in the brain and cognition in schizophrenia. Neuroscience and Biobehavioral Reviews, 67, 57–78. [DOI] [PubMed] [Google Scholar]
- Meyer U (2013). Developmental neuroinflammation and schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry, 42, 20–34. [DOI] [PubMed] [Google Scholar]
- Meyer U (2014). Prenatal poly(i:C) exposure and other developmental immune activation models in rodent systems. Biol Psychiatry, 75, 307–315. [DOI] [PubMed] [Google Scholar]
- Mocco J, Mack WJ, Ducruet AF, Sosunov SA, Sughrue ME, Hassid BG, et al. (2006). Complement component c3 mediates inflammatory injury following focal cerebral ischemia. Circ Res, 99, 209–217. [DOI] [PubMed] [Google Scholar]
- Modinos G, Iyegbe C, Prata D, Rivera M, Kempton MJ, Valmaggia LR, et al. (2013). Molecular genetic gene-environment studies using candidate genes in schizophrenia: A systematic review. Schizophr Res, 150, 356–365. [DOI] [PubMed] [Google Scholar]
- Mokhtari R, & Lachman HM (2016). The major histocompatibility complex (mhc) in schizophrenia: A review. J Clin Cell Immunol, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen PB, Pedersen CB, Hougaard DM, Norgaard-Petersen B, Mors O, Borglum AD, et al. (2010). A danish national birth cohort study of maternal hsv-2 antibodies as a risk factor for schizophrenia in their offspring. Schizophrenia Research, 122, 257–263. [DOI] [PubMed] [Google Scholar]
- Muller N (2014). Immunology of schizophrenia. Neuroimmunomodulation, 21, 109–116. [DOI] [PubMed] [Google Scholar]
- Murphy K, Travers P, Walport M, & Janeway C (2012). Janeway’s Immunobiology (8th ed.). New York: Garland Science. [Google Scholar]
- Needleman LA, Liu XB, El-Sabeawy F, Jones EG, & McAllister AK (2010). Mhc class i molecules are present both pre- and postsynaptically in the visual cortex during postnatal development and in adulthood. Proc Natl Acad Sci U S A, 107, 16999–17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimgaonkar VL, Prasad KM, Chowdari KV, Severance EG, & Yolken RH (2017). The complement system: A gateway to gene-environment interactions in schizophrenia pathogenesis. Mol Psychiatry, 22, 1554–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissila E, Korpela K, Lokki AI, Paakkanen R, Jokiranta S, de Vos WM, et al. (2017). C4b gene influences intestinal microbiota through complement activation in patients with paediatric-onset inflammatory bowel disease. Clin Exp Immunol, 190, 394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri O, Bossa F, Valvano MR, Corritore G, Latiano T, Martino G, et al. (2017). Crohn’s disease localization displays different predisposing genetic variants. PLoS One, 12, e0168821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks S, Avramopoulos D, Mulle J, McGrath J, Wang R, Goes FS, et al. (2018). Hla typing using genome wide data reveals susceptibility types for infections in a psychiatric disease enriched sample. Brain Behav Immun. [DOI] [PubMed] [Google Scholar]
- Pedersen MG, Stevens H, Pedersen CB, Norgaard-Pedersen B, & Mortensen PB (2011). Toxoplasma infection and later development of schizophrenia in mothers. The American Journal of Psychiatry, 168, 814–821. [DOI] [PubMed] [Google Scholar]
- Pollak TA, Drndarski S, Stone JM, David AS, McGuire P, & Abbott NJ (2018). The blood-brain barrier in psychosis. Lancet Psychiatry, 5, 79–92. [DOI] [PubMed] [Google Scholar]
- Prasad KM, Chowdari KV, D’Aiuto LA, Iyengar S, Stanley JA, & Nimgaonkar VL (2018). Neuropil contraction in relation to complement c4 gene copy numbers in independent cohorts of adolescent-onset and young adult-onset schizophrenia patients-a pilot study. Translational Psychiatry, 8, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presumey J, Bialas AR, & Carroll MC (2017). Complement system in neural synapse elimination in development and disease. Advances in Immunology, 135, 53–79. [DOI] [PubMed] [Google Scholar]
- Ransohoff RM, & Stevens B (2011). Neuroscience. How many cell types does it take to wire a brain? Science, 333, 1391–1392. [DOI] [PubMed] [Google Scholar]
- Rothermundt M, Arolt V, & Bayer TA (2001). Review of immunological and immunopathological findings in schizophrenia. Brain Behav Immun, 15, 319–339. [DOI] [PubMed] [Google Scholar]
- Saito H, Hirayama A, Umemura T, Joshita S, Mukawa K, Suga T, et al. (2018). Association between kir-hla combination and ulcerative colitis and crohn’s disease in a japanese population. PLoS One, 13, e0195778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, & Mazmanian SK (2015). Control of brain development, function, and behavior by the microbiome. Cell Host & Microbe, 17, 565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhya P, Danda D, Sharma D, & Scaria V (2016). Does the buck stop with the bugs?: An overview of microbial dysbiosis in rheumatoid arthritis. International Journal of Rheumatic Diseases, 19, 8–20. [DOI] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, et al. (2012). Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron, 74, 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepp-Berglind J, Atkinson C, Elvington M, Qiao F, Mannon P, & Tomlinson S (2012). Complement-dependent injury and protection in a murine model of acute dextran sulfate sodium-induced colitis. J Immunol, 188, 6309–6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics, C. (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature, 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz E, Maukonen J, Hyytiainen T, Kieseppa T, Oresic M, Sabunciyan S, et al. (2018). Analysis of microbiota in first episode psychosis identifies preliminary associations with symptom severity and treatment response. Schizophr Res, 192, 398–403. [DOI] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, et al. (2016). Schizophrenia risk from complex variation of complement component 4. Nature, 530, 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance EG, Alaedini A, Yang S, Halling M, Gressitt KL, Stallings CR, et al. (2012a). Gastrointestinal inflammation and associated immune activation in schizophrenia. Schizophr Res, 138, 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance EG, Dickerson FB, & Yolken RH (2018). Autoimmune phenotypes in schizophrenia reveal novel treatment targets. Pharmacol Ther. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance EG, Gressitt KL, Buka SL, Cannon TD, & Yolken RH (2014). Maternal complement c1q and increased odds for psychosis in adult offspring. Schizophr Res, 159, 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance EG, Gressitt KL, Halling M, Stallings CR, Origoni AE, Vaughan C, et al. (2012b). Complement c1q formation of immune complexes with milk caseins and wheat glutens in schizophrenia. Neurobiol Dis, 48, 447–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance EG, Gressitt KL, Stallings CR, Katsafanas E, Schweinfurth LA, Savage CL, et al. (2016a). Candida albicans exposures, sex specificity and cognitive deficits in schizophrenia and bipolar disorder. NPJ Schizophr, 2, 16018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance EG, Gressitt KL, Stallings CR, Origoni AE, Khushalani S, Leweke FM, et al. (2013). Discordant patterns of bacterial translocation markers and implications for innate immune imbalances in schizophrenia. Schizophr Res, 148, 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance EG, Prandovszky E, Castiglione J, & Yolken RH (2015). Gastroenterology issues in schizophrenia: Why the gut matters. Current Psychiatry Reports, 17, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance EG, Yolken RH, & Eaton WW (2016b). Autoimmune diseases, gastrointestinal disorders and the microbiome in schizophrenia: More than a gut feeling. Schizophr Res, 176, 23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatz CJ (2009). Mhc class i: An unexpected role in neuronal plasticity. Neuron, 64, 40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Xu J, Li Z, Huang Y, Yuan Y, Wang J, et al. (2018). Analysis of gut microbiota diversity and auxiliary diagnosis as a biomarker in patients with schizophrenia: A cross-sectional study. Schizophr Res. [DOI] [PubMed] [Google Scholar]
- Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe’er I, et al. (2009). Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature, 460, 753–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto A, & Rappeneau V (2017). Sex-dependent mental illnesses and mitochondria. Schizophr Res, 187, 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirts BH, Prasad KM, Pogue-Geile MF, Dickerson F, Yolken RH, & Nimgaonkar VL (2008). Antibodies to cytomegalovirus and herpes simplex virus 1 associated with cognitive function in schizophrenia. Schizophr Res, 106, 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobue A, Ito N, Nagai T, Shan W, Hada K, Nakajima A, et al. (2018). Astroglial major histocompatibility complex class i following immune activation leads to behavioral and neuropathological changes. Glia, 66, 1034–1052. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, et al. (2009). Common variants conferring risk of schizophrenia. Nature, 460, 744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan AH, Madison DV, Mateos JM, Fraser DA, Lovelett EA, Coutellier L, et al. (2013). A dramatic increase of c1q protein in the cns during normal aging. J Neurosci, 33, 13460–13474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan KE, Baldeweg T, & Friston KJ (2006). Synaptic plasticity and dysconnection in schizophrenia. Biol Psychiatry, 59, 929–939. [DOI] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, et al. (2007). The classical complement cascade mediates cns synapse elimination. Cell, 131, 1164–1178. [DOI] [PubMed] [Google Scholar]
- Stilling RM, Dinan TG, & Cryan JF (2014). Microbial genes, brain & behaviour - epigenetic regulation of the gut-brain axis. Genes, Brain, and Behavior, 13, 69–86. [DOI] [PubMed] [Google Scholar]
- Thelemann C, Eren RO, Coutaz M, Brasseit J, Bouzourene H, Rosa M, et al. (2014). Interferon-gamma induces expression of mhc class ii on intestinal epithelial cells and protects mice from colitis. PLoS One, 9, e86844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thion MS, Low D, Silvin A, Chen J, Grisel P, Schulte-Schrepping J, et al. (2018). Microbiome influences prenatal and adult microglia in a sex-specific manner. Cell, 172, 500–516 e516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrente F, Anthony A, Heuschkel RB, Thomson MA, Ashwood P, & Murch SH (2004). Focal-enhanced gastritis in regressive autism with features distinct from crohn’s and helicobacter pylori gastritis. Am J Gastroenterol, 99, 598–605. [DOI] [PubMed] [Google Scholar]
- Torrey EF, Bartko JJ, & Yolken RH (2012). Toxoplasma gondii and other risk factors for schizophrenia: An update. Schizophr Bull, 38, 642–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrey EF, & Peterson MR (1976). The viral hypothesis of schizophrenia. Schizophr Bull, 2, 136–146. [DOI] [PubMed] [Google Scholar]
- Tremlett H, Bauer KC, Appel-Cresswell S, Finlay BB, & Waubant E (2017). The gut microbiome in human neurological disease: A review. Ann Neurol, 81, 369–382. [DOI] [PubMed] [Google Scholar]
- Tsuang M (2000). Schizophrenia: Genes and environment. Biol Psychiatry, 47, 210–220. [DOI] [PubMed] [Google Scholar]
- Tsujita T, Niikawa N, Yamashita H, Imamura A, Hamada A, Nakane Y, et al. (1998). Genomic discordance between monozygotic twins discordant for schizophrenia. Am J Psychiatry, 155, 422–424. [DOI] [PubMed] [Google Scholar]
- Ursini G, Punzi G, Chen Q, Marenco S, Robinson JF, Porcelli A, et al. (2018). Convergence of placenta biology and genetic risk for schizophrenia. Nature Medicine, 24, 792–801. [DOI] [PubMed] [Google Scholar]
- Vaknin A, Eliakim R, Ackerman Z, & Steiner I (2004). Neurological abnormalities associated with celiac disease. Journal of Neurology, 251, 1393–1397. [DOI] [PubMed] [Google Scholar]
- Vasek MJ, Garber C, Dorsey D, Durrant DM, Bollman B, Soung A, et al. (2016). A complement-microglial axis drives synapse loss during virus-induced memory impairment. Nature, 534, 538–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu J, Kushnir V, Cassell B, Gyawali CP, & Sayuk GS (2014). The impact of psychiatric and extraintestinal comorbidity on quality of life and bowel symptom burden in functional gi disorders. Neurogastroenterology and Motility : the official journal of the European Gastrointestinal Motility Society, 26, 1323–1332. [DOI] [PubMed] [Google Scholar]
- Wekerle H (2017). Brain autoimmunity and intestinal microbiota: 100 trillion game changers. Trends Immunol, 38, 483–497. [DOI] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science, 334, 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Dalman C, Karlsson H, Lewis G, Osborn DPJ, Gardner R, et al. (2018). Childhood and parental asthma, future risk of bipolar disorder and schizophrenia spectrum disorders: A population-based cohort study. Schizophr Bull. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Buka SL, Cannon TD, Suzuki Y, Viscidi RP, Torrey EF, et al. (2009). Serological pattern consistent with infection with type i toxoplasma gondii in mothers and risk of psychosis among adult offspring. Microbes Infect, 11, 1011–1018. [DOI] [PubMed] [Google Scholar]
- Xiao J, Li Y, Gressitt KL, He H, Kannan G, Schultz TL, et al. (2016). Cerebral complement c1q activation in chronic toxoplasma infection. Brain Behav Immun, 58, 52–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav SK, Boppana S, Ito N, Mindur JE, Mathay MT, Patel A, et al. (2017). Gut dysbiosis breaks immunological tolerance toward the central nervous system during young adulthood. Proc Natl Acad Sci U S A, 114, E9318–E9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yolken RH, Severance EG, Sabunciyan S, Gressitt KL, Chen O, Stallings C, et al. (2015). Metagenomic sequencing indicates that the oropharyngeal phageome of individuals with schizophrenia differs from that of controls. Schizophr Bull, 41, 1153–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yolken RH, & Torrey EF (2008). Are some cases of psychosis caused by microbial agents? A review of the evidence. Mol Psychiatry, 13, 470–479. [DOI] [PubMed] [Google Scholar]
- Yuan X, Zhang P, Wang Y, Liu Y, Li X, Kumar BU, et al. (2018). Changes in metabolism and microbiota after 24-week risperidone treatment in drug naive, normal weight patients with first episode schizophrenia. Schizophr Res. [DOI] [PubMed] [Google Scholar]