Abstract
Over the past two decades, progress in understanding human infections with hepatitis A virus (HAV) and hepatitis E virus (HEV) has been eclipsed by the priority of combating persistent hepatitis B virus (HBV) and hepatitis C virus (HCV) infections. During that time, the global burden of liver disease caused by enteric hepatitis viruses has not abated. Because of vaccines, hepatitis A has become increasingly a disease of adults instead of early childhood in many regions of the world, resulting in an age-related shift toward more severe disease. HEV has remained endemic in many developing countries, and in well-developed, economically advanced countries it is now recognized as a cause of chronic, progressive liver disease in individuals with compromised immunity. The goal of this collection of articles is to review recent progress and to shine a bright light on gaps in our understanding of how these viruses replicate, cause disease, interact with the liver and host immune system, and are transmitted, along with prospects for improved control in human populations. Renewed efforts to study and compare HAV and HEV biology in humans and animal models have high potential to enhance our understanding of host–pathogen balance in the liver, and may contribute ultimately to the control of other infectious diseases of the liver.
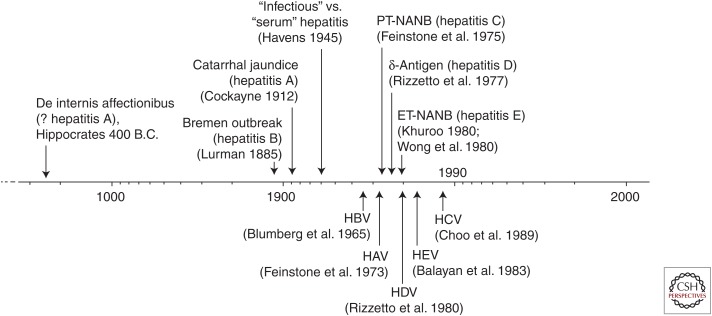
Hepatitis A and hepatitis E serve as bookends for the five types of viral hepatitis now known to afflict humans, not only in name but also in terms of when they were first recognized as distinct disease entities. Reports of epidemic jaundice extend back to the time of Hippocrates in the 5th century BC (Fig. 1). These disease outbreaks have often been attributed to hepatitis A virus (HAV), but in light of today’s knowledge could equally well have been caused by hepatitis E virus (HEV). Iatrogenic transmission of what was most likely hepatitis B was first described in a report of an outbreak in a Bremen shipyard in 1883 (Lurman 1885), whereas a clear description of what can be assumed to be person-to-person, household transmission of acute hepatitis A appeared in the medical literature ∼20 years later (Cockayne 1912). Despite this, hepatitis A and B were not distinguished as distinct disease entities, “infectious hepatitis” and “homologous serum hepatitis,” until several decades later, during the Second World War (Havens 1945). It was not until 1953, 65 years ago, that the terms “hepatitis A” and “hepatitis B” came into use (MacCallum 1953). With the discovery of the hepatitis B virus (HBV) surface antigen in the mid-1960s (Blumberg et al. 1965), followed by the identification of the HAV particle at the beginning of the 1970s (Feinstone et al. 1973), it became evident that most cases of posttransfusion hepatitis were caused by neither virus (Feinstone et al. 1975; Feinstone 2018). This led directly to recognition of what we now call hepatitis C. The existence of a second, enterically transmitted type of “non-A, non-B” hepatitis, hepatitis E, became apparent at the end of the decade, when serologic testing showed that persons involved in hepatitis outbreaks in India were not infected with HAV (Khuroo 1980; Wong et al. 1980). The responsible agent, HEV, was identified several years later (Balayan et al. 1983). The discovery of HAV can thus be said to have paved the way toward recognition of hepatitis C and E.
Figure 1.
A 2500-year timeline extending from an early description of four types of jaundice possibly written by Hippocrates to more recent recognition of different forms of transmissible hepatitis with distinct clinical or epidemiologic features (above) and identification of specific viral particles or virus-associated antigens (below) (see text for details). Hepatitis delta virus (HDV) infection is limited to persons infected with hepatitis B virus (HBV) (Wong et al. 1980). HCV, Hepatitis C virus; PT-NANB, posttransfusion non-A, non-B hepatitis; ET-NANB, enterically transmitted non-A, non-B hepatitis; HEV, hepatitis E virus; HAV, hepatitis A virus.
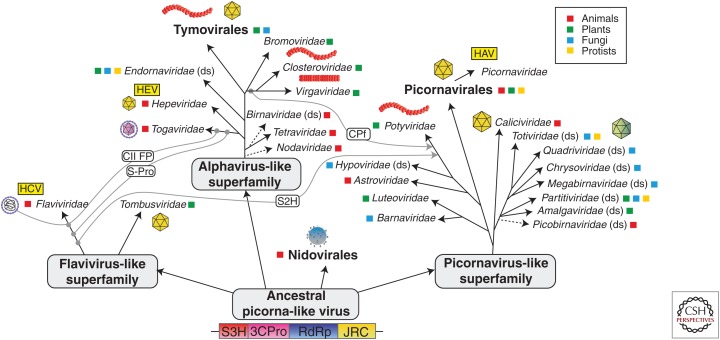
There are numerous parallels between HAV and HEV, both of which were discovered using immune electron microscopy in fecal material from experimentally infected humans (in the case of HEV, the lead investigator himself) (Feinstone et al. 1973; Balayan et al. 1983). Both are hepatotropic, positive-strand RNA viruses that usually cause acute, self-limited inflammatory disease of the liver. Although they have distinct evolutionary histories, they have common ancestral roots (Fig. 2) (Koonin et al. 2015). HAV is a member of the picornavirus-like superfamily of viruses, while HEV resides within the alphavirus-like superfamily, consistent with substantial differences in the organization of their genomes and in molecular aspects of their replication cycles (see Kenney and Meng 2018; McKnight and Lemon 2018). Amazingly, both viruses have evolved, almost certainly independently, to share a unique life cycle. In both cases, virus is shed in feces as naked, nonenveloped virions optimized for transmission through the environment, but circulates in the blood of infected persons as quasi-enveloped virions cloaked in host membranes that protect it from neutralizing antibodies (Feng et al. 2014). Not surprisingly, HAV and HEV share a common mode of fecal–oral transmission, and geographic variations in endemicity of both can be related to economic development and sanitation practices (see Jacobsen 2018; Nelson et al. 2018).
Figure 2.
Evolutionary relationships of the positive-strand RNA hepatitis viruses. Hepatitis A virus (HAV), hepatitis E virus (HEV), and hepatitis C virus (HCV) emerged from picornavirus-like, alphavirus-like, and flavivirus-like superfamilies that evolved from an ancestral picorna-like virus with superfamily 3 helicase (S3H), 3C-like protease (3Cpro), RNA-dependent RNA polymerase (RdEp), and jelly-roll capsid (JRC) modules. Gray lines represent horizontal gene transfers of superfamily 2 helicase (S2H), class II fusion protein (CF II), and filamentous virus capsid protein (CPf). (Image modified from Koonin et al. 2015; modified, with permission, from Elsevier in conjunction with Creative Commons Attribution License, CC BY.)
These similarities between HAV and HEV justify the dual focus on these two infectious agents in this collection of articles. Nonetheless, fundamental differences in the biology of these viruses abound—not only in terms of their molecular features, but also in terms of their interactions with the host: the diseases they cause, how they evoke and evade innate immune responses, and probably also how they are ultimately controlled by host immunity (see Feng and Lemon 2018; Wedemeyer 2018). One of these viruses (HEV) is zoonotic, and the other is not. Much of this is poorly understood and a target of current research. One key difference with substantial immediate medical consequence is the high mortality rate seen in women infected with genotype (gt)1 HEV during the second and third trimesters of pregnancy (Khuroo et al. 1981). A second key difference is the recent recognition that individuals with compromised T-cell function, such as recipients of solid organ transplants, are at risk for persistent gt3 HEV infection and progressive liver disease (Kamar et al. 2008). In sharp contrast, such persistent infections have never been recognized with HAV, even in patients with advanced acquired immunodeficiency syndrome (AIDS) caused by uncontrolled human immunodeficiency virus (HIV) infection. The lack of HAV persistence remains unexplained but suggests that cell-intrinsic antiviral responses may serve as a backstop for control of HAV, but not HEV infection.
Two previous published monographs have focused on hepatitis A, one edited by Robert Gerety (1984) and the other written by Gust and Feinstone (1988). To our knowledge, none has been published concerning hepatitis E. Much has been learned about both diseases over the past two decades, and HEV has in particular emerged as a significant threat to human health.
Effective vaccines have been developed for both viruses (Werzberger et al. 1992; Shrestha et al. 2007) and, in the case of HAV, are now in wide use globally, preventing millions of infections annually (see Innis and Lynch 2018; Shouval 2018). Nonetheless, HAV was estimated to have caused 47 million (95% confidence interval [CI]; 14–111 million) infections globally in 2010, resulting in 94,000 (95% CI; 30,000–222,000) deaths (Kirk et al. 2015). Most of these infections occur in economically underdeveloped regions or areas plagued by humanitarian crisis (see Jacobsen 2018). HAV infection is much less common today in well-developed Western countries. The incidence of acute hepatitis A among persons of all ages in the United States plummeted with the introduction of universal childhood immunization against HAV in 1999 (Advisory Committee on Immunization Practices 2006). However, successful public health interventions are often followed by complacency on the part of both the public and health authorities, and the past few years have noted a resurgence of hepatitis A, culminating most notably in a large sustained outbreak involving more than 500 persons and at least 20 HAV-related deaths in San Diego County in 2017.
In contrast to what might be considered a relatively minor reemergence of hepatitis A in the United States, we are witnessing the emergence of hepatitis E on a global scale. However, it is likely to be more our perceptions of hepatitis E that are emerging, not the virus itself or the disease it causes. Global estimates of HEV infection are difficult to come by, but the numbers are similar in magnitude to HAV infections as vaccines have had no impact outside China. An estimated 20 million (95% CI; 2.8–37 million) incident gt1 and gt2 HEV infections occurred in 2005, resulting in an estimated 70,000 (95% CI; 12,000–133,000) deaths (Rein et al. 2012). The number of zoonotic infections with gt3 and g4 HEV are even more difficult to estimate. However, with better diagnostic methods has come the realization that zoonotic infections with HEV gt3 and gt4 are much more prevalent in Western Europe than was considered to be case previously (see Dalton and Izopet 2018). Coupled with the discovery of chronic liver disease associated with persistent gt3 HEV in immunocompromised individuals, this is fueling interest in HEV infection among clinicians and basic scientists alike.
Interest in the science of hepatitis A largely waned following the licensure of effective HAV vaccines in the United States and Europe in the early 1990s, in part because of the perception that the hepatitis A threat was “fixed,” and in part because it was preempted by the discovery of hepatitis C virus (HCV) in 1989 (Choo et al. 1989) and recognition of the substantial long-term risk to health HCV infection poses. To some extent, this situation has reversed in recent years with the development of potent direct acting antivirals for HCV, and the discovery of quasi-enveloped eHAV virions and their role in human disease (Feng et al. 2013). Other major findings with HAV have included the realization that HAV elicits an incredibly paltry interferon response in the liver of infected chimpanzees (Lanford et al. 2011); that the immune control of HAV may be linked more directly to CD4+ than CD8+ T-cell responses (Zhou et al. 2012); that numerous closely related viruses (some with antigenic relatedness to human HAV) exist among bats and other small mammalian species, with evidence suggesting multiple shifts among host species (Drexler et al. 2015); that the structure of the HAV capsid differs dramatically from that of other common mammalian picornaviruses, in some ways resembling that of primitive insect dicistroviruses (Wang et al. 2015); and that the disease we know as hepatitis A can be reproduced by HAV infection of Ifnar1−/− mice, in which HAV-induced hepatocellular apoptosis and inflammation result from activation of innate immunity (Hirai-Yuki et al. 2016). Many other long-standing questions remain only partly answered, however, such as the reason for and consequences of the bizarre codon bias evident in the HAV genome that sets it apart from all other mammalian picornaviruses (Aragones et al. 2010; see Pintó et al. 2018).
HEV also has quirks that only now are coming to light and driving scientific enquiry. For example, how can HEV gt1 and gt2 viruses be so similar in genome structure, sequence, and antigenicity to gt3 and gt4 viruses, but differ so greatly in host range, apparent severity of acute hepatitis, and ability to persist in humans? And what accounts for the heightened morbidity and mortality of gt1 and gt2 infections in pregnant women (Khuroo et al. 1981)? Can an animal model be developed that recapitulates this enhanced pathology? There are numerous other HEV-specific questions at a molecular level. What is the significance of the ORF1 hypervariable region (HVR) and the rather striking ability of the virus to integrate host genetic sequences into its genome (see Dao Thi et al. 2018; Kenney and Meng 2018)? How does this happen, and does it have biological significance in terms of disease potential or continued viral evolution? Is this phenomenon unique to HEV, or is it potentially an attribute of all positive-strand viruses? Host genetic sequences have also been found incorporated into the genome of HCV (Li et al. 2011), and one wonders whether that could explain the origin of the unique yet diverse pX sequences found in HAV and other hepatoviruses (see Sander et al. 2018). Another question is whether the HEV ORF1 protein product actually undergoes proteolytic processing, like all other known positive-strand virus polyproteins, or does it somehow function as a large, single connected polyprotein? Finally, what is the significance of the putative amino-terminal signal sequence and glycosylation sites in the ORF2 capsid protein? How is this protein trafficked following its synthesis, and how does HEV assemble and gain egress from the hepatocyte?
Then, there are questions common to both HAV and HEV. How do they reach the liver once ingested with contaminated food or water? How important is replication within the gastrointestinal tract? And how do vaccines work, because both HAV and HEV circulate in the blood in a quasi-enveloped state resistant to the actions of neutralizing antibodies (Feng et al. 2013, 2014; Okamoto 2013)? Quasi-enveloped HAV and HEV virions are infectious, yet we have only a most rudimentary understanding of how they are produced and how they gain entry into hepatocytes (Yin et al. 2016). These questions are common to both HAV and HEV, although perhaps more urgent with HEV given the current limited state of prophylactic or therapeutic options for hepatitis E.
CONCLUDING REMARKS
Apart from the insight that studying HAV and HEV can provide concerning the evolution of RNA viruses generally, and how these two viruses specifically have evolved to evade host-protective responses to successfully parasitize the liver, continued research is likely to have both direct and indirect practical benefits. Therapeutics remain primitive for control of chronic HEV infection; more specific, better targeted, and more active antivirals than ribavirin and sofosbuvir are surely feasible (Dao Thi et al. 2016). Moreover, immune responses that provide spontaneous control of these infections and prevent viral persistence have yet to be identified despite the global impact of HAV and HEV on public health. A better understanding of the host immune responses that successfully control HAV and HEV infections in most persons may well provide novel insight into what goes wrong in the majority of acute HCV infections, leading to lifelong persistence of HCV. The development of an effective HCV vaccine remains an elusive goal, perhaps the holy grail of the hepatitis field in 2018. A better understanding of HAV and HEV immunity can only advance that goal.
Footnotes
Editors: Stanley M. Lemon and Christopher Walker
Additional Perspectives on Enteric Hepatitis Viruses available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Advisory Committee on Immunization Practices. 2006. Prevention of hepatitis A through active or passive immunization: Recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm Rep 55: 1–23. [PubMed] [Google Scholar]
- Aragones L, Guix S, Ribes E, Bosch A, Pinto RM. 2010. Fine-tuning translation kinetics selection as the driving force of codon usage bias in the hepatitis A virus capsid. PLoS Pathog 6: e1000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balayan MS, Andzhaparidze AG, Savinskaya SS, Ketiladze ES, Braginsky DM, Savinov AP, Poleschuk VF. 1983. Evidence for a virus in non-A, non-B hepatitis transmitted via the fecal–oral route. Intervirology 20: 23–31. [DOI] [PubMed] [Google Scholar]
- Blumberg BS, Alter HJ, Visnich S. 1965. A “new” antigen in leukemia sera. JAMA 191: 541–546. [DOI] [PubMed] [Google Scholar]
- Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244: 359–362. [DOI] [PubMed] [Google Scholar]
- Cockayne EA. 1912. Catarrhal jaundice, sporadic and epidemic, and its relation to acute yellow atrophy of the liver. QJM 6: 1–29. [Google Scholar]
- *.Dalton HR, Izopet J. 2018. Transmission and epidemiology of hepatitis E virus genotype 3 and 4 infections. Cold Spring Harb Perspect Med 10.1101/cshperspect.a032144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao Thi VL, Debing Y, Wu X, Rice CM, Neyts J, Moradpour D, Gouttenoire J. 2016. Sofosbuvir inhibits hepatitis E virus replication in vitro and results in an additive effect when combined with ribavirin. Gastroenterology 150: 82–85.e84. [DOI] [PubMed] [Google Scholar]
- *.Dao Thi VL, Wu X, Rice CM. 2018. Stem cell–derived culture models of hepatitis E virus infection. Cold Spring Harb Perspect Med 10.1101/cshperspect.a031799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler JF, Corman VM, Lukashev AN, van den Brand JMA, Gmyl A, Brunink S, Rasche A, Seggewiss N, Feng H, Leijten LM, et al. 2015. Evolutionary origins of hepatitis A virus in small mammals. Proc Natl Acad Sci 112: 15190–15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Feinstone SM. 2018. History of the discovery of hepatitis A virus. Cold Spring Harb Perspect Med 10.1101/cshperspect.a031740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstone SM, Kapikian AZ, Purcell RH. 1973. Hepatitis A: Detection by immune electron microscopy of a viruslike antigen associated with acute illness. Science 182: 1026–1028. [DOI] [PubMed] [Google Scholar]
- Feinstone SM, Kapikian AZ, Purcell RH, Alter HJ, Holland PV. 1975. Transfusion-associated hepatitis not due to viral hepatitis type A or B. N Engl J Med 292: 767–770. [DOI] [PubMed] [Google Scholar]
- *.Feng Z, Lemon SM. 2018. Innate immunity to enteric hepatitis viruses. Cold Spring Harb Perspect Med 10.1101/cshperspect.a033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM. 2013. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496: 367–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Hirai-Yuki A, McKnight KL, Lemon SM. 2014. Naked viruses that aren’t always naked: Quasi-enveloped agents of acute hepatitis. Annu Rev Virol 1: 539–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerety RJ. 1984. Hepatitis A. Academic, Orlando, FL. [Google Scholar]
- Gust ID, Feinstone SM. 1988. Hepatitis A. CRC, Boca Raton, FL. [Google Scholar]
- Havens WP Jr. 1945. Experiment in cross immunity between infectious hepatitis and homologous serum jaundice. Proc Soc Exp Biol Med 59: 148–150. [Google Scholar]
- Hirai-Yuki A, Hensley L, McGivern DR, Gonzalez-Lopez O, Das A, Feng H, Sun L, Wilson JE, Hu F, Feng Z, et al. 2016. MAVS-dependent host species range and pathogenicity of human hepatitis A virus. Science 353: 1541–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Innis BL, Lynch JA. 2018. Immunization against hepatitis E. Cold Spring Harb Perspect Med 10.1101/cshperspect.a032573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Jacobsen KH. 2018. Globalization and the changing epidemiology of hepatitis A virus. Cold Spring Harb Perspect Med 10.1101/cshperspect.a031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamar N, Selves J, Mansuy JM, Ouezzani L, Peron JM, Guitard J, Cointault O, Esposito L, Abravanel F, Danjoux M, et al. 2008. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N Engl J Med 358: 811–817. [DOI] [PubMed] [Google Scholar]
- *.Kenney SP, Meng X-J. 2018. Hepatitis E virus genome structure and replication strategy. Cold Spring Harb Perspect Med. 10.1101/cshperspect.a031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuroo MS. 1980. Study of an epidemic of non-A, non-B hepatitis: Possibility of another human hepatitis virus distinct from post-transfusion non-A, non-B type. Am J Med 68: 818–824. [DOI] [PubMed] [Google Scholar]
- Khuroo MS, Teli MR, Skidmore S, Sofi MA, Khuroo MI. 1981. Incidence and severity of viral hepatitis in pregnancy. Am J Med 70: 252–255. [DOI] [PubMed] [Google Scholar]
- Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Dopfer D, Fazil A, Fischer-Walker CL, Hald T, et al. 2015. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: A data synthesis. PLoS Med 12: e1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Dolja VV, Krupovic M. 2015. Origins and evolution of viruses of eukaryotes: The ultimate modularity. Virology 479–480: 2–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford RE, Feng Z, Chavez D, Guerra B, Brasky KM, Zhou Y, Yamane D, Perelson AS, Walker CM, Lemon SM. 2011. Acute hepatitis A virus infection is associated with a limited type I interferon response and persistence of intrahepatic viral RNA. Proc Natl Acad Sci 108: 11223–11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YP, Gottwein JM, Scheel TK, Jensen TB, Bukh J. 2011. MicroRNA-122 antagonism against hepatitis C virus genotypes 1–6 and reduced efficacy by host RNA insertion or mutations in the HCV 5′ UTR. Proc Natl Acad Sci 108: 4991–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurman A. 1885. Eine icterus epidemic. Berl Klin Woschenschr 22: 20–23. [Google Scholar]
- MacCallum FO. 1953. Hepatitis. Br Med Bull 9: 221–225. [DOI] [PubMed] [Google Scholar]
- *.McKnight KL, Lemon SM. 2018. Hepatitis A virus genome organization and replication strategy. Cold Spring Harb Perspect Med 10.1101/cshperspect.a033480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Nelson KE, Labrique AB, Kmush BL. 2018. Epidemiology of genotype 1 and 2 hepatitis E virus infections. Cold Spring Harb Perspect Med 10.1101/cshperspect.a031732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H. 2013. Culture systems for hepatitis E virus. J Gastroenterol 48: 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Pintó RM, Pérez-Rodríguez F-J, D’Andrea L, de Castellarnau M, Guix S, Bosch A. 2018. Hepatitis A virus codon usage: Implications for translation kinetics and capsid folding. Cold Spring Harb Perspect Med 10.1101/cshperspect.a031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein DB, Stevens GA, Theaker J, Wittenborn JS, Wiersma ST. 2012. The global burden of hepatitis E virus genotypes 1 and 2 in 2005. Hepatology 55: 988–997. [DOI] [PubMed] [Google Scholar]
- Rizzetto M, Canese MG, Arico S, Crivelli O, Trepo C, Bonino F, Verme G. 1977. Immunofluorescence detection of new antigen-antibody system (δ/anti-δ) associated to hepatitis B virus in liver and in serum of HBsAg carriers. Gut 18: 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzetto M, Hoyer B, Canese MG, Shih JWK, Purcell RH, Gerin JL. 1980. δ Agent: Association of δ antigen with hepatitis B surface antigen and RNA in serum of δ-infected chimpanzees. Proc Natl Acad Sci 77: 6124–6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Sander A-L, Corman VM, Lukashev AN, Drexler JF. 2018. Evolutionary origins of enteric hepatitis viruses. Cold Spring Harb Perspect Med 10.1101/cshperspect.a031690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Shouval D. 2018. Immunization against hepatitis A. Cold Spring Harb Perspect Med 10.1101/cshperspect.a031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha MP, Scott RM, Joshi DM, Mammen MP Jr, Thapa GB, Thapa N, Myint KS, Fourneau M, Kuschner RA, Shrestha SK, et al. 2007. Safety and efficacy of a recombinant hepatitis E vaccine. N Engl J Med 356: 895–903. [DOI] [PubMed] [Google Scholar]
- Wang X, Ren J, Gao Q, Hu Z, Sun Y, Li X, Rowlands DJ, Yin W, Wang J, Stuart DI, et al. 2015. Hepatitis A virus and the origins of picornaviruses. Nature 517: 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Wedemeyer H. 2018. Adaptive immunity to enteric hepatitis viruses. Cold Spring Harb Perspect Med 10.1101/cshperspect.a033472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werzberger A, Mensch B, Kuter B, Brown L, Lewis J, Sitrin R, Miller W, Shouval D, Wiens B, Calandra G, et al. 1992. A controlled trial of a formalin-inactivated hepatitis A vaccine in healthy children. N Engl J Med 327: 453–457. [DOI] [PubMed] [Google Scholar]
- Wong DC, Purcell RH, Sreenivasan MA, Prasad SR, Pavri KM. 1980. Epidemic and endemic hepatitis A in India: Evidence for a non-A, non-B hepatitis virus etiology. Lancet 2: 876–879. [DOI] [PubMed] [Google Scholar]
- Yin X, Ambardekar C, Lu Y, Feng Z. 2016. Distinct entry mechanisms for nonenveloped and quasi-enveloped hepatitis E viruses. J Virol 90: 4232–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Callendret B, Xu D, Brasky KM, Feng Z, Hensley LL, Guedj J, Perelson AS, Lemon SM, Lanford RE, et al. 2012. Dominance of the CD4+ T helper cell response during acute resolving hepatitis A virus infection. J Exp Med 209: 1481–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]