Abstract
Cordyceps, a type of Chinese herbal medicine that exhibits anti-angiogenesis and tumor growth suppression effects, has recently gained increasing popularity. However, high-quality, natural Cordyceps, such as Ophiocordyceps sinensis, is very rare and difficult to obtain in large amounts. Cordyceps is cultured instead of harvested from natural sources, but the quality with respect to the ingredients has not been fully studied. In this study, we performed an NMR metabolic profiling of aqueous extracts of Cordyceps without any sample treatment to evaluate the proper species and medium and influence of two different disinfection methods. It was discovered that Cordyceps militaris fungus and silkworm chrysalis medium were suitable for cultivation of Cordyceps. Furthermore, cordycepin, a Cordyceps-specific functional compound, was produced at different growth stages during different cultivation processes, even at the mycelial stage, and was found at three times higher concentrations in cultured C. militaris compared to that in naturally occurring C. militaris.
Subject terms: Metabolomics, Metabolomics
Introduction
Cordyceps is one of the most famous plant worms, and it has been used as one of the key materials in various traditional Chinese medicines for almost 2000 years1. In recent decades, Cordyceps has gained increasing popularity in the world, and several lines of evidence have shown that Cordyceps exhibits bioactivities such as anti-angiogenesis effects, tumor growth suppression and the regulation of apoptotic homeostasis2,3. Among approximately 400 different species of Cordyceps4, Ophiocordyceps sinensis, Cordyceps militaris, Cordyceps nutans and a related species, Paecilomyces tenuipes, are widely consumed as health foods and effective traditional medicines in Asia1,2,5,6.
O. sinensis, one of the most famous and expensive fungal species in the world, thrives in the cold and grassy alpine meadows of Tibet, which is approximately 3000 to 5000 meters high7. Therefore, naturally occurring O. sinensis is rare and hard to obtain. In recent studies, it has also been shown that O. sinensis has anti-oxidant and anti-apoptotic effects, but it is difficult to cultivate8,9. C. militaris, which is also widely known as a rare caterpillar fungus that is found throughout the northern hemisphere, has similar biological activities to O. sinensis, including anti-oxidant and anti-apoptotic effects1. Various cultured C. militaris are commonly sold as drug materials and health food products in China and South East Asia10,11. C. nutans is an entomoparasitic ascomycete that parasitizes hemipteran insects, and it has been found in Japan, Taiwan and China5. Compared to O. sinensis and C. militaris, there are only few studies regarding the effects of C. nutans. Paecilomyces is considered to be an anamorph of Cordyceps2. P. tenuipes, an entomogenous fungi that parasitizes mainly pupae, is a parasitic fungus in the larvae of lepidoptera. Due to its various biological activities, such as the cytotoxic activities of acetoxyscirpenediol and ergosterol peroxide, P. tenuipes is highly valuable as a health food and has been used as an effective, traditional, nutritious medicine in China for many years6,12.
In association with the rise in popularity, the demands for Cordyceps as an herbal supplement and health food have also increased. However, since high-quality, natural Cordyceps, such as O. sinensis, is very rare and difficult to obtain in large amounts, consumer demand has not been met using natural Cordyceps13. Although there are some studies on the cultivation process of Cordyceps, the actual quality of the cultured Cordyceps has not been fully studied in terms of the active compounds14,15. This is one of the major barriers to the development of cultivation methods to obtain high-quality cultured Cordyceps that can act as a replacement for naturally grown Cordyceps. In addition, many components have been found from natural and cultured Cordyceps by liquid chromatography (LC), gas chromatography (GC), and capillary electrophoresis (CE) combined with mass spectrometry (MS) and UV-visible spectrophotometry16–19, such as cordycepin, adenosine, proteins, amino acids, carbohydrates, carboxylic acids, lipids, glycosides and minerals. However, these conventional methods are all compound-targeted for the specific chemical characteristics of Cordyceps16–19.
NMR is an analytical technique with a high quantitative ability and reproducibility20. NMR spectra are taken in a nondestructive way, and therefore, provides comprehensive information regarding the chemical components directly from complex mixtures in a fast and simple way21,22. For the past decade, NMR has been recognized as a powerful technique for discerning the chemical properties of complex mixtures, and it has been widely applied to identify organic compounds in foods, such as milk, yogurt, coffee, mango juice, black garlic23–27, and many others. In the present study, solution NMR analysis of water extracts of Cordyceps without any separation and purification because consumers usually drink water extracts of Cordyceps were performed. Moreover, NMR-based metabolic profiling to investigate and evaluate newly improved cultivation processes of Cordyceps, which may bring potential commercial value to the industry of Cordyceps cultivation in the future were further performed.
Results and Discussion
NMR analysis of natural Cordyceps and related species
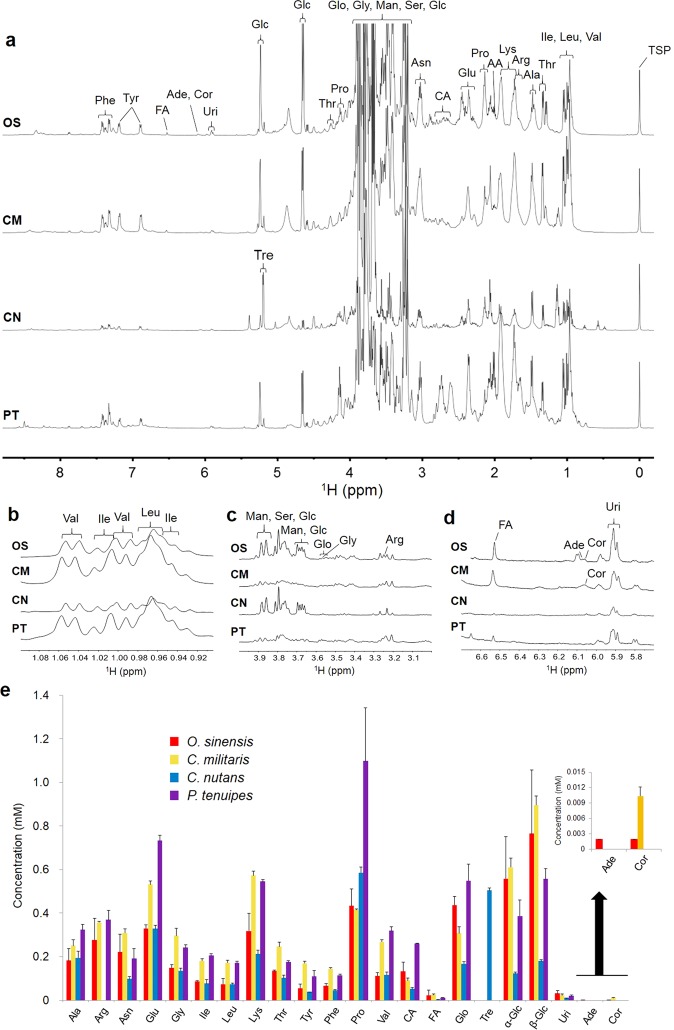
According to 1D and 2D NMR analysis and spiking experiments, 24 components, including amino acids, organic acids, glycerol and nucleosides, were identified in the extracts from natural Cordyceps and P. tenuipes (Tables S1–S3). From these, 21 compounds were quantified based on the 1H NMR spectra (Fig. 1), while serine, acetic acid and mannitol were not able to be quantified accurately due to heavy signal overlaps. Arginine signals were not detected in extracts of C. nutans, whereas trehalose signals were only detected in the extracts of C. nutans. Adenosine was only found in the extracts of O. sinensis, and cordycepin was not detected in the extracts of C. nutans and P. tenuipes. This observation suggests that O. sinensis and C. militaris are the major sources of cordycepin.
Figure 1.
NMR spectral analysis of natural Cordyceps and related species. (a‒d) 1H NMR spectra (a, −0.2 to 8.8 ppm; b, 0.9 to 1.1 ppm; c, 3.0 to 4.0 ppm; and d, 5.85 to 6.6) of the extracts of O. sinensis (OS), C. militaris (CM), C. nutans (CN), and P. tenuipes (PT). (e) Concentrations of organic compounds in the extracts of natural Cordyceps and P. tenuipes determined by NMR spectroscopy. Abbreviations: Ala, alanine; Arg, arginine; Asn, asparagine; Glu, glutamic acid; Gly, glycine; Ile, isoleucine; Leu, leucine; Lys, lysine; Ser, serine; Thr, threonine; Tyr, tyrosine; Phe, phenylalanine; Pro, proline; Val, valine; AA, acetic acid; CA, citric acid; FA, fumaric acid; Glo, glycerol; Man, mannitol; Tre, trehalose; Glc, glucose; Uri, uridine; Ade, adenosine; and Cor, cordycepin. Data are expressed as the means ± standard deviations (SD; n = 3).
Amino acids are regarded as biologically important compounds. For example, muscle protein synthesis is stimulated by amino acid supplementation28. 14 types of amino acids were quantified by NMR spectroscopy in the extracts of natural Cordyceps and P. tenuipes. Citric acid, fumaric acid, α/β-glucose, and glycerol have not been quantified in the extracts of natural Cordyceps and P. tenuipes in previous studies. Uridine, adenosine and cordycepin were observed by NMR spectroscopy without any separation. Adenosine, which plays a role in sleep control29, was specific to the extracts of O. sinensis. Cordycepin 3′-deoxyadenosine acts as a nucleoside antagonist, inhibits RNA synthesis30, and shows an antitumor effect by inducing apoptosis and cell cycle arrest in human tumor cells31. The concentrations of most of the NMR-observed amino acids in C. militaris extracts were higher than those in O. sinensis. The amounts of glycerol were lower in C. militaris than those in O. sinensis. Cordycepin, one of the most important biochemicals in Cordyceps, showed a 5-fold higher concentration in C. militaris than in O. sinensis. The concentrations of many of the components in the C. nutans extracts were obviously lower than those in O. sinensis. Trehalose, which can protect proteins and cellular membranes from inactivation or denaturation caused by a variety of stress conditions in Cordyceps32, was only found in the extracts of C. nutans. Arginine, adenosine and cordycepin were not detected in the extracts of C. nutans in the present study. The concentrations of most of the amino acids in the P. tenuipes extracts were higher than those in the O. sinensis extracts, except for asparagine. Furthermore, the amounts of the four amino acids were also higher in the P. tenuipes extracts than the levels found in C. militaris, including alanine, glutamic acid, proline, and valine. Generally, natural O. sinensis has been proven to be a natural Cordyceps with a high quality, but it is hard to obtain in large amounts. We have found that natural C. militaris is a good substitute for natural O. sinensis in terms of the major ingredients, including cordycepin, since the concentrations of most amino acids were higher than those in O. sinensis. Additionally, one of the most important biochemicals in Cordyceps, cordycepin, showed a 5-fold higher concentration than that in O. sinensis, which was not detected in C. nutans and P. tenuipes extracts.
Multivariate statistical analysis of natural Cordyceps and P. tenuipes
The 3 types of natural Cordyceps (O. sinensis, C. militaris, and C. nutans) and P. tenuipes were clearly distinguished by the principal component analysis (PCA). The loading plot for PC1 (Fig. 2b) showed that most of the bins had a negative value along the PC1 axis, which means that most of the signals had relatively higher intensities in the extracts from C. militaris and P. tenuipes than those from O. sinensis and C. nutans. On the other hand, among the 3 types of natural Cordyceps and P. tenuipes, trehalose was only detected in C. nutans (Fig. 1e) and the signals from mannitol showed high intensities in the extracts from O. sinensis and C. nutans (Fig. 1c). In Fig. 2c, the results indicate that sugar signals detected in the extracts of C. militaris and O. sinensis have relatively higher intensities than those in P. tenuipes and C. nutans, which is consistent with the quantitative data (Fig. 1e). On the other hand, the signals of alanine, glutamic acid, proline and citric acid have relatively higher intensities in P. tenuipes and C. nutans than in C. militaris and O. sinensis. Since the signal intensities of C. militaris were relatively higher than those of the other natural Cordyceps and P. tenuipes, along both the PC1 and PC2, C. militaris demonstrates great potential as a substitute for natural O. sinensis. C. militaris was found to contain relatively high concentrations of amino acids, saccharides and nucleosides, including cordycepin, compared to the other types of natural Cordyceps and P. tenuipes used in the present study.
Figure 2.
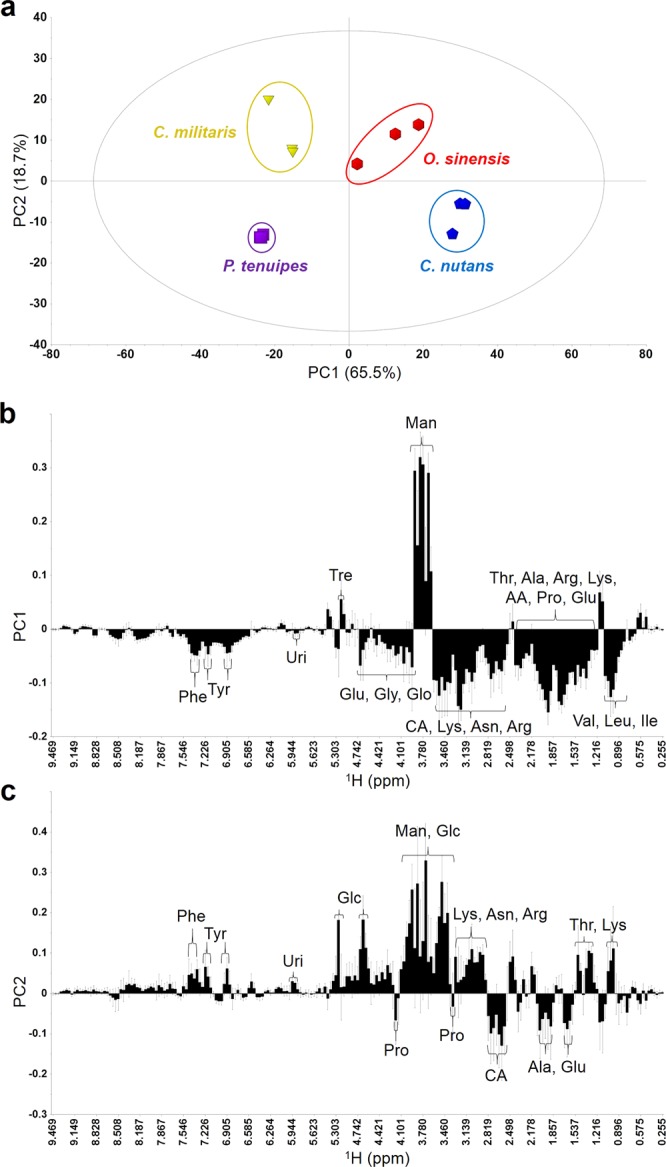
PCA analysis of natural Cordyceps and P. tenuipes. (a) PCA score plot of the extracts of 3 types of natural Cordyceps and P. tenuipes (n = 3). (b,c) Loading plots of (b) PC1 and (c) PC2 of natural Cordyceps and P. tenuipes samples (n = 3). Abbreviations: Man, mannitol; and AA, acetic acid. The other abbreviations are the same as those in Fig. 1.
NMR spectral analysis of cultured Cordyceps
Twenty-four components were identified in the extracts of cultured C. militaris based on a comparison with the data shown in Tables S1–S3. Then, 21 compounds out of the 24 compounds were also quantified by the 1H NMR spectra on the basis of the signal assignments, which are shown in Fig. 3a‒d. The quantitative results for the organic compounds in the cultured C. militaris are summarized in Fig. 3e. Trehalose was specific to C. militaris cultured in a rice medium. The C. militaris cultured in a silkworm chrysalis medium possessed higher concentrations of α- and β-glucose than that cultured in a rice medium. Trehalose biosynthesis from glucose 6-phosphate is a two-step enzymatic process that involves trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase, suggesting that the rice medium specifically activates this process33. The rice medium is rich in starch, which can be degraded to glucose and induce the synthesis of trehalose by C. militaris fungus during the cultivation process.
Figure 3.
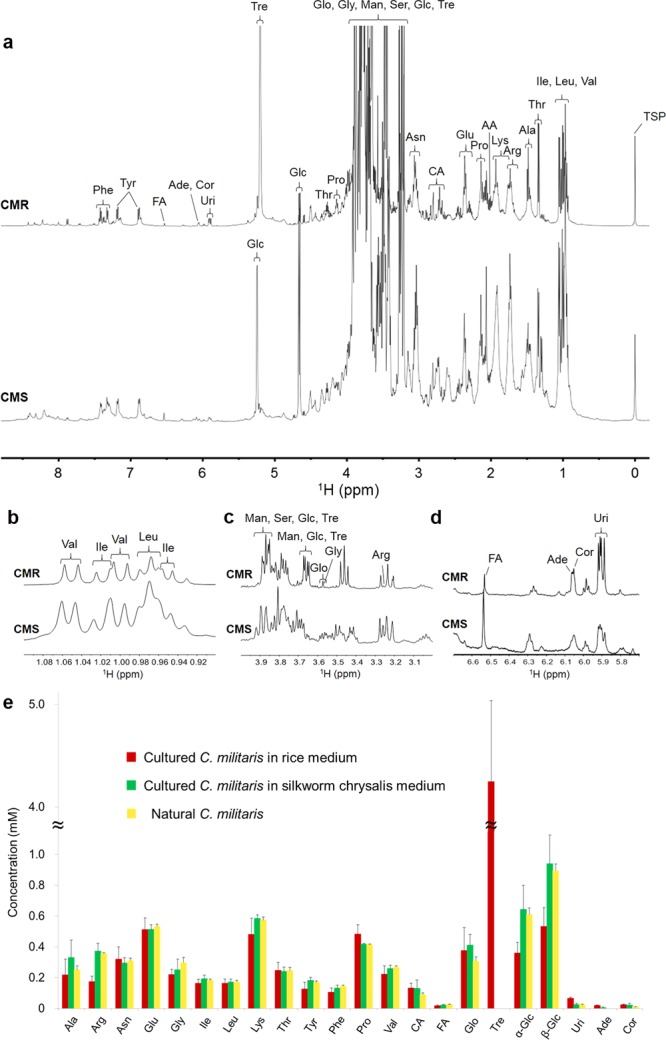
NMR spectral analysis of cultured Cordyceps. (a‒d) 1H NMR spectra (a, −0.2 to 8.8 ppm; b, 0.9 to 1.1 ppm; c, 3.0 to 4.0 ppm; and d, 5.85 to 6.6) of the extracts of C. militaris cultured in a silkworm chrysalis medium (CMS) and in a rice medium (CMR). (c) Concentrations of the organic compounds in extracts from C. militaris cultured in a silkworm chrysalis medium and a rice medium determined by NMR spectroscopy. The extracts of natural C. militaris (n = 3) were compared with those of C. militaris cultured in a silkworm chrysalis medium and a rice medium (n = 5). The other abbreviations are the same as those in Fig. 1. Data are represented as the means ± SD.
Multivariate statistical analysis of natural and cultured C. militaris
A PCA was applied to distinguish the 5 types of natural and cultured Cordyceps and P. tenuipes and was used to verify whether the two types of cultured C. militaris were similar to natural C. militaris compared with other related species. As shown in Fig. 4, the samples were clearly distinguished from one another by the PCA score plots. C. militaris cultured in a silkworm chrysalis medium showed a distribution more similar to that of natural C. militaris on the PCA score plot than C. militaris cultured in a rice medium and other natural Cordyceps. P. tenuipes was also distributed in close proximity to natural C. militaris, but the distribution did not overlap with the C. militaris cultured in a silkworm chrysalis medium. In general, the silkworm chrysalis medium should be chosen to cultivate C. militaris, because the C. militaris cultured in a silkworm chrysalis medium possessed almost the same contents as that cultured in a rice medium and it was the most similar to natural C. militaris. Thus, the C. militaris fungus and silkworm chrysalis medium were suitable for the cultivation of Cordyceps.
Figure 4.
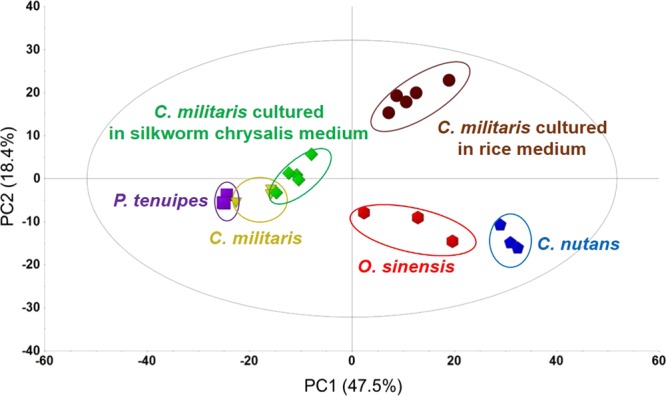
PCA score plot of the extracts of 3 types of natural Cordyceps, P. tenuipes (n = 3) and 2 types of cultured C. militaris (n = 5).
Chemical changes in cultured C. militaris extracts during cultivation processes
Natural Cordyceps may be affected by other contaminated microorganisms, such as fungi and bacteria, in the host worm, which may cause the changes in metabolic processes. However, it remains unclear how the interaction with the contaminated organisms influences the components of the cultured Cordyceps. To evaluate it, the chemical changes were monitored during cultivation processes with different disinfection steps. Figures 5 and 6 display the typical 1H NMR spectra of the extracts from the C. militaris samples culture using two different cultivation processes. The observed compounds include 14 amino acids (alanine, valine, isoleucine, leucine, threonine, arginine, asparagine, lysine, proline, glutamic acid, phenylalanine, tyrosine, glycine, and serine), 3 other organic acids (citric acid, acetic acid, and fumaric acid), 2 saccharides (glucose and mannitol), glycerol and 3 nucleosides (cordycepin, adenosine, and uridine). Among the detectable components, 21 components were quantitatively analyzed because their signals could be separately observed.
Figure 5.
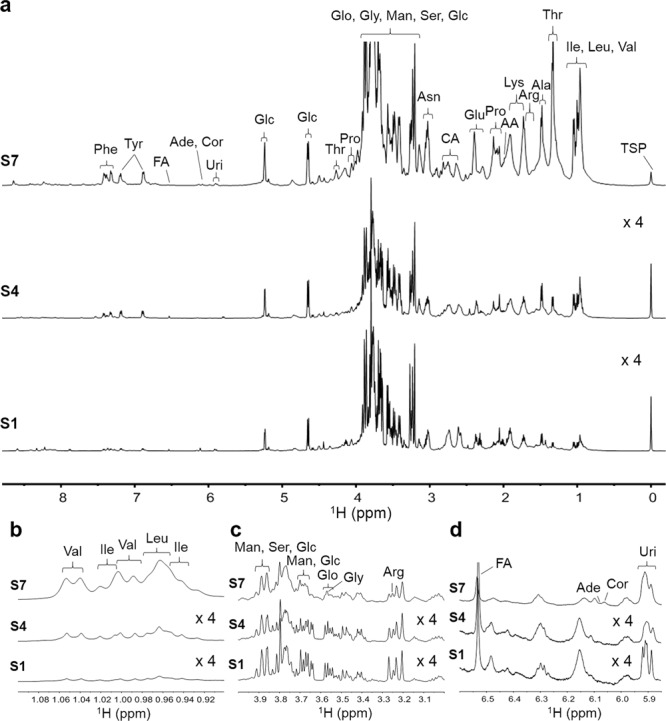
Chemical changes in cultured C. militaris extracts during cultivation processes 1. (a‒d) 1H NMR spectra (a, −0.2 to 8.8 ppm; b, 0.9 to 1.1 ppm; c, 3.0 to 4.0 ppm; and d, 5.85 to 6.6) of the extracts of the cultured C. militaris samples at stage 1 (S1), stage 4 (S4) and stage 7 (S7) by cultivation process 1. The other abbreviations are the same as those in Fig. 1.
Figure 6.
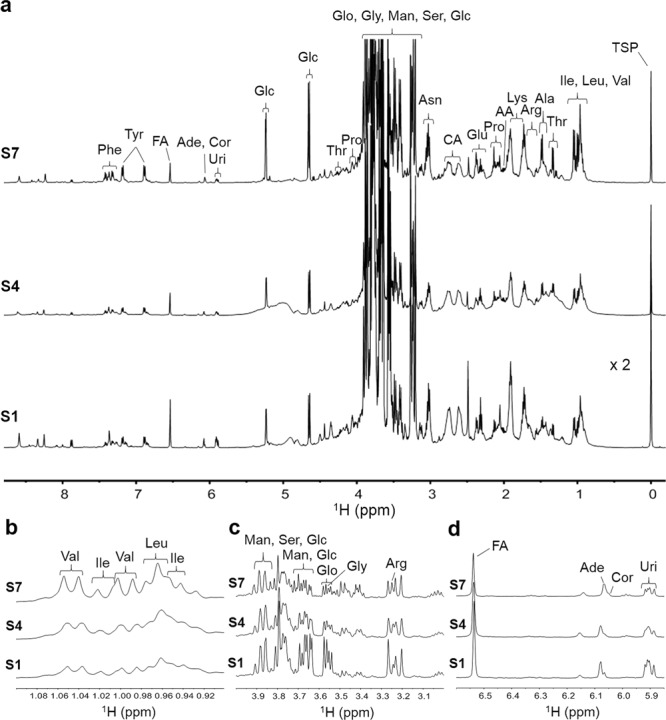
Chemical changes in cultured C. militaris extracts during cultivation processes 2. (a‒d) 1H NMR spectra (a, −0.2 to 8.8 ppm; b, 0.9 to 1.1 ppm; c, 3.0 to 4.0 ppm; and d, 5.85 to 6.6) of the extracts of the cultured C. militaris samples at stage 1 (S1), stage 4 (S4) and stage 7 (S7) by cultivation process 2. The other abbreviations are the same as those in Fig. 1.
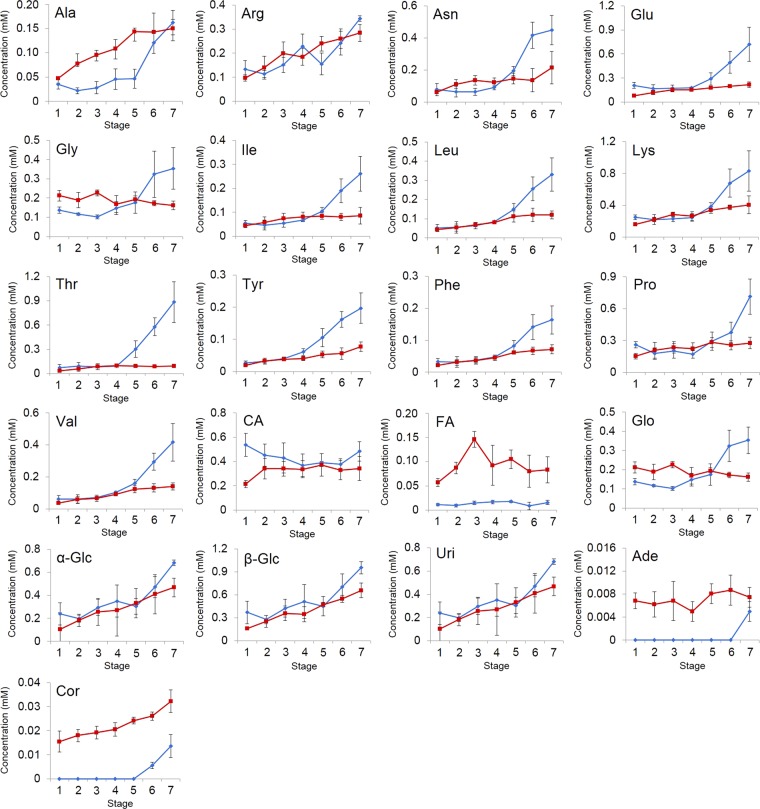
As shown in Fig. 7, the amounts of the 13 types of amino acids observed by NMR, especially from stage 4 to stage 7 during cultivation process 1, were found to be greater. The changes of amino acids were highly correlated with one another (Fig. 8a). Both the concentrations of α- and β-glucose increased during the cultivation processes, maintaining an α/β-anomer ratio of approximately 2:3, showing that α/β-anomer ratio is not affected by the biological processes. Glycerol increased rapidly from stage 5 to stage 7. Uridine tended to increase from stage 5 to stage 7. Adenosine was only detected at stage 7. The signals of cordycepin began to be detected during stage 6, and an increase was observed from stage 6 to stage 7. The pattern of cordycepin was similar to that of adenosine. Compared with the data from cultivation process 1, most of the observed amino acids increased slowly during cultivation process 2. Similar to the chemical changes in cultivation process 1, both the concentrations of α- and β-glucose increased during cultivation process 2, maintaining an α/β-anomer ratio of approximately 2:3. Glycerol decreased slightly during the cultivation process 2. The change of glycerol was negatively correlated with other detected components except for glycine (Fig. 8b). The adenosine signals were detected at stage 1, but the concentration hardly changed over time. The signals of cordycepin were first detected at stage 1, which was a mycelium state, and continued increasing during cultivation process 2 with a 2-fold greater amount compared to that from cultivation process 1. Thus, cordycepin can be synthesized at different growth stages in the cultivation processes with different disinfection steps, even at the stage of mycelia.
Figure 7.
Evolution of the NMR-detected components in the extracts from cultured C. militaris samples during two different cultivation processes. Blue lines represent cultivation process 1, and red lines represent cultivation process 2. Abbreviations are the same as those in Fig. 1. Data are represented as the means ± SD (n = 5).
Figure 8.
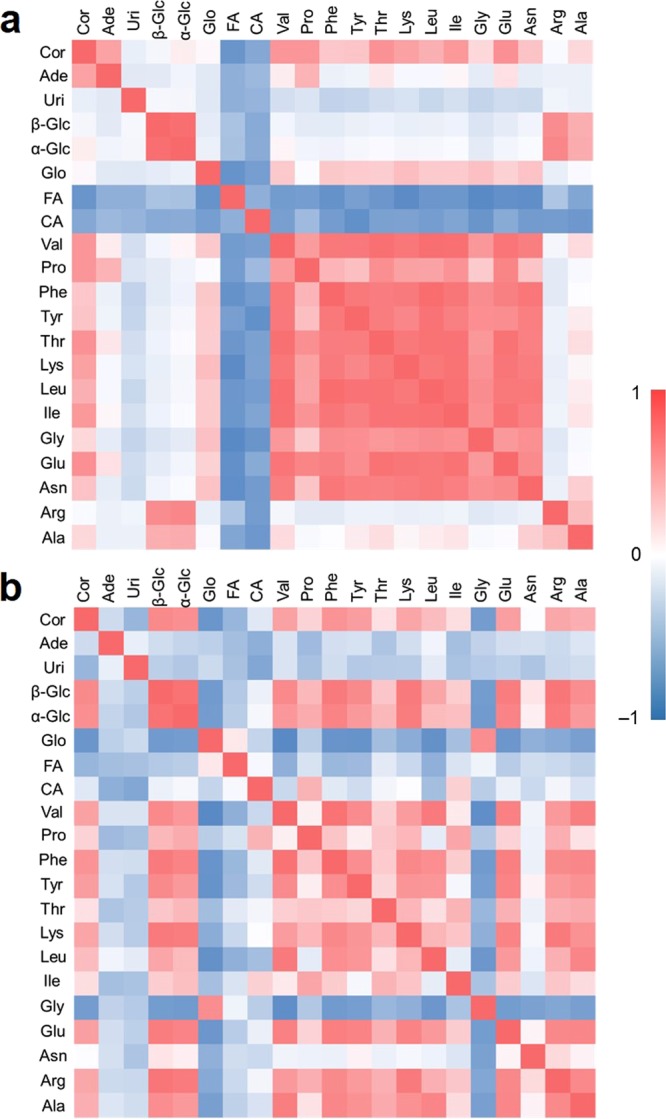
Correlation heatmaps among 21 compounds of the cultured C. militaris samples during (a) cultivation process 1 and (b) cultivation process 2. Abbreviations are the same as those in Fig. 1.
Reasons for the chemical changes during cultivation processes 1 and 2
The mycelium samples were directly disinfected in cultivation process 1, whereas the silkworm chrysalis media were disinfected before seeding of C. militaris in cultivation process 2. Cultivation processes 1 and 2 showed different patterns for the production of the amino acids. Based on the results shown in Figs 7 and 8, the amino acids are thought to be obtained via the decomposition of proteins. During cultivation process 1, the relatively higher concentrations of the amino acids were confirmed by comparing the results of cultivation process 2, which indicated that increased consumption of proteins for the production of amino acids occurred in cultivation process 1. Since the silkworm chrysalis medium was not disinfected in cultivation process 1, this observation suggests that the interaction between the C. militaris fungus and other microorganisms may cause more consumption of proteins, producing more amino acids during the cultivation process. Glucose was increased during both cultivation processes 1 and 2. Although glucose was thought to be consumed and decomposed into amino acids and organic acids, it can also be produced by polysaccharide degradation, which cannot be observed by NMR spectroscopy. Glycerol is also produced by polysaccharide degradation; however, the concentration of glycerol was not significantly increased in cultivation process 2. Glycerol may be effectively consumed as one of the carbon sources for the growth and/or metabolism of C. militaris without the interaction with other microorganisms.
Uridine and adenosine are hypothesized to be produced by phosphoribosyl diphosphate (PRPP), which can be obtained from glucose through the oxidative branch of the pentose phosphate pathway34. However, the cordycepin biosynthetic pathway has not been fully revealed. It is thought that cordycepin may be synthesized from adenosine, according to a previous study35, and thus the relatively higher amount of cordycepin in cultivation process 2 may be due to the higher amount of adenosine. The relatively lower amount of adenosine in cultivation process 1 may be caused by the influence of other microorganisms, which might consume adenosine as a nutrient source. Since cultivation process 2 included a disinfection method before seeding C. militaris, the C. militaris fungus could produce cordycepin more efficiently with less interaction with other microorganisms.
The difference between natural and cultured C. militaris products obtained by cultivation processes 1 and 2
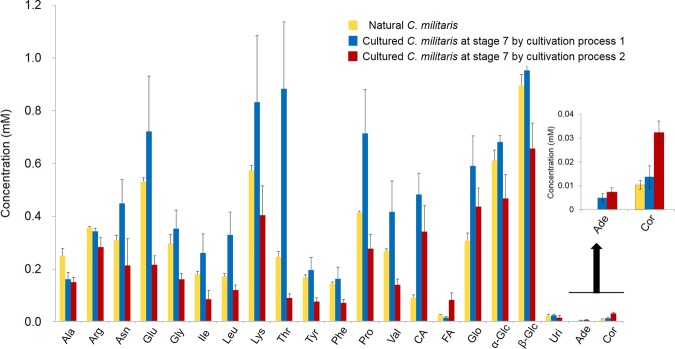
Figure 9 shows the concentrations of components in the extracts from the cultured C. militaris at stage 7 in cultivation processes 1 and 2 and the natural C. militaris samples. In the extracts of C. militaris obtained from cultivation process 1, the concentrations of the NMR-detected components, except for alanine, arginine and fumaric acid, were higher than those in natural C. militari. In the C. militaris extracts produced by cultivation process 2, the concentration of cordycepin was 3-fold higher than that in natural C. militaris. These results indicate that the C. militaris cultured using cultivation process 1 may be a good substitute for natural C. militaris and that cultivation process 2 could be applied to obtain larger amounts of cordycepin, instead of isolation from natural Cordyceps.
Figure 9.
Concentrations of the components in the extracts from natural (n = 3) and cultured C. militaris samples at stage 7 by cultivation processes 1 and 2 (n = 5). Abbreviations are the same as those in Fig. 1. Data are represented as the means ± SD.
Conclusions
The metabolic profiling of natural and cultured Cordyceps by NMR spectroscopy to investigate the cultivation processes was performed. It was discovered that the C. militaris fungus and silkworm chrysalis medium were suitable for the cultivation of Cordyceps. Furthermore, cordycepin can be biosynthesized at different growth stages in different cultivation processes, even at the stage of mycelia, and the C. militaris cultured in a silkworm medium that was disinfected before seeding contained three times more cordycepin than natural C. militaris. These findings will help further develop cultivation methods for Cordyceps for industrial uses and food engineering. The changes in the chemical composition in regarding functional components, such as cordycepin, may also be used to evaluate the influence of Cordyceps on human health.
Materials and Methods
Samples
Three types of natural Cordyceps (O. sinensis, C. militaris and C. nutans) and P. tenuipes samples were provided from 3 lots by Cordyce Co. Furthermore, 2 types of cultured C. militaris samples, cultured in a rice medium (Shanghai Jiao Tong University) and a silkworm chrysalis medium (Cordyce Co.), were provided from five lots of each type of Cordyceps sample from different cultivation processes. The mycelium samples were directly disinfected in cultivation process 1, whereas the silkworm chrysalis media were disinfected before the seeding of C. militaris in cultivation process 2. Then, they were transferred into sterilized moisturizing media. The samples of stage 1 were collected soon after disinfection of the mycelium samples, which were referred to as the 0-day state in the present study. The mycelia were incubated at 22 °C, and samples for stages 2 to 7 were collected every 5 days. Samples of the 7 growth stages were freeze-dried before the NMR measurements were taken. The 5 lots were provided for each stage of the C. militaris samples.
The Cordyceps samples were powdered using a Multi-beads Shocker (Yasui Kikai) with a MSH003(S) holder at 3000 rpm for 10 s. The samples (0.5 g) were soaked in 1.5 mL of deuterium oxide (99.7% Deuterium), and the mixture was shaken for 1 h using a vortex mixer at room temperature. The extracts were centrifuged at 15,000 × g at 20°C for 30 min, and 580 microliters supernatants were transferred to other tubes. Then, 20 μL of 1 mM 3-(trimethylsilyl)propanoic-2,2,3,3-d4 acid sodium salt (TSP-d4) in deuterium oxide was added to reach in the final concentration of 33 μM. The pH of the solution was measured with a pH meter. Then the solutions were transferred into 5-mm NMR tubes.
NMR spectroscopy
NMR measurements were performed at 293 K on an Agilent Unity INOVA-500 NMR spectrometer operating at 11.7 Tesla, observing 1H and 13C at 499.87 and 125.71 MHz, respectively and was equipped with a 5 mm triple resonance probe. 1H and 13C NMR chemical shifts are given in ppm related to TSP-d4 signal at 0.00 ppm as internal reference and coupling constants in Hz.
The quantitative 1H NMR spectra were acquired with pre-saturation of HDO signal using a PRESAT pulse sequence as follows: 64K datapoints distributed on a 6,000 Hz spectral width, providing a spectral resolution of 0.183 Hz; acquisition time, 5.461 s; recycle delay, 20 s; and 32 scans. To obtain a proper recycle delay time value, the spin-lattice relaxation time (T1) was determined using the partial relaxation method36,37. The 13C{1H} NMR spectra were acquired with aid of S2PUL pulse sequence as follows: 64K datapoints distributed on a 32,000 Hz spectral width, providing a spectral resolution of 0.977 Hz; acquisition time, 1.024 s; recycle delay, 2 s; and 100,000 scans. The quantification of the compounds has been performed by directly comparison of the signal area of a no overlapped signal for each compound in relation to the area of TSP-d4 signal at the concentration of 33 μM. NMR measurements for natural Cordyceps and P. tenuipes quantitation were performed in triplicate while those for cultured Cordyceps in quintuplet. The results in the figures and tables are expressed as the mean ± standard deviation (SD) based on the NMR data. The statistical analysis was performed using SAS University Edition (SAS Institute Inc.). Differences between the means were assessed using analysis of variance (ANOVA) followed by the Ryan-Einot-Gabriel-Welsch (REGWQ) multiple-range test. Differences were considered significant at p <0.05.
The 1H-1H DQF-COSY experiments were obtained by suppressing the HDO signal using the pre-saturation method, and the acquisition parameters were as follows: 32 scans in F2 (2K datapoints) per each 256 increments in F1; spectral widths, 6,000 Hz (F1 and F2); acquisition time, 0.171 s; and recycle delay, 2 s. The 1H-13C HSQC experiments were generated in the phase-sensitive mode with the following acquisition parameters: 64 scans in F2 (2K datapoints) per each 256 increments in F1; spectral widths, 6,000 Hz for 1H and 24,510 Hz for 13C; acquisition time, 0.171 s; and recycle delay, 2 s. The 1H-13C HMBC experiments were measured in the absolute mode with the following parameters: 64 scans in F2 (2K datapoints) per each 256 increment in F1; spectral widths, 6,000 Hz for 1H and 32,000 Hz for 13C; acquisition time, 0.171 s; and recycle delay, 2 s.
NMR signal assignments
The preprocessing of the free-induction decays (FIDs) and the subsequent Fourier transformations were performed using the MestReNova software (Mestrelab Research). To assign the signals, some NMR signals were analyzed by comparison with previously published data including NMR assignment data and composition data38–40. Although the 1H and 13C signals of the extracts from Cordyceps and P. tenuipes were shifted slightly relative to those in the published data due to the use of a different solvent, their correlations were not expected to change in the1H-1H DQF-COSY, 1H-13C HSQC and 1H-13C HMBC correlation maps. The signals were then confirmed and assigned to the candidate compounds on the basis of the 1H-1H DQF-COSY, 1H-13C HSQC and 1H-13C HMBC spectra.
Multivariate statistical analysis
The 1H NMR spectra from 0.20 to 9.40 ppm except HDO signal (4.75‒5.00 ppm) were data reduced by binning process in to equal 0.04 ppm buckets. The buckets area was then calculated and normalized to average sum method. For the multivariate statistical analysis, the resulting datasets were imported into SIMCA-P, version 13 (Umetrics). The quality of the model was described by its R2X and Q2 values. R2X was defined as the proportion of variance explained by the model, indicating the goodness of fit, and Q2 represented the predictability of the model26.
Supplementary information
Acknowledgements
We thank Y. Kondo (Cordyceps Corporation) for providing Cordyceps samples. This work was supported by a Grant-in-Aid for Scientific Research (S) from the Japan Society for the Promotion of Science (JSPS) (Grant No. 23228003) and Laboratory of Basic Science on Healthy Longevity.
Author Contributions
M.T. and Y.Z. designed the research. Y.L. performed the experiments. Y.L., T.M. and M.T. wrote the paper. M.T. edited the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44154-x.
References
- 1.Zhang Y, Li E, Wang C, Li Y, Liu X. Ophiocordyceps sinensis, the flagship fungus of China: terminology, life strategy and ecology. Mycology. 2012;3:2–10. [Google Scholar]
- 2.Paterson RRM. Cordyceps-A traditional Chinese medicine and another fungal therapeutic biofactory. Phytochemistry. 2009;69:1469–1495. doi: 10.1016/j.phytochem.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shrestha B, Zhang W, Zhang Y, Liu X. The medicinal fungus Cordyceps militaris: research and development. Mycol. Prog. 2012;11:599–614. doi: 10.1007/s11557-012-0825-y. [DOI] [Google Scholar]
- 4.Stensrud Ø, Hywel-Jones NL, Schumacher T. Towards a phylogenetic classification of Cordyceps: ITS nrDNA sequence data confirm divergent lineages and paraphyly. Mycol. Res. 2005;109:41–56. doi: 10.1017/S095375620400139X. [DOI] [PubMed] [Google Scholar]
- 5.Sasaki F, Miyamoto T, Yamamoto A, Tamai Y, Yajima T. Relationship between intraspecific variations and host insects of Ophiocordyceps nutans collected in Japan. Mycoscience. 2012;53:85–91. doi: 10.1007/S10267-011-0137-0. [DOI] [Google Scholar]
- 6.Hong IP, et al. Chemical components of Paecilomyces tenuipes (Peck) samson. Mycobiology. 2007;35:215–218. doi: 10.4489/MYCO.2007.35.4.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuo YC, et al. Regulation of bronchoalveolar lavage fluids cell function by the immunomodulatory agents from Cordyceps sinensis. Life Sci. 2001;68:1067–1082. doi: 10.1016/S0024-3205(00)01011-0. [DOI] [PubMed] [Google Scholar]
- 8.Cheng YJ, et al. Cordyceps sinensis prevents apoptosis in mouse liver with D-galactosamine/lipopolysaccharide-induced fulminant hepatic failure. Am. J. Chin. Med. 2014;42:427–441. doi: 10.1142/S0192415X14500281. [DOI] [PubMed] [Google Scholar]
- 9.Buenz EJ, Weaver JG, Bauer BA, Chalpin SD, Badley AD. Cordyceps sinensis extracts do not prevent Fas-receptor and hydrogen peroxide-induced T-cell apoptosis. J. Ethnoparmacol. 2004;90:57–62. doi: 10.1016/j.jep.2003.09.025. [DOI] [PubMed] [Google Scholar]
- 10.Wang BS, Lee CP, Chen ZT, Yu HM, Duh PD. Comparison of the hepatoprotective activity between cultured Cordyceps militaris and natural Cordyceps sinensis. J. Funct. Foods. 2012;4:489–495. doi: 10.1016/j.jff.2012.02.009. [DOI] [Google Scholar]
- 11.Yu R, et al. Structural elucidation and biological activity of a novel polysaccharide by alkaline extraction from cultured Cordyceps militaris. Carbohydr. Polym. 2009;75:166–171. doi: 10.1016/j.carbpol.2008.07.023. [DOI] [Google Scholar]
- 12.Nam KS, Jo YS, Kim YH, Hyun JW, Kim HW. Cytotoxic activities of acetoxyscirpenediol and ergosterol peroxide from Paecilomyces tenuipes. Life Sci. 2001;69:229–237. doi: 10.1016/S0024-3205(01)01125-0. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura K, et al. Combined effects of Cordyceps sinensis and methotrexate on hematogenic lung metastasis in mice. Receptors Channels. 2003;9:329–334. doi: 10.3109/713745176. [DOI] [PubMed] [Google Scholar]
- 14.Mao XB, Zhong JJ. Hyperproduction of cordycepin by two‐stage dissolved oxygen control in submerged cultivation of medicinal mushroom Cordyceps militaris in bioreactors. Biotechnol. Prog. 2004;20:1408–1413. doi: 10.1021/bp049765r. [DOI] [PubMed] [Google Scholar]
- 15.Mao XB, Eksriwong T, Chauvatcharin S, Zhong JJ. Optimization of carbon source and carbon/nitrogen ratio for cordycepin production by submerged cultivation of medicinal mushroom Cordyceps militaris. Process Biochem. 2005;40:1667–1672. doi: 10.1016/j.procbio.2004.06.046. [DOI] [Google Scholar]
- 16.Yang FQ, Feng K, Zhao J, Li SP. Analysis of sterols and fatty acids in natural and cultured Cordyceps by one-step derivatization followed with gas chromatography–mass spectrometry. J. Pharm. Biomed. Anal. 2009;49:1172–1178. doi: 10.1016/j.jpba.2009.02.025. [DOI] [PubMed] [Google Scholar]
- 17.Yang FQ, Ge L, Yong JWH, Tan SN, Li SP. Determination of nucleosides and nucleobases in different species of Cordyceps by capillary electrophoresis–mass spectrometry. J. Pharm. Biomed. Anal. 2009;50:307–314. doi: 10.1016/j.jpba.2009.04.027. [DOI] [PubMed] [Google Scholar]
- 18.Fan H, et al. Qualitative and quantitative determination of nucleosides, bases and their analogues in natural and cultured Cordyceps by pressurized liquid extraction and high performance liquid chromatography–electrospray ionization tandem mass spectrometry (HPLC–ESI–MS/MS) Anal. Chim. Acta. 2006;567:218–228. doi: 10.1016/j.aca.2006.03.032. [DOI] [Google Scholar]
- 19.Dong JZ, et al. Composition and characterization of cordyxanthins from Cordyceps militaris fruit bodies. J. Funct. Foods. 2013;5:1450–1455. doi: 10.1016/j.jff.2013.06.002. [DOI] [Google Scholar]
- 20.Pan Z, Raftery D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal. Bioanal. Chem. 2013;387:525–527. doi: 10.1007/s00216-006-0687-8. [DOI] [PubMed] [Google Scholar]
- 21.Defernez M, Colquhoun IJ. Factors affecting the robustness of metabolite fingerprinting using 1H NMR spectra. Phytochemistry. 2003;62:1009–1017. doi: 10.1016/S0031-9422(02)00704-5. [DOI] [PubMed] [Google Scholar]
- 22.Antalek B. Using PGSE NMR for chemical mixture analysis: quantitative aspects. Concepts Magn. Reson. 2007;30:219–235. doi: 10.1002/cmr.a.20093. [DOI] [Google Scholar]
- 23.Hu F, Furihata K, Ito-Ishida M, Kaminogawa S, Tanokura M. Nondestructive observation of bovine milk by NMR spectroscopy: analysis of existing states of compounds and detection of new compounds. J. Agric. Food Chem. 2004;52:4969–4974. doi: 10.1021/jf049616o. [DOI] [PubMed] [Google Scholar]
- 24.Liang T, et al. Comprehensive NMR analysis of compositional changes of black garlic during thermal processing. J. Agric. Food Chem. 2015;63:683–691. doi: 10.1021/jf504836d. [DOI] [PubMed] [Google Scholar]
- 25.Koda M, Furihata K, Wei F, Miyakawa T, Tanokura M. Metabolic discrimination of mango juice from various cultivars by band-selective NMR spectroscopy. J. Agric. Food Chem. 2012;60:1158–1166. doi: 10.1021/jf2041438. [DOI] [PubMed] [Google Scholar]
- 26.Wei F, Furihata K, Miyakawa T, Tanokura M. A pilot study of NMR-based sensory prediction of roasted coffee bean extracts. Food Chem. 2014;152:363–369. doi: 10.1016/j.foodchem.2013.11.161. [DOI] [PubMed] [Google Scholar]
- 27.Lu Y, Hu F, Miyakawa T, Tanokura M. Complex Mixture Analysis of Organic Compounds in Yogurt by NMR Spectroscopy. Metabolites. 2016;6:19. doi: 10.3390/metabo6020019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu G. Amino acids: metabolism, functions, and nutrition. Amino Acids. 2009;37:1–17. doi: 10.1007/s00726-009-0269-0. [DOI] [PubMed] [Google Scholar]
- 29.Basheer R, Strecker RE, Thakkar MM, McCarley RW. Adenosine and sleep-wake regulation. Prog. Neurobiol. 2004;73:379–396. doi: 10.1016/j.pneurobio.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura K, et al. Antitumor effect of cordycepin (3′-deoxyadenosine) on mouse melanoma and lung carcinoma cells involves adenosine A3 receptor stimulation. Anticancer Res. 2006;26:43–47. [PubMed] [Google Scholar]
- 31.Beach LR, Ross J. Cordycepin. An inhibitor of newly synthesized globin messenger RNA. J. Biol. Chem. 1978;253:2628–2632. [PubMed] [Google Scholar]
- 32.Wang S, et al. Simultaneous determination of nucleosides, myriocin, and carbohydrates in Cordyceps by HPLC coupled with diode array detection and evaporative light scattering detection. J. Sep. Sci. 2009;32:4069–4076. doi: 10.1002/jssc.200900570. [DOI] [PubMed] [Google Scholar]
- 33.François J, Parrou JL. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. Fems Microbiol. Rev. 2001;25:125–145. doi: 10.1111/j.1574-6976.2001.tb00574.x. [DOI] [PubMed] [Google Scholar]
- 34.Fan DD, Wang W, Zhong JJ. Enhancement of cordycepin production in submerged cultures of Cordyceps militaris by addition of ferrous sulfate. Biochem. Eng. J. 2012;60:30–35. doi: 10.1016/j.bej.2011.09.014. [DOI] [Google Scholar]
- 35.Aman S, Anderson DJ, Connolly TJ, Crittall AJ, Ji G. From adenosine to 3′-deoxyadenosine: development and scale up. Process Res. Dev. 2000;4:601–605. doi: 10.1021/op000209x. [DOI] [Google Scholar]
- 36.Hu F, Furihata K, Kato Y, Tanokura M. Nondestructive quantification of organic compounds in whole milk without pretreatment by two-dimensional NMR spectroscopy. J. Agric. Food Chem. 2007;55:4307–4311. doi: 10.1021/jf062803x. [DOI] [PubMed] [Google Scholar]
- 37.Lu Y, et al. Real-time monitoring of chemical changes in three kinds of fermented milk products during fermentation using quantitative difference nuclear magnetic resonance spectroscopy. J. Agric. Food Chem. 2018;66:1479–1487. doi: 10.1021/acs.jafc.7b05279. [DOI] [PubMed] [Google Scholar]
- 38.Park SJ, et al. Biochemical characterization of cultivated Cordyceps bassiana mycelia and fruiting bodies by 1H nuclear magnetic resonance spectroscopy. Metabolomics. 2013;9:236–246. doi: 10.1007/s11306-012-0442-4. [DOI] [Google Scholar]
- 39.Biological Magnetic Resonance Data Bank. http://www.bmrb.wisc.edu.
- 40.Spectral Database for Organic Compounds, SDBS. http://sdbs.db.aist.go.jp/sdbs/cgi-bin/direct_frame_top.cgi.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.