Brucella species cause brucellosis, a global zoonosis. A gene encoding a conserved DUF1849-family protein, which we have named EipB, is present in all sequenced Brucella and several other genera in the class Alphaproteobacteria. The manuscript provides the first functional and structural characterization of a DUF1849 protein. We show that EipB is secreted to the periplasm where it forms a spiral-shaped antiparallel β protein that is a determinant of cell envelope integrity in vitro and virulence in an animal model of disease. eipB genetically interacts with ttpA, which also encodes a periplasmic protein. We propose that EipB and TtpA function as part of a system required for cell envelope homeostasis in select Alphaproteobacteria.
KEYWORDS: Alphaproteobacteria, Brucella, DUF1849, PF08904, TPR, stress response
ABSTRACT
The Gram-negative cell envelope is a remarkable structure with core components that include an inner membrane, an outer membrane, and a peptidoglycan layer in the periplasmic space between. Multiple molecular systems function to maintain integrity of this essential barrier between the interior of the cell and its surrounding environment. We show that a conserved DUF1849 family protein, EipB, is secreted to the periplasmic space of Brucella species, a monophyletic group of intracellular pathogens. In the periplasm, EipB folds into an unusual 14-stranded β-spiral structure that resembles the LolA and LolB lipoprotein delivery system, though the overall fold of EipB is distinct from LolA/LolB. Deletion of eipB results in defects in Brucella cell envelope integrity in vitro and in maintenance of spleen colonization in a mouse model of Brucella abortus infection. Transposon disruption of ttpA, which encodes a periplasmic protein containing tetratricopeptide repeats, is synthetically lethal with eipB deletion. ttpA is a reported virulence determinant in Brucella, and our studies of ttpA deletion and overexpression strains provide evidence that this gene also contributes to cell envelope function. We conclude that eipB and ttpA function in the Brucella periplasmic space to maintain cell envelope integrity, which facilitates survival in a mammalian host.
IMPORTANCE Brucella species cause brucellosis, a global zoonosis. A gene encoding a conserved DUF1849-family protein, which we have named EipB, is present in all sequenced Brucella and several other genera in the class Alphaproteobacteria. The manuscript provides the first functional and structural characterization of a DUF1849 protein. We show that EipB is secreted to the periplasm where it forms a spiral-shaped antiparallel β protein that is a determinant of cell envelope integrity in vitro and virulence in an animal model of disease. eipB genetically interacts with ttpA, which also encodes a periplasmic protein. We propose that EipB and TtpA function as part of a system required for cell envelope homeostasis in select Alphaproteobacteria.
INTRODUCTION
Brucella spp. are the causative agents of brucellosis, which afflicts wildlife and livestock on a global scale and can occur in humans through contact with infected animals or animal products (1, 2). These intracellular pathogens are members of the class Alphaproteobacteria, a group of Gram-negative species that exhibit tremendous diversity in metabolic capacity, cell morphology, and ecological niches (3). In their mammalian hosts, Brucella cells must contend with the host immune system (4) and adapt to stresses, including oxidative assault from immune cells, acidic pH in the phagosomal compartment, and nutrient shifts during intracellular trafficking (5). Molecular components of the cell envelope play a key role in the ability of Brucella spp. to survive these stresses and to replicate in the intracellular niche (6, 7). As part of a systematic experimental survey of conserved Alphaproteobacterial protein domains of unknown function (DUFs), we recently described envelope integrity protein A (EipA). This periplasmic protein confers resistance to cell envelope stressors and determines Brucella abortus virulence in a mouse model of infection (8). In this study, we report a functional and structural analysis of envelope integrity protein B (EipB), a member of the uncharacterized gene family DUF1849.
DUF1849 (Pfam PF08904 [9]) is widespread among the Rhizobiales, Rhodospirillales, and Rhodobacterales (Fig. 1). To our knowledge, no functional data have been reported for this gene family other than results from a recent multispecies transposon sequencing (Tn-seq) study that showed stress sensitivity in Sinorhizobium meliloti DUF1849 (locus SMc02102) mutant strains (10). Here we show that the Brucella DUF1849 protein, EipB (locus tag bab1_1186; RefSeq locus BAB_RS21600), is a 280-residue periplasmic protein that folds into a 14-stranded, open β-barrel structure containing a conserved disulfide bond. We term this novel barrel structure a β-spiral and show that it resembles the lipoprotein chaperone LolB, though its overall fold is distinct. Replication and survival of a B. abortus strain from which we deleted eipB were attenuated in a mouse infection model, and deletion of eipB in both Brucella abortus and Brucella ovis enhanced sensitivity to compounds that affect the integrity of the cell envelope. We have further shown that B. abortus eipB deletion is synthetically lethal with transposon disruption of gene locus bab1_0430, which encodes a periplasmic tetratricopeptide-repeat (TPR)-containing protein that we have named TtpA. The Brucella melitensis ortholog of TtpA (locus tag BMEI1531) has been previously described as a molecular determinant of mouse spleen colonization (11), while a Rhizobium leguminosarum TtpA homolog (locus tag RL0936) is required for proper cell envelope function (12). We propose that TtpA and EipB coordinately function in the Brucella periplasm to ensure cell envelope integrity and to enable cell survival in the mammalian host niche.
FIG 1.
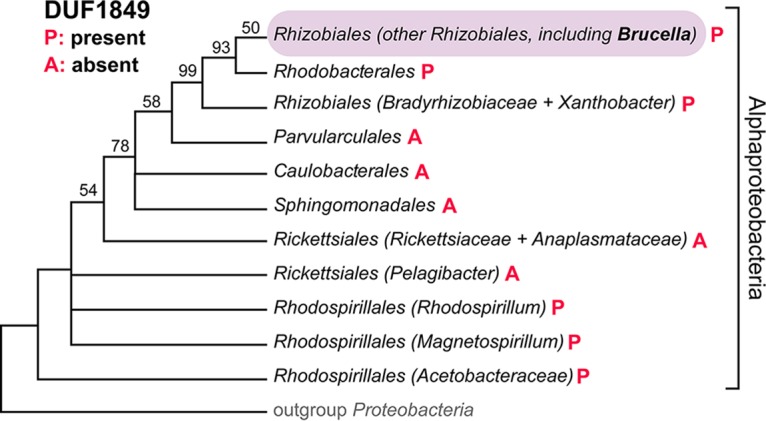
The DUF1849 sequence family is restricted to Alphaproteobacteria. A Bayesian phylogenetic tree shows the distribution of DUF1849 genes in different orders within the class Alphaproteobacteria (P, present; A, absent). Bayesian support values are shown when <100%; nodes were collapsed when support was <50% (adapted from the work of Williams et al. [50]). In Brucella abortus (order Rhizobiales), DUF1849 is encoded by gene locus bab1_1186 (i.e., eipB).
RESULTS
B. abortus eipB is required for maintenance of mouse spleen colonization.
As part of a screen to evaluate the role of conserved Alphaproteobacterial genes of unknown function in B. abortus infection biology, we infected THP-1 macrophage-like cells with wild-type (WT) B. abortus, an eipB deletion (ΔeipB) strain, and a genetically complemented ΔeipB strain. Infected macrophages were lysed, and CFU were enumerated on tryptic soy agar (TSA) plates at 1, 24 and 48 h postinfection. We observed no significant differences between strains at 1, 24 or 48 h postinfection, indicating that eipB was not required for entry, replication or intracellular survival in vitro (Fig. 2A).
FIG 2.
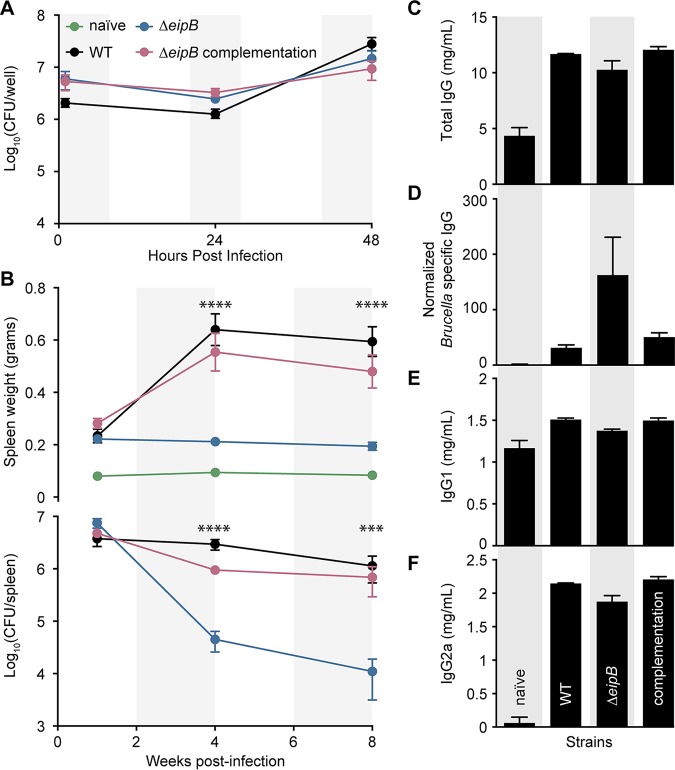
eipB is a genetic determinant of B. abortus virulence. (A) In vitro macrophage infection assay: infection of THP-1 cells with wild-type B. abortus 2308 (WT), the ΔeipB strain, and the eipB complementation strain. The numbers of B. abortus CFU recovered from the THP-1 cells at 1, 24, and 48 h postinfection are plotted. Each data point (n = 3 per strain) indicates the mean ± the standard error of the mean. (B) In vivo mouse infection assay: female BALB/c mice were injected intraperitoneally with the wild-type, ΔeipB, or ΔeipB complementation strain. Spleen weights (upper graph) and bacterial burdens (lower graph) were measured at 1, 4, and 8 weeks postinfection. Naïve, uninfected, naive mice. Data are means ± the standard errors of the mean (n = 5 mice per strain per time point). One-way analysis of variance (ANOVA), followed by Dunnett’s posttest (to wild type), supports the conclusion that spleens infected with the eipB deletion strain were significantly smaller at 4 (****, P < 0.0001) and 8 (****, P < 0.0001) weeks and had fewer CFU than spleens infected with the wild type at 4 (****, P < 0.0001) and 8 (***, P < 0.0007) weeks. (C to F) Antibody quantification in mouse sera harvested at 8 weeks postinfection from naive control mice or from mice infected with the wild-type, ΔeipB, or complementation strain. The amounts of total IgG (C), Brucella-specific IgG (D), IgG1 (E), and IgG2a (F) at 8 weeks were determined by ELISA. Each data point (naive, n = 3; WT, n = 2, ΔeipB; complementation, n = 4) indicates the mean ± the standard error of the mean.
We further evaluated the role of eipB in a BALB/c mouse infection model. Mice infected with the ΔeipB strain had no significant differences in spleen weight or bacterial load compared to mice infected with wild-type B. abortus strain 2308 at 1 week postinfection (Fig. 2B). However, at 4 and 8 weeks postinfection, mice infected with the wild type or with the complemented eipB deletion strains had pronounced splenomegaly and a bacterial load of approximately 5 × 106 CFU/spleen. In contrast, mice infected with the ΔeipB strain had smaller spleens, with approximately 2 orders of magnitude fewer bacteria (∼1 × 104 CFU/spleen) (Fig. 2B). We conclude that eipB is not required for initial spleen colonization but is necessary for full virulence and persistence in the spleen over an 8-week time course.
To assess the pathology of mice infected with wild-type and ΔeipB strains, we harvested spleens at 8 weeks postinfection and fixed, mounted, and subjected the samples to hematoxylin and eosin (H&E) staining (see Fig. S1 in the supplemental material). Compared to naive (uninfected) mice (Fig. S1A), we observed higher extramedullary hematopoiesis, histiocytic proliferation, granulomas, and the presence of Brucella immunoreactivities in spleens of mice infected with wild-type B. abortus 2308 and the genetically complemented mutant strain (Fig. S1B and D). Both the wild type and the complemented strain caused spleen inflammation with a reduced white to red pulp ratio as a result of lymphoid follicle disruption and red pulp expansion, which typically correlates with infiltration of inflammatory cells; these spleens also had increased marginal zones (Fig. S1B and D). As expected from the CFU enumeration data, mice infected with the ΔeipB strain had reduced pathological features: there was minimal change in the white/red pulp ratio and a minimal increase in marginal zones (Fig. S1C). There was no evidence of extramedullary hematopoiesis in mice infected with the ΔeipB strain, though histiocytic proliferation was mildly increased. Granulomas and Brucella immunoreactivities were rare in the ΔeipB strain (Fig. S1C). These results are consistent with a model in which eipB is required for full B. abortus virulence in a mouse model of infection. A summary of spleen pathology scores is presented in Table S1.
We further measured antibody responses in mice infected with ΔeipB and wild-type strains. The serum levels of total IgG, Brucella-specific IgG, subclass IgG1, and subclass IgG2a were measured by enzyme-linked immunosorbent assays (ELISA) (Fig. 2C to F). Antibody subclasses IgG2a and IgG1 were measured as markers of T helper 1 (Th1)- and Th2-specific immune responses, respectively. At 8 weeks postinfection, the total serum IgG level was higher in all infected mice relative to the uninfected control (Fig. 2C). The level of Brucella-specific IgG was approximately 5 times higher in ΔeipB strain-infected mice than in mice infected with wild type or the complemented mutant strain (Fig. 2D). Uninfected mice and mice infected with wild-type, ΔeipB, and ΔeipB-complemented strains showed no significant differences in IgG1 levels after 8 weeks (Fig. 2E). All infected mice had highly increased levels of IgG2a at 8 weeks postinfection relative to naive mice, though there was no difference between B. abortus strains (Fig. 2F). We conclude that ΔeipB strain infection results in the production of more B. abortus-specific antibodies than does infection with the wild type. Subclasses IgG1 and IgG2a do not apparently account for the higher levels of these specific antibodies. Large induction of IgG2a by all B. abortus strains is consistent with the known ability of B. abortus to promote a strong Th1 response (13, 14). However, the ΔeipB strain does not induce a more robust Th1 response than does the wild type based on our IgG2a measurements. We did not test whether antibodies contribute to clearance of the ΔeipB strain. Enhanced Brucella-specific antibody production may simply be a consequence of antigen release triggered by host clearance of the ΔeipB strain by other immune mechanisms.
The ΔeipB strain is sensitive to cell envelope stressors.
To test whether reduced virulence of the ΔeipB strain correlates with an increased sensitivity to stress in vitro, we evaluated B. abortus ΔeipB strain growth on TSA plates supplemented with known cell membrane/envelope stressors, including EDTA, ampicillin, and deoxycholate. The ΔeipB strain had 1.5 to 3 orders of magnitude fewer CFU compared to the wild type when titers were determined on TSA plates containing these compounds. All phenotypes were complemented by restoring the ΔeipB locus to the wild type (Fig. 3A). Together, these data provide evidence that eipB contributes to the resistance to compounds that compromise the integrity of the B. abortus cell membrane/envelope.
FIG 3.
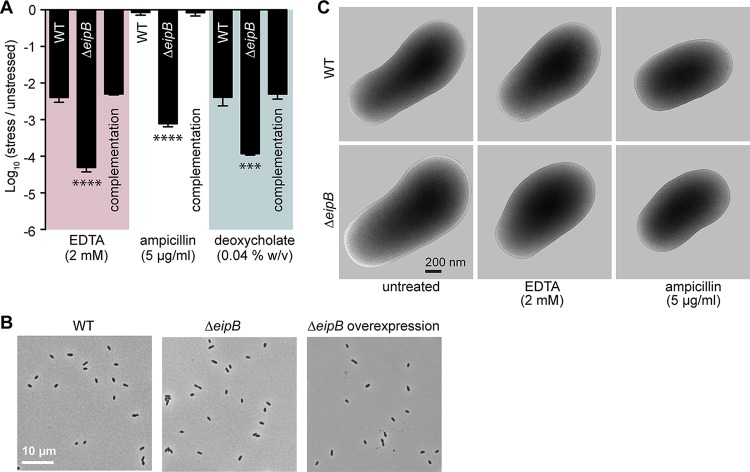
Assessing the effect of cell envelope stressors on B. abortus ΔeipB growth and survival. (A) Envelope stress survival assays. Serially diluted cultures of B. abortus wild-type, ΔeipB, and complementation strains were spotted onto plain TSA plates or TSA plates containing EDTA (2 mM), deoxycholate (0.04%, wt/vol), or ampicillin (5 μg/ml). After 3 to 5 days of growth at 37°C and 5% CO2, CFU for each condition were enumerated and plotted. This experiment was repeated four times; each data point indicates the mean ± the standard error of the mean. One-way ANOVA, followed by Dunnett’s posttest (to wild-type), supported the conclusion that the eipB deletion strain had significantly fewer CFU than the wild type in the presence of EDTA (****, P < 0.0001), ampicillin (****, P < 0.0001), and deoxycholate (***, P < 0.0003). (B) Light micrograph of B. abortus wild-type (left), ΔeipB (middle), and overexpression (right; induced with 5 mM IPTG) liquid cultures grown overnight in brucella broth. (C) Cryo-EM images of B. abortus wild-type and ΔeipB cells cultivated in liquid broth that either remained untreated or were treated with 2 mM EDTA or 5 μg/ml ampicillin for 4 h.
Although ΔeipB CFU were reduced relative to the wild type on agar plates containing all three envelope stressors that we assayed, we observed no apparent defects in ΔeipB cell morphology by light microscopy or cryo-electron microscopy when the strain was cultivated in liquid broth (Fig. 3B and C). Incubation of the ΔeipB strain with 2 mM EDTA or 5 μg/ml ampicillin (final concentration) in brucella broth for 4 h also had no apparent effect on cell structure, nor did eipB overexpression (Fig. 3B and C). Longer periods of growth in the presence of stressors may be required for differences in cell morphology/structure to be evident in broth. It may also be the case that the envelope stress phenotypes we observe are particular to growth on solid medium.
B. abortus ΔeipB agglutination phenotypes indicate the presence of smooth LPS.
In B. abortus, smooth lipopolysaccharide (LPS; containing O-polysaccharide) is an important virulence determinant (15). Smooth LPS can also act as a protective layer against treatments that compromise the integrity of the cell envelope (16). Loss of smooth LPS in the B. abortus ΔeipB strain could therefore explain the phenotypes we observe for this strain. To test this hypothesis, we assayed wild-type and ΔeipB agglutination in the presence of serum from a B. abortus-infected mouse. A major serological response to smooth Brucella species is to O-polysaccharide (17), and thus agglutination can provide an indirect indication of the presence or absence of smooth LPS on the surface of the cell. Both wild-type and ΔeipB strains agglutinated in the presence of serum from a B. abortus-infected mouse, providing evidence for the presence of O-polysaccharide in ΔeipB (Fig. S2A). As a negative control, we incubated the naturally rough species B. ovis with the same serum; B. ovis did not agglutinate in the presence of this serum (Fig. S2A). We further assayed agglutination of B. abortus wild-type and ΔeipB strains in the presence of acriflavine, which is demonstrated to agglutinate rough strains such as B. ovis (18, 19). After 2 h of incubation, we observed no agglutination of wild-type or ΔeipB B. abortus (Fig. S2B). We treated B. ovis with acriflavine as a positive control and observed agglutination, as expected (Fig. S2B). Together, these data indicate that deletion of eipB does not result in a loss of smooth LPS. However, we cannot rule out the possibility that the chemical structure of O-polysaccharide is altered in the ΔeipB strain.
EipB is a monomeric protein that is secreted to the periplasm.
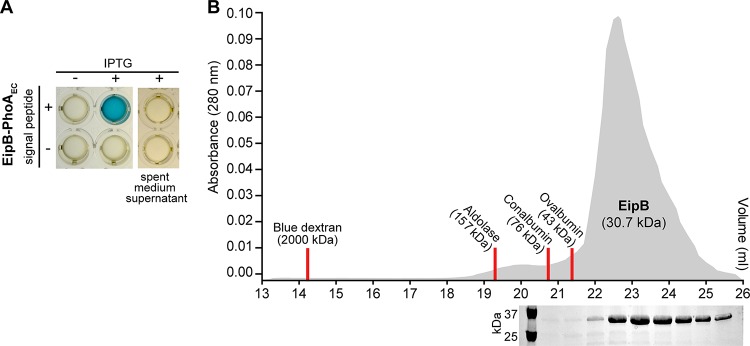
The N terminus (residues M1 to A30 [M1-A30]) of Brucella EipB contains a predicted signal peptide based on SignalP 4.2 analysis (20). EipB (DUF1849) homologs in other Alphaproteobacteria also have a predicted N-terminal secretion signal (Fig. S3). We note that EipB in our wild-type B. abortus 2308 strain has a methionine instead of a leucine at position 250. These two amino acids are interchangeable at this position in DUF1849 (Fig. S4). To test the prediction that EipB is a periplasmic protein, we fused the Escherichia coli periplasmic alkaline phosphatase gene (phoA) to B. abortus eipB and expressed fusions from a lac promoter in B. ovis. We generated (i) the full-length EipB protein (M1-K280) fused at its C terminus to E. coli PhoA (EipB-PhoAEc) and (ii) an EipB-PhoA fusion lacking the hypothetical EipB signal peptide sequence (EipBS29-K280-PhoAEc). After overnight growth in brucella broth in presence or absence of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), we adjusted each culture to the same density and loaded into a 96-well plate containing BCIP (5-bromo-4-chloro-3-indolylphosphate; final concentration, 200 μg/ml). BCIP is hydrolyzed to a blue pigment by PhoA, which can be measured colorimetrically. BCIP diffusion through the inner membrane is inefficient, and thus this reagent can be used to specifically detect PhoA activity in the periplasmic space or in the extracellular medium (21). After a 2-h incubation at 37°C, the well containing the B. ovis cells expressing the EipBM1-K280-PhoAEc fusion turned dark blue. We observed no color change in the well containing the B. ovis strain expressing the EipBS29-K280-PhoAEc protein fusion (Fig. 4A). As expected, no color change was observed in the absence of induction with 1 mM IPTG (Fig. 4A). To test whether EipB is secreted from the cell into the growth medium, we performed a similar experiment on spent medium supernatants from the different cultures. We observed no color change in these samples after 2 h of incubation, providing evidence that EipBM1-K280-PhoAEc is not secreted from the cell.
FIG 4.
EipB is monomeric in solution and is secreted to the Brucella periplasm. (A) Alkaline phosphatase assay. Overnight cultures of B. ovis expressing EipB with (+) or without (–) its signal peptide and fused to E. coli PhoA were grown in the presence (+) or absence (–) of 1 mM IPTG inducer. In a 96-well plate, these cultures were mixed with BCIP (200 μg/ml, final concentration) and developed for 2 h at 37°C and 5% CO2. Only the strain expressing EipB-PhoAEc with a signal peptide turned blue, providing evidence that the protein is located in the periplasm. As a control, spent medium supernatants were mixed with BCIP to test whether EipB-PhoAEc is secreted into the medium. After 2 h of incubation, no color change was observed, indicating that EipB-PhoAEc is not exported outside the cell. These experiments were performed at least three times with independent clones. A representative image is shown. (B) Size exclusion chromatography elution profile of purified EipB (in gray). Elution fractions were subjected to SDS-PAGE, and a single band migrating at ∼30 kDa was visible. Elution peaks of the molecular weight standards are shown as red lines. This experiment was performed twice and yielded similar elution profiles.
We further assayed the oligomeric state of affinity-purified B. abortus EipB in solution by size exclusion chromatography. The calculated molecular mass of His6-EipB (V31-K280) is 30.7 kDa. This protein eluted from a sizing column at a volume with an apparent molecular mass of ∼23 kDa, which is consistent with a monomer (Fig. 4B). There was no evidence of larger oligomers determined by size exclusion chromatography. From these data, we conclude that EipB is a monomeric periplasmic protein.
EipB folds into a spiral-like β-sheet that resembles PA1994, LolA, and LolB.
We postulated that the three-dimensional structure of EipB may provide molecular-level insight into its function in the cell. As such, we solved an X-ray crystal structure of B. abortus EipB (residues A30 to K280; PDB ID 6NTR). EipB lacking its signal peptide formed triclinic crystals (a = 47.4 Å, b = 69.2 Å, c = 83.2 Å, α = 90.1°, β = 90.0°, γ = 78.7°) that diffracted to 2.1-Å resolution; we refined this structure to Rwork= 0.195 and Rfree= 0.245. Crystallographic data and refinement statistics are summarized in Table S2. Four EipB molecules (chains A to D) are present in the crystallographic asymmetric unit.
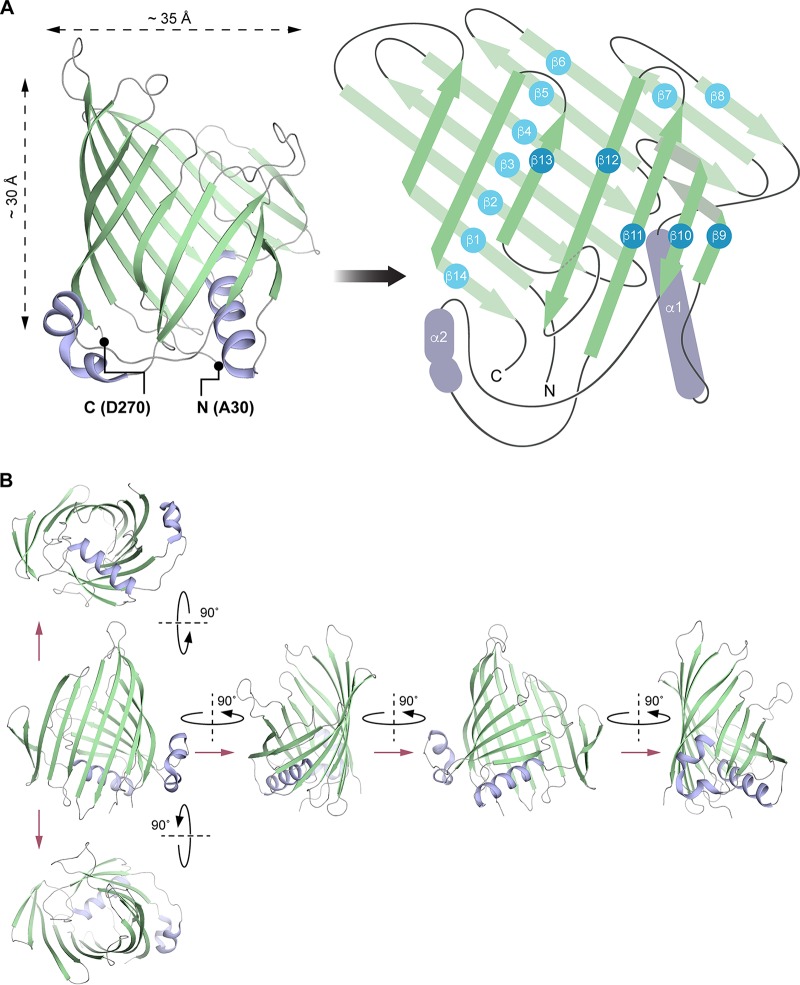
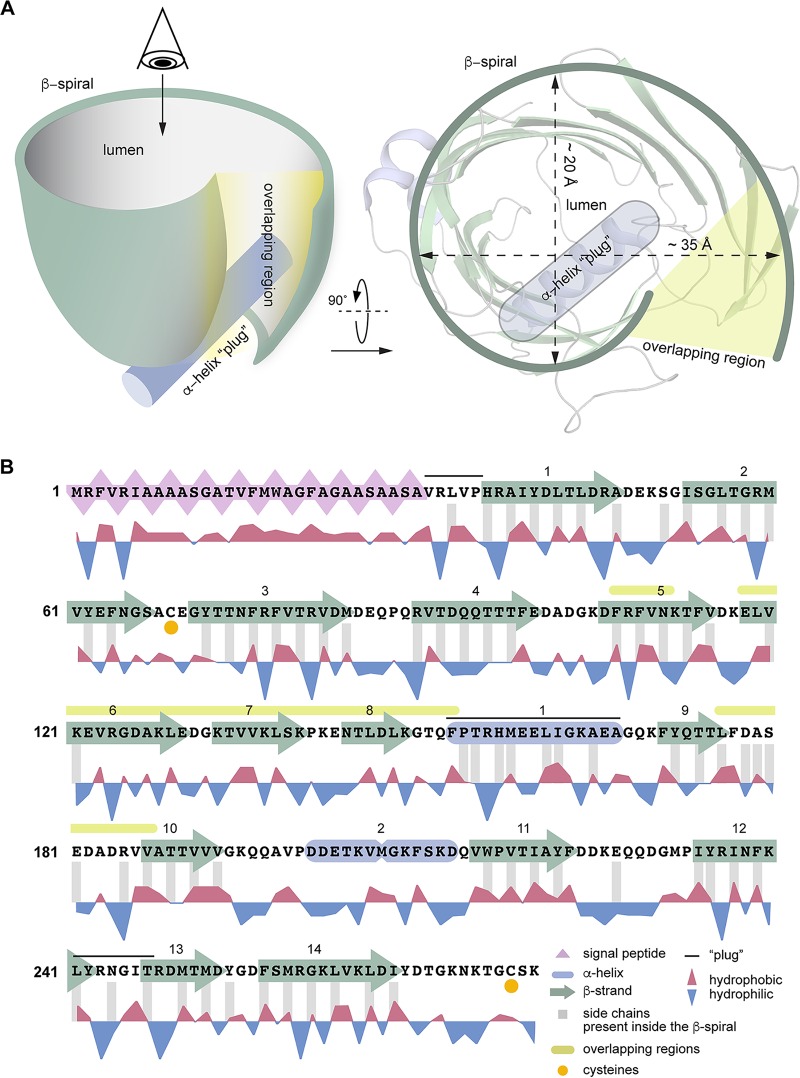
Each EipB monomer consists of 14 antiparallel β-strands (β1 to β14) forming an oval, spiral-like β-sheet (minor axis diameter, ∼25 Å; major axis diameter, ∼35 Å). Two regions of this β-spiral, involving β5, β6, β7, and β8, and the hairpin loop connecting β9 and β10 overlap (Fig. 5). Interactions between these two overlapping portions of structure are mostly hydrophobic, though polar contacts are also found in these regions (Fig. 5 and 6). One side of the spiral is occluded by the N terminus, a loop connecting β-strands 12 and 13, and α-helix 1, which form the bottom of this “cup” shaped protein (Fig. 5 and 6A). The external surface of EipB is positively and negatively charged and also presents small hydrophobic patches (Fig. S5); one helix, α2, is kinked and positioned at the surface of the cylindrical β-spiral (Fig. 5). The lumen of EipB is solvent accessible and is partially filled with the side chains of hydrophobic or acidic residues. Hydrophobic residues represent ∼66% of the residues present inside the EipB cavity (Fig. 5 and 6B). The size of this cavity suggests that EipB, in this conformation, can accommodate small molecules or ligands in its lumen.
FIG 5.
EipB adopts a β-spiral fold. (A, left) X-ray structure of EipB. EipB consist of 14 β-strands (in green) and 2 α-helices (in violet). The N terminus (A30) and the C terminus (D270) are reported on this structure. (Right) Simplified representation of EipB. The color code is the same as for the left panel. (B) Different orientations of EipB structure. The color code is the same as for panel A.
FIG 6.
Simplified representation of EipB structure. (A) EipB adopts a cuplike structure, 14 β-strands (in green) form an overlapping β-spiral (β5-β6-β7-β8 overlap with the β9-β10 connecting loop, highlighted in yellow in panels A and B). α1 (in violet) and the loop connecting β12 and β13 form the bottom of this “cup.” (B) Amino acid sequence of EipB. The sequence corresponding to the predicted signal peptide is highlighted in pink. Hydrophobic and hydrophilic residues are reported below the sequence. Residues with side chains present inside EipB cavity are highlighted with gray bars. Cysteines C69 and C278 are highlighted with orange dots. Structural elements forming the bottom of the β-spiral are highlighted with a black line.
We searched the EipB structure against the protein structure database using Dali (22) but failed to identify clear structural homologs. Pseudomonas aeruginosa PA1994 (PDB ID 2H1T) (23) was the closest structural match to EipB (root mean square deviation, ∼3.5; Z-score, ∼11) (Fig. S6A). Despite very low sequence identity (∼8%), PA1994 has noticeable structural similarities to EipB: it adopts a spiral-like β-fold involving 15 β-strands, which is occluded at one end with a long α-helix. Unlike EipB, PA1994 lacks a signal peptide and is predicted to be a cytoplasmic protein. Structural parallels between PA1994 and the periplasmic lipoprotein chaperones LolA/LolB have been noted and a role for PA1994 in glycolipid metabolism has been postulated (23), though this prediction remains untested. Like PA1994, EipB has structural similarities to LolA and LolB, in particular the antiparallel and curved β-sheet scaffold that engulfs a central α-helical plug (Fig. S6B). Whether Brucella EipB or DUF1849 proteins more generally function in trafficking lipoproteins or other molecules in the periplasm remains to be tested.
EipB has a conserved disulfide bond.
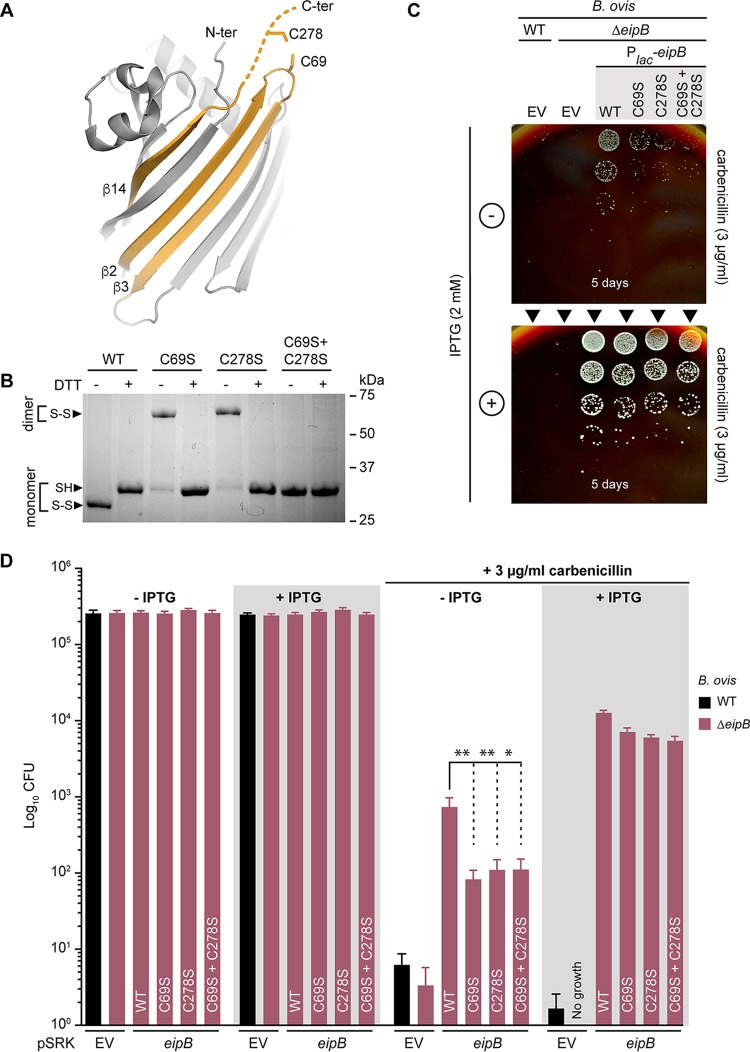
We identified two cysteines in EipB, C69 and C278, which are the two most conserved residues in the DUF1849 sequence family (Fig. S3 and S4). C69 is solvent exposed in Brucella EipB and positioned in a loop connecting β2 and β3. C278 is present at the C terminus of the protein, which immediately follows β14. β14 interacts with β13 and β1 and is spatially proximal to β2 and β3 (Fig. 7A). Given the proximity of these two cysteines in the EipB structure, we hypothesized that C69 and C278 form an internal disulfide bond. However, electron density for the 10 C-terminal residues (containing C278) is not well resolved in the EipB crystal structure, and a disulfide bond is not evident, likely because the protein was dialyzed against a buffer containing 2 mM 1,4-dithiothreitol (DTT) prior to crystallization.
FIG 7.
EipB has an internal disulfide bond. (A) Cysteines C69 and C278 are spatially proximal in the EipB structure and form a disulfide bond. C278 is present at the EipB C terminus that follows β14, and C69 is present in a loop connecting β2 and β3. (B) His-tagged wild-type EipB and EipB cysteine mutant proteins (C69S, C278S, and C69S+C278S) were purified and mixed with a protein loading buffer with (+) or without (−) 1 mM DTT. Protein samples were resolved by SDS–12% PAGE. This experiment was performed three times. The picture of a representative gel is presented. (C) Growth on SBA plates containing 3 μg/ml of carbenicillin with (+) or without (–) 2 mM IPTG of a serially diluted (10-fold dilution) B. ovis ΔeipB strain ectopically expressing wild-type EipB (Plac-eipB), C69S mutant (Plac-eipBC69S), C278S mutant (Plac-eipBC278S), or C69S+C278S mutant (Plac-eipBC69S+C278S). B. ovis wild-type (WT) and ΔeipB carrying the pSRK empty vector (EV) were used as a control. The days of growth at 37°C and 5% CO2 are reported for each plate. A representative picture of the different plates is presented. (D) Enumerated CFU after growth on SBA plates containing 3 μg/ml of carbenicillin with (+) or without (–) 2 mM IPTG of serially diluted (10-fold dilution) B. ovis ΔeipB strains expressing different versions of eipB from a plasmid (wild type and cysteine mutants; see panel C legend). EV strains and SBA plates with no carbenicillin, with or without IPTG, were used as controls. This experiment was independently performed twice, with two different clones each time, and all plate assays were done in triplicate. Each data point indicates the mean ± the standard error of the mean. One-way ANOVA, followed by Dunnett’s posttest (to the wild type), supports the conclusion that eipB-dependent protection against the cell wall antibiotic, carbenicillin, is significantly diminished when disulfide-forming residues C69 (**, P < 0.005) and C278 (**, P < 0.003) are individually or both (*, P < 0.01) mutated to serine. This is effect is evident with leaky eipB expression from Plac but diminished when the expression of wild-type and mutant eipB alleles is induced by IPTG.
To biochemically test whether these two cysteines form a disulfide bond, we purified B. abortus EipB under nonreducing conditions and mixed the protein with sodium dodecyl sulfate (SDS) gel loading dye with or without 1 mM DTT. We observed two bands that migrated differently in the 30-kDa region when the protein was resolved by 12% SDS-PAGE. EipB without DTT migrated farther than the DTT-treated protein, suggesting the presence of a disulfide bond (Fig. 7B). We performed this same experiment with three different EipB cysteine mutant proteins in which C69, C278, or both were mutated to serine. In the absence of DTT, EipBC69S and EipBC278S migrated at an apparent molecular weight of ∼60 kDa, corresponding to a dimeric EipB interacting through a S-S bond. After DTT treatment, these mutant proteins migrated the same as the reduced wild-type protein (Fig. 7B). As expected, the double cysteine mutant (EipBC69S+C278S) did not form an apparent dimer and was unaffected by DTT (Fig. 7B). From these data, we conclude that an internal disulfide bond can form between C69 and C278 in EipB and is likely present in vivo, since EipB resides in the oxidizing environment of the periplasm.
To test whether this disulfide bond affects EipB function, we measured CFU of a Brucella ovis ΔeipB (Δbov_1121) strain expressing wild-type B. abortus EipB or cysteine disulfide mutants on agar plates containing 3 μg/ml carbenicillin. B. ovis is a closely related biosafety level 2 (BSL2) surrogate for B. abortus. B. ovis and B. abortus EipB are identical with the exception of one amino acid at position 250 (Fig. S4). In this carbenicillin assay (Fig. 7C and D), B. abortus EipB complemented a B. ovis ΔeipB strain, suggesting that the substitution at residue 250 does not impair EipB function. We placed four different versions of eipB under the control of a lac promoter (Plac): Plac-eipBWT, Plac-eipBC69S, Plac-eipBC278S, and Plac-eipBC69S+C278S; the empty vector (EV) was used as a control. After 5 to 6 days of growth on Schaedler blood agar (SBA) plates containing 3 μg/ml of carbenicillin and no IPTG, we observed poor growth at only the lowest dilution for wild-type and ΔeipB strains carrying the empty vector control (also see Fig. S7A for an example of growth on 2 μg/ml carbenicillin plates). Corresponding colonies for the strains carrying the different Plac-eipB overexpression plasmids were more abundant though very small in the absence of IPTG induction. However, the strain harboring the wild-type eipB plasmid systematically grew at a 1-log-higher dilution than the cysteine mutant strains, indicating that the presence of the disulfide bond in eipB contributes to carbenicillin resistance on solid medium (Fig. 7C and D; see also Fig. S7A). These results indicate some level of leaky expression from the multicopy Plac-eipB plasmids. When induced with IPTG, overexpression of the different EipB variants enhanced growth in all strains (Fig. 7C and D). As expected, strains grown on control plates without carbenicillin had no growth defect, with or without IPTG induction (Fig. 7D). The morphology of B. ovis ΔeipB strains expressing the different variants of eipB appeared normal by phase-contrast microscopy (see Fig. S7B). These results provide evidence that EipB is necessary for full carbenicillin resistance in B. ovis and that cysteines 69 and 278 contribute to EipB function in vivo.
To evaluate the effect of these two cysteines on EipB stability in vitro, we measured the thermal stability of purified wild-type B. abortus EipB (EipBWT) and double cysteine mutant (EipBC69S+C278S) in presence or absence of 2 mM DTT. EipBWT melted at ∼46°C in the absence of DTT and at ∼41.5°C in the presence of DTT. EipBC69S+C278S melted at ∼42.3°C in the presence or absence of DTT (see Fig. S8). We conclude that an internal disulfide bond stabilizes EipB structure in vitro. Reduced stability of EipB lacking its conserved disulfide bond may contribute to the 1-log relative growth defect of ΔeipB strains expressing EipB cysteine mutants on SBA carbenicillin plates (Fig. 7C and D).
eipB deletion is synthetically lethal with bab1_0430 (ttpA) disruption and synthetically sick with disruption of multiple genes with cell envelope functions.
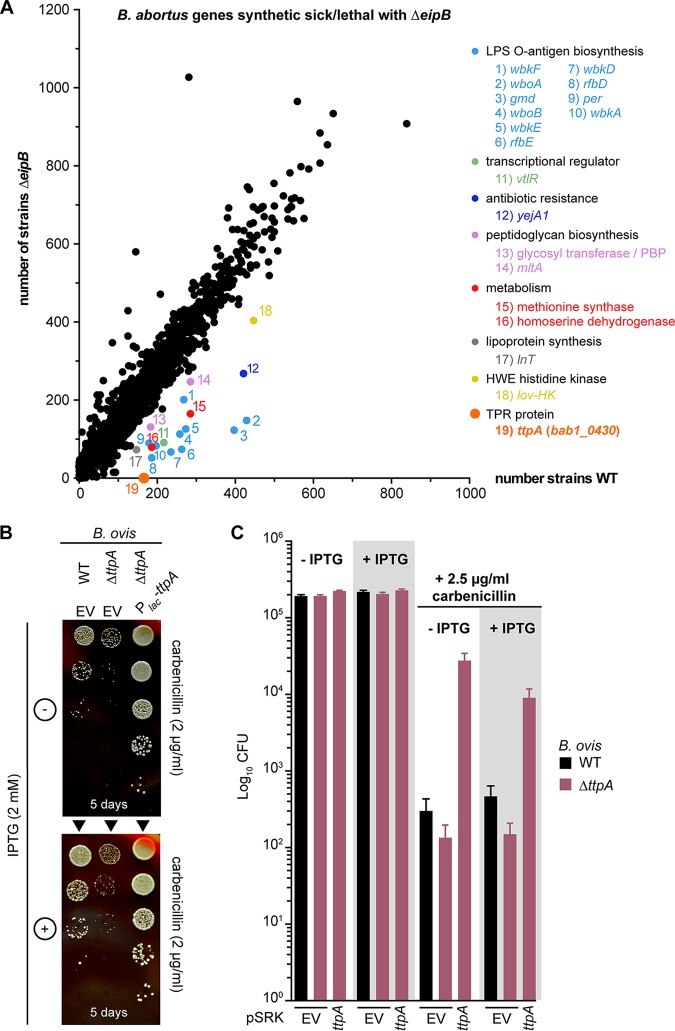
To further characterize how eipB functions in the Brucella cell, we aimed to identify transposon (Tn) insertion mutations that are synthetically lethal with eipB deletion in B. abortus (see Tables S3 and S4). In other words, we sought to discover genes that are dispensable in a wild-type genetic background but that cannot be disrupted in a ΔeipB background. By sequencing a Tn-Himar insertion library generated in B. abortus ΔeipB (NCBI Sequence Read Archive accession number SRR8322167) and a Tn-Himar library generated in wild-type B. abortus (NCBI Sequence Read Archive accession number SRR7943723), we uncovered evidence that disruption of bab1_0430 (RefSeq locus BAB_RS17965) is synthetically lethal with eipB deletion. Specifically, reproducible reads corresponding to insertions in the central 10 to 90% of bab1_0430 were not evident in ΔeipB but were present in wild-type (Fig. 8A). bab1_0430 encodes a 621-residue tetratricopeptide repeat-containing (TPR) protein with a predicted signal peptide and signal peptidase site at its N terminus. This protein was previously detected by mass spectrometry analyses of B. abortus extracts and was described as a cell envelope-associated protein (24) or periplasmic protein (25). Here, we refer to this gene as ttpA (tetratricopeptide repeat protein A gene) based on its similarity to Rhizobium leguminosarum ttpA (12).
FIG 8.
B. abortus eipB deletion is synthetically lethal with Tn-Himar disruption of bab1_0430, which encodes a tetratricopeptide repeat (TPR) protein. (A) Identification of B. abortus genes that are synthetically lethal or sick with eipB deletion. Tn-Himar insertion strains per gene (black dots) obtained in a B. abortus ΔeipB background are plotted as a function of strains per gene in a wild-type background. bab1_0430, for which we observed significantly fewer insertions in the ΔeipB strain than in the wild-type strain, is represented as an orange dot. Other synthetic sick genes are also evident in the plot, including the following genes involved in LPS O-antigen synthesis: wbkF (locus bab1_0535), wboA (bab1_0999), gmd (bab1_0545), wboB (bab1_1000), wbkE (bab1_0563), rfbE (bab1_0542), wbkD (bab1_0534), rfbD (bab1_0543), per (bab1_0544), and wbkA (bab1_0553). Also shown are genes related to peptidoglycan synthesis, mltA (bab1_2076) and a penicillin-binding protein gene (bab1_607), apolipoprotein N-acyltransferase gene lnt (bab1_2158), LysR transcriptional regulator gene vtlR (bab1_1517), extracellular solute binding protein gene yejA1 (bab1_0010), general stress response kinase gene lovhK (bab2_0652), and the metabolic genes encoding methionine synthase (bab1_0188) and homoserine dehydrogenase (bab1_1293). (B) Growth on SBA plates containing 2 μg/ml of carbenicillin with or without 2 mM IPTG serially diluted (10-fold dilution) B. ovis ΔttpA strains carrying the pSRK EV or ectopically expressing wild-type TtpA (Plac-ttpA). The wild-type (WT) B. ovis pSRK empty vector (EV) strain was used as a control. The days of growth at 37°C and 5% CO2 are reported for each plate. A representative picture of the different plates is presented. (C) Enumerated CFU, after growth on SBA plates containing 2.5 μg/ml of carbenicillin with or without 2 mM IPTG, of serially diluted (10-fold dilution) B. ovis wild-type and ΔttpA strains. The ΔttpA strain was either transformed with the empty vector (EV) or with pSRK-ttpA. EV wild-type strain and SBA plates with no carbenicillin and with or without IPTG were used as controls. This experiment was independently performed twice with two different clones each time, and all plate assays were done in triplicate. Each data point indicates the mean ± the standard error of the mean.
Genes involved in LPS O-antigen synthesis and previously described as synthetic lethal with eipA (bab1_1612) deletion in B. abortus (8) were synthetic sick with eipB deletion (Fig. 8A), as were genes involved in peptidoglycan synthesis: mltA (bab1_2076, lytic murein transglycosylase A) and bab1_0607 (glycosyl transferase/penicillin-binding protein 1A) (26) (Fig. 8A). There were reduced transposon insertions in solute binding protein gene yejA1 (bab1_0010) (Fig. 8A), which is involved in B. melitensis resistance to polymyxin (27). lnt (bab1_2158) and vtlR (bab1_1517) were also synthetic sick with the ΔeipB strain. lnt is an apolipoprotein N-acyltransferase gene involved in lipoprotein synthesis (28); vtlR encodes a LysR transcriptional regulator required for full B. abortus virulence (29) (Fig. 8A). Finally, the general stress sensor kinase gene lovHK (bab2_0652) (30), bab1_1293 (homoserine dehydrogenase), and bab1_0188 (methionine synthase) had fewer Tn insertions in the ΔeipB background relative to the wild type (Fig. 8A).
ttpA contributes to carbenicillin resistance.
As ttpA disruption is synthetic lethal with eipB deletion, we postulated that these two genes have complementary functions or are involved in a common physiological process (i.e., envelope integrity). Thus, to characterize ttpA and the nature of its connection to eipB, we deleted ttpA in B. ovis and evaluated its sensitivity to carbenicillin. All efforts to delete B. ovis ttpA (locus tag bov_0411) using a classic crossover recombination and sacB counterselection approach were unsuccessful, though hundreds of clones were screened. Efforts to delete the chromosomal copy by expressing a copy of ttpA from a plasmid also failed. This result is surprising considering that transposon insertions in B. abortus ttpA (NCBI Sequence Read Archive accession number SRR7943723) and B. ovis ttpA (NCBI Sequence Read Archive accession number SRR7943724) are tolerated in wild-type backgrounds (8). As an alternative approach to study the function of this gene, we inactivated ttpA using a single crossover recombination strategy. The resulting strain expressed a truncated version of TtpA containing the first 205 amino acids (including the signal peptide), immediately followed by 22 amino acids from the suicide plasmid. The corresponding B. ovis strain (ΔttpA) was then transformed with a plasmid-borne IPTG-inducible copy of ttpA (pSRK-ttpA) or with an empty plasmid vector (EV). We evaluated the sensitivity of these strains to carbenicillin by plating a dilution series on SBA plates containing 2 or 2.5 μg/ml carbenicillin, with or without IPTG inducer (Fig. 8B and C). Compared to the wild-type strain with an empty vector, the B. ovis ΔttpA strain with an empty vector had ∼0.5-log-unit-reduced CFU on carbenicillin SBA. The corresponding colonies of the B. ovis ΔttpA strain were noticeably smaller than the wild-type colonies. Genetic complementation of the ΔttpA strain with pSRK-ttpA restored growth on carbenicillin plates. B. ovis ΔttpA/pSRK-ttpA had ∼1.5-log-units more colonies than did the wild type in the presence of carbenicillin, with or without IPTG induction. Thus, leaky expression of ttpA from the lac promoter on pSRK-ttpA is apparently sufficient to protect this strain from carbenicillin on solid medium. The morphology of the B. ovis ΔttpA strains appeared normal by phase-contrast microscopy at ×630 magnification (Fig. S9).
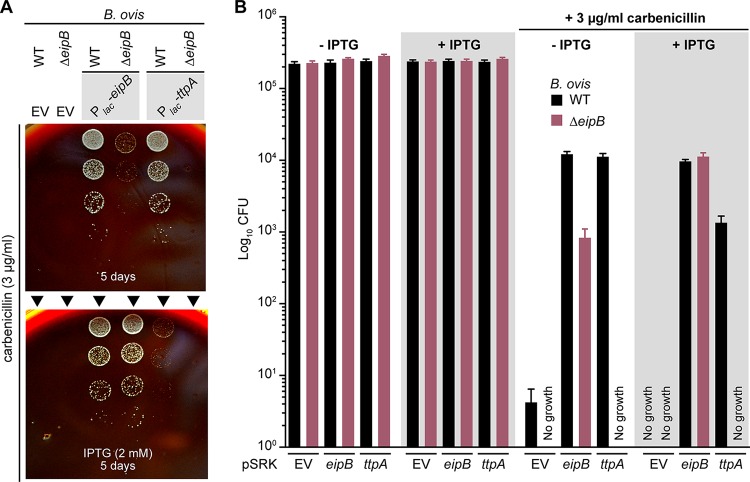
To further evaluate the effect of ttpA overexpression, we assayed B. ovis wild-type and ΔeipB strains carrying pSRK-ttpA. As before, we tested sensitivity of these inducible expression strains to carbenicillin by plating a dilution series on SBA plates containing 3 μg/ml of carbenicillin, with or without 2 mM IPTG inducer (Fig. 9). Wild-type B. ovis/pSRK-ttpA and wild-type B. ovis/pSRK-eipB strains had equivalent CFU in the absence of carbenicillin, with or without IPTG. ttpA or eipB provided an ∼3-log-unit protective effect without IPTG induction in the presence of carbenicillin compared to the wild-type empty vector strain (Fig. 9). Surprisingly, inducing ttpA expression with IPTG reduced its ability to protect in the presence of carbenicillin by 1 log unit (relative to uninduced), and the corresponding colonies were very small, suggesting slower growth when ttpA was induced (Fig. 9). This may be an effect of IPTG, based on reduced CFU counts of wild-type empty vector control under this condition. As expected, induced expression of eipB from Plac-eipB rescued the carbenicillin viability defect of ΔeipB. However, induced expression of ttpA from Plac-ttpA was not sufficient to rescue the ΔeipB carbenicillin phenotype (Fig. 9). As before, we observed highly reduced CFU for B. ovis wild-type or ΔeipB control strains carrying the pSRK empty vector (EV), when challenged with 3 μg/ml of carbenicillin. The morphology of wild-type or ΔeipB B. ovis strains overexpressing ttpA appeared normal by phase-contrast microscopy at ×630 magnification (Fig. S10).
FIG 9.
Overexpression of TtpA protects against carbenicillin treatment; protection requires EipB. (A) Growth on SBA plates containing 3 μg/ml of carbenicillin with or without 2 mM IPTG of serially diluted (10-fold dilution) B. ovis wild-type (WT) and ΔeipB strains expressing wild-type EipB (Plac-eipB) or TtpA (Plac-ttpA). B. ovis strains carrying the pSRK empty vector were used as a control. The days of growth at 37°C and 5% CO2 are reported for each plate. A representative picture of the different plates is presented. (B) Enumerated CFU after growth on SBA plates containing 3 μg/ml of carbenicillin with or without 2 mM IPTG of serially diluted (10-fold dilution) B. ovis wild-type and ΔeipB strains ectopically expressing eipB or ttpA. EV strains and SBA plates with no carbenicillin and with or without IPTG were used as controls. This experiment was independently performed twice with two different clones each time, and all plate assays were done in triplicate. Each data point indicates the mean ± the standard error of the mean.
The observed genetic interaction between eipB and ttpA, the fact that both single mutants have envelope phenotypes, and the fact that both gene products are secreted to the periplasm raised the possibility that EipB and TtpA physically interact. We tested interaction between EipB and TtpA proteins using bacterial two-hybrid and biochemical pulldown assays. We further evaluated whether a possible EipB-TtpA interaction is influenced by the presence or absence of the EipB internal disulfide bond using a biochemical pulldown. For our bacterial two-hybrid assay, EipBV31-K280 was fused to the T25 adenylate cyclase fragment, and TtpAK31-D621 was fused to the T18 or T18C adenylate cyclase fragments. For the pulldown assay, MBP-tagged TtpA (K31-D621) and His-tagged EipB (V31-K280; wild type and the different cysteine mutants) were copurified in the presence or absence of DTT. We found no evidence for direct interaction between EipB and TtpA, suggesting that the function of these two proteins in Brucella envelope stress adaptation is not achieved through direct interaction (Fig. S11).
DISCUSSION
Bacterial genome sequencing efforts over the past 2 decades have revealed thousands of protein domains of unknown function (DUFs). The DUF1849 sequence family is prevalent in the orders Rhizobiales, Rhodobacterales, and Rhodospirillales. To date, the function of DUF1849 has remained undefined. We have shown that a DUF1849 gene in Brucella spp., which we have named eipB, encodes a 14-stranded β-spiral protein that is secreted to the periplasm. eipB is required for maintenance of B. abortus spleen colonization in a mouse model of infection (Fig. 2), and eipB deletion in B. abortus and in B. ovis results in sensitivity to treatments that compromise the integrity of the cell envelope in vitro (Fig. 3). The envelope stress sensitivity of the B. abortus ΔeipB mutant likely contributes to its reduced virulence in a mouse. We further demonstrate that EipB contains a conserved disulfide bond that contributes to protein stability and function in vitro; the importance of this conserved disulfide to EipB function in vivo remains to be determined (Fig. 6 and 7; see also Fig. S3 and S4 in the supplemental material).
A lipoprotein connection?
An X-ray crystal structure of EipB shows that this periplasmic protein adopts an unusual β-spiral fold that shares structural similarity (DALI Z-score, 11.0) with a functionally uncharacterized P. aeruginosa protein, PA1994, despite low sequence identity (Fig. S6). It was previously noted (23) that PA1994 has structural features that resemble the lipoprotein carrier and chaperone proteins LolA and LolB, which have a central role in lipoprotein localization in select Gram-negative bacteria (31). Like LolA, LolB, and PA1994, Brucella EipB forms a curved hydrophobic β-sheet that is wrapped around an α-helix (Fig. S6B). Homologs of LolA are present in Brucella and other Alphaproteobacteria, but homologs of LolB are missing (28). Given the EipB structure, its periplasmic localization, and the phenotypes of a ΔeipB deletion strain, it is tempting to speculate that EipB (DUF1849) has a LolB-like function in the Brucella cell. However, it seems unlikely that LolB and EipB function in a structurally or biochemically equivalent manner. Certainly, we observe surface level similarity between LolA/LolB and EipB structures (Fig. S6), particularly in the antiparallel β-sheet region, but these proteins have topological differences that distinguish their folds. Moreover, LolB is a membrane anchored lipoprotein that facilitates lipoprotein targeting at the inner leaflet of the outer membrane. In contrast, Brucella EipB does not have a predicted site for lipidation (i.e., a lipobox) and is therefore unlikely to function as a membrane-anchored protein.
The number of unique barcoded Tn-Himar insertions in the apolipoprotein N-acyltransferase gene lnt (bab1_2158; lnt conserved domain database score < e−173) is lower than expected in a ΔeipB background relative to the wild type (Fig. 8A). This provides indirect evidence for a link between eipB and lipoproteins. Lnt catalyzes the final acylation step in lipoprotein biogenesis (32), which is often considered to be an essential cellular process. However, like Francisella tularensis and Neisseria gonorrhoeae (33), B. abortus lnt is dispensable (26) (Fig. 8A and Table S4). The data presented here suggest that transposon insertions are less tolerated in B. abortus lnt when eipB is missing. Additional experimentation is required to test a possible functional relationship between lnt and eipB. However, it is notable that we did not observe a synthetic genetic interaction between lnt and the gene encoding a structurally unrelated periplasmic envelope integrity protein, EipA, in a parallel Tn-seq experiment (8). Whether eipB actually influences lipoprotein biogenesis or localization remains to be tested.
TtpA: a periplasmic determinant of cell envelope function in Rhizobiaceae.
Transposon disruption of ttpA (bab1_0430) is not tolerated when eipB is deleted in B. abortus. ttpA, like eipB, contributes to carbenicillin resistance in vitro (Fig. 8 and 9). Though we observed a genetic interaction between eipB and ttpA, we found no evidence for a direct physical interaction between the two periplasmic proteins encoded by these genes (Fig. S11). TtpA is named for its tetratricopeptide repeat (TPR) motif; proteins containing TPR motifs are known to function in many different pathways in bacteria, including cell envelope biogenesis, and are often molecular determinants of virulence (34, 35). Indeed, deletion of ttpA has been reported to attenuate B. melitensis virulence in a mouse infection model of infection (11) and to increase R. leguminosarum membrane permeability and sensitivity to SDS and hydrophobic antibiotics (12). A genetic interaction between ttpA and the complex media growth deficient (cmdA-cmdD) operon has been reported in R. leguminosarum. Mutations in this operon result in envelope dysfunction and defects in cell morphology (12, 36). While B. abortus contains a predicted cmd operon (bab1_1573, bab1_1574, bab1_1575, and bab1_1576), these genes remain uncharacterized. We found no evidence for a synthetic genetic interaction between eipB and cmd in B. abortus.
Leaky expression of either eipB or ttpA from a plasmid strongly protected B. ovis from a cell wall antibiotic (carbenicillin). Surprisingly, inducing ttpA expression from a plasmid with IPTG did not protect as well as uninduced (i.e., leaky) ttpA expression (Fig. 9). IPTG induction of eipB expression from a plasmid did not have this same parabolic effect on cell growth/survival in the face of carbenicillin treatment. Considering that EipB and TtpA confer resistance to β-lactam antibiotics, which perturb peptidoglycan synthesis, one might hypothesize that these proteins influence the structure or synthesis of the cell wall. This hypothesis is reinforced by the fact that a lytic murein transglycosylase and a class A PBP/glycosyl transferase are synthetic sick with eipB deletion (Fig. 8A). In Escherichia coli, the TPR-containing protein LpoA is proposed to reach from the outer membrane through the periplasm to interact with the peptidoglycan synthase PBP1A (37). Models in which EipB and TtpA influence lipoprotein biosynthesis and/or cell wall metabolism are important to test as we work toward understanding the mechanisms by which these genes ensure Brucella cell envelope integrity and survival in a mammalian host.
MATERIALS AND METHODS
Agglutination assays, mouse and macrophage infection assays, antibody measurements, and the transposon sequencing experiments for this study were performed in parallel with our recent studies of eipA (8).
All experiments using live B. abortus 2308 were performed in biosafety level 3 facilities according to U.S. Centers for Disease Control and Prevention (CDC) select agent regulations at the University of Chicago Howard Taylor Ricketts Laboratory. All the B. abortus and B. ovis strains were cultivated at 37°C with 5% CO2; primer and strain information are available in Table S5 in the supplemental material.
Chromosomal deletions in B. abortus and in B. ovis.
The B. abortus and B. ovis ΔeipB deletion strains were generated using a double-crossover recombination strategy as previously described (8). Briefly, fragments corresponding to the 500-bp region upstream of the eipB start codon and the 500-bp region downstream of the eipB stop codon were ligated into the suicide plasmid pNPTS138, which carries the nptI gene for initial kanamycin selection and the sacB gene for counterselection on sucrose. Genetic complementation of the B. abortus deletion strain was carried out by transforming this strain with a pNPTS138 plasmid carrying the wild-type allele. The B. ovis ΔeipB strain was complemented with the pSRK-eipB plasmid (IPTG inducible).
To inactivate ttpA in B. ovis (bov_0411), a 527-nucleotide internal fragment was cloned into pNPTS138-cam (a suicide plasmid that we engineered to carry a chloramphenicol resistance marker) and used to disrupt the target gene by single crossover insertion. The recombinant clones were selected on SBA plates supplemented with 3 μg/ml chloramphenicol. The corresponding strain expresses the first 205 amino acids (including the signal peptide) of TtpA, plus 22 extra amino acids from the plasmid sequence, followed by a stop signal. This ΔttpA strain was complemented with pSRK-ttpA (kanamycin resistant).
Brucella EipB and TtpA overexpression strains.
For ectopic expression of B. ovis TtpA and the different versions of B. abortus EipB (wild-type, cysteine mutants, and the EipB-PhoAEc fusion with or without the signal peptide), the pSRKKm (Kanr) IPTG inducible plasmid was used (38). An overlapping PCR strategy was used to introduce cysteine mutations and to stitch the different DNA fragments to the E. coli alkaline phosphatase phoA (lacking its signal peptide). A Gibson assembly cloning strategy was then used to insert the different DNA fragments in the linearized pSRK plasmid. After sequencing, plasmids were introduced in B. abortus or B. ovis by overnight mating with E. coli WM3064 in the presence of 300 μM diaminopimelic acid (DAP) and plated on SBA plates supplemented with kanamycin.
Building and mapping the wild-type B. abortus and B. abortus ΔeipB Tn-Himar insertion libraries.
To build and map the different Tn-Himar insertion libraries, we used a barcoded transposon mutagenesis strategy developed by Wetmore and colleagues (39). A full and detailed protocol can be found in our previous paper (8). Statistics for the two different transposon insertion libraries are reported in Table S3.
Cell culture and macrophage infection assays.
Infection of inactivated macrophages differentiated from human monocytic THP-1 cells was performed as previously described (8). Briefly, for infection assays, 5 × 106 B. abortus cells were used to infect 5 × 104 THP-1 cells (multiplicity of infection of 1:100). To determine the numbers of intracellular bacteria at 1, 24, and 48 h postinfection, the infected cells were lysed, and the lysate was then serially diluted (10-fold serial dilution) and plated on TSA plates to enumerate the CFU.
Mouse infection assay.
All mouse studies were approved by the University of Chicago Institutional Animal Care and Use Committee (IACUC) and were performed as previously published (8). Briefly, 100 μl of a 5 × 105 CFU/ml B. abortus suspension were intraperitoneally injected into 6-week-old female BALB/c mice (Harlan Laboratories, Inc.). At 1, 4, and 8 weeks postinfection, five mice per strain were sacrificed, and the spleens were removed for weighing and CFU counting. At week 8, blood was also collected by cardiac puncture, and serum from each mouse was separated from blood using a serum separation tube (Sarstedt). Sera were subsequently used for ELISAs.
Determination of antibody responses at 8 weeks postinfection.
Total mouse serum IgG, IgG1, and IgG2a titers were measured using mouse-specific ELISA kits by following manufacturer’s instructions (eBioscience). Brucella-specific IgG titers were determined as previously published (8).
Spleen histology.
At 8 weeks postinfection, spleens (n = 1 per strain) were prepared for histology as previously described (8). Briefly, spleens were first fixed with formalin and submitted for tissue embedding, H&E staining, and immunohistochemistry to Nationwide Histology (Veradale, WA). For immunohistochemistry, goat anti-Brucella IgG was used (Tetracore, Inc.). Pictures of fixed mouse spleen slides were subsequently analyzed and scored.
Plate stress assays.
Stress assays were performed as previously published (8). Briefly, the different B. abortus and B. ovis strains were resuspended in sterile phosphate-buffered saline (PBS) or brucella broth to an optical density at 600 nm (OD600) of ∼0.015 (∼1 × 108 CFU/ml) and serially diluted (10-fold serial dilution). Then, 5 μl of each dilution were spotted onto TSA or SBA plates containing the different membrane stressors (2 to 5 μg/ml of ampicillin or carbenicillin, 0.04% (wt/vol) deoxycholate, or 2 mM EDTA [final concentration]).
To grow B. ovis strains containing pSRK-derived plasmids, all liquid cultures and plates were supplemented with 50 μg/ml kanamycin. When necessary, 2 mM IPTG (final concentration) was added to the plates to induce expression of EipB or TtpA from pSRK. We note that the B. ovis ΔttpA strains carry the pNPTS138 suicide plasmid (used for gene disruption), which results in chloramphenicol resistance. However, no chloramphenicol was added to the overnight cultures or the stress plates. For carbenicillin growth/survival assays, B. ovis strains were grown for 3 days at 37°C and 5% CO2 on SBA plates without carbenicillin and for 5 to 6 days when these plates contained 2, 2.5, or 3 μg/ml of carbenicillin.
Cryo-electron microscopy.
Cryo-electron microscopy was performed as previously described (8). Briefly, B. abortus cultures in brucella broth (OD600 of ∼0.015) were prepared with 2 mM EDTA or ampicillin (5 μg/ml) (final concentrations). After 4 h of incubation in the presence of EDTA or ampicillin, cells were harvested and fixed in PBS plus 4% formaldehyde. After 1 h, the cells were pelleted and resuspended in 500 μl EM buffer (40). Per CDC guidelines, cell killing was confirmed before sample removal for imaging. Fixed Brucella cells were vitrified on glow-discharged 200 mesh copper EM grids with extra-thick R2/2 holey carbon film (Quantifoil). Per grid, 3 μl of the sample was applied and automatically blotted and plunged into liquid ethane with the Leica EM GP plunge-freezer. Images were collected on a Talos L120C TEM (Thermo Fisher) using a Gatan cryo-TEM(626) holder. The images were acquired at a defocus of between 8 and 10 μm, with a pixel size of 0.458 nm.
Light microscopy images.
Phase-contrast images of B. abortus and B. ovis cells from plates or liquid broth (with or without 1 mM IPTG) were collected using a Leica DM 5000B microscope with an HCX PL APO 63×/1.4-numerical-aperture Ph3 objective. Images were acquired with a mounted Orca-ER digital camera (Hamamatsu) controlled by the Image-Pro software suite (Media Cybernetics). To prepare the different samples, cells were resuspended in PBS containing 4% formaldehyde.
Agglutination assay.
Agglutination assays were performed as previously described (8). The different Brucella strains (B. ovis and B. abortus) were harvested and resuspended in sterile PBS at an OD600 of ∼0.5. One milliliter of each cell suspension was loaded in a spectrophotometer cuvette and mixed with 20 μl of wild-type B. abortus-infected mouse serum or with acriflavine (final concentration, 5 mM), and the OD600 was measured at time zero and after 2 h. As a control, 1 ml of each cell suspension was also kept in a spectrophotometer cuvette without serum or acriflavine.
Alkaline phosphatase cell localization assay.
To determine the cellular localization of EipB, we used a B. ovis strain transformed with the pSRK plasmid carrying B. abortus eipB C-terminally fused to E. coli phoA. Two versions of this plasmid were built: one carrying the full-length eipB, which expressed the protein with its signal peptide, and one carrying a short version of eipB, which expressed the protein lacking the signal peptide. Alkaline phosphatase assays were performed as previously described (8). Briefly, aliquots of overnight culture of B. ovis (grown in presence or absence of 1 mM IPTG) were mixed with BCIP (final concentration, 200 μg/ml). After 2 h of incubation, the color change was visually assessed, and pictures were taken. The same experiment was performed with spent medium supernatants.
Size exclusion chromatography.
A DNA fragment corresponding to B. abortus eipB lacking the signal peptide (residues 31 to 280) was cloned into pET28a and transformed into the protein overexpression the E. coli Rosetta (DE3)/pLysS strain. Protein expression and purification was conducted using a Ni2+ affinity purification protocol as previously published (8). The purified protein was then dialyzed against a Tris-NaCl buffer (10 mM Tris [pH 7.4], 150 mM NaCl). The EipB oligomeric state was analyzed by size exclusion chromatography as previously described (8). Briefly, after concentration, a protein sample (500 μl at 5 mg/ml) was injected onto a GE Healthcare Superdex 200 10/300 GL column (flow rate, 0.5 ml/min). The elution profile was measured at 280 nm, and 500-μl fractions were collected during the run; the dialysis buffer described above was used for all runs. Protein standards (blue dextran, aldolase, conalbumin, and ovalbumin) injected onto the column were used to construct a calibration curve to estimate the molecular weight of purified EipB.
EipB expression, purification, and crystallization.
The DNA fragment corresponding to the B. abortus EipB protein (residues 31 to 280) was cloned into the pMCSG68 plasmid using a protocol previously published (8). For protein expression, the E. coli BL21-Gold(DE3) strain was used. Selenomethionine (Se-Met) protein expression and purification was performed as previously described (8). The purified protein was then dialyzed against 20 mM HEPES (pH 8), 250 mM NaCl, and 2 mM DTT buffer, and its concentration was determined. The purified Se-Met EipB protein was concentrated to 160 mg/ml for crystallization. Initial crystallization screening was carried out using the sitting-drop, vapor-diffusion technique. After a week, EipB crystallized in the triclinic space group P1 from the condition 70 (F10) of the MCSG-2 crystallization kit, which contains 24% polyethylene glycol (PEG) 1500 and 20% glycerol. Prior to flash freezing in liquid nitrogen, crystals were cryoprotected by briefly washing them in the crystallization solution containing 25% glycerol.
Crystallographic data collection and data processing.
Se-Met crystal diffraction was measured at a temperature of 100 K using a 2-s exposure/degree of rotation over 260°. The crystals diffracted to a resolution of 2.1 Å, and the corresponding diffraction images were collected on the ADSC Q315r detector with an X-ray wavelength near the selenium edge of 12.66 keV (0.97929 Å) for SAD phasing at the 19-ID beamline (SBC-CAT; Advanced Photon Source, Argonne, IL). Diffraction data were processed using the HKL3000 suite (41). B. abortus EipB crystals were twinned, and the data had to be reprocessed and scaled from the P21 space group to the lower symmetry space group P1 with the following cell dimensions: a = 47.36 Å, b = 69.24 Å, c = 83.24 Å, α = 90.09°, β = 90.02°, and γ = 78.66° (see Table S2). The structure was determined by SAD phasing using SHELX C/D/E, mlphare, and dm, and initial automatic protein model building with Buccaneer software, all implemented in the HKL3000 software package (41). The initial model was manually adjusted using COOT (42) and iteratively refined using COOT, PHENIX (43), and REFMAC (44); 5% of the total reflections were kept out of the refinement in both REFMAC and PHENIX throughout the refinement. The final structure converged to an Rwork of 19.5% and Rfree of 24.5% and includes 4 protein chains (A, 30-270; B, 31-271; C, 30-271; and D, 30-270), 9 ethylene glycol molecules, 2 glycerol molecules, and 129 ordered water molecules. The EipB protein contained three N-terminal residues (Ser-Asn-Ala) that remain from the cleaved tag. The stereochemistry of the structure was checked using PROCHECK (45) and a Ramachandran plot and was validated using the PDB validation server.
Disulfide bond reduction assays.
DNA fragments corresponding to B. abortus eipB cysteine mutants (C69S, C278S, and C69S+C278S) and lacking the signal peptide (residues M1 to A30) were cloned into pET28a and transformed into the protein overexpression E. coli Rosetta (DE3)/pLysS strain. Protein expression and Ni2+ affinity purification were conducted using protocols previously published (8). Briefly, for each protein, a pellet corresponding to a 250-ml culture was resuspended in 1.5 ml of BugBuster master mix (MD Millipore) supplemented with 50 μl of DNase I (5 mg/ml). After 20 min on ice, cell debris was pelleted, and the supernatant was mixed with 200 μl of Ni-NTA Superflow resin (Qiagen). Beads were washed with 8 ml of a 10 mM imidazole Tris-NaCl buffer (10 mM Tris [pH 7.4], 150 mM NaCl) and 5 ml of a 75 mM imidazole Tris-NaCl buffer. Proteins were eluted with 200 μl of a 500 mM imidazole Tris-NaCl buffer. Then, 50 μl of each purified protein (at 0.5 mg/ml) was mixed with 12.5 μl of a 4× protein loading dye containing or not containing 1 mM DTT. Samples were boiled for 5 min, and 10 μl was loaded onto a SDS–12% PAGE gel.
Thermal shift protein stability assay.
A thermal shift assay to assess protein stability was performed on 20-μl samples containing 25 μM purified B. abortus EipBWT or EipBC69S+C278S, 50× Sypro Orange (Invitrogen), and 2 mM DTT when needed. Each protein sample and solution was prepared with the same dialysis buffer (10 mM Tris [pH 7.4], 150 mM NaCl, 1 mM EDTA). Plates (96 well; MicroAmp EnduratePlate optical 96-well fast clear reaction plates; Applied Biosystems) were heated from 25 to 95°C with a ramp rate of 0.05°C/s and read by a thermocycler (QuantumStudio 5 real-time PCR system; Applied Biosystems/Thermo Fisher Scientific) using excitation and emission wavelengths of 470 ± 15 nm and 558 ± 11 nm, respectively. Protein Thermal Shift software v1.3 (Applied Biosystems/Thermo Fisher Scientific) was used to calculate the first derivative of the curve to determine the melting temperature.
Bacterial two-hybrid protein interaction assay.
To assay EipB interaction with TtpA, we used a bacterial two-hybrid system (46). Briefly, a B. abortus eipB DNA fragment (lacking the signal peptide) was cloned into pKT25 vector and a B. abortus ttpA fragment (lacking the signal peptide) was cloned into pUT18 or pUT18C vectors. The different pUT18, pUT18C, and pKT25 combinations were then cotransformed into a chemically competent E. coli reporter strain BTH101 and spotted onto Luria-Bertani agar plates (ampicillin [100 μg/ml] plus kanamycin [50 μg/ml]) supplemented with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 40 μg/ml).
Pulldown assay between EipB and TtpA.
To evaluate the interaction between B. abortus wild-type and cysteine mutant EipB and TtpA, the different genes were cloned into pET28a and pMAL-c2G expression plasmids and transformed in the E. coli Rosetta (DE3)/pLysS expression strain. The corresponding proteins (His6-EipBWT or His6-EipB cysteine mutants and MBP-TtpA) were overexpressed and purified using nickel affinity and amylose affinity gravity columns, respectively. Two milliliters of amylose resin was saturated with 10 ml of a clarified cell lysate corresponding to a 500-ml culture pellet of IPTG-induced Rosetta pMAL-c2G-ttpA. Beads were thoroughly washed with 50 ml of a Tris-NaCl buffer (10 mM Tris [pH 7.4], 150 mM NaCl), and 200-μl portions of these beads were mixed with 500 μl of nickel-purified EipB at ∼0.5 mg/ml (see reference 8 for a detailed nickel-affinity purification protocol). After 30 min of incubation in the presence or absence of 1 mM DTT, the flowthrough was saved, and the beads were thoroughly washed with a Tris-NaCl buffer supplemented or not supplemented with 1 mM DTT. The protein was eluted with 200 μl of the same buffer containing 20 mM maltose. The different protein samples (elutions and flowthroughs) were subjected SDS–12% PAGE and Coomassie stained.
Bioinformatics.
Figures of the structures, structural alignments, electrostatic potential representations, and root mean square deviation calculations were performed using PyMOL (PyMOL Molecular Graphics System, version 1.7.4; Schrödinger, LLC). Surface hydrophobicity was evaluated using the YRB python script (47). The XtalPred server (48) and Dali server (49) were used to identify proteins with the highest structural and sequence relatedness. The BLAST server (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to identify homologs of B. abortus EipB in different taxa within the Alphaproteobacteria. The EipB weblogo was generated by aligning 447 DUF1849 protein sequences of Alphaproteobacteria retrieved from the EMBL-EBI website (https://www.ebi.ac.uk/interpro/entry/IPR015000/proteins-matched). Alignment was generated with Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). When necessary, the C termini of sequences were realigned by hand. The Clustal alignment file was converted to a fasta file using http://sequenceconversion.bugaco.com/converter/biology/sequences/clustal_to_fasta.php. This file was then submitted to the skylign server (http://skylign.org/) to generate a weblogo. The alignment was processed with the following options: remove mostly empty columns/alignment sequences are full length/score.
Data availability.
For each Himar insertion library, Tn-seq read data have been deposited in the NCBI Sequence Read Archive under the following accession numbers: B. abortus 2308 wild type, SRR7943723 (BioProject PRJNA493942); B. abortus ΔeipB strain (Δbab1_1186), SRR8322167 (BioProject PRJNA510139). The coordinates of EipB have been deposited in the Protein Data Bank under 6NTR. Crystallographic data and refined model statistics are presented in Table S2. Diffraction images have been uploaded to the SBGrid diffraction data server (data doi:10.15785/SBGRID/445).
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Crosson laboratory for helpful discussions. We thank members of the SBC at Argonne National Laboratory for their help with data collection at the 19-ID beamline.
This study was supported by National Institutes of Health grants U19AI107792 and R01AI107159 to S.C.
J.H., J.W.W., and S.C. contributed to the design and conceptualization of the study. J.H., J.W.W., A.F., D.M.C., J.X.C., E.U., A.B., L.B., G.B., Y.K., and S.C. performed the experiments and acquired and analyzed the data. J.H., J.W.W., A.F., and S.C. interpreted the data. J.H. and S.C. wrote the original draft of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00134-19.
REFERENCES
- 1.Gorvel JP, Moreno E. 2002. Brucella intracellular life: from invasion to intracellular replication. Vet Microbiol 90:281–297. doi: 10.1016/S0378-1135(02)00214-6. [DOI] [PubMed] [Google Scholar]
- 2.Pappas G, Papadimitriou P, Akritidis N, Christou L, Tsianos EV. 2006. The new global map of human brucellosis. Lancet Infect Dis 6:91–99. doi: 10.1016/S1473-3099(06)70382-6. [DOI] [PubMed] [Google Scholar]
- 3.Batut J, Andersson SG, O’Callaghan D. 2004. The evolution of chronic infection strategies in the alpha-proteobacteria. Nat Rev Microbiol 2:933–945. doi: 10.1038/nrmicro1044. [DOI] [PubMed] [Google Scholar]
- 4.Atluri VL, Xavier MN, de Jong MF, den Hartigh AB, Tsolis RM. 2011. Interactions of the human pathogenic Brucella species with their hosts. Annu Rev Microbiol 65:523–541. doi: 10.1146/annurev-micro-090110-102905. [DOI] [PubMed] [Google Scholar]
- 5.Roop RM II, Gaines JM, Anderson ES, Caswell CC, Martin DW. 2009. Survival of the fittest: how Brucella strains adapt to their intracellular niche in the host. Med Microbiol Immunol 198:221–238. doi: 10.1007/s00430-009-0123-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byndloss MX, Tsolis RM. 2016. Brucella spp. virulence factors and immunity. Annu Rev Anim Biosci 4:111–127. doi: 10.1146/annurev-animal-021815-111326. [DOI] [PubMed] [Google Scholar]
- 7.Lamontagne J, Butler H, Chaves-Olarte E, Hunter J, Schirm M, Paquet C, Tian M, Kearney P, Hamaidi L, Chelsky D, Moriyon I, Moreno E, Paramithiotis E. 2007. Extensive cell envelope modulation is associated with virulence in Brucella abortus. J Proteome Res 6:1519–1529. doi: 10.1021/pr060636a. [DOI] [PubMed] [Google Scholar]
- 8.Herrou J, Willett JW, Fiebig A, Varesio LM, Czyz DM, Cheng JX, Ultee E, Briegel A, Bigelow L, Babnigg G, Kim Y, Crosson S. 2018. Periplasmic protein EipA determines envelope stress resistance and virulence in Brucella abortus. Mol Microbiol doi: 10.1111/mmi.14178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price MN, Wetmore KM, Waters RJ, Callaghan M, Ray J, Liu H, Kuehl JV, Melnyk RA, Lamson JS, Suh Y, Carlson HK, Esquivel Z, Sadeeshkumar H, Chakraborty R, Zane GM, Rubin BE, Wall JD, Visel A, Bristow J, Blow MJ, Arkin AP, Deutschbauer AM. 2018. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557:503–509. doi: 10.1038/s41586-018-0124-0. [DOI] [PubMed] [Google Scholar]
- 11.Lestrate P, Dricot A, Delrue RM, Lambert C, Martinelli V, De Bolle X, Letesson JJ, Tibor A. 2003. Attenuated signature-tagged mutagenesis mutants of Brucella melitensis identified during the acute phase of infection in mice. Infect Immun 71:7053–7060. doi: 10.1128/IAI.71.12.7053-7060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neudorf KD, Vanderlinde EM, Tambalo DD, Yost CK. 2015. A previously uncharacterized tetratricopeptide-repeat-containing protein is involved in cell envelope function in Rhizobium leguminosarum. Microbiology 161:148–157. doi: 10.1099/mic.0.082420-0. [DOI] [PubMed] [Google Scholar]
- 13.Street NE, Schumacher JH, Fong TA, Bass H, Fiorentino DF, Leverah JA, Mosmann TR. 1990. Heterogeneity of mouse helper T cells: evidence from bulk cultures and limiting dilution cloning for precursors of Th1 and Th2 cells. J Immunol 144:1629–1639. [PubMed] [Google Scholar]
- 14.Svetic A, Jian YC, Lu P, Finkelman FD, Gause WC. 1993. Brucella abortus induces a novel cytokine gene expression pattern characterized by elevated IL-10 and IFN-γ in CD4+ T cells. Int Immunol 5:877–883. doi: 10.1093/intimm/5.8.877. [DOI] [PubMed] [Google Scholar]
- 15.Lapaque N, Moriyon I, Moreno E, Gorvel JP. 2005. Brucella lipopolysaccharide acts as a virulence factor. Curr Opin Microbiol 8:60–66. doi: 10.1016/j.mib.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Papo N, Shai Y. 2005. A molecular mechanism for lipopolysaccharide protection of Gram-negative bacteria from antimicrobial peptides. J Biol Chem 280:10378–10387. doi: 10.1074/jbc.M412865200. [DOI] [PubMed] [Google Scholar]
- 17.Palmer DA, Douglas JT. 1989. Analysis of Brucella lipopolysaccharide with specific and cross-reacting monoclonal antibodies. J Clin Microbiol 27:2331–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alton GG, Jones LM, Pietz DE. 1975. Laboratory techniques in brucellosis. Monogr Ser World Health Organ 1975:1–163. [PubMed] [Google Scholar]
- 19.Turse JE, Pei J, Ficht TA. 2011. Lipopolysaccharide-deficient Brucella variants arise spontaneously during infection. Front Microbiol 2:54. doi: 10.3389/fmicb.2011.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen H. 2017. Predicting secretory proteins with SignalP. Methods Mol Biol 1611:59–73. doi: 10.1007/978-1-4939-7015-5_6. [DOI] [PubMed] [Google Scholar]
- 21.Marrichi M, Camacho L, Russell DG, DeLisa MP. 2008. Genetic toggling of alkaline phosphatase folding reveals signal peptides for all major modes of transport across the inner membrane of bacteria. J Biol Chem 283:35223–35235. doi: 10.1074/jbc.M802660200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm L, Laakso LM. 2016. Dali server update. Nucleic Acids Res 44:W351–W355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakolitsa C, Kumar A, McMullan D, Krishna SS, Miller MD, Carlton D, Najmanovich R, Abdubek P, Astakhova T, Chiu HJ, Clayton T, Deller MC, Duan L, Elias Y, Feuerhelm J, Grant JC, Grzechnik SK, Han GW, Jaroszewski L, Jin KK, Klock HE, Knuth MW, Kozbial P, Marciano D, Morse AT, Nigoghossian E, Okach L, Oommachen S, Paulsen J, Reyes R, Rife CL, Trout CV, van den Bedem H, Weekes D, White A, Xu Q, Hodgson KO, Wooley J, Elsliger MA, Deacon AM, Godzik A, Lesley SA, Wilson IA. 2010. The structure of the first representative of Pfam family PF06475 reveals a new fold with possible involvement in glycolipid metabolism. Acta Crystallogr Sect F Struct Biol Cryst Commun 66:1211–1217. doi: 10.1107/S1744309109022684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchesini MI, Connolly J, Delpino MV, Baldi PC, Mujer CV, DelVecchio VG, Comerci DJ. 2011. Brucella abortus choloylglycine hydrolase affects cell envelope composition and host cell internalization. PLoS One 6:e28480. doi: 10.1371/journal.pone.0028480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connolly JP, Comerci D, Alefantis TG, Walz A, Quan M, Chafin R, Grewal P, Mujer CV, Ugalde RA, DelVecchio VG. 2006. Proteomic analysis of Brucella abortus cell envelope and identification of immunogenic candidate proteins for vaccine development. Proteomics 6:3767–3780. doi: 10.1002/pmic.200500730. [DOI] [PubMed] [Google Scholar]
- 26.Sternon JF, Godessart P, Goncalves de Freitas R, Van der Henst M, Poncin K, Francis N, Willemart K, Christen M, Christen B, Letesson JJ, De Bolle X. 2018. Transposon sequencing of Brucella abortus uncovers essential genes for growth in vitro and inside macrophages. Infect Immun 86:e00312–e00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Bie P, Cheng J, Lu L, Cui B, Wu Q. 2016. The ABC transporter YejABEF is required for resistance to antimicrobial peptides and the virulence of Brucella melitensis. Sci Rep 6:31876. doi: 10.1038/srep31876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goolab S, Roth RL, van Heerden H, Crampton MC. 2015. Analyzing the molecular mechanism of lipoprotein localization in Brucella. Front Microbiol 6:1189. doi: 10.3389/fmicb.2015.01189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheehan LM, Budnick JA, Blanchard C, Dunman PM, Caswell CC. 2015. A LysR-family transcriptional regulator required for virulence in Brucella abortus is highly conserved among the alpha-proteobacteria. Mol Microbiol 98:318–328. doi: 10.1111/mmi.13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HS, Willett JW, Jain-Gupta N, Fiebig A, Crosson S. 2014. The Brucella abortus virulence regulator, LovhK, is a sensor kinase in the general stress response signalling pathway. Mol Microbiol 94:913–925. doi: 10.1111/mmi.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okuda S, Tokuda H. 2011. Lipoprotein sorting in bacteria. Annu Rev Microbiol 65:239–259. doi: 10.1146/annurev-micro-090110-102859. [DOI] [PubMed] [Google Scholar]
- 33.LoVullo ED, Wright LF, Isabella V, Huntley JF, Pavelka MS Jr. 2015. Revisiting the Gram-negative lipoprotein paradigm. J Bacteriol 197:1705–1715. doi: 10.1128/JB.02414-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeytuni N, Zarivach R. 2012. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure 20:397–405. doi: 10.1016/j.str.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 35.Cerveny L, Straskova A, Dankova V, Hartlova A, Ceckova M, Staud F, Stulik J. 2013. Tetratricopeptide repeat motifs in the world of bacterial pathogens: role in virulence mechanisms. Infect Immun 81:629–635. doi: 10.1128/IAI.01035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vanderlinde EM, Magnus SA, Tambalo DD, Koval SF, Yost CK. 2011. Mutation of a broadly conserved operon (RL3499-RL3502) from Rhizobium leguminosarum biovar viciae causes defects in cell morphology and envelope integrity. J Bacteriol 193:2684–2694. doi: 10.1128/JB.01456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jean NL, Bougault CM, Lodge A, Derouaux A, Callens G, Egan AJ, Ayala I, Lewis RJ, Vollmer W, Simorre JP. 2014. Elongated structure of the outer-membrane activator of peptidoglycan synthesis LpoA: implications for PBP1A stimulation. Structure 22:1047–1054. doi: 10.1016/j.str.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khan SR, Gaines J, Roop RM II, Farrand SK. 2008. Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl Environ Microbiol 74:5053–5062. doi: 10.1128/AEM.01098-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wetmore KM, Price MN, Waters RJ, Lamson JS, He J, Hoover CA, Blow MJ, Bristow J, Butland G, Arkin AP, Deutschbauer A. 2015. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. mBio 6:e00306–e00315. doi: 10.1128/mBio.00306-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willett JW, Herrou J, Briegel A, Rotskoff G, Crosson S. 2015. Structural asymmetry in a conserved signaling system that regulates division, replication, and virulence of an intracellular pathogen. Proc Natl Acad Sci U S A 112:E3709–E3718. doi: 10.1073/pnas.1503118112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. 2006. HKL-3000: the integration of data reduction and structure solution - from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58:1948–1954. doi: 10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 44.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. 1993. Procheck: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 46.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hagemans D, van Belzen IAEM, Morán Luengo T, Rüdiger SGD. 2015. A script to highlight hydrophobicity and charge on protein surfaces. Front Mol Biosci 2:56. doi: 10.3389/fmolb.2015.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slabinski L, Jaroszewski L, Rychlewski L, Wilson IA, Lesley SA, Godzik A. 2007. XtalPred: a web server for prediction of protein crystallizability. Bioinformatics 23:3403–3405. doi: 10.1093/bioinformatics/btm477. [DOI] [PubMed] [Google Scholar]
- 49.Holm L, Rosenstrom P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res 38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams KP, Sobral BW, Dickerman AW. 2007. A robust species tree for the Alphaproteobacteria. J Bacteriol 189:4578–4586. doi: 10.1128/JB.00269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For each Himar insertion library, Tn-seq read data have been deposited in the NCBI Sequence Read Archive under the following accession numbers: B. abortus 2308 wild type, SRR7943723 (BioProject PRJNA493942); B. abortus ΔeipB strain (Δbab1_1186), SRR8322167 (BioProject PRJNA510139). The coordinates of EipB have been deposited in the Protein Data Bank under 6NTR. Crystallographic data and refined model statistics are presented in Table S2. Diffraction images have been uploaded to the SBGrid diffraction data server (data doi:10.15785/SBGRID/445).