Herein the crystal structure of the VapBC-1 complex from nontypeable Haemophilus influenzae (NTHi) is described. Our results show that some of the mutations in the PIN domain of the VapC-1 toxin were associated with decreased toxicity in E. coli, but the mutants retained the ability to homodimerize and to heterodimerize with the wild-type cognate antitoxin, VapB-1. A new system was designed and constructed to quantify the effects of these mutations on NTHi survival during infections of primary human tissues ex vivo. Any mutation to a conserved amino acid in the PIN domain significantly decreased the number of survivors compared to that of the in cis wild-type toxin under the same conditions.
KEYWORDS: PIN domain, crystal structure, persistence, protein-protein interactions, ribonuclease, toxin-antitoxin
ABSTRACT
Toxin-antitoxin (TA) gene pairs have been identified in nearly all bacterial genomes sequenced to date and are thought to facilitate persistence and antibiotic tolerance. TA loci are classified into various types based upon the characteristics of their antitoxins, with those in type II expressing proteic antitoxins. Many toxins from type II modules are ribonucleases that maintain a PilT N-terminal (PIN) domain containing conserved amino acids considered essential for activity. The vapBC (virulence-associated protein) TA system is the largest subfamily in this class and has been linked to pathogenesis of nontypeable Haemophilus influenzae (NTHi). In this study, the crystal structure of the VapBC-1 complex from NTHi was determined to 2.20 Å resolution. Based on this structure, aspartate-to-asparagine and glutamate-to-glutamine mutations of four conserved residues in the PIN domain of the VapC-1 toxin were constructed and the effects of the mutations on protein-protein interactions, growth of Escherichia coli, and pathogenesis ex vivo were tested. Finally, a novel model system was designed and utilized that consists of an NTHi ΔvapBC-1 strain complemented in cis with the TA module containing a mutated or wild-type toxin at an ectopic site on the chromosome. This enabled the analysis of the effect of PIN domain toxin mutants in tandem with their wild-type antitoxin under the control of the vapBC-1 native promoter and in single copy. This is the first report of a system facilitating the study of TA mutant operons in the background of NTHi during infections of primary human tissues ex vivo.
IMPORTANCE Herein the crystal structure of the VapBC-1 complex from nontypeable Haemophilus influenzae (NTHi) is described. Our results show that some of the mutations in the PIN domain of the VapC-1 toxin were associated with decreased toxicity in E. coli, but the mutants retained the ability to homodimerize and to heterodimerize with the wild-type cognate antitoxin, VapB-1. A new system was designed and constructed to quantify the effects of these mutations on NTHi survival during infections of primary human tissues ex vivo. Any mutation to a conserved amino acid in the PIN domain significantly decreased the number of survivors compared to that of the in cis wild-type toxin under the same conditions.
INTRODUCTION
Toxin-antitoxin (TA) loci were first identified on plasmids as modules that aided in plasmid maintenance and stability by mediating the killing of plasmid-free cells and thereby ensuring stable plasmid inheritance (1, 2). Following the advent of whole-genome sequencing, TAs were found to be maintained in the chromosomes of numerous Gram-negative and Gram-positive bacterial genera as well as in the Archaea (3–5). Chromosomally located TAs have been implicated as important metabolic regulators in many human pathogens, including Escherichia coli, nontypeable Haemophilus influenzae (NTHi), Mycobacterium tuberculosis, and Staphylococcus aureus (6–9). Stressful environmental conditions, such as nutrient limitation, heat, pH, and oxidative stress from the host immune response or antibiotic therapy (10–13), initiate degradation of the labile antitoxin via bacterial proteases such as Lon and ClpXP (14, 15). After the antitoxin is degraded, the toxin is free to inhibit essential cellular processes, such as ATP and protein synthesis, DNA replication, peptidoglycan synthesis, and cell division (16–20), resulting in growth arrest and facilitating a persister state.
TA modules are operons typically organized with the antitoxin located directly upstream of the toxin. Six different TA families have been identified and characterized by the nature of the antitoxin. In type I and type III systems, the antitoxin is an RNA. The type I antitoxin is antisense RNA that binds the toxin mRNA under normal cellular conditions, inhibiting its translation (21). Following degradation of the type I antitoxin, the toxin is translated and depolarizes the cell membrane, thereby inhibiting ATP synthesis (21, 22). In the type III TA loci, the antitoxin is a pseudoknot RNA that binds to the protein toxin and prevents it from cleaving RNA (23, 24). Type II, IV, V, and VI systems all contain protein antitoxins. In type II systems, both the antitoxin and the toxin are proteins that form a nontoxic complex upon translation (25). Many toxins of the type II TA modules have been characterized as RNases (17, 26). Type IV systems are unique in that the antitoxin inhibits toxin activity by interacting directly with the targets of the toxin, which are cytoskeletal proteins necessary for proper cell division (20, 27). The only type V TA locus described to date is ghoST, in which the antitoxin inhibits the activity of the toxin by cleaving its mRNA (28). There is one identified type VI locus currently, designated socAB. In this module, the antitoxin SocA does not inhibit the activity of the SocB toxin directly but rather guides it to the protease ClpXP (29). When SocB is not degraded by ClpXP, it binds to DnaN and inhibits DNA replication and elongation (29). Free-living prokaryotes tend to have more TA loci than obligate intracellular microorganisms, and pathogenic bacteria often have more TA loci than their nonpathogenic relatives (30). Maintaining numerous TA loci in their genomes may increase the ability of microorganisms to rapidly respond to stresses in their microenvironments, and there is growing interest in studying the contributions of these systems to microbial pathogenesis and persistence.
Type II modules are currently the best characterized, and the vapBC (virulence-associated protein) system is the largest subfamily in this class, with VapB being the antitoxin and VapC the RNase toxin. The VapB antitoxin has two roles in the VapBC system: a region near its C terminus binds to and neutralizes the toxin, while its N terminus binds DNA and increases the TA complex stability in autoregulating transcription of its cognate operon (31). The VapC toxin contains a conserved PilT N-terminal (PIN) (32) domain that is crucial for its activity. While it has been shown that some VapC homologues are able to target specific RNA in bacterial cells (26, 33, 34), it is important to note that a VapC toxin has not yet been cocrystallized with its specific RNA target (31).
NTHi is a human-adapted bacterial commensal of the upper respiratory tract and a leading cause of recurrent otitis media in the pediatric population (35). This organism maintains two vapBC modules in its chromosome, designated vapBC-1 and vapBC-2. We have previously shown that the vapBC-1 locus significantly increases survival during infection, both in vivo and ex vivo (7). Despite low primary sequence homology among the VapC toxins from different microorganisms, the PIN domain is ubiquitous and characterized by highly conserved acidic, polar, and hydrophobic residues in the active site (31, 36). Based on the VapBC-1 crystal structure, we introduced directed mutations to four PIN domain residues and evaluated the resulting mutant’s protein-protein interactions, effects on the growth of E. coli, and ability to survive during infections of primary human tissues ex vivo.
RESULTS
Structural analysis of VapBC-1.
Although VapBC structures from other organisms have been determined (37), the structure of VapBC-1 from NTHi has not been reported. Therefore, the crystal structure of VapBC-1 was determined from a polyhistidine-tagged construct that allowed the VapB-1 antitoxin to be coexpressed with the VapC-1 toxin. The primary sequences of the purified toxin and antitoxin used for crystallization are shown in Fig. S1 in the supplemental material.
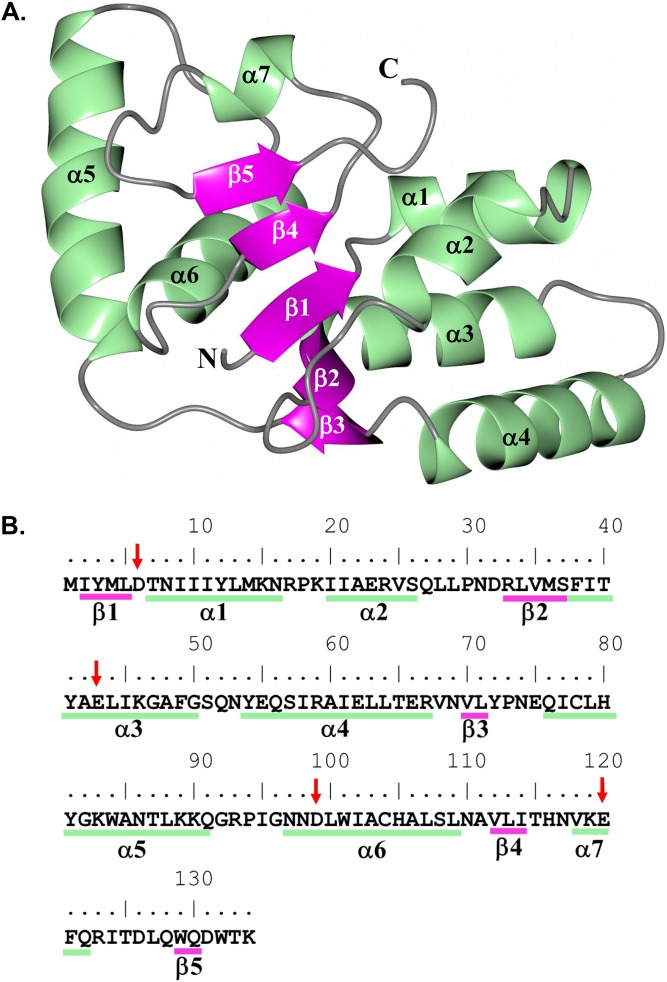
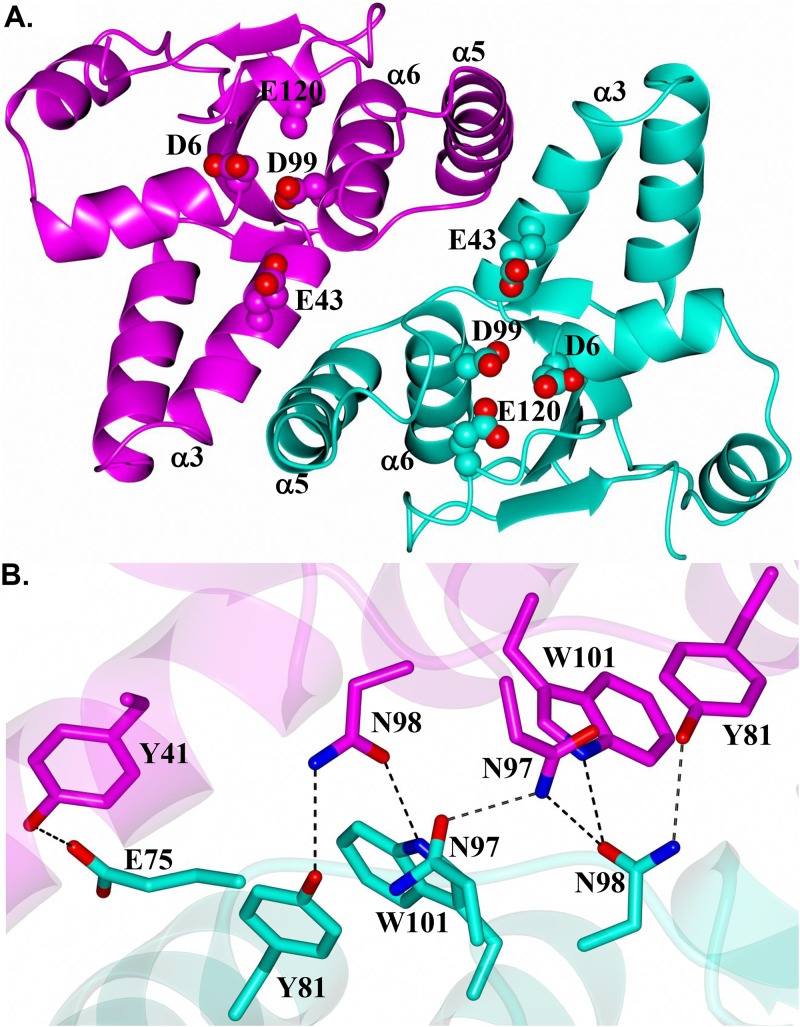
The overall structure of NTHi VapC-1 adopts an α-β-α sandwich fold in which five central β-strands are flanked by two α-helical regions typically observed for PIN-like domains (Fig. 1A). Subunits of VapC-1 form a dimer in which the hydrophobic surfaces of helices α3 and α5 are positioned approximately 10 Å from one another (Fig. 2A). The active-site residues D6, E43, D99, and E120, depicted as spheres in Fig. 2A, are conserved among various VapC species and adopt a spatial arrangement similar to that of previously determined structures (Fig. S2). The main network of hydrogen bonds is formed between dimer subunits through residues Y81 in helix α5 and N97, N98, and W101 of helix α6 (Fig. 2B). An additional hydrogen bond is present between E75 of subunit B and Y41 in helix α3 of subunit A. The reciprocal interaction, E75 in subunit A with Y41 of subunit B, is not observed, as the side chains are positioned 5 Å apart. Thus, this hydrogen bond interaction is not likely a critical interaction between dimer subunits. Similar to previously determined structures (38, 39), the VapC-1 dimer has an interface area of 1,024 Å2 and a solvation free-energy gain from dimer formation (ΔGint) of −21.6 kcal/mol, and 13% of the accessible surface area is buried, as determined with the Proteins, Interfaces, Structures and Assemblies program (PISA) (40). As noted above, there is minimal hydrogen bonding between the dimer subunits and no salt bridge interactions are present.
FIG 1.
Structure of the H. influenzae VapC-1 monomer subunit. (A) Colored by secondary structure with α-helices (green) and β-sheets (magenta). (B) Primary and secondary structural elements of VapC-1. Conserved active-site residues are indicated by arrows.
FIG 2.
Structure of the H. influenzae VapC-1 dimer showing subunits A and B in magenta and cyan, respectively. (A) View along the 2-fold axis of the dimer. Helices that form the interacting surfaces between subunits are indicated. Conserved active-site residues are rendered as spheres. Note that E120 (chain A) was partially disordered, so not all of the side chain atoms were modeled. (B) Hydrogen bond interactions (dashed lines) between residues of the dimer.
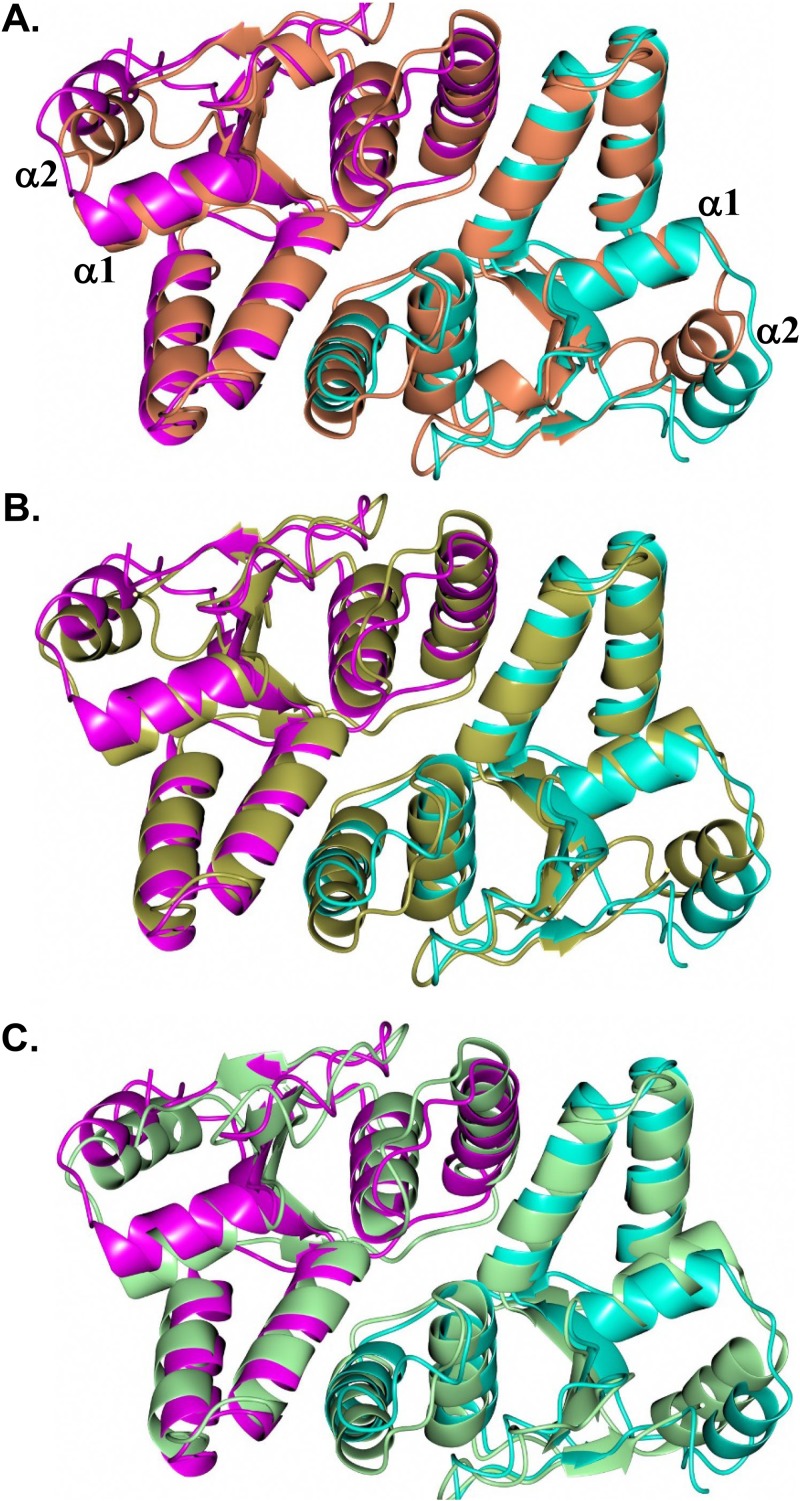
The VapC-1 dimer adopts an arrangement similar to that of structures from other organisms (Fig. 3). Comparison of the NTHi VapC-1 dimer with homologous structures, identified from a PDBeFold search (41), yielded root mean square (RMS) deviations between Cα atoms of 1.85 Å (251 residues, VapC D98N from Shigella flexneri, PDB no. 5ECD) (42), 1.85 Å (253 residues, VapBC from S. flexneri, PDB no. 3TND) (43) and 1.79 Å (256 residues, VapBC from Rickettsia felis in complex with DNA, PDB no. 3ZVK) (44). The main difference is observed in helix α2, which is moved further away from helix α1 in the NTHi VapC-1 dimer, as noted in Fig. 3A. As shown below, this relative difference in position of helices α1 and α2 is necessary to accommodate VapB-1 binding. Slightly lower RMS deviations were observed when comparing single subunits, and superposition with VapC-1 (subunit A) yielded proteins with the following PDB identification numbers: 5ECD (subunit A, 1.48 Å, 126 residues), 3TND (subunit C, 1.37 Å, 129 residues), and 3ZVK (subunit A, 1.54 Å, 131 residues). The main conformational differences occur in the relative positions of helices α1 and α2, as was observed for superposition of the dimers noted above.
FIG 3.
Comparison of the H. influenzae VapC-1 dimer (magenta/cyan) with the VapC dimer from homologous structures. (A) VapC D98N (coral) from S. flexneri (PDB no. 5ECD); (B) VapBC (gold) from S. flexneri (PDB no. 3TND); (C) VapBC (green) from R. felis in complex with DNA (DNA not shown) (PDB no. 3ZVK).
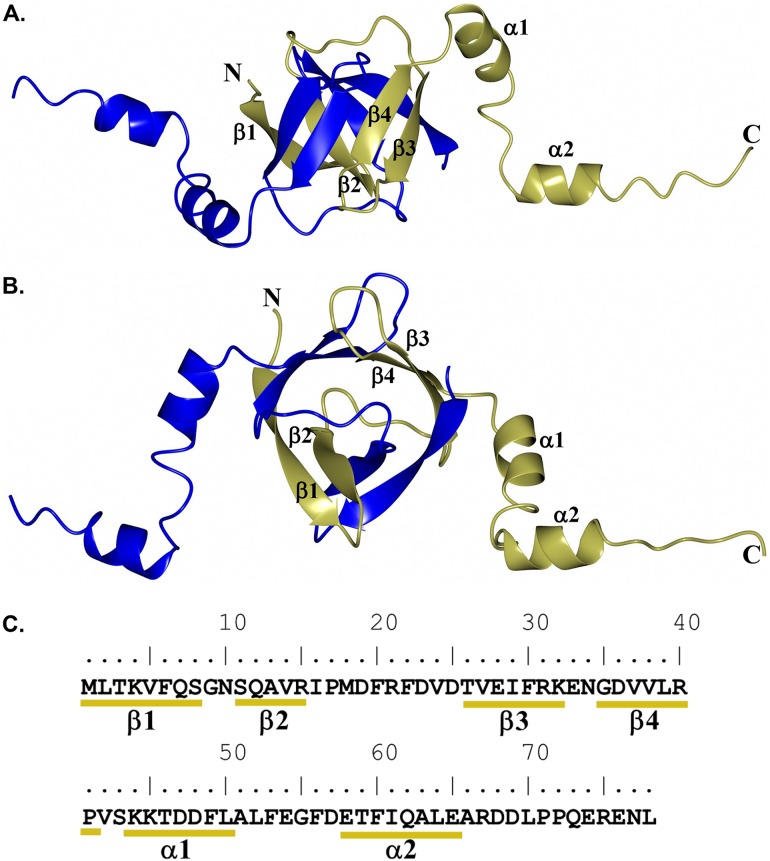
The VapB-1 portion of the NTHi VapBC-1 complex could be modeled from residues M1 to P72 (subunit C) and residues M1 to D69 (subunit D). The overall structure of VapB-1 consists of a ribbon-helix-helix motif in which the four β-sheets of the N terminus dimerize with another subunit to form a β-barrel type of architecture (Fig. 4). As determined using PISA (40), extensive hydrogen bonding and salt bridge interactions are present between the dimer subunits (27 and 11, respectively), forming an interface area of 1,662 Å2 and a solvation free-energy gain of −17.0 kcal/mol. Notably, 25% of the accessible surface area is buried to form the dimer interface. The C terminus contains a long stretch of flexible residues occupied by two α-helices that bind to the VapC-1 toxin, forming a 1:1 interaction between VapB-1 and VapC-1 subunits. A similar architecture is observed for other VapBC structures, such as VapBC from R. felis in complex with DNA (PDB no. 3ZVK). Superposition yielded RMS deviations between Cα atoms of 1.89 Å (90 residues). As shown in Fig. S3, the dimerization core regions are quite similar. However, the VapC-1 binding region of the C terminus adopts a markedly different orientation relative to the VapB-1 β-barrel dimer core. This is not too surprising, given that VapBC complexes often adopt various quaternary structural arrangements that produce higher-order oligomers (31).
FIG 4.
Structure of the H. influenzae VapB-1 dimer showing subunits C (gold) and D (blue). Secondary structure elements are annotated for subunit C. (A) Perpendicular view; (B) view along the β-barrel axis; (C) primary and secondary structural elements of VapB-1.
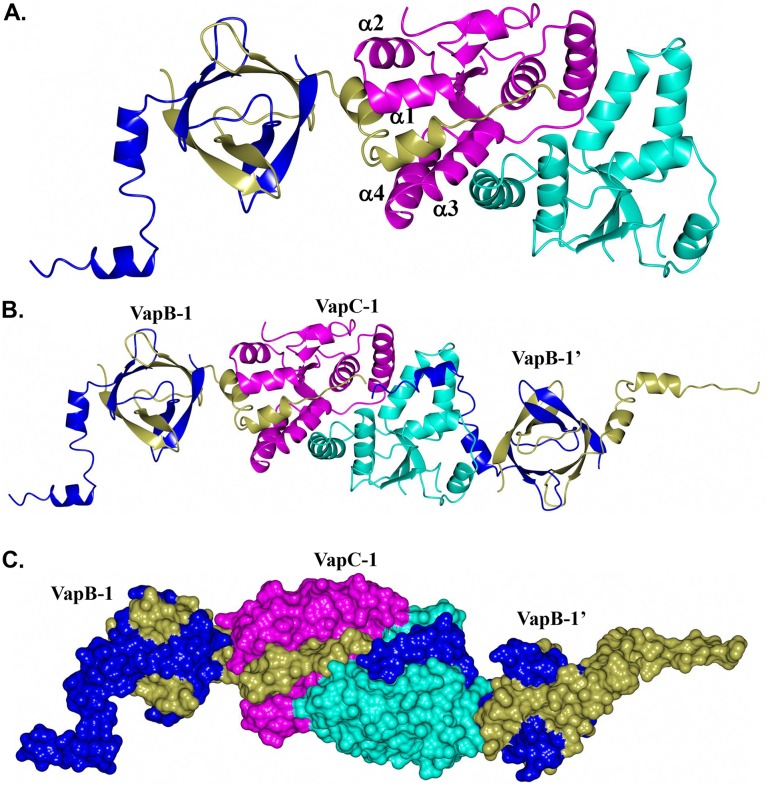
Although the toxin-antitoxin form commonly observed consists of 1:1 interactions between subunits, the quaternary arrangement resulting from the binding of NTHi VapB-1 to VapC-1 in this structure appears to be unique. As shown in Fig. 5A, the asymmetric unit consists of a VapB-1 and VapC-1 dimer in which the C-terminal helices of VapB-1 (subunit C) occupy a cleft formed by α1 to α4 of VapC-1 (subunit A). Subunit B of VapC-1 forms a similar interaction with VapB-1 (subunit D). However, this interaction is with a VapB-1 molecule related by crystallographic symmetry (Fig. 5B) and creates tightly packed pairs of VapB-1 and VapC-1 dimers (Fig. 5C). This arrangement results in a row of alternating VapB-1 and VapC-1 dimers that propagate (21 screw) along the crystallographic c axis, as shown in Fig. S4.
FIG 5.
Structure of VapBC-1 with VapB-1 subunits C/D (gold/blue) and VapC-1 subunits A/B (magenta/cyan). (A) Asymmetric unit showing the positioning of the C-terminal tail of VapB-1 (subunit C) occupying the cleft formed by α1 to α4 of VapC-1 (subunit A); (B) asymmetric unit with an extra VapB-1 dimer related by crystallographic symmetry (VapB-1’); (C) same as that shown in panel B but rendered as a surface.
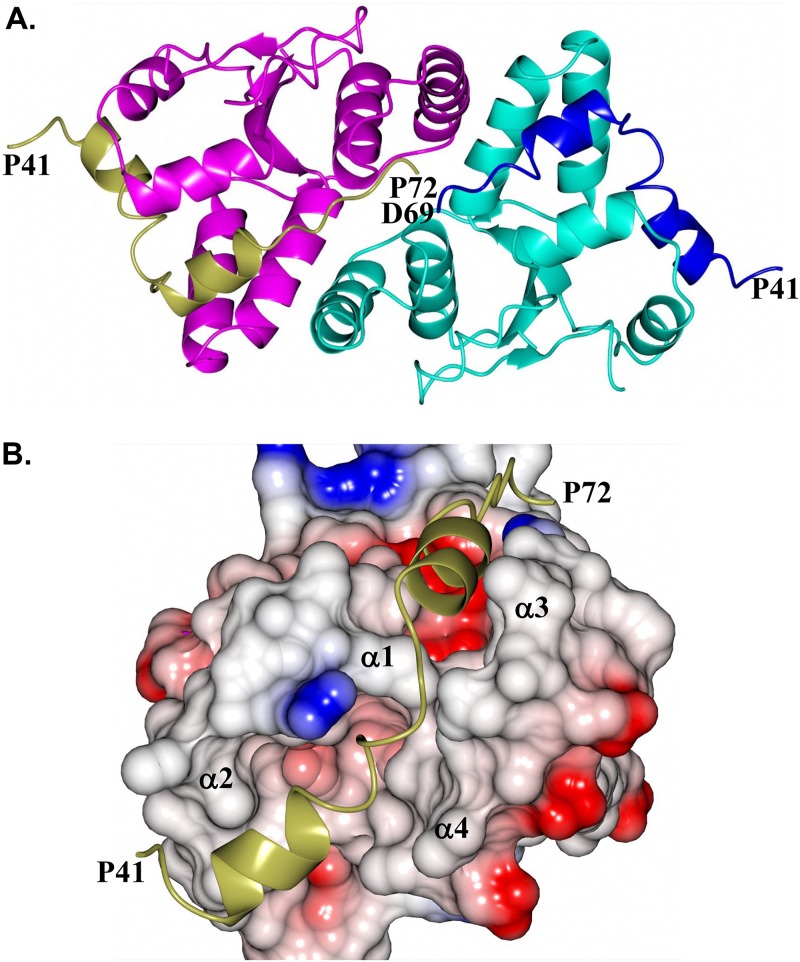
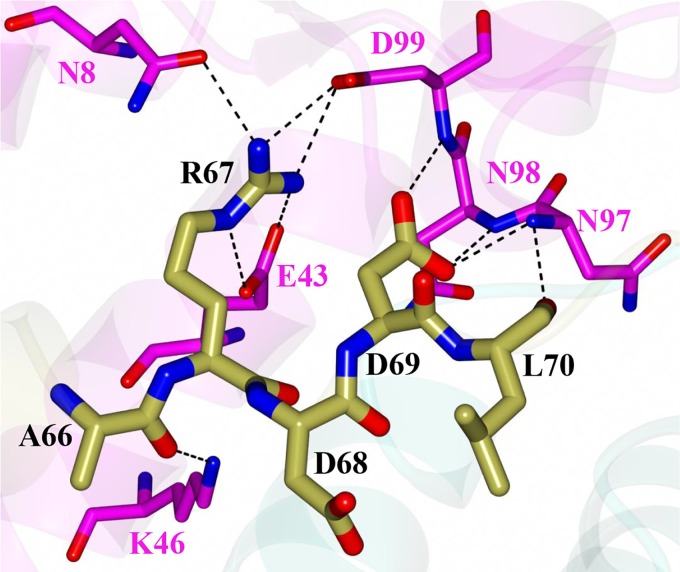
As noted above, the C-terminal tails of VapB-1 are positioned within a cleft formed by α1 to α4 of a VapC-1 dimer. Specifically, the C-terminal tail spanning P41 to P72 (subunit C) and P41 to D69 (subunit D) of VapB-1 (Fig. 6A) position the α-helices within a complementary cleft of VapC-1 subunits A and B, respectively (Fig. 6B). The binding interface between VapB-1 and VapC-1 is 1,385 Å2 with a solvation free-energy gain of −21.8 kcal/mol. The majority of the hydrogen bond and salt bridge interactions occur at the C terminus of VapB-1, spanning residues A66 to L70 (Fig. 7). Notably, R67 of VapB-1 interacts with E43 and D99 of VapC-1, which are conserved catalytic residues along with D6 and E120, among various VapC species (45). Similar interactions were observed for the S. flexneri VapBC structure (PDB no. 3TND), suggesting a similar mode of VapC-1 inhibition. Additional hydrogen bond interactions are observed between G55 and N52 of VapB-1 and N56 and N52 of VapC-1 (not shown).
FIG 6.
Binding of VapB-1 to VapC-1. (A) Zoomed-in view of Fig. 5B showing the binding of the C-terminal tail of VapB-1 subunit C residues P41 to P72 (gold) and subunit D residues P41 to D69 (blue) bound to the VapC-1 dimer; (B) electrostatic surface of VapC-1 subunit A showing the binding of the C-terminal α-helices of VapB-1 (P41 to P72, gold) positioned within the cleft formed by α1 to α4 of VapC-1.
FIG 7.
Hydrogen bond interactions between the C-terminal end of VapB-1 subunit C (gold) and VapC-1 subunit A (magenta).
In an effort to determine the oligomeric state of VapBC-1 in solution, a sample was analyzed by size exclusion chromatography and SDS-PAGE. A peak was observed at an elution volume of 13.70 ml, which corresponds to an approximate molecular weight of 100,786 Da (Fig. S5A, peak F). The calculated molecular weights of the VapB-1 and VapC-1 dimers are 21,722 Da and 33,742 Da, respectively, and the molecular weight of the two dimers of both the antitoxin and the toxin is 110,928 Da. Therefore, this peak most likely arises from the T4A4 oligomer. Notably, this peak contains both VapB-1 and VapC-1, as determined by SDS-PAGE (Fig. S5B). However, additional peaks were observed at larger elution volumes that correspond to species with molecular weights lower than those of the VapB-1 and VapC-1 subunits. Surprisingly, each of these peaks contains both VapB-1 and VapC-1, as observed by SDS-PAGE. It is possible that this phenomenon arises from nonspecific interactions between the column matrix and the protein complex, retarding its elution and resulting in both proteins appearing in each peak fraction.
Protein-protein interactions in vivo.
Proteins encoded by type II TA modules form nontoxic heterocomplexes of the antitoxin and toxin homodimers under normal growth conditions in vivo (37). In order to examine antitoxin and toxin homodimer formation and heterodimerization, a bacterial LexA-based system (46) was employed. The LexA repressor consists of two domains, a DNA-binding domain (DBD) and a dimerization domain. The DBD can function as a transcriptional repressor only in dimeric form. Other domains can be fused in-frame to the DBD and will restore the repressor’s function only if they interact (47). If the fused proteins interact, the lacZ reporter gene is repressed, reducing β-galactosidase activity, which is quantitated biochemically as dimensionless Miller units (48). Strong protein-protein interactions result in minimal β-galactosidase activity, whereas weak interactions result in significant activity.
This system was applied to evaluate the ability of VapC-1 single and double mutants to interact with wild-type (WT) VapB-1 antitoxin (heterodimerization) as well as with themselves (homodimerization) in vivo. The homodimerization of the wild-type VapC-1 toxin was not evaluated due to its substantial toxicity when overexpressed in a high-copy-number plasmid. Table 1 shows that the VapC-1 mutants retained the ability to homodimerize with comparative strengths of interaction of E120Q > D6N > E43Q D99N > E43Q > D99N > D6N D99N mutants. Mutant or wild-type VapC-1 toxins could heterodimerize with wild-type VapB-1 at relative levels of D6N > D6N D99N > E43Q D99N > E120Q > WT > E43Q > D99N mutants (Table 2).
TABLE 1.
Homodimerization of VapC-1 mutantsa
| Construct | Homodimerization of LexA DBD fusion in SU101 (Miller units) |
|
|---|---|---|
| Avg | ±SD | |
| pSR658 (control) | 1,302.6 | 78.2 |
| VapC-1 D6N | 38.5 | 12.9 |
| VapC-1 D99N | 59.5 | 9.4 |
| VapC-1 D6N D99N | 148.3 | 24.8 |
| VapC-1 E43Q | 55.5 | 9.8 |
| VapC-1 E43Q D99N | 39.2 | 2.2 |
| VapC-1 E120Q | 27.6 | 8.4 |
The homodimerization of the wild-type VapC-1 toxin was not evaluated due to its substantial toxicity when overexpressed in high copy number.
TABLE 2.
Heterodimerization of VapC-1 mutants with the wild-type VapB-1 antitoxin
| Construct | Heterodimerization of LexA DBD fusion in SU202 (Miller units) |
|
|---|---|---|
| Avg | ±SD | |
| Control vector | 1,813.4 | 130.0 |
| VapC-1 D6N | 37.9 | 11.5 |
| VapC-1 D99N | 360.5 | 30.6 |
| VapC-1 D6N D99N | 56.0 | 6.0 |
| VapC-1 E43Q | 202.2 | 18.9 |
| VapC-1 E43Q D99N | 113.3 | 5.5 |
| VapC-1 E120Q | 167.3 | 11.6 |
| VapC-1 wild type | 190.4 | 6.4 |
VapC-1 activity in vivo.
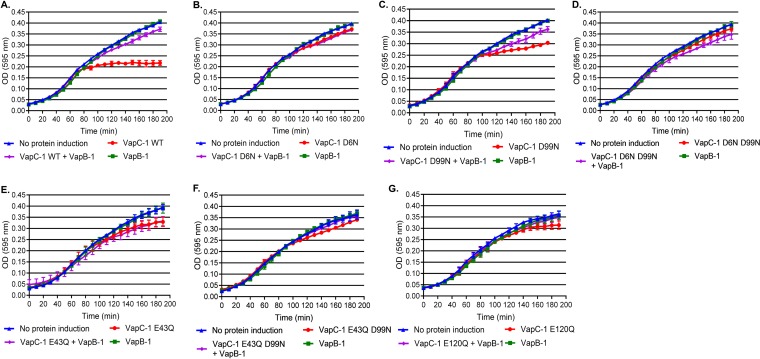
A growth recovery assay was used to evaluate VapC-1 activity in E. coli. Mutant or wild-type VapC-1 toxins were individually cloned under the control of the PBAD promoter and induced with 3.3 mM (0.05%, wt/vol) arabinose (49). The wild-type VapB-1 antitoxin was under the control of the Ptrc promoter, and its expression was induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) (50). Thus, VapC-1 and VapB-1 could be induced either independently or simultaneously from compatible plasmids that are cotransformed and expressed in the same E. coli strain, LMG 194. To initiate the assay, a culture in early growth phase was split and treated with (i) medium only, (ii) arabinose only, (iii) IPTG only, or (iv) both arabinose and IPTG. Typically, significant growth arrest occurred at approximately 70 min after wild-type VapC-1 was induced, but this was not observed if the antitoxin was expressed alone (Fig. 8A, red and green symbols, respectively). If VapB-1 and VapC-1 were coinduced, the antitoxin inhibited the toxin RNase activity, and growth was similar to that of the noninduced control (Fig. 8A, purple and blue symbols, respectively). The results of experiments with the VapC-1 mutants are presented in Fig. 8B to G. Significantly attenuated growth was observed upon induction of VapC-1 in strains with the wild-type and D99N and E120Q mutant toxins (red symbols in Fig. 8A, C, and G, respectively), compared to the growth observed when VapB-1 was coinduced (purple diamonds). However, the VapC-1 D6N, D6N D99N, E43Q, and E43Q D99N mutants did not cause significant growth arrest when induced in vivo (Fig. 8B and D to F, red symbols).
FIG 8.
E. coli growth recovery assays with inducible wild-type or mutant VapC-1 toxins. (A) Noninduced E. coli growth (measured by absorbance at 595 nm) or growth following induction of wild-type (WT) VapC-1, VapB-1, or both wild-type VapC-1 and VapB-1. (B to G) Similar growth experiments were performed with the VapC-1 D6N (B), D99N (C), D6N D99N (D), E43Q (E), E43Q D99N (F), and E120Q (G) mutants. All data are plotted as the mean ± SD of the results of three biological replicates in triplicate. Statistical comparisons of growth following wild-type or mutant VapC-1 inductions with that of the corresponding noninduced controls were made using the repeated-measures ANOVA. The significance of the results were as follows: VapC-1 WT, P < 0.0001 (A); D6N mutant, P = 0.21 (B); D99N mutant, P = 0.0015 (C); D6N D99N mutant, P = 0.15 (D); E43Q mutant, P = 0.39 (E); E43Q D99N mutant, P = 0.28 (F); E120Q mutant, P = 0.017 (G).
Construction of an in cis ectopic system to study TA modules.
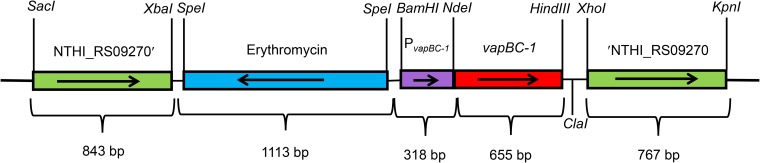
The growth recovery assay in E. coli strain LMG 194 is a valuable tool to evaluate the effects of toxin and antitoxin expression. However, the antitoxin and toxin proteins are cloned into separate multicopy vectors to allow independent expression and are controlled by artificial promoters. Therefore, the assay does not allow either expression via the native transcriptional regulation of the locus or the characterization of a mutant locus in single copy on the chromosome. To evaluate these aspects, a novel system was developed that consisted of the native vapBC-1 promoter controlling an operon containing the wild-type vapB-1 gene, followed by either the wild-type or each mutant vapC-1 gene (Fig. 9). This system mimics the natural sequential arrangement and copy number of the operon. The module is flanked by homologous regions of the metE pseudogene from NTHi strain 86-028NP (NTHI_RS09270; an authentic frameshift resulting in a deletion of over 100 amino acids), which targets the reconstructed TA locus to a chromosomal region that is not in use. Briefly, fusions are amplified by PCR, used to transform NTHi, and homologously recombined into the chromosome at the NTHI_RS09270 pseudogene site in single copy. NTHi is an obligate human parasite with natural deletions in many genes involving metabolism. Since it scavenges a number of essential molecules from its host, this results in the requirement to supplement the growth medium in vitro but not in vivo or ex vivo (51–53). The well-characterized 86-028NP ΔvapBC-1 strain (7) was chosen as the background to analyze each tandem fusion to eliminate contributions by the wild-type vapBC-1 locus. This novel approach is the most biologically relevant system currently available to study the effects of TA locus mutants in the background of NTHi.
FIG 9.
NTHi delivery vector for single-copy in cis expression of cloned genes. This construct is targeted to NTHI_RS09270, the metE pseudogene in strain 86-028NP. The native vapBC-1 promoter controls either the wild-type vapBC-1 operon or an engineered operon consisting of the wild-type vapB-1 antitoxin and one of the vapC-1 mutated toxins.
To ensure that the in cis mutant modules had no effect on the replication of strains, the growth of the reconstructed wild type, the vapBC-1 mutants, and the parent strain 86-028NP ΔvapBC-1 was measured over 10 h. There were no significant differences in growth among the strains (data not shown), supporting the use of this model to investigate variations in pathogenesis attributable to mutations in VapC-1.
Effect of VapC-1 mutations on survival of NTHi in primary human tissues.
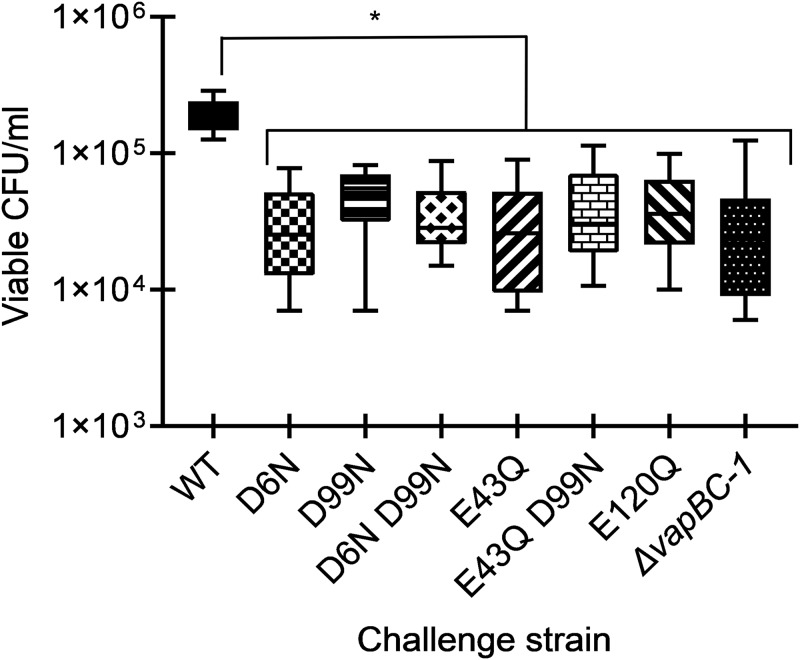
The EpiAirway model (AIR 100 ABF; MatTek, Ashland, MA, USA), which consists of antibiotic-free primary human respiratory epithelial tissue at the air-liquid interface, was used to evaluate the impact of VapC-1 mutations on NTHi survival during infections. After coculturing the in cis-complemented strains with EpiAirway tissues in antibiotic-free medium for 48 h, the tissues were treated with 100 μg/ml gentamicin for 90 min to kill any NTHi not internalized, as this antibiotic does not enter the tissues. The surviving internalized bacteria were recovered, diluted, and plated for viable colony counts. This experiment was performed with six tissues originating from at least two different EpiAirway kits. The survival of all strains containing VapC-1 mutants was significantly reduced compared to that of the strain containing wild-type toxin (Fig. 10). The survival of the parent strain, 86-028NP ΔvapBC-1, was also significantly decreased, as previously reported (7).
FIG 10.
Number of viable CFU/milliliter of NTHi in cis-complemented mutants or the wild-type operon in primary human tissues at the air-liquid interface. EpiAirway tissues were infected at ∼1.0 × 104 CFU per insert with either the 86-028NP ΔvapBC-1 deletion strain, the 86-028NP ΔvapBC-1 NTHI_RS09270::PvapBC-1::vapB-1vapC-1 (wild-type) strain, the 86-028NP ΔvapBC-1 NTHI_RS09270::PvapBC-1::vapB-1vapC-1 (D6N) strain, the 86-028NP ΔvapBC-1 NTHI_RS09270::PvapBC-1::vapB-1vapC-1 (D99N) strain, the 86-028NP ΔvapBC-1 NTHI_RS09270::PvapBC-1::vapB-1vapC-1 (D6N D99N) strain, the 86-028NP ΔvapBC-1 NTHI_RS09270::PvapBC-1::vapB-1vapC-1 (E43Q) strain, the 86-028NP ΔvapBC-1 NTHI_RS09270::PvapBC-1::vapB-1vapC-1 (E43Q D99N) strain, the 86-028NP ΔvapBC-1 NTHI_RS09270::PvapBC-1::vapB-1vapC-1 (E120Q) strain, or the 86-028NP ΔvapBC-1 parent strain. At 48 h after infection, gentamicin-resistant bacteria were harvested for colony counts. Data are expressed as the mean ± SD (n = 6). All in cis mutants and the parent strain were significantly attenuated for survival compared to the strain carrying the wild-type VapC-1 in cis. An asterisk denotes a P of ≤0.05.
DISCUSSION
The VapC-1 toxin of NTHi is an efficient and highly active RNase, and the vapBC-1 locus is crucial for survival during infection, both ex vivo in primary human tissues and in vivo in the chinchilla model of acute otitis media (7). The PIN domain of VapC homologues has been implicated in the binding and cleavage of RNA substrates (42), and many acidic residues in this domain are highly conserved (45). Other studies have evaluated various mutations to the PIN domain (54, 55), and we previously demonstrated that the VapC-1 D99N mutant had significantly reduced RNase activity, while retaining the ability to bind VapB-1 (56).
The NTHi VapBC-1 structure provides insight into heterodimeric interactions and enables comparisons with VapBC complexes from other organisms. Overall, the VapBC-1 homodimer subunits are similar to other reported VapB and VapC structures, adopting similar protein-protein interactions with the VapB C-terminal helices occupying clefts within VapC dimers. The homodimer subunits in VapBC complexes often form higher-order oligomers, such as heterotetramers, heterohexamers, and heteroctamers (31). Interestingly, the VapBC-1 complex does not adopt any of these higher-order quaternary structures but instead forms a unique linear array of alternating VapB and VapC dimers. It should be noted that the VapB construct used for crystallization contains 18 additional residues at the N terminus from the cloning vector pET24b, which were mostly disordered. Therefore, it is unclear if this unique quaternary structure arrangement is an artifact resulting from these additional residues.
Based on the crystal structure of VapBC-1, two highly conserved aspartate residues and two glutamate residues in the toxin PIN domain were mutated to their beta-amino derivatives, asparagine and glutamine. Analyses of these mutant constructs indicated decreased RNase activity in vivo measured by growth recovery assays and decreased survival during ex vivo infections of primary human tissues compared to that of the wild-type toxin when expressed in the background of NTHi using a novel single-copy in cis system. We also found that the effects of these mutations were not necessarily additive, since although a VapC-1 D99N mutant was significantly attenuated when overexpressed in E. coli growth recovery assays, a D6N D99N double mutant was not. Interestingly, all VapC-1 mutants were able to strongly homodimerize as well as to bind wild-type VapB-1, suggesting that the module’s quaternary structure is somewhat decoupled from the enzymatic activity of the toxin.
The results reported here support the notion that VapBC-1 (and TA modules in general) have evolved a number of mechanisms to maintain their activity and that their presence confers a significant advantage to the microorganism. Many studies have linked TA modules to the survival of bacterial pathogens in host tissues (7, 56–60). Achieving and maintaining persistence during an infection is central for both successfully evading the immune response and for nonspecific antibiotic tolerance, leading to a pathogen’s survival of clinical therapy. This characteristic can substantially contribute to recurrent or chronic infections and implicates TA modules as therapeutic targets. Indeed, when we tested the effects of the toxin mutations on NTHi survival inside primary human respiratory tissues ex vivo, we found that any perturbation of the acidic residues in the conserved PIN domain negatively affected survival in comparison to the wild-type toxin under the same conditions. Our data suggest that targeting any of these individual residues in the toxin could potentially result in a significant decrease in survival of NTHi during infection and imply that small-molecule inhibitors could be most effective in this approach.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in these studies are listed in Table S1 in the supplemental material. E. coli strains were grown in LB broth or agar with or without 100 μg/ml ampicillin, 250 μg/ml erythromycin, 20 μg/ml chloramphenicol, 30 μg/ml of kanamycin, or 12 μg/ml tetracycline, as required. NTHi strains were grown in brain heart infusion broth or agar supplemented with 10 μg/ml heme-histidine and 10 μg/ml β-NAD (sBHI) or on chocolate agar with 5 U/ml bacitracin. NTHi was routinely cultured at 37°C with 5% CO2. To construct the ectopic strains, transformants were selected on chocolate agar plates with 5 μg/ml erythromycin, and single colonies were passaged for 3 days on selection plates before being confirmed by DNA sequencing of PCR-amplified purified genomic DNA.
Site-directed mutagenesis of VapC-1.
VapC-1 D6N and D99N mutants cloned in tandem with a wild-type VapB-1 in the vector pET24b (Millipore Sigma, Burlington, MA) were previously described (61). These constructs were used as the templates for plasmid expression sets. Synthetic constructs of VapC-1 with E43Q, E120Q, and E43Q D99N mutations were generated in tandem with wild-type VapB-1 by Eurofins (Eurofins Genomics, Louisville, KY). The VapC-1 D6N D99N construct, also in tandem with the wild-type VapB-1, was produced by GenScript (Piscataway, NJ). Each construct was confirmed by DNA sequencing (Eurofins Genomics, Louisville, KY) following subcloning into the vectors listed in Table S1.
Purification of VapC-1 for crystallization studies.
To overexpress VapBC-1 for purification, pDD686 in BL21(DE3) (17) was grown to logarithmic phase at 37°C with shaking, induced for 18 h with 1 mM IPTG at 26°C, and harvested at 2,500 × g into pellets from 25-ml aliquots. After one freeze-thaw cycle, VapBC-1 was isolated in accordance with the MagneHis protein purification system (Promega, Madison, WI) with the following exceptions: the lysis solution was 1× BugBuster protein extraction reagent (Millipore Sigma, Burlington, MA) with 100 mM HEPES (pH 7.2), 300 mM NaCl, 1× ProteCEASE-50 (EDTA free), and 2 μl/ml RNase-free DNase (Thermo Fisher Scientific, Waltham, MA); the wash buffer contained 100 mM HEPES (pH 7.2), 300 mM NaCl, and 20 mM imidazole; and the elution buffer contained 100 mM HEPES (pH 7.2), 300 mM NaCl, and 500 mM imidazole. One microgram of purified VapBC-1 was separated on a Bolt precast 4 to 12% polyacrylamide gel and analyzed via Western blotting using the iBlot and iBind system (Thermo Fisher Scientific, Waltham, MA). The nitrocellulose blot was probed with a monoclonal anti-His tag primary antibody (THE anti-His; GenScript, Piscataway, NJ), goat anti-mouse IgG–horseradish peroxidase (HRP) secondary antibody (GenScript, Piscataway, NJ), detected with SuperSignal West Dura chemiluminescence substrate (Thermo Fisher Scientific, Waltham, MA) and imaged on a UVP BioSpectrum 815 system (UVP LLC, Upland, CA).
Crystallization and data collection.
A purified preparation of the NTHi VapBC-1 protein complex was concentrated to 20 mg/ml in 20 mM NaCl, 10 mM HEPES (pH 7.2) for crystallization screening. All crystallization experiments were set up using an NT8 drop setting robot (Formulatrix, Inc.) and UVXPO MRC (Molecular Dimensions) sitting drop vapor diffusion plates at 18°C. One hundred nanoliters of protein and 100 nl of crystallization solution were dispensed and equilibrated against 50 μl of the latter. Crystals approximately 100 μm long that displayed a needle morphology (Fig. S6) were observed within 1 week from the Proplex screen (Molecular Dimensions) condition C3 (20% [wt/vol] PEG 4000, 100 mM sodium acetate [pH 5.0], 200 mM ammonium acetate). A cryoprotectant solution composed of 80% crystallization solution and 20% (vol/vol) PEG 200 was dispensed (2 μl) onto the drop, and crystals were harvested with a cryoloop immediately and stored in liquid nitrogen. X-ray diffraction data were collected at the Advanced Photon Source beamline 17-ID using a Dectris Pilatus 6 M pixel array detector.
Structure solution and refinement.
Intensities were integrated using XDS (62, 63) via Autoproc (64), and the Laue class analysis and data scaling, using two data sets to increase the multiplicity, were performed with Aimless (65), which indicated that the crystals belong to the mmm Laue class. Structure solution was conducted by molecular replacement with Morda (66) to place a VapC-1 dimer using Shigella flexneri VapC (PDB no. 5ECD) as the template, and the top solution was obtained in the space group P212121. The VapC-1 coordinates from Morda were input as a fixed solution into Phaser (67), and a VapB monomer from the Rickettsia felis VapBC structure (PDB no. 3ZVK) was used as a search model. The final solution from Phaser consisted of a VapB-1 and VapC-1 dimer in the asymmetric unit, and the model was improved by automated building with Arp/wARP (68). Further refinement and manual model building were conducted with Phenix (69) and Coot (70), respectively. Disordered side chains were truncated to the point at which electron density could be observed. Structure validation was conducted with Molprobity (71), and figures were prepared using the CCP4MG package (72). Structure superposition was carried out with GESAMT (73). Crystallographic data are provided in Table 3.
TABLE 3.
Crystallographic data for VapBC-1
| Parameter | Value(s) for VapBC-1 |
|---|---|
| Data collection statistics | |
| Unit cell parameters (Å, °) | a = 43.88, b = 57.32, c = 175.75 |
| Space group | P212121 |
| Resolution (Å)a | 48.01–2.20 (2.28–2.20) |
| Wavelength (Å) | 1.0000 |
| Temp (K) | 100 |
| No. of observed reflections | 303,859 |
| No. of unique reflections | 23,388 |
| <l/σ(l)>a | 10.9 (2.1) |
| Completeness (1%)a | 100 (100) |
| Multiplicitya | 13.0 (13.7) |
| Rmerge (%)a ,b | 16.8 (161.3) |
| Rmeas (%)a ,d | 17.5 (167.7) |
| Rpim (%)a ,d | 4.9 (45.2) |
| CC1/2a ,e | 0.998 (0.784) |
| Refinement statistics | |
| Resolution (Å)a | 48.01–2.20 |
| No. of reflections (working/test)a | 22,146/1,166 |
| Rfactor/Rfree (%)a ,c | 18.6/23.4 |
| No. of atoms (VapB-1/VapC-1/water) | 1,085/2,103/58 |
| Model quality | |
| RMS deviations | |
| Bond length (Å) | 0.009 |
| Bond angle (°) | 0.981 |
| Avg B-factor (Å2) | |
| All atoms | 43.5 |
| VapB-1 | 48.0 |
| VapC-1 | 41.3 |
| Water | 41.8 |
| Coordinate error (maximum likelihood) (Å) | 0.24 |
| Ramachandran plot | |
| Most favored (%) | 96.8 |
| Additionally allowed (%) | 3.0 |
Values in parenthesis are for the highest-resolution shell.
Rmerge = ΣhklΣi |Ii(hkl) – <I(hkl)>|/ΣhklΣi Ii(hkl), where Ii(hkl) is the intensity measured for the ith reflection and <I(hkl)> is the average intensity of all reflections with indices hkl.
Rfactor = Σhkl ǁFobs (hkl) | − |Fcalc (hkl)ǁ/Σhkl |Fobs (hkl)|; Rfree is calculated in an identical manner using 5% of randomly selected reflections that were not included in the refinement.
Size exclusion chromatography.
A 0.5-ml sample (5.8 mg/ml) of the coexpressed and purified VapBC-1 construct was loaded onto a Superdex 200 10/300 GL column (GE Healthcare) equilibrated with 200 mM NaCl, 50 mM HEPES (pH 7.2). The sample was applied at a flow rate of 0.5 ml using an AKTA pure fast protein liquid chromatograph, and 1-ml fractions were collected. Chromatography data were analyzed using the Unicorn 7 software package (GE Healthcare). The column was calibrated using protein standard mixture containing thyroglobulin (670,000 Da), gamma globulin (158,000 Da), ovalbumin (44,000 Da), myoglobin (17,000 Da), and vitamin B12 (1,350 Da). The molecular weights of the VapBC-1 peak fractions were estimated relative to the least-squares fit of the calibration standards.
Cloning and protein-protein interactions of VapC-1 mutants.
The VapC-1 D99N, E43Q, E43Q D99N, and E120Q mutants were cloned by amplifying the vapC-1 D99N gene from pDD758, the vapC-1 E43Q gene from pDD1165, the vapC-1 E43Q D99N gene from pDD1196, or the vapC-1 E120Q gene from pDD1166 using high-fidelity Phusion Flash DNA polymerase (Thermo Fisher Scientific, Waltham, MA, USA) and the primers 322LexFor and 322LexRev. The PCR products, pSR658, and pSR659 were digested with SacI and KpnI prior to ligation, resulting in pDD1120, pDD1123, pDD1170, pDD1178, pDD1199, pDD1200, pDD1177, and pDD1179. VapC-1 D6N and D6N D99N were cloned by amplifying the vapC-1 D6N gene from pDD757 and the vapC-1 D6N D99N gene from pDD1118, using high-fidelity Phusion Flash DNA Polymerase with pETD6NC1SacFor and 322LexRev primers. The PCR products, pSR660, and pSR661 were digested with SacI and KpnI prior to ligation, resulting in pDD1125, pDD1126, pDD1129, and pDD1128. The wild-type VapC-1 toxin was cloned into pSR659 with the above-described method from NTHi strain 86-028NP genomic DNA (pDD1150). All clones were confirmed by DNA sequencing. The pSR659 or pSR661 constructs were cotransformed with pDD866 into SU202 to perform heterodimerization assays. The pSR658 or pSR660 constructs were individually transformed into SU101 for homodimerization assays.
Expression of the LexA fusions was induced by IPTG, and if the fusion proteins interacted, a competent repressor that could bind the operator site and decrease transcription of the lacZ reporter gene was formed in the reporter strain. This resulted in a measurable decrease in β-galactosidase activity compared to a strain carrying the control unfused vectors. However, because β-galactosidase is a long-lived enzyme, our reporter strains were grown overnight in the presence of 1 mM IPTG so that any enzyme expressed prior to the LexA chimera induction was degraded. This strategy results in a more reliable and accurate quantification of homodimerization and heterodimerization. Following overnight incubation on LB agar plates with 1 mM IPTG and appropriate antibiotics, reporter strains carrying the control vectors (pSR658 and pSR659 or pSR660 and pSR661) (47) or the LexA DBD fusions were inoculated and grown to an optical density at 600 nm (OD600) of ∼0.5 in LB broth with 1 mM IPTG and appropriate antibiotics. Protein-protein interactions were quantified by β-galactosidase activity assays and compared to the activity of the reporter strain carrying the unfused vectors, which has the highest activity due to the lack of repressor formation. The algorithm for quantifying β-galactosidase activity was as follows: [OD420 – (1.75 × OD550)/(t × v × OD600)] × 1,000, where t is time of reaction development in minutes and v is volume of sample in milliliters (48). This equation allows for the normalization of different culture densities for comparison purposes.
Cloning and growth-recovery assays with VapC-1.
To clone wild-type and mutant VapC-1 genes for the growth recovery assays, the wild-type vapC-1 gene was amplified from NTHi strain 86-028NP genomic DNA, the vapC-1 D6N gene was amplified from pDD757, the vapC-1 D99N gene was amplified from pDD758, the vapC-1 D6N D99N gene was amplified from pDD1118, the vapC-1 E43Q gene was amplified from pDD1165, the vapC-1 E43Q D99N gene was amplified from a synthetic gene produced by Eurofins, and the vapC-1 E120Q gene was amplified from pDD1166 using high-fidelity Phusion Flash DNA polymerase with pBAD86C1SacFor and 322Rev primers. The PCR products and pBAD33 were digested with SacI and HindIII prior to ligation, resulting in pDD946, pDD1132, pDD1133, pDD1119, pDD1175, pDD1194, and pDD1174. The forward primer was designed to include the putative Shine-Dalgarno sequence for VapC-1. The vapB-1 gene was amplified from 86-028NP genomic DNA using high-fidelity Phusion Flash DNA polymerase with 321LexFor and 321LexRev primers. After digestion with SacI and KpnI the PCR fragment was ligated into pTrcHisA, resulting in pDD935. All clones were verified by DNA sequencing. Wild-type vapC-1 and each of the six mutants were cotransformed with pDD935 into E. coli strain LMG 194.
To perform the growth recovery assays, strains were grown at 37°C to early log phase (OD600, ∼0.2 to 0.3) in LB broth with 20 μg/ml chloramphenicol and 100 μg/ml ampicillin before being diluted in a 1:1 (vol/vol) ratio to a final volume of 150 μl of medium in a 96-well Falcon polystyrene microplate (Corning, Corning, NY) containing LB broth with 20 μg/ml chloramphenicol and 100 μg/ml ampicillin, with or without 1 mM IPTG, 6.6 mM (0.1%, wt/vol) arabinose, or both. The final induction concentrations were 0.5 mM IPTG and 3.3 mM (0.05%, wt/vol) arabinose. The plate was covered with a gas-permeable membrane, loaded into a Multiskan FC incubating plate reader (Thermo Fisher Scientific, Waltham, MA), and incubated at 37°C with a shaking protocol of 5 s on and 5 s off, with measurements taken every 10 min at OD595 for 190 min. Each assay included three biological replicates with three technical replicates.
Construction of the in cis delivery system and reconstituted NTHi strains.
Based on a previously published ectopic delivery vector in NTHi (74), the metE pseudogene in the chromosome of strain 86-028NP (NTHI_RS09270) was targeted as the site for gene delivery and single-copy expression in cis. The delivery vector was constructed by amplifying 843 bp of the 5′ end of NTHI_RS09270 from 86-028NP genomic DNA using high-fidelity Phusion Flash DNA polymerase with 2009-1SacFor and 2009-1XbaRev primers and ligating the product into pBluescript SK+ digested with SacI and XbaI, resulting in pDD1058. The second homologous region of 767 bp of the 3′ end of NTHI_RS09270 was amplified from 86-028NP genomic DNA using high-fidelity Phusion Flash DNA polymerase with 2009-2XhoFor and 2009-2KpnRev primers and ligated into pDD1058 cut with XhoI and KpnI, resulting in pDD1063. The last step was amplifying an erythromycin resistance cassette from pEJ18 (75) with PermCSpeFor and ErmCSpeRev primers and ligating into pDD1063 cut with SpeI, resulting in the delivery vector, pDD1140. The native vapBC-1 promoter from 86-028NP was cloned into the multiple cloning site of the delivery vector using the primers BC1PromBamFor and BC1PromNdeRev. The reverse primer contains an engineered NdeI site (CATATG) in which the ATG of the restriction site (underlined) serves as the start codon for the fused vapBC-1 genes. This allowed us to clone all constructs with their native transcriptional regulation and organization. The vapBC-1 promoter was ligated to pDD1140 cut with BamHI and EcoRV, resulting in pDD1155. Because the vapC-1 mutants are in tandem with the wild-type vapB-1 gene, each was cloned into pDD1155 for delivery into the 86-028NP chromosome. The vapBC-1 D6N locus was amplified from pDD757, vapBC-1 D99N was amplified from pDD758, vapBC-1 D6N D99N was amplified from pDD1118, vapBC-1 E43Q was amplified from pDD1165, vapBC-1 E43Q D99N was amplified from a synthetic gene produced by Eurofins, vapBC-1 E120Q was amplified from pDD1166, and wild-type vapBC-1 was amplified from pDD686 using high-fidelity Phusion Flash DNA polymerase with 86B1NdeFor and 322Rev primers. Each amplicon was ligated to pDD1155 digested with NdeI and HindIII, resulting in pDD1159, pDD1160, pDD1161, pDD1167, pDD1204, pDD1168, and pDD1169. This resulted in vapBC-1 with the desired mutations in vapC-1 under the control of its native promoter (Fig. 9).
The strains of NTHi 86-028NP ΔvapBC-1 NTHI_RS09270::PvapBC-1::vapB-1vapC-1 with wild-type vapC-1 and the six mutants were constructed by homologous recombination into the chromosome of 86-028NP ΔvapBC-1 by PCR products containing both the flanking regions of NTHI_RS09270 and each promoter-tandem gene clone using MIV transformation (53). After selection on chocolate agar with 5 μg/ml erythromycin, a single colony of each transformant was passaged three times on antibiotic-containing plates. DNA sequencing of PCR-amplified genomic DNA preparations confirmed each correct strain.
To ensure that the in cis fusions did not affect the growth dynamics of NTHi, strains were grown to early log phase (OD600, ∼0.2 to 0.3) in sBHI broth before dilution into a 96-well Falcon polystyrene microplate, and absorbance was measured by following the same protocol as that of the growth recovery assays over the course of 10 h. Each growth assay included three biological replicates with three technical replicates.
NTHi infections of primary human tissues.
EpiAirway tissues (AIR-100-ABF; MatTek, Ashland, MA) were maintained, inoculated, and harvested as previously described (76), with the following exception: inserts were inoculated with approximately 25 μl of a 1.0 × 104 CFU/ml bacterial suspension of the desired strain. Inserts were cocultured with the parent, wild-type, or mutant strains for 48 h before harvesting and quantification by counting viable colonies from a plated dilution series on chocolate agar plates.
Statistical analyses.
Differences between means were determined using Student's t test. Differences among multiple-group treatments were determined by the repeated-measures analysis of variance (ANOVA) or two-way ANOVA as appropriate using Prism 8.0.1 (GraphPad Software, La Jolla, CA). A P value of ≤0.05 was considered statistically significant.
Data availability.
Coordinates and structure factors were deposited in the worldwide Protein Data Bank (wwPDB) under accession code 6NKL.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the National Institute on Deafness and Other Communication Disorders cooperative agreement U01 DC014756 to D.A.D. N.P.C. and M.S. were supported by the National Center for Advancing Translational Sciences (NCATS) Division of Pre-Clinical Innovation Intramural Program. Use of the University of Kansas Protein Structure Laboratory was supported by a grant from the National Institute of General Medical Sciences (P30 GM110761) at the National Institutes of Health. Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Hauptman-Woodward Medical Research Institute. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02-06CH11357.
We thank L. C. Zavada for technical support.
The National Institute on Deafness and Other Communication Disorders (NIDCD) had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00026-19.
REFERENCES
- 1.Gerdes K, Rasmussen PB, Molin S. 1986. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci U S A 83:3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogura T, Hiraga S. 1983. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc Natl Acad Sci U S A 80:4784–4788. doi: 10.1073/pnas.80.15.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masuda Y, Miyakawa K, Nishimura Y, Ohtsubo E. 1993. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J Bacteriol 175:6850–6856. doi: 10.1128/jb.175.21.6850-6856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerdes K. 2000. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J Bacteriol 182:561–572. doi: 10.1128/JB.182.3.561-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamaguchi Y, Park JH, Inouye M. 2011. Toxin-antitoxin systems in bacteria and archaea. Annu Rev Genet 45:61–79. doi: 10.1146/annurev-genet-110410-132412. [DOI] [PubMed] [Google Scholar]
- 6.Correia FF, D'Onofrio A, Rejtar T, Li L, Karger BL, Makarova K, Koonin EV, Lewis K. 2006. Kinase activity of overexpressed HipA is required for growth arrest and multidrug tolerance in Escherichia coli. J Bacteriol 188:8360–8367. doi: 10.1128/JB.01237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren D, Walker AN, Daines DA. 2012. Toxin-antitoxin loci vapBC-1 and vapXD contribute to survival and virulence in nontypeable Haemophilus influenzae. BMC Microbiol 12:263. doi: 10.1186/1471-2180-12-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramage HR, Connolly LE, Cox JS. 2009. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet 5:e1000767. doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weaver KE, Reddy SG, Brinkman CL, Patel S, Bayles KW, Endres JL. 2009. Identification and characterization of a family of toxin-antitoxin systems related to the Enterococcus faecalis plasmid pAD1 par addiction module. Microbiology 155:2930–2940. doi: 10.1099/mic.0.030932-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen SK, Mikkelsen M, Pedersen K, Gerdes K. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc Natl Acad Sci U S A 98:14328–14333. doi: 10.1073/pnas.251327898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tachdjian S, Kelly RM. 2006. Dynamic metabolic adjustments and genome plasticity are implicated in the heat shock response of the extremely thermoacidophilic archaeon Sulfolobus solfataricus. J Bacteriol 188:4553–4559. doi: 10.1128/JB.00080-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemos JA, Brown TA, Abranches J, Burne RA. 2005. Characteristics of Streptococcus mutans strains lacking the MazEF and RelBE toxin-antitoxin modules. FEMS Microbiol Lett 253:251–257. doi: 10.1016/j.femsle.2005.09.045. [DOI] [PubMed] [Google Scholar]
- 13.Fernández-García L, Blasco L, Lopez M, Bou G, García-Contreras R, Wood T, Tomas M. 2016. Toxin-antitoxin systems in clinical pathogens. Toxins (Basel) 8:E227. doi: 10.3390/toxins8070227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Melderen L, Thi MH, Lecchi P, Gottesman S, Couturier M, Maurizi MR. 1996. ATP-dependent degradation of CcdA by Lon protease. Effects of secondary structure and heterologous subunit interactions. J Biol Chem 271:27730–27738. doi: 10.1074/jbc.271.44.27730. [DOI] [PubMed] [Google Scholar]
- 15.Aizenman E, Engelberg-Kulka H, Glaser G. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3',5'-bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci U S A 93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogel J, Argaman L, Wagner EG, Altuvia S. 2004. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr Biol 14:2271–2276. doi: 10.1016/j.cub.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Daines DA, Wu MH, Yuan SY. 2007. VapC-1 of nontypeable Haemophilus influenzae is a ribonuclease. J Bacteriol 189:5041–5048. doi: 10.1128/JB.00290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simic M, De Jonge N, Loris R, Vesnaver G, Lah J. 2009. Driving forces of gyrase recognition by the addiction toxin CcdB. J Biol Chem 284:20002–20010. doi: 10.1074/jbc.M109.014035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mutschler H, Gebhardt M, Shoeman RL, Meinhart A. 2011. A novel mechanism of programmed cell death in bacteria by toxin-antitoxin systems corrupts peptidoglycan synthesis. PLoS Biol 9:e1001033. doi: 10.1371/journal.pbio.1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masuda H, Tan Q, Awano N, Wu KP, Inouye M. 2012. YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol Microbiol 84:979–989. doi: 10.1111/j.1365-2958.2012.08068.x. [DOI] [PubMed] [Google Scholar]
- 21.Fozo EM, Hemm MR, Storz G. 2008. Small toxic proteins and the antisense RNAs that repress them. Microbiol Mol Biol Rev 72:579–589. doi: 10.1128/MMBR.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerdes K, Thisted T, Martinussen J. 1990. Mechanism of post-segregational killing by the hok/sok system of plasmid R1: sok antisense RNA regulates formation of a hok mRNA species correlated with killing of plasmid-free cells. Mol Microbiol 4:1807–1818. doi: 10.1111/j.1365-2958.1990.tb02029.x. [DOI] [PubMed] [Google Scholar]
- 23.Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP. 2009. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci U S A 106:894–899. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blower TR, Pei XY, Short FL, Fineran PC, Humphreys DP, Luisi BF, Salmond GP. 2011. A processed noncoding RNA regulates an altruistic bacterial antiviral system. Nat Struct Mol Biol 18:185–190. doi: 10.1038/nsmb.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leplae R, Geeraerts D, Hallez R, Guglielmini J, Drèze P, Van Melderen L. 2011. Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res 39:5513–5525. doi: 10.1093/nar/gkr131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winther KS, Gerdes K. 2011. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc Natl Acad Sci U S A 108:7403–7407. doi: 10.1073/pnas.1019587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan Q, Awano N, Inouye M. 2011. YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeleton proteins, FtsZ and MreB. Mol Microbiol 79:109–118. doi: 10.1111/j.1365-2958.2010.07433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Lord DM, Cheng HY, Osbourne DO, Hong SH, Sanchez-Torres V, Quiroga C, Zheng K, Herrmann T, Peti W, Benedik MJ, Page R, Wood TK. 2012. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat Chem Biol 8:855–861. doi: 10.1038/nchembio.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aakre CD, Phung TN, Huang D, Laub MT. 2013. A bacterial toxin inhibits DNA replication elongation through a direct interaction with the β sliding clamp. Mol Cell 52:617–628. doi: 10.1016/j.molcel.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandey DP, Gerdes K. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res 33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bendtsen KL, Brodersen DE. 2017. Higher-order structure in bacterial VapBC toxin-antitoxin complexes. Subcell Biochem 83:381–412. doi: 10.1007/978-3-319-46503-6_14. [DOI] [PubMed] [Google Scholar]
- 32.Wall D, Kaiser D. 1999. Type IV pili and cell motility. Mol Microbiol 32:1–10. doi: 10.1046/j.1365-2958.1999.01339.x. [DOI] [PubMed] [Google Scholar]
- 33.Liu M, Zhang Y, Inouye M, Woychik NA. 2008. Bacterial addiction module toxin Doc inhibits translation elongation through its association with the 30S ribosomal subunit. Proc Natl Acad Sci U S A 105:5885–5890. doi: 10.1073/pnas.0711949105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masuda H, Inouye M. 2017. Toxins of prokaryotic toxin-antitoxin systems with sequence-specific endoribonuclease activity. Toxins (Basel) 9:E140. doi: 10.3390/toxins9040140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casey JR, Pichichero ME. 2004. Changes in frequency and pathogens causing acute otitis media in 1995-2003. Pediatr Infect Dis J 23:824–828. doi: 10.1097/01.inf.0000136871.51792.19. [DOI] [PubMed] [Google Scholar]
- 36.Arcus VL, McKenzie JL, Robson J, Cook GM. 2011. The PIN-domain ribonucleases and the prokaryotic VapBC toxin-antitoxin array. Protein Eng Des Sel 24:33–40. doi: 10.1093/protein/gzq081. [DOI] [PubMed] [Google Scholar]
- 37.Coussens NP, Daines DA. 2016. Wake me when it's over–bacterial toxin-antitoxin proteins and induced dormancy. Exp Biol Med (Maywood) 241:1332–1342. doi: 10.1177/1535370216651938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bendtsen KL, Brodersen DE. 2017. HIgher-order structure in bacterial VapBC toxin-antitoxin complexes In Harris J, Marles-Wright J (ed), Macromolecular protein complexes, vol 83 Springer, Cham, Switzerland. [DOI] [PubMed] [Google Scholar]
- 39.Deep A, Kaundal S, Agarwal S, Singh R, Thakur KG. 2017. Crystal structure of Mycobacterium tuberculosis VapC20 toxin and its interactions with cognate antitoxin, VapB20, suggest a model for toxin-antitoxin assembly. FEBS J 284:4066–4082. doi: 10.1111/febs.14289. [DOI] [PubMed] [Google Scholar]
- 40.Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 41.Krissinel E, Henrick K. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 42.Xu K, Dedic E, Brodersen DE. 2016. Structural analysis of the active site architecture of the VapC toxin from Shigella flexneri. Proteins 84:892–899. doi: 10.1002/prot.25002. [DOI] [PubMed] [Google Scholar]
- 43.Dienemann C, Boggild A, Winther KS, Gerdes K, Brodersen DE. 2011. Crystal structure of the VapBC toxin-antitoxin complex from Shigella flexneri reveals a hetero-octameric DNA-binding assembly. J Mol Biol 414:713–722. doi: 10.1016/j.jmb.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mate MJ, Vincentelli R, Foos N, Raoult D, Cambillau C, Ortiz LM. 2012. Crystal structure of the DNA-bound VapBC2 antitoxin/toxin pair from Rickettsia felis. Nucleic Acids Res 40:3245–3258. doi: 10.1093/nar/gkr1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Senissar M, Manav MC, Brodersen DE. 2017. Structural conservation of the PIN domain active site across all domains of life. Protein Sci 26:1474–1492. doi: 10.1002/pro.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daines DA, Silver RP. 2000. Evidence for multimerization of Neu proteins involved in polysialic acid synthesis in Escherichia coli K1 using improved LexA-based vectors. J Bacteriol 182:5267–5270. doi: 10.1128/JB.182.18.5267-5270.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daines DA, Granger-Schnarr M, Dimitrova M, Silver RP. 2002. Use of LexA-based system to identify protein-protein interactions in vivo. Methods Enzymol 358:153–161. doi: 10.1016/S0076-6879(02)58087-3. [DOI] [PubMed] [Google Scholar]
- 48.Miller JH. 1972. Experiments in molecular genetics, p 352–355. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 49.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brosius J, Erfle M, Storella J. 1985. Spacing of the –10 and –35 regions in the tac promoter. Effect on its in vivo activity. J Biol Chem 260:3539–3541. [PubMed] [Google Scholar]
- 51.Klein RD, Luginbuhl GH. 1979. Simplified media for the growth of Haemophilus influenzae from clinical and normal flora sources. J Gen Microbiol 113:409–411. doi: 10.1099/00221287-113-2-409. [DOI] [PubMed] [Google Scholar]
- 52.Herriott RM, Meyer EY, Vogt M, Modan M. 1970. Defined medium for growth of Haemophilus influenzae. J Bacteriol 101:513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herriott RM, Meyer EM, Vogt M. 1970. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J Bacteriol 101:517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hamilton B, Manzella A, Schmidt K, DiMarco V, Butler JS. 2014. Analysis of non-typeable Haemophilous influenzae VapC1 mutations reveals structural features required for toxicity and flexibility in the active site. PLoS One 9:e112921. doi: 10.1371/journal.pone.0112921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zaychikova MV, Mikheecheva NE, Belay YO, Alekseeva MG, Melerzanov AV, Danilenko VN. 2018. Single nucleotide polymorphisms of Beijing lineage Mycobacterium tuberculosis toxin-antitoxin system genes: their role in the changes of protein activity and evolution. Tuberculosis (Edinb) 112:11–19. doi: 10.1016/j.tube.2018.06.011. [DOI] [PubMed] [Google Scholar]
- 56.Daines DA, Jarisch J, Smith AL. 2004. Identification and characterization of a nontypeable Haemophilus influenzae putative toxin-antitoxin locus. BMC Microbiol 4:30. doi: 10.1186/1471-2180-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Agarwal S, Tiwari P, Deep A, Kidwai S, Gupta S, Thakur KG, Singh R. 2018. System-wide analysis unravels the differential regulation and in vivo essentiality of virulence-associated proteins B and C toxin-antitoxin systems of Mycobacterium tuberculosis. J Infect Dis 217:1809–1820. doi: 10.1093/infdis/jiy109. [DOI] [PubMed] [Google Scholar]
- 58.Wen W, Liu B, Xue L, Zhu Z, Niu L, Sun B. 2018. Autoregulation and virulence control by the toxin-antitoxin system SavRS in Staphylococcus aureus. Infect Immun 86:e00032-18. doi: 10.1128/IAI.00032-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gotfredsen M, Gerdes K. 1998. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol Microbiol 29:1065–1076. doi: 10.1046/j.1365-2958.1998.00993.x. [DOI] [PubMed] [Google Scholar]
- 60.Deep A, Tiwari P, Agarwal S, Kaundal S, Kidwai S, Singh R, Thakur KG. 2018. Structural, functional and biological insights into the role of Mycobacterium tuberculosis VapBC11 toxin-antitoxin system: targeting a tRNase to tackle mycobacterial adaptation. Nucleic Acids Res doi: 10.1093/nar/gky924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cline SD, Saleem S, Daines DA. 2012. Regulation of the vapBC-1 toxin-antitoxin locus in nontypeable Haemophilus influenzae. PLoS One 7:e32199. doi: 10.1371/journal.pone.0032199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kabsch W. 1988. Automatic indexing of rotation diffraction patterns. J Appl Crystallogr 21:67–72. doi: 10.1107/S0021889887009737. [DOI] [Google Scholar]
- 63.Kabsch W. 2010. Xds. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, Bricogne G. 2011. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr 67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Evans PR. 2011. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr 67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vagin A, Lebedev A. 2015. MoRDa, an automatic molecular replacement pipeline. Acta Crystallogr A Found Adv 71:s19. doi: 10.1107/S2053273315099672. [DOI] [Google Scholar]
- 67.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langer G, Cohen SX, Lamzin VS, Perrakis A. 2008. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc 3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. 2004. Developments in the CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr 60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 73.Krissinel E. 2012. Enhanced fold recognition using efficient short fragment clustering. J Mol Biochem 1:76–85. [PMC free article] [PubMed] [Google Scholar]
- 74.Daines DA, Smith AL. 2004. Construction of a nontypeable Haemophilus influenzae-specific ectopic delivery vector. J Microbiol Methods 57:421–424. doi: 10.1016/j.mimet.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 75.Robinson E, Juhas M, Hood D, Crook D. 2010. Construction of a novel shuttle vector for use in Haemophilus influenzae and H. parainfluenzae. J Microbiol Methods 83:330–334. doi: 10.1016/j.mimet.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ren D, Daines DA. 2011. Use of the EpiAirway model for characterizing long-term host-pathogen interactions. J Vis Exp. doi: 10.3791/3261 55 e3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 78.Diederichs K, Karplus PA. 1997. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol 4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- 79.Weiss MS. 2001. Global indicators of X-ray data quality. J Appl Crystallogr 34:130–135. doi: 10.1107/S0021889800018227. [DOI] [Google Scholar]
- 80.Karplus PA, Diederichs K. 2012. Linking crystallographic model and data quality. Science 336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Evans P. 2012. Biochemistry. Resolving some old problems in protein crystallography. Science 336:986–987. doi: 10.1126/science.1222162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors were deposited in the worldwide Protein Data Bank (wwPDB) under accession code 6NKL.