Bacteria use weapons to deliver effectors into target cells. One of these weapons, called the type VI secretion system (T6SS), could be compared to a nano-spear gun using a springlike mechanism for effector injection. By targeting bacteria and eukaryotic cells, the T6SS reshapes bacterial communities and hijacks host cell defenses. In enteroaggregative Escherichia coli, the T6SS is a multiprotein machine that comprises a cytoplasmic tail and a peptidoglycan-anchored trans-envelope channel. In this work, we show that TagL comprises an N-terminal domain that mediates contact with the channel and a peptidoglycan-binding domain that binds the cell wall. We then determine at which stage of T6SS biogenesis TagL is recruited and how TagL absence impacts the assembly pathway.
KEYWORDS: protein transport, assembly pathway, domain swapping, membrane complex, peptidoglycan, protein secretion
ABSTRACT
The type VI secretion system (T6SS) is an injection apparatus that uses a springlike mechanism for effector delivery. The contractile tail is composed of a needle tipped by a sharpened spike and wrapped by the sheath that polymerizes in an extended conformation on the assembly platform, or baseplate. Contraction of the sheath propels the needle and effectors associated with it into target cells. The passage of the needle through the cell envelope of the attacker is ensured by a dedicated trans-envelope channel complex. This membrane complex (MC) comprises the TssJ lipoprotein and the TssL and TssM inner membrane proteins. MC assembly is a hierarchized mechanism in which the different subunits are recruited in a specific order: TssJ, TssM, and then TssL. Once assembled, the MC serves as a docking station for the baseplate. In enteroaggregative Escherichia coli, the MC is accessorized by TagL, a peptidoglycan-binding (PGB) inner membrane-anchored protein. Here, we show that the PGB domain is the only functional domain of TagL and that the N-terminal transmembrane region mediates contact with the TssL transmembrane helix. Finally, we conduct fluorescence microscopy experiments to position TagL in the T6SS biogenesis pathway, demonstrating that TagL is recruited to the membrane complex downstream of TssL and is not required for baseplate docking.
IMPORTANCE Bacteria use weapons to deliver effectors into target cells. One of these weapons, called the type VI secretion system (T6SS), could be compared to a nano-spear gun using a springlike mechanism for effector injection. By targeting bacteria and eukaryotic cells, the T6SS reshapes bacterial communities and hijacks host cell defenses. In enteroaggregative Escherichia coli, the T6SS is a multiprotein machine that comprises a cytoplasmic tail and a peptidoglycan-anchored trans-envelope channel. In this work, we show that TagL comprises an N-terminal domain that mediates contact with the channel and a peptidoglycan-binding domain that binds the cell wall. We then determine at which stage of T6SS biogenesis TagL is recruited and how TagL absence impacts the assembly pathway.
INTRODUCTION
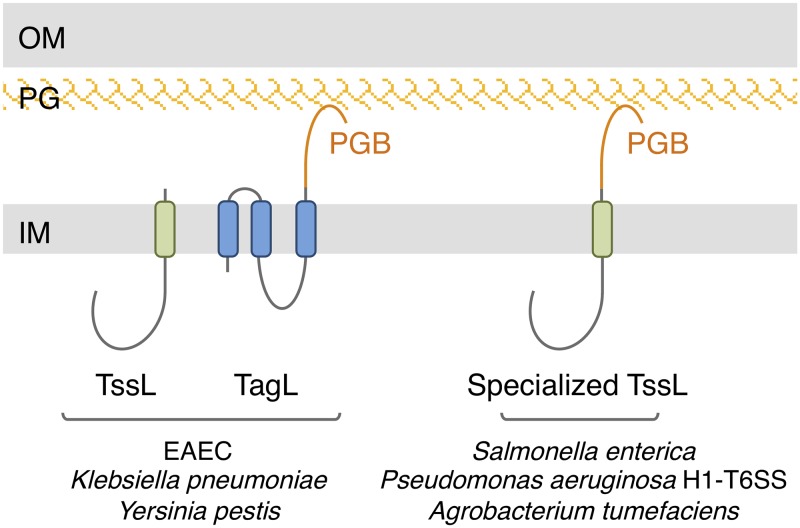
Bacteria have evolved sophisticated mechanisms to attack or defend against competitors that share the same environment, such as different bacterial species, amoeba, other unicellular microorganisms, or fungi (1, 2). Among these mechanisms, the type VI secretion system (T6SS) is a multiprotein complex that is widespread in Gram-negative bacteria (3–9). The T6SS is capable of delivering effectors in bacteria and eukaryotic cells and is therefore an important player in interbacterial competition and pathogenesis (10–14). At the molecular level, the T6SS can be architecturally and mechanistically compared to a nanocrossbow or -spear gun (6, 15). It is related to a broad family of contractile injection systems (CIS) that include bacteriophages, R-pyocins, and anti-feeding prophages (16–19). A CIS comprises a springlike structure, called the tail, composed of an inner tube terminated by a spike, wrapped by a contractile sheath and built on an assembly platform (18, 19). Once in contact with the target cell, the sheath contracts and hence propels the inner tube/spike needle. The T6SS tail tube is made of Hcp hexamers stacked on each other and tipped by a trimer of the VgrG spike protein, which is further sharpened by the metal-bound PAAR protein (20–24). The contractile sheath that wraps the inner tube is composed of two proteins, TssB and TssC (15, 25–28). The T6SS tail assembly platform, or baseplate (BP), comprises the TssEFGK subunits that form a complex similar to bacteriophage baseplate wedges (29–32). Similar to baseplate assembly in bacteriophages, six TssEFGK wedges polymerize around the VgrG spike to form the functional baseplate (31–32). Once the baseplate is assembled, the TssA protein will control the polymerization of the tube and of the sheath by adding tube and sheath building blocks from the distal end until the tail contacts the TagA stopper at the opposite membrane (33–37). In addition to the conserved CIS tail, T6SSs comprise an additional transenvelope complex that anchors the tail to the cell envelope, positions the tail for firing toward the exterior, and serves as a channel for the passage of the inner tube (38, 39). This membrane complex (MC) is composed of three proteins: TssJ, TssL, and TssM. TssJ is an outer membrane lipoprotein, whereas TssL and TssM are two inner membrane proteins, inserted by one and three transmembrane helices (TMHs), respectively (40–43). TssL dimerizes and interacts with TssM by its single TMH (42, 44, 45), whereas the C-terminal region of the TssM periplasmic domain binds to TssJ (39, 46). The negative-stain and cryo-electron microscopy and cryo-electron tomography structures of the 1.7-MDa T6SS MC from enteroaggregative Escherichia coli (EAEC) have been reported, defining a rocket-like shape, with a large cytoplasmic base followed by arches likely corresponding to the TssL and TssM TMHs and by pillars crossing the periplasm and corresponding to the TssM periplasmic domain and TssJ lipoprotein (39, 47, 48). The central portion of the MC defines a channel that is narrowed at the periplasmic entrance and closed at its outer membrane extremity, suggesting that the MC undergoes significant conformational changes during firing to allow the passage of the needle (39, 47). The cytoplasmic base of the membrane complex represents the docking station for the assembled baseplate, mainly by contacts between the C-terminal domain of the baseplate TssK subunit with the cytoplasmic domains of TssL and TssM (30, 32, 43, 49–52). The assembly of the membrane complex starts with the positioning of the TssJ lipoprotein and the ordered recruitment of TssM and TssL (39). Polymerization of the TssJLM complex to assemble the MC requires local remodeling of the peptidoglycan, which is ensured by a dedicated or housekeeping transglycosylase (53, 54). In most T6SSs, the MC is stably anchored to the cell wall by a peptidoglycan-binding (PGB) domain (38, 55). In most cases, the PGB domain is fused to the C terminus of the TssL TMH (Fig. 1). However, other T6SSs have evolved different strategies to be anchored to the peptidoglycan layer, such as accessory inner membrane or periplasmic proteins bearing a PGB domain (55). In EAEC, the PGB domain is carried by the accessory protein TagL (formerly known as SciZ) that associates with the membrane complex by directly interacting with TssL (38). TagL is constituted of three TMHs and the periplasmic PGB domain (Fig. 1) (38). Mutations within the PGB domain that prevent peptidoglycan binding in vivo and in vitro abolish T6SS activity, demonstrating that TagL-mediated anchoring to the cell wall is necessary for T6SS function (38). Here, we show that fusing the TagL PGB domain to the C terminus of TssL is sufficient to compensate for the absence of tagL, demonstrating that the PGB domain is the only functional domain of TagL. We further demonstrate that TagL-TssL complex formation is mediated by interactions between their TMHs. We then generated a functional superfolder green fluorescent protein (sfGFP)-TagL fusion protein and showed by fluorescence microscopy that it colocalizes with TssL at the base of the sheath and is recruited downstream of TssL but upstream of baseplate docking. Finally, microscopy recordings of functional GFP fusions to T6SS subunits in the absence of tagL demonstrate that TagL is not necessary for recruitment of TssA and of the baseplate but is required for sheath polymerization.
FIG 1.
Schematic representations of selected T6SS subunits that bind the cell wall. In EAEC and other species, T6SS gene clusters encode TssL and TagL, a three-TMH protein with a periplasmic peptidoglycan-binding (PGB) domain (orange). In the vast majority of cases, the PGB domain is fused to TssL to yield the TssL-PGB specialized TssL protein. Additional cases have been described by Aschtgen et al. (55). IM, inner membrane; PG, peptidoglycan; OM, outer membrane.
RESULTS
TagL PGB domain fusion to TssL bypasses the requirements for full-length TagL.
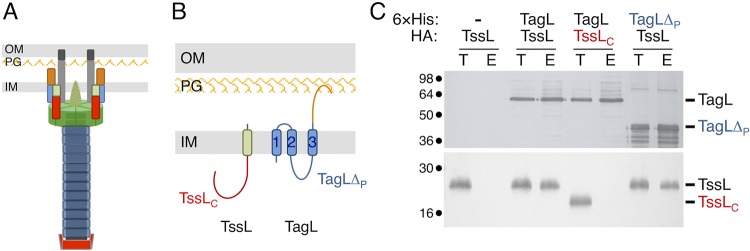
The EAEC T6SS sci1 gene cluster encodes TagL, a three-TMH inner membrane protein that bears a periplasmic peptidoglycan-binding (PGB) domain (38) (Fig. 1). Binding to the cell wall is required for T6SS function as TagL PGB domain amino acid substitutions that prevent in vitro and in vivo interaction with the peptidoglycan abolish T6SS activity (38). However, although TagL is not broadly conserved in T6SSs, T6SS gene clusters usually encode a PGB domain that can be either carried by an accessory protein or fused to the TssL core component (55) (Fig. 1). We therefore asked whether the N-terminal transmembrane portion of TagL is necessary for T6SS function. For this, we engineered a strain deleted of both tssL and tagL. ΔtssL-tagL cells, as well as ΔtssL-tagL cells expressing tssL or tagL only, were unable to release Hcp in the culture supernatant and to outcompete E. coli K-12 cells in antibacterial assays (Fig. 2), in agreement with the essential roles of TssL and TagL in the EAEC T6SS (38, 42). However, a plasmid-borne fragment encoding both TssL and TagL complemented the phenotypes associated with the ΔtssL-tagL deletion to wild-type levels (Fig. 2). Interestingly, fusion of the TagL periplasmic PGB domain at the C terminus of TssL to yield the TssL-PGB domain chimeric protein was sufficient to promote T6SS activity in ΔtssL-tagL cells (Fig. 2). This result demonstrates that the C-terminal PGB domain is the only functional domain of TagL. In agreement with this observation, production of the TssL-PGB domain chimeric protein bearing substitutions that prevent anchorage of the PGB domain to the cell wall (TssL-PGB*, with substitutions N494L/L497N/S498A/R501Q/A502D [38]) was unable to restore T6SS activity in ΔtssL-tagL cells (Fig. 2).
FIG 2.
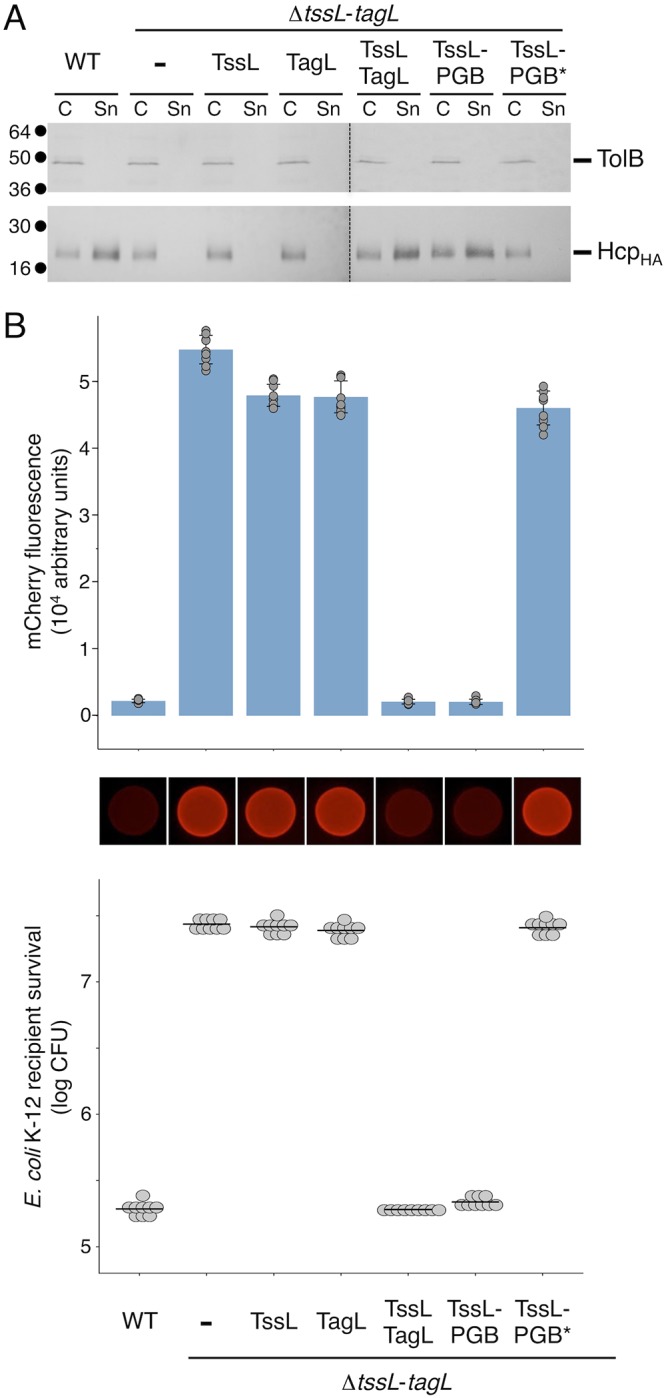
The PGB domain is the only functional domain of TagL. T6SS functional assays were performed with wild-type (WT) cells and ΔtssL-tagL cells bearing the pASK-IBA37(+) empty vector (−) or the pASK-IBA37(+) vector encoding TssL, TagL, both TssL and TagL, the TagL PGB fused to TssL (TssL-PGB), or the mutated TagL PGB fused to TssL (TssL-PGB*). (A) Hcp release was assessed by separating whole cells (C) and supernatant (Sn) fractions from the indicated strains producing HA-tagged Hcp from the pOK-HcpHA vector. A total of 2 × 108 cells and the trichloroacetic acid-precipitated material of the supernatant from 5 × 108 cells were subjected to 12.5% acrylamide SDS-PAGE and immunodetected using the anti-HA monoclonal antibody and the anti-TolB polyclonal antibody (lysis control). The positions of HcpHA and TolB are indicated on the right. Molecular weight markers (in kilodaltons) are indicated on the left. (B) E. coli K-12 recipient cells (W3110 expressing mCherry; Ampr) were mixed with the indicated attacker cells, spotted onto SIM agar plates, and incubated for 4 h at 37°C. The image of a representative bacterial spot is shown below the graph reporting the fluorescence level of the spots. The average fluorescence and standard deviation are indicated, as well as the values from nine independent measurements (gray dots). The lower graph reports the number of surviving E. coli recipient cells counted on selective ampicillin medium. The gray dots correspond to the nine values from three independent biological replicates. The average is indicated by a horizontal bar.
The TagL N-terminal region mediates interaction with the TssL TMH.
The observation that the PGB domain of TagL (orange in Fig. 3A and B) is necessary and sufficient for T6SS activity raised the question of the role of its N-terminal region that comprises three TMHs, a short periplasmic linker connecting TMH1 and TMH2, and a cytoplasmic domain located between TMH2 and TMH3 (blue in Fig. 3A and B). We hypothesized that this region might engage in the interaction with TssL. To test whether the TagL N-terminal region is important for TagL-TssL complex formation, we performed coprecipitation analyses. As previously shown (38), precipitation of 6×His-tagged TagL on nickel magnetic beads coprecipitated TssL (Fig. 3C, third and fourth lanes). However, TagL did not coprecipitate TssLC, a TssL variant with a deletion of its C-terminal TMH (Fig. 3A and B, red, and Fig. 3C, fifth and sixth lanes), suggesting that TagL-TssL complex formation does not require interactions between their cytoplasmic domains and, thus, that it likely involves their TMHs. In agreement with this hypothesis, a TagL variant deleted of its periplasmic domain but retaining its three TMHs (TagLΔP) (Fig. 3A and B, blue) coprecipitated TssL at levels comparable to those of full-length TagL (Fig. 3C, seventh and eighth lanes). From these results, we conclude that the TagL periplasmic region, which includes the PGB domain, and the TssL cytoplasmic domain do not contribute to TagL-TssL complex formation and, hence, that the interaction of the two partners is likely mediated by their TMHs. Taken together, the results shown in Fig. 2 and 3 suggest that the TagL PGB domain is a functional module for T6SS activity whereas its TMHs are required for recruitment to the T6SS MC via direct interactions with the TssL TMH.
FIG 3.
TssL and TagL interact via their TMHs. (A) Schematic representation of the EAEC T6SS. The membrane complex comprises TssJ (dark gray), TssM (light gray), TssL (red, N-terminal cytoplasmic domain [TssLC]; green, C-terminal TMH), and TagL (blue, N-terminal membrane domain [TagLΔP]; orange, C-terminal peptidoglycan-binding domain). The baseplate is shown in green. The contractile tail is shown in blue. The TssA protein, located at the distal end of the tail, is shown in red. IM, inner membrane; PG, peptidoglycan; OM, outer membrane. (B) Schematic representation of EAEC TssL and TagL highlighting their topologies (38, 42) and different domains, using the scheme described for panel A. The three TagL TMHs are numbered from the N terminus. (C) Coprecipitation assay. Igepal CA-630-solubilized extracts of E. coli cells producing the indicated proteins or protein variants were mixed with nickel magnetic beads to precipitate 6×His-tagged TagL and interacting partners. The total lysates (T) and eluted (E) material were subjected to 12.5% acrylamide SDS-PAGE and immunodetected with anti-His (TagL) and anti-HA (TssL) monoclonal antibodies. The positions of TssL, TagL, and their variants are indicated on the right. Molecular weight markers (in kilodaltons) are indicated on the left.
TagL colocalizes with the membrane complex at the base of the tail.
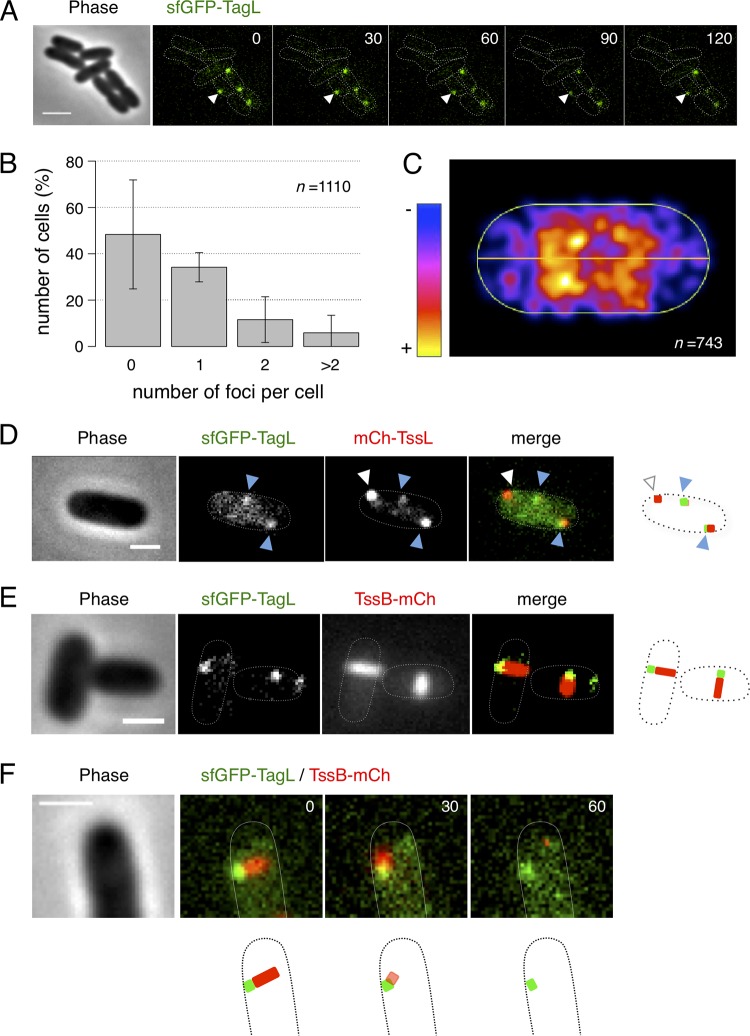
To define the localization and dynamics of TagL in wild-type EAEC cells, we engineered a chromosomally encoded superfolder green fluorescent protein (sfGFP) fusion to TagL by inserting the sfGFP-encoding fragment in frame, downstream of the tagL ATG start codon. Hence, sfGFP-TagL is produced from the native locus, under the control of the expression signals of tagL. Antibacterial assays demonstrated that this sfGFP-TagL fusion is functional as sfGFP-TagL-producing cells outcompete E. coli K-12 competitor cells (see Fig. S1 in the supplemental material). Fluorescence microscopy analyses showed that sfGFP-TagL clusters in foci in EAEC cells (Fig. 4A). Time-lapse recordings further showed that these foci remain static (Fig. 4A). Quantitative and distribution analyses demonstrated that most cells contain fewer than 2 foci per cell (Fig. 4B) and that sfGFP-TagL foci localize throughout the cell body with an underrepresentation at the cell poles (Fig. 4C). The number of foci per cell and their spatial distribution are comparable to what has been previously reported for sfGFP-TssL and sfGFP-TssM in EAEC (39). In agreement with this observation, and with previous protein-protein interaction data showing that TagL interacts with TssL (38) (Fig. 3C), fluorescence microscopy analyses of EAEC cells producing both sfGFP-TagL and TssL fused to mCherry (mCh-TssL) demonstrated that, in most cases, TagL and TssL colocalize (Fig. 4D). In some cases, only an mCh-TssL focus was visible without evidence for the presence of sfGFP-TagL (Fig. 4D, white arrowheads). While we cannot rule out that some MCs may assemble without TagL, we hypothesize that these cases represent assembly intermediates in which TagL has not been recruited yet to the MC. Nevertheless, these results suggest that TagL colocalizes with the MC and, hence, should be located at the base of the extended sheath. Indeed, sfGFP-TagL foci localized at one extremity of the T6SS sheath (Fig. 4E). Time-lapse recordings further showed that the sheaths contract on sfGFP-TagL foci (Fig. 4F), demonstrating that TagL localizes at the base of the sheath.
FIG 4.
Localization, dynamics, and distribution of TagL. (A) Representative fluorescence microscopy time-lapse recording of wild-type EAEC cells producing sfGFP-TagL. Individual images were taken every 30 s. The arrowheads highlight the position of one sfGFP-TagL focus, showing that it is static over time. Scale bar, 2 μm. (B) Percentage of cells with 0, 1, 2, or >2 sfGFP-TagL foci (n = 1,110 cells from four biological replicates). The mean number of foci per cell is 0.67 ± 0.86. (C) Spatial distribution of sfGFP-TagL foci. Shown is a projection of the foci from n = 743 cells on a single cell (from blue to yellow, according to the color chart). (D and E) Representative cells producing sfGFP-TagL and mCherry-TssL or TssB-mCherry, as indicated. The merged images show overlays of the sfGFP and mCherry channels, with schematic representations at right. White and blue arrowheads in panel D indicate isolated TssL foci and TssL/TagL foci, respectively. Scale bars, 1 μm. (F) Representative fluorescence microscopy time-lapse recording of a wild-type EAEC cell producing sfGFP-TagL and TssB-mCherry. Individual images were taken every 30 s. A schematic representation is shown below. Scale bar, 1 μm.
Recruitment and role of TagL during T6SS biogenesis in EAEC.
T6SS biogenesis is a hierarchized pathway in which subunits or complexes are recruited in a strict order. T6SS biogenesis is initiated by the assembly of the MC (39) and followed by the docking of the BP (30, 32) and the extension of the tail tube/sheath structure (30, 39). TssA is recruited to the TssJM complex and helps the recruitment of the BP complex prior to tail tube/sheath polymerization (33). Finally, TagA stops tail tube/sheath polymerization and maintains the sheath under the extended conformation (37). We therefore wished to determine at which step TagL is recruited and how TagL absence impacts the T6SS biogenesis pathway.
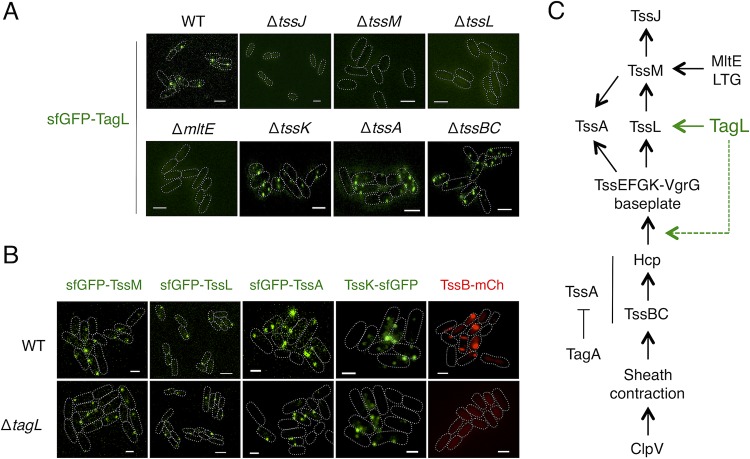
To define the requirements for TagL recruitment at the T6SS, we fused sfGFP to TagL in various mutant backgrounds deleted of T6SS genes. Figure 5A shows that sfGFP-TagL localizes in foci in the absence of TssK, TssA, or the TssBC sheath subunit. In addition, statistical analyses showed that the absence of these T6SS subunits does not significantly impact the localization and distribution of sfGFP-TagL foci (Fig. S2). These results suggest that TagL is recruited to the apparatus prior to BP assembly and docking to the MC. Biogenesis of the MC starts with the positioning of TssJ and the subsequent recruitment of TssM and TssL (39). Then, TssJLM complexes polymerize to assemble the MC once the cell wall has been locally degraded by MltE (54). Fluorescence microscopy experiments showed that TssJ, TssM, TssL, and MltE are necessary for sfGFP-TagL recruitment (Fig. 5A), suggesting that TagL binds to the MC once this complex has been fully assembled.
FIG 5.
Recruitment and impact of TagL on T6SS biogenesis. (A) Representative fluorescence microscopy fields of wild-type (WT), ΔtssJ, ΔtssM, ΔtssL, ΔmltE, ΔtssK, ΔtssA, and ΔtssBC cells producing sfGFP-TagL. Statistical analyses are shown in Fig. S2 in the supplemental material. Scale bars, 2 μm. (B) Representative fluorescence microscopy fields of wild-type (WT) or ΔtagL cells producing sfGFP-TssM, sfGFP-TssL, sfGFP-TssA, TssK-sfGFP, or TssB-mCh. Statistical analyses are shown in Fig. S3. Scale bars, 1 μm. (C) Schematic representation of the EAEC T6SS biogenesis pathway based on previous studies (30, 32, 33, 39, 54), highlighting the position of TagL (green). TagL is recruited after completion of the MC and prior to BP docking (solid green line). TssA recruitment, baseplate positioning (represented by the TssK subunit in this study), and tail tube/sheath polymerization (represented by TssBC in this study) are not required for TagL localization, but TagL is necessary for tail tube/sheath polymerization (dotted green line). LTG, lytic transglycosylase.
Based on these results, we hypothesized that TagL should not be required for MC assembly but will be necessary for BP docking. To verify this hypothesis, we deleted the tagL gene in EAEC cells producing sfGFP-TssM, sfGFP-TssL, sfGFP-TssA, TssK-sfGFP, or TssB-mCh from their native loci. As expected, the absence of TagL did not impact the recruitment of TssM or TssL (Fig. 5B). However, in contrast to our initial hypothesis, TssA and TssK formed foci in the absence of TagL (Fig. 5B). Statistical analyses showed that the number of sfGFP-TssM, sfGFP-TssL, sfGFP-TssA, and TssK-sfGFP foci per cell was not impacted by the absence of TagL (Fig. S3), suggesting that TagL does not participate in the stability of the T6SS MC and BP. Taken together, these results demonstrate that TagL binding to the MC is not a prerequisite for BP docking. Interestingly, although the absence of TagL does not prevent TssA recruitment to the MC, TssA forms static foci which do not move to the opposite membrane in ΔtagL cells (Fig. S4A), suggesting that TagL is necessary for sheath extension. Indeed, no sheath structure was observable in the absence of TagL (Fig. 5B). Finally, because previous data have shown that the MC is static over time (39), we asked whether TagL-mediated interaction with the cell wall stabilizes the MC and prevents its diffusion in the cell envelope. This hypothesis is, however, unlikely as time-lapse recordings showed that sfGFP-TssM foci remain static in the absence of TagL (Fig. S4B).
Taken together, our fluorescence microscopy observations demonstrate that TagL is recruited once the MC is fully assembled, is dispensable for the assembly of the MC and for the docking of the BP, but is necessary for tail sheath extension (Fig. 5C).
DISCUSSION
In this work, we provide details on the role of the TagL peptidoglycan-binding protein for T6SS function and during T6SS biogenesis in EAEC.
TagL comprises two domains: an N-terminal domain constituted of three TMHs and a C-terminal domain that binds the cell wall. Our results show that the PGB domain is the only functional domain of TagL as a fusion protein between TssL and the TagL PGB domain is sufficient to support T6SS activity. Based on this observation, we hypothesize that the TagL N-terminal transmembrane domain is only necessary to bind TssL and hence to accessorize the T6SS with the PGB domain. Interestingly, previous analyses have shown that most T6SS gene clusters encode proteins with peptidoglycan-binding motifs. However, these proteins come in many flavors: inner membrane-anchored accessory proteins such as TagL or TagP, predicted periplasmic proteins such as TagN or TagW, or a protein with the PGB domain fused to the TssL core component (“specialized” TssL) (55). A detailed phylogenetic study of these proteins would be required to understand how these genes have been acquired and how they have evolved to accessorize the T6SS. One interesting question is whether specialized TssL arose from a reduction of the gene cluster by fusing the PGB domain of TagL/TagN/TagP/TagW to TssL or whether TagL/TagN/TagP/TagW evolved from the partition of the TssL core and PGB domains. Our observations that a TagL PGB domain fusion to TssL bypasses the requirement for full-length TagL and that its TMHs are only necessary to mediate contact with TssL tend to support the reduction hypothesis.
By using deletion variants, we have shown that TssL-TagL complex formation is mediated by their TMHs. While we have not defined which of the three TagL TMHs is implicated in the interaction with the TssL C-terminal TMH, previous studies have reported that the TssL TMH is engaged in interaction with itself and with the TssM TMHs (39, 43, 45). Because there is no conserved sequence homology or helical motifs between the TagL and TssM TMHs, we suggest that TagL and TssM do not share the same face of interaction on the TssL TMHs and, hence, that TssL-TagL and TssL-TssM interactions are not exclusive. This hypothesis is in agreement with previous data reporting the coprecipitation of a ternary TssM-TssL-TagL complex (38). These observations suggest that in addition to the tryptophan residues engaged in π-π interactions in the TssL TMH dimer (45), two additional faces of the TssL TMH are involved in TssM and TagL interactions. Further studies need to be performed to better understand the organization of the TMHs of the TssM-TssL-TagL complex in the inner membrane.
In this work, we performed a comprehensive fluorescence microscopy analysis to position TagL in the T6SS biogenesis pathway. By using a functional sfGFP-TagL fusion, we showed that TagL colocalizes with TssL at the base of the extended sheath, in agreement with TagL association with the MC. From our time-lapse fluorescence recordings of cells producing both sfGFP-TagL and TssB-mCherry, one can notice that TagL remains at the MC during sheath contraction, suggesting that TagL is associated with TssL throughout the T6SS mechanism of action. We can hypothesize that TagL-mediated PG anchoring is important to stabilize the MC and to prevent its dissociation by the mechanical strength generated by sheath contraction (15). Our analyses of sfGFP-TagL focus formation in various T6SS mutant backgrounds revealed that positioning TagL at the site of T6SS assembly requires TssJ, TssM, TssL, and MltE but does not require TssK, TssA, or TssBC. Therefore, TagL is recruited to the MC prior to BP docking. Because the MltE lytic transglycosylase enables full assembly of the MC by locally remodeling the cell wall (54), these results suggest that TagL is recruited to the MC when its assembly is completed (Fig. 5C). Indeed, TagL and TagL-mediated PG anchoring are not required for TssJLM MC assembly, in agreement with the facts that TagL is an accessory protein that is not conserved in all T6SSs (38, 55) and that the MC can be purified from E. coli K-12 cells lacking TagL (39). However, although TagL is recruited upstream of the BP, it is not necessary for BP docking to the MC (Fig. 5C). This result clearly shows that the T6SS biogenesis pathway is not a strict linear pathway, with the ordered recruitment of one subunit after the other, but, rather, that independent branches exist. However, while the BP docks to the MC in ΔtagL cells, extension of the tail/tube cannot proceed in the absence of TagL. This observation hence suggests that once the BP docks to the MC, TagL mediates a structural modification of the MC or BP that is required to initiate sheath extension. Electron microscopy models of the MC bound to TagL or of the MC-BP complex would likely provide further information on the impact of TagL on the conformation of the MC or BP.
MATERIALS AND METHODS
Bacterial strains, media, growth conditions, and chemicals.
Bacterial strains are listed in Table S1 in the supplemental material. Escherichia coli K-12 DH5α and W3110 strains were used for cloning procedures and coimmunoprecipitations, respectively. Escherichia coli K-12 W3110 cells carrying the pFPV-mCherry plasmid (mCherry under the control of the constitutive and strong ribosomal rpsM promoter; Ampr [56]) were used as recipients for the antibacterial competition assays. The enteroaggregative E. coli (EAEC) wild-type strain 17-2, its ΔtagL, ΔtssM, ΔtssL, ΔtssJ, ΔtssA, and ΔtssK derivatives, and 17-2 strains producing the TssB-mCh, TssB-sfGFP, sfGFP-TssA, sfGFP-TssM, sfGFP-TssL, mCh-TssL, and TssK-sfGFP fusions from their native chromosomal loci have been previously described (30, 33, 38–40, 49). Strains were routinely grown in LB broth at 37°C with aeration. For induction of the T6SS sci1 genes, strains were grown in Sci1 inducing medium (SIM; M9 minimal medium supplemented with 0.25% glycerol 1 μg/ml vitamin B1, 40 μg/ml Casamino Acids, and 10% [vol/vol] LB) (57). Plasmids and mutations were maintained by the addition of ampicillin (100 μg/ml for K-12; 200 μg/ml for EAEC), kanamycin (50 μg/ml for K-12, 50 μg/ml for chromosomal insertion on EAEC, and 100 μg/ml for plasmid-bearing EAEC). Gene expression from pASK-IBA vectors was induced for 30 min with 0.05 μg/ml anhydrotetracycline (AHT) (IBA Technologies).
Strain construction.
The 17-2 derivative strains were engineered by lambda Red recombination (58) using plasmid pKOBEG (59). For construction of the 17-2 strain deleted of tagL or of both tssL and tagL (ΔtssL-tagL strain), a kanamycin cassette was amplified from plasmid pKD4 (58) with primers carrying 50-bp extensions corresponding to the 5′ and 3′ sequences of the region to be deleted. For construction of 17-2 strain derivatives producing sfGFP-TagL, a kanamycin-sfGFP cassette was amplified from the pKD4-Nter-sfGFP vector (39) with primers carrying 50-bp extensions corresponding to the upstream and downstream regions of the tagL initiation codon. PCR products were electroporated into target cells, and lambda Red-mediated recombination at the proper locus was verified by PCR. The excision of the kanamycin cassette, when possible, was then obtained by production of the Flp recombinase using pCP20 (58). The final strains were verified by PCR.
Plasmid construction.
Plasmids used in this study are listed in Table S1. PCRs were performed with a Biometra thermocycler, using Pfu Turbo DNA polymerase (Stratagene). Custom oligonucleotides were synthesized by Sigma-Aldrich and are listed in Table S1. The pASK-IBA37(+) vector producing N-terminally 6×His-tagged TagL (initially called pIBA-28 [38]), the FLAG-tagged TssL or TssL fused to the periplasmic domain (amino acids 360 to 576) of TagL (TssL-PGB, initially named pIBA-SciP and pIBA-SciP-PG [42]), and the pOK12 derivatives producing N-terminally hemagglutinin (HA)-tagged Hcp (HcpHA) (38) or TssL (initially named pOK-SciP-HA [38]) have been previously described. The pOK12 plasmid producing the TssL cytoplasmic domain (TssLC; amino acids 1 to 186) was constructed by standard restriction/ligation cloning by inserting an EcoRI-tssLC-XhoI fragment into the EcoRI-XhoI-digested pOK12 derivative plasmid pMS600 (40). Plasmid producing TagL deleted of the periplasmic region (TagLΔP) was constructed by replacing tagL codon 366 by a TAA stop codon on pIBA-TagL, using site-directed mutagenesis and primers carrying the desired substitutions. The pIBA-TssL-TagL plasmid, producing both TssL and TagL, and pIBA-TssL-PGB*, producing TssL fused to the TagL mutated PGB domain (PGB*; Asn494Leu-Leu497Asn-Ser498Ala-Arg501Gln-Ala502Asp substitutions [38]), were constructed by restriction-free cloning (38, 60). Briefly, the sequence encoding the TssL protein or the mutated PGB domain of TagL was amplified from 17-2 DNA or plasmid pIBA-TagL-PG* (38), respectively, with oligonucleotides carrying 5′ extensions annealing upstream and downstream of the site of insertion on the target plasmid. The products of the first PCR were then used as oligonucleotides for a second PCR using the target vector as the template. The tssL fragment was inserted into plasmid pIBA-TagL, whereas the PGB*-encoding fragment was inserted into plasmid pIBA-TssL. All constructs were verified by restriction analyses and DNA sequencing (Eurofins Genomics).
T6SS function reporter assays.
An Hcp release assay was performed as described previously (40), using plasmid pOK-HcpHA, except that cells were grown in SIM to an absorbance at a wavelength (λ) of 600 nm (A600) of 0.6. The periplasmic TolB protein was used as a control for cell integrity. An interbacterial competition assay was performed as described previously (61) with modifications as follows: (i) E. coli W3110 cells constitutively expressing mCherry from plasmid pFPV-mCherry (Ampr) (56) were used as recipients; (ii) fluorescence measurements with the Tecan microplate reader were performed by exciting mCherry at a λ of 575 nm and recording its emission at a λ of 610 nm; and (iii) survival of recipient cells was calculated by counting CFU (CFU on recipient-selective LB plates supplemented with ampicillin).
Coprecipitation on nickel magnetic beads.
A total of 5 × 109 E. coli W3110 cells producing the proteins of interest was harvested and resuspended in CelLytic B lysis reagent (Sigma-Aldrich) supplemented with 100 μg/ml lysozyme, 100 μg/ml DNase, protease inhibitors (Complete; Roche), and 1.65 mM octylphenoxy poly(ethyleneoxy)ethanol (Igepal CA-630; Sigma-Aldrich). After a 30-min incubation at 25°C with strong agitation, unsolubilized material was discarded by centrifugation for 20 min at 20,000 × g. The cleared lysates were supplemented with 10 mM imidazole and incubated with nickel magnetic beads (PureProteome; Millipore) for 2 h at room temperature on a wheel. Magnetic beads were then washed three times with CelLytic B lysis reagent supplemented with Igepal CA-630 and 20 mM imidazole, resuspended in nonreducing Laemmli loading dye, and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analyses using monoclonal anti-His and anti-HA antibodies.
Fluorescence microscopy and data analysis.
Cells producing the chromosomal sfGFP or mCherry fusion proteins were grown in SIM to an A600 of 0.6 to 0.8, harvested, and resuspended in SIM to an A600 of 8. Cells were then spotted on a thin pad of SIM supplemented with 2% agarose, covered with a coverslip, and incubated for 20 to 30 min at room temperature before microscopy acquisition. Fluorescence microscopy recordings were performed with a Nikon Eclipse Ti microscope equipped with an Orcaflash 4.0 LT digital camera (Hamamatsu) and a perfect focus system (PFS) to automatically maintain focus so that the point of interest within the specimen was always kept in sharp focus at all times despite mechanical or thermal perturbations. All fluorescence images were acquired with a minimal exposure time to minimize bleaching and phototoxicity effects. Exposure times were typically 30 ms for phase contrast, 250 ms for sfGFP fusion proteins, and 200 ms for TssB-mCh and TssL-mCh. The images shown in the figures are representative regions cropped from large fields and are from at least triplicate experiments. Images were analyzed using ImageJ (62) and the MicrobeJ plug-in (63). Statistical data set analysis was performed using the R software environment (https://www.r-project.org/).
Miscellaneous.
SDS-PAGE and transfer onto nitrocellulose membranes (Amersham Protran, 0.2-μm pore size; GE Healthcare) were performed with standard procedures using Mini-Protean II systems (Bio-Rad). After protein transfer, membranes were probed with primary antibodies and goat secondary antibodies coupled to alkaline phosphatase. Immunodetections were performed in alkaline buffer in the presence of 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium. The anti-FLAG (clone M2; Sigma-Aldrich), anti-HA (clone HA-7; Sigma-Aldrich), and antihistidine tag (clone AD1.1.10; Bio-Rad) monoclonal antibodies and alkaline phosphatase-conjugated goat anti-mouse and anti-rabbit secondary antibodies (Beckman Coulter) are commercially available. The anti-TolB polyclonal antibodies were from the laboratory collection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Leon Espinosa for helpful discussions and advice on fluorescence microscopy, Laure Journet, Dukas Jurėnas, and Yassine Cherrak for critical reading of the manuscript, laboratory members for discussions, Moly Ba, Isabelle Bringer, Annick Brun, and Oliver Uderso for technical assistance, and Jean-Phil Monce-Lipp for encouragement.
This work was supported by the Centre National de la Recherche Scientifique (CNRS), the Aix-Marseille Université (AMU), and grants from the Agence Nationale de la Recherche (ANR-10-JCJC-1303, ANR-14-CE14-0006, and ANR-17-CE11-0039) and Fondation pour la Recherche Médicale (FRM; DEQ20180339165) to E.C., Y.G.S., A.Z., and M.-S.A. were supported by doctoral fellowships from the French Ministère de l'Education Supérieure et de la Recherche. M.-S.A. also received end-of-thesis support from an Attaché Temporaire d'Enseignement et de Recherche (ATER) fellowship from the Aix-Marseille Université. A.Z. received end-of-thesis support from a fellowship from the FRM (FDT20140931060).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00173-19.
REFERENCES
- 1.Chassaing B, Cascales E. 2018. Antibacterial weapons: targeted destruction in the microbiota. Trends Microbiol 26:329–338. doi: 10.1016/j.tim.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Coyne MJ, Comstock LE. 2019. Type VI secretion systems and the gut microbiota. Microbiol Spectr 7:2. doi: 10.1128/microbiolspec.PSIB-0009-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bingle LE, Bailey CM, Pallen MJ. 2008. Type VI secretion: a beginner's guide. Curr Opin Microbiol 11:3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Cascales E. 2008. The type VI secretion toolkit. EMBO Rep 9:735–741. doi: 10.1038/embor.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. 2009. Dissecting the bacterial type VI secretion system by a genome-wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104. doi: 10.1186/1471-2164-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zoued A, Brunet YR, Durand E, Aschtgen MS, Logger L, Douzi B, Journet L, Cambillau C, Cascales E. 2014. Architecture and assembly of the type VI secretion system. Biochim Biophys Acta 1843:1664–1673. doi: 10.1016/j.bbamcr.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 7.Ho BT, Dong TG, Mekalanos JJ. 2014. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cianfanelli FR, Monlezun L, Coulthurst SJ. 2016. Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol 24:51–62. doi: 10.1016/j.tim.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Basler M. 2015. Type VI secretion system: secretion by a contractile nanomachine. Philos Trans R Soc B 370:20150021. doi: 10.1098/rstb.2015.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russell AB, Peterson SB, Mougous JD. 2014. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol 12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durand E, Cambillau C, Cascales E, Journet L. 2014. VgrG, Tae, Tle, and beyond: the versatile arsenal of type VI secretion effectors. Trends Microbiol 22:498–507. doi: 10.1016/j.tim.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Alcoforado Diniz J, Liu YC, Coulthurst SJ. 2015. Molecular weaponry: diverse effectors delivered by the type VI secretion system. Cell Microbiol 17:1742–1751. doi: 10.1111/cmi.12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hachani A, Wood TE, Filloux A. 2016. Type VI secretion and anti-host effectors. Curr Opin Microbiol 29:81–93. doi: 10.1016/j.mib.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Trunk K, Peltier J, Liu YC, Dill BD, Walker L, Gow NAR, Stark MJR, Quinn J, Strahl H, Trost M, Coulthurst SJ. 2018. The type VI secretion system deploys antifungal effectors against microbial competitors. Nat Microbiol 3:920–931. doi: 10.1038/s41564-018-0191-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brackmann M, Nazarov S, Wang J, Basler M. 2017. Using force to punch holes: mechanics of contractile nanomachines. Trends Cell Biol 27:623–632. doi: 10.1016/j.tcb.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Bönemann G, Pietrosiuk A, Mogk A. 2010. Tubules and donuts: a type VI secretion story. Mol Microbiol 76:815–821. doi: 10.1111/j.1365-2958.2010.07171.x. [DOI] [PubMed] [Google Scholar]
- 17.Sarris PF, Ladoukakis ED, Panopoulos NJ, Scoulica EV. 2014. A phage tail-derived element with wide distribution among both prokaryotic domains: a comparative genomic and phylogenetic study. Genome Biol Evol 6:1739–1747. doi: 10.1093/gbe/evu136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cascales E. 2017. Microbiology: and Amoebophilus invented the machine gun! Curr Biol 27:R1170–R1173. doi: 10.1016/j.cub.2017.09.025. [DOI] [PubMed] [Google Scholar]
- 19.Taylor NMI, van Raaij MJ, Leiman PG. 2018. Contractile injection systems of bacteriophages and related systems. Mol Microbiol 108:6–15. doi: 10.1111/mmi.13921. [DOI] [PubMed] [Google Scholar]
- 20.Ballister ER, Lai AH, Zuckermann RN, Cheng Y, Mougous JD. 2008. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc Natl Acad Sci U S A 105:3733–3738. doi: 10.1073/pnas.0712247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, Burley SK, Almo SC, Mekalanos JJ. 2009. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A 106:4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. 2013. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brunet YR, Hénin J, Celia H, Cascales E. 2014. Type VI secretion and bacteriophage tail tubes share a common assembly pathway. EMBO Rep 15:315–321. doi: 10.1002/embr.201337936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Renault MG, Zamarreno Beas J, Douzi B, Chabalier M, Zoued A, Brunet YR, Cambillau C, Journet L, Cascales E. 2018. The gp27-like hub of VgrG serves as adaptor to promote Hcp tube assembly. J Mol Biol 430:3143–3156. doi: 10.1016/j.jmb.2018.07.018. [DOI] [PubMed] [Google Scholar]
- 25.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kudryashev M, Wang RY, Brackmann M, Scherer S, Maier T, Baker D, DiMaio F, Stahlberg H, Egelman EH, Basler M. 2015. Structure of the type VI secretion system contractile sheath. Cell 160:952–962. doi: 10.1016/j.cell.2015.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Brackmann M, Castaño-Díez D, Kudryashev M, Goldie KN, Maier T, Stahlberg H, Basler M. 2017. Cryo-EM structure of the extended type VI secretion system sheath-tube complex. Nat Microbiol 2:1507–1512. doi: 10.1038/s41564-017-0020-7. [DOI] [PubMed] [Google Scholar]
- 28.Salih O, He S, Planamente S, Stach L, MacDonald JT, Manoli E, Scheres SHW, Filloux A, Freemont PS. 2018. Atomic structure of type VI contractile sheath from Pseudomonas aeruginosa. Structure 26:329–336.e3. doi: 10.1016/j.str.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.English G, Byron O, Cianfanelli FR, Prescott AR, Coulthurst SJ. 2014. Biochemical analysis of TssK, a core component of the bacterial type VI secretion system, reveals distinct oligomeric states of TssK and identifies a TssK-TssFG subcomplex. Biochem J 461:291–304. doi: 10.1042/BJ20131426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brunet YR, Zoued A, Boyer F, Douzi B, Cascales E. 2015. The type VI secretion TssEFGK-VgrG phage-like baseplate is recruited to the TssJLM membrane complex via multiple contacts and serves as assembly platform for tail tube/sheath polymerization. PLoS Genet 11:e1005545. doi: 10.1371/journal.pgen.1005545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nazarov S, Schneider JP, Brackmann M, Goldie KN, Stahlberg H, Basler M. 2018. Cryo-EM reconstruction of type VI secretion system baseplate and sheath distal end. EMBO J 37:e97103. doi: 10.15252/embj.201797103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cherrak Y, Rapisarda C, Pellarin R, Bouvier G, Bardiaux B, Allain F, Malosse C, Rey M, Chamot-Rooke J, Cascales E, Fronzes R, Durand E. 2018. Biogenesis and structure of a type VI secretion baseplate. Nat Microbiol 3:1404–1416. doi: 10.1038/s41564-018-0260-1. [DOI] [PubMed] [Google Scholar]
- 33.Zoued A, Durand E, Brunet YR, Spinelli S, Douzi B, Guzzo M, Flaugnatti N, Legrand P, Journet L, Fronzes R, Mignot T, Cambillau C, Cascales E. 2016. Priming and polymerization of a bacterial contractile tail structure. Nature 531:59–63. doi: 10.1038/nature17182. [DOI] [PubMed] [Google Scholar]
- 34.Zoued A, Durand E, Santin YG, Journet L, Roussel A, Cambillau C, Cascales E. 2017. TssA: the cap protein of the type VI secretion system tail. Bioessays 39:10. doi: 10.1002/bies.201600262. [DOI] [PubMed] [Google Scholar]
- 35.Vettiger A, Winter J, Lin L, Basler M. 2017. The type VI secretion system sheath assembles at the end distal from the membrane anchor. Nat Commun 8:16088. doi: 10.1038/ncomms16088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dix SR, Owen HJ, Sun R, Ahmad A, Shastri S, Spiewak HL, Mosby DJ, Harris MJ, Batters SL, Brooker TA, Tzokov SB, Sedelnikova SE, Baker PJ, Bullough PA, Rice DW, Thomas MS. 2018. Structural insights into the function of type VI secretion system TssA subunits. Nat Commun 9:4765. doi: 10.1038/s41467-018-07247-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santin YG, Doan T, Lebrun R, Espinosa L, Journet L, Cascales E. 2018. In vivo TssA proximity labelling during type VI secretion biogenesis reveals TagA as a protein that stops and holds the sheath. Nat Microbiol 3:1304–1313. doi: 10.1038/s41564-018-0234-3. [DOI] [PubMed] [Google Scholar]
- 38.Aschtgen MS, Gavioli M, Dessen A, Lloubès R, Cascales E. 2010. The SciZ protein anchors the enteroaggregative Escherichia coli type VI secretion system to the cell wall. Mol Microbiol 75:886–899. doi: 10.1111/j.1365-2958.2009.07028.x. [DOI] [PubMed] [Google Scholar]
- 39.Durand E, Nguyen VS, Zoued A, Logger L, Péhau-Arnaudet G, Aschtgen MS, Spinelli S, Desmyter A, Bardiaux B, Dujeancourt A, Roussel A, Cambillau C, Cascales E, Fronzes R. 2015. Biogenesis and structure of a type VI secretion membrane core complex. Nature 523:555–560. doi: 10.1038/nature14667. [DOI] [PubMed] [Google Scholar]
- 40.Aschtgen MS, Bernard CS, de Bentzmann S, Lloubès R, Cascales E. 2008. SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J Bacteriol 190:7523–7531. doi: 10.1128/JB.00945-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma LS, Lin JS, Lai EM. 2009. An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its Walker a motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J Bacteriol 191:4316–4329. doi: 10.1128/JB.00029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aschtgen MS, Zoued A, Lloubès R, Journet L, Cascales E. 2012. The C-tail anchored TssL subunit, an essential protein of the enteroaggregative Escherichia coli Sci-1 type VI secretion system, is inserted by YidC. Microbiologyopen 1:71–82. doi: 10.1002/mbo3.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Logger L, Aschtgen MS, Guérin M, Cascales E, Durand E. 2016. Molecular dissection of the interface between the type VI secretion TssM cytoplasmic domain and the TssG baseplate component. J Mol Biol 428:4424–4437. doi: 10.1016/j.jmb.2016.08.032. [DOI] [PubMed] [Google Scholar]
- 44.Durand E, Zoued A, Spinelli S, Watson PJ, Aschtgen MS, Journet L, Cambillau C, Cascales E. 2012. Structural characterization and oligomerization of the TssL protein, a component shared by bacterial type VI and type IVb secretion systems. J Biol Chem 287:14157–14168. doi: 10.1074/jbc.M111.338731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zoued A, Duneau JP, Durand E, España AP, Journet L, Guerlesquin F, Cascales E. 2018. Tryptophan-mediated dimerization of the TssL transmembrane anchor is required for type VI secretion system activity. J Mol Biol 430:987–1003. doi: 10.1016/j.jmb.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 46.Felisberto-Rodrigues C, Durand E, Aschtgen MS, Blangy S, Ortiz-Lombardia M, Douzi B, Cambillau C, Cascales E. 2011. Towards a structural comprehension of bacterial type VI secretion systems: characterization of the TssJ-TssM complex of an Escherichia coli pathovar. PLoS Pathog 7:e1002386. doi: 10.1371/journal.ppat.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rapisarda C, Cherrak Y, Kooger R, Schmidt V, Pellarin R, Logger L, Cascales E, Pilhofer M, Durand E, Fronzes R. 15 March 2019. In situ and high-resolution cryo-EM structure of the type VI secretion membrane complex. EMBO J doi: 10.15252/embj.2018100886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yin M, Yan Z, Li X. 2019. Architecture of type VI secretion system membrane core complex. Cell Res 29:251–253. doi: 10.1038/s41422-018-0130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zoued A, Durand E, Bebeacua C, Brunet YR, Douzi B, Cambillau C, Cascales E, Journet L. 2013. TssK is a trimeric cytoplasmic protein interacting with components of both phage-like and membrane anchoring complexes of the type VI secretion system. J Biol Chem 288:27031–27041. doi: 10.1074/jbc.M113.499772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerc AJ, Diepold A, Trunk K, Porter M, Rickman C, Armitage JP, Stanley-Wall NR, Coulthurst SJ. 2015. Visualization of the Serratia type VI secretion system reveals unprovoked attacks and dynamic assembly. Cell Rep 12:2131–2142. doi: 10.1016/j.celrep.2015.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zoued A, Cassaro CJ, Durand E, Douzi B, España AP, Cambillau C, Journet L, Cascales E. 2016. Structure-function analysis of the TssL cytoplasmic domain reveals a new interaction between the type VI secretion baseplate and membrane complexes. J Mol Biol 428:4413–4423. doi: 10.1016/j.jmb.2016.08.030. [DOI] [PubMed] [Google Scholar]
- 52.Nguyen VS, Logger L, Spinelli S, Legrand P, Huyen Pham TT, Nhung Trinh TT, Cherrak Y, Zoued A, Desmyter A, Durand E, Roussel A, Kellenberger C, Cascales E, Cambillau C. 2017. Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex. Nat Microbiol 2:17103. doi: 10.1038/nmicrobiol.2017.103. [DOI] [PubMed] [Google Scholar]
- 53.Weber BS, Hennon SW, Wright MS, Scott NE, de Berardinis V, Foster LJ, Ayala JA, Adams MD, Feldman MF. 2016. Genetic dissection of the type VI secretion system in Acinetobacter and identification of a novel peptidoglycan hydrolase, TagX, required for its biogenesis. mBio 7:e01253-16. doi: 10.1128/mBio.01253-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Santin YG, Cascales E. 2017. Domestication of a housekeeping transglycosylase for assembly of a Type VI secretion system. EMBO Rep 18:138–149. doi: 10.15252/embr.201643206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aschtgen MS, Thomas MS, Cascales E. 2010. Anchoring the type VI secretion system to the peptidoglycan: TssL, TagL, TagP… what else? Virulence 1:535–540. doi: 10.4161/viru.1.6.13732. [DOI] [PubMed] [Google Scholar]
- 56.Drecktrah D, Levine-Wilkinson S, Dam T, Winfree S, Knodler LA, Schroer TA, Steele-Mortimer O. 2008. Dynamic behavior of Salmonella-induced membrane tubules in epithelial cells. Traffic 9:2117–2129. doi: 10.1111/j.1600-0854.2008.00830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brunet YR, Bernard CS, Gavioli M, Lloubès R, Cascales E. 2011. An epigenetic switch involving overlapping Fur and DNA methylation optimizes expression of a type VI secretion gene cluster. PLoS Genet 7:e1002205. doi: 10.1371/journal.pgen.1002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chaveroche MK, Ghigo JM, d'Enfert C. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res 28:E97. doi: 10.1093/nar/28.22.e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van den Ent F, Löwe J. 2006. RF cloning: a restriction-free method for inserting target genes into plasmids. J Biochem Biophys Methods 67:67–74. doi: 10.1016/j.jbbm.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 61.Flaugnatti N, Le TT, Canaan S, Aschtgen MS, Nguyen VS, Blangy S, Kellenberger C, Roussel A, Cambillau C, Cascales E, Journet L. 2016. A phospholipase A1 antibacterial type VI secretion effector interacts directly with the C-terminal domain of the VgrG spike protein for delivery. Mol Microbiol 99:1099–1118. doi: 10.1111/mmi.13292. [DOI] [PubMed] [Google Scholar]
- 62.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ducret A, Quardokus EM, Brun YV. 2016. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol 1:16077. doi: 10.1038/nmicrobiol.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.