Abstract
Improving the stability of antibodies for manufacture and shelf life is one of the main focuses of antibody engineering. One stabilization strategy is to perform specific mutations in human antibodies based on highly stable antibodies in other species. To identify the key residues for mutagenesis, it is necessary to understand the roles of these residues in stabilizing the antibody. Here, we use molecular dynamics simulations to study the molecular origin of the four shark immunoglobulin new antigen receptors constant domains (C1–C4). According to the unfolding pathways and the conformational free energy surfaces in 8 M urea at 380 K, the C2 domain is the most stable, followed by C4, C1, and C3, which agrees with the experimental findings. The C1 and C3 domains follow a common unfolding pathway in which the unfolding starts from the edge strands, particularly strand g, and then gradually progresses to the inner strands. Detailed structural analysis of the C2 domain reveals a “sandwich-like” R339-E322-R341 salt-bridge cluster on strand g, which grants ultrahigh stability to the C2 domain. We further design two sets of mutations by mutating E322 to alanine or setting all atomic charges in E322 to zero to break the salt-bridge cluster in the C2 domain, which confirms the importance of the salt bridges in stability. In the C4 domain, the D80-K104 salt bridge on strand g also strengthens the stability. On the other hand, in the C1 and C3 domains, there is no salt bridge on strand g. In addition to the salt bridges, the overall hydrophobicity score of the hydrophobic core is also positively correlated with the domain stability. Our findings provide a detailed microscopic picture of the molecular origin of the four shark immunoglobulin new antigen receptors constant domains that not only explains the differences in their structural stability but also provides important insights into future antibody design.
Introduction
Diverged from a common ancestor with other jawed vertebrates ∼500 million years ago (1), the cartilaginous fish comprise more than 700 extant species, including sharks, skates, and rays (2, 3, 4). These members are the earliest known vertebrates on Earth with the basic components of the vertebrate adaptive immune system (5, 6). Specifically, shark antibodies function naturally in the harsh environment of shark sera, which consists of 350 mM of urea and 1000 of mOsmol (7, 8, 9), exhibiting extremely high stability. Attracted by the potential for therapeutic antibody design, an amount of studies on shark antibodies have been designed by academic and industrial laboratories worldwide to gain more insights into the structural features rendering the high stability (5, 10, 11, 12, 13, 14, 15, 16).
There are three isotypes of shark antibodies (17, 18), immunoglobulin (Ig) M (19), IgW (20), and IgNAR (21). IgM is an isotype of the Ig and is regarded as the primordial in vertebrate evolution. IgW is orthologous to mammalian IgD (22). Ig new antigen receptor (IgNAR) exists in shark sera at a concentration of 0.1–1.0 mg/mL (23) and is the major antibody of sharks’ adaptive immune system. Without light chains, IgNAR is a homodimer that comprises only heavy chains (2, 18, 24, 25), and each heavy chain consists of five constant domains and one variable domain that mediate antigen binding (2). The variable domain is a soluble single domain and is tethered to the constant domains via flexible hinge-like regions (19). The constant domains consist of two-layer sandwich-like β-sheets that are linked by a buried disulfide bridge (26). Several studies on the IgNAR structure have been published since it was first isolated from the nurse shark serum in 1995 (2). The atomic resolution structure of the IgNAR constant domains was resolved by x-ray crystallography, and the stability of four constant domains C1–C4 were studied by thermochemical denaturation experiments (26). It was shown that the C2 and C4 domains were very tolerant to irreversible chemical and thermal denaturation, whereas the C1 and C3 domains were less resistant to the perturbation (26). However, it remains unknown what the underlying molecular mechanism is that contributes to the difference in the stability of the four constant domains.
In this study, we investigated the structural stability of the IgNAR C1–C4 domains at the molecular level by all-atom molecular dynamics simulations and provided a microscopic picture of the unfolding dynamics and the underlying molecular mechanisms. We found that the salt bridges on the edge strand, strand g, including R339-E322-R341 salt-bridge cluster and D80-K104 salt bridge, were essential for the high stability of the C2 and C4 domains, respectively. Mutation(s) on E322 and K104 disrupted the salt bridges and destabilized the C2 and C4 domains, respectively. In addition, the hydrophobicity of the hydrophobic core also has a strong impact on the domain stability. Taken all together, the structural analysis of the unfolding pathways revealed the key elements governing the high stability of the shark IgNAR C1–C4 domains and provided insights into the stability engineering of the human antibody.
Methods
The initial structures of the four IgNAR constant domains C1–C4 were obtained from the crystal structures deposited in the Protein Data Bank as 4Q97 (C1), 4Q9B (C2), 4Q9C (C3), and 2MKL (C4) (26). Fig. 1 shows the sequence alignment and the initial structures of the four constant domains. The multiple sequence similarity of the four domains was 44.72% as measured by using DNAMAN software (Lynnon Corporation). Moreover, the structural alignment analysis demonstrated that the four domains also showed a very high structural similarity (Fig. S1). Given that the IgNARs innately function in the harsh environment of shark sera, exhibiting extremely high stability (26), all denaturation simulations were performed in 8 M urea at 380 K to accelerate the unfolding process, following the system setup protocols in previous studies (27, 28). Please see Table S1 and Fig. S2 for more details on the system setup.
Figure 1.
(A) Sequence alignment of the IgNAR constant domains C1–C4. (B–E) Initial structures of the IgNAR constant domains are shown. Each constant domain consists of seven β-sheets arranged in a two-layered sandwich structure linked by a buried disulfide bond colored in purple. The seven β-sheets (a–g) are shown in cartoon representation in blue, cyan, green, lime, yellow, orange, and red, respectively. To see this figure in color, go online.
All simulations were carried out using the NAMD program (29), and all systems were built and visualized using the VMD software (30). The CHARMM27 (parameter set c32b1) force field (31) was used for the proteins and urea. The CHARMM-modified TIP3P model (32) was applied for water. The particle mesh Ewald method (33) was used for the long-range electrostatic interactions. The typical 12 Å cutoff was used for the van der Waals interactions. All production runs were performed under NPT ensemble, with both temperature and pressure controlled by the Langevin equation (34). Given that the shark antibodies are particularly stable at the physiological temperature, the simulations were performed in 8 M urea at 380 K. A time step of 2 fs was used, and the bond lengths were constrained in all simulations. The standard equilibration procedures were adapted for the systems. Each solvated system was minimized for 40,000 steps and equilibrated for 80 ps at 380 K and 1 atm with position restraints on all heavy atoms of the proteins. Three independent 300 ns trajectories of each constant domain were collected for the analysis.
To testify whether the force fields have an effect on the simulation results, the AMBER-03 force field (35) was also adopted to redo the simulations of C2 domain to compare the structural features of C2 domain under different force fields.
Results and Discussions
To characterize the unfolding processes of the four IgNAR constant domains, we analyzed the time dependence of the root mean-square deviation (RMSD) of the backbone atoms and the fraction of the native contacts (Q value) as compared to the crystal structures. This analysis has been used successfully to evaluate the stability and the degree of retention of the native structure of proteins in previous studies (27, 36). Following the typical cutoff distance of 6.5 Å for a residue-residue contact (27, 37, 38, 39), we defined a native contact between residues i and j, where j − i > 3, when the distance between any heavy atom of i and any heavy atom of j was less than 6.5 Å in the crystal structures. Therefore, for the folded state, Q = 1, and for the fully unfolded state, Q = 0.
Based on the time evolution of the RMSD and Q values, the C1 and C3 domains were completely unfolded by 300 ns, whereas the C2 and C4 domains were remarkably more stable. As shown in Fig. 2 A, the RMSD value of the C1 domain increased from 0 to 16.0 Å within 90 ns and that of the C3 domain increased to 8.0 Å in 75 ns. Meanwhile, the Q values of the C1 and C3 domains decreased dramatically from 1 to 0.45 and 0.52 (Fig. 2 B), respectively. Following the trend, the RMSD values continued to increase, and the Q values continued to decrease, but at a relatively slower rate. At ∼225 ns, the RMSD values of the C1 and C3 domains increased to around 22.5 and 20.5 Å, respectively. Similarly, the Q values of the two domains decreased further to 0.04 and 0.07, respectively. After 230 ns, the RMSD and Q values fluctuated slightly around these values, indicating that the two domains had been fully unfolded. Whereas, the RMSD values of the C2 and C4 domains remained nearly constant at ∼3.5 and ∼6.5 Å, respectively, for ∼200 ns after the initial slow growth (< ∼40 ns). The RMSD values remained stable until 250 ns. In the last 50 ns, the RMSD value of the C4 domain showed a slight increase to ∼10.0 Å due to the fluctuation of strand d. Moreover, we further tested the convergence of the simulations of all the four domains in all three independent 300 ns trajectories, each using the “all-to-all RMSD analysis” (40, 41, 42). In general, simulations of C2 and C4 domains converged better than that of C1 and C3 domains (Fig. S3). This was reasonable because the structure of C2 and C4 domains was highly stable during the entire simulation in all three trajectories, whereas C1 and C3 domains mostly unfolded after 200 ns of simulations and then underwent significant conformational fluctuations, thus leading to a relatively poor convergence of the simulations. The observed differences in the structural stability between the four domains were consistent with the experimental results (26).
Figure 2.
(A) The backbone root mean-square deviation (RMSD) and (B) the fraction of the native contacts (Q value) of the IgNAR C1–C4 domains as a function of the simulation time to show the stability of the four domains in 8 M urea at 380 K. (C) The relative conformational free energy surfaces (FESs) for C1–C4 domains that constructed along with the reaction coordinates of solvent accessible surface area (y-axis) and Q (x-axis) are shown. The free energy color bar (in kJ/mol) is provided on the right side of each figure. The typical configuration that corresponds to the free energy minimum in FESs is connected with an arrow. To see this figure in color, go online.
On the other hand, we further constructed the conformational free energy landscapes (FESs) of all the four domains to study their relative thermodynamic stability using two reaction coordinates, the solvent accessible surface area and the fraction of the native contacts (Q) (Fig. 2 C). Typical configurations that correspond to the local/global energy minimum in FES basins were extracted and analyzed. In general, the FES distributions of C2 and C4 domains were very similar from each other, both containing only one free energy basin, with a compact (and well-converged) “native-like” state around the basin. Near the basins, the overall structures of the C2 and C4 domains were very stable and native-like even under these conditions. The overall (un)folding free energies were estimated to be −15.0 and −13.2 kJ/mol for C2 and C4 domains, respectively (Fig. 2 C). On the other hand, C1 and C3 domains also shared a similar FES landscape, quite different from that of C2 and C4 domains, with a much broader and less well-converged surface. Both displayed a double free energy basin in the FES. For C1, the first free energy minimum was located at (0.61, 8443 Å2), with a free energy of −11.0 kJ/mol and a typical structure of its strand g unfolded. At the second minimum (0.01, 1200 Å2), the C1 domain was significantly more unfolded, with much less native contacts and a slightly lower free energy of −11.5 kJ/mol, indicating that under these conditions (high temperature and high urea concentration), the unfolded structure is more favored for C1. The C3 domain displayed similar features, with its first minimum located at (0.52, 7981 Å2), showing a free energy of −10.5 kJ/mol and a similar partially unfolded structure (strand d and g unfolded), and its second minimum located at (0.01, 11,505 Å2) showing a more significantly unfolded structure and a slightly lower free energy of −10.6 kJ/mol. Overall, these additional analyses of thermodynamics further support our original conclusion from the analyses of unfolding kinetics.
To obtain a microscopic view of the differences in the unfolding dynamics between different domains and to reveal the underlying mechanisms, we investigated the unfolding pathways of the C1–C4 domains at residue level. We calculated the time evolution of the fraction of native contacts for every residue (Qres) of each domain (Fig. 3) and examined some important intermediates along the pathways (Fig. 4).
Figure 3.
Time evolution of the fraction of the native contacts for each residue (Qres) of the (A) C1, (B) C2, (C) C3, and (D) C4 domains in one of the three independent trajectories. The color scale is provided on the right side, for the folded state, Q = 1, and for the fully unfolded state, Q = 0. To see this figure in color, go online.
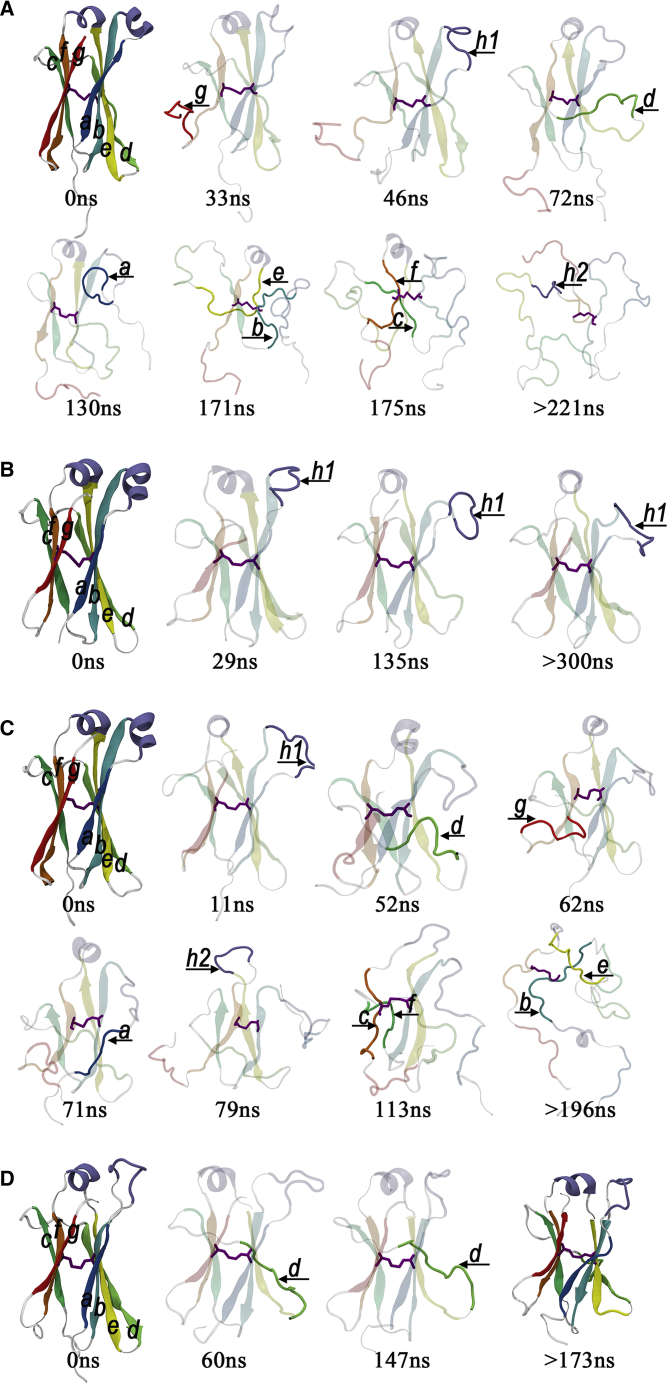
Figure 4.
Snapshots from the representative trajectories at critical time points for the (A) C1, (B) C2, (C) C3, and (D) C4 domains to show the important intermediate structures in the unfolding pathways. Unfolded β-strands along the simulations are indicated by an arrow. To see this figure in color, go online.
For the C1 domain, we took run1 as the example for the detailed analysis, and after only 33 ns, the Qres value and the secondary structure of strand g quickly disappeared. The unfolding of strand g was followed by that of helix-1, of which the Qres value and the secondary structure were mostly lost by 46 ns. At 72 ns, strand d lost most of the native contacts, and at 130 ns, strand a also lost most of its native contacts. The two pairs of the adjacent stands that form the hydrophobic core of the constant domains, strands b and e and strands c and f, lost their native contacts completely at 171 and 175 ns, respectively. Finally, at 221 ns, the helix-2 also lost its native contacts. We found that the helix-2 began to unfold only when the salt bridge between E215 on the helix-2 and K181 on strand c was destroyed (Fig. S4). After this point, the secondary structure of the C1 domain was completely lost. As expected, the observed unfolding sequence in run1 was also observed in the other two runs, despite the exact unfolding kinetics showing some variations. For instance, in all three trajectories of C1 domain, the unfolding initiated at the edge strands (particularly strand g), gradually progressed to the inner strands (i.e., the hydrophobic core strands), and ended with the unfolding of helix-2 (Fig. S5). As for the variations in the unfolding kinetics, strand g unfolded at 33, 19, and 14 ns for run1, run2, and run3, respectively (Fig. S6); helix-2 fully unfolded at 221, 168, and 253 ns for run1, run2, and run3, respectively. In contrast to the C1 domain, the C2 domain was much more stable throughout the simulation in all three trajectories (Figs. S5 and S7). The domain remained folded except for the helix-1, which became unfolded in every simulation. The poor stability of the helix-1 was due to its direct exposure to water.
The unfolding dynamics of the C3 domain were very similar to that of the C1 domain, in which the unfolding process always started from the edge strands (strand g, strand a, or strand d) or helix-1 or helix-2 because of the direct exposure of those components to solvent (Figs. S5 and S8). The only difference was that helix-2 of the C3 domain disappeared much earlier (at t = 73 ± 8 ns) than that of the C1 domain (at t = 214 ± 43 ns) because the former was the lack of the protection from the salt bridge between E215 and K181 in the C1 domain. Similar to the C2 domain, the C4 domain is also very stable. Only the edge strand d disappeared at around 60 ns. These unfolding pathways were very robust. We observed similar unfolding sequences in the other two trajectories of each domain. See Fig. S3 for the analysis of the additional trajectories.
Together, we propose a general unfolding pathway for the C1 and C3 domains. The unfolding process started from the regions that are directly accessible to the solvent, including the edge strands (particularly strand g) and helix-1, and then gradually progressed to the inner strands that form the hydrophobic core. This unfolding process was very similar to zipper unfolding, in which the edge strand was completely unzipped once the hydrogen bonds between the edge strand and its neighboring strand was destroyed.
Based on the unfolding dynamics illustrated above, we further analyzed the local structural properties of one of the edge strands, strand g, and uncovered the molecular origins of the four domains that resulted in the different stabilities.
There are some common structural features of strand g of the four constant domains, such as locating at the C-terminal end, directly exposing to water, forming antiparallel β-sheets with strand f, and preserving a dry interface with strand a by the internal-facing side chains. On the other hand, there are distinct structural features of the strand g of the four domains. In the C2 domain, we found a “sandwich-like” salt-bridge cluster formed by R339 and R341 in strand g and E322 in strand f as R339-E322-R341 (Fig. 5 B). In the C4 domain, a salt bridge was formed between D80 in strand g and K104 in helix-2 (Fig. 5 D). However, in the C1 and C3 domains, there were no salt bridges in strand g (Fig. 5, A and C). We then evaluated the stabilities of the salt bridges by calculating the distances between the Cβ atoms of the corresponding residues in the salt bridges. In the C2 domain, the Cβ-Cβ distances between E322 and R339 and between E322 and R341 remained stable throughout the simulation (Fig. 5 E). In the C4 domain, the Cβ-Cβ distance between D80 and K104 was also quite stable despite some fluctuations at the end of the simulation (Fig. 5 F). These results suggested that the salt bridges formed by the residues in strand g of the C2 and C4 domains were stable. Based on the unfolding pathways we demonstrated above, the C2 and C4 domains were much more stable than the C1 and C3 domains, in which the unfolding process started from strand g. We therefore propose that the formation of intermolecular salt bridges between strand g and the other strand is highly associated with the structural stability of strand g itself and even the whole domain.
Figure 5.
(A–D) The local structure of strand g of the C1–C4 domains. The side chains of all amino acids in strand g (red) and strand a (blue) are shown in bonds. The three salt bridges formed by the residues in strand g and strand f (orange) in the C2 domain and by the residues in strand g and the loop connecting helix-2 in the C4 domain are displayed in bold bonds. (E and F) Shown are the distances between the Cβ-Cβ atoms of the salt bridges in the C2 (E) and C4 (F) domains as a function of the simulation time, respectively. To see this figure in color, go online.
To validate our hypothesis, we mutated E322 in strand f to an alanine (E322A) to break the native R339-E322-R341 salt-bridge cluster in the C2 domain (Fig. 6 A). The reason that we selected the C2 domain for the mutation study was because it was more stable than the C4 domain. The time-dependent backbone RMSD value was used again to measure the structural stability of the mutant protein. As shown in Fig. 6 B, the RMSD value of the mutant C2 domain increased linearly to ∼9.0 Å along the simulation, whereas that of the wild-type C2 domain remained nearly constant at ∼3.5 Å. The Cβ-Cβ distances between A322 with R339 and R341 also increased from ∼6.0 to ∼25.0 Å as compared with the wild-type protein, in which the distances remained constant (Fig. 6 C). By the end of the 300 ns simulation, the structure of the mutant C2 domain had been largely unfolded in all three independent trajectories. These results demonstrated that the stability of the C2 domain was strongly impaired by the E322A mutation. Furthermore, we characterized the unfolding dynamics of the three independent trajectories of the mutant C2 domain by the time evolution of Qres. In all three trajectories, the unfolding process started from the edge strand g. To further validate the important role of the salt bridge in protecting the protein structure, we generated an “idealized” mutation, with all atomic charges in E322 residue set to zero (denoted as E322(e0) mutant) (Fig. S10). As expected, the stabilities of the C2 domain was considerably impaired by the E322(e0) mutation—within a very short period of time, E322(e0) mutant C2 domain totally unfolded in all three repeats. Moreover, without the protection of the wild-type R339-E322-R341 salt-bridge cluster, the unfolding of E322(e0) mutant C2 domain also initiated at strand g, consistent with the unfolding sequence of E322A mutant. These results agreed with our proposed hypothesis that strand g was like a gate that plays a significant role in protecting the structural stability of the C2 domain, and the “sandwich-like” R339-E322-R341 salt-bridge cluster was like a very solid lock that controls the opening and closing of the gate. Furthermore, we also have performed a similar idealized mutation for C4 domain by setting all atomic charges in K104 residue to zero to destruct the wild-type K104-D80 salt bridge (denoted by K104(e0) mutant, Fig. S11). Clearly, the stabilities of the C4 domain was also severely impaired by the K104(e0) mutation in all three trajectories. Again similarly, with the lack of protection of the K104-D80 salt bridge, the unfolding of K104(e0) mutant C4 domain also started from strand g and d.
Figure 6.
(A) Mutation of Glu322 to Ala (E322A) in the C2 domain. (B) Time-dependent RMSD values of the E322A mutant and the wild-type C2 domains are shown. (C) Time-dependent Cβ-Cβ distances between A322 (or E322) with R339 and R341 of the E322A mutant (or the wild-type) C2 domain are shown. (D) Final structures of the three independent 300 ns trajectories for the E322A mutant are shown. (E) Time-dependent Qres of the three trajectories for the mutant, the color scale is on the right-hand side of each panel. To see this figure in color, go online.
On the other hand, we noticed that the salt-bridge strength could be overestimated in many popular force fields, and this overvaluation in the AMBER-03 force field was milder than in others (43). Therefore, we adopted AMBER-03 force fields to redo simulations of C2 domain systems to compare the structural behaviors of the C2 domain under different force fields. Here, the C2 domain was selected for comparison because of its highest stability among the four domains. As shown in Fig. S12 A, the C2 domain still displayed excellent stability throughout the entire simulation in all of the three independent trajectories under the AMBER-03 force field, with a (un)folding free energy of −11.6 kJ/mol (Fig. S13), which is slightly weaker than that of −15.0 kJ/mol in the CHARMM27 force field, indicating less of an overestimation of the salt-bridge strengths indeed. Similarly, when the atomic charges in E322 were set to zero (i.e., E322(e0) mutant), the native salt-bridge cluster of “R339-E322-R341” was destructed, and the overall stability of the C2 domain was significantly weakened (Fig. S12 B). Interestingly, the mutant C2 domain also initiated unfolding at strand g in all three repeats. Overall, these results obtained from the simulations using the AMBER-03 force field were consistent with those from CHARMM27. Thus, despite the potential overestimation of salt-bridge strengths in CHARMM27, we believe the overall conclusion on the relative stability among all four domains still remains consistent across two different force fields. Interestingly, the important impact of a single charged residue at the critical position on the overall stability of a protein has also been observed in many other systems, including human γD-crystallins (27) and lysozyme (44).
We also studied the correlation between the hydrophobicity of the hydrophobic core (Fig. 7) and the structural stability of the wild-type protein. The hydrophobic core was defined as the region where the internal-facing side chains were within 6.0 Å of the center of mass of the inner strands b, c, e, and f. The hydrophobic score of the hydrophobic core was calculated by the sum of the hydropathy indexes of all residues at the hydrophobic core (45, 46). Similar to previous literature (47, 48, 49, 50, 51, 52, 53), here, the hydropathy index was assigned according to the water-vapor transfer free energies (ΔG°transfer) and the interior-exterior distribution of amino acid side chains determined by Jack Kyte and Russell Doolittle (45). This hydropathy index has been widely used in many previous studies to estimate the hydrophobicity of residues despite some limitations still existing, such as less accuracy for some specific residues like Cys and Pro (45, 46). The higher the hydrophobic score, the stronger the hydrophobicity of the core. The C2 and C4 domains have relative higher hydrophobic scores than the C1 and C3 domains. The hydrophobic scores are 16.2, 21.1, 13.1, and 20.7 for the C1 to C4 domains, respectively, which were positively correlated with the structural stabilities of the C1 to C4 domains. The E322A mutant in the C2 domain did not directly damage the hydrophobic core of the protein but might have triggered some allosteric effects on the hydrophobic core and, to some extent, lowered the domain stability.
Figure 7.
Hydrophobic scores for the hydrophobic cores of the C1–C4 domains. (A–D) The hydrophobic cores of the C1–C4 domains represented in transparent surface mode are shown. (E) The hydropathy indexes for the residues at the hydrophobic core are shown. (F) The hydrophobic scores for the hydrophobic cores of the C1–C4 domains are shown. To see this figure in color, go online.
Conclusions
In this study, we systematically evaluated the stability of the shark antibody IgNAR constant domains C1–C4 in relation to their molecular structures using all-atom molecular dynamics simulations. We performed denaturation simulations and demonstrated that the C2 and C4 domains were much more stable than the C1 and C3 domains, which is highly consistent with the experimental observations (26). We observed a common unfolding pathway for the C1 and C3 domains, in which the unfolding process started from the edge strands (particularly strand g) and helix-1 and then gradually progressed to the inner strands that formed the hydrophobic core. The edge strands and the loop helix were directly exposed to the solvent and were easier to be attacked or disrupted by water molecules. Detailed structural analyses suggested that the salt bridges formed by the residues on strand g and the residue(s) on the other components of the C2 and C4 domains contributed to the high stability of these two domains. In contrast, strand g of the C1 and C3 domains did not form salt bridges. In addition, we dissected the important role of the salt bridges on strand g in preserving the structure of the C2 domain by two sets of single mutations (E322A and an idealized E322(e0)) and using different force fields. The protecting role of the salt bridge in strand g on the C4 domain was also verified. As the hydrophobicity of the hydrophobic core was also positively correlated with the structural stability of the domain, in addition to the salt-bridge disruption, the E322A mutation might cause some long-range allosteric effects on the hydrophobic core of the C2 domain as well and thus further decrease the stability of the mutant C2 domain. This study not only supports the general conclusion of the experiment but also provides a molecular picture of the underlying mechanism and unfolding dynamics. Moreover, the revealed intrinsic molecular mechanism in controlling the stability of the four IgNAR constant domains provides unique insights into antibody research, which is useful for the antibody design in the biomedical engineering field.
Author Contributions
Z.Y. and R.Z. conceived and designed the research. H.Z., S.L., X.Y., and Z.L. performed the molecular dynamics simulations. H.Z., S.L., X.Y., Z.L., and Z.Y analyzed the data. H.Z., Z.Y. and R.Z. co-wrote the manuscript. All authors discussed the results and commented on the manuscript.
Acknowledgments
We thank Bruce Berne, Hongsuk Kang, Qiwen Shao, and Xuanyu Meng for helpful discussions.
This work was partially supported by the National Natural Science Foundation of China (grants 11574224, 11404233, and 11374221) and the Natural Science Foundation of Jiangsu Province (grant BK20161213). R.Z. acknowledges the support from IBM Blue Gene Science Program, A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions and Jiangsu Provincial Key Laboratory of Radiation Medicine and Protection.
Editor: Alan Grossfield.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.04.013.
Contributor Information
Zaixing Yang, Email: zxyang@suda.edu.cn.
Ruhong Zhou, Email: ruhongz@us.ibm.com.
Supporting Material
References
- 1.Dooley H., Flajnik M.F. Antibody repertoire development in cartilaginous fish. Dev. Comp. Immunol. 2006;30:43–56. doi: 10.1016/j.dci.2005.06.022. [DOI] [PubMed] [Google Scholar]; Dooley, H., and M. F. Flajnik. 2006. Antibody repertoire development in cartilaginous fish. Dev. Comp. Immunol. 30:43-56. [DOI] [PubMed]
- 2.Greenberg A.S., Avila D., Flajnik M.F. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature. 1995;374:168–173. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]; Greenberg, A. S., D. Avila, …, M. F. Flajnik. 1995. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature. 374:168-173. [DOI] [PubMed]
- 3.Litman G.W., Anderson M.K., Rast J.P. Evolution of antigen binding receptors. Annu. Rev. Immunol. 1999;17:109–147. doi: 10.1146/annurev.immunol.17.1.109. [DOI] [PubMed] [Google Scholar]; Litman, G. W., M. K. Anderson, and J. P. Rast. 1999. Evolution of antigen binding receptors. Annu. Rev. Immunol. 17:109-147. [DOI] [PubMed]
- 4.Flajnik M.F. Comparative analyses of immunoglobulin genes: surprises and portents. Nat. Rev. Immunol. 2002;2:688–698. doi: 10.1038/nri889. [DOI] [PubMed] [Google Scholar]; Flajnik, M. F. 2002. Comparative analyses of immunoglobulin genes: surprises and portents. Nat. Rev. Immunol. 2:688-698. [DOI] [PubMed]
- 5.Criscitiello M.F. What the shark immune system can and cannot provide for the expanding design landscape of immunotherapy. Expert Opin. Drug Discov. 2014;9:725–739. doi: 10.1517/17460441.2014.920818. [DOI] [PubMed] [Google Scholar]; Criscitiello, M. F. 2014. What the shark immune system can and cannot provide for the expanding design landscape of immunotherapy. Expert Opin. Drug Discov. 9:725-739. [DOI] [PubMed]
- 6.Flajnik M.F., Rumfelt L.L. The immune system of cartilaginous fish. Curr. Top. Microbiol. Immunol. 2000;248:249–270. doi: 10.1007/978-3-642-59674-2_11. [DOI] [PubMed] [Google Scholar]; Flajnik, M. F., and L. L. Rumfelt. 2000. The immune system of cartilaginous fish. Curr. Top. Microbiol. Immunol. 248:249-270. [DOI] [PubMed]
- 7.Kovaleva M., Ferguson L., Barelle C. Shark variable new antigen receptor biologics - a novel technology platform for therapeutic drug development. Expert Opin. Biol. Ther. 2014;14:1527–1539. doi: 10.1517/14712598.2014.937701. [DOI] [PubMed] [Google Scholar]; Kovaleva, M., L. Ferguson, …, C. Barelle. 2014. Shark variable new antigen receptor biologics - a novel technology platform for therapeutic drug development. Expert Opin. Biol. Ther. 14:1527-1539. [DOI] [PubMed]
- 8.England J.L., Haran G. Role of solvation effects in protein denaturation: from thermodynamics to single molecules and back. Annu. Rev. Phys. Chem. 2011;62:257–277. doi: 10.1146/annurev-physchem-032210-103531. [DOI] [PMC free article] [PubMed] [Google Scholar]; England, J. L., and G. Haran. 2011. Role of solvation effects in protein denaturation: from thermodynamics to single molecules and back. Annu. Rev. Phys. Chem. 62:257-277. [DOI] [PMC free article] [PubMed]
- 9.Saerens D., Ghassabeh G.H., Muyldermans S. Single-domain antibodies as building blocks for novel therapeutics. Curr. Opin. Pharmacol. 2008;8:600–608. doi: 10.1016/j.coph.2008.07.006. [DOI] [PubMed] [Google Scholar]; Saerens, D., G. H. Ghassabeh, and S. Muyldermans. 2008. Single-domain antibodies as building blocks for novel therapeutics. Curr. Opin. Pharmacol. 8:600-608. [DOI] [PubMed]
- 10.Clem L.W. Phylogeny of immunoglobulin structure and function. IV. Immunoglobulins of the giant grouper, Epinephelus itaira. J. Biol. Chem. 1971;246:9–15. [PubMed] [Google Scholar]; Clem, L. W. 1971. Phylogeny of immunoglobulin structure and function. IV. Immunoglobulins of the giant grouper, Epinephelus itaira. J. Biol. Chem. 246:9-15. [PubMed]
- 11.Müller M.R., O’Dwyer R., Barelle C.J. Generation and isolation of target-specific single-domain antibodies from shark immune repertoires. Methods Mol. Biol. 2012;907:177–194. doi: 10.1007/978-1-61779-974-7_9. [DOI] [PubMed] [Google Scholar]; Muller, M. R., R. O’Dwyer, …, C. J. Barelle. 2012. Generation and isolation of target-specific single-domain antibodies from shark immune repertoires. Methods Mol. Biol. 907:177-194. [DOI] [PubMed]
- 12.Liu J.L., Anderson G.P., Goldman E.R. Selection of cholera toxin specific IgNAR single-domain antibodies from a naïve shark library. Mol. Immunol. 2007;44:1775–1783. doi: 10.1016/j.molimm.2006.07.299. [DOI] [PubMed] [Google Scholar]; Liu, J. L., G. P. Anderson, …, E. R. Goldman. 2007. Selection of cholera toxin specific IgNAR single-domain antibodies from a naive shark library. Mol. Immunol. 44:1775-1783. [DOI] [PubMed]
- 13.Walsh R., Nuttall S., Locarnini S. Targeting the hepatitis B virus precore antigen with a novel IgNAR single variable domain intrabody. Virology. 2011;411:132–141. doi: 10.1016/j.virol.2010.12.034. [DOI] [PubMed] [Google Scholar]; Walsh, R., S. Nuttall, …, S. Locarnini. 2011. Targeting the hepatitis B virus precore antigen with a novel IgNAR single variable domain intrabody. Virology. 411:132-141. [DOI] [PubMed]
- 14.Jena B., Moyes J.S., Cooper L.J. Driving CAR-based T-cell therapy to success. Curr. Hematol. Malig. Rep. 2014;9:50–56. doi: 10.1007/s11899-013-0197-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jena, B., J. S. Moyes, …, L. J. Cooper. 2014. Driving CAR-based T-cell therapy to success. Curr. Hematol. Malig. Rep. 9:50-56. [DOI] [PMC free article] [PubMed]
- 15.Luo M., Kim H., Ohta Y. Construction of a nurse shark (Ginglymostoma cirratum) bacterial artificial chromosome (BAC) library and a preliminary genome survey. BMC Genomics. 2006;7:106. doi: 10.1186/1471-2164-7-106. [DOI] [PMC free article] [PubMed] [Google Scholar]; Luo, M., H. Kim, …, Y. Ohta. 2006. Construction of a nurse shark (Ginglymostoma cirratum) bacterial artificial chromosome (BAC) library and a preliminary genome survey. BMC Genomics. 7:106. [DOI] [PMC free article] [PubMed]
- 16.Könning D., Zielonka S., Kolmar H. Camelid and shark single domain antibodies: structural features and therapeutic potential. Curr. Opin. Struct. Biol. 2017;45:10–16. doi: 10.1016/j.sbi.2016.10.019. [DOI] [PubMed] [Google Scholar]; Konning, D., S. Zielonka, …, H. Kolmar. 2017. Camelid and shark single domain antibodies: structural features and therapeutic potential. Curr. Opin. Struct. Biol. 45:10-16. [DOI] [PubMed]
- 17.Rumfelt L.L., Lohr R.L., Flajnik M.F. Diversity and repertoire of IgW and IgM VH families in the newborn nurse shark. BMC Immunol. 2004;5:8. doi: 10.1186/1471-2172-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rumfelt, L. L., R. L. Lohr, …, M. F. Flajnik. 2004. Diversity and repertoire of IgW and IgM VH families in the newborn nurse shark. BMC Immunol. 5:8. [DOI] [PMC free article] [PubMed]
- 18.Hamers-Casterman C., Atarhouch T., Hamers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]; Hamers-Casterman, C., T. Atarhouch, …, R. Hamers. 1993. Naturally occurring antibodies devoid of light chains. Nature. 363:446-448. [DOI] [PubMed]
- 19.Roux K.H., Greenberg A.S., Flajnik M.F. Structural analysis of the nurse shark (new) antigen receptor (NAR): molecular convergence of NAR and unusual mammalian immunoglobulins. Proc. Natl. Acad. Sci. USA. 1998;95:11804–11809. doi: 10.1073/pnas.95.20.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]; Roux, K. H., A. S. Greenberg, …, M. F. Flajnik. 1998. Structural analysis of the nurse shark (new) antigen receptor (NAR): molecular convergence of NAR and unusual mammalian immunoglobulins. Proc. Natl. Acad. Sci. USA. 95:11804-11809. [DOI] [PMC free article] [PubMed]
- 20.Rumfelt L.L., Diaz M., Flajnik M.F. Unprecedented multiplicity of Ig transmembrane and secretory mRNA forms in the cartilaginous fish. J. Immunol. 2004;173:1129–1139. doi: 10.4049/jimmunol.173.2.1129. [DOI] [PubMed] [Google Scholar]; Rumfelt, L. L., M. Diaz, …, M. F. Flajnik. 2004. Unprecedented multiplicity of Ig transmembrane and secretory mRNA forms in the cartilaginous fish. J. Immunol. 173:1129-1139. [DOI] [PubMed]
- 21.Berstein R.M., Schluter S.F., Marchalonis J.J. A new high molecular weight immunoglobulin class from the carcharhine shark: implications for the properties of the primordial immunoglobulin. Proc. Natl. Acad. Sci. USA. 1996;93:3289–3293. doi: 10.1073/pnas.93.8.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]; Berstein, R. M., S. F. Schluter, …, J. J. Marchalonis. 1996. A new high molecular weight immunoglobulin class from the carcharhine shark: implications for the properties of the primordial immunoglobulin. Proc. Natl. Acad. Sci. USA. 93:3289-3293. [DOI] [PMC free article] [PubMed]
- 22.Ohta Y., Flajnik M. IgD, like IgM, is a primordial immunoglobulin class perpetuated in most jawed vertebrates. Proc. Natl. Acad. Sci. USA. 2006;103:10723–10728. doi: 10.1073/pnas.0601407103. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ohta, Y., and M. Flajnik. 2006. IgD, like IgM, is a primordial immunoglobulin class perpetuated in most jawed vertebrates. Proc. Natl. Acad. Sci. USA. 103:10723-10728. [DOI] [PMC free article] [PubMed]
- 23.Dooley H., Stanfield R.L., Flajnik M.F. First molecular and biochemical analysis of in vivo affinity maturation in an ectothermic vertebrate. Proc. Natl. Acad. Sci. USA. 2006;103:1846–1851. doi: 10.1073/pnas.0508341103. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dooley, H., R. L. Stanfield, …, M. F. Flajnik. 2006. First molecular and biochemical analysis of in vivo affinity maturation in an ectothermic vertebrate. Proc. Natl. Acad. Sci. USA. 103:1846-1851. [DOI] [PMC free article] [PubMed]
- 24.Zielonka S., Empting M., Kolmar H. Structural insights and biomedical potential of IgNAR scaffolds from sharks. MAbs. 2015;7:15–25. doi: 10.4161/19420862.2015.989032. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zielonka, S., M. Empting, …, H. Kolmar. 2015. Structural insights and biomedical potential of IgNAR scaffolds from sharks. MAbs. 7:15-25. [DOI] [PMC free article] [PubMed]
- 25.Flajnik M.F., Deschacht N., Muyldermans S. A case of convergence: why did a simple alternative to canonical antibodies arise in sharks and camels? PLoS Biol. 2011;9:e1001120. doi: 10.1371/journal.pbio.1001120. [DOI] [PMC free article] [PubMed] [Google Scholar]; Flajnik, M. F., N. Deschacht, and S. Muyldermans. 2011. A case of convergence: why did a simple alternative to canonical antibodies arise in sharks and camels? PLoS Biol. 9:e1001120. [DOI] [PMC free article] [PubMed]
- 26.Feige M.J., Gräwert M.A., Buchner J. The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins. Proc. Natl. Acad. Sci. USA. 2014;111:8155–8160. doi: 10.1073/pnas.1321502111. [DOI] [PMC free article] [PubMed] [Google Scholar]; Feige, M. J., M. A. Grawert, …, J. Buchner. 2014. The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins. Proc. Natl. Acad. Sci. USA. 111:8155-8160. [DOI] [PMC free article] [PubMed]
- 27.Yang Z., Xia Z., Zhou R. Dissecting the contributions of β-hairpin tyrosine pairs to the folding and stability of long-lived human γD-crystallins. Nanoscale. 2014;6:1797–1807. doi: 10.1039/c3nr03782g. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yang, Z., Z. Xia, …, R. Zhou. 2014. Dissecting the contributions of β-hairpin tyrosine pairs to the folding and stability of long-lived human γD-crystallins. Nanoscale. 6:1797-1807. [DOI] [PMC free article] [PubMed]
- 28.Xia Z., Yang Z., Zhou R. UV-radiation induced disruption of dry-cavities in human γD-crystallin results in decreased stability and faster unfolding. Sci. Rep. 2013;3:1560. doi: 10.1038/srep01560. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xia, Z., Z. Yang, …, R. Zhou. 2013. UV-radiation induced disruption of dry-cavities in human γD-crystallin results in decreased stability and faster unfolding. Sci. Rep. 3:1560. [DOI] [PMC free article] [PubMed]
- 29.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]; Phillips, J. C., R. Braun, …, K. Schulten. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781-1802. [DOI] [PMC free article] [PubMed]
- 30.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38, 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]; Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33-38, 27-28. [DOI] [PubMed]
- 31.Brooks B.R., Brooks C.L., 3rd, Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]; Brooks, B. R., C. L. Brooks 3rd, …, M. Karplus. 2009. CHARMM: the biomolecular simulation program. J. Comput. Chem. 30:1545-1614. [DOI] [PMC free article] [PubMed]
- 32.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]; Jorgensen, W. L., J. Chandrasekhar, …, M. L. Klein. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926-935.
- 33.Essmann U., Perera L., Pedersen L.G. A smooth particle Mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]; Essmann, U., L. Perera, …, L. G. Pedersen. 1995. A smooth particle Mesh Ewald method. J. Chem. Phys. 103:8577-8593.
- 34.Langevin P. On the theory of Brownian motion. Cr Hebd Acad Sci. 1908;146:530–533. [Google Scholar]; Langevin, P. 1908. On the theory of Brownian motion. Cr Hebd Acad Sci. 146:530-533.
- 35.Duan Y., Wu C., Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]; Duan, Y., C. Wu, …, P. Kollman. 2003. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 24:1999-2012. [DOI] [PubMed]
- 36.Chen S.H., Elber R. The energy landscape of a protein switch. Phys. Chem. Chem. Phys. 2014;16:6407–6421. doi: 10.1039/c3cp55209h. [DOI] [PubMed] [Google Scholar]; Chen, S. H., and R. Elber. 2014. The energy landscape of a protein switch. Phys. Chem. Chem. Phys. 16:6407-6421. [DOI] [PubMed]
- 37.Miyazawa S., Jernigan R.L. Residue-residue potentials with a favorable contact pair term and an unfavorable high packing density term, for simulation and threading. J. Mol. Biol. 1996;256:623–644. doi: 10.1006/jmbi.1996.0114. [DOI] [PubMed] [Google Scholar]; Miyazawa, S., and R. L. Jernigan. 1996. Residue-residue potentials with a favorable contact pair term and an unfavorable high packing density term, for simulation and threading. J. Mol. Biol. 256:623-644. [DOI] [PubMed]
- 38.Betancourt M.R., Thirumalai D. Pair potentials for protein folding: choice of reference states and sensitivity of predicted native states to variations in the interaction schemes. Protein Sci. 1999;8:361–369. doi: 10.1110/ps.8.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]; Betancourt, M. R., and D. Thirumalai. 1999. Pair potentials for protein folding: choice of reference states and sensitivity of predicted native states to variations in the interaction schemes. Protein Sci. 8:361-369. [DOI] [PMC free article] [PubMed]
- 39.Chen S.H., Kang S.G., Zhou R. Charging nanoparticles: increased binding of Gd@C82(OH)22 derivatives to human MMP-9. Nanoscale. 2018;10:5667–5677. doi: 10.1039/c8nr00127h. [DOI] [PubMed] [Google Scholar]; Chen, S. H., S. G. Kang, …, R. Zhou. 2018. Charging nanoparticles: increased binding of Gd@C82(OH)22 derivatives to human MMP-9. Nanoscale. 10:5667-5677. [DOI] [PubMed]
- 40.Grossfield A., Zuckerman D.M. Quantifying uncertainty and sampling quality in biomolecular simulations. Annu. Rep. Comput. Chem. 2009;5:23–48. doi: 10.1016/S1574-1400(09)00502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Grossfield, A., and D. M. Zuckerman. 2009. Quantifying uncertainty and sampling quality in biomolecular simulations. Annu. Rep. Comput. Chem. 5:23-48. [DOI] [PMC free article] [PubMed]
- 41.García A.E. Large-amplitude nonlinear motions in proteins. Phys. Rev. Lett. 1992;68:2696–2699. doi: 10.1103/PhysRevLett.68.2696. [DOI] [PubMed] [Google Scholar]; Garcia, A. E. 1992. Large-amplitude nonlinear motions in proteins. Phys. Rev. Lett. 68:2696-2699. [DOI] [PubMed]
- 42.Grossfield A., Patrone P.N., Zuckerman D.M. Best practices for quantification of uncertainty and sampling quality in molecular simulations [Article v1.0] Living J. Comput. Mol. Sci. 2018;1:5067. doi: 10.33011/livecoms.1.1.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]; Grossfield, A., P. N. Patrone, …, D. M. Zuckerman. 2018. Best practices for quantification of uncertainty and sampling quality in molecular simulations [Article v1.0]. Living J. Comput. Mol. Sci. 1: 5067 [DOI] [PMC free article] [PubMed]
- 43.Debiec K.T., Gronenborn A.M., Chong L.T. Evaluating the strength of salt bridges: a comparison of current biomolecular force fields. J. Phys. Chem. B. 2014;118:6561–6569. doi: 10.1021/jp500958r. [DOI] [PMC free article] [PubMed] [Google Scholar]; Debiec, K. T., A. M. Gronenborn, and L. T. Chong. 2014. Evaluating the strength of salt bridges: a comparison of current biomolecular force fields. J. Phys. Chem. B. 118:6561-6569. [DOI] [PMC free article] [PubMed]
- 44.Zhou R., Eleftheriou M., Berne B.J. Destruction of long-range interactions by a single mutation in lysozyme. Proc. Natl. Acad. Sci. U.S.A. 2007;104:5824–5829. doi: 10.1073/pnas.0701249104. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhou, R., M. Eleftheriou, …, B. J. Berne. 2007. Destruction of long-range interactions by a single mutation in lysozyme. Proc. Natl. Acad. Sci. U.S.A. 104:5824-5829. [DOI] [PMC free article] [PubMed]
- 45.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]; Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed]
- 46.Chothia C. The nature of the accessible and buried surfaces in proteins. J. Mol. Biol. 1976;105:1–12. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]; Chothia, C. 1976. The nature of the accessible and buried surfaces in proteins. J. Mol. Biol. 105:1-12. [DOI] [PubMed]
- 47.Uversky V.N., Gillespie J.R., Fink A.L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]; Uversky, V. N., J. R. Gillespie, and A. L. Fink. 2000. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 41:415-427. [DOI] [PubMed]
- 48.Ashbaugh H.S., Hatch H.W. Natively unfolded protein stability as a coil-to-globule transition in charge/hydropathy space. J. Am. Chem. Soc. 2008;130:9536–9542. doi: 10.1021/ja802124e. [DOI] [PubMed] [Google Scholar]; Ashbaugh, H. S., and H. W. Hatch. 2008. Natively unfolded protein stability as a coil-to-globule transition in charge/hydropathy space. J. Am. Chem. Soc. 130:9536-9542. [DOI] [PubMed]
- 49.Hirokawa T., Boon-Chieng S., Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]; Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 14:378-379. [DOI] [PubMed]
- 50.Kosinski J., von Appen A., Beck M. Xlink Analyzer: software for analysis and visualization of cross-linking data in the context of three-dimensional structures. J. Struct. Biol. 2015;189:177–183. doi: 10.1016/j.jsb.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kosinski, J., A. von Appen, …, M. Beck. 2015. Xlink Analyzer: software for analysis and visualization of cross-linking data in the context of three-dimensional structures. J. Struct. Biol. 189:177-183. [DOI] [PMC free article] [PubMed]
- 51.Lee D.W., Whittaker G.R. Use of AAScatterPlot tool for monitoring the evolution of the hemagglutinin cleavage site in H9 avian influenza viruses. Bioinformatics. 2017;33:2431–2435. doi: 10.1093/bioinformatics/btx203. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lee, D. W., and G. R. Whittaker. 2017. Use of AAScatterPlot tool for monitoring the evolution of the hemagglutinin cleavage site in H9 avian influenza viruses. Bioinformatics. 33:2431-2435. [DOI] [PMC free article] [PubMed]
- 52.Sanz-Bravo A., Martín-Esteban A., López de Castro J.A. Allele-specific alterations in the peptidome underlie the joint association of HLA-A∗29:02 and endoplasmic reticulum aminopeptidase 2 (ERAP2) with birdshot chorioretinopathy. Mol. Cell. Proteomics. 2018;17:1564–1577. doi: 10.1074/mcp.RA118.000778. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sanz-Bravo, A., A. Martin-Esteban, …, J. A. Lopez de Castro. 2018. Allele-specific alterations in the peptidome underlie the joint association of HLA-A∗29:02 and endoplasmic reticulum aminopeptidase 2 (ERAP2) with birdshot chorioretinopathy. Mol. Cell. Proteomics. 17:1564-1577. [DOI] [PMC free article] [PubMed]
- 53.Sanz M.A., Madan V., Nieva J.L. Interfacial domains in Sindbis virus 6K protein. Detection and functional characterization. J. Biol. Chem. 2003;278:2051–2057. doi: 10.1074/jbc.M206611200. [DOI] [PubMed] [Google Scholar]; Sanz, M. A., V. Madan, …, J. L. Nieva. 2003. Interfacial domains in Sindbis virus 6K protein. Detection and functional characterization. J. Biol. Chem. 278:2051-2057. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.