Abstract
Objective
Activation of the Wnt-signaling pathway is known to inhibit differentiation in adipocytes. However, there is a gap in our understanding of the transcriptional network regulated by components of the Wnt-signaling pathway during adipogenesis and in adipocytes during postnatal life. The key intracellular effectors of the Wnt-signaling pathway occur through TCF transcription factors such as TCF7L2 (transcription factor-7-like 2). Several genetic variants in proximity to TCF7L2 have been linked to type 2 diabetes through genome-wide association studies in various human populations. Our work aims to functionally characterize the adipocyte specific gene program regulated by TCF7L2 and understand how this program regulates metabolism.
Methods
We generated Tcf7l2F/F mice and assessed TCF7L2 function in isolated adipocytes and adipose specific knockout mice. ChIP-sequencing and RNA-sequencing was performed on the isolated adipocytes with control and TCF7L2 knockout cells. Adipose specific TCF7L2 knockout mice were challenged with high fat diet and assessed for body weight, glucose tolerance, and lipolysis.
Results
Here we report that TCF7L2 regulates adipocyte size, endocrine function, and glucose metabolism. Tcf7l2 is highly expressed in white adipose tissue, and its expression is suppressed in genetic and diet-induced models of obesity. Genome-wide distribution of TCF7L2 binding and gene expression analysis in adipocytes suggests that TCF7L2 directly regulates genes implicated in cellular metabolism and cell cycle control. When challenged with a high-fat diet, conditional deletion of TCF7L2 in adipocytes led to impaired glucose tolerance, impaired insulin sensitivity, promoted weight gain, and increased adipose tissue mass. This was accompanied by reduced expression of triglyceride hydrolase, reduced fasting-induced free fatty acid release, and adipocyte hypertrophy in subcutaneous adipose tissue.
Conclusions
Together our studies support that TCF7L2 is a central transcriptional regulator of the adipocyte metabolic program by directly regulating the expression of genes involved in lipid and glucose metabolism.
Keywords: Diabetes, Adipose tissue, Obesity, Wnt signaling, Lipolysis
Highlights
-
•
Type 2 diabetes risk gene Tcf7l2 is downregulated in adipose tissue of obese mice.
-
•
TCF7L2 occupies genes involved in metabolic control and cell cycle.
-
•
Inverse correlation between Tcf7l2 expression and glucose control.
-
•
Conditional deletion of Tcf7l2 in adipocytes leads to glucose intolerance and adipocyte hypertrophy.
-
•
Tcf7l2 regulates adipose tissue lipolysis and lipogenesis.
1. Introduction
The Wnt signaling pathway is an integral regulator of cellular proliferation and development and has more recently been shown to regulate cellular metabolism [1], [2]. Canonical Wnt signaling is initiated when extracellular wnt ligands activate cell surface receptors on target cells. This leads to stabilization and subsequent translocation of β-catenin to the nucleus where it activates TCF/LEF transcription factors [3], [4]. In the absence of Wnt ligand, TCF transcription factors are bound to TLEs, a family of transcriptional repressors that silence Wnt-mediated gene expression [5]. Several components of the Wnt signaling pathway are known to regulate metabolic disease in humans including the TCF/LEF family member, TCF7L2 [6]. Genome-wide association studies in humans aimed at uncovering novel genetic variants associated with type 2 diabetes have identified polymorphisms (rs12255372 and rs7903146) in Tcf7l2 that correlate with type 2 diabetes across multiple populations [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54], [55], [56], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72], [73], [74], [75], [76], [77], [78], [79], [80], [81], [82], [83], [84], [85], [86], [87], [88], [89], [90], [91], [92], [93], [94], [95], [96], [97], [98], [99], [100], [101], [102], [103], [104], [105], [106], [107], [108], [109], [110], [111], [112], [113], [114], [115], [116], [117], [118], [119], [120], [121], [122], [123], [124], [125], [126], [127], [128], [129], [130], [131]. Remarkably, polymorphisms in Tcf7l2 have a greater impact on risk for developing type 2 diabetes than any other risk allele that has been identified in humans, including those found in PPARγ, a transcription factor that regulates adipocyte function [10], [19], [132].
A causal relationship between Tcf7l2 and type 2 diabetes has been explored in mice expressing multiple copies of Tcf7l2 using a BAC transgenic system. This led to overexpression of the human Tcf7l2 gene in multiple tissues, which resulted in impaired glucose handling [133]. Efforts to understand how TCF7L2 functions in various tissues to regulate systemic glycemia have yielded mixed results. Mice lacking TCF7L2 in pancreatic β-cells show normal β-cell function and insulin secretion, which might be expected since patients who harbor Tcf7l2 risk alleles have increased expression of TCF7L2 in the pancreas [134]. In contrast, mice with deletion of TCF7L2 in the liver have improved glucose tolerance and reduced gluconeogenesis, while mice overexpressing TCF7L2 in the liver are glucose intolerant [134]. Additional studies are needed to characterize the role of TCF7L2 in other metabolically active tissues to gain a full understanding of how TCF7L2 contributes to the complex etiology of type 2 diabetes.
One tissue that regulates both carbohydrate and lipid metabolism is the adipose tissue. White adipocytes synthesize and store triglycerides from newly synthesized fatty acids or from dietary sources via chylomicrons. Adipocytes also operate as endocrine cells, secreting bioactive peptides that regulate glucose and lipid metabolism including leptin, adiponectin, resistin, and adipsin [135], [136], [137], [138]. Previous studies show that TCF7L2 regulates adipocyte differentiation. Expression of a dominant negative form of TCF7L2 in fibroblasts blocks Wnt-signaling and stimulates cells to differentiate into adipocytes [139]. During adipocyte differentiation Wnt-responsive genes are actively repressed by TLE3 through a direct interaction with TCF7L2 [140]. Given the central role of adipose tissue in type 2 diabetes pathology and the role of TCF7L2 in adipogenesis, defining the direct transcriptional targets of TCF7L2 in adipocytes will provide opportunities to understanding how this pathway drives metabolic control.
Here we show that Tcf7l2 is highly expressed in adipocytes, and its expression is reduced in adipose tissue of mice fed high-fat diet and in genetic models of obesity. We demonstrate that conditional deletion of Tcf7l2 in adipocytes leads to adipocyte hypertrophy, and impaired glucose handling on a high-fat diet. Genome-wide analysis of TCF7L2 binding and gene expression analysis supports that TCF7L2 is a key metabolic regulator in adipocytes. TCF7L2 directly regulates genes involved in triglyceride hydrolysis and lipogenesis, and adipocyte depletion of TCF7L2 ultimately promotes lipid storage and adipocyte hypertrophy. These studies highlight that TCF7L2 has a role beyond development that extends to metabolic control in adipocytes. Together our findings suggest that targeted therapies that enhance Tcf7l2 expression in adipocytes are likely to improve glucose metabolism in individuals carrying this susceptibility gene.
2. Materials and methods
2.1. Animals
Mice were maintained in 12 h light/12 h dark with free access to water and standard chow (Tekland Global 2920X-030818). For high fat diet (HFD) studies, mice were fed rodent diet with 60 kcal% fat diet (D12492, Research Diets, Inc). Tcf7l2F/F mice were generated from JM8.F6 ES cells originally derived from C57BL/6N mice through the EUCOMM collection, Tcf7l2tm1a(EUCOMM)Wtsi (EM:06158). The L1L2_Bact_P cassette was inserted at position 55907842 of Chromosome 19 upstream of exon5 (Build GRCm38) (Exon5: TCGAACAAAG TACCGGTGGT GCAACACCCC CACCATGTCC ACCCACTCAC GCCTCTCATC ACGTACAGCA ATGAACACTT CACCCCGGGA AATCCACCTC CGCACTTACC AGCTGACGTA GACCCCAAAA CAGG). The cassette is composed of an FRT flanked lacZ/neomycin cassette followed by a loxP site. An additional loxP site is inserted downstream of the targeted exon5 at position 55908926 (Supplemental Figure 2A). Exon 5 is thus flanked by loxP sites (Supplemental Figure 2B). A “conditional ready” (floxed) allele was created by crossing mice with Actin-flp mice to induce recombinase expression in the germline. Removal of LacZ and Neomycin cassettes were confirmed by PCR using primers flanking FRT sites, forward (CTGCCCCAATACAACATACACA), and reverse (CTCATAAATTAACCGGACAATGATG). Flp induced recombination leads to a 306 bp PCR product, which was observed. Subsequent breeding of Tcf7l2F/+ mice was completed using adiponectin-Cre mice [141] to induce deletion of exon5 in adipocytes. The Tcf7l2F allele was genotyped for the presence of the terminal loxP site primers forward (TCCTAAGCAAACTAGGTAGCAATGA) and reverse (TTCAGTTTGTACATCTGTAAAATGGG) as well as for the presence of the FRT site primers forward (CTGCCCCAATACAACATACACA) and reverse (CTCATAAATTAACCGGACAATGATG) (Supplemental Figure 2C).
For glucose tolerance tests (GTTs) mice were fasted for 6 h, then given an intraperitoneal injection of glucose (1 mg glucose/g body weight). Insulin tolerance tests (ITT) was performed by intraperitoneal injection of 0.75U insulin/kg body weight (Lilly). In both of these experiments, blood glucose was monitored via tail vein bleed using a glucometer (Contour).
2.2. Body composition
Non-anesthetized mice were weighed and then placed in Bruker Minispec LF50 NMR to measure lean body mass, fat mass, and water mass. Measurements were made from mice fed chow or high-fat diets as indicated in figure legends.
2.3. In vivo lipolysis
Tcf7l2F/F;Adp-Cre and littermate Tcf7l2F/F control mice received an intraperitoneal injection of CL-316,243 administered at a dose of 1 mg/kg body weight or a volume matched vehicle control injection of PBS. Food was removed at T0 and the mice were allowed free access to water. Blood was taken by tail vein immediately before the injection for T0 and then taken at 30 min, 1 h, 3 h, and 6 h post-injection. Blood was spun at 2,000× g for 20 min in a refrigerated centrifuge and serum was collected and transferred to a new tube. Glycerol measurements were performed by colorimetric assay following manufacturer instructions (Cayman). Free fatty acids were measured using a Free Fatty Acid Quantitation Kit (Sigma) according to manufacturer's instructions. Insulin measurements were performed on the serum of mice fasted for 6 h by ELISA (Crystal Chemical).
2.4. Cell culture
Preadipocytes were isolated from minced inguinal white adipose tissue of Tcf7l2F/F mice by digestion with Collagenase type II (Sigma). Cells from the stromal vascular fraction were then resuspended in HBSS and stained with Biotin CD31 (Biolegend), Biotin CD45 (Biolegend), Biotin Ter-119 (Biolegend), FITC Streptavidin (Biolegend), PE CD34 (BD Biosciences), V500 Sca1 (BD Biosciences), PE/Cy7 CD29 (Biolegend), and Sytox Blue (Thermo Fisher). Dead cells were excluded by uptake of Sytox blue and lineage positive cells were excluded by FITC Streptavidin. Preadipocytes were positively selected for by CD34, Sca1, and CD29 staining. Cells were sorted on a BD FACS Aria cell sorter and analyzed with BD FACSDiva software. Isolated preadipocytes were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10%FBS(RMBIO) and maintained in a 5% CO2 atmosphere and were then immortalized by retroviral expression of the SV40 large T antigen using a hygromycin selection marker. To generate an inducible knockout cell line immortalized cells were then infected with retroviral vector pMSCV CreERT2 using a puromycin selection marker. For differentiation cells were plated, grown to confluence, and then stimulated with differentiation media (1 μM dexamethasone, 0.5 mM isobutylmethylxanthine, and 5 μg/mL insulin) for two days. The cells were maintained in 10% FBS DMEM with 5 μg/mL insulin.
2.5. Histology
Tissues were excised and fixed in a 4% buffered formaldehyde for 24 h then transferred to 70% ethanol. White adipose tissue was embedded in paraffin and sliced into 10 μM sections and stained for hematoxylin and eosin.
2.6. RNA isolation
Adipose tissue was excised and then placed in 1 mL Trizol reagent (Thermo Fisher Scientific). Tissue was lysed using a tissuelyzerII (Qiagen). Homogenates were then spun at 12,000 g for 10 min at 4 °C and top lipid bilayer removed. RNA extraction was then carried out by manufacturer's instructions for Trizol. cDNA was synthesized with SuperScript IV VILO Master Mix (Thermo Fisher Scientific) and qPCR was performed on ABI Quant Studio 6 flex with KAPA SYBR FAST (Kapa Biosystems).
2.7. Gene expression
Human gene expression data was acquired from GTEx database. The most abundant transcript of Tcf7l2 in white adipose tissue corresponds to TCF7L2v-003 which encodes for 3802 bp transcript corresponding to NM_030756 and genecode ID ENST00000369397. This transcript includes the following exons, 1,2,3,4,8,9,10,11,12,13,14,15,16,19, that encode for a 596 amino acid protein. The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 05/17/17.
2.8. Western blot analysis
2.8.1. Tissue
Epididymal white adipose tissue was excised from Tcf7l2F/F and Tcf7l2F/F;+Adp-Cre mice and processed using RIPA and glass dounce. Adipose tissue lysate was spun at 12,000 rpm, and top lipid layer was removed. This was repeated twice to ensure that lipids were removed from lysate. Protein was quantified using a BCA Protein Assay Kit (Pierce) and 50 ug of protein was diluted into Laemmli sample buffer, heated at 95 °C for 5 min, loaded on 4–12% TPX gel (Biorad), and transferred to Protran Premium 0.45 nitrocellulose (GE-Amersham). The membrane was blocked with 5% BSA in PBS-T. TCF7L2 was detected using antibody TCF4 (C48H11) Rabbit mAB #2569 (Cell Signaling) in 5%BSA in PBS-T overnight at 4 °C. HMGB1 was detected using antibody HMGB1 antibody #ab18256 (Abcam) in 5% milk in PBS-T overnight at 4 °C. Blots were probed with secondary antibody Goat Anti-Rabbit IgG (H + L) secondary antibody, HRP #656120 (Thermo Fisher Scientific).
2.8.2. Cells
Cells were homogenized using RIPA buffer plus proteinase inhibitor and incubated on ice for 20 min. Cell homogenate was spun at 12,000×g for 10 min at 4 °C. Protein was quantified using a BCA Protein Assay Kit (Pierce). Samples were diluted in Laemmli sample buffer, heated at 95 °C for 5 min and loaded on 4–12% TPX gel (Biorad) and transferred to Protran Premium 0.45 nitrocellulose (GE-Amersham). The membrane was blocked with 5% milk in PBS-T. TCF7L2 was detected using antibody TCF4 (C48H11) Rabbit mAB #2569 (Cell Signaling) in 5%BSA in PBS-T overnight at 4 °C. HMGB1 was detected using antibody HMGB1 antibody #ab18256 (Abcam) in 5% milk in PBS-T overnight at 4 °C. Blots were probed with secondary antibody Goat Anti-Rabbit IgG (H + L) secondary antibody, HRP #656120 (Thermo Fisher Scientific) and run on SDS page.
2.9. Chromatin immunoprecipitation and next generation sequencing
Immortalized Tcf72F/F preadipocytes expressing pMSCV CreERT2 were treated with either 500 nM of (z) 4-hydoxytamoxifen (Sigma) or vehicle and then induced to differentiate with DMEM containing 10% FBS, 1 mM dexamethasone, 0.5 mM isobutylmethylxanthine, 20 nM GW1929, and 5 mg/ml insulin for 2 days, followed by 5 mg/ml insulin and 20 nM GW1929 alone. After 8 days of differentiation, cells were fixed with 1% formaldehyde for 10 min at RT. Fixation was quenched with the addition of 125 mM glycine for 5 min at RT. Cells were washed 2 times in cold PBS and harvested in Farnham Lysis Buffer and nuclei isolation was performed using a dounce. Homogenates were then spun at 4000 rpm for 5 min at 4 °C. Nuclei were resuspended in ChIP RIPA buffer (1xPBS, 1%NP-40, 0.5% NaDoc, 0.1%SDS, protease inhibitor cocktail (Roche)) and sonicated in Diagenode Pico according to manufacturer's instructions. Samples were then spun at 10,000 g for 5 min at 4 °C and supernatant was incubated with antibody TCF4 (C48H11) Rabbit mAB #2569 (Cell Signaling) or Rabbit IgG #2729 (Cell Signaling) overnight at 4 °C. Protein A/G magnetic beads #88802 (Thermo Fisher Scientific) were then added to Ab-Ag complexes for 4 h at 4 °C. ChIP reactions were washed in ChIP wash buffer 1 (0.1% SDS, 0.1% NaDoc, 1% Triton, 0.15M NaCl, 1 mM EDTA, 20 mM Tris pH 8.0, protease inhibitor cocktail (Roche)) for 5 min at 4 °C, ChIP wash buffer 2 (0.1% SDS, 0.1% NaDoc, 1% Triton, 0.5M NaCl, 1 mM EDTA, 10 mM Tris pH 8.0, protease inhibitor cocktail (Roche)) for 5 min at 4 °C, LiCl wash buffer (100 mM Tris pH 7.5, 500 mM LiCl, 1% NP-40, 1% NaDoc, protease inhibitor cocktail (Roche)) for 3 min at 4 °C 5 times, and TE 1 min at 4 °C. Chromatin was eluted with Elution buffer (1% SDA, 0.1M NaHCO3) at 65 °C for 1 h. Crosslinks were reversed by incubated samples in 0.2M NaCl overnight at 65 °C. Samples were then incubated with 10 ug of RNase A for 1 h at 37 °C followed by 20ug of Proteinase K for 2 h at 45 °C. Samples were then purified using Zymo Chip DNA purification and concentrator kit (Zymo). ChIP-seq Library was prepared with DNA SMART ChIP-seq Kit (Takara) according to manufacturer's instructions and purified using AMPure XP beads #A63880 (Beckman Coulter). Sequencing libraries (25 pM) were chemically denatured and applied to an Illumina HiSeq v4 single read flow cell using an Illumina cBot. Hybridized molecules were clonally amplified and annealed to sequencing primers with reagents from an Illumina HiSeq SR Cluster Kit v4-cBot (GD-401-4001). Following transfer of the flowcell to an Illumina HiSeq 2500 instrument (HCSv2.2.38 and RTA v1.18.61), a 50-cycle single-read sequence run was performed using HiSeq SBS Kit v4 sequencing reagents (FC-401-4002). RNA-seq and ChIP-seq data is deposited in GEO DataSets.
2.10. Integration of ChIP- seq and RNA-seq analysis
FASTQ sequences were aligned to the mouse genome (UCSC build mm10) with Novocraft Novoalign (version 3.7, http://www.novocraft.com), allowing for one random alignment for reads with multiple alignments. Alignments had moderately high duplication levels; therefore, alignments at duplicate positions were subsampled to an overall duplication rate of 15% and maximum duplicate depth of 30 using bam_partial_dedup (https://github.com/tjparnell/HCI-Scripts/blob/master/BamFile/bam_partial_dedup.pl) to avoid false peaks while preserving natural signal strength. Fragment coverage bedGraph files were generated with BioToolBox (version 1.52, https://github.com/tjparnell/biotoolbox) bam2wig using 200 bp as an extension, excluding mitochondrial chromosome and Encode black list regions, depth-normalizing to 1 million reads, and taking the mean of replicates. Peaks were called manually with Macs2 (version 2.1.1.20160309, https://github.com/taoliu/MACS) using a lambda control bedGraph generated from the input with bam2wig (small and local lambda set to 1 kb and 10 kb, respectively) and peak calling thresholds of q-value of 2 (1% FDR), minimum peak length 250 bp, and peak gap of 100 bp. ChIP peaks were annotated with the R package ChIPSeeker (version 1.12.1) using UCSC knownGene transcript and Ensembl annotation tables. Promoters were defined as ±3 kb from the TSS. Unique transcript start sites were collected using BioToolBox get_gene_regions. Heat maps were generated with deepTools (version 2.5.4, http://deeptools.readthedocs.io/en/develop/) computeMatrix and plotHeatMap. Transcription factor site motif was performed using Homer (version 4.9, http://homer.ucsd.edu/homer/) findMotifsGenome, and Jaspar http://159.149.160.88/pscan_chip_dev/.
RNA was purified using Qiagen RNeasy mini Kit and library preps were made with Illumina TruSeq Stranded Total RNA Sample Prep Kit with Ribo-Zero Gold. Samples were sequenced on Illumina HiSeq 2000 with HiSeq 125 Cycle paired-end Sequencing v4. Reads were aligned to mm10,M_musculus_Dec_2011, GRCm38 using Bioconductor RNA-Seq workflow to find differentially expressed genes using DESeq2 and the hciR package on Github to simplify the R code.
2.11. HMDP analysis
The Hybrid Mouse Diversity Panel arrays and clinical traits were generated as previously described [142], [143]. Gene expression and PC vector correlations were assessed using the BicorAndPvalue function in the R package WGCNA [144]. Principle Component coordinates for each mouse was extracted from the base R function prcomp and plotted using the package facoextra as previously described [145]. Pathway enrichment was performed on the top 500 genes, either positively or negatively correlated with Tcf7l2 within the adipose tissue expression arrays using The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 [146], [147].
2.12. In vitro lipolysis assay
Immortalized Tcf72F/F preadipocytes expressing pMSCV CreERT2 were treated with either 500 nM of (z) 4-hydoxytamoxifen (Sigma) or vehicle and then induced to differentiate with DMEM containing 10% FBS, 1 mM dexamethasone, 0.5 mM isobutylmethylxanthine, 20 nM GW1929, and 5 mg/ml insulin for 2 days, followed by 5 mg/ml insulin and 20 nM GW1929 alone. After 10 days of differentiation cells were washed with PBS and incubated in Krebs Ringer buffer for two hours. Cells were then treated with 100 nM CL 316,243 for four hours. Fatty acids from media were then quantified using a Free Fatty Acid Quantitation Kit (Sigma) according to manufacturer's instructions.
2.13. Statistical analysis
Student T-test or ANOVA followed by Tukey posthoc analysis. Analysis was completed using excel or Prism 7.
3. Results
3.1. Tcf7l2 expression is reduced in adipose tissue of high fat fed mice and genetic models of obesity
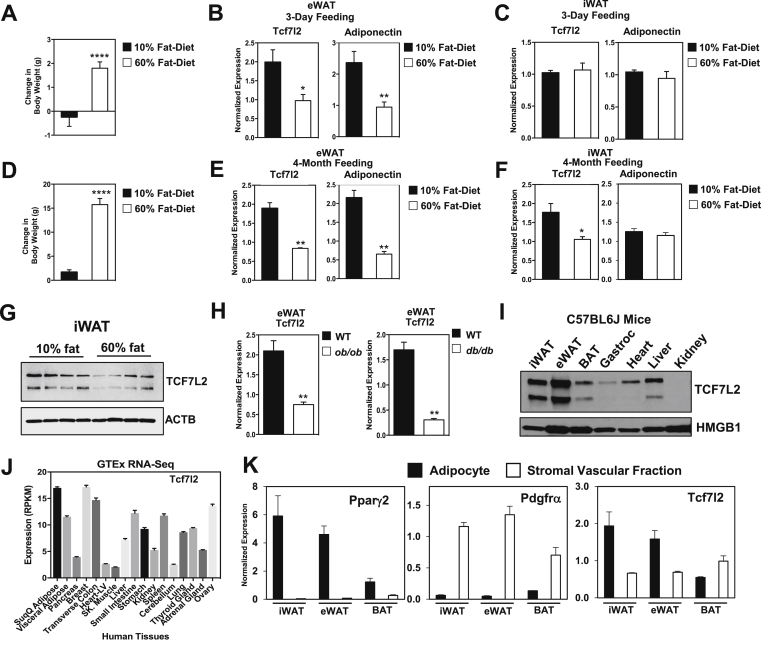
The strong association of Tcf7l2 with type 2 diabetes prompted us to explore the regulation of TCF7L2 in adipose tissue. To test whether Tcf7l2 expression changes with high-fat diet, mice were placed on 10% or 60% high-fat diet for 3-days. When compared to 10% high-fat diet, mice fed 60% high-fat diet had increased body weight (Figure 1A) and reduced mRNA of Tcf7l2 and adiponectin in epididymal white adipose tissue (eWAT) (Figure 1B), while mRNA of Tcf7l2 and adiponectin was similar in inguinal white adipose tissue (iWAT) (Figure 1C). To determine whether Tcf7l2 expression remains low with prolonged high-fat diet, we measured expression of Tcf7l2 in eWAT and iWAT after 4-months of high-fat diet, and found reduced gene expression of Tcf7l2, Adiponectin and Cfd (adipsin) (Figure 1E,F). The decreased TCF7L2 expression in HFD was also observed by western blot analysis (Figure 1G). To test whether genetic models of obesity had altered expression of Tcf7l2, we measured gene expression in eWAT of ob/ob and db/db mice and found reduced expression of Tcf7l2 (Figure 1H). To determine the normal distribution of TCF7L2, we completed western blot analysis on multiple tissues, and found that TCF7L2 had the highest expression in epididymal and inguinal white adipose tissue in mice (Figure 1I). To develop a broader view of Tcf7l2 expression in humans, we queried the Genotype-Tissue Expression (GTEx) portal, where gene expression data from RNA sequencing is available from multiple human tissues post mortem. Expression data is shown as RPKMs (number Reads Per Kilobase gene model and Million mapped reads) using 0.5 RPKM as threshold for detection. In humans, we found that Tcf7l2 expression was high in subcutaneous and visceral adipose tissue, while expression in pancreas, heart and skeletal muscle was relatively low (Figure 1J). To understand where Tcf7l2 is expressed, we fractionated adipose tissue, and found that Tcf7l2 expression is highest in white adipocytes, with lower expression in the stromal vascular fraction (Figure 1K). The decrease in expression in both diet-induced and genetic models of obesity, and the high expression in the adipocyte fraction, raised the possibility that reduced Tcf7l2 expression in adipocytes might lead to impaired glucose or lipid metabolism.
Figure 1.
TCF7L2 expression is reduced in white adipose tissue with high-fat diet and genetic models of obesity. (A) Change in body weight of C57Bl/6J mice placed on 10% or 60% HFD for 3 days (males, n = 5, 3-months of age). (B) Tcf7l2 and adipoq (adiponectin) gene expression in eWAT and (C) iWAT. (D) Change in body weight of C57Bl/76J mice placed on 60% HFD for 4 months. (males, n = 5 for each group, 60% HFD started at 14 weeks old) (E) Tcf7l2 and adipoq gene expression in eWAT and (F) in iWAT, gene expression normalized to Rps3 expression.(G) Western blot analysis in iWAT of TCF7L2 and ACTB (β-actin) after 4-month HFD. (H) Tcf7l2 expression in eWAT of control, ob/ob mice and db/db mice, gene expression was normalized to Rps3 expression. (I) Western blot analysis of TCF7L2 and HMGB1 expression in mouse tissues. (J) Tcf7l2 expression in human tissues from publicly available GTEx RNA-Seq data set. (K) Gene expression in fractionated adipose tissue depots from C57BL6/J 3-month old male mice. P-values were determined using Student's t-test, *p < 0.05.
3.2. Inducible deletion of Tcf7l2 promotes adipocyte differentiation and lipid accumulation
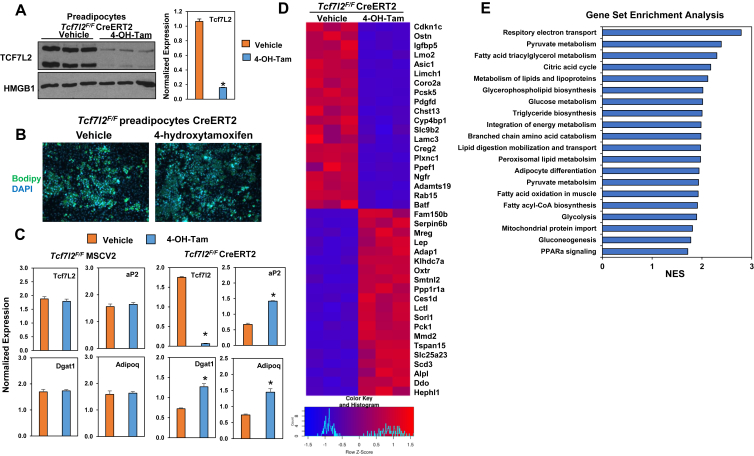
To test the in vitro role of TCF7L2 on adipocyte differentiation and lipid storage, we generated mice from embryonic stem cells engineered by EMMA for C57BL/6NTac-Tcf7l2<tm1a (EUCOMM)Wtsi>/Wtsi (EM:06158). Cells were injected into fertilized embryos and chimeras were bred with C57BL6J mice. Pups from the F1 cross were genotyped and those carrying the targeted allele were bred with Actin-Flpe mice to remove the neomycin cassette. Tcf7l2F/F mice have loxP sites flanking exon 5, which is expressed in all Tcf7l2 transcripts. Tcf7l2F/F preadipocytes were immortalized by retroviral expression of LargeT antigen and subsequently infected with viral vectors for CreERT2 [148], [149]. Western blot analysis of cells treated with (z)-4-hydroxy tamoxifen resulted in greatly diminished TCF7L2 protein (Figure 2A) and mRNA (Figure 2C). Subsequent differentiation of TCF7L2 null preadipocytes led to enhanced differentiation as supported by increased expression of differentiation markers, including Adiponectin, aP2, and Dgat1 expression (Figure 2C) as well as increased lipid accumulation as measured by bodipy staining (Figure 2B). To develop a global assessment of the role of TCF7L2 in adipocytes, we measured differential gene expression by RNA-sequencing analysis (Figure 2D). Gene set enrichment analysis (GSEA) on differentially expressed genes from control (Vehicle) or TCF7L2 null cells (4-OH-Tam) identified several metabolic pathways, including those involved in the respiratory electron transport chain, pyruvate metabolism, fatty acid and triglyceride metabolism, glucose metabolism, amino acid metabolism, and adipocyte differentiation (Figure 2E).
Figure 2.
Inducible deletion of TCF7L2 results in enhanced adipocyte differentiation and lipid accumulation. (A) Western blot of TCF7L2 and HMGB1 expression in preadipocytes harvested from Tcf7l2F/F mice. CreERT2 expressing preadipocytes were treated with vehicle (ethanol) or 50 nM 4-hydroxytamoxifen, n = 3. Corresponding changes in Tcf7l2 transcript (bar graph). (B) Bodipy and DAPI staining of differentiated Tcf7l2F/F preadipocytes expressing CreERT2. (C) Gene expression changes were measured by real-time PCR and normalized to Rps3 expression. (D) Heat map of differential gene expression changes in control and TCF7L2-null cells (E) Gene Set Enrichment Analysis of differentially expressed genes in control and TCF7L2-null cells. n = 3. P-values were determined using Student's t-test, *p < 0.05.
3.3. Genome-wide analysis of TCF7L2 binding sites suggests direct transcriptional regulation of metabolic and proliferative pathways
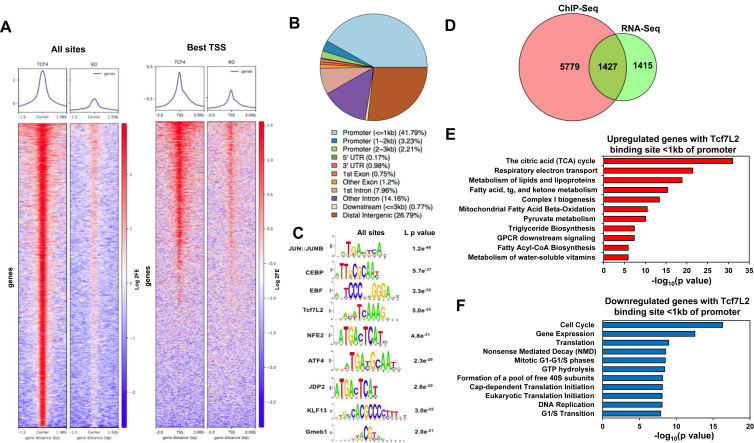
To define the transcriptional pathways that are directly regulated by TCF7L2, we performed ChIP-seq analysis using preadipocytes from Tcf7l2F/F mice. Cells were engineered to express CreERT2 enabling conditional deletion of TCF7L2 with a single tamoxifen treatment. Genome-wide occupancy of TCF7L2 showed 12,073 total binding sites which mapped to 7,207 genes. As expected, occupancy was greatly diminished in Tcf7l2F/F cells receiving tamoxifen (Figure 2A,C). Notably, TCF7L2 binding sites were largely localized near the transcriptional start site (Figure 3A). Although the majority of TCF7L2 binding sites were located within promoter regions, we observed binding in intronic, 5′ UTR, 3′UTR, and distal intergenic regions (Figure 3B). To identify direct targets of TCF7L2, we completed integrated analysis of RNA-seq and ChIP-seq data and found that 1,427 genes that were differentially regulated had a TCF7L2 binding site nearby (Figure 3D). We then performed GO analysis on transcripts that were differentially regulated and found that upregulated genes were enriched in metabolic pathways such as the TCA cycle, respiratory electron transport chain, lipid and triacylglycerol metabolism, while downregulated genes were highly enriched in cell cycle, DNA replication, translation and transcriptional pathways, pathways involved in proliferation (Figure 3F). To determine whether other transcription factors associate with TCF7L2 binding sites, we performed motif analysis on the ChIP fragments using Jaspar. Through this analysis we identified transcription factor binding motifs of Jun and ATF4, which are known to regulate proliferation. We also identified motifs for transcription factors that regulate various aspects of metabolism such as CEBP, EBF, and KLF13 (Figure 3C). These findings provide evidence for a direct role for TCF7L2 in regulating gene expression of metabolic pathways.
Figure 3.
Genome wide analysis of TCF7L2 binding sites shows direct regulation of metabolic and proliferative pathways. (A) Heat map of ChIP-seq data of TCF7L2 binding in proximity to the transcriptional start site. (B) Genomic distribution of TCF7L2 occupancy. (C) Motif analysis of TCF7L2 ChIP fragments using JASPAR. (D) Venn diagram showing overlap between differential gene expression and genes occupied by TCF7L2. (E) GO analysis of upregulated genes with TCF7L2 binding within 1 kb of promoter region. (F) GO analysis of downregulated genes with TCF7L2 binding within 1 kb of promoter region. n = 3.
3.4. Tcf7l2 expression correlates with HOMA-IR, fat mass, and metabolic genes across Hybrid Mouse Diversity Panel
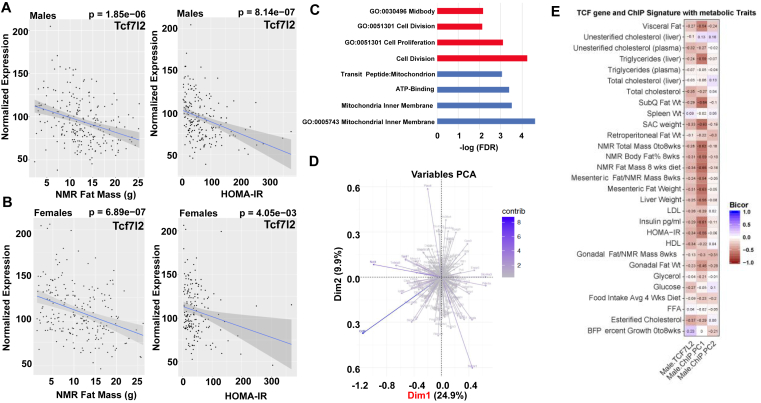
Given that we observed a negative correlation with Tcf7l2 expression and adiposity in C57BL6/J male mice, and this mouse strain is susceptible to obesity, we next asked if similar patterns of variation persisted across genetically diverse populations. We interrogated the Hybrid Mouse Diversity Panel (HMDP) in order to assess how natural variation in Tcf7l2 expression relates to metabolic phenotypes. This panel consists of over a hundred classical and recombinant inbred mouse strains whereby environmental influence can be tightly controlled, thus maximizing our ability to see how genetic architecture plays a role in complex phenotypes [150], [151]. We specifically analyzed a population which was fed a high-fat/high-sucrose diet for 8 weeks as this seemed particularly relevant for studies of metabolism and diet-induced metabolic dysfunction [142], [143]. Here, we observed striking negative correlations between Tcf7l2 expression in adipose tissue and both fat mass and HOMA-IR (Figure 4A, B). Furthermore, we analyzed the strongest negatively or positively correlated genes with Tcf7l2 in adipose tissue where we observed strong enrichments in cell cycle and mitochondrial processes, respectively (Figure 4C, D). These data show that the patterns of natural variation of adipose tissue Tcf7l2 recapitulate a significant role in regulating pertinent gene expression pathways in fat and consequent changes in metabolic homeostasis, consistent with our observations generated through targeted deletion of Tcf7l2.
Figure 4.
Tcf7l2 expression correlates with HOMA-IR, fat mass, and metabolic genes across mouse populations (A) Analysis of Hybrid Mouse Diversity Panel (HMDP) fed a high-fat/high-sucrose diet for 8 weeks for correlation of Tcf7l2 expression in male mice with HOMA-IR and fat mass in female mice (B). ((C) Gene ontology of principle component analysis prioritized two vectors which explained the total HMDP variance in gene expression of the top 500-enriched genes from the ChIP-Seq profiles and Gene Ontology (D). Tcf7l2 gene and ChIP signature integrated with metabolic traits across HMDP.
We next asked if integration of the data generated from ChIP-sequencing experiments could further inform the mechanism by which Tcf7l2 was exerting its functions across a population. We therefore implemented a principal component approach used to capture patterns of variation of larger gene sets across genetically diverse individuals [145]. The basic intuition underlying this approach is as follows: ChIP-seq experiments informed direct binding regions and downstream genes being regulated directly by TCF7L2. Given that TCF7L2 is likely exerting its effects on metabolism through its regulations on gene expression, those transcripts identified in the ChIP-seq should also vary in a manner consistent with Tcf7l2. Therefore, capturing a metric of the variance in the genes across a population should further identify key regulatory nodes allowing Tcf7l2 to exert robust effects and thus show stronger correlations across relevant traits. Along these lines, principle component analysis prioritized two vectors which explained the maximal HMDP variance in gene expression from the ChIP-seq profiles (Figure 4C, D). We note that several genes, such as Ccl2, Ncf4 and Slc44a3 showed substantial individual contribution to the first PC vector. Importantly, this vector which explained the most total variance in gene expression (PC1) showed patterns similar, but substantially more significant than Tcf7l2 itself to clinical traits (Figure 4E). This integrative analysis not only validated the physiologic implications of the Tcf7l2 ChIP-seq data but additionally prioritized several key genes in which TCF7L2 could regulate to exert its effects.
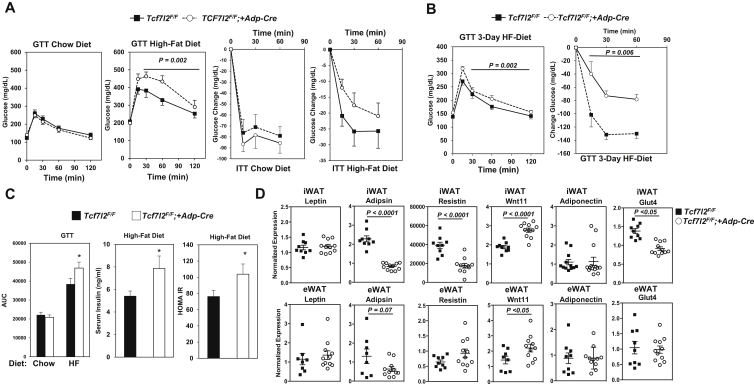
3.5. Selective deletion of TCF7L2 in adipocytes leads to glucose intolerance and insulin resistance on a high-fat diet
Because of the striking negative correlations between Tcf7l2 expression in adipose tissue with both fat mass and HOMA-IR (Figure 4A, B), we hypothesized that functional role of TCF7L2 in adipocytes is important in the etiology of type 2 diabetes associated with obesity. To test this hypothesis, we generated adipose specific knockout mice by breeding Tcf7l2F/F mice with adiponectin-cre (Adp-Cre), an adipocyte selective Cre driver. Glucose tolerance tests showed similar glucose excursion curves between Tcf7l2F/F and Tcf7l2F/F;+Adp-Cre mice on a chow diet. In contrast, when mice were challenged with a high-fat diet, Tcf7l2F/F;+Adp-Cre mice showed impaired glucose tolerance relative to Tcf7l2F/F controls (Figure 5A). To assess the insulin responsiveness of these mice, we performed an insulin tolerance test on mice fed a chow and high-fat diet. Mice had a similar drop in glucose in response to 0.75U/kg of insulin with chow, while on a high-fat diet Tcf7l2F/F;+Adp-Cre mice had a greater resistance to insulin's glucose lowering effects when compared to Tcf7l2F/F controls (Figure 5A). The observed glucose intolerance found in Tcf7l2F/F;+Adp-Cre mice was accompanied by increased serum insulin levels (Figure 5C). The impaired glucose tolerance and elevated insulin levels suggest that the TCF7L2 knockout mice developed insulin resistance, which is supported by calculation of the HOMA-IR (Figure 5C). The impaired glucose tolerance was seen within 3-days of high-fat feeding, and mice became insulin resistant during this period as well, suggesting that the insulin resistance is driving the impairment in glucose handling. After 30 min from the initial administration of insulin, Tcf7l2F/F control mice showed a decrease in blood glucose levels by 132 mg/dL, while Tcf7l2F/F;+Adp-Cre mice had a 73 mg/dL drop. In response to this short 3-day high-fat diet feeding, insulin lost its glucose lowering ability by almost 2-fold Tcf7l2F/F;+Adp-Cre mice (Figure 5B). Glucose intolerance can result from increased gluconeogenesis in the liver; however, we did not see changes in PEPCK or G6Pase gene expression (Supplemental Figure 5C).
Figure 5.
Adipocyte specific deletion of TCF7L2 leads to glucose intolerance, insulin resistance and altered adipokine expression on HFD. (A) Glucose tolerance test (GTT) and insulin tolerance test (ITT) of Tcf7l2F/F and Tcf7l2F/F;+Adp-Cre mice on control and HFD for 12 weeks (males n = 9 for contols and n = 11 for Tcf7l2F/F;+Adp-Cre at age 6 months) (B) GTT and ITT for Tcf7l2F/F and Tcf7l2F/F;+Adp-Cre mice on control and HFD for 3 days. (males n = 9 for contols and n = 11 for Tcf7l2F/F;+Adp-Cre at age 3 months) (C) AUC for GTT, fasting insulin, and HOMA IR. (D) gene expression in iWAT and eWAT of adipokines and Glut4, gene expression normalized to Rps3 expression. (males n = 9 for contols and n = 11 for Tcf7l2F/F;+Adp-Cre at age 6 months) P-values were determined using ANOVA o student T-test, *p < 0.05.
Adipose tissue is able to regulate systemic metabolism through secretion of adipokines, to assess the impact of TCF7L2 loss on adipokine transcripts we measured gene expression by real-time PCR in adipose tissue depots. In eWAT, we found similar expression of several adipokines including Leptin, Resistin, and adiponectin in Tcf7l2F/F and Tcf7l22F/F;+Adp-Cre mice, while components of the Wnt pathway such as Wnt11 were increased in TCF7L2 knockout mice. In contrast, in the subcutaneous iWAT depot, adipsin and resistin were decreased, while Wnt11 was elevated, and leptin expression remained the same between Tcf7l2F/F and Tcf7l2F/F;+Adp-Cre mice. Similarly, Glut4 expression was reduced in iWAT of Tcf7l2F/F;+Adp-Cre mice and not in eWAT (Figure 5D). These results supported that the loss of Tcf7l2 in adipocytes leads to impaired glucose metabolism and altered expression of adipokines in mice.
3.6. TCF7L2 regulates adipose tissue mass and adipocyte size
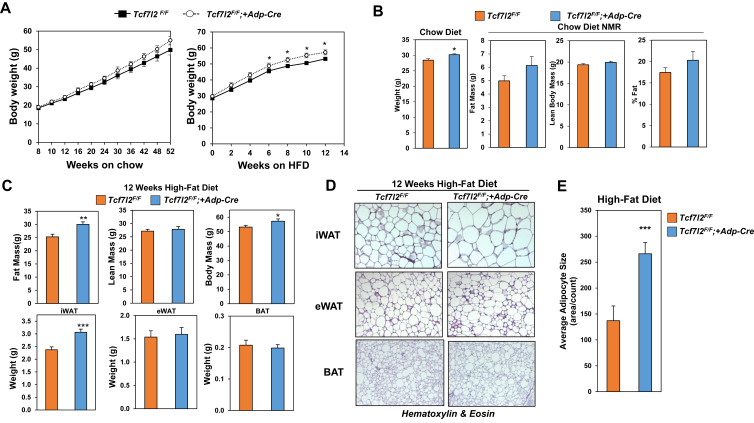
Due to the negative correlations between Tcf7l2 expression in adipose tissue with body fat mass (Figure 4B), we wanted to assess the importance of TCF7L2 in weight gain. We measured body weights of Tcf7l2F/F mice and Tcf7l2F/F;+Adp-Cre mice on chow and high-fat diet. On chow diet, both groups had similar gains in body weight and only started to diverge as the mice aged, at 1 year Tcf7l2F/F;+Adp-Cre mice started to gain more than Tcf7l2F/F control mice (Figure 6A). To observe subtle changes in body weight, we increased the n number of mice monitored for body weight on chow diet and observed significant increases in body weight but little changes in body composition as measured by Nuclear Magnetic Resonance (NMR) (Figure 6B) For high-fat diet experiments, mice were placed on a 60% high-fat diet and body weights were measured over a 12-week period. Body weight curves diverged after 8-weeks of 60% high-fat diet and showed that selective deletion of TCF7L2 increased weigh gain in response to a high-fat diet (Figure 6A). To determine whether loss of TCF7L2 increased adiposity on a high-fat diet, we measured whole body composition using NMR, and found increased fat mass and body mass in Tcf7l2F/F;+Adp-Cre mice, while lean body mass was unchanged (Figure 6C). To determine which adipose tissue depots were contributing to the differences in fat mass on a high-fat diet, the fat pads were excised and weighed. We observed increased iWAT weight, however eWAT and BAT were unchanged (Figure 6C). To test if adipocyte size was altered in iWAT, we completed histological analysis using hematoxylin and eosin (H&E) staining of iWAT and found that TCF7L2 null adipose tissue had increased adipocyte size relative to controls (Figure 6D). Adipocyte size was quantified and found to be twice as large in TCF7L2 null adipocytes (Figure 6E). These findings suggest that during conditions of high fat diet TCF7L2 regulates adipocyte size and adipose tissue mass in a depot specific manner.
Figure 6.
Selective deletion of TCF7L2 in adipocytes promotes weight gain and subcutaneous adipocyte hypertrophy. (A) Body weights of Tcf7l2F/F and Tcf7l2F/F;+Adp-Cre mice on Chow (males, n = 6 per group) and HFD (males n = 9 for controls and n = 11 for Tcf7l2F/F;Adp-Cre mice at age 6 months). (B) NMR on chow diet (males n = 9 for controls and n = 11 for Tcf7l2F/F;Adp-Cre mice at age 3 months). (C) NMR and tissue weight on 12 week HFD. (D) H + E staining of eWAT, iWAT and BAT after 12 weeks HFD. (E) Average adipocyte size in iWAT of mice fed HFD. P-values were determined using Student's t-test, *p < 0.05.
3.7. TCF7L2 regulates lipogenic and lipolytic gene expression programs
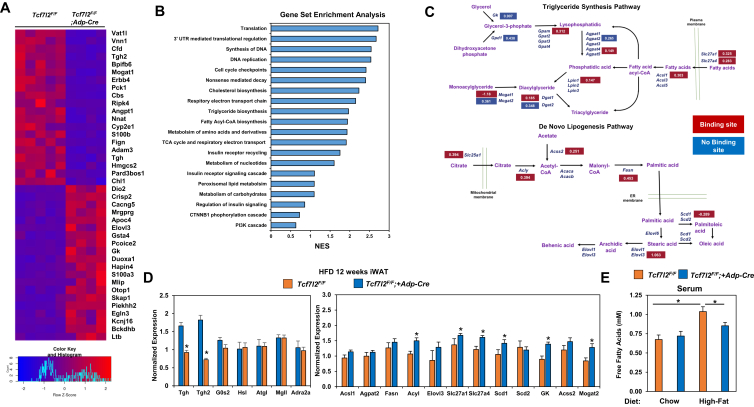
To identify transcriptional targets of TCF7L2 that alter adipocyte size we performed RNA-seq analysis on subcutaneous adipose tissue from both Tcf7l2F/F and Tcf7l2F/F;+Adp-Cre mice. Because adiponectin is expressed after the initiation of differentiation in adipocytes, the RNA-seq from the Tcf7l2F/F;+Adp-Cre iWAT will inform on a subset of TCF7L2 targets separate from those in the isolated Tcf7l2F/F adipocytes where TCF7L2 was knocked-out before differentiation (Figure 2). Similar to the RNA-seq in the isolated Tcf7l2F/F adipocytes, we found altered expression of metabolic pathways. Notably lipolytic genes, triglyceride hydrolases (Tgh) and Tgh2 expression were downregulated in the hypertrophic tissue of Tcf7l2F/F;+Adp-Cre mice (Figure 7A). Gene set enrichment analysis showed altered gene expression in translational control, DNA synthesis, cell cycle, cholesterol metabolism, and triglyceride biosynthesis and fatty acyl-CoA biosynthesis (Figure 7B). Review of our ChIP-seq data in isolated adipocytes assessing genes in the lipogenic pathways showed TCF7L2 binding sites within 1 kb of the promoter region in genes that encode for enzymes involved in triglyceride synthesis and de novo lipogenesis (Figure 7C). These results were confirmed by real-time PCR analysis where we found that Tgh and Tgh2 expression were reduced in iWAT of Tcf7l2F/F;+Adp-Cre, while several transcripts implicated in lipogenesis were increased, an outcome that may drive the larger adipocyte size (Figure 7D). The decreased Tgh and Tgh2 expression prompted further exploration of lipolysis and plasma free fatty acid levels. When compared to Tcf7l2F/F controls, Tcf7l2F/F;+Adp-Cre mice showed reduced free fatty acid levels, which suggested that lipolysis was decreased (Figure 7E).
Figure 7.
TCF7L2 regulates lipogenic and lipolytic gene expression programs. (A) Heat map of differential gene expression changes in iWAT of Tcf7l2F/F and Tcf7l2F/F;+Adp-Cre mice on 12 week HFD (males, n = 5 for control and n = 4 for Tcf7l2F/F;+Adp-Cre mice at age 6 months). (B) Gene Set Enrichment Analysis of gene expression changes in iWAT. (C) Schematic of triglyceride synthesis and de novo lipogeneis pathway with gene expression changes from RNA-Seq analysis. Red boxes indicate TCF7L2 occupancy within 1 kb of the promoter, while blue boxes indicate that there is a lack of nearby TC7L2 binding site. (D) Gene expression of lipolytic and lipogenic genes in iWAT, gene expression normalized to Rps3 expression. (E) Free fatty acid levels in serum. P-values were determined using student's t-test, *p < 0.05.
3.8. Tcf7l2 regulates fasting-induced weight loss and adipose tissue lipolysis
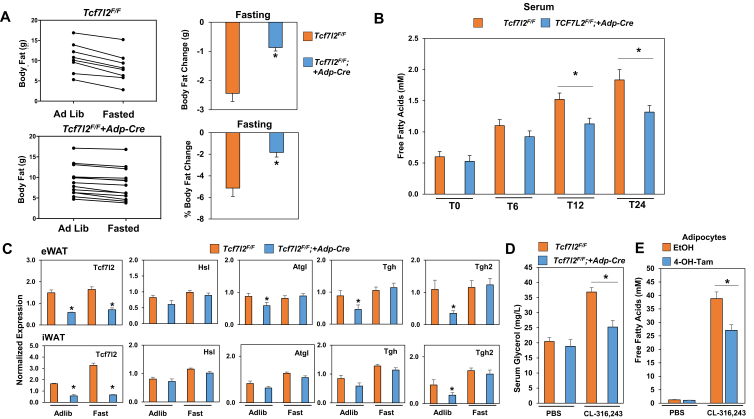
Tcf7l2F/F;+Adp-Cre mice on a HFD had reduced levels of Tgh and Tgh2 expression in iWAT and serum free fatty acid levels were decreased, which suggested that TCF7L2 regulates adipose tissue lipolysis. These results let us to challenged Tcf7l2F/F and Tcf7l2F/F;+Adp-Cre mice with 24 h of fasting to stimulate adipose tissue lipolysis. We observed that Tcf7l2F/F;+Adp-Cre mice lost less body fat as measured by NMR, than Tcf7l2F/F mice (Figure 8A). In this paradigm, we also measured free fatty acid (FFA) levels in the serum throughout the 24 h fast and saw a rise in FFAs in both Tcf7l2F/F and Tcf7l2F/F;+Adp-Cre mice, however the induction in Tcf7l2F/F;+Adp-Cre mice was lower compared to control Tcf7l2F/F mice (Figure 8B). We measured expression of lipolytic genes such as Lipe (encodes hormone sensitive lipase) and Atgl (adipose triglyceride lipase) and saw minimal differences only in the adlib state. We measured Tgh and Tgh2 expression and again saw reduced expression in Tcf7l2F/F;+Adp-Cre mice, however only in the ad libitum state (Figure 8C). Utilizing the ChIP-seq data from the isolated Tcf7l2F/Fadipocytes, we were able to observe TCF7L2 binding in close proximity to the genes that encode Tgh and Tgh2 (Supplemental Figure 8A). To test whether response to a lipolytic stimulus was impaired with the loss of Tcf7l2, mice were administered a single dose of CL-316,243, and glycerol was measured in the blood. We found a 1.5 fold reduction in glycerol levels in Tcf7l2F/F; +Adp-Cre mice when compared to Tcf7l2F/F controls (Figure 8D). Together, these findings suggest that TCF7L2 regulates lipolysis and directly regulates Tgh and Tgh2 expression.
Figure 8.
Loss of TCF7L2 leads to impaired adipocyte lipolysis. (A) Body weight of TCF7L2F/F and TCF7L2F/F;+Adp-Cre mice fasted for 24 h compares to Ad Lib controls. (B) Fasting time course of serum free fatty acid levels. (C) Gene expression of lipolytic genes in eWAT and iWAT, gene expression normalized to Rps3 expression. (males n = 8 for controls and n = 12 for TCF7L2F/F;+Adp-Cre mice at age 5 months). (D) TCF7L2F/F and TCF7L2F/F;+Adp-Cre were administered 1 mg/kg of CL-316,243 or vehicle control. Serum glycerol was measured 30 min after treatment. (E) Immortalized TCF7L2F/F preadipocytes retrovirally expressing CreERT2 were treated with vehicle or 500 nM 4-hydroxytamoxifen for 24 h 10 days post differentiation. Cells were then treated with either PBS or 100 nM CL316,243 for 4 h and media was measured for concentration of free fatty acids. P-values were determined using Student's t-test or in the case of multiple group comparison an ANOVA with a Tukey Posthoc Analysis, *p < 0.05.
4. Discussion
One of the strongest genetic predispositions for type 2 diabetes in humans is a variant of the Tcf7l2 gene [7]. Polymorphisms in Tcf7l2 have also been linked to body weight, triglyceride metabolism, and gestational diabetes [152], [153], [154]. In this study, we report that the Wnt-effector TCF7L2 is enriched in both human and mouse white adipose tissue and its expression is reduced in white adipose tissue of both genetic and diet-induced models of obesity. The conditional deletion of TCF7L2 in adipocytes leads to impaired glucose metabolism, insulin resistance, altered adipokine and GLUT4 expression, and adipocyte hypertrophy on a high-fat diet. Additionally, loss of TCF7L2 in preadipocytes leads to enhanced differentiation and lipid accumulation. Genome-wide analysis of TCF7L2 binding suggests a direct role in regulation of genes involved in both lipogenesis and lipolysis, an outcome that likely results in adipocyte hypertrophy. Knockout of TCF7L2 in adipocytes led to reduced expression of triglyceride hydrolases (Tgh)1 and 2 as well as increased expression in lipogenic genes. The in vivo impact of this impaired lipid processing was observed in adipose specific TCF7L2 knockout mice during fasting, where the knockout mice had reduced weight loss and reduced serum free fatty acid levels compared to the control. These studies underscore a direct role for TCF7L2 in the regulation of glucose and lipid metabolism in the mature adipocyte.
Mechanisms that lead to activation of Wnt-signaling have been shown to block differentiation of adipocytes [139]. While loss of function studies targeting β-catenin in preadipocytes show enhanced adipocyte differentiation [155]. Similarly, expression of a dominant negative form of TCF7L2 in fibroblasts results in enhanced adipocyte differentiation [139]. Our findings are consistent with these previous studies where we observed that the loss of Tcf7l2 in preadipocytes led to enhanced differentiation and lipid accumulation. Combined ChIP-seq and RNA-seq analysis shows that loss of TCF7L2 led to an induction of genes that are enriched in metabolic pathways such as the TCA cycle, respiratory electron transport chain, lipid and triacylglycerol metabolism. Downregulated genes were highly enriched in pathways that are involve in proliferation such as cell cycle, DNA replication, and translation. These data suggest that the block in adipocyte differentiation likely occurs through direct regulation of transcriptional targets involved in proliferation and a shift in metabolism. Perhaps a shift in the metabolic program that favors a switch from glycolysis towards oxidative respiration [156]. However, our findings are in contrast with recently published results from Chen et al. (2018) in which knock-down of Tcf7l2 led to impaired adipocyte differentiation [157]. This may be due to the use of different genetic models, they employed shRNA technology in 3T3-L1 cells while we employed Tcf7l2F/F preadipocytes engineered with retroviral expression of tamoxifen inducible Cre recombinase, CreERT2. These two genetic models may differ in the timing of TCF7L2 knockout or knockdown and TCF7L2 may be required for generation of preadipocytes and inhibition of subsequent differentiation. In other tissues, TCF7L2 is required for the maintenance of Lgr5+ stem cells in the adult small intestine [158]. The essential role of TCF7L2 in maintaining stemness, may be apparent in adipocyte progenitors, an outcome that needs to be further explored.
Unlike other anti-adipogenic factors like Wnt10b and Wnt6, TCF7L2 expression is maintained in the mature adipocyte, leading to the possibility that it may have separate roles in adipogenesis and in differentiated adipocytes [159]. Other components of the Wnt signaling pathway have been shown to have roles in both differentiation and in the mature adipocyte, an example of this is found in the TCF/LEF repressor TLE3 which also coactivates PPARγ [140]. Previous studies on TCF7L2 in the mature adipocyte have focused on assessing the impact of metabolic disease conditions on TCF7L2 expression in adipocytes. Kuzmicki et al.(2011) observed significant reduction in TCF7L2 expression in visceral white adipose tissue in gestational diabetes [160]. However, when corrected for BMI, the changes in expression are no longer apparent, suggesting that adiposity is driving the reduced expression of TCF7L2 in human adipose tissue and not diabetes. In our studies we have observed reduced Tcf7l2 expression during high fat diet and genetic models of obesity in mice on the C57BL6J background. Typically, metabolic studies are often carried out in the inbred strain C57BL6J mice which are somewhat susceptible to obesity. However, one inbred mouse strain does not adequately reflect the genetic diversity seen in human populations, which has thought to be a major reason for poor translation of a substantial number of metabolic studies. The Hybrid Mouse Diversity Panel (HMDP) which consists of a similar variation of SNPs to most human populations were therefore used to assess how natural variation in Tcf7l2 expression relates to traits such as HOMA-IR, fat mass, liver triglycerides and liver weight. We also observed trends consistent with observations from direct modification of Tcf7l2 with respect to metabolic phenotypes and hypothesize that we would see similar correlations in diverse human populations.
Our work characterized the role of TCF7L2 in mature adipocytes during obesogenic growth. We pursued these questions with use of Tcf7l2F/F mice where cre expression was driven by the adiponectin promoter which is activated post-adipocyte commitment. These adipose specific TCF7L2 knockout mice were phenotyped using both chow and high fat diet. In our studies, loss of TCF7L2 in adipocytes enhanced weight gain associated with high-fat diet and aging. On a high-fat diet loss of TCF7L2 leads to adipocyte hypertrophy, an outcome that may be driven by an increase in lipogenesis and impaired lipolysis. In our ChIP-seq data TCF7L2 was found in close proximity to genes that encode for enzymes involved in lipogenesis. Furthermore, we observed increased lipid accumulation in preadipocytes where Tcf7l2 was knocked-out before differentiation, suggesting that TCF7L2 normally has an inhibitory role in adipocyte differentiation through direct regulation of lipogenesis. Adipocyte hypertrophy may also be driven by impaired lipolysis. In addition to reduced expression of Tgh1 and Tgh2 with the loss of Tcf7l2, ChIP-Seq analysis showed TCF7L2 occupancy in the genomic loci that includes the Ces1 family that contain Ces1f,d (Tgh1 and Tgh2). Similar to TCF7L2F/F;+Adp-Cre mice, mice lacking TGH develop adipocyte hypertrophy and have lower serum free fatty acid levels, suggesting that decreased TGH expression promotes adipocyte hypertrophy [161]. Although TGH1 and 2 are triglyceride hydrolases, they do not associate with the lipid droplet in adipocytes but are located in the endoplasmic reticulum and they are not activated by adrenergic stimulation such as isoproterenol [162]. In Drosophila, there is evidence that activation of wnt-signaling using an axin gain of function allele leads to reduced lipid droplet size, which is associated with elevated free fatty acid levels [168]. Based on these observations, Wnt-signaling may have a conserved role in regulating lipolysis. Recent studies on a human prospective cohort of spontaneous weight change, showed reduced adipose tissue lipolysis in subcutaneous white adipose tissue. This showed that disturbed lipolysis increased the risk for impaired glucose metabolism [169]. This leads to the possibility that the changes in adipose tissue lipolysis seen with loss of Tcf7l2 in adipocytes, may drive the changes in glucose metabolism that we observe. Our data suggest that the Wnt effector, TCF7L2 directly regulates lipid handling through direct inhibition of lipogenic programs and activation of lipolytic gene expression program.
The glucose intolerance and reduced insulin sensitivity that we observed with the loss of TCF7L2 occurred at 3-days and 3-months of high-fat feeding. These findings would suggest that the loss of TCF7L2 manifests early and predisposes mice to diabetes when challenged with a high-fat diet. These findings are consistent with prior observations that adipocytes are key regulators of systemic glucose metabolism. Other components of the Wnt-signaling pathway have been reported to affect glucose metabolism as well. Gain of function studies in transgenic mice overexpressing Wnt10b in adipocytes show protection against high-fat diet induced obesity, and obesity in ob/ob and Agouti mice [163]. These mice also show improvements in glucose handling [163]. These findings suggest that the reduced levels of TCF7L2 that are observed in white adipose tissue in genetic and diet-induced models of obesity contributes to the development of diabetes.
One mechanism through which TCF7L2 in the adipocyte could be contributing to the development of type 2 diabetes is through regulation of adipokine expression. Adipokines released from adipocytes have pleiotropic effects on distant tissues to ultimately regulate glucose, lipid, and energy balance. Adipokines like adipsin are reduced in genetic and diet-induced models of obesity, and, when levels are restored to normal, there are whole body improvements in glucose control. Recently adipsin was shown to improve β-cell function in diabetes by enhancing glucose stimulated insulin secretion [137]. In our studies, we found that loss of TCF7L2 in adipocytes leads to reduced expression of adipsin, which in the long-term may lead to impaired β-cell function. These changes were most apparent in inguinal white adipose tissue in high-fat fed mice. It has been well established that TCF7L2 regulates β-cell function and insulin secretion in mice and humans [32], [164]. Humans who carry the T risk allele rs7903146 have increased expression of Tcf7L2 in the pancreas and have a higher proinsulin to insulin ratio in their plasma resulting from higher expression of proinsulin processing genes [165]. Previous studies observed no changes in Tcf7L2 expression in adipose tissue in humans carrying risk T risk allele rs7903146 but only a subset of Tcf7L2 transcripts were assayed [166]. More recent studies have found educed expression of some isoforms in subcutaneous adipose tissue that also inversely correlated with BMI in non-carriers [167]. Although Tcf7L2 seems to have a direct role in β-cell function and insulin secretion, our studies suggest that TCF7L2 activity in adipose tissue also may affect pancreatic β-cells indirectly through adipsin.
Our work describes the functional role of TCF7L2 in adipogenesis, mature adipocytes, and in obesogenic growth. We observed that the loss of TCF7L2 in adipocytes leads to glucose intolerance and insulin resistance. Adipose specific knockout of Tcf7l2, led to adipocyte hypertrophy that was accompanied by impaired lipolysis in response to fasting. This inability to mobilize the triglyceride pool is likely a driving factor leading to adipocyte hypertrophy. Developing therapies that modulate TCF7L2 expression may provide opportunities to treat type 2 diabetes in individuals that carry a risk allele in the TCF7L2 locus.
Acknowledgements
The authors are grateful to members of the Diabetes and Metabolism Research Center and the Biochemistry Department at the University of Utah for useful discussion and feedback. This study was supported by NIDDK KO1DK097285, NIDDK RO3DK103089, NIDDK RO1DK103930, NIDDK 2P30DK020579, NIDDK 5T32DK007115, AHA Postdoctoral Fellowship, and Margolis Research Foundation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.03.003.
Conflict of interest
No conflict of interests to report.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Morin P.J., Sparks A.B., Korinek V., Barker N., Clevers H., Vogelstein B. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275(5307):1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 2.Nusse R., Varmus H.E. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982;31(1):99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 3.Behrens J., von Kries J.P., Kuhl M., Bruhn L., Wedlich D., Grosschedl R. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382(6592):638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 4.Li V.S., Ng S.S., Boersema P.J., Low T.Y., Karthaus W.R., Gerlach J.P. Wnt signaling through inhibition of beta-catenin degradation in an intact Axin1 complex. Cell. 2012;149(6):1245–1256. doi: 10.1016/j.cell.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Cavallo R.A., Cox R.T., Moline M.M., Roose J., Polevoy G.A., Clevers H. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395(6702):604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- 6.Miravet S., Piedra J., Miro F., Itarte E., Garcia de Herreros A., Dunach M. The transcriptional factor Tcf-4 contains different binding sites for beta-catenin and plakoglobin. Journal of Biological Chemistry. 2002;277(3):1884–1891. doi: 10.1074/jbc.M110248200. [DOI] [PubMed] [Google Scholar]
- 7.Grant S.F., Thorleifsson G., Reynisdottir I., Benediktsson R., Manolescu A., Sainz J. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nature Genetics. 2006;38(3):320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 8.Cauchi S., Meyre D., Dina C., Choquet H., Samson C., Gallina S. Transcription factor TCF7L2 genetic study in the French population: expression in human beta-cells and adipose tissue and strong association with type 2 diabetes. Diabetes. 2006;55(10):2903–2908. doi: 10.2337/db06-0474. [DOI] [PubMed] [Google Scholar]
- 9.Palmer N.D., Hester J.M., An S.S., Adeyemo A., Rotimi C., Langefeld C.D. Resequencing and analysis of variation in the TCF7L2 gene in African Americans suggests that SNP rs7903146 is the causal diabetes susceptibility variant. Diabetes. 2011;60(2):662–668. doi: 10.2337/db10-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chauhan G., Spurgeon C.J., Tabassum R., Bhaskar S., Kulkarni S.R., Mahajan A. Impact of common variants of PPARG, KCNJ11, TCF7L2, SLC30A8, HHEX, CDKN2A, IGF2BP2, and CDKAL1 on the risk of type 2 diabetes in 5,164 Indians. Diabetes. 2010;59(8):2068–2074. doi: 10.2337/db09-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Damcott C.M., Pollin T.I., Reinhart L.J., Ott S.H., Shen H., Silver K.D. Polymorphisms in the transcription factor 7-like 2 (TCF7L2) gene are associated with type 2 diabetes in the Amish: replication and evidence for a role in both insulin secretion and insulin resistance. Diabetes. 2006;55(9):2654–2659. doi: 10.2337/db06-0338. [DOI] [PubMed] [Google Scholar]
- 12.Groves C.J., Zeggini E., Minton J., Frayling T.M., Weedon M.N., Rayner N.W. Association analysis of 6,736 U.K. subjects provides replication and confirms TCF7L2 as a type 2 diabetes susceptibility gene with a substantial effect on individual risk. Diabetes. 2006;55(9):2640–2644. doi: 10.2337/db06-0355. [DOI] [PubMed] [Google Scholar]
- 13.Saxena R., Gianniny L., Burtt N.P., Lyssenko V., Giuducci C., Sjogren M. Common single nucleotide polymorphisms in TCF7L2 are reproducibly associated with type 2 diabetes and reduce the insulin response to glucose in nondiabetic individuals. Diabetes. 2006;55(10):2890–2895. doi: 10.2337/db06-0381. [DOI] [PubMed] [Google Scholar]
- 14.Scott L.J., Bonnycastle L.L., Willer C.J., Sprau A.G., Jackson A.U., Narisu N. Association of transcription factor 7-like 2 (TCF7L2) variants with type 2 diabetes in a Finnish sample. Diabetes. 2006;55(9):2649–2653. doi: 10.2337/db06-0341. [DOI] [PubMed] [Google Scholar]
- 15.Bodhini D., Radha V., Dhar M., Narayani N., Mohan V. The rs12255372(G/T) and rs7903146(C/T) polymorphisms of the TCF7L2 gene are associated with type 2 diabetes mellitus in Asian Indians. Metabolism. 2007;56(9):1174–1178. doi: 10.1016/j.metabol.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 16.Chang Y.C., Chang T.J., Jiang Y.D., Kuo S.S., Lee K.C., Chiu K.C. Association study of the genetic polymorphisms of the transcription factor 7-like 2 (TCF7L2) gene and type 2 diabetes in the Chinese population. Diabetes. 2007;56(10):2631–2637. doi: 10.2337/db07-0421. [DOI] [PubMed] [Google Scholar]
- 17.Helgason A., Palsson S., Thorleifsson G., Grant S.F., Emilsson V., Gunnarsdottir S. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nature Genetics. 2007;39(2):218–225. doi: 10.1038/ng1960. [DOI] [PubMed] [Google Scholar]
- 18.Amoli M.M., Amiri P., Tavakkoly-Bazzaz J., Charmchi E., Hafeziyeh J., Keramatipour M. Replication of TCF7L2 rs7903146 association with type 2 diabetes in an Iranian population. Genetics and Molecular Biology. 2010;33(3):449–451. doi: 10.1590/S1415-47572010005000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Florez J.C., Jablonski K.A., Bayley N., Pollin T.I., de Bakker P.I., Shuldiner A.R. TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. New England Journal of Medicine. 2006;355(3):241–250. doi: 10.1056/NEJMoa062418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burwinkel B., Shanmugam K.S., Hemminki K., Meindl A., Schmutzler R.K., Sutter C. Transcription factor 7-like 2 (TCF7L2) variant is associated with familial breast cancer risk: a case-control study. BMC Cancer. 2006;6:268. doi: 10.1186/1471-2407-6-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humphries S.E., Gable D., Cooper J.A., Ireland H., Stephens J.W., Hurel S.J. Common variants in the TCF7L2 gene and predisposition to type 2 diabetes in UK European Whites, Indian Asians and Afro-Caribbean men and women. Journal of Molecular Medicine (Berlin) 2006;84(12):1005–1014. doi: 10.1007/s00109-006-0108-7. [DOI] [PubMed] [Google Scholar]
- 22.Munoz J., Lok K.H., Gower B.A., Fernandez J.R., Hunter G.R., Lara-Castro C. Polymorphism in the transcription factor 7-like 2 (TCF7L2) gene is associated with reduced insulin secretion in nondiabetic women. Diabetes. 2006;55(12):3630–3634. doi: 10.2337/db06-0574. [DOI] [PubMed] [Google Scholar]
- 23.Zhang C., Qi L., Hunter D.J., Meigs J.B., Manson J.E., van Dam R.M. Variant of transcription factor 7-like 2 (TCF7L2) gene and the risk of type 2 diabetes in large cohorts of U.S. women and men. Diabetes. 2006;55(9):2645–2648. doi: 10.2337/db06-0643. [DOI] [PubMed] [Google Scholar]
- 24.Barber T.M., Bennett A.J., Groves C.J., Sovio U., Ruokonen A., Martikainen H. Disparate genetic influences on polycystic ovary syndrome (PCOS) and type 2 diabetes revealed by a lack of association between common variants within the TCF7L2 gene and PCOS. Diabetologia. 2007;50(11):2318–2322. doi: 10.1007/s00125-007-0804-z. [DOI] [PubMed] [Google Scholar]
- 25.Chandak G.R., Janipalli C.S., Bhaskar S., Kulkarni S.R., Mohankrishna P., Hattersley A.T. Common variants in the TCF7L2 gene are strongly associated with type 2 diabetes mellitus in the Indian population. Diabetologia. 2007;50(1):63–67. doi: 10.1007/s00125-006-0502-2. [DOI] [PubMed] [Google Scholar]
- 26.Dahlgren A., Zethelius B., Jensevik K., Syvanen A.C., Berne C., Cohort U. Variants of the TCF7L2 gene are associated with beta cell dysfunction and confer an increased risk of type 2 diabetes mellitus in the ULSAM cohort of Swedish elderly men. Diabetologia. 2007;50(9):1852–1857. doi: 10.1007/s00125-007-0746-5. [DOI] [PubMed] [Google Scholar]
- 27.Guo T., Hanson R.L., Traurig M., Muller Y.L., Ma L., Mack J. TCF7L2 is not a major susceptibility gene for type 2 diabetes in Pima Indians: analysis of 3,501 individuals. Diabetes. 2007;56(12):3082–3088. doi: 10.2337/db07-0621. [DOI] [PubMed] [Google Scholar]
- 28.Hayashi T., Iwamoto Y., Kaku K., Hirose H., Maeda S. Replication study for the association of TCF7L2 with susceptibility to type 2 diabetes in a Japanese population. Diabetologia. 2007;50(5):980–984. doi: 10.1007/s00125-007-0618-z. [DOI] [PubMed] [Google Scholar]
- 29.Horikoshi M., Hara K., Ito C., Nagai R., Froguel P., Kadowaki T. A genetic variation of the transcription factor 7-like 2 gene is associated with risk of type 2 diabetes in the Japanese population. Diabetologia. 2007;50(4):747–751. doi: 10.1007/s00125-006-0588-6. [DOI] [PubMed] [Google Scholar]
- 30.Kimber C.H., Doney A.S., Pearson E.R., McCarthy M.I., Hattersley A.T., Leese G.P. TCF7L2 in the Go-DARTS study: evidence for a gene dose effect on both diabetes susceptibility and control of glucose levels. Diabetologia. 2007;50(6):1186–1191. doi: 10.1007/s00125-007-0661-9. [DOI] [PubMed] [Google Scholar]
- 31.Lehman D.M., Hunt K.J., Leach R.J., Hamlington J., Arya R., Abboud H.E. Haplotypes of transcription factor 7-like 2 (TCF7L2) gene and its upstream region are associated with type 2 diabetes and age of onset in Mexican Americans. Diabetes. 2007;56(2):389–393. doi: 10.2337/db06-0860. [DOI] [PubMed] [Google Scholar]
- 32.Lyssenko V., Lupi R., Marchetti P., Del Guerra S., Orho-Melander M., Almgren P. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. Journal of Clinical Investigation. 2007;117(8):2155–2163. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marzi C., Huth C., Kolz M., Grallert H., Meisinger C., Wichmann H.E. Variants of the transcription factor 7-like 2 gene (TCF7L2) are strongly associated with type 2 diabetes but not with the metabolic syndrome in the MONICA/KORA surveys. Hormone and Metabolic Research. 2007;39(1):46–52. doi: 10.1055/s-2007-957345. [DOI] [PubMed] [Google Scholar]
- 34.Mayans S., Lackovic K., Lindgren P., Ruikka K., Agren A., Eliasson M. TCF7L2 polymorphisms are associated with type 2 diabetes in northern Sweden. European Journal of Human Genetics. 2007;15(3):342–346. doi: 10.1038/sj.ejhg.5201773. [DOI] [PubMed] [Google Scholar]
- 35.Parra E.J., Cameron E., Simmonds L., Valladares A., McKeigue P., Shriver M. Association of TCF7L2 polymorphisms with type 2 diabetes in Mexico City. Clinical Genetics. 2007;71(4):359–366. doi: 10.1111/j.1399-0004.2007.00780.x. [DOI] [PubMed] [Google Scholar]
- 36.Pearson E.R., Donnelly L.A., Kimber C., Whitley A., Doney A.S., McCarthy M.I. Variation in TCF7L2 influences therapeutic response to sulfonylureas: a GoDARTs study. Diabetes. 2007;56(8):2178–2182. doi: 10.2337/db07-0440. [DOI] [PubMed] [Google Scholar]
- 37.Sale M.M., Smith S.G., Mychaleckyj J.C., Keene K.L., Langefeld C.D., Leak T.S. Variants of the transcription factor 7-like 2 (TCF7L2) gene are associated with type 2 diabetes in an African-American population enriched for nephropathy. Diabetes. 2007;56(10):2638–2642. doi: 10.2337/db07-0012. [DOI] [PubMed] [Google Scholar]
- 38.Schafer S.A., Tschritter O., Machicao F., Thamer C., Stefan N., Gallwitz B. Impaired glucagon-like peptide-1-induced insulin secretion in carriers of transcription factor 7-like 2 (TCF7L2) gene polymorphisms. Diabetologia. 2007;50(12):2443–2450. doi: 10.1007/s00125-007-0753-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Vliet-Ostaptchouk J.V., Shiri-Sverdlov R., Zhernakova A., Strengman E., van Haeften T.W., Hofker M.H. Association of variants of transcription factor 7-like 2 (TCF7L2) with susceptibility to type 2 diabetes in the Dutch Breda cohort. Diabetologia. 2007;50(1):59–62. doi: 10.1007/s00125-006-0477-z. [DOI] [PubMed] [Google Scholar]
- 40.Wang J., Kuusisto J., Vanttinen M., Kuulasmaa T., Lindstrom J., Tuomilehto J. Variants of transcription factor 7-like 2 (TCF7L2) gene predict conversion to type 2 diabetes in the Finnish Diabetes Prevention Study and are associated with impaired glucose regulation and impaired insulin secretion. Diabetologia. 2007;50(6):1192–1200. doi: 10.1007/s00125-007-0656-6. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe R.M., Allayee H., Xiang A.H., Trigo E., Hartiala J., Lawrence J.M. Transcription factor 7-like 2 (TCF7L2) is associated with gestational diabetes mellitus and interacts with adiposity to alter insulin secretion in Mexican Americans. Diabetes. 2007;56(5):1481–1485. doi: 10.2337/db06-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agalliu I., Suuriniemi M., Prokunina-Olsson L., Johanneson B., Collins F.S., Stanford J.L. Evaluation of a variant in the transcription factor 7-like 2 (TCF7L2) gene and prostate cancer risk in a population-based study. The Prostate. 2008;68(7):740–747. doi: 10.1002/pros.20732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alsmadi O., Al-Rubeaan K., Mohamed G., Alkayal F., Al-Saud H., Al-Saud N.A. Weak or no association of TCF7L2 variants with Type 2 diabetes risk in an Arab population. BMC Medical Genetics. 2008;9:72. doi: 10.1186/1471-2350-9-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bielinski S.J., Pankow J.S., Folsom A.R., North K.E., Boerwinkle E. TCF7L2 single nucleotide polymorphisms, cardiovascular disease and all-cause mortality: the Atherosclerosis Risk in Communities (ARIC) study. Diabetologia. 2008;51(6):968–970. doi: 10.1007/s00125-008-1004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gonzalez-Sanchez J.L., Martinez-Larrad M.T., Zabena C., Perez-Barba M., Serrano-Rios M. Association of variants of the TCF7L2 gene with increases in the risk of type 2 diabetes and the proinsulin:insulin ratio in the Spanish population. Diabetologia. 2008;51(11):1993–1997. doi: 10.1007/s00125-008-1129-2. [DOI] [PubMed] [Google Scholar]
- 46.Hazra A., Fuchs C.S., Chan A.T., Giovannucci E.L., Hunter D.J. Association of the TCF7L2 polymorphism with colorectal cancer and adenoma risk. Cancer Causes & Control. 2008;19(9):975–980. doi: 10.1007/s10552-008-9164-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huertas-Vazquez A., Plaisier C., Weissglas-Volkov D., Sinsheimer J., Canizales-Quinteros S., Cruz-Bautista I. TCF7L2 is associated with high serum triacylglycerol and differentially expressed in adipose tissue in families with familial combined hyperlipidaemia. Diabetologia. 2008;51(1):62–69. doi: 10.1007/s00125-007-0850-6. [DOI] [PubMed] [Google Scholar]
- 48.Kang E.S., Kim M.S., Kim Y.S., Hur K.Y., Han S.J., Nam C.M. A variant of the transcription factor 7-like 2 (TCF7L2) gene and the risk of posttransplantation diabetes mellitus in renal allograft recipients. Diabetes Care. 2008;31(1):63–68. doi: 10.2337/dc07-1005. [DOI] [PubMed] [Google Scholar]
- 49.Mahurkar S., Bhaskar S., Reddy D.N., Prakash S., Rao G.V., Singh S.P. TCF7L2 gene polymorphisms do not predict susceptibility to diabetes in tropical calcific pancreatitis but may interact with SPINK1 and CTSB mutations in predicting diabetes. BMC Medical Genetics. 2008;9:80. doi: 10.1186/1471-2350-9-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyake K., Horikawa Y., Hara K., Yasuda K., Osawa H., Furuta H. Association of TCF7L2 polymorphisms with susceptibility to type 2 diabetes in 4,087 Japanese subjects. Journal of Human Genetics. 2008;53(2):174–180. doi: 10.1007/s10038-007-0231-5. [DOI] [PubMed] [Google Scholar]
- 51.Rees S.D., Bellary S., Britten A.C., O'Hare J.P., Kumar S., Barnett A.H. Common variants of the TCF7L2 gene are associated with increased risk of type 2 diabetes mellitus in a UK-resident South Asian population. BMC Medical Genetics. 2008;9:8. doi: 10.1186/1471-2350-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ren Q., Han X.Y., Wang F., Zhang X.Y., Han L.C., Luo Y.Y. Exon sequencing and association analysis of polymorphisms in TCF7L2 with type 2 diabetes in a Chinese population. Diabetologia. 2008;51(7):1146–1152. doi: 10.1007/s00125-008-1039-3. [DOI] [PubMed] [Google Scholar]
- 53.Saadi H., Nagelkerke N., Carruthers S.G., Benedict S., Abdulkhalek S., Reed R. Association of TCF7L2 polymorphism with diabetes mellitus, metabolic syndrome, and markers of beta cell function and insulin resistance in a population-based sample of Emirati subjects. Diabetes Research and Clinical Practice. 2008;80(3):392–398. doi: 10.1016/j.diabres.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 54.Cho Y.M., Kim T.H., Lim S., Choi S.H., Shin H.D., Lee H.K. Type 2 diabetes-associated genetic variants discovered in the recent genome-wide association studies are related to gestational diabetes mellitus in the Korean population. Diabetologia. 2009;52(2):253–261. doi: 10.1007/s00125-008-1196-4. [DOI] [PubMed] [Google Scholar]
- 55.Cornelis M.C., Qi L., Kraft P., Hu F.B. TCF7L2, dietary carbohydrate, and risk of type 2 diabetes in US women. American Journal of Clinical Nutrition. 2009;89(4):1256–1262. doi: 10.3945/ajcn.2008.27058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goode E.L., Szabo C., Prokunina-Olsson L., Vierkant R.A., Fredericksen Z.S., Collins F.S. No association between a candidate TCF7L2 variant and risk of breast or ovarian cancer. BMC Cancer. 2009;9:312. doi: 10.1186/1471-2407-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kabagambe E.K., Glasser S.P., Ordovas J.M., Warodomwichit D., Tsai M.Y., Hopkins P.N. TCF7L2 polymorphisms and inflammatory markers before and after treatment with fenofibrate. Diabetology & Metabolic Syndrome. 2009;1(1):16. doi: 10.1186/1758-5996-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo Y., Wang H., Han X., Ren Q., Wang F., Zhang X. Meta-analysis of the association between SNPs in TCF7L2 and type 2 diabetes in East Asian population. Diabetes Research and Clinical Practice. 2009;85(2):139–146. doi: 10.1016/j.diabres.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 59.Nordman S., Ostenson C.G., Efendic S., Gu H.F. Loci of TCF7L2, HHEX and IDE on chromosome 10q and the susceptibility of their genetic polymorphisms to type 2 diabetes. Experimental and Clinical Endocrinology & Diabetes. 2009;117(4):186–190. doi: 10.1055/s-0028-1100419. [DOI] [PubMed] [Google Scholar]
- 60.Prokunina-Olsson L., Kaplan L.M., Schadt E.E., Collins F.S. Alternative splicing of TCF7L2 gene in omental and subcutaneous adipose tissue and risk of type 2 diabetes. PLoS One. 2009;4(9):e7231. doi: 10.1371/journal.pone.0007231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prokunina-Olsson L., Welch C., Hansson O., Adhikari N., Scott L.J., Usher N. Tissue-specific alternative splicing of TCF7L2. Human Molecular Genetics. 2009;18(20):3795–3804. doi: 10.1093/hmg/ddp321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stolerman E.S., Manning A.K., McAteer J.B., Fox C.S., Dupuis J., Meigs J.B. TCF7L2 variants are associated with increased proinsulin/insulin ratios but not obesity traits in the Framingham Heart Study. Diabetologia. 2009;52(4):614–620. doi: 10.1007/s00125-009-1266-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tabara Y., Osawa H., Kawamoto R., Onuma H., Shimizu I., Miki T. Replication study of candidate genes associated with type 2 diabetes based on genome-wide screening. Diabetes. 2009;58(2):493–498. doi: 10.2337/db07-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thorsby P.M., Midthjell K., Gjerlaugsen N., Holmen J., Hanssen K.F., Birkeland K.I. Comparison of genetic risk in three candidate genes (TCF7L2, PPARG, KCNJ11) with traditional risk factors for type 2 diabetes in a population-based study--the HUNT study. Scandinavian Journal of Clinical & Laboratory Investigation. 2009;69(2):282–287. doi: 10.1080/00365510802538188. [DOI] [PubMed] [Google Scholar]
- 65.Tong Y., Lin Y., Zhang Y., Yang J., Zhang Y., Liu H. Association between TCF7L2 gene polymorphisms and susceptibility to type 2 diabetes mellitus: a large Human Genome Epidemiology (HuGE) review and meta-analysis. BMC Medical Genetics. 2009;10:15. doi: 10.1186/1471-2350-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Warodomwichit D., Arnett D.K., Kabagambe E.K., Tsai M.Y., Hixson J.E., Straka R.J. Polyunsaturated fatty acids modulate the effect of TCF7L2 gene variants on postprandial lipemia. Journal of Nutrition. 2009;139(3):439–446. doi: 10.3945/jn.108.096461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu J., Steck A.K., Babu S., Yu L., Miao D., McFann K. Single nucleotide transcription factor 7-like 2 (TCF7L2) gene polymorphisms in antiislet autoantibody-negative patients at onset of diabetes. Journal of Clinical Endocrinology & Metabolism. 2009;94(2):504–510. doi: 10.1210/jc.2007-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boccardi V., Ambrosino I., Papa M., Fiore D., Rizzo M.R., Paolisso G. Potential role of TCF7L2 gene variants on cardiac sympathetic/parasympathetic activity. European Journal of Human Genetics. 2010;18(12):1333–1338. doi: 10.1038/ejhg.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cruz M., Valladares-Salgado A., Garcia-Mena J., Ross K., Edwards M., Angeles-Martinez J. Candidate gene association study conditioning on individual ancestry in patients with type 2 diabetes and metabolic syndrome from Mexico City. Diabetes Metabolism Research and Reviews. 2010;26(4):261–270. doi: 10.1002/dmrr.1082. [DOI] [PubMed] [Google Scholar]
- 70.Haupt A., Thamer C., Heni M., Ketterer C., Machann J., Schick F. Gene variants of TCF7L2 influence weight loss and body composition during lifestyle intervention in a population at risk for type 2 diabetes. Diabetes. 2010;59(3):747–750. doi: 10.2337/db09-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Neuman R.J., Wasson J., Atzmon G., Wainstein J., Yerushalmi Y., Cohen J. Gene-gene interactions lead to higher risk for development of type 2 diabetes in an Ashkenazi Jewish population. PLoS One. 2010;5(3):e9903. doi: 10.1371/journal.pone.0009903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Potapov V.A., Shamkhalova M.N., Smetanina S.A., Bel'chikova L.N., Suplotova L.A., Shestakova M.V. Polymorphic markers TCF7L2 rs12255372 and SLC30A8 rs13266634 confer susceptibility to type 2 diabetes in a Russian population. Genetika. 2010;46(8):1123–1131. [PubMed] [Google Scholar]
- 73.Zampetti S., Spoletini M., Petrone A., Capizzi M., Arpi M.L., Tiberti C. Association of TCF7L2 gene variants with low GAD autoantibody titre in LADA subjects (NIRAD Study 5) Diabetic Medicine. 2010;27(6):701–704. doi: 10.1111/j.1464-5491.2010.02997.x. [DOI] [PubMed] [Google Scholar]
- 74.Bonetti S., Trombetta M., Malerba G., Boselli L., Trabetti E., Muggeo M. Variants and haplotypes of TCF7L2 are associated with beta-cell function in patients with newly diagnosed type 2 diabetes: the Verona Newly Diagnosed Type 2 Diabetes Study (VNDS) 1. Journal of Clinical Endocrinology & Metabolism. 2011;96(2):E389–E393. doi: 10.1210/jc.2010-1677. [DOI] [PubMed] [Google Scholar]
- 75.Dabelea D., Dolan L.M., D'Agostino R., Jr., Hernandez A.M., McAteer J.B., Hamman R.F. Association testing of TCF7L2 polymorphisms with type 2 diabetes in multi-ethnic youth. Diabetologia. 2011;54(3):535–539. doi: 10.1007/s00125-010-1982-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Franco L.F., Crispim F., Pereira A.C., Moises R.S., Japanese-Brazilian Diabetes Study G. Variants of transcription factor 7-like 2 (TCF7L2) gene and incident glucose intolerance in Japanese-Brazilians. Brazilian Journal of Medical and Biological Research. 2011;44(3):240–244. doi: 10.1590/s0100-879x2011007500010. [DOI] [PubMed] [Google Scholar]
- 77.Klunder-Klunder M., Mejia-Benitez M.A., Flores-Huerta S., Burguete-Garcia A.I., Garcia-Mena J., Cruz M. rs12255372 variant of TCF7L2 gene is protective for obesity in Mexican children. Archives of Medical Research. 2011;42(6):495–501. doi: 10.1016/j.arcmed.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 78.Kurzawski M., Dziewanowski K., Kedzierska K., Wajda A., Lapczuk J., Drozdzik M. Association of transcription factor 7-like 2 (TCF7L2) gene polymorphism with posttransplant diabetes mellitus in kidney transplant patients medicated with tacrolimus. Pharmacological Reports. 2011;63(3):826–833. doi: 10.1016/s1734-1140(11)70595-3. [DOI] [PubMed] [Google Scholar]
- 79.Martinez-Gomez L.E., Cruz M., Martinez-Nava G.A., Madrid-Marina V., Parra E., Garcia-Mena J. A replication study of the IRS1, CAPN10, TCF7L2, and PPARG gene polymorphisms associated with type 2 diabetes in two different populations of Mexico. Annals of Human Genetics. 2011;75(5):612–620. doi: 10.1111/j.1469-1809.2011.00668.x. [DOI] [PubMed] [Google Scholar]
- 80.Mohan V., Goldhaber-Fiebert J.D., Radha V., Gokulakrishnan K. Screening with OGTT alone or in combination with the Indian diabetes risk score or genotyping of TCF7L2 to detect undiagnosed type 2 diabetes in Asian Indians. Indian Journal of Medical Research. 2011;133:294–299. [PMC free article] [PubMed] [Google Scholar]
- 81.Muendlein A., Saely C.H., Geller-Rhomberg S., Sonderegger G., Rein P., Winder T. Single nucleotide polymorphisms of TCF7L2 are linked to diabetic coronary atherosclerosis. PLoS One. 2011;6(3):e17978. doi: 10.1371/journal.pone.0017978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Papadopoulou A., Lynch K.F., Shaat N., Hakansson R., Ivarsson S.A., Berntorp K. Gestational diabetes mellitus is associated with TCF7L2 gene polymorphisms independent of HLA-DQB1*0602 genotypes and islet cell autoantibodies. Diabetic Medicine. 2011;28(9):1018–1027. doi: 10.1111/j.1464-5491.2011.03359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Alami F.M., Ahmadi M., Bazrafshan H., Tabarraei A., Khosravi A., Tabatabaiefar M.A. Association of the TCF7L2 rs12255372 (G/T) variant with type 2 diabetes mellitus in an Iranian population. Genetics and Molecular Biology. 2012;35(2):413–417. doi: 10.1590/S1415-47572012005000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Connor A.E., Baumgartner R.N., Baumgartner K.B., Kerber R.A., Pinkston C., John E.M. Associations between TCF7L2 polymorphisms and risk of breast cancer among Hispanic and non-Hispanic white women: the Breast Cancer Health Disparities Study. Breast Cancer Research and Treatment. 2012;136(2):593–602. doi: 10.1007/s10549-012-2299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guinan K.J. Worldwide distribution of type II diabetes-associated TCF7L2 SNPs: evidence for stratification in Europe. Biochemical Genetics. 2012;50(3–4):159–179. doi: 10.1007/s10528-011-9456-2. [DOI] [PubMed] [Google Scholar]
- 86.Gupta V., Vinay D.G., Rafiq S., Kranthikumar M.V., Janipalli C.S., Giambartolomei C. Association analysis of 31 common polymorphisms with type 2 diabetes and its related traits in Indian sib pairs. Diabetologia. 2012;55(2):349–357. doi: 10.1007/s00125-011-2355-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kalnina I., Geldnere K., Tarasova L., Nikitina-Zake L., Peculis R., Fridmanis D. Stronger association of common variants in TCF7L2 gene with nonobese type 2 diabetes in the Latvian population. Experimental and Clinical Endocrinology & Diabetes. 2012;120(8):466–468. doi: 10.1055/s-0032-1306298. [DOI] [PubMed] [Google Scholar]
- 88.Mattei J., Qi Q., Hu F.B., Sacks F.M., Qi L. TCF7L2 genetic variants modulate the effect of dietary fat intake on changes in body composition during a weight-loss intervention. American Journal of Clinical Nutrition. 2012;96(5):1129–1136. doi: 10.3945/ajcn.112.038125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nemr R., Turki A., Echtay A., Al-Zaben G.S., Daher H.S., Irani-Hakime N.A. Transcription factor-7-like 2 gene variants are strongly associated with type 2 diabetes in Lebanese subjects. Diabetes Research and Clinical Practice. 2012;98(3):e23–e27. doi: 10.1016/j.diabres.2012.09.044. [DOI] [PubMed] [Google Scholar]
- 90.Vcelak J., Vejrazkova D., Vankova M., Lukasova P., Bradnova O., Halkova T. T2D risk haplotypes of the TCF7L2 gene in the Czech population sample: the association with free fatty acids composition. Physiological Research. 2012;61(3):229–240. doi: 10.33549/physiolres.932272. [DOI] [PubMed] [Google Scholar]
- 91.Yalamanchi S.K., Sam S., Cardenas M.O., Holaday L.W., Urbanek M., Dunaif A. Association of fibrillin-3 and transcription factor-7-like 2 gene variants with metabolic phenotypes in PCOS. Obesity (Silver Spring) 2012;20(6):1273–1278. doi: 10.1038/oby.2011.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ciccacci C., Di Fusco D., Cacciotti L., Morganti R., D'Amato C., Novelli G. TCF7L2 gene polymorphisms and type 2 diabetes: association with diabetic retinopathy and cardiovascular autonomic neuropathy. Acta Diabetologica. 2013;50(5):789–799. doi: 10.1007/s00592-012-0418-x. [DOI] [PubMed] [Google Scholar]
- 93.Dou H., Ma E., Yin L., Jin Y., Wang H. The association between gene polymorphism of TCF7L2 and type 2 diabetes in Chinese Han population: a meta-analysis. PLoS One. 2013;8(3):e59495. doi: 10.1371/journal.pone.0059495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ling Q., Xie H., Lu D., Wei X., Gao F., Zhou L. Association between donor and recipient TCF7L2 gene polymorphisms and the risk of new-onset diabetes mellitus after liver transplantation in a Han Chinese population. Journal of Hepatology. 2013;58(2):271–277. doi: 10.1016/j.jhep.2012.09.025. [DOI] [PubMed] [Google Scholar]
- 95.Pang D.X., Smith A.J., Humphries S.E. Functional analysis of TCF7L2 genetic variants associated with type 2 diabetes. Nutrition, Metabolism, and Cardiovascular Diseases. 2013;23(6):550–556. doi: 10.1016/j.numecd.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Peng S., Zhu Y., Lu B., Xu F., Li X., Lai M. TCF7L2 gene polymorphisms and type 2 diabetes risk: a comprehensive and updated meta-analysis involving 121,174 subjects. Mutagenesis. 2013;28(1):25–37. doi: 10.1093/mutage/ges048. [DOI] [PubMed] [Google Scholar]
- 97.Turki A., Al-Zaben G.S., Mtiraoui N., Marmmuoch H., Mahjoub T., Almawi W.Y. Transcription factor-7-like 2 gene variants are strongly associated with type 2 diabetes in Tunisian Arab subjects. Gene. 2013;513(2):244–248. doi: 10.1016/j.gene.2012.10.086. [DOI] [PubMed] [Google Scholar]
- 98.Uma Jyothi K., Jayaraj M., Subburaj K.S., Prasad K.J., Kumuda I., Lakshmi V. Association of TCF7L2 gene polymorphisms with T2DM in the population of Hyderabad, India. PLoS One. 2013;8(4):e60212. doi: 10.1371/journal.pone.0060212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang J., Zhang J., Li L., Wang Y., Wang Q., Zhai Y. Association of rs12255372 in the TCF7L2 gene with type 2 diabetes mellitus: a meta-analysis. Brazilian Journal of Medical and Biological Research. 2013;46(4):382–393. doi: 10.1590/1414-431X20132677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang C., Bao W., Rong Y., Yang H., Bowers K., Yeung E. Genetic variants and the risk of gestational diabetes mellitus: a systematic review. Human Reproduction Update. 2013;19(4):376–390. doi: 10.1093/humupd/dmt013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Barros C.M., Araujo-Neto A.P., Lopes T.R., Barros M.A., Motta F.J., Canalle R. Association of the rs7903146 and rs12255372 polymorphisms in the TCF7L2 gene with type 2 diabetes in a population from northeastern Brazil. Genetics and Molecular Research. 2014;13(3):7889–7898. doi: 10.4238/2014.September.29.1. [DOI] [PubMed] [Google Scholar]
- 102.Ben-Salem A., Ajina M., Suissi M., Daher H.S., Almawi W.Y., Mahjoub T. Polymorphisms of transcription factor-7-like 2 (TCF7L2) gene in Tunisian women with polycystic ovary syndrome (PCOS) Gene. 2014;533(2):554–557. doi: 10.1016/j.gene.2013.09.104. [DOI] [PubMed] [Google Scholar]
- 103.Pagan A., Sabater-Molina M., Olza J., Prieto-Sanchez M.T., Blanco-Carnero J.E., Parrilla J.J. A gene variant in the transcription factor 7-like 2 (TCF7L2) is associated with an increased risk of gestational diabetes mellitus. European Journal of Obstetrics & Gynecology and Reproductive Biology. 2014;180:77–82. doi: 10.1016/j.ejogrb.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 104.Reyes-Lopez R., Perez-Luque E., Malacara J.M. Metabolic, hormonal characteristics and genetic variants of TCF7L2 associated with development of gestational diabetes mellitus in Mexican women. Diabetes Metab Res Rev. 2014;30(8):701–706. doi: 10.1002/dmrr.2538. [DOI] [PubMed] [Google Scholar]
- 105.Shi X., Cai Q., Zou M., Shen Y. Correlation between TCF7L2 gene polymorphism and genetic susceptibility in women with gestational diabetes mellitus. Zhonghua Fu Chan Ke Za Zhi. 2014;49(8):588–593. [PubMed] [Google Scholar]
- 106.Shokouhi S., Delpisheh A., Haghani K., Mahdizadeh M., Bakhtiyari S. Association of rs7903146, rs12255372, and rs290487 polymorphisms in TCF7L2 gene with type 2 diabetes in an Iranian Kurdish ethnic group. Clinical Laboratory. 2014;60(8):1269–1276. doi: 10.7754/clin.lab.2013.130809. [DOI] [PubMed] [Google Scholar]
- 107.Tsai M.K., Wang H.M., Shiang J.C., Chen I.H., Wang C.C., Shiao Y.F. Sequence variants of ADIPOQ and association with type 2 diabetes mellitus in Taiwan Chinese Han population. ScientificWorld Journal. 2014;2014:650393. doi: 10.1155/2014/650393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Acharya S., Al-Elq A., Al-Nafaie A., Muzaheed M., Al-Ali A. Type 2 diabetes mellitus susceptibility gene TCF7L2 is strongly associated with hyperglycemia in the Saudi Arabia Population of the eastern province of Saudi Arabia. European Review for Medical and Pharmacological Sciences. 2015;19(16):3100–3106. [PubMed] [Google Scholar]
- 109.Bodhini D., Chidambaram M., Liju S., Prakash V.G., Gayathri V., Shanthirani C.S. Association of TCF7L2 polymorphism with diabetic nephropathy in the South Indian population. Annals of Human Genetics. 2015 doi: 10.1111/ahg.12122. [DOI] [PubMed] [Google Scholar]
- 110.de Melo S.F., Frigeri H.R., dos Santos-Weiss I.C., Rea R.R., de Souza E.M., Alberton D. Polymorphisms in FTO and TCF7L2 genes of Euro-Brazilian women with gestational diabetes. Clinical Biochemistry. 2015;48(16–17):1064–1067. doi: 10.1016/j.clinbiochem.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 111.Gammoh E., Mahmood N.A., Madan S., Ebrahim B.H., Mustafa F.E., Almawi W.Y. Transcription factor-7-like 2 gene variants affect the metabolic phenotypes of polycystic ovary syndrome. Annals of Nutrition & Metabolism. 2015;67(4):228–235. doi: 10.1159/000439546. [DOI] [PubMed] [Google Scholar]
- 112.Ibrahim A.T., Hussain A., Salih M.A., Ibrahim O.A., Jamieson S.E., Ibrahim M.E. Candidate gene analysis supports a role for polymorphisms at TCF7L2 as risk factors for type 2 diabetes in Sudan. Journal of Diabetes and Metabolic Disorders. 2015;15:4. doi: 10.1186/s40200-016-0225-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Moran Y., Labrador L., Camargo M.E., Fernandez D., Chiurillo M.A. Design of an allele-specific PCR assay to genotype the rs12255372 SNP in a pilot study of association between common TCF7L2 polymorphisms and type 2 diabetes in Venezuelans. Arch Endocrinol Metab. 2015;60(3):246–251. doi: 10.1590/2359-3997000000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nanfa D., Sobngwi E., Atogho-Tiedeu B., Noubiap J.J., Donfack O.S., Mofo E.P. Association between the TCF7L2 rs12255372 (G/T) gene polymorphism and type 2 diabetes mellitus in a Cameroonian population: a pilot study. Clinical and Translational Medicine. 2015;4:17. doi: 10.1186/s40169-015-0058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ngwa E.N., Sobngwi E., Atogho-Tiedeu B., Noubiap J.J., Donfack O.S., Guewo-Fokeng M. Association between the rs12255372 variant of the TCF7L2 gene and obesity in a Cameroonian population. BMC Research Notes. 2015;8:717. doi: 10.1186/s13104-015-1661-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pourahmadi M., Erfanian S., Moradzadeh M., Jahromi A.S. Non-association between rs7903146 and rs12255372 polymorphisms in transcription factor 7-like 2 gene and type 2 diabetes mellitus in Jahrom City, Iran. Diabetes & Metabolism Journal. 2015;39(6):512–517. doi: 10.4093/dmj.2015.39.6.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rafati R., Jalal R., Asoodeh A., Matin M.M. Association of rs12255372 (TCF7L2) and D76N (PDX-1) polymorphisms with type 2 diabetes in a population living in Northeast Iran. Archives of Iranian Medicine. 2015;18(6):376–378. [PubMed] [Google Scholar]
- 118.Yako Y.Y., Madubedube J.H., Kengne A.P., Erasmus R.T., Pillay T.S., Matsha T.E. Contribution of ENPP1, TCF7L2, and FTO polymorphisms to type 2 diabetes in mixed ancestry ethnic population of South Africa. African Health Sciences. 2015;15(4):1149–1160. doi: 10.4314/ahs.v15i4.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang Y., Xu J.R., Wang Y.J., Liu X.M. Association of TCF7L2 gene polymorphisms with susceptibility to type 2 diabetes mellitus in a Chinese Hui population. Genetics and Molecular Research. 2015;14(3):10064–10071. doi: 10.4238/2015.August.21.13. [DOI] [PubMed] [Google Scholar]
- 120.Yao H., Wang Z., Wang T., Ma Y., Su Y., Ma Q. Association of TCF7L2 genetic polymorphisms with type 2 diabetes mellitus in the Uygur population of China. International Journal of Environmental Research and Public Health. 2015;12(9):11797–11814. doi: 10.3390/ijerph120911797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chang S., Wang Z., Wu L., Lu X., Shangguan S., Xin Y. Association between TCF7L2 polymorphisms and gestational diabetes mellitus: a meta-analysis. Journal of Diabetes Investigation. 2016 doi: 10.1111/jdi.12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chidambaram M., Liju S., Saboo B., Sathyavani K., Viswanathan V., Pankratz N. Replication of genome-wide association signals in Asian Indians with early-onset type 2 diabetes. Acta Diabetologica. 2016;53(6):915–923. doi: 10.1007/s00592-016-0889-2. [DOI] [PubMed] [Google Scholar]
- 123.Dhawan D., Padh H. Genetic variations in TCF7L2 influence therapeutic response to sulfonylureas in Indian diabetics. Diabetes Research and Clinical Practice. 2016;121:35–40. doi: 10.1016/j.diabres.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 124.Han L., Li Y., Tang L., Chen Z., Zhang T., Chen S. IGF2BP2 rs11705701 polymorphisms are associated with prediabetes in a Chinese population: a population-based case-control study. Experimental and Therapeutic Medicine. 2016;12(3):1849–1856. doi: 10.3892/etm.2016.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hindy G., Mollet I.G., Rukh G., Ericson U., Orho-Melander M. Several type 2 diabetes-associated variants in genes annotated to WNT signaling interact with dietary fiber in relation to incidence of type 2 diabetes. Genes & Nutrition. 2016;11:6. doi: 10.1186/s12263-016-0524-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.InterAct C. Investigation of gene-diet interactions in the incretin system and risk of type 2 diabetes: the EPIC-InterAct study. Diabetologia. 2016;59(12):2613–2621. doi: 10.1007/s00125-016-4090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lopez-Ortiz M.M., Garay-Sevilla M.E., Tejero M.E., Perez-Luque E.L. Analysis of the interaction between transcription factor 7-like 2 genetic variants with nopal and wholegrain fibre intake: effects on anthropometric and metabolic characteristics in type 2 diabetes patients. British Journal of Nutrition. 2016;116(6):969–978. doi: 10.1017/S0007114516002798. [DOI] [PubMed] [Google Scholar]
- 128.Erkoc Kaya D., Arikoglu H., Kayis S.A., Ozturk O., Gonen M.S. Transcription factor 7-like 2 (TCF7L2) gene polymorphisms are strong predictorsof type 2 diabetes among nonobese diabetics in the Turkish population. Turkish Journal of Medical Sciences. 2017;47(1):22–28. doi: 10.3906/sag-1507-160. [DOI] [PubMed] [Google Scholar]
- 129.Gallardo-Blanco H.L., Villarreal-Perez J.Z., Cerda-Flores R.M., Figueroa A., Sanchez-Dominguez C.N., Gutierrez-Valverde J.M. Genetic variants in KCNJ11, TCF7L2 and HNF4A are associated with type 2 diabetes, BMI and dyslipidemia in families of Northeastern Mexico: a pilot study. Experimental and Therapeutic Medicine. 2017;13(2):523–529. doi: 10.3892/etm.2016.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hou Z., Li M., Cao Y. TCF7L2, CAPN10 polymorphisms are associated with gestational diabetes mellitus (GDM) risks: a meta-analysis. Gynecological Endocrinology. 2017:1–6. doi: 10.1080/09513590.2017.1290066. [DOI] [PubMed] [Google Scholar]
- 131.Miranda-Lora A.L., Cruz M., Molina-Diaz M., Gutierrez J., Flores-Huerta S., Klunder-Klunder M. Associations of common variants in the SLC16A11, TCF7L2, and ABCA1 genes with pediatric-onset type 2 diabetes and related glycemic traits in families: a case-control and case-parent trio study. Pediatric Diabetes. 2017 doi: 10.1111/pedi.12497. [DOI] [PubMed] [Google Scholar]
- 132.Zeggini E., McCarthy M.I. TCF7L2: the biggest story in diabetes genetics since HLA? Diabetologia. 2007;50(1):1–4. doi: 10.1007/s00125-006-0507-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Savic D., Ye H., Aneas I., Park S.Y., Bell G.I., Nobrega M.A. Alterations in TCF7L2 expression define its role as a key regulator of glucose metabolism. Genome Research. 2011;21(9):1417–1425. doi: 10.1101/gr.123745.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Boj S.F., van Es J.H., Huch M., Li V.S., Jose A., Hatzis P. Diabetes risk gene and Wnt effector Tcf7l2/TCF4 controls hepatic response to perinatal and adult metabolic demand. Cell. 2012;151(7):1595–1607. doi: 10.1016/j.cell.2012.10.053. [DOI] [PubMed] [Google Scholar]
- 135.Halaas J.L., Gajiwala K.S., Maffei M., Cohen S.L., Chait B.T., Rabinowitz D. Weight-reducing effects of the plasma protein encoded by the obese gene. Science. 1995;269(5223):543–546. doi: 10.1126/science.7624777. [DOI] [PubMed] [Google Scholar]
- 136.Scherer P.E., Williams S., Fogliano M., Baldini G., Lodish H.F. A novel serum protein similar to C1q, produced exclusively in adipocytes. Journal of Biological Chemistry. 1995;270(45):26746–26749. doi: 10.1074/jbc.270.45.26746. [DOI] [PubMed] [Google Scholar]
- 137.Lo J.C., Ljubicic S., Leibiger B., Kern M., Leibiger I.B., Moede T. Adipsin is an adipokine that improves beta cell function in diabetes. Cell. 2014;158(1):41–53. doi: 10.1016/j.cell.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Steppan C.M., Bailey S.T., Bhat S., Brown E.J., Banerjee R.R., Wright C.M. The hormone resistin links obesity to diabetes. Nature. 2001;409(6818):307–312. doi: 10.1038/35053000. [DOI] [PubMed] [Google Scholar]
- 139.Ross S.E. Inhibition of adipogenesis by wnt signaling. Science. 2000;289(5481):950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 140.Villanueva C.J., Waki H., Godio C., Nielsen R., Chou W.L., Vargas L. TLE3 is a dual-function transcriptional coregulator of adipogenesis. Cell Metabolism. 2011;13(4):413–427. doi: 10.1016/j.cmet.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Eguchi J., Wang X., Yu S., Kershaw E.E., Chiu P.C., Dushay J. Transcriptional control of adipose lipid handling by IRF4. Cell Metabolism. 2011;13(3):249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Parks B.W., Nam E., Org E., Kostem E., Norheim F., Hui S.T. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metabolism. 2013;17(1):141–152. doi: 10.1016/j.cmet.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Parks B.W., Sallam T., Mehrabian M., Psychogios N., Hui S.T., Norheim F. Genetic architecture of insulin resistance in the mouse. Cell Metabolism. 2015;21(2):334–347. doi: 10.1016/j.cmet.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yu J., Seldin M.M., Fu K., Li S., Lam L., Wang P. Topological arrangement of cardiac fibroblasts regulates cellular plasticity. Circulation Research. 2018;123(1):73–85. doi: 10.1161/CIRCRESAHA.118.312589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 147.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ali S.H., DeCaprio J.A. Cellular transformation by SV40 large T antigen: interaction with host proteins. Seminars in Cancer Biology. 2001;11(1):15–23. doi: 10.1006/scbi.2000.0342. [DOI] [PubMed] [Google Scholar]
- 149.Metzger D., Clifford J., Chiba H., Chambon P. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(15):6991–6995. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ghazalpour A., Rau C.D., Farber C.R., Bennett B.J., Orozco L.D., van Nas A. Hybrid mouse diversity panel: a panel of inbred mouse strains suitable for analysis of complex genetic traits. Mammalian Genome. 2012;23(9–10):680–692. doi: 10.1007/s00335-012-9411-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lusis A.J., Seldin M.M., Allayee H., Bennett B.J., Civelek M., Davis R.C. The Hybrid Mouse Diversity Panel: a resource for systems genetics analyses of metabolic and cardiovascular traits. The Journal of Lipid Research. 2016;57(6):925–942. doi: 10.1194/jlr.R066944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Engelbrechtsen L., Hansen T.H., Mahendran Y., Pyl P., Andersson E., Jonsson A. Homozygous carriers of the TCF7L2 rs7903146 T-allele show altered postprandial response in triglycerides and triglyceride-rich lipoproteins. Scientific Reports. 2017;7:43128. doi: 10.1038/srep43128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cauchi S., Choquet H., Gutierrez-Aguilar R., Capel F., Grau K., Proenca C. Effects of TCF7L2 polymorphisms on obesity in European populations. Obesity (Silver Spring) 2008;16(2):476–482. doi: 10.1038/oby.2007.77. [DOI] [PubMed] [Google Scholar]
- 154.Chang Shaoyan, Wang Zhen, Wu Lihua, Lu Xiaolin, Shangguan Shaofang, Xin Yu, Li Li, Wang Li. Association between TCF7L2 polymorphisms and gestational diabetes mellitus: a meta-analysis. Journal of Diabetes Investigation. 2017;8(4):560–570. doi: 10.1111/jdi.12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kennell J.A., MacDougald O.A. Wnt signaling inhibits adipogenesis through beta-catenin-dependent and -independent mechanisms. Journal of Biological Chemistry. 2005;280(25):24004–24010. doi: 10.1074/jbc.M501080200. [DOI] [PubMed] [Google Scholar]
- 156.Shyh-Chang N., Ng H.H. The metabolic programming of stem cells. Genes & Development. 2017;31(4):336–346. doi: 10.1101/gad.293167.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chen X., Ayala I., Shannon C., Fourcaudot M., Acharya N.K., Jenkinson C.P. The diabetes gene and wnt pathway effector TCF7L2 regulates adipocyte development and function. Diabetes. 2018;67(4):554–568. doi: 10.2337/db17-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.van Es J.H., Haegebarth A., Kujala P., Itzkovitz S., Koo B.K., Boj S.F. A critical role for the wnt effector Tcf4 in adult intestinal homeostatic self-renewal. Molecular and Cellular Biology. 2012;32(10):1918–1927. doi: 10.1128/MCB.06288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cawthorn W.P., Bree A.J., Yao Y., Du B., Hemati N., Martinez-Santibanez G. Wnt6, Wnt10a and Wnt10b inhibit adipogenesis and stimulate osteoblastogenesis through a beta-catenin-dependent mechanism. Bone. 2012;50(2):477–489. doi: 10.1016/j.bone.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kuzmicki M., Telejko B., Wawrusiewicz-Kurylonek N., Kalejta K., Lemancewicz A., Zdrodowski M. The expression of transcription factor 7-like 2 (TCF7L2) in fat and placental tissue from women with gestational diabetes. Diabetes Research and Clinical Practice. 2011;94(2):e43–e46. doi: 10.1016/j.diabres.2011.07.033. [DOI] [PubMed] [Google Scholar]
- 161.Wei E., Ben Ali Y., Lyon J., Wang H., Nelson R., Dolinsky V.W. Loss of TGH/Ces3 in mice decreases blood lipids, improves glucose tolerance, and increases energy expenditure. Cell Metabolism. 2010;11(3):183–193. doi: 10.1016/j.cmet.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 162.Wei E., Gao W., Lehner R. Attenuation of adipocyte triacylglycerol hydrolase activity decreases basal fatty acid efflux. Journal of Biological Chemistry. 2007;282(11):8027–8035. doi: 10.1074/jbc.M605789200. [DOI] [PubMed] [Google Scholar]
- 163.Wright W.S., Longo K.A., Dolinsky V.W., Gerin I., Kang S., Bennett C.N. Wnt10b inhibits obesity in ob/ob and agouti mice. Diabetes. 2007;56(2):295–303. doi: 10.2337/db06-1339. [DOI] [PubMed] [Google Scholar]
- 164.Shu L., Sauter N.S., Schulthess F.T., Matveyenko A.V., Oberholzer J., Maedler K. Transcription factor 7-like 2 regulates beta-cell survival and function in human pancreatic islets. Diabetes. 2008;57(3):645–653. doi: 10.2337/db07-0847. [DOI] [PubMed] [Google Scholar]
- 165.Zhou Y., Park S.-Y., Su J., Bailey K., Ottosson-Laakso E., Shcherbina L. TCF7L2 is a master regulator of insulin production and processing. Human Molecular Genetics. 2014;23(24):6419–6431. doi: 10.1093/hmg/ddu359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Elbein S.C., Chu W.S., Das S.K., Yao-Borengasser A., Hasstedt S.J., Wang H. Transcription factor 7-like 2 polymorphisms and type 2 diabetes, glucose homeostasis traits and gene expression in US participants of European and African descent. Diabetologia. 2007;50(8):1621–1630. doi: 10.1007/s00125-007-0717-x. [DOI] [PubMed] [Google Scholar]
- 167.Mondal A.K., Das S.K., Baldini G., Chu W.S., Sharma N.K., Hackney O.G. Genotype and tissue-specific effects on alternative splicing of the transcription factor 7-like 2 gene in humans. Journal of Clinical Endocrinology & Metabolism. 2010;95(3):1450–1457. doi: 10.1210/jc.2009-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhang T., Hsu F.N., Xie X.J., Li X., Liu M., Gao X. Reversal of hyperactive Wnt signaling-dependent adipocyte defects by peptide boronic acids. Proc Natl Acad Sci USA. 2017 Sep 5;114(36):E7469–E7478. doi: 10.1073/pnas.1621048114. Epub 2017 Aug 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Arner P., Andersson D.P., Bäckdahl J., Dahlman I., Rydén M. Weight gain and impaired glucose metabolism in women are predicted by inefficient subcutaneous fat cell lipolysis. Cell Metab. 2018 Jul 3;28(1):45–54.e3. doi: 10.1016/j.cmet.2018.05.004. Epub 2018 May 31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.