Abstract
Objective
The ferritin heavy/heart chain (FTH) gene encodes the ferroxidase component of the iron (Fe) sequestering ferritin complex, which plays a central role in the regulation of cellular Fe metabolism. Here we tested the hypothesis that ferritin regulates organismal Fe metabolism in a manner that impacts energy balance and thermal homeostasis.
Methods
We developed a mouse strain, referred herein as FthR26 fl/fl, expressing a tamoxifen-inducible Cre recombinase under the control of the Rosa26 (R26) promoter and carrying two LoxP (fl) sites: one at the 5′end of the Fth promoter and another the 3' end of the first Fth exon. Tamoxifen administration induces global deletion of Fth in adult FthR26Δ/Δ mice, testing whether FTH is required for maintenance of organismal homeostasis.
Results
Under standard nutritional Fe supply, Fth deletion in adult FthR26Δ/Δ mice led to a profound deregulation of organismal Fe metabolism, oxidative stress, inflammation, and multi-organ damage, culminating in death. Unexpectedly, Fth deletion was also associated with a profound atrophy of white and brown adipose tissue as well as with collapse of energy expenditure and thermogenesis. This was attributed mechanistically to mitochondrial dysfunction, as assessed in the liver and in adipose tissue.
Conclusion
The FTH component of ferritin acts as a master regulator of organismal Fe homeostasis, coupling nutritional Fe supply to organismal redox homeostasis, energy expenditure and thermoregulation.
Keywords: Iron metabolism, Redox homeostasis, Adipose tissue, Energy expenditure, Adipose tissue, Mitochondria
Highlights
-
•
Ferritin acts as a master regulator of organismal iron metabolism.
-
•
Regulation of Fe metabolism by Ferritin sustains organismal redox homeostasis.
-
•
Ferritin is essential to support organismal energy expenditure and thermogenesis.
-
•
Ferritin is essential to support mitochondrial function and integrity in parenchymal tissues.
1. Introduction
As a divalent metal, Fe can exchange electrons with a variety of acceptor and donor molecules. This property was likely co-opted early through evolution to catalyze vital redox-based reactions in most forms of life. As an evolutionary trade-off, when Fe exchanges electrons with superoxide (O2•-) or hydrogen peroxide (H2O2), via the Haber–Weiss/Fenton reactions, it produces hydroxyl radicals (HO−) and other damaging reactive oxygen species (ROS) [1], [2]. Therefore, intracellular Fe content and redox activity must be tightly regulated to avoid oxidative damage to cellular macromolecules and organelles [2].
Intracellular Fe content is regulated by the relative rate of cellular import, via the divalent metal transporter 1 (DMT1) [3] and transferrin receptor 1 (TFR1) [4], versus export, via the solute carrier family 40 (SLC40A1; ferroportin) [5]. Intracellular Fe redox activity is controlled by ferritin, a multimeric protein complex composed of variable ratios of liver/light chain (FTL) and FTH [6], [7].
FTH and FTL are encoded by distinct genes, with germline Fth deletion being embryonically lethal in mice [8] and insects [9]. Ftl deletion however, is not embryonically lethal in mice [10], suggesting that these are not functionally redundant genes. FTL is thought to provide the scaffold supporting the ferritin multimeric complex, while the ferroxidase activity of FTH converts redox-active Fe2+ into redox-inactive Fe3+ via a process referred to as Fe nucleation [6], [7]. When intracellular Fe content is low, the Fe-responsive element-binding protein (IRP) 2 represses FTH and FTL mRNA translation, while intracellular Fe accumulation promotes IRP2 degradation and allows for FTH and FTL mRNA translation [2], [11], [12], [13]. The ferritin complex acquires Fe from the Fe-chaperone poly (rC)–binding protein 1 (PCBP1) [14] and can convert up to 4500 Fe2+ atoms into Fe3+, per ferritin complex [6], [7]. This allows to reduce intracellular Fe2+ content and stabilize IRP2 repressing FTH and FTL mRNA translation [2], [11], [12], [13]. When this occurs, ferritin releases Fe via an autophagy disassembly process involving the nuclear receptor co-activator 4 (NCOA4), termed ferritinophagy [15], [16].
Induction of FTH expression is essential to prevent oxidative tissue damage in response to infection [17], [18], [19] or tissue injury [20], [21]. Whether FTH regulates organismal Fe metabolism and redox homeostasis to prevent oxidative tissue damage under steady state conditions, to the best of our knowledge, has not been established. Several studies in mice have shown a relatively minor impact of FTH on organismal Fe metabolism and redox homeostasis, for example, when Fth is deleted specifically in intestinal epithelium [22], hepatocytes and leukocytes [23], [24] or in the kidney [20]. While this may suggest that ferritin plays a minor role in the regulation of organismal Fe metabolism and redox homeostasis under steady state conditions, one should take into consideration that a significant proportion of ferritin is secreted, possibly acting in a non-cell autonomous manner [16], [25]. It is possible therefore that ferritin secretion contributes to the regulation of organismal Fe metabolism and redox homeostasis, including in mice carrying tissue-specific Fth deletions [20], [22], [23], [24].
Here we report that global Fth deletion in adult mice leads to a profound disruption of organismal Fe metabolism and redox homeostasis, associated with failure to sustain energy expenditure and thermal homeostasis. This is attributed mechanistically to intracellular Fe accumulation and deregulation of mitochondrial function in parenchyma tissues that are critical to control energy expenditure and thermal homeostasis, as determined in the liver and fat. We conclude that ferritin acts as a gatekeeper of organismal Fe metabolism, coupling nutritional Fe supply to organismal energy expenditure and thermoregulation.
2. Results
2.1. FTH is essential to maintain organismal Fe homeostasis
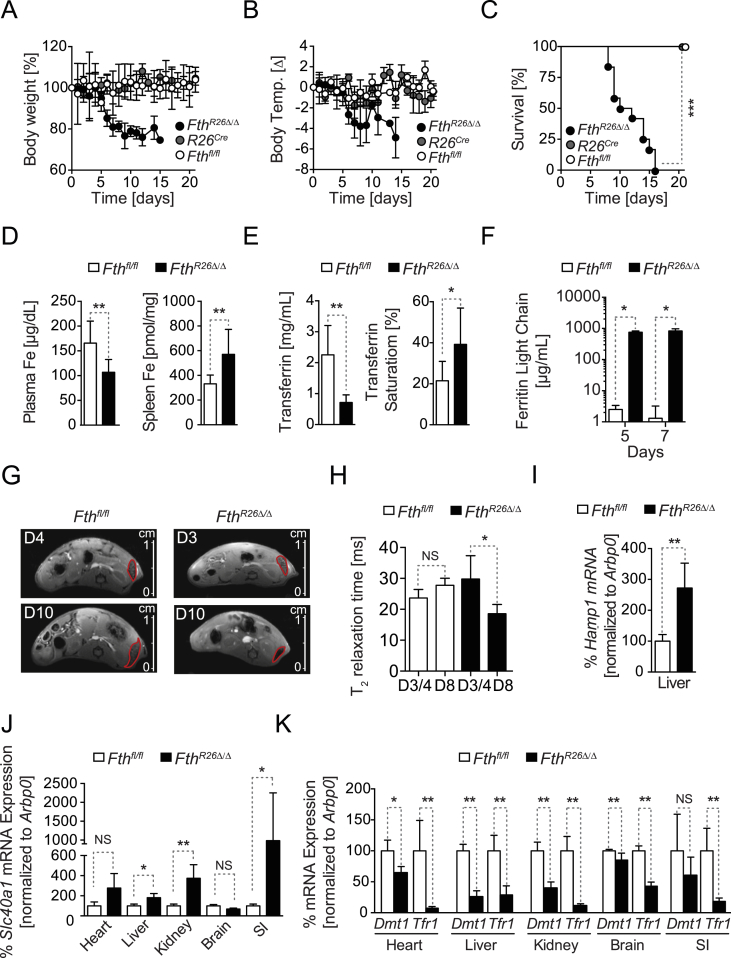
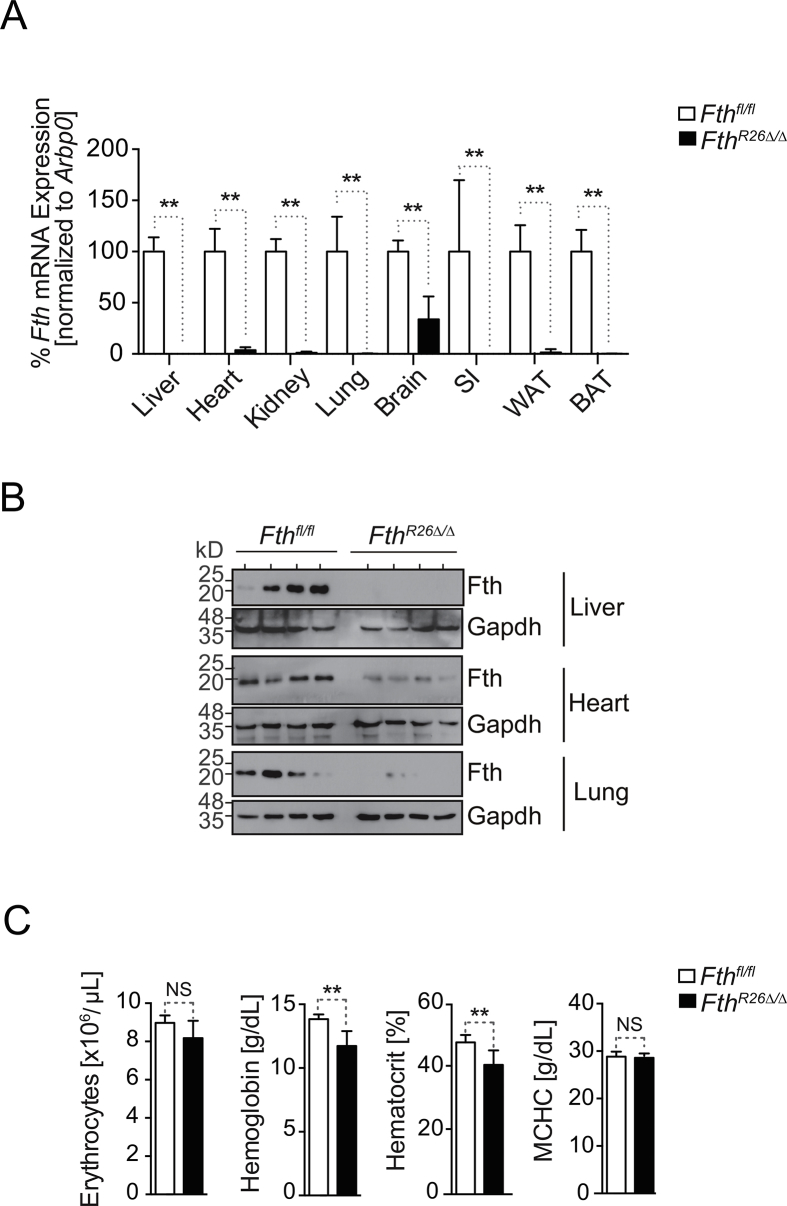
We reasoned that global and inducible deletion of Fth in FthR26Δ/Δ mice should reveal whether FTH regulates organismal Fe metabolism. In contrast to previous studies using tissue-specific Fth deletions [20], [22], [23], [24], global Fth deletion (Figure S1A, B) resulted in rapid loss of body weight (Figure 1A) and temperature (Fig. 1B) and death of all FthR26Δ/Δ mice (Fig. 1C). This was not observed in control R26CreERT2 or Fthfl/fl mice, receiving tamoxifen at the same dosage and schedule (Figure 1A–C).
Figure 1.
FTH regulates organismal Fe and redox homeostasis. Relative (A) body weight, (B) temperature, and (C) survival of FthR26Δ/Δ (n = 12), R26CreERT2 (n = 5) and Fthfl/fl (n = 7) mice, after tamoxifen administration at day 0. Data in (A) and (B) are represented as mean ± SD, pooled from 4 experiments with similar trend. Data in (C) are from the same 4 experiments as (A, B). (D) Concentration of non-heme Fe in plasma and spleen of Fthfl/fl mice (n = 9) and FthR26Δ/Δ (n = 8) mice, 7 days after tamoxifen administration. Data represented as mean ± SD, pooled from 3 experiments with similar trend. (E) Transferrin concentration and saturation in blood of same mice as (D). (F) Ferritin (i.e. light chain) concentration in plasma of FthR26Δ/Δ (n = 4) and Fthfl/fl mice (n = 4), at the days indicated, after tamoxifen administration. Data represented as mean ± SD, pooled from 2 experiments with similar trend. (G) Representative MRI of the abdomen in axial view at day 4 and 10 after tamoxifen administration for the same Fthfl/fl mouse and at day 3 and 8 after tamoxifen administration for the same FthR26Δ/Δ mouse. (H) Quantification of the T2 relaxation time in the spleen of Fthfl/fl (n = 4) and FthR26Δ/Δ (n = 4) mice at day 3/4 and day 8 after tamoxifen administration. Relative expression of (I) Hamp1 (J) ferroportin (Slc40a1), (K) Dmt1 and Tfr1 mRNA in Fthfl/fl (n = 4) and FthR26Δ/Δ (n = 6) mice, 7 days after tamoxifen administration. Data are represented as mean ± SD, pooled from 2 independent experiments, with similar trend. Small intestine (SI). NS: non-significant, *P < 0.05, **P < 0.01, ***P < 0.001. P values in (C) were determined using Log-rank (Mantel–Cox) test. Mann–Whitney test was used for comparison between two groups and One-Way ANOVA with Bonferroni Post-hoc Test was used for comparison between multiple groups.
FthR26Δ/Δ mice developed hypoferremia, i.e. lower than normal Fe concentration in plasma (Figure 1D), low transferrin concentration, and high transferrin saturation in plasma (Figure 1E). There was also an accumulation of ferritin, i.e. Ftl, in plasma, as assessed by ELISA (Figure 1F) and Fe in the spleen, as assessed by magnetic resonance imaging (MRI), using T2 relaxation time as a measure of Fe-induced signal cancellation (Figure 1G,H), respectively. This suggests that, under steady state nutritional Fe supply, FthR26Δ/Δ mice develop a profound deregulation of organismal Fe metabolism, associated with a decrease in body temperature (Figure 1B), reminiscent of Fe deficiency in rodents [26] and humans [27], [28], [29].
Development of hypoferremia in FthR26Δ/Δ mice was associated with the induction of hepcidin (Hamp1) mRNA expression in the liver (Figure 1I), a central regulator of organismal Fe homeostasis that prevents Fe cellular export [30]. Moreover, FthR26Δ/Δ mice induced ferroportin mRNA expression in parenchyma tissues, including the liver, kidneys and small intestine (Figure 1J), while repressing Dmt1 and Tfr1 mRNA expression in the heart, liver, kidneys, brain and small intestine (Figure 1K). This suggests that FthR26Δ/Δ mice favor cellular Fe export from parenchyma tissues in detriment of Fe import, probably disrupting organismal Fe compartmentalization.
The largest pool of Fe in vertebrates is contained in the heme groups of hemoglobin, with 80% of circulating Fe being used for heme biosynthesis during erythropoiesis [2], [31]. While hypoferremia impairs erythropoiesis and leads to anemia [32], FthR26Δ/Δ mice did not develop overt anemia (Fig. S1C), suggesting that FthR26Δ/Δ mice succumb to an impairment of other vital functions not related directly to erythropoiesis.
2.2. FTH regulates organismal redox homeostasis
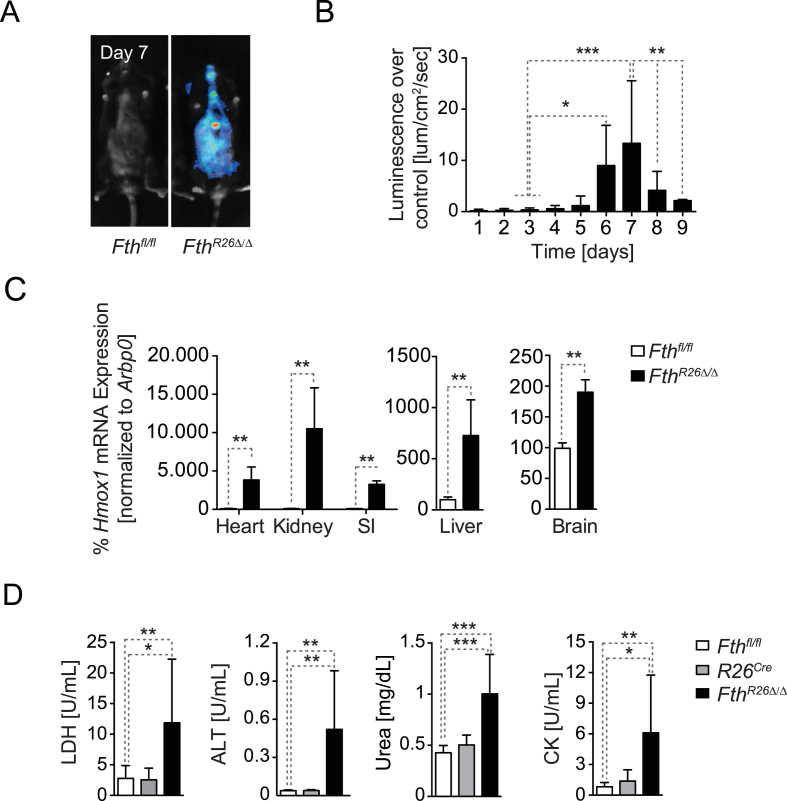
It is well established that the ferroxidase activity of FTH can prevent Fe2+ from partaking in the production of ROS via the Haber–Weiss and Fenton reactions [1]. However, to what extent this property of FTH acts under physiologic conditions to regulate organismal redox homeostasis was not formally established. To test this hypothesis FthR26fl/fl mice were crossed with OKD48Luc mice, expressing ubiquitously a luciferase reporter under the control of a synthetic oxidative-stress responsive promoter activated by the transcription factor nuclear factor (erythroid-derived 2)-like 2 (NRF2) [33]. Fth deletion in FthR26Δ/ΔOKD48Luc mice led to a robust induction of luciferase expression in different organs, i.e. oxidative stress, not observed in control Fthfl/flOKD48Luc mice (Figure 2A). This oxidative stress response peaked at day 7 after tamoxifen administration (Figure 2B), consistent with the onset of mortality (Figure 1C). Fth deletion was also associated with induction of endogenous NRF2-responsive genes, as illustrated for heme oxygenase 1 (Hmox1) mRNA expression in the heart, kidneys, small intestine, liver and brain of FthR26Δ/Δ vs. Fthfl/fl mice (Figure 2C). This shows that under steady state nutritional Fe supply, FTH is essential to regulate Fe metabolism in a manner that maintains organismal redox homeostasis.
Figure 2.
FTH is essential to maintain organismal redox homeostasis (A) Luciferase activity in whole body of OKD48lucFthfl/fl and OKD48lucFthR26Δ/Δ mice, 7 days after tamoxifen administration. (B) Quantification of luciferase activity in OKD48lucFthR26Δ/Δ mice (n = 7) relative to OKD48lucFthfl/fl (n = 8), shown as mean ± SD, pooled from 2 experiments with similar trend. (C) Hmox1 mRNA quantified by qRT-PCR in different organs from Fthfl/fl (n = 4) and FthR26Δ/Δ (n = 6) mice, harvested 7 days after tamoxifen administration. Data are shown as mean ± SD, pooled from 2 independent experiments with similar trend. SI: small intestine. (D) Serological markers of organ dysfunction in FthR26Δ/Δ (n = 10), R26Cre (n = 5) and Fthfl/fl (n = 10) mice, 7 days after tamoxifen administration. Data are shown as mean ± SD, pooled from 4 experiments with similar trend. NS: non-significant, *P < 0.05, **P < 0.01, ***P < 0.001. Mann–Whitney test was used for comparison between two groups. One-Way ANOVA with Bonferroni Post-hoc Test was used for comparison between multiple groups.
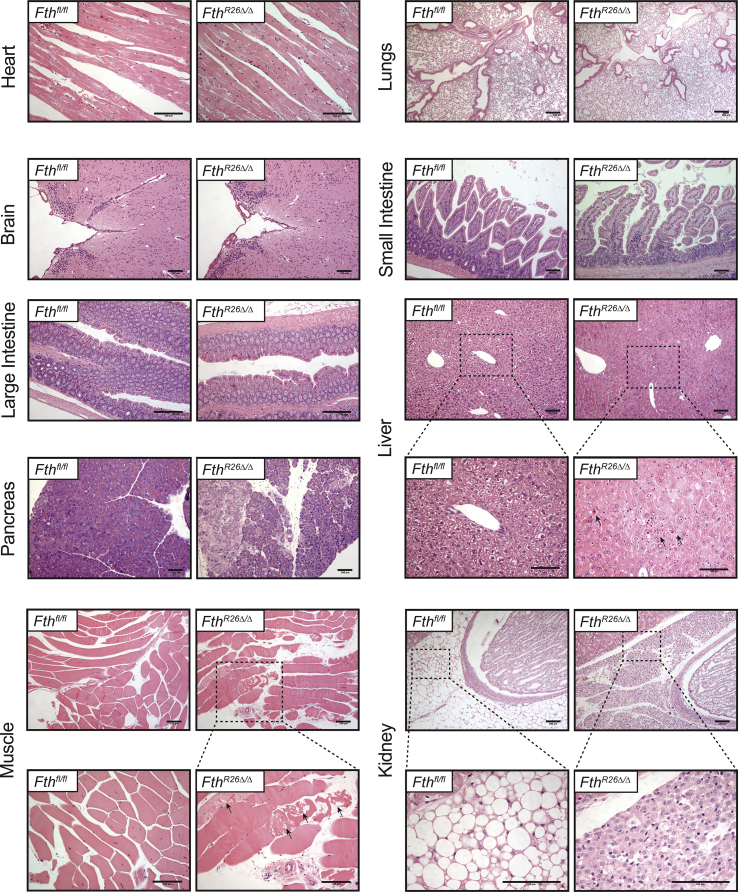
Consistent with the occurrence of oxidative stress, FthR26Δ/Δ mice developed multi-organ damage, as illustrated by lactate dehydrogenase (LDH), aminotransferase (ALT; liver damage), urea (kidney damage) and creatinine kinase (CK; muscle damage) accumulation in plasma (Figure 2D). Liver injury was characterized histologically by hypertrophic and multinucleated hepatocytes containing acidophilic granular cytoplasm as well as by apoptotic-foci (Fig. S2). FthR26Δ/Δ mice also developed skeletal muscle necrosis (Fig. S2) and focal lesions in the pancreas without other apparent histopathologic alterations in heart, lungs, brain, small intestine and large intestine and (Fig. S2). We noticed, however, an apparent atrophy of perirenal adipose tissue associated with extensive leukocyte infiltration (Fig. S2). This suggests that FTH acts under steady state conditions to sustain organismal redox homeostasis and prevent organ damage.
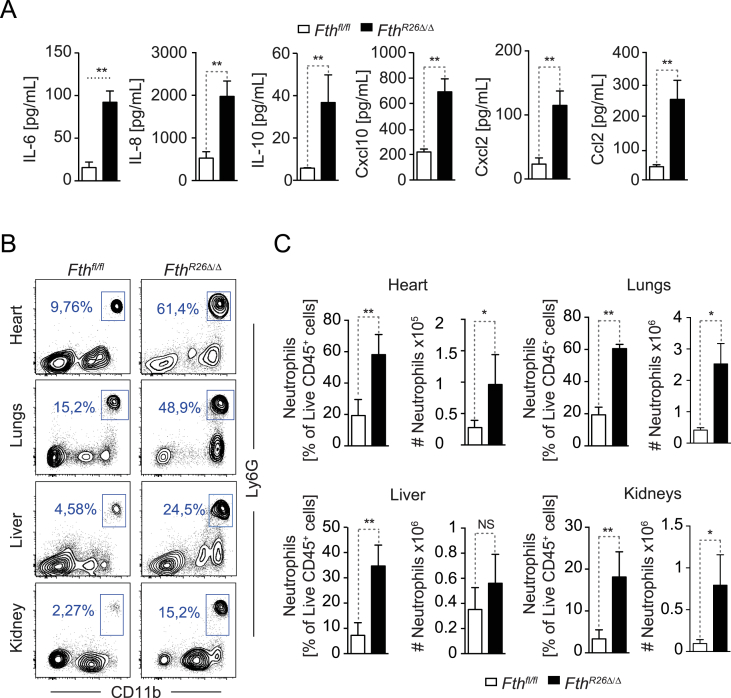
FthR26Δ/Δ mice developed a systemic inflammatory response characterized by the accumulation of interleukin (IL)-6, IL-8, and IL-10 and chemokines Ccl2, Cxcl2, and Cxcl10 in plasma (Figure 3A) and associated with neutrophil infiltration in parenchyma tissues, such as the heart, liver, lungs, and kidneys (Figure 3B,C). There was no induction of pro-inflammatory cytokines typically associated with macrophage activation, such as Tumor Necrosis Factor (TNF) or IL-1β (data not shown). This suggests that macrophages are not driving the systemic inflammatory response observed in FthR26Δ/Δ mice, which is probably triggered by sterile tissue damage.
Figure 3.
FTH protects against inflammation-driven tissue damage. (A) Cytokine and chemokine concentration in the sera of FthR26Δ/Δ (n = 9) and Fthfl/fl (n = 5) mice. Data are shown as mean ± SD, pooled from 3 experiments with similar trend. (B) Representative flow cytometry staining of live CD45+CD11bhiLy6-Ghi neutrophils in different organs from FthR26Δ/Δ and Fthfl/fl mice. (C) Quantification of live CD45+CD11bhiLy6-Ghi neutrophils in different organs from FthR26Δ/Δ (n = 7) and Fthfl/fl (n = 6) mice. Percentage (%) and total numbers (#) are represented as mean ± SEM, pooled from 3 experiments with similar trend. NS: non-significant, *P < 0.05, **P < 0.01. Mann–Whitney test was used for comparison between two groups. Sera and organs in (A, B, C) were harvested 7 days after tamoxifen administration.
2.3. FTH regulates energy expenditure
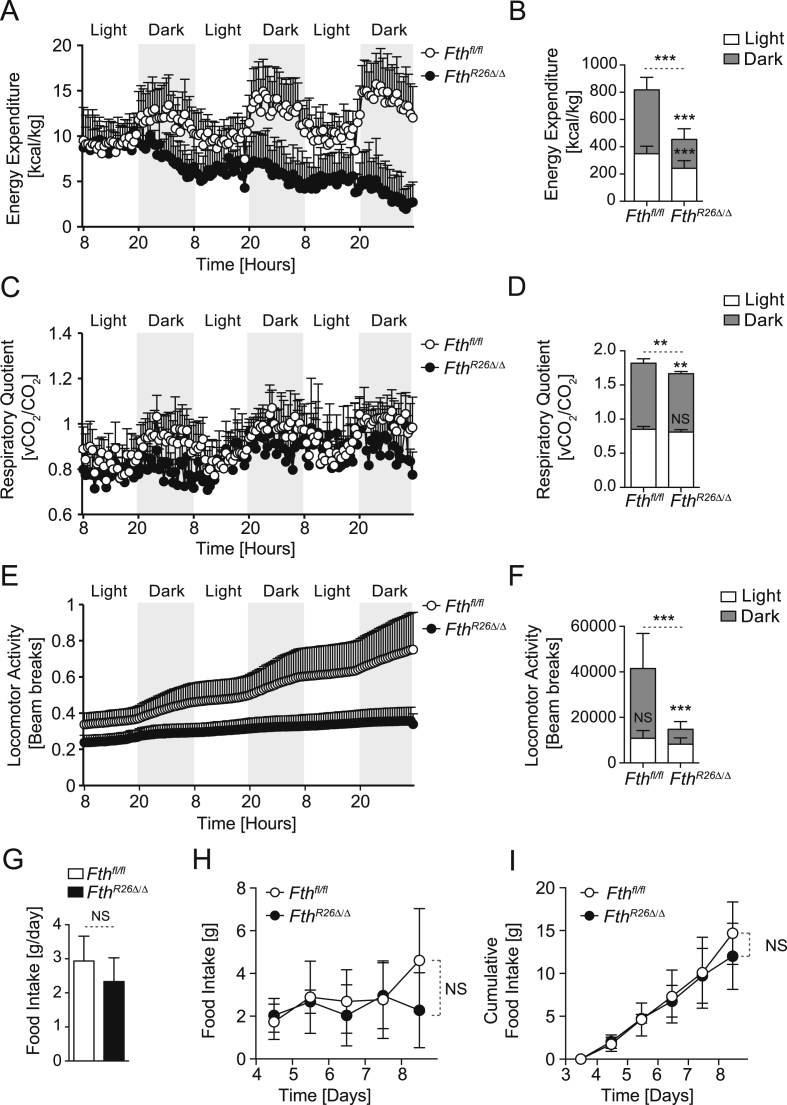
Body weight and temperature loss in FthR26Δ/Δ mice suggested that regulation of Fe metabolism by FTH controls organismal energy balance. Accordingly, FthR26Δ/Δ mice had a marked reduction in energy expenditure (Figure 4A,B) and a subtle reduction of respiratory quotient (RQ) (Figure 4C,D), as well as a drastic reduction in locomotor activity (Figure 4E,F), when compared to control Fthfl/fl mice. Decreased energy expenditure in FthR26Δ/Δ mice was not associated with changes in food intake, when compared to control Fthfl/fl mice (Figure 4G–I). This suggests that FTH plays a fundamental role in the control of energetic balance.
Figure 4.
FTH is essential to support energy expenditure. Cumulative (A,B) Energy expenditure, (C,D) Respiratory Quotient, and (E,F) Locomotor Activity of Fthfl/fl (n = 14) and FthR26Δ/Δ (n = 8), monitored from days 6–8, after the first tamoxifen administration. Data are represented as mean ± SD, pooled from 3 independent experiments. Average (G), daily (H), and cumulative (I) food intake of Fthfl/fl (n = 8) and FthR26Δ/Δ (n = 6), monitored from days 6–8, after the first tamoxifen administration. Data is represented as mean ± SD, from 2 independent experiments. NS: non-significant, **P < 0.01, ***P < 0.001.
2.4. FTH is essential to maintain adipose tissue homeostasis
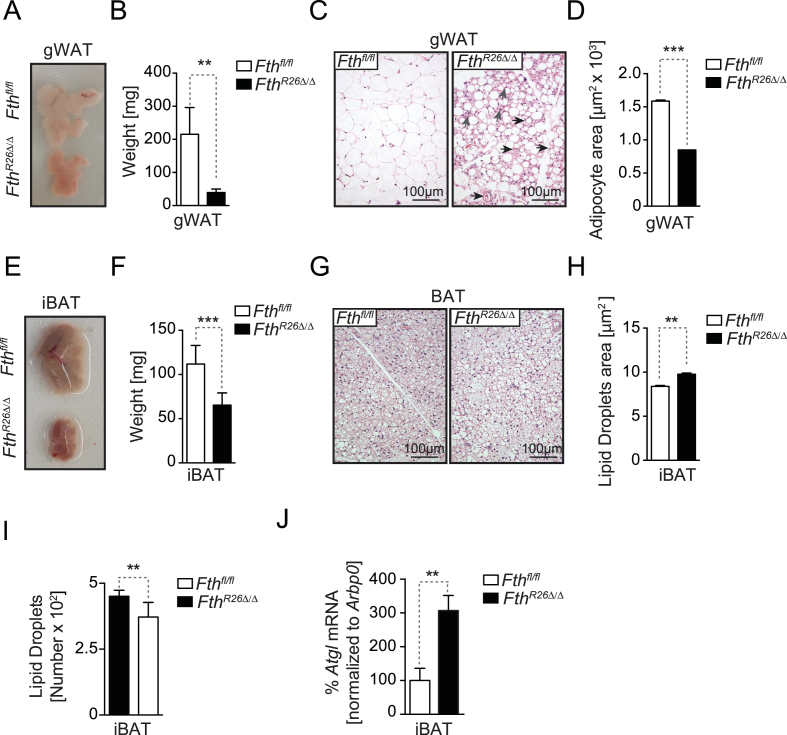
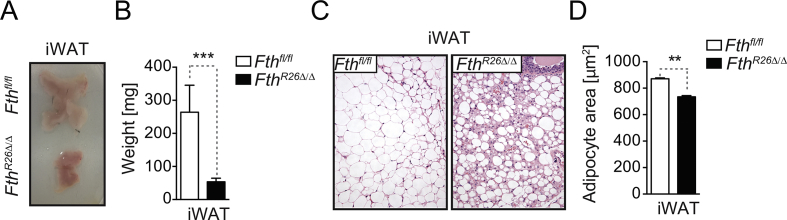
The adipose tissue plays a central role in maintenance of energy balance [34], [35] and regulation of Fe metabolism can impact adipose tissue function [36], for example via mechanisms linked to lipid peroxidation [37]. Therefore, we hypothesized that regulation of Fe metabolism by FTH might impact adipose tissue homeostasis. In strong support of this hypothesis, we found that FthR26Δ/Δ mice had a 75–80% reduction in white adipose tissue (WAT) mass, when compared to control Fthfl/fl mice (Figure 5A,B, S3A,B). This was associated with WAT leukocyte infiltration and the emergence of vascularized structures (Figure 5C, S3C), as well as with decreased adipocyte area (Figure 5D, S3D).
Figure 5.
FTH is essential to maintain adipose tissue homeostasis. (A) Representative picture of gonadal fat (gWAT) pads. (B) Mean of the gWAT gross mass ± SD (n = 7–8, mice per genotype), from 3 experiments with similar trend. (C) Representative H&E staining of gWAT. (D) Mean of gWAT adipocyte area ± SD (n = 3, mice per genotype). (E) Representative picture of interscapular BAT (iBAT). (F) Mean iBAT gross mass ± SD (n = 7–8 per genotype), from 3 experiments with similar trend. (G) Representative H&E staining of iBAT. (H) Mean iBAT lipid droplet area ± SD (n = 3, mice per genotype). (I) Mean iBAT lipid droplet numbers in ± SD (n = 3, mice per genotype). (J) Mean of the relative Atgl mRNA expression ± SD in iBAT (n = 3, mice per genotype) from one experiment. NS: non-significant, **P < 0.01, ***P < 0.001. Mann–Whitney test was used for comparison between two groups. All experiments were performed 7 days after tamoxifen administration.
FthR26Δ/Δ mice had a 40% reduction of brown adipose tissue (BAT) mass, when compared to control Fthfl/fl mice (Figure 5E,F). This was not associated with major histological alterations (Figure 5G) or reduced adipocyte area, despite a slight increase in lipid droplet area (Figure 5H) and a decrease in lipid droplet number (Figure 5I). The BAT from FthR26Δ/Δ mice also showed a marked up-regulation of adipose triglyceride lipase (Atgl) mRNA expression, when compared to control Fthfl/fl mice (Figure 5J). Overall, this suggests that FTH affects lipid mobilization in WAT and BAT, presumably therefore impacting on energy balance.
2.5. FTH is essential to support adaptive thermogenesis
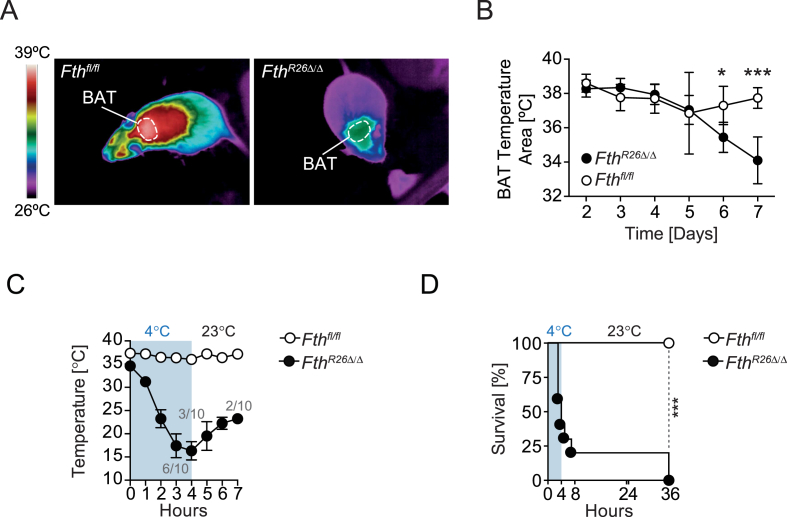
One of the major pathologic outcomes associated with Fth deletion is the loss of thermal homeostasis (Figure 1B). Considering that BAT acts as a thermogenic organ essential to provide adaptive thermoregulation [38], [39], we asked whether FTH impacts on BAT thermogenesis. In strong support of this hypothesis, FthR26Δ/Δ mice showed a marked reduction in BAT temperature, when compared to control Fthfl/fl mice maintained under standard husbandry conditions, i.e. 20–22 °C (Figure 6A,B).
Figure 6.
FTH is essential to support thermogenesis. (A) Representative thermal images at day 7 after Tamoxifen administration. (B) Longitudinal quantification of skin temperature surrounding BAT. (C) Mean rectal temperature ± SD in mice (n = 9–10 per genotype), challenged with environmental cold (4 °C; blue), pooled from 3 independent experiments with similar trend. (D) Survival of Fthfl/fl (n = 10), FthR26Δ/Δ (n = 9) mice challenged as in (C). P values in (B) were calculated using Mann–Whitney test and in (D) using Log-rank (Mantel-Cox) test. *P < 0.05, ***P < 0.001.
We then asked whether FTH is required to provide adaptive thermogenesis in response to cold stress (4 °C). Unlike control Fthfl/fl mice that maintained body temperature and survived when exposed to cold stress, FthR26Δ/Δ mice lost body temperature and succumbed to cold stress (Figure 6C,D). This demonstrates that FTH is essential to maintain BAT thermogenic capacity and sustain organismal bioenergetics, both at steady state and in response to cold stress.
2.6. Regulation of cellular Fe content by FTH is required to support mitochondrial function
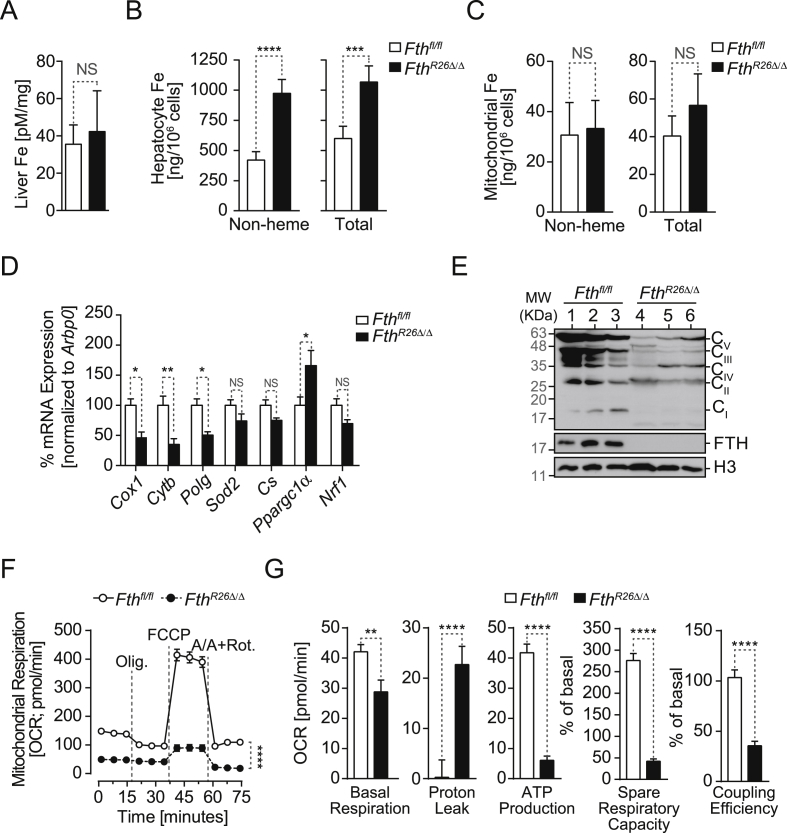
Regulation of cellular Fe metabolism exerts a major impact on mitochondrial function, presumably because mitochondria rely on Fe import to generate Fe-sulfur clusters and heme used by a variety of mitochondrial proteins, including those supporting the electron transport chain (ETC) [2], [40], [41], [42]. We used primary mouse hepatocytes as an experimental model to address whether FTH regulates cellular Fe metabolism in a manner that impacts on mitochondrial function. While the overall liver Fe content was similar in FthR26Δ/Δ and Fthfl/fl mice (Figure 7A), hepatocytes from FthR26Δ/Δ mice had a two-fold higher Fe content, as compared to hepatocytes from Fthfl/fl mice (Figure 7B). This suggests that Fe accumulates in hepatocytes from FthR26Δ/Δ mice probably transiting from another liver cell compartment, most likely Kupffer cells [43].
Figure 7.
Regulation of cellular Fe metabolism by FTH is essential to sustain mitochondrial function in hepatocytes. (A) Fe concentration in the liver of Fthfl/fl (n = 8) and FthR26Δ/Δ (n = 9) mice, 7 days after tamoxifen administration. Data are represented as mean ± SD, pooled from 3 independent experiments. (B) Fe concentration in primary hepatocytes isolated from Fthfl/fl (n = 5) and FthR26Δ/Δ (n = 5), 7 days after tamoxifen administration. Data are represented as mean ± SD, pooled from 3 independent experiments. (C) Mitochondrial Fe concentration in primary hepatocytes isolated from the same mice as in (B). (D) Mean relative mRNA expression ± SD in the liver of Fthfl/fl and FthR26Δ/Δ mice (n = 6–7 per genotype), 7 days after tamoxifen administration. Data pooled from 2 independent experiments with similar trend. (E) Relative expression of ETC complex I (i.e. NADH dehydrogenase), II (i.e. succinate dehydrogenase), III (i.e. subunit Core 2/Uqcrc2), IV (subunit I/Cox1), V (i.e. ATP synthase) proteins and FTH detected by Western Blot in the liver of Fthfl/fl and FthR26Δ/Δ mice (n = 3 per genotype), 7 days after tamoxifen administration. Histone H3 was used as loading control. (F) Oxidative consumption rate (OCR) in hepatocytes isolated from Fthfl/fland FthR26Δ/Δ mice. Data are representative of 3 independent experiments. (G) Basal respiration, proton leak and ATP production, spare respiratory capacity (SRC) and coupling efficiency in the same hepatocytes as in (F). Oligomycin (Olig.); carbonilcyanide p-triflouromethoxyphenylhydrazone (FCCP); Antimycin A/Rotenone (Rot.). Mann–Whitney test was used for comparison between two groups. Data analyzed 7 days after tamoxifen administration. NS: non-significant, ***P < 0.001, ****P < 0.0001.
The mitochondrial Fe content of hepatocytes from FthR26Δ/Δ mice was similar to that of hepatocytes from Fthfl/fl mice (Figure 7C). While this suggests that FTH does not regulate mitochondrial Fe import we asked whether regulation of hepatocyte Fe content by FTH regulates mitochondrial function. In keeping with this notion, expression of mitochondrial genes coding different ETC components, i.e. cytochrome c oxidase I (Cox1), cytochrome b (Cytb; apocytochrome b of ubiquinol:ferricytochrome c oxidoreductase) and polymerase-γ (Polg), was reduced in the liver of FthR26Δ/Δ vs. Fthfl/fl mice (Figure 7D). Expression of nuclear-encoded genes regulating mitochondrial function was similar in the liver of FthR26Δ/Δ vs. Fthfl/fl mice, as illustrated for mitochondrial superoxide dismutase (Sod2) or citrate synthase (Cs), which catalyzes the first reaction of the tricarboxylic acid (TCA) cycle. This was also the case for the transcriptional factor nuclear respiratory factor 1 (Nrf1), which regulates heme biosynthesis and mitochondrial DNA transcription. In contrast, expression of nuclear coded peroxisome proliferator-activated receptor gamma co-activator 1-alpha (Ppargc1α/Pgc1α) mRNA, a master regulator of mitochondrial biogenesis, was higher in the liver of FthR26Δ/Δ vs. control Fthfl/fl mice (Figure 7D).
Expression of mitochondrial ETC proteins in the liver of FthR26Δ/Δ mice was strongly decreased, when compared to control Fthfl/fl mice, as illustrated for nicotinamide adenine dinucleotide (NADH) dehydrogenase (complex I), succinate dehydrogenase (complex II), cytochrome b-c1 complex subunit 2 (Uqcrc2; complex III), cytochrome c oxidase subunit 1 (Cox1; complex IV), and ATP synthase (complex V) (Figure 7E). This suggests that Fe accumulation in hepatocytes from FthR26Δ/Δ mice impacts the expression of mitochondrial as well as nuclear genes that are critical to support mitochondrial function.
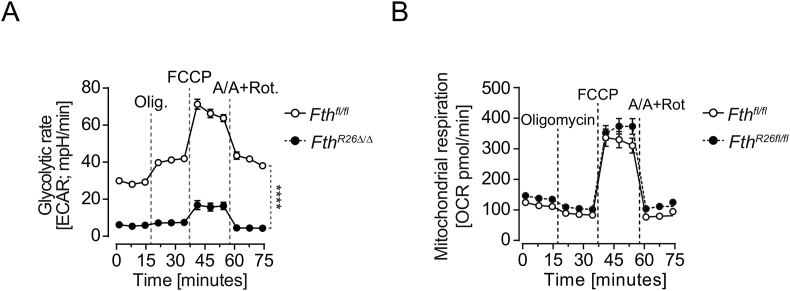
To assess whether FTH regulates mitochondrial function, we monitored oxygen consumption rate (OCR). Hepatocytes from FthR26Δ/Δ mice had lower basal respiration rate, increased proton leakage, and reduced ATP production, respiratory capacity, and coupling efficiency (Figure 7F,G), as well as reduced glycolytic rate (Fig. S4A), when compared to Fthfl/fl hepatocytes. This was not observed in hepatocytes from control Fthfl/fl and FthR26fl/fl mice not receiving tamoxifen (Fig. S4B). Taken together, these observations show that regulation of Fe metabolism by FTH is essential to support mitochondrial function in hepatocytes.
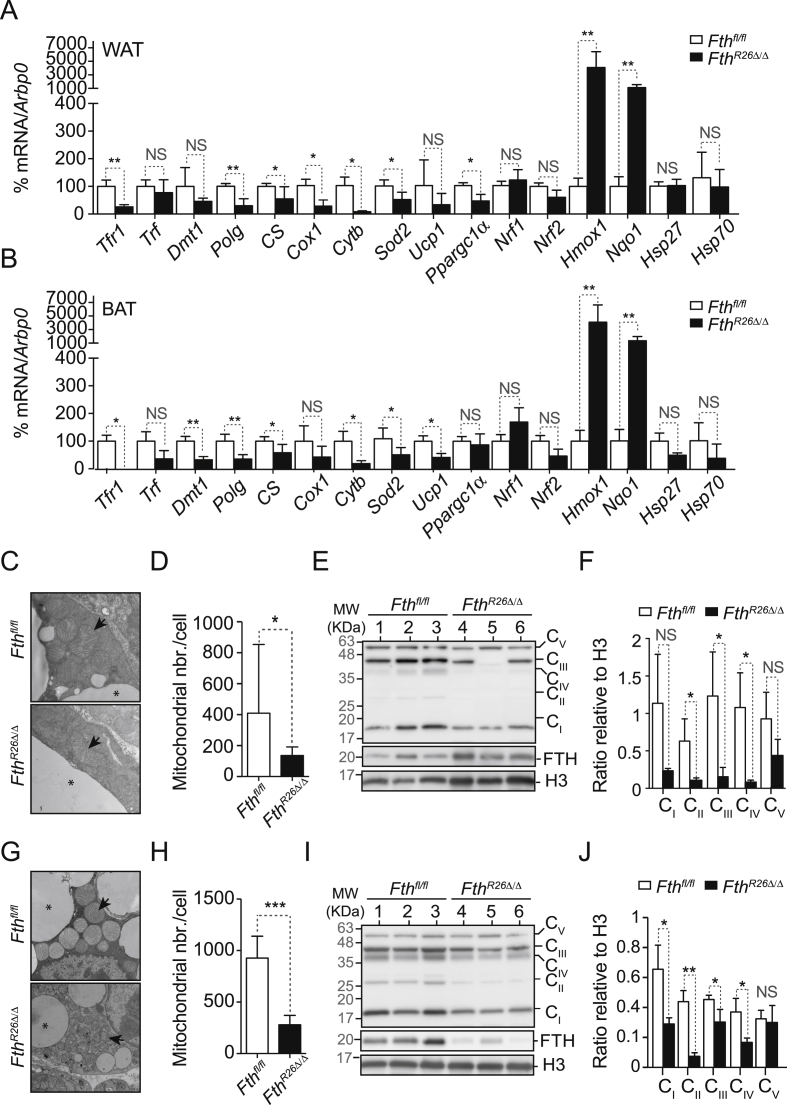
We then asked whether FTH regulates mitochondrial function in other tissues, such as the WAT and BAT, which play a critical role in supporting organismal energy expenditure and thermoregulation. The WAT from FthR26Δ/Δ mice had lower Tfr1, but not Dmt1 mRNA expression, as compared to the WAT from control Fthfl/fl mice (Figure 8A), while both genes were reduced in the BAT from FthR26Δ/Δ vs. control Fthfl/fl mice (Figure 8B).
Figure 8.
FTH is essential to sustain mitochondrial function in fat. Mean relative mRNA expression ± SD in (A) WAT and (B) BAT from Fthfl/fl (n = 5) and FthR26Δ/Δ mice (n = 7) mice, 7 days after tamoxifen treatment. Data are represented as mean ± SD, pooled from 2 independent experiments. (C) Representative electron micrograph of inguinal WAT from FthR26Δ/Δ and Fthfl/fl mice on day 7 post tamoxifen administration. (D) Relative number of mitochondria in inguinal WAT from Fthfl/fl (n = 8) and FthR26Δ/Δ (n = 11) mice on day 7 after tamoxifen administration. Data are represented as mean ± SD, pooled from 3 independent experiments. (E) Relative expression of ETC complex I (i.e. Nadh dehydrogenase), II (i.e. succinate dehydrogenase), III (i.e. subunit Core 2/Uqcrc2), IV (subunit I/Cox1), V (i.e. ATP synthase) proteins and FTH, detected by western blot in WAT from Fthfl/fl and FthR26Δ/Δ mice (n = 3 per genotype), 7 days after tamoxifen administration. Histone H3 was used as loading control. (F) Quantification of different ETC complex proteins detected in (E). Data are represented as mean ± SD. (G) Representative electron micrograph of the BAT from FthR26Δ/Δ and Fthfl/fl mice on day 7 after tamoxifen administration. (H) Relative number of mitochondria in BAT from Fthfl/fl (n = 10) and FthR26Δ/Δ (n = 11) mice. Data represented as mean ± SD, pooled from 3 independent experiments. (I) Relative expression of ETC complex I (i.e. Nadh dehydrogenase), II (i.e. succinate dehydrogenase), III (i.e. subunit Core 2/Uqcrc2), IV (subunit I/Cox1), V (i.e. ATP synthase) proteins, and FTH detected by western blot in the BAT of Fthfl/fl and FthR26Δ/Δ mice (n = 3 per genotype), 7 days after tamoxifen administration. Histone H3 was used as loading control. (J) Quantification of different ETC complex (C) proteins, detected in (I). Data are represented as mean ± SD. Mann–Whitney test was used for comparison between two groups. NS: non-significant. *P < 0.05, **P < 0.01, ***P < 0.001. Arrows in (C) and (G) highlight mitochondria and * highlights fat reservoir.
Expression of mitochondrial Polg, Cs, Cox1, Cytb and Sod2 mRNA was reduced in the WAT from FthR26Δ/Δ vs. control Fthfl/fl mice (Figure 8A,B). This was also observed in the BAT, with the exception of Cox1 (Figure 8B).
We then asked whether Fth impacts on the expression of uncoupling protein 1 (UCP1), a nuclear-encoded gene highly expressed in the BAT, which dissipates energy produced by the ETC, in the form of heat [38]. Expression of Ucp1 mRNA was markedly reduced in BAT from FthR26Δ/Δ vs. Fthfl/fl mice (Figure 8B). Expression of Ucp1 mRNA in WAT was similar in FthR26Δ/Δ vs. Fthfl/fl mice (Figure 8A). Expression of Ppargc1α/Pgc1α mRNA, a nuclear encoded gene controlling mitochondrial biogenesis, was repressed in WAT (Figure 8A) but not in BAT (Figure 8B) from FthR26Δ/Δ vs. control Fthfl/fl mice. This was not the case for other nuclear encoded genes controlling mitochondrial function, such as Nrf1 or Nrf2 (Figure 8A,B). However, the expression of Nrf2-regulated Hmox1 or NAD(P)H Quinone Dehydrogenase 1 (Nqo1) mRNA was highly induced in WAT and BAT from FthR26Δ/Δ vs. control Fthfl/fl mice (Figure 8A,B). This was not observed for mRNA encoding heat shock proteins, such as heat shock protein 27 (Hsp27) and 70 (Hsp70) (Figure 8A,B). This suggests that regulation of Fe metabolism by FTH impacts the expression of mitochondrial and nuclear genes supporting mitochondrial function in WAT and BAT.
While the morphological structure of the mitochondria in WAT adipocytes from FthR26Δ/Δ mice was indistinguishable from that of controls Fthfl/fl mice (Figure 8C), the number of mitochondria per cell in the WAT from FthR26Δ/Δ was decreased, when compared to control Fthfl/fl mice (Figure 8D). This was corroborated by a marked decrease in the expression of mitochondrial ETC proteins in the WAT from FthR26Δ/Δ vs. control Fthfl/fl mice, as illustrated for the complex II component succinate dehydrogenase, Uqcrc2 in complex III and Cox1 in complex IV (Figure 8E,F). Albeit not significant, a similar trend was observed for the complex I component Nadh dehydrogenase and the complex V component ATP synthase (Figure 8E,F).
In contrast to mitochondria in the WAT, mitochondria in the BAT from FthR26Δ/Δ mice had a profoundly disrupted morphological structure, as compared to control Fthfl/fl mice (Figure 8G). Namely, mitochondria in the BAT from FthR26Δ/Δ mice had smaller size and irregular shape, containing fewer cristae and lower matrix density, as compared to Fthfl/fl mice where BAT mitochondria were abundant, large sized and in most cases spherical with large parallel cristae and a dense matrix (Figure 8G). Mitochondria in the BAT from FthR26Δ/Δ mice also had primary and secondary lysosomes with dense bodies and inclusions (Figure 8G), suggesting that autophagy is a conspicuous event. In keeping with this notion, the number of mitochondria per cell was markedly reduced in the BAT from FthR26Δ/Δ vs. control Fthfl/fl mice (Figure 8H). Accordingly, the expression of mitochondrial ETC proteins, including Nadh dehydrogenase (complex I), succinate dehydrogenase (complex II), Uqcrc2 (complex III), and Cox1 (complex IV) were also reduced, while this was not the case for complex V ATP synthase (Figure 8I,J).
Overall, these observations suggest that regulation of Fe metabolism by FTH is strictly required to preserve the mitochondrial function in WAT and BAT, which should contribute to explain why FTH exerts such a major impact on organismal energy balance and thermogenesis.
3. Discussion
Analyses of loss-of-function mutations or deletion of genes regulating Fe metabolism in animals have proven invaluable to gain further understanding of how Fe metabolism is regulated at an organismal level [2], [44]. Because most Fe-regulatory genes are evolutionarily conserved, this approach informs critically on how orthologous human genes impact on human Fe metabolism [2], [44]. This same approach, however, had not been used to determine whether ferritin regulates organismal Fe metabolism, initially due to the embryonic lethality associated with constitutive Fth deletion in mice [8]. Several studies used tissue-specific Fth deletion instead, to infer a rather minor impact on organismal Fe homeostasis in mice [20], [22], [23], [24]. This is, however, at odds with a large body of literature suggesting that ferritin acts as a central regulator of cellular Fe metabolism [2], [44]. A possible explanation for this apparent discrepancy may be that a significant proportion of ferritin is secreted [16], [25]. It is possible therefore, that extracellular ferritin compensates for the pathophysiologic impact of Fth deletion in specific cell compartments. Consistent with this notion we found that global Fth deletion in adult mice leads to a profound disruption of organismal Fe homeostasis (Figure 1).
The observation that global Fth deletion leads to the development of oxidative stress in different organs suggests that ferritin is a critical regulator of organismal redox homeostasis, preventing oxidative organ damage (Figure 2A–C) and, therefore, presumably ensuing systemic inflammation (Figure 3). Whether deregulation of redox homeostasis, associated with Fth deletion, is owed to unfettered ROS production via the Haber–Weiss/Fenton reactions [1], [2], to dysfunctional mitochondrial function (Figure 7, Figure 8), or to both remains to be established.
Deletion of genes regulating organismal Fe metabolism in animals is most often associated with the development of anemia [2], [32], [44], which is not the case for Fth deletion (Fig. S1C). One possible explanation for this is that deregulation of Fe metabolism upon Fth deletion impacts on vital functions leading to the rapid onset of lethality (Figure 1), before overt anemia is allowed to develop (Fig. S1C). In strong support of this notion Fth deletion leads to a severe impairment of energy expenditure (Figure 4) and thermal homeostasis (Figure 4), presumably due to loss of mitochondrial function in liver, WAT, and BAT (Figure 7, Figure 8). This is reminiscent of impaired thermoregulation caused by nutritional Fe-deficiency in rodents [26] and humans [27], [28], [29], suggesting that Fth deletion causes a “functional state” of Fe-deficiency, despite adequate Fe supply through diet (Figure 1). Presumably, ferritin is required to couple nutritional Fe supply with organismal Fe availability, supporting energy balance and thermogenesis. The question then becomes: How does ferritin impact on energy balance and thermal homeostasis?
One possible explanation is that ferritin regulates Fe metabolism in a manner that impacts mitochondrial function in organs controlling energy balance and thermal homeostasis, such as the liver (Figure 7), WAT and BAT (Figure 8). This is consistent with previous observations showing that other master regulators of cellular Fe metabolism, such as IRPs, control mitochondrial integrity and function [41]. In contrast to IRPs [41], however, ferritin does not regulate mitochondrial Fe content (Figure 7A–C). Instead, it controls cellular redox homeostasis, likely preventing oxidative mitochondrial damage (Figure 8) [42]. This is particularly relevant in the liver as well as in the BAT, where mitochondria sustain thermogenesis (Figure 6) [38].
Another possible explanation for how ferritin impacts on energy balance and thermal homeostasis it that this occurs via regulation of WAT function, as suggested by the profound WAT atrophy associated with Fth deletion (Figure 5A–C). While this is in keeping with a proposed functional relationship between Fe and fat metabolism [36], [45], whether regulation of Fe metabolism by ferritin acts directly or indirectly on WAT is not clear.
It is possible that regulation of Fe metabolism by ferritin impacts on WAT lipolysis, consistent with the induction of Atgl in WAT from Fth-deleted mice (Figure 5J). One of the mechanisms via which WAT lipolysis might be induced is through the production of norepinephrine in Fth-deleted mice, a lipolytic catecholamine released by the sympathetic nervous system [46]. Consistent with this notion, nutritional Fe deficiency is associated with norepinephrine accumulation in blood and urine [26], as well as with loss of adaptive thermoregulation [26], [28], [29].
The consequences of Fth deletion are likely multi-factorial and it is therefore difficult to disentangle the causes and consequences of the lethal outcome associated with Fth deletion. While the data obtained demonstrate that FTH impacts a number of vital homeostatic parameters, it is likely that other cellular functions not addressed in this study may also be affected, impacting organismal homeostasis.
In conclusion, we unveiled an unsuspected functional relationship between organismal Fe metabolism, energy expenditure, and adaptive thermoregulation with implications to the current understanding of energy expenditure and thermoregulation in mammals. Further dissection of the molecular mechanisms regulating this metabolic crosstalk should uncover molecular targets for the treatment of diseases associated with metabolic deregulation.
4. Experimental models
4.1. Animals
R26CreERT2Fthfl/fl mice were generated by intercrossing Fthfl/fl mice (obtained originally from Prof. Lukas Kuhn; ETH, Switzerland) [23] with Rosa26CreERT2 (Jackson Laboratory; stock 008463). OKD48 transgenic mice (RIKEN BioResource Center) were crossed with Fthfl/fl, R26CreERT2 and R26CreERT2Fthfl/fl to generate OKD48Fthfl/fl, OKD48R26CreERT2 and OKD48R26CreERT2Fthfl/fl, respectively. Germ free R26CreERT2Fthfl/fl mice were generated by embryo transfer into the germ-free facility of the Instituto Gulbenkian de Ciência. All animal protocols were approved by the Instituto Gulbenkian de Ciência ethical committee and the “Órgão Responsável pelo Bem-estar dos Animais (ORBEA).” These were consequently licensed by the Direccão Geral de Alimentação e Veterinária (DGAV). All experimental procedures follow the Portuguese (Portaria n° 1005/92, Decreto-Lei n° 113/2013) and European (Directive 2010/63/EU) legislations, concerning housing, husbandry and animal welfare. MRI experiments were approved by a governmental committee on animal welfare and were performed according to the national animal protection guidelines (North Rhine-Westphalia Agency for Nature, Environment, and Consumer Protection: 84–02.04.2012.A293).
4.2. Primary hepatocytes
Isolation of primary hepatocytes from livers of fed mice was performed essentially as described [47]. Livers were perfused in anesthetized mice (i.p. Ketamine/Xylazine). Briefly, a peritoneal incision was performed to expose the inferior vena cava and hepatic portal vein, a 23G butterfly needle connected to a peristaltic pump (MINIPULS 3, Gilson) was inserted into the inferior vena cava, and the hepatic portal vein was cut open. Livers were perfused (25 mL Liver Perfusion buffer; Gibco™), digested (40 mL collagenase digestion buffer; HBSS, 6.67 mM CaCl2, 50 mM HEPES and 240 ng/mL Type IV collagenase; sigma), collected, dissected free of the hepatic serous membrane, and stirred in liver perfusion medium to suspend the parenchymal cells. Cell suspension was filtered (70 μM strainer) and centrifuged (50×g, 2 min, RT). The supernatant was discarded, hepatocytes were washed (2 × 25mL William's E medium, 5% FBS, 1% Penicillin-Streptomycin) and centrifuged (50×g, 2 min). Hepatocyte purity was typically >98% as assessed via flow cytometry (CD45-, Albumin+; data not shown). Hepatocytes were plated onto collagen-coated Seahorse XF96 plates (7000 cells/well) in 180 μL William's E medium (5% FBS, 1% Penicillin-Streptomycin). Plates, were coated using collagen coating solution (70 μL; 20 mM acetic acid, 50 μg/mL rat tail collagen I; Thermo Fischer scientific) incubated (37 °C, 1 h), washed (3 x PBS) and air-dried.
4.3. Inducible gene deletion
Conditional deletion of the Fthfl/fl allele in R26CreERT2FthΔ/Δ mice was achieved at 7–9 weeks of age by tamoxifen (Sigma–Aldrich, Sintra, Portugal) gavage (225 mg/kg BW in 100 μL Corn Oil, 5% EtOH; 3 times every other day). Weight and temperature were monitored daily (Rodent Thermometer BIO-TK8851, Bioseb, France). Fth deletion in germ-free R26CreERT2FthΔ/Δ mice was achieved using sterile tamoxifen food (Sniff) with control R26CreERT2FthΔ/Δ maintained under specific pathogen free conditions also receiving tamoxifen food.
4.4. qRT-PCR
RNA was isolated from organs using the NucleoSpin RNA kit (Macherey–Nagel). Briefly, mice were euthanized and perfused (PBS; ice cold) by cardiac puncture. Organs were collected into Eppendorf tubes and snap frozen in liquid nitrogen. For RNA isolation organs were disrupted using a mortar and pestle under constant freezing in liquid nitrogen. Disrupted tissues were re-suspended in Trizol and passed 10 times through a 23 g needle and appropriate volume of chloroform was added (200uL per mL of Trizol). After centrifugation, the aqueous phase was transferred to the column of the above-mentioned kit and processed according to the manufacturer instructions. cDNA was transcribed from total RNA with transcriptor first strand cDNA synthesis kit (Roche). Quantitative real-time PCR (qRT-PCR) was performed using 1 μg cDNA and SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA) in duplicate on a 7500 Fast Real-Time PCR System (Applied Biosystems) under the following conditions: 95 °C/10 min, 40 cycles/95 °C/15 s, annealing at 60 °C/30 s, and elongation 72 °C/30 s. Primers were designed using Primer Blast [48] and are listed in the Table 1.
Table 1.
Key resource table.
| Reagent or Resource | Source | Identifier | |
|---|---|---|---|
| Mouse Strains | |||
| B6.C57BL/6 Fthfl/fl | Lukas Kuhn, ETH, Switzerland | [23] | |
| B6.129 Gt(ROSA)26Sortm1(cre/ERT2)Tyj/J (ROSA26CreERT2) | Jackson Laboratory | JAX stock: 008463 | |
| B6.C57BL/6 OKD48 | RIKEN BioResource Center | RBRC05704 [33] | |
| Critical Commercial Kit Assays | |||
| NucleoSpin RNA kit | Macherey & Nagel | 740955 | |
| Transcriptor first strand cDNA synthesis kit | Roche | ||
| MILLIPLEX MAP Mouse Cytokine/Chemokine Magnetic Bead Panel - Premixed 25 Plex - Immunology Multiplex Assay | Millipore (Merk) | MCYTOMAG-70K-PMX | |
| Free Glycerol Reagent (F6428); Glycerol Standard Solution |
Sigma | G7793 | |
| Free Fatty Acid Quantification Kit | Sigma | MAK044 | |
| Seahorse XF Cell Mito Stress Test Kit | Agilent Technologies | 103015–100 | |
| Mitochondria Isolation Kit for Cultured Cells | Thermo Scientific™ | 89874 | |
| Ferritin (Mouse) ELISA Kit | Abnova | KA1941 | |
| Chemicals | |||
| Tamoxifen® | Sigma–Aldrich | T5648 | |
| Corn Oil | Sigma–Aldrich | C8267 | |
| Tamoxifen Food | Sniff | ||
| Luciferin | Promega | PROME1605 | |
| Antibodies (Company/Clone, Ref) | |||
| CD11b FITC | BD Biosciences | M1/70, 553310 | |
| Ly6G PE | BD Biosciences | 1A8, 551461 | |
| CD45 APC-Cy7 | Invitrogen | 30-F11, 4331936 | |
| CD16/CD32 | BioLegend | 93, 101331 | |
| FTH1 | Cell Signaling | D1D4, #4393 | |
| Histone H3 | Cell Signaling | 9715 | |
| GAPDH | SciGen | AB0049-200 | |
| MitoProfile® OXPHOS WB |
Abcam |
ab110413 |
|
| Oligonucleotides |
Sequences |
||
| Arbp0 Fwd | 5′ CTTTGGGCATCACCACGAA 3′ | ||
| Arbp0 Rev | 5′ GCTGGCTCCCACCTTGTCT 3′ | ||
| Fth Fwd | 5′ CCATCAACCGCCAGATCAAC 3′ | ||
| Fth Rev | 5′ GCCACATCATCTCGGTCAAA 3′ | ||
| Hmox1 Fwd | 5′ TGACACCTGAGGTCAAGCAC 3′ | ||
| Hmox1 Rev | 5′ TCTCTGCAGGGGCAGTATCT 3′ | ||
| Hamp1 Fwd | 5′GAGAGACACCAACTTCCCCA 3′ | ||
| Hamp1 Rev | 5′TCAGGATGTGGCTCTAGGCT 3′ | ||
| Fpn Fwd | 5′TGCCTTAGTTGTCCTTTGGG 3′ | ||
| Fpn Rev | 5′GTGGAGAGAGAGTGGCCAAG 3′ | ||
| Tfr1 Fwd | 5′GTTTTTGTGAGGATGCAGACTATCC 3′ | ||
| Tfr1 Rev | 5′GCTGAGGAACTTTCTGAGTCAATG 3′ | ||
| Trf Fwd | 5′TGTGACCTGTGTATTGGCCC 3′ | ||
| Trf Rev | 5′GCAGGGTTCTTTCCTTCGGT 3′ | ||
| Dmt1 Fwd | 5′ GCAGTGGTTAGCGTGGCTTATT 3′ | ||
| Dmt1 Rev | 5′ AGACAGACCCAATGCAATCAAA 3′ | ||
| Cox1 Fwd | 5′ TTCGGAGCCTGAGCGGGAAT 3′ | ||
| Cox1 Rev | 5′ ATGCCTGCGGCTAGCACTGG 3′ | ||
| Cytb Fwd | 5′ CTTAGCCATACACTACACATCAG 3′ | ||
| Cytb Rev | 5′ ATCCATAATATAAGCCTCGTCC 3′ | ||
| Ucp1 Fwd | 5′ ACTGCCACACCTCCAGTCATT 3′ | ||
| Ucp1 Rev | 5′ CTTTGCCTCACTCAGGATTGG 3′ | ||
| Ppargc1a Fwd | 5′ CCCTGCCATTGTTAAGAC 3′ | ||
| Ppargc1a Rev | 5′ TGCTGCTGTTCCTGTTTTC 3′ | ||
| Nrf1 Fwd | 5′ CCAGVAAGTCCAGCAGGTCC 3′ | ||
| Nrf1 Rev | 5′ TTCCCTGTTGCCACAGCAGC 3′ | ||
| Polg Fwd | 5′ GCCCCACTGTAGAATCCGCTG 3′ | ||
| Polg Rev | 5′ AGCAGCAGGCAGAACTAGAGG 3′ | ||
| Sod2 Fwd | 5′ CCCAAAGGAGAGTTGCTGGAGG 3′ | ||
| Sod2 Rev | 5′ GCTCCCACACGTCAATCCCC 3′ | ||
| Cs Fwd | 5′ TGGGGTGCTGCTCCAGTACTAT 3′ | ||
| Cs Rev | 5′ AGTCTTAAAGGCCCCTGAAACAA 3′ | ||
| AtgL Fwd | 5′ TGGTTCAGTAGGCCATTCCT 3′ | ||
| AtgL Rev | 5′ CACTTTAGCTCCAADDATGA 3′ | ||
| Hk2 Fwd | 5′ GCCAGCCTCTCCTGATTTTAGTGT3′ | ||
| Hk2 Rev | 5′ GGGAACACAAAAGACCTCTTCTGG 3′ | ||
| Nd1 Fwd | 5′ CTAGCAGAAACAAACCGGGC 3′ | ||
| Nd1 Rev | 5′ CCGGCTGCGTATTCTACGTT 3′ | ||
| Reagents | |||
| Liver Perfusion buffer | Gibco® | 17701–038 | |
| Type IV collagenase | Sigma | C5138 | |
| Type D Collagenase | Roche | 11088866001 | |
| William's E medium | Gibco® | 32551–087 | |
| Rat Tail Collagen I | Gibco® | A1048301 | |
| SuperSignal™ West Pico PLUS Chemiluminescent Substrate | ThermoFisher Scientific | 34577 | |
4.5. In vivo luciferase assay
Luciferase activity was measured in live anesthetized (Ketamine/Xylazine, i.p.) mice, essentially as described [49]. Briefly, the abdomen was shaved, and mice received luciferin (2mg/mouse in 100 μL PBS, i.v.). Luciferase signal was acquired in a Hamamatsu Aequoria, equipped with an electron multiplying CCD (EMCCD) camera, used at highest sensitivity (255) and maximum gain (5) for 10, 30, 60, 120, and 240 s. Signal quantification was performed using Fiji software (ImageJ).
4.6. Fe quantification in organs
Spleen samples were dried (24 h, 99 °C), dissolved (3M HCl, 10% trichloracetic acid; TCA; overnight at 65 °C) and diluted (10 μL) in water (590 μL). β-mercaptoethanol (10 μL), sodium acetate (pH 4.5; 500 μL) and bathophenanthroline-disulfonic acid (80 μL) were added (37 °C, 1 h) and absorbance (λ = 535 nm) was measured using a microplate reader (Bio-Rad 3550-UV). Magnetic Resonance Imaging was used to detect signal cancellation induced by iron and to quantify the corresponding T2 relaxation time. Briefly, longitudinal MRI was performed on days 3–4 and 8–9, after the first tamoxifen administration. Images were acquired at 9.4 T on a Bruker BioSpec 94/20 system (Ettlingen, Germany) equipped with a 0.7 T/m gradient system and ParaVision 5.1 operating software (Bruker BioSpin, Ettlingen, Germany). A 72-mm volume resonator coil was used for signal transmission, while a 4 channel receive coil array was used for signal reception. During MRI, the animals were anesthetized with isoflurane (1.5–2.5% isoflurane and 0.7/0.3 air/O2 mixture), and core body temperature and respiration rates were monitored using an MRI compatible monitoring system (SA Instruments, Stony Brook, NY). Respiratory-gated anatomical MRI of the abdomen was performed in axial view using a self-gated cine FLASH sequence (IntraGate, Bruker BioSpin MRI, Ettlingen, Germany) with the following parameters: TR: 8 ms, TE: 2.9 ms, FA: 10°, number of repetitions 100, FOV: 30 × 30 mm2, Matrix: 256 × 256, slice thickness: 1 mm. T2 relaxation maps were acquired in axial view, using a multi-spin multi-echo sequence (MSME; TR: 2500 ms, 30 echoes, FOV: 30 × 30 mm2, matrix: 128 × 128, averages: 2, slice thickness: 1.5 mm) to determine corresponding T2 relaxation times.
4.7. Mitochondria isolation
Mitochondria were isolated from purified mouse hepatocytes using the Mitochondria Isolation Kit for Cultured Cells (ThermoFisher Scientific), according to manufacturer instructions and using the chemical disruption option for cell membrane lysis. Mitochondria isolations were performed in duplicate for each set of purified hepatocytes, with 3 × 106 cells used per mitochondrial isolation. Isolated mitochondria were immediately snap frozen in liquid nitrogen and kept at −20 °C prior to further use.
4.8. Cellular and mitochondrial Fe quantification
Non-heme and total liver and mitochondrial iron levels were measured as previously described [50], [51], with the following modifications. Briefly, cell pellets or isolated mitochondria from 3 × 106 primary mouse hepatocytes were resuspended (200 μl PBS) split (100 μL duplicates) and hydrolyzed (50 μl 26% HCl, 1,8M trichloroacetic acid; 65 °C; overnight). To measure total iron, one of hydrolyzed samples was boiled (120 °C, 1 h) to release heme-iron under acidic conditions. Both samples were clarified by centrifugation (3.000 g) and clarified sample (60 μL) was transferred to a 96-well plate and incubated (5min. RT) in 160 μL of 3.8M sodium acetate, 575 μM bathophenanthroline disulfonic acid, and 2.5 mM ascorbic acid. Absorbance was measured at λ539nm and iron concentration was calculated against standard samples containing known concentrations of iron, and verified using the formula , where As is the sample absorbance, Ab is the blank absorbance, V is the reaction volume (0.22), MW is the molecular weight of iron (56 g·mol−1), e is the milimolar absorptivity of bathophenanthroline disulfonic acid (22.14 mM−1·cm−1), () is the path length (0.6 cm) and c is the number of cells used (3 × 106). Each mitochondrial isolation was performed in technical duplicates and the measured values averaged before analysis.
4.9. Western blot
Proteins were extracted, electrophoresed, and electrotransferred essentially as described (Gozzelino et al., 2012). Briefly, organs were collected after euthanasia and perfusion (ice cold PBS). Organs were snap frozen in liquid nitrogen. For protein extraction tissues were extracted in RIPA buffer using a tissue douncer kit, sonicated and centrifuged. Supernatant was collected and total protein was quantified using Bradford assay. Anti-FTH1 (clone D1D4; 1:1000; Cell Signaling) and anti-β-actin (Sigma; A5441, 1:5000) were detected using peroxidase conjugated secondary antibodies (1 h; RT) and developed with SuperSignal™ West Pico PLUS Chemiluminescent Substrate (ThermoFisher Scientific). For detection of ETC complexes, samples were processed according to manufacture instructions (Mitosciences, Abcam). Briefly, livers, WAT and BAT were directly lysed in 2x SDS page sample buffer (20% glycerol, 4% SDS, 100 mM Tris pH6.8, 0.002% bromophenol blue, 100 mM dithiothreitol) and homogenized in a tissue lyser (Qiagen) using tungsten carbide beads. Samples were then sonicated, heated (10min; 50 °C) to ensure preservation of mitochondrial complexes, and centrifuged. Supernatant was collected and total protein was quantified using NanoDrop™ 1000. MitoProfile® Total OXPHOS Rodent WB Antibody Cocktail (Mitosciences, Abcam, Catalog ab110413; 1:1000), anti-FTH1 (Cell Signaling; clone D1D4; 1:1000), anti-Histone H3 (Cell Signaling; catalog 9715; 1:1000) were detected using peroxidase conjugated secondary antibodies (1 h; room temperature) and developed with SuperSignal™ West Pico PLUS Chemiluminescent Substrate (ThermoFisher Scientific). For WAT and BAT, western blots were developed using Amersham Imager 680 (GEHealthcare), equipped with a Peltier cooled Fujifilm Super CCD. Densitometry analysis was performed with ImageJ (Rasband, W.S., ImageJ, U.S. NIH, Bethesda, Maryland, USA, https://imagej.nih.gov/ij/,1997-2014), using images without saturated pixels.
4.10. Serology
Mice were euthanized using CO2 inhalation, and blood was collected by cardiac puncture. Blood was transferred into an EDTA containing tube for hemogram analyses or heparin tubes for serology (ALT, AST, Urea, Creatinin, Creatinine Kinase, LDH, Iron, Transferrin and Transferrin saturation), performed by DNAtech (Lisbon). Ferritin levels in Plasma were determined with the Ferritin (Mouse) ELISA kit from Abnova. Briefly, mice were euthanized and blood collected by cardiac puncture using a heparinized syringe. Blood samples were stored 30 min in the fridge and subsequently centrifuged at 2000g, 10 min, 4 °C. The plasma was then collected into a fresh 1.5 mL Eppendorf tube and stored at −80 °C for further analysis. For ferritin detection samples were diluted 1:40 (Fthlox/lox mice) and 1:4000–1:5000 (R26FthΔ/Δ mice) and the assay was conducted according to the manufacturer's instructions. Absorbance λ600 were measured on microplate spectrophotometer (Thermo Scientific™ Multiskan™ GO). Plasma cytokine levels (IL-1α, IL-1β, IL-6, TNF-α, IL-10, IL-12, IP-10, KC, MCP1, MIP-1α, MIP-1β, MIP2, VIG, RANTES, VEGF) were determined by using the MILLIPLEX® MAP magnetic bead-based multi-analyte panels according to the manufacturer instructions and analyzed on a MAGPIX® system. Data analysis was performed by MILLIPLEX® Analyst 5.1 software.
4.11. Leukocyte isolation and flow cytometry
To quantify the numbers of tissue resident leukocytes organs were perfused and digested (Collagenase D; 1 mg/mL and DNase I; 10 μg/mL). Briefly, liver, kidney, lung and heart were cut into small pieces, digested in 10 mL digestion medium (HBSS supplemented with Proteinase and DNase) under shaking (220 rpm, 37 °C for 45 min), passed through a cell strainer (100 μM), and washed with 5 mL of medium (5 mL RPMI complete medium; 10% FCS, PenStrep, l-Glutamine, HEPES, β-mercaptoethanol). Cells were pelleted (300 g, 4 °C, 10 min), and RBC were lyzed (5 mL RBC lysis buffer; 5 min, RT). Lysis was stopped by adding 5 mL of medium, and cells were passed through a 40 μM cell strainer, centrifuged (300 g, 4 °C, 10 min) and re-suspended in 3 mL (liver and kidney) or 1 mL (heart and lung) medium. Leukocytes were stained with anti-CD45, CD11b, and Ly6G monoclonal antibodies. Cell acquisition was performed using either a CyanADP (Beckman Coulter) or a BD LSRFortessa X-20 (BD Biosciences) flow cytometer. Samples were analyzed using FlowJo software.
4.12. Histology
Organs were harvested, fixed in 10% formalin, embedded in paraffin, sectioned into 3 μm-thick sections, and stained with Hematoxylin and Eosin (H&E). Whole sections were analyzed and images acquired with a Leica DMLB2 microscope (Leica) and NanoZoomer-SQ Digital slide scanner (Hamamatsu). For WAT adipocytes area and BAT lipid droplets measurements tissues were processed to paraffin-embedded sections (3 μm sections), stained with H&E and scanned into digital images (NanoZoomer-SQ Digital slide scanner -Hamamatsu). The average WAT adipocyte size in adipose tissue sections [expressed as the average cross-sectional area per cell (μm2)] was determined using Fiji software, as described elsewhere [52]. Briefly, a slide scanned picture was captured at 2.5x. An average of 1500 adipocytes were measured per sample (n = 3 mice). The following macro was applied: run (“Set Scale...”, “distance = 560 known = 250 pixel = 1 unit = um global”); run (“Duplicate...”, “ ”); run (“Subtract Background...”, “rolling = 50 light separate sliding”); run (“Despeckle”); run (“8-bit”); setAutoThreshold(“Mean dark”);//run (“Threshold...”);//setThreshold(250, 255); setOption(“BlackBackground”, false); run (“Convert to Mask”); run (“Make Binary”); run (“Dilate”); run (“Close-”); run (“Invert”); run (“Analyze Particles...”, “size = 330–15000 circularity = 0.50–1.00 display exclude clear summarize add”). The average BAT lipid droplet size [expressed as the average cross-sectional area per lipid droplet (μm2)], and the number of lipid droplets were quantified in 3 non-overlapping 40x fields for each mouse (n = .3 mice). An average of 11000 lipid droplets per mouse were measured. The following macro was applied: run (“Set Scale...”, “distance = 452 known = 100 pixel = 1 unit = um global”); run (“Duplicate...”, “ ”); run (“Subtract Background...”, “rolling = 50 light separate sliding”); run (“Despeckle”); run (“8-bit”); setAutoThreshold(“Mean dark”);//run (“Threshold...”);//setThreshold(245, 255); setOption(“BlackBackground”, false); run (“Convert to Mask”); run (“Make Binary”); run (“Dilate”); run (“Close-"); run (“Invert”); run (“Watershed”); run (“Analyze Particles...”, “size = 1–5000 circularity = 0.4–1.00 display exclude clear summarize add”).
4.13. Calorimetric system
Animals were analyzed for EE, RQ, LA, and FI using a calorimetric system (LabMaster; TSE Systems; Bad Homburg, Germany). Animals were placed in a temperature-controlled (24 °C) box through which air was pumped. After calibrating the system with the reference gases (20.9% O2, 0.05% CO2 and 79.05% N2), metabolic rate was measured for 6 days, as previously described [53], [54]. Energy expenditure (EE), respiratory coefficient (RQ), locomotor activity (LA), and food intake (FI9 were recorded every 30 min). Animals were placed for adaptation for 2 days before starting the measurements. Animals were anesthetized and no other special preparation was performed before measurements.
4.14. BAT temperature measurements
Skin temperature surrounding BAT was recorded with an infrared camera (B335: Compact-Infrared-Thermal-Imaging-Camera; FLIR; West Malling, Kent, UK) and analyzed with a specific software package (FLIR-Tools-Software, FLIR; West Malling, Kent, UK) as previously shown [53]. For each animal and day, area surrounding BAT was delimited in the infrared picture recorded and average skin temperature area was calculated.
4.15. Electron microscopy
Animals were anesthetized and fixed using 2% (v/v) formaldehyde (®EMS), 2.5% (v/v) glutaraldehyde (®Polysciences) in 0.1M Phosphate Buffer (PB) (pH 7.4) by perfusion using a peristaltic pump. Organs were dissected and immersed in the same primary fixative (1 h; RT). Further processing was achieved using a PELCO BioWave Microwave Processor at 23 °C, restricted by a PELCO SteadyTemp Pro. Samples were additionally fixed in the primary fixative using a time-sequence of 7 × 2 min with ON and OFF sequential cycles of 0 and 100 W irradiating power in vacuum and rinsed with PB before post-fixation in 1% (v/v) osmium tetroxide (®EMS) with 1% (w/v) potassium ferrocyanide (®Sigma Aldrich) in PB for 8 × 2 min also with ON and OFF sequential cycles of 100 W in vacuum. Subsequently, samples were washed with PB and dH2O twice and immersed in 1% (w/v) tannic acid (®EMS) followed by en-bloc staining with 0.5% (w/v) uranyl acetate. Both steps were made using a time-sequence of 7 × 2 min with ON and OFF sequential cycles of 0 and 150 W irradiating power in vacuum. Between the steps, samples were rinsed with dH2O. Dehydration was done in a graded ethanol series of 30%, 50%, 75%, 90% and 100%, for 40 s at 150 W each. EPON resin (®EMS), 25%, 50%, 75% and 100% was infiltrated, for 3 min at 250 W in vacuum each step, and cured Over-Night, at 60 °C. Sections of 70 nm were obtained on a Leica UC7 and mounted on palladium-copper grids coated with 1% (w/v) formvar (®Agar Scientific) in chloroform (®VWR). Sections were stained with 1% (w/v) uranyl acetate and Reynolds lead citrate for 5 min each and imaged on Hitachi H-7650 Transmission Electron Microscope operating at 100 keV.
4.16. DNA extraction and mtDNA/nDNA qPCR
BAT and WAT were harvested directly into lysis buffer (EDTA solution (100 mM NaCl, 10 mM EDTA, 0.5% SDS and 20 mM Tris-HCl, pH 7.4)) and homogenized in a tissue lyser (Qiagen) using tungsten carbide beads. Upon homogenization, an equal volume of Phenol:Chloroform:Isoamyl Alcohol (25:24:1, v/v) was added. DNA, present in the aqueous phase, was precipitated using 1 vol isopropanol and 0.3 M sodium acetate for 3 h at −20 °C. The isolated DNA was used to perform the quantification of mitochondrial DNA (mtDNA) in comparison to nuclear DNA (nDNA) using a qRT-PCR -based method, similar to what was previously described [55]. Briefly, qRT-PCR was performed using 20 ng of DNA and SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA), in duplicate on a 7500 Fast Real-Time PCR System (Applied Biosystems), under the following conditions: 50 °C/2min and 95 °C/5 min (Hold stage), 45 cycles/95 °C/10 s, annealing at 60 °C/30 s, and elongation 72 °C/20 s, followed by melting curve: 95 °C for 15 s, 60 °C for 1 min, and gradual increase in temperature up to 95 °C. Primers for NADH-ubiquinone oxidoreductase chain 1 encoded by the mitochondrial gene MT-Nd1 (Nd1) and for the nuclear encoded hexokinase 2 gene (Hk2) [55] are listed in the Table 1. Mitochondria number per cell was calculated by the ratio of mRNA expression of the single copy mitochondrial gene Nd1 and the single copy nuclear gene Hk2.
4.17. Seahorse assays
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured using a Seahorse XFe96 analyzer (Agilent Tech.) and the Seahorse XF Cell Mito Stress Test Kit according to instructions from the manufacturer. Specifically, purified primary mouse hepatocytes were plated on collagen-coated Seahorse XF96 plates (7,000 cells per well) in Williams E medium supplemented with 5% FBS and 1% Pen/Strep, and allowed to adhere (37 °C; 3 h). Cells were washed twice with warm Seahorse XF Assay Medium (pH 7.4, 1 mM sodium pyruvate, 2 mM l-glutamine, 10 mM glucose), and XF Assay Medium (180 μL) was added per well. Cells were incubated in a non-CO2 incubator (37 °C; 1 h) prior to assay. The analyzer was programmed to calibrate and equalize samples, followed by 3 baseline measurements (3 min each) and mixing (3 min) between measurements prior to inhibitor injection. The inhibitors were injected in the following order: Oligomycin (2 μM); FCCP (0.5 μM); Antimycin A/Rotenone (0.5 μM); and 3 measurements (3 min each) were made following each injection with 3min. mixing between measurements.
4.18. Quantification and statistical analysis
Statistical analysis was conducted using GraphPad Prism 6 software. All distributed data are displayed as means ± standard deviation of the mean (SD) unless otherwise noted. Measurements between two groups were performed with a Mann–Whitney U test. Groups of three or more were analyzed by one-way analysis of variance (ANOVA) or the Kruskal–Wallis test. Survival was assessed using a log-rank (Mantel–Cox) test. Statistical parameters for each experiment can be found within the corresponding figure legends.
Author contribution
MPS conceived the research and wrote the manuscript with BB and FB, who designed and carried out most of the experimental work. RM performed experiments analyzing mitochondrial function and intracellular Fe content. PBA assisted FB in cold stress experiments. IGG and ML performed mouse metabolic analysis, JMN analyzed electron microscopy and adipose tissue with PBA. ARC performed and analyzed experiments to quantify mitochondria and mitochondrial ETC protein expression. IM and AD contributed to different aspects of metabolic analyses related to adipose tissue. PV performed western blot analyzes, RF histological analysis. SW, VH, and JG performed MRI. SC bred all the mice used and helped with experiments. All authors provided critical feedback and contributed critically to shaping experimental work, analysis and manuscript writing.
Acknowledgements
The authors thank the Inflammation group (IGC) for insightful technical and intellectual contributions and Erin M. Tranfield at the Electron Microscopy Unit (IGC). MPS is supported by Fundação Calouste Gulbenkian and by Fundação para a Ciência e a Tecnologia, Portugal (PTDC/IMI-IMU/5723/2014 and 02/SAICT/2017). FCT support to ARC (SFRH/BPD/101608/2014) and PBA (PTDC/IMI-IMU/5723/2014) is acknowledged. BB and SC were supported in part by European Community 7th Framework 294709-DAMAGECONTROL ERC-2011-AdG to MPS, FB by Marie Skłodowska-Curie Research Fellowship (REGDAM 707998), RM by EMBO long-term fellowship (ALTF 290-2017), ML by Xunta de Galicia (2015-CP079) and MINECO from EU FEDER Program (SAF2015-71026-R), SW and JG by the German Ministry of Education and Research (BMBF; grant 01 EO 1502) via the Jena Center of Sepsis Control and Care.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.03.008.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
References
- 1.Winterbourn C.C. Toxicity of iron and hydrogen peroxide: the Fenton reaction. Toxicology Letters. 1995;82–83:969–974. doi: 10.1016/0378-4274(95)03532-x. [DOI] [PubMed] [Google Scholar]
- 2.Muckenthaler M.U., Rivella S., Hentze M.W., Galy B. A red carpet for iron metabolism. Cell. 2017;168:344–361. doi: 10.1016/j.cell.2016.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gunshin H., Mackenzie B., Berger U.V., Gunshin Y., Romero M.F., Boron W.F. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 4.Kuhn L.C., McClelland A., Ruddle F.H. Gene transfer, expression, and molecular cloning of the human transferrin receptor gene. Cell. 1984;37:95–103. doi: 10.1016/0092-8674(84)90304-0. [DOI] [PubMed] [Google Scholar]
- 5.Donovan A., Lima C.A., Pinkus J.L., Pinkus G.S., Zon L.I., Robine S. The iron exporter ferroportin/Slc40a1 is essential for iron homeostasis. Cell Metabolism. 2005;1:191–200. doi: 10.1016/j.cmet.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Harrison P.M., Arosio P. Ferritins - molecular properties, iron storage function and cellular regulation. Biochimica et Biophysica Acta Bioenergetics. 1996;1275:161–203. doi: 10.1016/0005-2728(96)00022-9. [DOI] [PubMed] [Google Scholar]
- 7.Gozzelino R., Soares M.P. Coupling heme and iron metabolism via ferritin H chain. Antioxidants and Redox Signaling. 2014;20:1754–1769. doi: 10.1089/ars.2013.5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreira C., Bucchini D., Martin M.E., Levi S., Arosio P., Grandchamp B. Early embryonic lethality of H ferritin gene deletion in mice. Journal of Biological Chemistry. 2000;275:3021–3024. doi: 10.1074/jbc.275.5.3021. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez-Morales N., Mendoza-Ortiz M.A., Blowes L.M., Missirlis F., Riesgo-Escovar J.R. Ferritin is required in multiple tissues during Drosophila melanogaster development. PLoS One. 2015;10:e0133499. doi: 10.1371/journal.pone.0133499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W., Garringer H.J., Goodwin C.B., Richine B., Acton A., VanDuyn N. Systemic and cerebral iron homeostasis in ferritin knock-out mice. PLoS One. 2015;10:e0117435. doi: 10.1371/journal.pone.0117435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hentze M.W., Caughman S.W., Rouault T.A., Barriocanal J.G., Dancis A., Harford J.B. Identification of the iron-responsive element for the translational regulation of human ferritin mRNA. Science. 1987;238:1570–1573. doi: 10.1126/science.3685996. [DOI] [PubMed] [Google Scholar]
- 12.Rouault T.A., Hentze M.W., Caughman S.W., Harford J.B., Klausner R.D. Binding of a cytosolic protein to the iron-responsive element of human ferritin messenger RNA. Science. 1988;241:1207–1210. doi: 10.1126/science.3413484. [DOI] [PubMed] [Google Scholar]
- 13.Meyron-Holtz E.G., Ghosh M.C., Iwai K., LaVaute T., Brazzolotto X., Berger U.V. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. The EMBO Journal. 2004;23:386–395. doi: 10.1038/sj.emboj.7600041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi H., Bencze K.Z., Stemmler T.L., Philpott C.C. A cytosolic iron chaperone that delivers iron to ferritin. Science. 2008;320:1207–1210. doi: 10.1126/science.1157643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mancias J.D., Wang X., Gygi S.P., Harper J.W., Kimmelman A.C. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105–109. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiou B., Connor J.R. Emerging and dynamic biomedical uses of ferritin. Pharmaceuticals (Basel) 2018;11 doi: 10.3390/ph11040124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weis S., Carlos A.R., Moita M.R., Singh S., Blankenhaus B., Cardoso S. Metabolic adaptation establishes disease tolerance to sepsis. Cell. 2017;169:1263–1275. doi: 10.1016/j.cell.2017.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gozzelino R., Andrade B.B., Larsen R., Luz N.F., Vanoaica L., Seixas E. Metabolic adaptation to tissue iron overload confers tolerance to malaria. Cell Host & Microbe. 2012;12:693–704. doi: 10.1016/j.chom.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 19.Reddy V.P., Chinta K.C., Saini V., Glasgow J.N., Hull T.D., Traylor A. Ferritin H deficiency in myeloid compartments dysregulates host energy metabolism and increases susceptibility to Mycobacterium tuberculosis infection. Frontiers in Immunology. 2018;9:860. doi: 10.3389/fimmu.2018.00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zarjou A., Bolisetty S., Joseph R., Traylor A., Apostolov E.O., Arosio P. Proximal tubule H-ferritin mediates iron trafficking in acute kidney injury. Journal of Clinical Investigation. 2013;123:4423–4434. doi: 10.1172/JCI67867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berberat P.O., Katori M., Kaczmarek E., Anselmo D., Lassman C., Ke B. Heavy chain ferritin acts as an antiapoptotic gene that protects livers from ischemia reperfusion injury. FASEB Journal. 2003;17:1724–1726. doi: 10.1096/fj.03-0229fje. [DOI] [PubMed] [Google Scholar]
- 22.Vanoaica L., Darshan D., Richman L., Schumann K., Kuhn L.C. Intestinal ferritin H is required for an accurate control of iron absorption. Cell Metabolism. 2010;12:273–282. doi: 10.1016/j.cmet.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Darshan D., Vanoaica L., Richman L., Beermann F., Kuhn L.C. Conditional deletion of ferritin H in mice induces loss of iron storage and liver damage. Hepatology. 2009;50:852–860. doi: 10.1002/hep.23058. [DOI] [PubMed] [Google Scholar]
- 24.Vanoaica L., Richman L., Jaworski M., Darshan D., Luther S.A., Kuhn L.C. Conditional deletion of ferritin h in mice reduces B and T lymphocyte populations. PLoS One. 2014;9:e89270. doi: 10.1371/journal.pone.0089270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen L.A., Gutierrez L., Weiss A., Leichtmann-Bardoogo Y., Zhang D.L., Crooks D.R. Serum ferritin is derived primarily from macrophages through a nonclassical secretory pathway. Blood. 2010;116:1574–1584. doi: 10.1182/blood-2009-11-253815. [DOI] [PubMed] [Google Scholar]
- 26.Dillmann E., Johnson D.G., Martin J., Mackler B., Finch C. Catecholamine elevation in iron deficiency. American Journal of Physiology. 1979;237:R297–R300. doi: 10.1152/ajpregu.1979.237.5.R297. [DOI] [PubMed] [Google Scholar]
- 27.Lukaski H.C., Hall C.B., Nielsen F.H. Thermogenesis and thermoregulatory function of iron-deficient women without anemia. Aviation Space & Environmental Medicine. 1990;61:913–920. [PubMed] [Google Scholar]
- 28.Brigham D., Beard J. Iron and thermoregulation: a review. Critical Reviews in Food Science and Nutrition. 1996;36:747–763. doi: 10.1080/10408399609527748. [DOI] [PubMed] [Google Scholar]
- 29.Beard J.L., Borel M.J., Derr J. Impaired thermoregulation and thyroid function in iron-deficiency anemia. American Journal of Clinical Nutrition. 1990;52:813–819. doi: 10.1093/ajcn/52.5.813. [DOI] [PubMed] [Google Scholar]
- 30.Nemeth E., Tuttle M.S., Powelson J., Vaughn M.B., Donovan A., Ward D.M. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 31.Soares M.P., Hamza I. Macrophages and iron metabolism. Immunity. 2016;44:492–504. doi: 10.1016/j.immuni.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camaschella C. Iron-deficiency anemia. New England Journal of Medicine. 2015;373:485–486. doi: 10.1056/NEJMc1507104. [DOI] [PubMed] [Google Scholar]
- 33.Oikawa D., Akai R., Tokuda M., Iwawaki T. A transgenic mouse model for monitoring oxidative stress. Scientific Reports. 2012;2:229. doi: 10.1038/srep00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosen E.D., Spiegelman B.M. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kajimura S., Saito M. A new era in brown adipose tissue biology: molecular control of brown fat development and energy homeostasis. Annual Review of Physiology. 2014;76:225–249. doi: 10.1146/annurev-physiol-021113-170252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hubler M.J., Peterson K.R., Hasty A.H. Iron homeostasis: a new job for macrophages in adipose tissue? Trends in Endocrinology and Metabolism. 2015;26:101–109. doi: 10.1016/j.tem.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rumberger J.M., Peters T., Jr., Burrington C., Green A. Transferrin and iron contribute to the lipolytic effect of serum in isolated adipocytes. Diabetes. 2004;53:2535–2541. doi: 10.2337/diabetes.53.10.2535. [DOI] [PubMed] [Google Scholar]
- 38.Cannon B., Nedergaard J. Brown adipose tissue: function and physiological significance. Physiological Reviews. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 39.Lowell B.B., S-Susulic V., Hamann A., Lawitts J.A., Himms-Hagen J., Boyer B.B. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature. 1993;366:740–742. doi: 10.1038/366740a0. [DOI] [PubMed] [Google Scholar]
- 40.Andrews N.C., Schmidt P.J. Iron homeostasis. Annual Review of Physiology. 2007;69:69–85. doi: 10.1146/annurev.physiol.69.031905.164337. [DOI] [PubMed] [Google Scholar]
- 41.Galy B., Ferring-Appel D., Sauer S.W., Kaden S., Lyoumi S., Puy H. Iron regulatory proteins secure mitochondrial iron sufficiency and function. Cell Metabolism. 2010;12:194–201. doi: 10.1016/j.cmet.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 42.Walter P.B., Knutson M.D., Paler-Martinez A., Lee S., Xu Y., Viteri F.E. Iron deficiency and iron excess damage mitochondria and mitochondrial DNA in rats. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:2264–2269. doi: 10.1073/pnas.261708798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sibille J.C., Kondo H., Aisen P. Interactions between isolated hepatocytes and Kupffer cells in iron metabolism: a possible role for ferritin as an iron carrier protein. Hepatology. 1988;8:296–301. doi: 10.1002/hep.1840080218. [DOI] [PubMed] [Google Scholar]
- 44.Fleming R.E., Feng Q., Britton R.S. Knockout mouse models of iron homeostasis. Annual Review of Nutrition. 2011;31:117–137. doi: 10.1146/annurev-nutr-072610-145117. [DOI] [PubMed] [Google Scholar]
- 45.Gao Y., Li Z., Gabrielsen J.S., Simcox J.A., Lee S.H., Jones D. Adipocyte iron regulates leptin and food intake. Journal of Clinical Investigation. 2015;125:3681–3691. doi: 10.1172/JCI81860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng W., Pirzgalska R.M., Pereira M.M., Kubasova N., Barateiro A., Seixas E. Sympathetic neuro-adipose connections mediate leptin-driven lipolysis. Cell. 2015;163:84–94. doi: 10.1016/j.cell.2015.08.055. [DOI] [PubMed] [Google Scholar]
- 47.Dentin R., Pegorier J.P., Benhamed F., Foufelle F., Ferre P., Fauveau V. Hepatic glucokinase is required for the synergistic action of ChREBP and SREBP-1c on glycolytic and lipogenic gene expression. Journal of Biological Chemistry. 2004;279:20314–20326. doi: 10.1074/jbc.M312475200. [DOI] [PubMed] [Google Scholar]
- 48.Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeney V., Ramos S., Bergman M.L., Bechmann I., Tischer J., Ferreira A. Control of disease tolerance to malaria by nitric oxide and carbon monoxide. Cell Reports. 2014;8:126–136. doi: 10.1016/j.celrep.2014.05.054. [DOI] [PubMed] [Google Scholar]
- 50.Grundy M.A., Gorman N., Sinclair P.R., Chorney M.J., Gerhard G.S. High-throughput non-heme iron assay for animal tissues. Journal of Biochemical and Biophysical Methods. 2004;59:195–200. doi: 10.1016/j.jbbm.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 51.Martins R., Maier J., Gorki A.D., Huber K.V., Sharif O., Starkl P. Heme drives hemolysis-induced susceptibility to infection via disruption of phagocyte functions. Nature Immunology. 2016;17:1361–1372. doi: 10.1038/ni.3590. [DOI] [PubMed] [Google Scholar]
- 52.Parlee S.D., Lentz S.I., Mori H., MacDougald O.A. Quantifying size and number of adipocytes in adipose tissue. Methods in Enzymology. 2014;537:93–122. doi: 10.1016/B978-0-12-411619-1.00006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martinez de Morentin P.B., Gonzalez-Garcia I., Martins L., Lage R., Fernandez-Mallo D., Martinez-Sanchez N. Estradiol regulates brown adipose tissue thermogenesis via hypothalamic AMPK. Cell Metabolism. 2014;20:41–53. doi: 10.1016/j.cmet.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martinez-Sanchez N., Seoane-Collazo P., Contreras C., Varela L., Villarroya J., Rial-Pensado E. Hypothalamic AMPK-ER stress-JNK1 Axis mediates the central actions of thyroid hormones on energy balance. Cell Metabolism. 2017;26:212–229 e212. doi: 10.1016/j.cmet.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quiros P.M., Goyal A., Jha P., Auwerx J. Analysis of mtDNA/nDNA ratio in mice. Current Protocols in Mouse Biology. 2017;7:47–54. doi: 10.1002/cpmo.21. [DOI] [PMC free article] [PubMed] [Google Scholar]