Abstract
Let-7, a gene firstly known to control the timing of Caenorhabditis elegans larval development does not code for a protein but instead produces small non-coding RNAs, microRNAs. Higher animals have multiple isoforms of mature let-7 microRNAs. Mature let-7 family members share the same “seed sequence” and distinct from each other slightly by ‘non-seed’ sequence region. Let-7 has emerged as a central regulator of systemic energy homeostasis and it displays remarkable plasticity in metabolic responses to nutrients availability and physiological activities. In this review, we discuss recent studies highlighting post-transcriptional mechanisms that govern metabolic reprogramming in distinct cells by let-7. We focus on the participation of the let-7 clusters in immune cells, and suggest that tissue-specific regulation of the let-7 clusters by engineered mouse models might impact metabolic homeostasis and will be required to elucidate their physiological and pathological roles in the in vivo disease models.
Introduction
Although initial observations in immune-metabolism were made nearly a century ago, recent work of cellular metabolism broadly and deeply permeated immunological research [1], [2]. Researches have been elucidating the molecular basis for how extracellular signals or molecules control the uptake and utilization of nutrients in quiescent and activated immune cells [3], [4]. Mounting evidences support the concept that deep understanding how metabolic programs regulates immune cell functional roles may provide new therapeutic opportunities for many pathological diseases, which are associated with dysregulation of immune system, such as type II diabetes (T2D) [5], [6], [7]. Different subtypes of immune cells can adopt distinct metabolic allocations that allow the immune cells to adjust its requirements for energy and maintain its physiological functions [8], [9]. The big challenge of immune cell metabolic network has always been its own complex, which derives from plentiful tiny metabolites and the complex regulation of the activities of key enzymes as well as transportations and accumulations of key metabolites.
MicroRNA (miRNA) is a small non-coding RNA that is highly conserved from worms to humans. It mediates multiple metabolic activities, depending on its binding capacity to the three prime untranslated region (3’-UTR) of the targeted mRNAs for repressing corresponding target gene expression by either translational inhibition or mRNA decay [10], [11], [12], [19], [20], [21]. The “seed sequence” is essential for the binding of the miRNA to the mRNA. The seed sequence (also called seed region) is a conserved short sequence that is mostly situated at positions 2–7 (or 2–6, or 2–8) from the 5′-end of miRNAs [13]. Even though base pairing of miRNA and its target mRNA does not need to match perfectly, the “seed sequence” has to be perfectly complementary in most cases [14].
Currently, a series of miRNAs have been recognized as biomarkers in clinical medicine, with high conserved expression across species, such as miR-155 in breast cancer and acute myeloid leukemia [15], [16], [17], [18]. They also make great contributions to the control of energy homeostasis and metabolic disorders such as T2D [19], [20], [21]. Although the sophisticated mechanisms of action and the multipronged influence of miRNAs on physiological and pathological activities have not been fully interpreted, recent progress has been made in deciphering the individual roles of certain miRNA such as the let-7 cluster in specific immunological contexts.
The let-7 family, highly conserved across multiple animal species, is one of the well-studied miRNA clusters [22], [23], [24]. The role of let-7 family members as a tumor suppressor has been very well documented but their physiological roles in the regulation of immune cell responses is slowly unfolding [24], [25], [26]. The dys-regulated expression of let-7 can lead to aberrant immune function, including the development of multiple sclerosis and asthma [27], [28]. In recent years, a couple of let-7 family members have implicated in metabolic reprogramming and immune system development. Let-7’s function has increased notably, and there has been collected discussion of their potential use as therapeutics for metabolic-related and immune-related diseases [29], [30], [31], [32], [33].
In this review, we discuss the general features of the let-7 family across different species, with a brief discussion on its regulation. We then discuss recent advances in let-7 regulation of physiological and pathological metabolic reprograms, focusing on current but limited information regarding the metabolic network and their mechanisms underlying this regulation in various mouse models in vivo and tissue cultures in vitro. In the next section, we explore the connection between dys-regulated let-7 cluster expression and metabolic regulation in different types of immune cells. With our current understanding of the functions and mechanisms of let-7, we find that there is great promise in the field for using let-7 both to understand the metabolic mechanisms of immune cell fate decisions and to develop diagnostic and therapeutic strategies for aberrant immune cell-related human diseases.
Let-7 Cluster
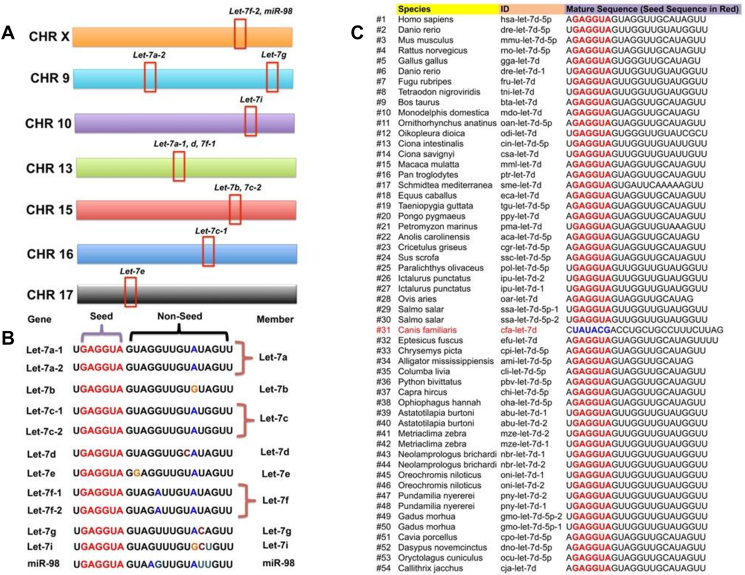
Some miRNAs are situated in polycistronic miRNA “clusters”, wherein majority of miRNA genes are generated only from a single primary transcript [34], [35]. The high conservation of miRNA clusters across species indicates evolutionary pressure to maintain such a complicated regulation network. One of the best-characterized polycistronic miRNA clusters is the let-7 cluster [23]. Forward genetics experiments initially identified let-7 as a heterochronic gene in Caenorhabditis elegans (C. elegans) [36], [37], [38]. In mice, twelve let-7 genes are present in the murine genome and disperse as eight clusters on seven different chromosomes, although some of let-7 genes are clustered together such as the let-7a-1/let-7d/let-7f-1 cluster (let-7adf) (Figure 1, A–B) [39]. In human, the let-7 family is composed of nine mature let-7 members, which are encoded by twelve different genomic loci [33]. Interestingly, the worm and the fly have only single member, named as cel-let-7 and dmel-let-7, respectively [40]. This might be because of the unique functional demands for let-7 or any other reasons, for example, positive Darwinian selection might be one of the driving forces underlying the formation and evolution of single let-7 miRNA clustering in worm and fly, which needs a mechanism to deepen our understanding of evolution between genomic contexts and novel let-7 clusters in higher species, supporting the “functional co-adaptation” for let-7 miRNA clustering. Further experiments, for instance, by transfecting cel-let-7 or dmel-let-7 miRNAs into human or mouse cells and extensively profiling the transcriptome alteration with deep-sequencing, so it could demonstrate the functional co-adaptation between lower and higher animal miRNAs in the let-7 clusters. Generally, higher animals have similar sets of let-7 family members, although slight differences may be observed, for example, let-7 h (dre-let-7 h) only exists in the zebrafish but not in the human and mouse genomes [41]. Different from other miRNA clusters, such as the miRNAs encoded by the miR-17-92 cluster which can be group into four different “seed” families, whose members are predicted to target distinct sets of genes, all let-7 members share the same “seed” [42], [43] in mouse and human genome and this seed region is generally the most critical portion of let-7 functional elements, suggesting that functional redundancy of let-7 may exist [23]. Intriguingly, over 50 species including C. elegans, Drosophila, Zebrafish, Mus musculus and Homo sapiens all express a same version of let-7d seed sequences (nucleotides 2–8 in mature miRNAs) “GAGGUA”, but interestingly, only Canis familiaris has its own version of “UAUACG” (Figure 1C) [23], [26]. This conserved feature of let-7d suggests that its targets and functions may be similar across diverse animal species. What is the difference between the bioprocessing of Canis familiaris let-7d from others such as mmu-let-7d? Is the Canis familiaris let-7d functional different or redundant in different type of cells, compared with other members? What is the physiological role of Canis familiaris let-7d in vivo? It is worthy to address all these questions and understand why Canis familiaris let-7d is unique in future.
Figure 1.
General features of mature let-7. (A) Chromosome location and distribution of let-7 genes in murine genome. (B) Mature sequence of murine let-7 genes. (C) Sequence comparison of mature let-7d across species.
To deep understanding the differences of biological functions among each let-7 cluster, identification of the miRNA recognizing element (MRE) is the initial step, because it is well known that miRNA target recognition is largely dictated by short “seed” sequences, and binding of miRNAs with their targets is strongly affected by the nucleotides in the 5′ end of the miRNA–MRE duplex, however, the flanking sequences (non-seed) around short “seed” also contribute to specific miRNA–target interactions [51], [52]. Despite the strong similarity in the sequences of let-7 clusters, tiny differences in their “non-seed” mature sequences may result in both overlapping and distinct messenger RNA targets. It has been reported that both the functional partnership between seed and non-seed sequences in mediating C. elegans temporal development and a diversity among miRNA effectors in the contributions of seed and non-seed regions to activity, thus, single point mutations on the non-seed sequences of each let-7 cluster would help to define the sequence requirements of let-7 single member or cluster's bio-functions [53], [54], [55]. We may thus hypothesize that advanced technologies such like (CRISPR)/Cas9 or transcription activator-like effector nuclease (TALEN) may be utilized to interrogate the functional relevance of non-seed region of different clusters to post-transcriptional silencing in several species including C. elegans, Zebrafish and Drosophila. Moreover, investigating how let-7 miRNA family members are expressed in clusters and how they control signaling pathways is likely to increase the knowledge of several biology processes. Let-7 is regulated by key transcriptional factors such as c-Myc that control the above-mentioned processes as well as RNA-binding proteins including lin28a/b, which has been reviewed widely [25], [48], [49], [50]. To further understanding of the half-life and stability of the pri-let-7, pre-let-7 and mature let-7 would help provide insight into each let-7 cluster's tissue-specific functions in various immune cells.
This combined mutual repression may therefore accelerate the understanding of the contribution of specific let-7 member–target interactions to the regulation of biological processes in vitro and in vivo. Future work will reveal the mechanisms that determine seed dependence and non-seed contributions for different let-7 member: target in distinct cells. It will be interesting to address whether it is evident that noncanonical let-7–target interactions where no match to the seed. For instance, Ago-RISC prepared from different immune cells could be used to understand how the stabilities of miRNAs in Ago proteins are influenced by non-canonical interactions. The non-canonical interactions of let-7 and targets may provide a novel means to fine-tune, refine and diversify miRNAs in different cells. A deeper understanding of the more precise contributions from interactions between let-7-mediated seed and/or non-seed with target may provide new avenues for using accurate let-7 member to alter the function of specific immune cell subsets.
It is unclear whether and how the multiple let-7 clusters are functionally equivalent or control different molecular pathways, and the majority of their functionally relevant targets also remain to be further identified. The analysis of genetically engineered mice carrying gain-of-function and loss-of-function mutations of each let-7 cluster will be essential to furthering our understanding of the biological functions of this important let-7 cluster.
Let-7 in Physiological and Pathological Cellular Metabolic Reprogramming
Recent findings reveal that miRNAs also contribute to insulin signaling and glucose homeostasis and highlight the potential pathological roles of aberrant miRNA expression in metabolic disorders such as obesity, insulin resistance (IR), and T2D [56]. Proper control of metabolic homeostasis is crucial to the maintenance of human health. Global expression analyses demonstrate that alterations in miRNA levels correlate with various metabolic diseases [57]. MiRNAs have been showed to regulate central metabolic pathways and thus play vital roles in maintaining organismal energy balance and metabolic homeostasis [58]. For example, miR-103/107 regulates insulin and glucose homeostasis [59], whereas miR-34a regulates hepatic lipid homeostasis [60].
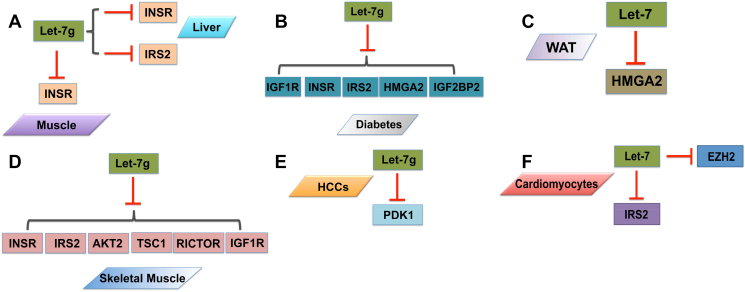
Let-7 was recently shown to be involved in the regulation of glucose metabolism [31], [44], [61], [62]. The emergence of let-7 as important regulators of metabolism has gained much interest from a clinical perspective, and let-7 target genes are associated with T2D in human a genome-wide association study (GWAS) [62]. Genetically engineered mouse models of let-7 are critical for evaluating the physiological roles of this miRNA cluster in metabolic diseases such as obesity and diabetes. For instance, global overexpression of let-7 g in mice results in growth retardation and impairs glucose tolerance [30]. Global knockdown of the let-7 family by anti-let-7 could be sufficient to prevent impaired glucose tolerance and enhance blood glucose levels and insulin resistance in obese mice [30]. Mechanistically, anti-let-7 improves insulin sensitivity in both liver and muscle, at least by partially restoring expression levels of insulin receptor (INSR) and insulin receptor substrate 2 (IRS2) (Figure 2, A–D) [30]. Thus, knockdown of let-7 by anti-let-7 may provide an approach to treat metabolic diseases such as T2D. Let-7 can also regulate glucose metabolism in multiple organs (Figure 2, B and C) [30]. Specific overexpression of let-7 in skeletal muscle is sufficient to promote growth [61]. Overexpression of let-7 affects not only organismal size and onset of puberty in mice but also human retardation [62]. Pancreas-specific overexpression of let-7 in mice results in impaired glucose tolerance and reduced glucose-induced pancreatic insulin secretion [30]. Inducible let-7 transgenic mice not only have reduced body size and growth retardation, but also have hyperglycemia and glucose intolerance [31]. The ability to closely recapitulate human physiology and pathology in mouse models, and the conservation between humans and mice suggest that studies of let-7 in mice could provide the most meaningful insights on the role of this clusters in the molecular pathogenesis of human diseases. Current evidence of let-7 is also being directed towards the development of useful mouse models to evaluate these miRNAs therapeutics pre-clinically. The CRISPR-Cas9 system is capable of producing multiple miRNA knock-in/out mouse models in few weeks and, therefore, has immense potential in let-7 clusters functional delineation in multiple human cellular contexts.
Figure 2.
Regulation of metabolic programs by let-7 in different tissues through targeting single or multiple genes.
Let-7 is also important to the regulation of metabolic programs in various cell lines. For example, in mouse embryonic stem cells (ESCs), inducing let-7 plays a critical role on the Threonine (Thr)-Glycine (Gly)-S-adenosyl-methionine (SAM) pathway [63]. In mouse embryonic fibroblasts (MEFs), suppression of let-7 alone is not sufficient to regulate the regenerative capacity but has no effect on cell migration [32]. In myoblasts, let-7 regulates Akt and S6 phosphorylation, which enhances the insulin-sensitivity and glucose uptake [62]. Let-7 is essential to nutrient homeostasis in higher organisms. In muscle and white fat, let-7 prevents mTORC1 activation to induce autophagy under nutrient deprivation [29]. In maturing cardiomyocytes (CMs), let-7 is an important mediator in augmenting metabolic energetics. Let-7 regulates cardiomyocytes metabolism by promoting maturity of stem cell derivatives through simultaneously repressing on two of its targets, IRS2 and EZH2 (Figure 2F) [46]. Let-7 repression by anti-let-7 is necessary but insufficient to enhance tissue repair, but interestingly, despite an efficient knockdown of let-7, neither systemic nor topical delivery of anti-let-7 enhances pinnal tissue repair whereas overexpression of let-7 after ear punch biopsy inhibits wound closure and pinnal repair [32].
Metabolism is not only regulated by let-7 in normal tissues but also in pathological cellular context. For example, let-7 family miRNAs, most notably let-7 g, repressed aerobic glycolysis in human hepatocellular carcinoma cancer cell lines (HCCs). Let-7 overexpression blocks glucose uptake in the examined HCCs, and let-7 specifically suppresses Pyruvate dehydrogenase kinase isozyme 1 (PDK1) expression but no other Oxidative phosphorylation (OxPhos) enzymes (Figure 2E) [64]. Up-regulation of let-7a contributes to the attenuation of endothelial cell migration and insulin signaling induced by acrolein [65]. The circulating levels of let-7 g display a female-specific elevation in individuals with metabolic syndrome, which is associated with the risk of developing cardiovascular disease and T2D [66], [67]. Let-7 also contributes to IL-13 synthesis and release in skeletal muscle from insulin-resistant T2D patients [68]. Interestingly, let-7 could couple with lin-4, which is another miRNA that was identified from a study of developmental timing in the C. elegans, to regulate intestinal mammalian target of rapamycin (mTOR) signaling so it regulates fat metabolism and vitellogenesis through the PQM-1 zinc finger transcription factor in C. elegans [69]. A number of open questions remain concerning the function of let-7 in metabolic control. For example, it is unclear whether let-7 has important regulatory impact on cholesterol and lipid metabolism in vitro and in vivo, and how widespread the contribution of individual let-7 member is to metabolic control.
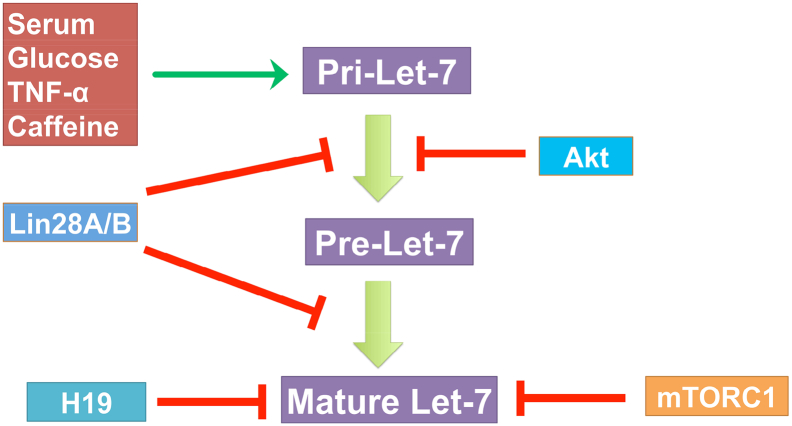
Other factors that regulate the expression of let-7 also might indirectly contribute to metabolic reprograms. For example, miR-107 has been reported to negatively regulate let-7 through a direct interaction. Deletion of miR-107 dramatically increases the stability of mature let-7 and down-regulates let-7 targets because miR-107 directly interacts with let-7 through internal loop of the let-7/miR-107 duplex is critical for repression of let-7 expression [70]. Does miR-107 affect metabolic programs in vivo by targeting let-7? The miR-107 iTg and KO mouse models as well as miR-107/let-7 double transgenic mouse models could be utilized to address these questions in future.
Different factors including serum, glucose, TNF-α and even caffeine can regulate the promoter activity and the let-7a-1/let-7d/let-7f-1 cluster (let-7adf) miRNA expression [71]. Ornithine decarboxylase inhibits glycolytic metabolism by regulating let-7 expression in neuroblastoma [72]. Acting as a molecular sponge, long noncoding RNA H19 inhibits let-7 (Figure 3) [73]. Acute hyper-insulinemia can down-regulate H19 through PI3K/AKT-dependent phosphorylation of the miRNA processing factor K-homology splicing regulatory protein (KSRP), which promotes let-7 biogenesis and also mediates H19 destabilization [74]. Could the influence of let-7 on growth and metabolism be mediated by any other interacting factors such as H19? The molecular details of the interaction on this miRNA in metabolic diseases await further elucidation. Future work should hone in on comprehensively analyzing the gene regulatory networks mediated by let-7 clusters that drive aberrant metabolic-related issues as well as factors controlling let-7 expression, with the aim of identifying key nodes for targeted therapeutic delivery of let-7 and its related molecules.
Figure 3.
Regulation of let-7 biogenesis by various factors.
Roles of Let-7 in Energy and Cellular Metabolism in Immune Cells
In comparison to their well-defined role in tumorigenesis as a tumor suppressor, let-7 is frequently expressed in a variety of immune cells, including B cells and macrophages [44], [45]. Moreover, the individual let-7 family member can regulate multiple components of both normal and disease pathways of metabolism and immune cell homeostasis [29], [44]. The development of some metabolic diseases such as diabetes involves a complex interaction between pancreatic β-cells and cells of both the innate and adaptive immune systems [75], [76]. Recently, Immune-metabolism has emerged as a chief breakthrough especially in the field of diabetes mellitus [77], [78]. The activation, growth, proliferation, and effector functions of different types of immune cells are intimately linked and dependent on dynamic changes in cellular metabolic programs [4]. Based on recent studies, let-7 is mostly linked to activation of B cells, macrophage responses, and proliferation and activation of T cell lineage [30], [44], [46], [47].
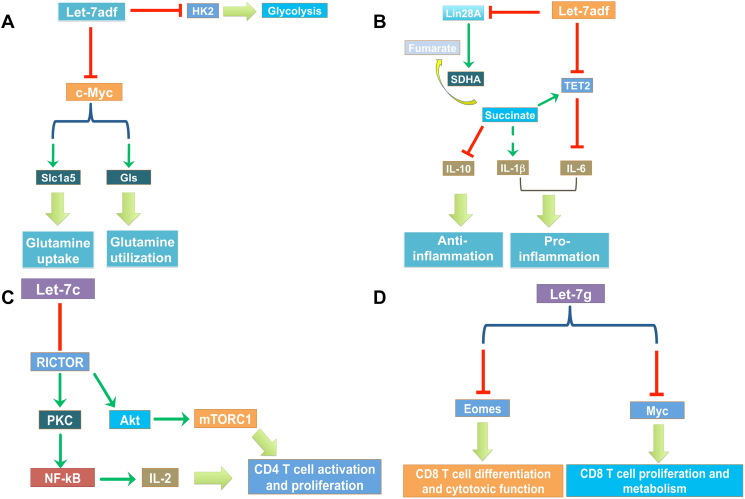
B cells promote metabolic disease by supporting T cell–mediated inflammation and insulin resistance and B cell–null mice are protected from pathogenic outcomes of obesity and inflammation [79], [80]. Upon activation of B cell by toll-like receptor (TLR) ligands, B cell can further proliferate and differentiate to generate antibody-producing cells [81]. The metabolic demands are high in the proliferative early B cell stages in the bone marrow (BM) and lower in the pre-B, immature, and transitional stages of spleen [82]. After generation of the mature B cell repertoires, energy needs be uptake and utilized for antigen-driven proliferation and antibody production [81]. Glucose, glutamine, and fatty acids are three necessary sources of nutrients for B cell growth and energy support [83]. Glucose uptake, which boosts dramatically after B cell activation, provides the substrate for glycolysis and pyruvate molecules for TCA cycle entry [84]. Uptake and utilization of glutamine also increases dramatically after B cell activation, followed by the induction of increased solute transporter proteins and is up-regulated in a manner dependent on PDK1 and AKT [85], [86]. Importantly, glutamine provides another potential source of nitrogen and enters mitochondria and feeds the TCA cycle after anaplerotic conversion into a-ketoglutarate (a-KG) through glutaminase (Gls)-mediated generation of glutamate [87]. In TLR4 agonist-activated B cells, analogous mTORC1-dependent increases in amino acid uptake and mTORC1 activation have been noted [88]. C-Myc, which is a canonical target of let-7 [89], is a master regulator of cell proliferation and growth [90], [91], and it promotes transcription and translation of key effectors in response to nutrient uptake and mTOR activation [92]. Our recent work shows that let-7adf specifically inhibits T cell-independent (TI) antigen-induced IgM antibody production and we find this miRNA cluster suppresses the acquisition and utilization of both glucose and glutamine, through directly targeting hexokinase 2 (Hk2) and repressing a glutamine transporter Slc1a5 and Gls, a mechanism mediated by regulation of c-Myc (Figure 4A). This study suggests a novel role of let-7adf as a “metabolic brake” on B cell antibody production [44]. Although it is clear that the let-7adf cluster acts as a negative regulator of TI-induced antibody production, a deeper understanding of whether and how let-7 clusters regulate metabolic phenotypes to support B cell functions, such as CSR and memory responses, and will shed light on how let-7 adapts to different physiological environments. In addition, further understanding the let-7adf cluster's pathological role in B cells requires long-term monitoring of aging let-7adf KO mice or using the pathological mouse models such as systemic lupus erythematosus (SLE) model. The potential for mechanistic links between intracellular metabolites to effect changes in the posttranscriptional networks mediated by miRNAs such as let-7 needs to be elucidated as well in future.
Figure 4.
Let-7 plays critical roles in different types of immune cells.
In the case of regulating B cell metabolism, the body of work with B cells is as yet far less extensive investigated than that with T cells or macrophages. For example, macrophages fulfill a broad range of functions in host defense, tissue homeostasis and pathology [93], [94]. Recently, a growing number of studies highlight the crucial role of metabolic reprogramming in macrophage activation [95], [96]. Metabolic pathways not only provide energy but also regulate the macrophage's phenotype and functions. While the induction of glycolysis is well accepted as a key feature of inflammatory macrophages (M1 macrophages), the mechanisms linking metabolic rewiring and macrophage function remained largely obscure. Literature elegantly demonstrates how particular glycolytic enzymes support pro-inflammatory macrophage functions [97], [98]. Succinate, a TCA cycle intermediate, is a critical metabolite that modulates macrophage inflammatory responses by enhancing expression of the key pro-inflammatory cytokine, IL-1β [99], [100]. Our recent work shows that using both KO and myeloid cell-specific transgenic mouse models, we find that a physiological role of the let-7adf cluster is to promote IL-6 secretion by LPS-activated macrophages through down-regulating Tet2 (Figure 4B). This let-7 cluster represses Tet2 using two different mechanisms: direct targeting and indirect enhancement of succinate accumulation. A further understanding of how let-7 clusters regulate metabolic phenotypes support macrophage activation, such as M2 macrophage responses and monocytes differentiation, and will shed light on how let-7 adapts to different innate immune systems.
Lately, endogenous metabolites including itaconate and succinate have emerged as key regulators of macrophage function [100], [101], but their precise regulatory mechanism of action remains unclear. Although the let-7adf cluster is critical regulator of macrophage inflammatory responses, whether and how they are involved in M1 macrophages needs to be further understood. In addition, it may be possible to target metabolic changes in macrophages therapeutically through anti-let-7. Moreover, approaches might include inhibition of isocitrate dehydrogenase (IDH) and/or SDH, which will prevent inflammatory macrophage activation while supporting anti-inflammatory pathways, including the induction of IL-10 and suppression of IL-1β. Further work will be needed to explore the utility of targeting specific metabolic events in macrophages for therapeutic gain and anti-let-7 treatment might have good potential.
CD4 T cells can respond to T cell receptor (TCR) signaling with full activation and the acquisition of effector functions [102]. Anergy CD4 T cells display the inability to proliferate and effector functions including cytokine secretion in response to secondary stimulation [103]. Profiling analysis of TCR-induced signaling showed elevated phosphorylation of AKT S473 and S6 ribosomal protein [104], indicating hyperactivity of mTORC1 and mTORC2 and overexpression of the key mTOR complex components Raptor and Rictor [105]. It has been reported that let-7c-mediated regulation of mTORC2 can facilitate discrimination between activation and anergy in CD4 T cells (Figure 4C) [106]. CD8 T cells undergo rapid clonal expansion after antigen recognition, followed by differentiation into effector cytotoxic T lymphocytes (CTLs) and memory T cells, which are both capable of producing effector cytokines and eliminate target cells [107]. This transition in CD8 T cells is accompanied with metabolic reprograming from oxidative phosphorylation to aerobic glycolysis, providing energy and larger amounts of the biomacromolecular intermediates needed to support growth [108]. Let-7 g acts as a molecular hub by converting the strength of TCR signaling into CD8 T cell function and it suppress the proliferation and metabolism of activated CD8 T cells through the suppression of c-Myc, and repression of effector function through both eomesodermin (Eomes)-dependent and -independent mechanisms (Figure 4D) [47]. It is interesting to investigate whether let-7 is involved in other T cell subtypes such as Th1 and Th17 cells. The T cell conditional KO or iTg mouse models might be utilized to answer these questions.
Regarding the influence of let-7 on the complex crosstalk among T cells, B cells, and macrophages, there have been no studies demonstrating this role in the pathological disease models such as murine systemic lupus erythematosus (SLE) model. In thinking of these similar clusters, however, it is important to consider that single or multiple clusters may be involved in regulating a single mRNA target that may be involved in regulating immune cell functions. It is now well recognized that let-7 controls metabolism in several subtypes of immune cells, thereby generating a functional link, which disrupts energy homeostasis in case of disconnection in the miRNA-metabolism interplays. A challenging road lies ahead for defining the role of let-7 cluster in other subtypes of immune cells such as dendritic cells (DCs) and regulatory T cells (Treg) cells, which play important roles in the pathogenesis of diseases such as T2D and for establishing their usefulness as new medications and clinically reliable biomarkers.
Current Opinion and Future Conceptual Perspective on Let-7
Despite great progress in understanding the let-7 cluster's roles in metabolic programming in different types of cells including immune cells, several key questions remain unanswered. Let-7 functions as efficient fine-tuners of protein expression, rather than as “on–off” switches and they represent a mechanism to balance the immune cell responses, which when left unchecked, can have physiological or pathological effects. Future studies should also be undertaken to find out whether other factors are utilized in most tissues to modulate expression of let-7 in various immune cells. Given the importance of let-7 in processes such as inflammation, and antibody production and understanding of these precisely controllers may translate in therapeutic strategies to treat human diseases.
A central issue regarding let-7 concerns the assessment of let-7 activity and function, both of which depend on which let-7 cluster or member functions and on the relative let-7 targets. When considering the therapeutic potential of let-7, it must be kept in mind that the factors influencing biological roles of let-7 should also be considered by its own concentration, in addition to its multiple targets. Certain mature let-7 might regulate the expression of bioprocessing of its own or other let-7 clusters in the cytoplasm, where they function in a noncanonical manner by directly regulating Dicer [109]. Extracellular let-7 are indeed involved in cell–cell communications, for instance, exosome-mediated transfer of let-7d from Treg cell to Th1 cells contributed to suppression and prevention of systemic disease, which would show promise for their use as potential therapeutic tools [110], [111]. Exosomes are predicted to have low antigenicity and toxicity, thus it is extremely important as carriers for targeted delivery of let-7. Such a novel experimental tool would reveal their possible utility in the treatment of diseases or as targets of therapeutic interventions. In addition, the possible clinical roles that let-7 might play in immune pathology could be understood by using engineered let-7 cluster mouse models. A very important question is that of identifying and validating the let-7 cluster's targetome in different types of immune cells, which is critical for our understanding of the regulatory networks governing biological processes mediated by let-7. It is also important to note that an increased number of validated target set might open up new avenues for therapeutic intervention in those settings, where one or more of the cluster's members are involved. Comparing different mechanisms by which individual let-7 cluster affects the immune system will be required to elucidate their roles in the in vivo disease models. Mouse disease models in vivo would shed light on the potentiality of targeting let-7 components therapeutically.
Although the use of intravenous delivery of anti-let-7 for the tumor treatment in immune-compromised mice has showed effective role in blockage of tumor growth, indicating let-7 as a potential therapeutic target [112], [113], [114], [115], there are still some issues need to be addressed, for instance, the selective delivery such as the blood–brain barrier (BBB) and side effects. Further studies are necessary to assess the impact of specific let-7 cluster-mediated therapies for prevention of off-target effects and improvement of delivery efficiency while preventing inflammatory responses. Additionally, elucidating the individual functions of let-7 clusters towards different physiological roles will be fundamental to understand the degree of functional overlap and interworking among all these miRNA let-7 clusters.
Given the importance of let-7 in regulating metabolism, it is tempting to speculate that at least one let-7 cluster might play a role in modulating metabolism in each cellular context. The generation of engineered mouse models to study each individual let-7 member is warranted to fully answer these essential questions, which we trust will provide new insights into the regulation of crucial metabolic programs by let-7 and indicate an additional level of regulation by this conserved class of RNA molecules in the form of functional overlap.
Acknowledgments
Acknowledgements
The author sincerely apologizes to those whose work was not cited due to time and space constraints.
Conflict of interest
The author declares that I have no conflicts of interests.
References
- 1.O'Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016;16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Neill L. Immunometabolism and the land of milk and honey. Nat Rev Immunol. 2017;17:217. doi: 10.1038/nri.2017.22. [DOI] [PubMed] [Google Scholar]
- 3.Buck MD, Sowell RT, Kaech SM, Pearce EL. Metabolic Instruction of Immunity. Cell. 2017;169:570–586. doi: 10.1016/j.cell.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganeshan K, Chawla A. Metabolic regulation of immune responses. Annu Rev Immunol. 2014;32:609–634. doi: 10.1146/annurev-immunol-032713-120236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Assmann N, Finlay DK. Metabolic regulation of immune responses: therapeutic opportunities. J Clin Invest. 2016;126:2031–2039. doi: 10.1172/JCI83005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bettencourt IA, Powell JD. Targeting metabolism as a novel therapeutic approach to autoimmunity, inflammation, and transplantation. J Immunol. 2017;198:999–1005. doi: 10.4049/jimmunol.1601318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Norata GD, Caligiuri G, Chavakis T, Matarese G, Netea MG, Nicoletti A, O'Neill LA, Marelli-Berg FM. The Cellular and Molecular Basis of Translational Immunometabolism. Immunity. 2015;43:421–434. doi: 10.1016/j.immuni.2015.08.023. [DOI] [PubMed] [Google Scholar]
- 8.Williams NC, O'Neill LAJ. A role for the Krebs cycle intermediate citrate in metabolic reprogramming in innate immunity and inflammation. Front Immunol. 2018;9:141. doi: 10.3389/fimmu.2018.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corcoran SE, O'Neill LA. HIF1alpha and metabolic reprogramming in inflammation. J Clin Invest. 2016;126:3699–3707. doi: 10.1172/JCI84431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 11.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 12.Mehta A, Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nat Rev Immunol. 2016;16:279–294. doi: 10.1038/nri.2016.40. [DOI] [PubMed] [Google Scholar]
- 13.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 14.Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7:147–154. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han JG, Jiang YD, Zhang CH, Yang YM, Pang D, Song YN, Zhang GQ. A novel panel of serum miR-21/miR-155/miR-365 as a potential diagnostic biomarker for breast cancer. Ann Surg Treat Res. 2017;92:55–66. doi: 10.4174/astr.2017.92.2.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu H, Liu MF, Wang ED. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70:3119–3127. doi: 10.1158/0008-5472.CAN-09-4250. [DOI] [PubMed] [Google Scholar]
- 17.Narayan N, Morenos L, Phipson B, Willis SN, Brumatti G, Eggers S, Lalaoui N, Brown LM, Kosasih HJ, Bartolo RC, Zhou L, Catchpoole D, Saffery R, Oshlack A, Goodall GJ, Ekert PG. Functionally distinct roles for different miR-155 expression levels through contrasting effects on gene expression, in acute myeloid leukaemia. Leukemia. 2017;31:808–820. doi: 10.1038/leu.2016.279. [DOI] [PubMed] [Google Scholar]
- 18.Joyce CE, Novina CD. miR-155 in acute myeloid leukemia: not merely a prognostic marker? J Clin Oncol. 2013;31:2219–2221. doi: 10.1200/JCO.2012.48.3180. [DOI] [PubMed] [Google Scholar]
- 19.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang LF, Jiang S, Liu MF. MicroRNA regulation and analytical methods in cancer cell metabolism. Cell Mol Life Sci. 2017;74:2929–2941. doi: 10.1007/s00018-017-2508-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dumortier O, Hinault C, Van Obberghen E. MicroRNAs and metabolism crosstalk in energy homeostasis. Cell Metab. 2013;18:312–324. doi: 10.1016/j.cmet.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Lee H, Han S, Kwon CS, Lee D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell. 2016;7:100–113. doi: 10.1007/s13238-015-0212-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Balzeau J, Menezes MR, Cao S, Hagan JP. The LIN28/let-7 Pathway in Cancer. Front Genet. 2017;8:31. doi: 10.3389/fgene.2017.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang S, Baltimore D. RNA-binding protein Lin28 in cancer and immunity. Cancer Lett. 2016;375:108–113. doi: 10.1016/j.canlet.2016.02.050. [DOI] [PubMed] [Google Scholar]
- 26.Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer. 2010;17:F19–F36. doi: 10.1677/ERC-09-0184. [DOI] [PubMed] [Google Scholar]
- 27.Guan H, Fan D, Mrelashvili D, Hao H, Singh NP, Singh UP, Nagarkatti PS, Nagarkatti M. MicroRNA let-7e is associated with the pathogenesis of experimental autoimmune encephalomyelitis. Eur J Immunol. 2013;43:104–114. doi: 10.1002/eji.201242702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polikepahad S, Knight JM, Naghavi AO, Oplt T, Creighton CJ, Shaw C, Benham AL, Kim J, Soibam B, Harris RA, Coarfa C, Zariff A, Milosavljevic A, Batts LM, Kheradmand F, Gunaratne PH, Corry DB. Proinflammatory role for let-7 microRNAS in experimental asthma. J Biol Chem. 2010;285:30139–30149. doi: 10.1074/jbc.M110.145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubinsky AN, Dastidar SG, Hsu CL, Zahra R, Djakovic SN, Duarte S, Esau CC, Spencer B, Ashe TD, Fischer KM, MacKenna DA, Sopher BL, Masliah E, Gaasterland T, Chau BN, Pereira de Almeida L, Morrison BE, La Spada AR. Let-7 coordinately suppresses components of the amino acid sensing pathway to repress mTORC1 and induce autophagy. Cell Metab. 2014;20:626–638. doi: 10.1016/j.cmet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frost RJ, Olson E. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci U S A. 2011;108:21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen LH, Zhu H. Lin28 and let-7 in cell metabolism and cancer. Transl Pediatr. 2015;4:4–11. doi: 10.3978/j.issn.2224-4336.2015.01.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shyh-Chang N, Zhu H, Yvanka de Soysa T, Shinoda G, Seligson MT, Tsanov KM, Nguyen L, Asara JM, Cantley LC, Daley GQ. Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell. 2013;155:778–792. doi: 10.1016/j.cell.2013.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su JL, Chen PS, Johansson G, Kuo ML. Function and regulation of let-7 family microRNAs. Microrna. 2012;1:34–39. doi: 10.2174/2211536611201010034. [DOI] [PubMed] [Google Scholar]
- 34.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Zhang R, Su B. Diversity and evolution of MicroRNA gene clusters. Sci China C Life Sci. 2009;52:261–266. doi: 10.1007/s11427-009-0032-5. [DOI] [PubMed] [Google Scholar]
- 36.Rougvie AE. Control of developmental timing in animals. Nat Rev Genet. 2001;2:690–701. doi: 10.1038/35088566. [DOI] [PubMed] [Google Scholar]
- 37.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 38.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 39.Hertel J, Bartschat S, Wintsche A, Otto C, Students of the Bioinformatics Computer L, Stadler PF. Evolution of the let-7 microRNA family. RNA Biol. 2012;9:231–241. doi: 10.4161/rna.18974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frasch M. A matter of timing: microRNA-controlled temporal identities in worms and flies. Genes Dev. 2008;22:1572–1576. doi: 10.1101/gad.1690608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aguirre A, Montserrat N, Zacchigna S, Nivet E, Hishida T, Krause MN, Kurian L, Ocampo A, Vazquez-Ferrer E, Rodriguez-Esteban C, Kumar S, Moresco JJ, Yates JR, 3rd, Campistol JM, Sancho-Martinez I, Giacca M, Izpisua Belmonte JC. In vivo activation of a conserved microRNA program induces mammalian heart regeneration. Cell Stem Cell. 2014;15:589–604. doi: 10.1016/j.stem.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuo G, Wu CY, Yang HY. MiR-17-92 cluster and immunity. J Formos Med Assoc. 2019;118(1 Pt 1):2–6. doi: 10.1016/j.jfma.2018.04.013. [DOI] [PubMed] [Google Scholar]
- 43.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang S, Yan W, Wang SE, Baltimore D. Let-7 Suppresses B Cell Activation through Restricting the Availability of Necessary Nutrients. Cell Metab. 2018;27:393–403 e394. doi: 10.1016/j.cmet.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 45.Banerjee S, Xie N, Cui H, Tan Z, Yang S, Icyuz M, Abraham E, Liu G. MicroRNA let-7c regulates macrophage polarization. J Immunol. 2013;190:6542–6549. doi: 10.4049/jimmunol.1202496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tolonen AM, Magga J, Szabo Z, Viitala P, Gao E, Moilanen AM, Ohukainen P, Vainio L, Koch WJ, Kerkela R, Ruskoaho H, Serpi R. Inhibition of Let-7 microRNA attenuates myocardial remodeling and improves cardiac function postinfarction in mice. Pharmacol Res Perspect. 2014;2:e00056. doi: 10.1002/prp2.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wells AC, Daniels KA, Angelou CC, Fagerberg E, Burnside AS, Markstein M, Alfandari D, Welsh RM, Pobezinskaya EL, Pobezinsky LA. Modulation of let-7 miRNAs controls the differentiation of effector CD8 T cells. Elife. 2017;6 doi: 10.7554/eLife.26398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z, Lin S, Li JJ, Xu Z, Yao H, Zhu X, Xie D, Shen Z, Sze J, Li K, Lu G, Chan DT, Poon WS, Kung HF, Lin MC. MYC protein inhibits transcription of the microRNA cluster MC-let-7a-1~let-7d via noncanonical E-box. J Biol Chem. 2011;286:39703–39714. doi: 10.1074/jbc.M111.293126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, Jung J, Gao P, Dang CV, Beer MA, Thomas-Tikhonenko A, Mendell JT. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci U S A. 2009;106:3384–3389. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kehl T, Backes C, Kern F, Fehlmann T, Ludwig N, Meese E, Lenhof HP, Keller A. About miRNAs, miRNA seeds, target genes and target pathways. Oncotarget. 2017;8:107167–107175. doi: 10.18632/oncotarget.22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang H, Artiles KL, Fire AZ. Functional relevance of "seed" and "non-seed" sequences in microRNA-mediated promotion of C. elegans developmental progression. RNA. 2015;21:1980–1992. doi: 10.1261/rna.053793.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye W, Lv Q, Wong CK, Hu S, Fu C, Hua Z, Cai G, Li G, Yang BB, Zhang Y. The effect of central loops in miRNA:MRE duplexes on the efficiency of miRNA-mediated gene regulation. PLoS One. 2008;3:e1719. doi: 10.1371/journal.pone.0001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steiman-Shimony A, Shtrikman O, Margalit H. Assessing the functional association of intronic miRNAs with their host genes. RNA. 2018;24(8):991–1004. doi: 10.1261/rna.064386.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rottiers V, Najafi-Shoushtari SH, Kristo F, Gurumurthy S, Zhong L, Li Y, Cohen DE, Gerszten RE, Bardeesy N, Mostoslavsky R, Naar AM. MicroRNAs in metabolism and metabolic diseases. Cold Spring Harb Symp Quant Biol. 2011;76:225–233. doi: 10.1101/sqb.2011.76.011049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hartig SM, Hamilton MP, Bader DA, McGuire SE. The miRNA Interactome in Metabolic Homeostasis. Trends Endocrinol Metab. 2015;26:733–745. doi: 10.1016/j.tem.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deiuliis JA. MicroRNAs as regulators of metabolic disease: pathophysiologic significance and emerging role as biomarkers and therapeutics. Int J Obes. 2016;40:88–101. doi: 10.1038/ijo.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Z, Wu S, Muhammad S, Ren Q, Sun C. miR-103/107 promote ER stress-mediated apoptosis via targeting the Wnt3a/beta-catenin/ATF6 pathway in preadipocytes. J Lipid Res. 2018;59:843–853. doi: 10.1194/jlr.M082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fu T, Choi SE, Kim DH, Seok S, Suino-Powell KM, Xu HE, Kemper JK. Aberrantly elevated microRNA-34a in obesity attenuates hepatic responses to FGF19 by targeting a membrane coreceptor beta-Klotho. Proc Natl Acad Sci U S A. 2012;109:16137–16142. doi: 10.1073/pnas.1205951109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shinoda G, Shyh-Chang N, Soysa TY, Zhu H, Seligson MT, Shah SP, Abo-Sido N, Yabuuchi A, Hagan JP, Gregory RI, Asara JM, Cantley LC, Moss EG, Daley GQ. Fetal deficiency of lin28 programs life-long aberrations in growth and glucose metabolism. Stem Cells. 2013;31:1563–1573. doi: 10.1002/stem.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, Urbach A, Thornton JE, Triboulet R, Gregory RI, Consortium D, Investigators M, Altshuler D, Daley GQ. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, Asara JM, Daley GQ, Cantley LC. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma X, Li C, Sun L, Huang D, Li T, He X, Wu G, Yang Z, Zhong X, Song L. Lin28/let-7 axis regulates aerobic glycolysis and cancer progression via PDK1. Nat Commun. 2014;5:5212. doi: 10.1038/ncomms6212. [DOI] [PubMed] [Google Scholar]
- 65.O'Toole TE, Abplanalp W, Li X, Cooper N, Conklin DJ, Haberzettl P, Bhatnagar A. Acrolein decreases endothelial cell migration and insulin sensitivity through induction of let-7a. Toxicol Sci. 2014;140:271–282. doi: 10.1093/toxsci/kfu087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang YT, Tsai PC, Liao YC, Hsu CY, Juo SH. Circulating microRNAs have a sex-specific association with metabolic syndrome. J Biomed Sci. 2013;20:72. doi: 10.1186/1423-0127-20-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaur J. A comprehensive review on metabolic syndrome. Cardiol Res Pract. 2014;2014:943162. doi: 10.1155/2014/943162. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68.Jiang LQ, Franck N, Egan B, Sjogren RJ, Katayama M, Duque-Guimaraes D, Arner P, Zierath JR, Krook A. Autocrine role of interleukin-13 on skeletal muscle glucose metabolism in type 2 diabetic patients involves microRNA let-7. Am J Physiol Endocrinol Metab. 2013;305:E1359–E1366. doi: 10.1152/ajpendo.00236.2013. [DOI] [PubMed] [Google Scholar]
- 69.Dowen RH, Breen PC, Tullius T, Conery AL, Ruvkun G. A microRNA program in the C. elegans hypodermis couples to intestinal mTORC2/PQM-1 signaling to modulate fat transport. Genes Dev. 2016;30:1515–1528. doi: 10.1101/gad.283895.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen PS. miR-107 promotes tumor progression by targeting the let-7 microRNA in mice and humans. J Clin Invest. 2017;127:1116. doi: 10.1172/JCI92099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katayama M, Sjogren RJ, Egan B, Krook A. miRNA let-7 expression is regulated by glucose and TNF-alpha by a remote upstream promoter. Biochem J. 2015;472:147–156. doi: 10.1042/BJ20150224. [DOI] [PubMed] [Google Scholar]
- 72.Lozier AM, Rich ME, Grawe AP, Peck AS, Zhao P, Chang AT, Bond JP, Sholler GS. Targeting ornithine decarboxylase reverses the LIN28/Let-7 axis and inhibits glycolytic metabolism in neuroblastoma. Oncotarget. 2015;6:196–206. doi: 10.18632/oncotarget.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gao Y, Wu F, Zhou J, Yan L, Jurczak MJ, Lee HY, Yang L, Mueller M, Zhou XB, Dandolo L. The H19/let-7 double-negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic Acids Res. 2014;42:13799–13811. doi: 10.1093/nar/gku1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shu CJ, Benoist C, Mathis D. The immune system's involvement in obesity-driven type 2 diabetes. Semin Immunol. 2012;24:436–442. doi: 10.1016/j.smim.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lehuen A, Diana J, Zaccone P, Cooke A. Immune cell crosstalk in type 1 diabetes. Nat Rev Immunol. 2010;10:501–513. doi: 10.1038/nri2787. [DOI] [PubMed] [Google Scholar]
- 77.Kornete M, Mason ES, Piccirillo CA. Immune Regulation in T1D and T2D: Prospective Role of Foxp3+ Treg Cells in Disease Pathogenesis and Treatment. Front Endocrinol (Lausanne) 2013;4:76. doi: 10.3389/fendo.2013.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.DeFuria J, Belkina AC, Jagannathan-Bogdan M, Snyder-Cappione J, Carr JD, Nersesova YR, Markham D, Strissel KJ, Watkins AA, Zhu M. B cells promote inflammation in obesity and type 2 diabetes through regulation of T-cell function and an inflammatory cytokine profile. Proc Natl Acad Sci U S A. 2013;110:5133–5138. doi: 10.1073/pnas.1215840110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Franchina DG, Grusdat M, Brenner D. B-Cell Metabolic Remodeling and Cancer. Trends Cancer. 2018;4:138–150. doi: 10.1016/j.trecan.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 80.Kunisawa J, Sugiura Y, Wake T, Nagatake T, Suzuki H, Nagasawa R, Shikata S, Honda K, Hashimoto E, Suzuki Y. Mode of Bioenergetic Metabolism during B Cell Differentiation in the Intestine Determines the Distinct Requirement for Vitamin B1. Cell Rep. 2015;13:122–131. doi: 10.1016/j.celrep.2015.08.063. [DOI] [PubMed] [Google Scholar]
- 81.Mauri C, Bosma A. Immune regulatory function of B cells. Annu Rev Immunol. 2012;30:221–241. doi: 10.1146/annurev-immunol-020711-074934. [DOI] [PubMed] [Google Scholar]
- 82.Boothby M, Rickert RC. Metabolic regulation of the immune humoral response. Immunity. 2017;46:743–755. doi: 10.1016/j.immuni.2017.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Teuwen LA, Geldhof V, Carmeliet P. How glucose, glutamine and fatty acid metabolism shape blood and lymph vessel development. Dev Biol. 2019;447(1):90–102. doi: 10.1016/j.ydbio.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 84.Olenchock BA, Rathmell JC, Vander Heiden MG. Biochemical underpinnings of immune cell metabolic phenotypes. Immunity. 2017;46:703–713. doi: 10.1016/j.immuni.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Baracho GV, Cato MH, Zhu Z, Jaren OR, Hobeika E, Reth M, Rickert RC. PDK1 regulates B cell differentiation and homeostasis. Proc Natl Acad Sci U S A. 2014;111:9573–9578. doi: 10.1073/pnas.1314562111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim S, Jee K, Kim D, Koh H, Chung J. Cyclic AMP inhibits Akt activity by blocking the membrane localization of PDK1. J Biol Chem. 2001;276:12864–12870. doi: 10.1074/jbc.M001492200. [DOI] [PubMed] [Google Scholar]
- 87.Yang C, Ko B, Hensley CT, Jiang L, Wasti AT, Kim J, Sudderth J, Calvaruso MA, Lumata L, Mitsche M. Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol Cell. 2014;56:414–424. doi: 10.1016/j.molcel.2014.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Raybuck AL, Cho SH, Li J, Rogers MC, Lee K, Williams CL, Shlomchik M, Thomas JW, Chen J, Williams JV. B Cell-Intrinsic mTORC1 Promotes Germinal Center-Defining Transcription Factor Gene Expression, Somatic Hypermutation, and Memory B Cell Generation in Humoral Immunity. J Immunol. 2018;200:2627–2639. doi: 10.4049/jimmunol.1701321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 90.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hsieh AL, Walton ZE, Altman BJ, Stine ZE, Dang CV. MYC and metabolism on the path to cancer. Semin Cell Dev Biol. 2015;43:11–21. doi: 10.1016/j.semcdb.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Luo W, Weisel F, Shlomchik MJ. B cell receptor and CD40 signaling are rewired for synergistic induction of the c-Myc transcription factor in germinal center B cells. Immunity. 2018;48:313–326 e315. doi: 10.1016/j.immuni.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Varol C, Mildner A, Jung S. Macrophages: development and tissue specialization. Annu Rev Immunol. 2015;33:643–675. doi: 10.1146/annurev-immunol-032414-112220. [DOI] [PubMed] [Google Scholar]
- 94.Gordon S, Pluddemann A. Tissue macrophages: heterogeneity and functions. BMC Biol. 2017;15:53. doi: 10.1186/s12915-017-0392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Minton K Immunometabolism. Stress-induced macrophage polarization. Nat Rev Immunol. 2017;17:277. doi: 10.1038/nri.2017.41. [DOI] [PubMed] [Google Scholar]
- 96.Van den Bossche J, O'Neill LA, Menon D Macrophage Immunometabolism. Where Are We (Going)? Trends Immunol. 2017;38:395–406. doi: 10.1016/j.it.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 97.Langston PK, Shibata M, Horng T. Metabolism supports macrophage activation. Front Immunol. 2017;8:61. doi: 10.3389/fimmu.2017.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Curi R, de Siqueira Mendes R, de Campos Crispin LA, Norata GD, Sampaio SC, Newsholme P. A past and present overview of macrophage metabolism and functional outcomes. Clin Sci (Lond) 2017;131:1329–1342. doi: 10.1042/CS20170220. [DOI] [PubMed] [Google Scholar]
- 99.Mills E, O'Neill LA. Succinate: a metabolic signal in inflammation. Trends Cell Biol. 2014;24:313–320. doi: 10.1016/j.tcb.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 100.Mills EL, Kelly B, Logan A, Costa ASH, Varma M, Bryant CE, Tourlomousis P, Dabritz JHM, Gottlieb E, Latorre I. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell. 2016;167:457–470. doi: 10.1016/j.cell.2016.08.064. [e413] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lampropoulou V. Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab. 2016;24:158–166. doi: 10.1016/j.cmet.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bantug GR, Galluzzi L, Kroemer G, Hess C. The spectrum of T cell metabolism in health and disease. Nat Rev Immunol. 2018;18:19–34. doi: 10.1038/nri.2017.99. [DOI] [PubMed] [Google Scholar]
- 104.Genot EM, Arrieumerlou C, Ku G, Burgering BM, Weiss A, Kramer IM. The T-cell receptor regulates Akt (protein kinase B) via a pathway involving Rac1 and phosphatidylinositide 3-kinase. Mol Cell Biol. 2000;20:5469–5478. doi: 10.1128/mcb.20.15.5469-5478.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12:325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Marcais A, Blevins R, Graumann J, Feytout A, Dharmalingam G, Carroll T, Amado IF, Bruno L, Lee K, Walzer T. microRNA-mediated regulation of mTOR complex components facilitates discrimination between activation and anergy in CD4 T cells. J Exp Med. 2014;211:2281–2295. doi: 10.1084/jem.20132059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang N, Bevan MJ. CD8(+) T cells: foot soldiers of the immune system. Immunity. 2011;35:161–168. doi: 10.1016/j.immuni.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.van der Windt GJ, Pearce EL. Metabolic switching and fuel choice during T-cell differentiation and memory development. Immunol Rev. 2012;249:27–42. doi: 10.1111/j.1600-065X.2012.01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu B, Liu M, Wang J, Zhang X, Wang X, Wang P, Wang H, Li W, Wang Y. DICER-dependent biogenesis of let-7 miRNAs affects human cell response to DNA damage via targeting p21/p27. Nucleic Acids Res. 2015;43:1626–1636. doi: 10.1093/nar/gku1368. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 110.Matsuura K, De Giorgi V, Schechterly C, Wang RY, Farci P, Tanaka Y, Alter HJ. Circulating let-7 levels in plasma and extracellular vesicles correlate with hepatic fibrosis progression in chronic hepatitis C. Hepatology. 2016;64:732–745. doi: 10.1002/hep.28660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Okoye IS, Coomes SM, Pelly VS, Czieso S, Papayannopoulos V, Tolmachova T, Seabra MC, Wilson MS. MicroRNA-Containing T-Regulatory-Cell-Derived Exosomes Suppress Pathogenic T Helper 1 Cells. Immunity. 2014;41:503. doi: 10.1016/j.immuni.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yang X, Rutnam ZJ, Jiao C, Wei D, Xie Y, Du J, Zhong L, Yang BB. An anti-let-7 sponge decoys and decays endogenous let-7 functions. Cell Cycle. 2012;11:3097–3108. doi: 10.4161/cc.21503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Price C, Chen J. MicroRNAs in cancer biology and therapy: current status and perspectives. Genes Dis. 2014;1:53–63. doi: 10.1016/j.gendis.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Moore KJ, Rayner KJ. Local anti-miR delivery: the latest in the arsenal of drug-eluting stents. Arterioscler Thromb Vasc Biol. 2015;35:1905–1906. doi: 10.1161/ATVBAHA.115.306187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim DG, Kim KH, Seo YJ, Yang H, Marcusson EG, Son E, Lee K, Sa JK, Lee HW, Nam DH. Anti-miR delivery strategies to bypass the blood-brain barrier in glioblastoma therapy. Oncotarget. 2016;7:29400–29411. doi: 10.18632/oncotarget.8837. [DOI] [PMC free article] [PubMed] [Google Scholar]