Abstract
Non-coding RNAs (ncRNAs) are crucial regulatory elements in most biological processes and reproduction is also controlled by them. The different types of ncRNAs, as well as the high complexity of these regulatory pathways, present a complex scenario; however, recent studies have shed some light on these questions, discovering the regulatory function of specific ncRNAs on concrete reproductive biology processes. This mini review will focus on the role of ncRNAs in spermatogenesis and oogenesis, and their potential use as biomarkers for reproductive diseases or for reproduction success.
Keywords: Non-coding RNAs, Spermatogenesis, Oogenesis, Reproductive transgenerational effects, Biomarkers
1. Introduction
Non-coding RNAs (ncRNAs) are regulatory elements of gene expression and chromatin structure [1]. It is well established that differential susceptibility to these non-coding RNAs contributes to tissue-specific gene expression. They are relevant from very early stages of germ line development, but their function is not only important in these early stages; non-coding RNAs are crucial players in postranscriptional gene control in spermatogenesis and oogenesis. In this review we will show the latest findings on the role of these molecules in such complex and highly regulated processes.
There are different classes of ncRNAs, but in this mini review we will focus mainly on microRNAs (miRNAs), and, in less detail, on Piwi interacting RNAs (piRNAS) and long non-coding RNAs (lnc RNAs) and their role in gametogenesis.
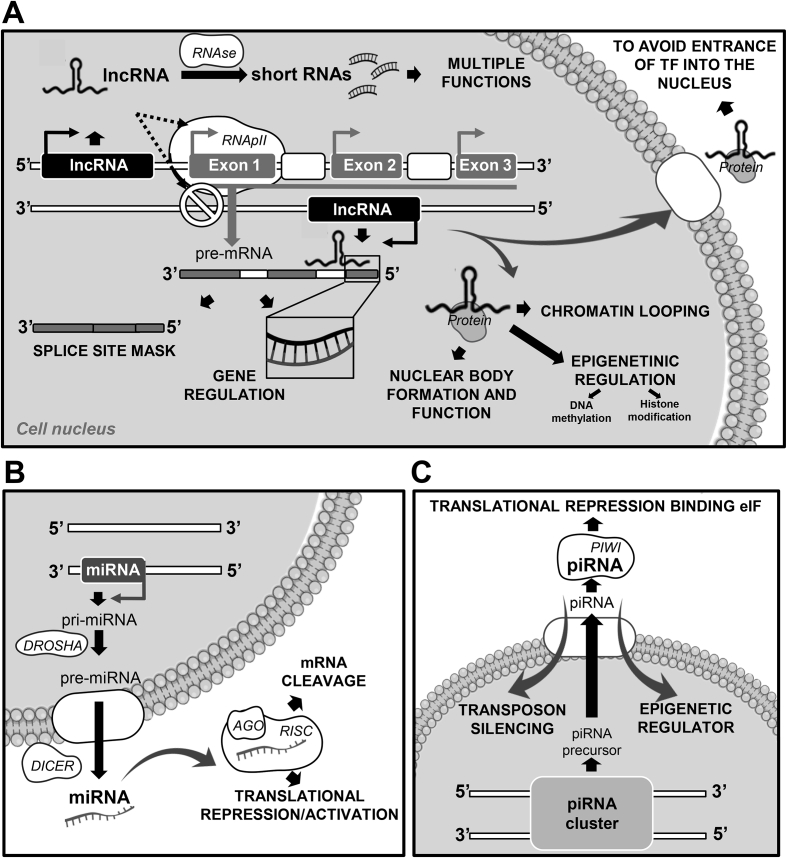
miRNA biogenesis is orchestrated at several levels: transcription (processing by Drosha and Dicer), RNA methylation and adenylation and uridylation. In the canonical pathway, miRNAs are transcribed as large primary miRNAs (pri-miRNAs) in the nucleus and after that these pri-miRNAs are methylated by methyltransferase 3 (METTL3) [2,3]. Subsequently, these pri-miRNAS are processed into mi-RNA precursor pre-miRNAs, (60–70 nt) by the DROSHA-DGCR8 complex and exported to the cytoplasm by exportin-5 (Exp-5). In the cytoplasm, pre-miRNAS are cleaved by DICER to form a double-stranded mature miRNA of 22 nucleotides which are incorporated into a ribonuclear vesicle forming the RNA-induced silencing complex (RISC) [2,3] (Fig. 1). In the non-canonical mode, several pathways are included such as ii) DICER-independent and DROSHA-DGCR8 dependent and ii) DICER-dependent and DROSHA-DGCR8 independent [4]. Small nucleolar RNAs (snoRNAs) are an example of the second pathway where the processing of snoRNAs to miRNAs requires Dicer activity but is independent of Drosha/DGCR8 [4]. Several miRNAs are crucial in different steps of gametogenesis, either for maintaining the undifferentiated state or inducing differentiation. The fact that one single miRNA could have different mRNA targets [5] and one specific mRNA 3′UTR could be regulated by several miRNAs, adds complexity to this regulatory pathway. Recently it was also discovered that a class of miRNA-recognition elements exist in protein-coding sequences (CDS) and CDS-targeted miRNAs are thought to be GW182 (glycine-tryptophan repeat-containing protein of 182 kDa) independent and induce transient ribosome stalling instead of mRNA destabilization [6]. In mammals, it is known that miRNAs control about 50% of all protein-coding genes and participate in most cellular processes [2]. Regarding piRNAs, this regulatory pathway is responsible for germline specification and gametogenesis and plays an important role in defending against transposable elements during the epigenetic reprogramming stage in the fetal testis [7] (Fig. 1). Two different pathways are involved in the piRNA biogenesis model: the primary processing pathway and a secondary amplification pathway (ping-pong amplification loop) that amplifies secondary piRNAs. In the germline, piRNA biogenesis involves both pathways [8]. In the first, piRNAs are transcribed from genomic regions (piRNA clusters), processed, and loaded onto Piwi or Aub. Piwi carries out transcriptional gene silencing in the nucleus [8]. The Aub–piRNA complex (some maternally inherited) serves as a trigger to start the ping-pong amplification pathway. The ping-pong pathway silences the target transposon sequence and amplifies the piRNA one [8].
Fig. 1.
Non coding RNAs. Mechanisms of action. A.- Long non coding RNAs and short RNAs. B.- miRNAs. C.- piRNAs.
Both types of molecules (miRNA and piRNA) require a specific group of protein regulator: miRNAs are always dependent on Argonaute proteins, which are essential components of the RISC, and piRNAs require PIWI proteins to be functional. However, lncRNAs, which represent the largest proportion of ncRNAs, do not require a specific group of proteins to be fully functional and their biogenesis has unique features [9]. Instead, they are able to interact with a wide variety of proteins or even with other ncRNAs to perform diverse functions in both the nucleus and cytoplasm (Fig. 1). They can inhibit DNA binding proteins and miRNA-induced mRNA degradation and translation, promote DNA methylation by binding to DNA methylation factors or promote histone modification by binding to histones (Fig. 1). They can act as precursors of short RNAs with different regulatory roles in the cell [10] and are also important in chromatin looping and nuclear body formation and function [11] (Fig. 1). Interestingly, some elements of these different pathways seem to converge in a germ-cell specific RNA processing centre known as the Cromatoid Body, which organizes and controls RNA processing in male germ cells [12]. This mini-review addresses recent findings on the role of these molecules in gametogenesis as well as their relationship with reproductive disorders (Table 1), transgenerational effects and their potential importance as biomarkers for reproductive success.
Table 1.
Summary of differential expression of ncRNAs linked to male reproductive disorders.
| Reproductive disorder | Sample | non-coding RNAs | Species | Ref. |
|---|---|---|---|---|
| Azoospermia | Seminal plasma | ↓miR-34c-5p, ↓miR-122, ↓miR-146b-5p, ↓miR-181a, | Human (Homo sapiens) | 101 |
| ↓miR-374b, ↓miR-509-5p, ↓miR-513a-5p | ||||
| Seminal plasma | ↓piR-31068, ↓piR-31925, ↓miR-43771, ↓piR-43773 | Human (Homo sapiens) | 114 | |
| ↓piR-31098 | ||||
| Asthenozoospermia | Seminal plasma | ↑miR-34c-5p, ↑miR-122, ↑miR-146b-5p, ↑miR-181a, | Human (Homo sapiens) | 101 |
| ↑miR-374b, ↑miR-509-5p, ↑miR-513a-5p | ||||
| Sperm | ↑miR-30a, ↑miR-363, ↑miR-26a, ↑miR-200a, ↑miR-141, | Human (Homo sapiens) | 102 | |
| ↑miR-429, ↑miR-193b | ||||
| ↓miR-122, ↓miR-34b, ↓miR-1973 | ||||
| Seminal plasma | ↓piR-31068, ↓piR-31925, ↓miR-43771, ↓piR-43773 | Human (Homo sapiens) | 114 | |
| Sperm | ↓Nrf2, ↓HOTAIR | Human (Homo sapiens) | 115 | |
| DNA damaged sperm | Seminal plasma | ↓miR-424/322 | Human (Homo sapiens) | 104 |
| Erectile dysfunction | Corpus cavernosum tissue | ↓miR-141 | Rat (Rattus norvegicus) | 113 |
| CP/CPPS | Expressed prostatic secretion (EPS) | ↑miR-21-5p, ↑miR-30a-5p, ↑miR-30d-5p, ↑miR103a-3p, | Human (Homo sapiens) | 111 |
| (Chronic prostatitis/Chronic pelvic pain syndrome) | ↑miR-141-3p | |||
| Non-obstructive azoospermia (NOA) | Testicular tissue | ↑miR-193a-3p, ↑miR-193a-5p, ↑miR-210, ↑miR-23a, | Human (Homo sapiens) | 99 |
| ↑miR-302a, ↑miR-371-5p, ↑miR-374a, ↑miR-423-3p, | ||||
| ↑miR-491-3p, ↑miR-554, ↑miR-557, ↑miR-565, | ||||
| ↑miR-572, ↑miR-638, ↑miR-654-5p, ↑miR-663, | ||||
| ↑miR-744, ↑miR-801 | ||||
| Testicular tissue (cryptorchid) | ↓miR-3663-5p, ↓miR-1233-3p, ↓miR-552-5p, ↓miR-449b-5p, | Human (Homo sapiens) | 100 | |
| ↓miR-7153-5p, ↓ miR-122-5p, ↓miR-552-3p, ↓miR-449a | ||||
| Testicular tissue | ↑miR-19b, ↑miR-let-7a | Human (Homo sapiens) | 110 | |
| Oligoasthenozoospermia | Sperm | ↑miR-141, ↑miR-193b, ↑miR-26a, ↑miR-200c, | Human (Homo sapiens) | 102 |
| ↑miR-29a, ↑miR-429, ↑miR-200a | ||||
| ↓miR-34b, ↓miR-15b, ↓miR-34c-5p,↓miR-449b, | ||||
| ↓miR-1973, ↓miR-122, ↓miR-16,↓miR-19a | ||||
| Seminal plasma | ↑miR-1275, ↑miR-4298, ↑miR-3675-3p, ↑miR-765, | Human (Homo sapiens) | 109 | |
| ↑miR-483-5p, ↑miR-1299, ↑miR-766, ↑miR-4306 | ||||
| ↓miR-26b, ↓miR-363, ↓miR-30b,↓miR-17, | ||||
| ↓miR-148a, ↓miR-21, ↓miR-20a,↓miR-23b | ||||
| Sperm | ↓Nrf2, ↓HOTAIR | Human (Homo sapiens) | 115 | |
| Prostate cancer | Blood | ↑miR-1825, ↑miR-484, ↑miR-141, | Human (Homo sapiens) | 112 |
| ↓miR-let-7b, ↓miR-205 | ||||
| SCO (Sertoli cell-only) syndrome | Testicular tissues | ↓miR-202-5p, ↓miR-34c-5p, ↓miR-10b, ↓miR-191, ↓miR-126 | Human (Homo sapiens) | 103 |
2. Primordial germ cell specification and spermatogenesis
2.1. PGC specification
Primordial germ cells (PGCs) are the embryonic precursors of germ cells that will generate the gametes (oocyte and spermatozoa) [13]. Germ cell specification, differentiation, and development involve complex regulatory networks at both transcriptional and post-transcriptional levels, including epigenetic mechanisms and the participation of small ncRNAs [14]. Different mechanisms of PGC specification have been described among diverse species. In mammals, germ cells are not specified by inheritance of maternal germplasm as occurs in teleost fish, but instead they are specified later by different inductive signals related to the bone morphogenetic protein ligands Bmp4 [15] and Bmp8b [16]. In this inductive mode, some miRNAs are selectively expressed in PGCs as miR-10b, −18a, −93, −106b, -126–3p, −127, −181a, −181b, and −301, all of them having an important function in these cells. These miRs are involved in differentiation, migration and apoptosis in PGCs in mice [17]. Specifically, miRNAs have been revealed as important in the PGC specification process, for instance the miR-290-295 cluster, whose deficiency results in an abnormal germ cell with deficient PGC migration in mice [18] or miR-29b, whose upregulation induces female PGC development by targeting DNA methyltransferase Dnmt3a and Dnmt3b [19].
In the inherited mode, the miRNA pathway plays a critical role in germ cell development as well, and some germ plasm-specific miRNAs like miR-202–5p, have recently been identified in zebrafish [20]. This miRNA was characterized as a PGC/germline marker in zebrafish due to its potential role in germ cell development. Moreover, other miRNAS have been related to PGC migration in this species, such as miR-430, regulating sdf1a and cxcr7 mRNAs key transcripts regulating migration [21]. In addition, it has been demonstrated in medaka that the interaction of dead end (dnd) transcript protects germ plasm RNAs from miR-430 mediated degradation to ensure PGC specification allowing PGC formation and maternal RNA turnover [22].
On the other hand, piRNAs are a class of sncRNAs that were discovered in germline cells. They safeguard the germline genome from retrotransposons and protect genomic stability [23,24]. For example, some functions have been described for piRNA pathway components in DNA methylation remodeling during early PGC specification in mammals [25]. In addition, although the effects of mutations on development differ among species, loss of Piwi function in mice or zebrafish, results in progressive loss of germ cells by apoptosis, thus demonstrating its importance in germ cell maintenance [26].
The role of lncRNAs in PGC specification has not been described. However, some reviews have suggested their possible implications in controlling transcription factors related to PGC specification such as BLIMP1/PRDM1 or DAZL [27,28]. Specifically, more than 300 binding sites of BLIMP1/PRDM1 in mouse PGCs, are associated with non-coding genes whose functions in PGCs specification are still unknown [27,29].
2.2. Spermatogenesis
Spermatogenesis is the process by which germ cells proliferate and differentiate into haploid male gametes. Post-transcriptional regulation is particularly important during the late steps of spermatogenesis when the compacting sperm nucleus becomes transcriptionally inhibited [30]. Non-coding RNAs have been shown to play a critical role during spermatogenesis in the control of gene expression, at the transcriptional level as components of chromatin remodeling complexes or post-transcriptional regulation [31]. This complex process is divided into three main phases and, interestingly, the miRNA profile is unique in each phase; (I) the first phase includes the mitotic proliferation and formation of spermatogonia from germ cells, (II) in the second phase spermatid formation occurs through spermatocyte meiosis and finally (III) spermiogenesis, this phase results in mature spermatozoa production from spermatids. In order to simplify, we will divide the process into early stages (phase I) and later stages of spermatogenesis (phases II and III):
2.2.1. The early stage of spermatogenesis
In this stage, different miR have been described in mammals as crucial for germ cell self-renewal and differentiation such as miR-34c. This miRNA promotes mouse spermatogonial stem cell (SSCs) differentiation by targeting Nanos2 [32]. Other key miRNAs are miR-293, 291a-5p, 290–5p and 294, whose targets are involved in cell cycle regulation [33]. In this sense, miR-21 inhibition increases the germ cell in the early stages of mouse spermatogenesis [34]. Other miRNAs, such as the Let-7 miR family, play an important role in mouse spermatogonial differentiation from undifferentiated spermatogonia to A1 spermatogonia through suppression of Lin28 [35] whereas others, such as miR-146, are crucial for keeping spermatogonia in an undifferentiated state in this species [36]. Additional miRNAs have been described as having a critical role in spermatogonial stem cell self-renewal and differentiation such as miR-20, miR-21 and miR-106 regulating spermatogonial homeostasis [37], miR-224 that promotes SSCs self-renewal via targeting DMRT1 in mouse [38], miR-202–3p involved in spermatogonial meiosis initiation and miR-10b related to SSC self-renewal via targeting KLF4 in mouse [39,40].
Some lncRNAs are known to carry out important functions in male germ cell development in mammals. Two spermatogonia specific lncRNA have been described, Spga-lncRNA1 and 2, which are crucial for maintaining SSC stemness [41]. Recently, lncRNA-033862 has been described as a molecular marker in SSC maintenance; this lncRNA, subjected to GDNF signaling, was highly expressed in mouse SSCs and could regulate the impaired self-renewal, survival and maintenance of SSCs [42].
2.2.2. The later stage of spermatogenesis
This stage includes meiosis phases and spermiogenesis. The role of miR has been largely described in mammals. Although miR 34-c has been identified in SSCs and its importance in germ cells previously described in the present review, this specific miR has an additional role in spermatocytes and round spermatids related to apoptosis events [43], and has been suggested as a regulator of the NOTCH signaling pathway that controls germ cell differentiation [44]. Interestingly, miR-34c in spermatozoa is also essential for first cell division of the mouse embryo [45]. Other miR involved in the regulation of meiotic and post meiotic events in later stages of spermatogenesis is the miR-449 cluster. Its upregulation is crucial in meiosis initiation during murine spermatogenesis [46]. This miR is also involved in germ cell apoptosis through its targets: BCL2 and AFT1 [40]. In the chromatin remodeling phase, some miRNAs such as miR-122 and 469 regulate chromatin condensation and protamine targeting [47].
In the zebrafish model, different miRNAs have been identified predominantly expressed in germ cells at different spermatogenetic stages (including spermatogonia and spermatozoa), for instance dre-miR-202–5p [48].
Recent studies reveal the importance of lncRNA in later stages of spermatogenesis. LncRNA-Tcam1 and lncRNA-HSVIII are described as crucial in pachytene spermatocytes, suggesting their participation in the transcriptional regulation of spermatocyte-specific gene expression. Specific lncRNAs are known to have other important functions, such as Tsx (testis-specific X-linked), which controls the apoptosis of pachytene spermatocytes in mouse [49]. Some additional functions related to post-transcriptional control have been determined for lncRNAS in this phase of spermatogenesis, for example, Tubulin cofactor A (TBCA), which interacts with tubulin during the microtubule rearrangement process [50]. However, the scenario in human is different, much less is known about lncRNAs in spermatogenesis. NLC1-C has been associated to male infertility through the control of miRNA expression via RNA-binding proteins in human spermatogenesis [51].
3. Oogenesis
Oogenesis is the process of creating the fertilizable ovum. It represents the most dramatic cellular differentiation pathway in females. It is a highly precise, orchestrated, and cyclic process. In folliculogenesis in mammals, small healthy primordial resting follicles (the oocyte and the surrounding somatic cells) progressively lead to the development and selection of dominant follicles (DFs) by extreme differentiation of both oocyte and somatic cells [52], especially granulose cells (GCs) [53]. Folliculogenesis is controlled by the luteinizing hormone (LH). Successive LH surge waves regulate the growth of a single follicle which becomes the dominant follicle (DF) while the rest, the subordinate follicles (SFs), undergo atresia [54]. The complex process from primordial follicles to mature follicles occurs by the differentiation, proliferation and apoptosis of granulose cells [55] and it requires a large network of gene interactions and tightly regulated genes. Communication between GCs and oocytes and vice versa [56,57] is crucial for oocyte growth. As expected, this differentiation and morphological transformations are complexly orchestrated by molecular mechanisms. Thus, dysregulation in the expression of key genes would be critical in determining the fate of DFs or SFs [[58], [59], [60]].
In recent years, post-transcriptional regulation by ncRNAs in this process has become a focus of attention. Many of the first related studies focused on identifying small RNA species within the organ, tissues and specific cell types of the gonad.
The vast majority of publications on ncRNAs in oogenesis and ovary function centre on miRNAs. These conserved tiny non-coding RNAs have been mainly studied using KO experiments in mice. Due to the importance of DROSHA-DGCR8 and DICER proteins in their biogenesis, conditional knockout (cKO) models have been developed to evaluate the general involvement of miRNAs in the ovary. Using these approaches, nowadays there is evidence of the key roles of miRNA in folliculogenesis, oocyte maturation and ovulation. Moreover, miRNAs are described as key elements in the assembly of primordial follicles, the transition from primordial to primary follicles, follicular growth, oocyte maduration, ovulation and the formation of the corpus luteum in mammals. As expected, miRNA expression within the ovary differs attending to cell type, function, and stage of the estrous cycle. miRNA screening analysis of mammalian ovarian tissues have defined the expression of these regulating molecules in the female gonads of several species [[61], [62], [63], [64], [65]].
Conditional knockout of Dicer1 from follicular GCs in mammals caused defects such as: abnormal oocyte maturation, disrupted follicular development and ovulation, increased follicular atresia, and infertility [[66], [67], [68]]. Interestingly, there are inklings about the regulation of bovine follicle development during the estrus cycle by a single miRNA via a canonical pathway, in which the target gene of miRNA plays a central role [69]. Predictably, numerous miRNAs target transcription factors (TFs) as well, such as TGF-β superfamily members, the luteinizing hormone receptor (LHR) and follicle stimulating hormone receptor (FSHR) closely linked to follicular development. Defects in these molecules similarly affect cellular communication and normal follicle development [[70], [71], [72]]. Furthermore, numerous publications have proven that specific miRNAs are involved in different aspects of GC processes [73], for instance, proliferation [38,[74], [75], [76]], survival [[77], [78], [79]], terminal differentiation [80], steroidogenesis [[81], [82], [83], [84]], and cumulus expansion [85] in mammals. As an example, miRNA-224 is proven to be involved in transforming growth factor-beta-mediated mouse GCs proliferation and GC function by targeting smad4 in mouse [83]. Thus, miRNAs may be involved in the selection of DFs. The difficult nature of miRNA target interaction, regulation, and function, nevertheless, represents a challenge for functional analysis. Some miRNAs regulate precise signaling pathways, others, the majority, perform in clusters and are fine regulators of cellular functions [86].
As a specific example of the regulatory role of miRNA in primordial follicle formation, the overexpression of miR-143 in murine 15.5 dpc ovaries inhibits the formation of primordial follicles by suppressing the proliferation of pre-granulosa cells. Transfected 18.5 dpc ovaries with miR-376a showed an increased number of primordial follicles. miR-320, miR-133, miR-383 modulate the secretion of E2, an inhibitor of primordial follicle formation, miR-145, miR-181a or the activity of activin, an inducer of primordial follicle formation (reviewed by Grossman and Shalgi [87])
The analysis of miRNAs in ovarian samples confirmed analogous expression patterns in ovaries in different mammalian species, including humans [61], mice [62,88,89], pigs [63], sheep [90], goats [64], and cows [65,[91], [92], [93]]. However, in other non-mammalian vertebrates such as teleost fish, the functional unit of the ovary is not the ovarian follicule. Although studied to a lesser extent, there is evidence of the role of ncRNAs in oogenesis in some fish species. For example, in zebrafish it was shown that a specific miRNA, miR-430, is responsible for maternal mRNA clearance during the embryonic development of zebrafish [94]. Bouchareb and colleagues in 2016, using microarray analysis, identified 66 miRNAs that are significantly overabundant in the ovary of the teleost model medaka [95]. miR-202 was among these sets of miRNAs. This molecule was previously described as being expressed in the gonads of other non-mammalian species like rainbow trout, frog or chicken [[96], [97], [98]]. Microarray revealed that miR-202 exhibited strict ovarian-predominance in medaka, suggesting the presence of an isomiR of miR-202–3p in Oryzias latipes [95]. In the same work, authors also described 8 novel ovarian-predominant miRNAs (miR-4785, miR-6352, miR-4653, miR-878, miR-487, miR-1288, miR-743, miR-729). Their analysis also revealed two miRNAs (miR-4785 and miR-6352) which exhibit strict ovarian expression suggesting key regulatory roles in oogenesis and/or early development, possibly involving a maternal effect since the predicted target of miR-4785 was fshr, and in the case of miR-6352 the predicted targets were smg8, ddx20 and ddx6 genes, all crucial in key ovarian regulation pathways.
4. The role of non-coding RNAs in some reproductive disorders: their potential use as biomarkers
The aberrant expression of non-coding RNAs could be associated with different disorders in humans [99]. Male infertility could be due to a complete lack of sperm (azoospermia), to low sperm production (oligospermia) or reduced sperm motility (asthenozoospermia), among other causes. A dysregulation of ncRNAs has been observed in all of the above-described pathological conditions. For example, the alteration of miRNA expression was detected in non-obstructive azoospermia (NOA) patients [100,101] and in asthenozoospermic patients [102]. A miRNA microarray analysis of spermatozoa from nine asthenozoospermic and nine oligoasthenozoospermic men revealed 50 upregulated, 27 downregulated miRNAs, and 42 upregulated, 44 downregulated miRNAs, respectively, when compared to normozoospermic men [103]. However, in males, the importance of miRNAs and their potential use as biomarkers are not limited to germ cells. An immunohistochemistry study performed on testicular tissue samples from azoospermic men with Sertoli cell-only syndrome (SCO syndrome), showed specific localization of miR-202–5p in Sertoli cells in normal fertile men, but not in those of infertile men with SCO [104]. Moreover, miRNAs are not only present within the cell. An analysis of seminal plasma from 94 infertile men with a high sperm DNA fragmentation index demonstrated that miR-424 is downregulated in this group, suggesting a role for miR-424 in repairing double-strand breaks during spermatogenesis [105].
The fact that these molecules can be found in extracellular fluids such as seminal plasma, follicular fluid, vaginal secretions, blood serum and saliva [106,107], and that they are stable and resistant to nuclease activity, enables then to be detected by noninvasive methods and could therefore be considered for possible use as non-invasive biomarkers [108]. In fact, miR-122–3p and miR-141–5p are proven to be stable in seminal plasma and are of certain value in the diagnosis of idiopathic asthenospermia [109]. Higher expression levels of miR-765 and miR-1275 and significantly lower expression level of miR-15a were also found in oligoasthenozoospermic subfertile men compared with normozoospermic men [110] and two miRNAs (miR-I9b and let-7a) have been proposed as good diagnostic molecular biomarkers for idiopathic infertile cases with NOA or oligozoospermia [111]. In a recent study, miR-21–5p was been clearly associated with chronic prostatitis patients with significant pelvic pain [112] and in another recent study, 5 miRNAs (miR-1825, miR-484, miR-205, miR-141, and let-7b) were described as potential biomarkers for Prostate cancer tumors [113]. Furthermore, some miRNAs for instance miR-141, have been associated with erectile dysfunction [114].
Regarding the influence of other classes of non-coding RNAs on male fertility, a study carried out in 211 infertile patients (asthenozoospermia and azoospermia) and 91 fertile controls, identified 61 altered piRNAs in the infertile patient group and found 5 piRNAs with high diagnostic potential to distinguish asthenozoospermic and azoospermic individuals from healthy controls [115]. Long non-coding RNAs have also been associated with sperm abnormalities. HOTAIR is a long non-coding RNA that shows lower levels in asthenozoospermic and oligoasthenozoospermic patients [116]. Authors suggest that the decrease in HOTAIR expression led to reactive oxygen species (ROS) related defects in sperm function [116].
In females, some common reproductive disorders are polycystic ovary syndrome (PCOS), endometriosis and ovarian cancer, and in all of them ncRNAs seem to have relevant roles or could be considered as biomarkers of reproductive pathogenesis. As an example, it has been suggested that miRNA could be used as a biomarker to distinguish polycystic ovaries patients from normal menstruating women [108]. A study carried out by Long et al. found that the expression levels of three miRNAs (miR-222, miR-146a and miR-30c) were significantly increased in PCOS patients with respect to the controls [117]. In the particular case of endometriosis, a chronic inflammatory disease, it has been shown that the loss of miR let-7b contributes to the pathophysiology of the disease. Interestingly, in a murine model, treatment using miRNA Let-7b reduced endometriosis lesion size, and authors propose local treatment with this miRNA as a promising therapy for endometriosis [118]. Circulating miRNAs have also been proposed as biomarkers of endometrial cancer [119] in which long non-coding RNAs also play a role. Altered expression of lncRNA in endometrial cancer has been observed although, to date, there are no studies on their profiles during physiological endometrial changes. It has been found that dysregulation of the lncRNAs MALAT1 and H19 are shared across endometrial cancer and endometriosis [119]. Both miRNAs and lncRNAs are thought to be promising molecular markers in ovarian cancers [120]. Apart from these pathogenic states, maternal aging is also a risk factor that correlates with human infertility, and is also associated with an altered miRNA profile. Some miRNAs seems to be differentially present in follicular fluid obtained from younger or older women [121].
To summarize the role of this wide variety of ncRNA in reproductive disorders, we can conclude with a statement by Fertilita et al. in their recent review “the complexity of ncRNA networks makes difficult to understand if deregulation of specific molecules represents the etiological cause of a disease or, alternatively, a secondary effect, depending on the perturbation of physiological pathways” [119]. Nevertheless, it has been clearly demonstrated that this group of molecules has a huge potential for use as biomarkers, more research being necessary to fully understand the complex interactions within these networks (Table 1).
5. Transgenerational effects mediated by non-coding RNAs
During recent years, several studies point to ncRNAs as important players in epigenetic inheritance of modifications mediated by environmental factors [1,122]. Nowadays it is generally accepted that the parental lifestyle can leave a mark on the progeny, and non-coding RNAs are also relevant players in that respect. Studies have been carried out to determine how important day-to-day factors such as diet, exercise and stress produce modifications in ncRNAs that can be transmitted to progeny. As an example, high fat diet modified sperm miRNAs abundance affecting sperm DNA damage, ROS levels and reduction in sperm capacitation and, more interestingly, metabolic disturbances are observed in two generations of mice [123]. It is also known that paternal stress modifiessperm miRNAs, which is reflected not only in changes in expression in the brains of offspring through the F3 generations [124,125], but also in anxiety and depressive phenotypes in the offspring [126]. Apart from diet and stress, exercise it is another important factor to take into account, because it can alter sperm small non-coding RNAs. Studies carried out in mice demonstrated that running had some effect on sperm miRNAs, three miRNAs in particular being altered (miR-19b, miR-455 and miR-133a). This study also associated paternal exercise with an anxiolytic behavioral phenotype of male offspring, ascribing this behavior to small non-coding RNAs modifications [127].
Taking into account that all these data suggest that alterations in sperm ncRNAs could have a clear effect on the progeny, it is interesting to consider that they could be also mediators in the transgenerational inheritance of undesirable effects caused by different toxics. Some studies show ncRNAs participation in epigenetic inheritance unrelated to DNA methylation or histone modification [1]. Evidence of direct participation of miRNA in the epigenetic transmission of effects caused by reprotoxicants has been confirmed in human [128]. Apart from endocrine disruptor compounds, there are other toxics that can modify the sperm ncRNAs population. As an example, quantitative RT-PCR confirmed lower levels for hsa-miR-296–5p, hsa-miR-3940, and hsa-miR-520d-3p in smokers compared to non-smokers [129]. Although it cannot be assumed that sperm ncRNAs modifications necessarily imply in all cases a real alteration in the progeny, it is clear from all existing data that ncRNAs could contribute to inheriting changes in successive generations through germ cells.
6. Conclusions
This review highlights the importance of ncRNAs in regulating PGC specification, spermatogenesis and oogenesis from fish to human. Interestingly, these molecules could be used as biomarkers either for reproductive disorders or for reproductive success and their presence in extracellular fluids allows evaluation by non-invasive methods. Although much research is needed to fully understand the regulatory functions of ncRNAs in reproduction and their potential involvement in transgenerational transmission, existing data support the idea that germ cell ncRNas could contribute to inheriting changes in successive generations.
Acknowledgments
The authors would like to acknowledge project AGL2015-68330-C2-1-R (MINECO/FEDER) PTA2016-11987-I contract (MINECO/FEDER) and AQUA-CIBUS international net 318RT0549 funded by CYTED.
References
- 1.Larriba E., del Mazo J. Role of non-coding RNAs in the transgenerational epigenetic transmission of the effects of reprotoxicants. Int. J. Mol. Sci. 2016;17:452. doi: 10.3390/ijms17040452. [DOI] [PMC free article] [PubMed] [Google Scholar]; E. Larriba, J. del Mazo, Role of Non-Coding RNAs in the Transgenerational Epigenetic Transmission of the Effects of Reprotoxicants, Int. J. Mol. Sci. 17 (2016) 452. doi:10.3390/ijms17040452. [DOI] [PMC free article] [PubMed]
- 2.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]; M. Ha, V.N. Kim, Regulation of microRNA biogenesis, Nat. Rev. Mol. Cell Biol. 15 (2014) 509-524. doi:10.1038/nrm3838. [DOI] [PubMed]
- 3.Alarcón C.R., Lee H., Goodarzi H., Halberg N., Tavazoie S.F. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]; C.R. Alarcon, H. Lee, H. Goodarzi, N. Halberg, S.F. Tavazoie, N6-methyladenosine marks primary microRNAs for processing, Nature. 519 (2015) 482-485. doi:10.1038/nature14281. [DOI] [PMC free article] [PubMed]
- 4.Miyoshi K., Miyoshi T., Siomi H. Many ways to generate microRNA-like small RNAs: non-canonical pathways for microRNA production. Mol. Genet. Genom. 2010;284:95–103. doi: 10.1007/s00438-010-0556-1. [DOI] [PubMed] [Google Scholar]; K. Miyoshi, T. Miyoshi, H. Siomi, Many ways to generate microRNA-like small RNAs: non-canonical pathways for microRNA production, Mol. Genet. Genomics. 284 (2010) 95-103. doi:10.1007/s00438-010-0556-1. [DOI] [PubMed]
- 5.Peterson S.M., Thompson J.A., Ufkin M.L., Sathyanarayana P., Liaw L., Congdon C.B. Common features of microRNA target prediction tools. Front. Genet. 2014;5:23. doi: 10.3389/fgene.2014.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]; S.M. Peterson, J.A. Thompson, M.L. Ufkin, P. Sathyanarayana, L. Liaw, C.B. Congdon, Common features of microRNA target prediction tools, Front. Genet. 5 (2014) 23. doi:10.3389/fgene.2014.00023. [DOI] [PMC free article] [PubMed]
- 6.Zhang K., Zhang X., Cai Z., Zhou J., Cao R., Zhao Y., Chen Z., Wang D., Ruan W., Zhao Q., Liu G., Xue Y., Qin Y., Zhou B., Wu L., Nilsen T., Zhou Y., Fu X.-D. A novel class of microRNA-recognition elements that function only within open reading frames. Nat. Struct. Mol. Biol. 2018 doi: 10.1038/s41594-018-0136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; K. Zhang, X. Zhang, Z. Cai, J. Zhou, R. Cao, Y. Zhao, Z. Chen, D. Wang, W. Ruan, Q. Zhao, G. Liu, Y. Xue, Y. Qin, B. Zhou, L. Wu, T. Nilsen, Y. Zhou, X.-D. Fu, A novel class of microRNA-recognition elements that function only within open reading frames, Nat. Struct. Mol. Biol. (2018). doi:10.1038/s41594-018-0136-3. [DOI] [PMC free article] [PubMed]
- 7.Russell S.J., LaMarre J. Transposons and the PIWI pathway: genome defense in gametes and embryos. Reproduction. 2018;156:R111–R124. doi: 10.1530/REP-18-0218. [DOI] [PubMed] [Google Scholar]; S.J. Russell, J. LaMarre, Transposons and the PIWI pathway: genome defense in gametes and embryos, Reproduction. 156 (2018) R111-R124. doi:10.1530/REP-18-0218. [DOI] [PubMed]
- 8.Iwasaki Y.W., Siomi M.C., Siomi H. PIWI-interacting RNA: its biogenesis and functions. Annu. Rev. Biochem. 2015;84:405–433. doi: 10.1146/annurev-biochem-060614-034258. [DOI] [PubMed] [Google Scholar]; Y.W. Iwasaki, M.C. Siomi, H. Siomi, PIWI-Interacting RNA: Its Biogenesis and Functions, Annu. Rev. Biochem. 84 (2015) 405-433. doi:10.1146/annurev-biochem-060614-034258. [DOI] [PubMed]
- 9.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]; J.J. Quinn, H.Y. Chang, Unique features of long non-coding RNA biogenesis and function, Nat. Rev. Genet. 17 (2016) 47-62. doi:10.1038/nrg.2015.10. [DOI] [PubMed]
- 10.Luk A.C.-S., Chan W.-Y., Rennert O.M., Lee T.-L. Long noncoding RNAs in spermatogenesis: insights from recent high-throughput transcriptome studies. Reproduction. 2014;147:R131–R141. doi: 10.1530/REP-13-0594. [DOI] [PubMed] [Google Scholar]; A.C.-S. Luk, W.-Y. Chan, O.M. Rennert, T.-L. Lee, Long noncoding RNAs in spermatogenesis: insights from recent high-throughput transcriptome studies, REPRODUCTION. 147 (2014) R131-R141. doi:10.1530/REP-13-0594. [DOI] [PubMed]
- 11.Kung J.T.Y., Colognori D., Lee J.T. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]; J.T.Y. Kung, D. Colognori, J.T. Lee, Long Noncoding RNAs: Past, Present, and Future, Genetics. 193 (2013) 651-669. doi:10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed]
- 12.Kotaja N., Sassone-Corsi P. The chromatoid body: a germ-cell-specific RNA-processing centre. Nat. Rev. Mol. Cell Biol. 2007;8:85–90. doi: 10.1038/nrm2081. [DOI] [PubMed] [Google Scholar]; N. Kotaja, P. Sassone-Corsi, The chromatoid body: a germ-cell-specific RNA-processing centre, Nat. Rev. Mol. Cell Biol. 8 (2007) 85-90. doi:10.1038/nrm2081. [DOI] [PubMed]
- 13.Ohinata Y., Ohta H., Shigeta M., Yamanaka K., Wakayama T., Saitou M. A signaling principle for the specification of the germ cell lineage in mice. Cell. 2009;137:571–584. doi: 10.1016/j.cell.2009.03.014. [DOI] [PubMed] [Google Scholar]; Y. Ohinata, H. Ohta, M. Shigeta, K. Yamanaka, T. Wakayama, M. Saitou, A Signaling Principle for the Specification of the Germ Cell Lineage in Mice, Cell. 137 (2009) 571-584. doi:10.1016/j.cell.2009.03.014. [DOI] [PubMed]
- 14.Tang W.W.C., Kobayashi T., Irie N., Dietmann S., Surani M.A. Specification and epigenetic programming of the human germ line. Nat. Rev. Genet. 2016;17:585–600. doi: 10.1038/nrg.2016.88. [DOI] [PubMed] [Google Scholar]; W.W.C. Tang, T. Kobayashi, N. Irie, S. Dietmann, M.A. Surani, Specification and epigenetic programming of the human germ line, Nat. Rev. Genet. 17 (2016) 585-600. doi:10.1038/nrg.2016.88. [DOI] [PubMed]
- 15.Lawson K.A., Dunn N.R., Roelen B.A., Zeinstra L.M., Davis A.M., V Wright C., Korving J.P., Hogan B.L. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. http://www.ncbi.nlm.nih.gov/pubmed/10049358 [DOI] [PMC free article] [PubMed] [Google Scholar]; K.A. Lawson, N.R. Dunn, B.A. Roelen, L.M. Zeinstra, A.M. Davis, C. V Wright, J.P. Korving, B.L. Hogan, Bmp4 is required for the generation of primordial germ cells in the mouse embryo., Genes Dev. 13 (1999) 424-436. http://www.ncbi.nlm.nih.gov/pubmed/10049358 (accessed November 5, 2018). [DOI] [PMC free article] [PubMed]
- 16.Ying Y., Liu X.-M., Marble A., Lawson K.A., Zhao G.-Q. Requirement of Bmp8b for the generation of primordial germ cells in the mouse. Mol. Endocrinol. 2000;14:1053–1063. doi: 10.1210/mend.14.7.0479. [DOI] [PubMed] [Google Scholar]; Y. Ying, X.-M. Liu, A. Marble, K.A. Lawson, G.-Q. Zhao, Requirement of Bmp8b for the Generation of Primordial Germ Cells in the Mouse, Mol. Endocrinol. 14 (2000) 1053-1063. doi:10.1210/mend.14.7.0479. [DOI] [PubMed]
- 17.Bhin J., Jeong H.-S., Kim J.S., Shin J.O., Hong K.S., Jung H.-S., Kim C., Hwang D., Kim K.-S. PGC-enriched miRNAs control germ cell development. Mol. Cell. 2015;38:895–903. doi: 10.14348/molcells.2015.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]; J. Bhin, H.-S. Jeong, J.S. Kim, J.O. Shin, K.S. Hong, H.-S. Jung, C. Kim, D. Hwang, K.-S. Kim, PGC-Enriched miRNAs Control Germ Cell Development., Mol. Cells. 38 (2015) 895-903. doi:10.14348/molcells.2015.0146. [DOI] [PMC free article] [PubMed]
- 18.Medeiros L.A., Dennis L.M., Gill M.E., Houbaviy H., Markoulaki S., Fu D., White A.C., Kirak O., Sharp P.A., Page D.C., Jaenisch R. Mir-290-295 deficiency in mice results in partially penetrant embryonic lethality and germ cell defects. Proc. Natl. Acad. Sci. Unit. States Am. 2011;108:14163–14168. doi: 10.1073/pnas.1111241108. [DOI] [PMC free article] [PubMed] [Google Scholar]; L.A. Medeiros, L.M. Dennis, M.E. Gill, H. Houbaviy, S. Markoulaki, D. Fu, A.C. White, O. Kirak, P.A. Sharp, D.C. Page, R. Jaenisch, Mir-290-295 deficiency in mice results in partially penetrant embryonic lethality and germ cell defects, Proc. Natl. Acad. Sci. 108 (2011) 14163-14168. doi:10.1073/pnas.1111241108. [DOI] [PMC free article] [PubMed]
- 19.Takada S., Berezikov E., Choi Y.L., Yamashita Y., Mano H. Potential role of miR-29b in modulation of Dnmt3a and Dnmt3b expression in primordial germ cells of female mouse embryos. RNA. 2009;15:1507–1514. doi: 10.1261/rna.1418309. [DOI] [PMC free article] [PubMed] [Google Scholar]; S. Takada, E. Berezikov, Y.L. Choi, Y. Yamashita, H. Mano, Potential role of miR-29b in modulation of Dnmt3a and Dnmt3b expression in primordial germ cells of female mouse embryos, RNA. 15 (2009) 1507-1514. doi:10.1261/rna.1418309. [DOI] [PMC free article] [PubMed]
- 20.Zhang J., Liu W., Jin Y., Jia P., Jia K., Yi M. MiR-202-5p is a novel germ plasm-specific microRNA in zebrafish. Sci. Rep. 2017;7:7055. doi: 10.1038/s41598-017-07675-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; J. Zhang, W. Liu, Y. Jin, P. Jia, K. Jia, M. Yi, MiR-202-5p is a novel germ plasm-specific microRNA in zebrafish, Sci. Rep. 7 (2017) 7055. doi:10.1038/s41598-017-07675-x. [DOI] [PMC free article] [PubMed]
- 21.Staton A.A., Knaut H., Giraldez A.J. miRNA regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration. Nat. Genet. 2011;43:204–211. doi: 10.1038/ng.758. [DOI] [PMC free article] [PubMed] [Google Scholar]; A.A. Staton, H. Knaut, A.J. Giraldez, miRNA regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration, Nat. Genet. 43 (2011) 204-211. doi:10.1038/ng.758. [DOI] [PMC free article] [PubMed]
- 22.Hong N., Li M., Yuan Y., Wang T., Yi M., Xu H., Zeng H., Song J., Hong Y. Dnd is a critical specifier of primordial germ cells in the medaka fish. Stem Cell Reports. 2016;6:411–421. doi: 10.1016/j.stemcr.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; N. Hong, M. Li, Y. Yuan, T. Wang, M. Yi, H. Xu, H. Zeng, J. Song, Y. Hong, Dnd Is a Critical Specifier of Primordial Germ Cells in the Medaka Fish., Stem Cell Reports. 6 (2016) 411-421. doi:10.1016/j.stemcr.2016.01.002. [DOI] [PMC free article] [PubMed]
- 23.Weick E.-M., Miska E.A. piRNAs: from biogenesis to function. Development. 2014;141:3458–3471. doi: 10.1242/dev.094037. [DOI] [PubMed] [Google Scholar]; E.-M. Weick, E.A. Miska, piRNAs: from biogenesis to function, Development. 141 (2014) 3458-3471. doi:10.1242/dev.094037. [DOI] [PubMed]
- 24.Ku H.-Y., Lin H. PIWI proteins and their interactors in piRNA biogenesis, germline development and gene expression. Natl. Sci. Rev. 2014;1:205–218. doi: 10.1093/nsr/nwu014. [DOI] [PMC free article] [PubMed] [Google Scholar]; H.-Y. Ku, H. Lin, PIWI proteins and their interactors in piRNA biogenesis, germline development and gene expression, Natl. Sci. Rev. 1 (2014) 205-218. doi:10.1093/nsr/nwu014. [DOI] [PMC free article] [PubMed]
- 25.von Meyenn F., Berrens R.V., Andrews S., Santos F., Collier A.J., Krueger F., Osorno R., Dean W., Rugg-Gunn P.J., Reik W. Comparative principles of DNA methylation reprogramming during human and mouse in vitro primordial germ cell specification. Dev. Cell. 2016;39:104–115. doi: 10.1016/j.devcel.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; F. von Meyenn, R.V. Berrens, S. Andrews, F. Santos, A.J. Collier, F. Krueger, R. Osorno, W. Dean, P.J. Rugg-Gunn, W. Reik, Comparative Principles of DNA Methylation Reprogramming during Human and Mouse In Vitro Primordial Germ Cell Specification, Dev. Cell. 39 (2016) 104-115. doi:10.1016/J.DEVCEL.2016.09.015. [DOI] [PMC free article] [PubMed]
- 26.Houwing S., Kamminga L.M., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., Plasterk R.H.A., Hannon G.J., Draper B.W., Ketting R.F. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]; S. Houwing, L.M. Kamminga, E. Berezikov, D. Cronembold, A. Girard, H. van den Elst, D. V. Filippov, H. Blaser, E. Raz, C.B. Moens, R.H.A. Plasterk, G.J. Hannon, B.W. Draper, R.F. Ketting, A Role for Piwi and piRNAs in Germ Cell Maintenance and Transposon Silencing in Zebrafish, Cell. 129 (2007) 69-82. doi:10.1016/J.CELL.2007.03.026. [DOI] [PubMed]
- 27.Taylor D.H., Chu E.T.-J., Spektor R., Soloway P.D. Long non-coding RNA regulation of reproduction and development. Mol. Reprod. Dev. 2015;82:932–956. doi: 10.1002/mrd.22581. [DOI] [PMC free article] [PubMed] [Google Scholar]; D.H. Taylor, E.T.-J. Chu, R. Spektor, P.D. Soloway, Long non-coding RNA regulation of reproduction and development., Mol. Reprod. Dev. 82 (2015) 932-956. doi:10.1002/mrd.22581. [DOI] [PMC free article] [PubMed]
- 28.Zhang C., Gao L., Xu E.Y. LncRNA, a new component of expanding RNA-protein regulatory network important for animal sperm development. Semin. Cell Dev. Biol. 2016;59:110–117. doi: 10.1016/j.semcdb.2016.06.013. http://www.ncbi.nlm.nih.gov/pubmed/27345292 [DOI] [PubMed] [Google Scholar]; C. Zhang, L. Gao, E.Y. Xu, LncRNA, a new component of expanding RNA-protein regulatory network important for animal sperm development, Semin. Cell Dev. Biol. 59 (2016) 110-117. http://www.ncbi.nlm.nih.gov/pubmed/27345292 (accessed October 16, 2018). [DOI] [PubMed]
- 29.Magnúsdóttir E., Surani M.A., McLaren A. How to make a primordial germ cell. Development. 2014;141:245–252. doi: 10.1242/dev.098269. [DOI] [PubMed] [Google Scholar]; E. Magnusdottir, M.A. Surani, A. McLaren, How to make a primordial germ cell., Development. 141 (2014) 245-252. doi:10.1242/dev.098269. [DOI] [PubMed]
- 30.Yadav R.P., Kotaja N. Small RNAs in spermatogenesis. Mol. Cell. Endocrinol. 2014;382:498–508. doi: 10.1016/j.mce.2013.04.015. [DOI] [PubMed] [Google Scholar]; R.P. Yadav, N. Kotaja, Small RNAs in spermatogenesis, Mol. Cell. Endocrinol. 382 (2014) 498-508. doi:10.1016/J.MCE.2013.04.015. [DOI] [PubMed]
- 31.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]; J.L. Rinn, H.Y. Chang, Genome Regulation by Long Noncoding RNAs, Annu. Rev. Biochem. 81 (2012) 145-166. doi:10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed]
- 32.Yu M., Mu H., Niu Z., Chu Z., Zhu H., Hua J. miR-34c enhances mouse spermatogonial stem cells differentiation by targeting Nanos2. J. Cell. Biochem. 2014;115:232–242. doi: 10.1002/jcb.24655. [DOI] [PubMed] [Google Scholar]; M. Yu, H. Mu, Z. Niu, Z. Chu, H. Zhu, J. Hua, miR-34c Enhances Mouse Spermatogonial Stem Cells Differentiation by Targeting Nanos2, J. Cell. Biochem. 115 (2014) 232-242. doi:10.1002/jcb.24655. [DOI] [PubMed]
- 33.McIver S.C., Stanger S.J., Santarelli D.M., Roman S.D., Nixon B., McLaughlin E.A. A unique combination of male germ cell miRNAs coordinates gonocyte differentiation. PLoS One. 2012;7:e35553. doi: 10.1371/journal.pone.0035553. [DOI] [PMC free article] [PubMed] [Google Scholar]; S.C. McIver, S.J. Stanger, D.M. Santarelli, S.D. Roman, B. Nixon, E.A. McLaughlin, A Unique Combination of Male Germ Cell miRNAs Coordinates Gonocyte Differentiation, PLoS One. 7 (2012) e35553. doi:10.1371/journal.pone.0035553. [DOI] [PMC free article] [PubMed]
- 34.Niu Z., Goodyear S.M., Rao S., Wu X., Tobias J.W., Avarbock M.R., Brinster R.L. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. Unit. States Am. 2011;108:12740–12745. doi: 10.1073/pnas.1109987108. [DOI] [PMC free article] [PubMed] [Google Scholar]; Z. Niu, S.M. Goodyear, S. Rao, X. Wu, J.W. Tobias, M.R. Avarbock, R.L. Brinster, MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells, Proc. Natl. Acad. Sci. 108 (2011) 12740-12745. doi:10.1073/pnas.1109987108. [DOI] [PMC free article] [PubMed]
- 35.Tong M.-H., Mitchell D., Evanoff R., Griswold M.D. Expression of Mirlet7 family microRNAs in response to retinoic acid-induced spermatogonial differentiation in mice. Biol. Reprod. 2011;85:189–197. doi: 10.1095/biolreprod.110.089458. [DOI] [PMC free article] [PubMed] [Google Scholar]; M.-H. Tong, D. Mitchell, R. Evanoff, M.D. Griswold, Expression of Mirlet7 family microRNAs in response to retinoic acid-induced spermatogonial differentiation in mice., Biol. Reprod. 85 (2011) 189-197. doi:10.1095/biolreprod.110.089458. [DOI] [PMC free article] [PubMed]
- 36.Huszar J.M., Payne C.J. MicroRNA 146 (Mir146) modulates spermatogonial differentiation by retinoic acid in Mice1. Biol. Reprod. 2013;88:15. doi: 10.1095/biolreprod.112.103747. [DOI] [PMC free article] [PubMed] [Google Scholar]; J.M. Huszar, C.J. Payne, MicroRNA 146 (Mir146) Modulates Spermatogonial Differentiation by Retinoic Acid in Mice1, Biol. Reprod. 88 (2013) 15. doi:10.1095/biolreprod.112.103747. [DOI] [PMC free article] [PubMed]
- 37.He Z., Jiang J., Kokkinaki M., Tang L., Zeng W., Gallicano I., Dobrinski I., Dym M. MiRNA-20 and mirna-106a regulate spermatogonial stem cell renewal at the post-transcriptional level via targeting STAT3 and Ccnd1. Stem Cell. 2013;31:2205–2217. doi: 10.1002/stem.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]; Z. He, J. Jiang, M. Kokkinaki, L. Tang, W. Zeng, I. Gallicano, I. Dobrinski, M. Dym, MiRNA-20 and mirna-106a regulate spermatogonial stem cell renewal at the post-transcriptional level via targeting STAT3 and Ccnd1., Stem Cells. 31 (2013) 2205-2217. doi:10.1002/stem.1474. [DOI] [PMC free article] [PubMed]
- 38.Cui N., Hao G., Zhao Z., Wang F., Cao J., Yang A. MicroRNA-224 regulates self-renewal of mouse spermatogonial stem cells via targeting DMRT1. J. Cell Mol. Med. 2016;20:1503–1512. doi: 10.1111/jcmm.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]; N. Cui, G. Hao, Z. Zhao, F. Wang, J. Cao, A. Yang, MicroRNA-224 regulates self-renewal of mouse spermatogonial stem cells via targeting DMRT1., J. Cell. Mol. Med. 20 (2016) 1503-1512. doi:10.1111/jcmm.12838. [DOI] [PMC free article] [PubMed]
- 39.Chen J., Cai T., Zheng C., Lin X., Wang G., Liao S., Wang X., Gan H., Zhang D., Hu X., Wang S., Li Z., Feng Y., Yang F., Han C. MicroRNA-202 maintains spermatogonial stem cells by inhibiting cell cycle regulators and RNA binding proteins. Nucleic Acids Res. 2016;45:gkw1287. doi: 10.1093/nar/gkw1287. [DOI] [PMC free article] [PubMed] [Google Scholar]; J. Chen, T. Cai, C. Zheng, X. Lin, G. Wang, S. Liao, X. Wang, H. Gan, D. Zhang, X. Hu, S. Wang, Z. Li, Y. Feng, F. Yang, C. Han, MicroRNA-202 maintains spermatogonial stem cells by inhibiting cell cycle regulators and RNA binding proteins, Nucleic Acids Res. 45 (2016) gkw1287. doi:10.1093/nar/gkw1287. [DOI] [PMC free article] [PubMed]
- 40.Chen X., Li X., Guo J., Zhang P., Zeng W. The roles of microRNAs in regulation of mammalian spermatogenesis. J. Anim. Sci. Biotechnol. 2017;8:35. doi: 10.1186/s40104-017-0166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; X. Chen, X. Li, J. Guo, P. Zhang, W. Zeng, The roles of microRNAs in regulation of mammalian spermatogenesis, J. Anim. Sci. Biotechnol. 8 (2017) 35. doi:10.1186/s40104-017-0166-4. [DOI] [PMC free article] [PubMed]
- 41.Luk A.C.-S., Chan W.-Y., Rennert O.M., Lee T.-L. Long noncoding RNAs in spermatogenesis: insights from recent high-throughput transcriptome studies. Reproduction. 2014;147:R131–R141. doi: 10.1530/REP-13-0594. [DOI] [PubMed] [Google Scholar]; A.C.-S. Luk, W.-Y. Chan, O.M. Rennert, T.-L. Lee, Long noncoding RNAs in spermatogenesis: insights from recent high-throughput transcriptome studies., Reproduction. 147 (2014) R131-R141. doi:10.1530/REP-13-0594. [DOI] [PubMed]
- 42.Hu K., Zhang J., Liang M. LncRNA AK015322 promotes proliferation of spermatogonial stem cell C18-4 by acting as a decoy for microRNA-19b-3p. Vitro Anim. Cell Dev. Biol. 2017;53:277–284. doi: 10.1007/s11626-016-0102-5. [DOI] [PubMed] [Google Scholar]; K. Hu, J. Zhang, M. Liang, LncRNA AK015322 promotes proliferation of spermatogonial stem cell C18-4 by acting as a decoy for microRNA-19b-3p, Vitr. Cell. Dev. Biol. - Anim. 53 (2017) 277-284. doi:10.1007/s11626-016-0102-5. [DOI] [PubMed]
- 43.Liang X., Zhou D., Wei C., Luo H., Liu J., Fu R., Cui S. MicroRNA-34c enhances murine male germ cell apoptosis through targeting ATF1. PLoS One. 2012;7 doi: 10.1371/journal.pone.0033861. [DOI] [PMC free article] [PubMed] [Google Scholar]; X. Liang, D. Zhou, C. Wei, H. Luo, J. Liu, R. Fu, S. Cui, MicroRNA-34c Enhances Murine Male Germ Cell Apoptosis through Targeting ATF1, PLoS One. 7 (2012) e33861. doi:10.1371/journal.pone.0033861. [DOI] [PMC free article] [PubMed]
- 44.Bouhallier F., Allioli N., Lavial F., Chalmel F., Perrard M.-H., Durand P., Samarut J., Pain B., Rouault J.-P. Role of miR-34c microRNA in the late steps of spermatogenesis. RNA. 2010;16:720–731. doi: 10.1261/rna.1963810. [DOI] [PMC free article] [PubMed] [Google Scholar]; F. Bouhallier, N. Allioli, F. Lavial, F. Chalmel, M.-H. Perrard, P. Durand, J. Samarut, B. Pain, J.-P. Rouault, Role of miR-34c microRNA in the late steps of spermatogenesis, RNA. 16 (2010) 720-731. doi:10.1261/rna.1963810. [DOI] [PMC free article] [PubMed]
- 45.Liu W.-M., Pang R.T.K., Chiu P.C.N., Wong B.P.C., Lao K., Lee K.-F., Yeung W.S.B. Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc. Natl. Acad. Sci. U.S.A. 2012;109:490–494. doi: 10.1073/pnas.1110368109. [DOI] [PMC free article] [PubMed] [Google Scholar]; W.-M. Liu, R.T.K. Pang, P.C.N. Chiu, B.P.C. Wong, K. Lao, K.-F. Lee, W.S.B. Yeung, Sperm-borne microRNA-34c is required for the first cleavage division in mouse., Proc. Natl. Acad. Sci. U. S. A. 109 (2012) 490-494. doi:10.1073/pnas.1110368109. [DOI] [PMC free article] [PubMed]
- 46.Bao J., Li D., Wang L., Wu J., Hu Y., Wang Z., Chen Y., Cao X., Jiang C., Yan W., Xu C. MicroRNA-449 and MicroRNA-34b/c function redundantly in murine testes by targeting E2F transcription factor-retinoblastoma protein (E2F-pRb) pathway. J. Biol. Chem. 2012;287:21686–21698. doi: 10.1074/jbc.M111.328054. [DOI] [PMC free article] [PubMed] [Google Scholar]; J. Bao, D. Li, L. Wang, J. Wu, Y. Hu, Z. Wang, Y. Chen, X. Cao, C. Jiang, W. Yan, C. Xu, MicroRNA-449 and MicroRNA-34b/c Function Redundantly in Murine Testes by Targeting E2F Transcription Factor-Retinoblastoma Protein (E2F-pRb) Pathway, J. Biol. Chem. 287 (2012) 21686-21698. doi:10.1074/jbc.M111.328054. [DOI] [PMC free article] [PubMed]
- 47.Yu Z., Raabe T., Hecht N.B. MicroRNA Mirn122a reduces expression of the posttranscriptionally regulated germ cell transition protein 2 (Tnp2) messenger RNA (mRNA) by mRNA Cleavage1. Biol. Reprod. 2005;73:427–433. doi: 10.1095/biolreprod.105.040998. [DOI] [PubMed] [Google Scholar]; Z. Yu, T. Raabe, N.B. Hecht, MicroRNA Mirn122a Reduces Expression of the Posttranscriptionally Regulated Germ Cell Transition Protein 2 (Tnp2) Messenger RNA (mRNA) by mRNA Cleavage1, Biol. Reprod. 73 (2005) 427-433. doi:10.1095/biolreprod.105.040998. [DOI] [PubMed]
- 48.Jia K.-T., Zhang J., Jia P., Zeng L., Jin Y., Yuan Y., Chen J., Hong Y., Yi M. Identification of MicroRNAs in zebrafish spermatozoa. Zebrafish. 2015;12:387–397. doi: 10.1089/zeb.2015.1115. [DOI] [PubMed] [Google Scholar]; K.-T. Jia, J. Zhang, P. Jia, L. Zeng, Y. Jin, Y. Yuan, J. Chen, Y. Hong, M. Yi, Identification of MicroRNAs in Zebrafish Spermatozoa, Zebrafish. 12 (2015) 387-397. doi:10.1089/zeb.2015.1115. [DOI] [PubMed]
- 49.Anguera M.C., Ma W., Clift D., Namekawa S., Kelleher R.J., Lee J.T., Lee J.T. Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PLoS Genet. 2011;7:e1002248. doi: 10.1371/journal.pgen.1002248. [DOI] [PMC free article] [PubMed] [Google Scholar]; M.C. Anguera, W. Ma, D. Clift, S. Namekawa, R.J. Kelleher, J.T. Lee, J.T. Lee, Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain., PLoS Genet. 7 (2011) e1002248. doi:10.1371/journal.pgen.1002248. [DOI] [PMC free article] [PubMed]
- 50.Nolasco S., Bellido J., Gonçalves J., Tavares A., Zabala J.C., Soares H. The expression of tubulin cofactor A (TBCA) is regulated by a noncoding antisense Tbca RNA during testis maturation. PLoS One. 2012;7:e42536. doi: 10.1371/journal.pone.0042536. [DOI] [PMC free article] [PubMed] [Google Scholar]; S. Nolasco, J. Bellido, J. Gonçalves, A. Tavares, J.C. Zabala, H. Soares, The expression of tubulin cofactor A (TBCA) is regulated by a noncoding antisense Tbca RNA during testis maturation., PLoS One. 7 (2012) e42536. doi:10.1371/journal.pone.0042536. [DOI] [PMC free article] [PubMed]
- 51.Lü M., Tian H., Cao Y., He X., Chen L., Song X., Ping P., Huang H., Sun F. Downregulation of miR-320a/383-sponge-like long non-coding RNA NLC1-C (narcolepsy candidate-region 1 genes) is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation. Cell Death Dis. 2015;6 doi: 10.1038/cddis.2015.267. e1960–e1960. [DOI] [PMC free article] [PubMed] [Google Scholar]; M. Lu, H. Tian, Y. Cao, X. He, L. Chen, X. Song, P. Ping, H. Huang, F. Sun, Downregulation of miR-320a/383-sponge-like long non-coding RNA NLC1-C (narcolepsy candidate-region 1 genes) is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation, Cell Death Dis. 6 (2015) e1960-e1960. doi:10.1038/cddis.2015.267. [DOI] [PMC free article] [PubMed]
- 52.Knight P.G., Glister C. Potential local regulatory functions of inhibins, activins and follistatin in the ovary. Reproduction. 2001;121:503–512. doi: 10.1530/rep.0.1210503. http://www.ncbi.nlm.nih.gov/pubmed/11277869 [DOI] [PubMed] [Google Scholar]; P.G. Knight, C. Glister, Potential local regulatory functions of inhibins, activins and follistatin in the ovary., Reproduction. 121 (2001) 503-512. http://www.ncbi.nlm.nih.gov/pubmed/11277869 (accessed November 20, 2018). [DOI] [PubMed]
- 53.Tu F., Pan Z.X., Yao Y., Liu H.L., Liu S.R., Xie Z., Li Q.F. miR-34a targets the inhibin beta B gene, promoting granulosa cell apoptosis in the porcine ovary. Genet. Mol. Res. 2014;13:2504–2512. doi: 10.4238/2014.January.14.6. [DOI] [PubMed] [Google Scholar]; F. Tu, Z.X. Pan, Y. Yao, H.L. Liu, S.R. Liu, Z. Xie, Q.F. Li, miR-34a targets the inhibin beta B gene, promoting granulosa cell apoptosis in the porcine ovary., Genet. Mol. Res. 13 (2014) 2504-2512. doi:10.4238/2014.January.14.6. [DOI] [PubMed]
- 54.Ginther O.J., Wiltbank M.C., Fricke P.M., Gibbons J.R., Kot K. Selection of the dominant follicle in cattle. Biol. Reprod. 1996;55:1187–1194. doi: 10.1095/biolreprod55.6.1187. http://www.ncbi.nlm.nih.gov/pubmed/8949873 [DOI] [PubMed] [Google Scholar]; O.J. Ginther, M.C. Wiltbank, P.M. Fricke, J.R. Gibbons, K. Kot, Selection of the dominant follicle in cattle., Biol. Reprod. 55 (1996) 1187-1194. http://www.ncbi.nlm.nih.gov/pubmed/8949873 (accessed November 20, 2018). [DOI] [PubMed]
- 55.Yada H., Hosokawa K., Tajima K., Hasegawa Y., Kotsuji F. Role of ovarian theca and granulosa cell interaction in hormone productionand cell growth during the bovine follicular maturation process. Biol. Reprod. 1999;61:1480–1486. doi: 10.1095/biolreprod61.6.1480. http://www.ncbi.nlm.nih.gov/pubmed/10569992 [DOI] [PubMed] [Google Scholar]; H. Yada, K. Hosokawa, K. Tajima, Y. Hasegawa, F. Kotsuji, Role of ovarian theca and granulosa cell interaction in hormone productionand cell growth during the bovine follicular maturation process., Biol. Reprod. 61 (1999) 1480-1486. http://www.ncbi.nlm.nih.gov/pubmed/10569992 (accessed November 20, 2018). [DOI] [PubMed]
- 56.Aerts J.M.J., Bols P.E.J. Ovarian follicular dynamics: a review with emphasis on the bovine species. Part I: folliculogenesis and pre-antral follicle development. Reprod. Domest. Anim. 2010;45:171–179. doi: 10.1111/j.1439-0531.2008.01302.x. [DOI] [PubMed] [Google Scholar]; J.M.J. Aerts, P.E.J. Bols, Ovarian follicular dynamics: a review with emphasis on the bovine species. Part I: Folliculogenesis and pre-antral follicle development., Reprod. Domest. Anim. 45 (2010) 171-179. doi:10.1111/j.1439-0531.2008.01302.x. [DOI] [PubMed]
- 57.Buccione R., Schroeder A.C., Eppig J.J. Interactions between somatic cells and germ cells throughout mammalian oogenesis. Biol. Reprod. 1990;43:543–547. doi: 10.1095/biolreprod43.4.543. http://www.ncbi.nlm.nih.gov/pubmed/2289008 [DOI] [PubMed] [Google Scholar]; R. Buccione, A.C. Schroeder, J.J. Eppig, Interactions between somatic cells and germ cells throughout mammalian oogenesis., Biol. Reprod. 43 (1990) 543-547. http://www.ncbi.nlm.nih.gov/pubmed/2289008 (accessed November 20, 2018). [DOI] [PubMed]
- 58.Canty M.J., Boland M.P., Evans A.C.O., Crowe M.A. Alterations in follicular IGFBP mRNA expression and follicular fluid IGFBP concentrations during the first follicle wave in beef heifers. Anim. Reprod. Sci. 2006;93:199–217. doi: 10.1016/j.anireprosci.2005.06.033. [DOI] [PubMed] [Google Scholar]; M.J. Canty, M.P. Boland, A.C.O. Evans, M.A. Crowe, Alterations in follicular IGFBP mRNA expression and follicular fluid IGFBP concentrations during the first follicle wave in beef heifers., Anim. Reprod. Sci. 93 (2006) 199-217. doi:10.1016/j.anireprosci.2005.06.033. [DOI] [PubMed]
- 59.Evans A.C.O., Ireland J.L.H., Winn M.E., Lonergan P., Smith G.W., Coussens P.M., Ireland J.J. Identification of genes involved in apoptosis and dominant follicle development during follicular waves in cattle. Biol. Reprod. 2004;70:1475–1484. doi: 10.1095/biolreprod.103.025114. [DOI] [PubMed] [Google Scholar]; A.C.O. Evans, J.L.H. Ireland, M.E. Winn, P. Lonergan, G.W. Smith, P.M. Coussens, J.J. Ireland, Identification of genes involved in apoptosis and dominant follicle development during follicular waves in cattle., Biol. Reprod. 70 (2004) 1475-1484. doi:10.1095/biolreprod.103.025114. [DOI] [PubMed]
- 60.Fayad T., Lévesque V., Sirois J., Silversides D.W., Lussier J.G. Gene expression profiling of differentially expressed genes in granulosa cells of bovine dominant follicles using suppression subtractive Hybridization1. Biol. Reprod. 2004;70:523–533. doi: 10.1095/biolreprod.103.021709. [DOI] [PubMed] [Google Scholar]; T. Fayad, V. Levesque, J. Sirois, D.W. Silversides, J.G. Lussier, Gene Expression Profiling of Differentially Expressed Genes in Granulosa Cells of Bovine Dominant Follicles Using Suppression Subtractive Hybridization1, Biol. Reprod. 70 (2004) 523-533. doi:10.1095/biolreprod.103.021709. [DOI] [PubMed]
- 61.Liang Y., Ridzon D., Wong L., Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]; Y. Liang, D. Ridzon, L. Wong, C. Chen, Characterization of microRNA expression profiles in normal human tissues, BMC Genomics. 8 (2007) 166. doi:10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed]
- 62.Ro S., Song R., Park C., Zheng H., Sanders K.M., Yan W. Cloning and expression profiling of small RNAs expressed in the mouse ovary. RNA. 2007;13:2366–2380. doi: 10.1261/rna.754207. [DOI] [PMC free article] [PubMed] [Google Scholar]; S. Ro, R. Song, C. Park, H. Zheng, K.M. Sanders, W. Yan, Cloning and expression profiling of small RNAs expressed in the mouse ovary, RNA. 13 (2007) 2366-2380. doi:10.1261/rna.754207. [DOI] [PMC free article] [PubMed]
- 63.Li M., Liu Y., Wang T., Guan J., Luo Z., Chen H., Wang X., Chen L., Ma J., Mu Z., Jiang A., Zhu L., Lang Q., Zhou X., Wang J., Zeng W., Li N., Li K., Gao X., Li X. Repertoire of porcine microRNAs in adult ovary and testis by deep sequencing. Int. J. Biol. Sci. 2011;7:1045–1055. doi: 10.7150/ijbs.7.1045. http://www.ncbi.nlm.nih.gov/pubmed/21927574 [DOI] [PMC free article] [PubMed] [Google Scholar]; M. Li, Y. Liu, T. Wang, J. Guan, Z. Luo, H. Chen, X. Wang, L. Chen, J. Ma, Z. Mu, A. Jiang, L. Zhu, Q. Lang, X. Zhou, J. Wang, W. Zeng, N. Li, K. Li, X. Gao, X. Li, Repertoire of porcine microRNAs in adult ovary and testis by deep sequencing., Int. J. Biol. Sci. 7 (2011) 1045-1055. http://www.ncbi.nlm.nih.gov/pubmed/21927574 (accessed November 20, 2018). [DOI] [PMC free article] [PubMed]
- 64.Ling Y.-H., Ren C.-H., Guo X.-F., Xu L.-N., Huang Y.-F., Luo J.-C., Zhang Y.-H., Zhang X.-R., Zhang Z.-J. Identification and characterization of microRNAs in the ovaries of multiple and uniparous goats (Capra hircus) during follicular phase. BMC Genomics. 2014;15:339. doi: 10.1186/1471-2164-15-339. [DOI] [PMC free article] [PubMed] [Google Scholar]; Y.-H. Ling, C.-H. Ren, X.-F. Guo, L.-N. Xu, Y.-F. Huang, J.-C. Luo, Y.-H. Zhang, X.-R. Zhang, Z.-J. Zhang, Identification and characterization of microRNAs in the ovaries of multiple and uniparous goats (Capra hircus) during follicular phase., BMC Genomics. 15 (2014) 339. doi:10.1186/1471-2164-15-339. [DOI] [PMC free article] [PubMed]
- 65.Hossain M.M., Ghanem N., Hoelker M., Rings F., Phatsara C., Tholen E., Schellander K., Tesfaye D. Identification and characterization of miRNAs expressed in the bovine ovary. BMC Genomics. 2009;10:443. doi: 10.1186/1471-2164-10-443. [DOI] [PMC free article] [PubMed] [Google Scholar]; M.M. Hossain, N. Ghanem, M. Hoelker, F. Rings, C. Phatsara, E. Tholen, K. Schellander, D. Tesfaye, Identification and characterization of miRNAs expressed in the bovine ovary., BMC Genomics. 10 (2009) 443. doi:10.1186/1471-2164-10-443. [DOI] [PMC free article] [PubMed]
- 66.Hong X., Luense L.J., McGinnis L.K., Nothnick W.B., Christenson L.K. Dicer1 is essential for female fertility and normal development of the female reproductive system. Endocrinology. 2008;149:6207–6212. doi: 10.1210/en.2008-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]; X. Hong, L.J. Luense, L.K. McGinnis, W.B. Nothnick, L.K. Christenson, Dicer1 Is Essential for Female Fertility and Normal Development of the Female Reproductive System, Endocrinology. 149 (2008) 6207-6212. doi:10.1210/en.2008-0294. [DOI] [PMC free article] [PubMed]
- 67.Nagaraja A.K., Andreu-Vieyra C., Franco H.L., Ma L., Chen R., Han D.Y., Zhu H., Agno J.E., Gunaratne P.H., DeMayo F.J., Matzuk M.M. Deletion of Dicer in somatic cells of the female reproductive tract causes sterility. Mol. Endocrinol. 2008;22:2336–2352. doi: 10.1210/me.2008-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]; A.K. Nagaraja, C. Andreu-Vieyra, H.L. Franco, L. Ma, R. Chen, D.Y. Han, H. Zhu, J.E. Agno, P.H. Gunaratne, F.J. DeMayo, M.M. Matzuk, Deletion of Dicer in somatic cells of the female reproductive tract causes sterility., Mol. Endocrinol. 22 (2008) 2336-2352. doi:10.1210/me.2008-0142. [DOI] [PMC free article] [PubMed]
- 68.Gonzalez G., Behringer R.R. Dicer is required for female reproductive tract development and fertility in the mouse. Mol. Reprod. Dev. 2009;76:678–688. doi: 10.1002/mrd.21010. [DOI] [PMC free article] [PubMed] [Google Scholar]; G. Gonzalez, R.R. Behringer, Dicer is required for female reproductive tract development and fertility in the mouse, Mol. Reprod. Dev. 76 (2009) 678-688. doi:10.1002/mrd.21010. [DOI] [PMC free article] [PubMed]
- 69.Gebremedhn S., Salilew-Wondim D., Ahmad I., Sahadevan S., Hossain M.M., Hoelker M., Rings F., Neuhoff C., Tholen E., Looft C., Schellander K., Tesfaye D. MicroRNA expression profile in bovine granulosa cells of preovulatory dominant and subordinate follicles during the late follicular phase of the estrous cycle. PLoS One. 2015;10:e0125912. doi: 10.1371/journal.pone.0125912. [DOI] [PMC free article] [PubMed] [Google Scholar]; S. Gebremedhn, D. Salilew-Wondim, I. Ahmad, S. Sahadevan, M.M. Hossain, M. Hoelker, F. Rings, C. Neuhoff, E. Tholen, C. Looft, K. Schellander, D. Tesfaye, MicroRNA Expression Profile in Bovine Granulosa Cells of Preovulatory Dominant and Subordinate Follicles during the Late Follicular Phase of the Estrous Cycle., PLoS One. 10 (2015) e0125912. doi:10.1371/journal.pone.0125912. [DOI] [PMC free article] [PubMed]
- 70.Bao B., Garverick H.A. Expression of steroidogenic enzyme and gonadotropin receptor genes in bovine follicles during ovarian follicular waves: a review. J. Anim. Sci. 1998;76:1903–1921. doi: 10.2527/1998.7671903x. http://www.ncbi.nlm.nih.gov/pubmed/9690647 [DOI] [PubMed] [Google Scholar]; B. Bao, H.A. Garverick, Expression of steroidogenic enzyme and gonadotropin receptor genes in bovine follicles during ovarian follicular waves: a review., J. Anim. Sci. 76 (1998) 1903-1921. http://www.ncbi.nlm.nih.gov/pubmed/9690647 (accessed November 20, 2018). [DOI] [PubMed]
- 71.Vitt U.A., Hayashi M., Klein C., Hsueh A.J.W. Growth differentiation factor-9 stimulates proliferation but suppresses the follicle-stimulating hormone-induced differentiation of cultured granulosa cells from small antral and preovulatory rat Follicles1. Biol. Reprod. 2000;62:370–377. doi: 10.1095/biolreprod62.2.370. [DOI] [PubMed] [Google Scholar]; U.A. Vitt, M. Hayashi, C. Klein, A.J.W. Hsueh, Growth Differentiation Factor-9 Stimulates Proliferation but Suppresses the Follicle-Stimulating Hormone-Induced Differentiation of Cultured Granulosa Cells from Small Antral and Preovulatory Rat Follicles1, Biol. Reprod. 62 (2000) 370-377. doi:10.1095/biolreprod62.2.370. [DOI] [PubMed]
- 72.Hayashi K.-G., Ushizawa K., Hosoe M., Takahashi T. Differential genome-wide gene expression profiling of bovine largest and second-largest follicles: identification of genes associated with growth of dominant follicles. Reprod. Biol. Endocrinol. 2010;8:11. doi: 10.1186/1477-7827-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]; K.-G. Hayashi, K. Ushizawa, M. Hosoe, T. Takahashi, Differential genome-wide gene expression profiling of bovine largest and second-largest follicles: identification of genes associated with growth of dominant follicles., Reprod. Biol. Endocrinol. 8 (2010) 11. doi:10.1186/1477-7827-8-11. [DOI] [PMC free article] [PubMed]
- 73.Donadeu F.X., Schauer S.N., Sontakke S.D. Involvement of miRNAs in ovarian follicular and luteal development. J. Endocrinol. 2012;215:323–334. doi: 10.1530/JOE-12-0252. [DOI] [PubMed] [Google Scholar]; F.X. Donadeu, S.N. Schauer, S.D. Sontakke, Involvement of miRNAs in ovarian follicular and luteal development, J. Endocrinol. 215 (2012) 323-334. doi:10.1530/JOE-12-0252. [DOI] [PubMed]
- 74.Yan G., Zhang L., Fang T., Zhang Q., Wu S., Jiang Y., Sun H., Hu Y. MicroRNA-145 suppresses mouse granulosa cell proliferation by targeting activin receptor IB. FEBS Lett. 2012;586:3263–3270. doi: 10.1016/j.febslet.2012.06.048. [DOI] [PubMed] [Google Scholar]; G. Yan, L. Zhang, T. Fang, Q. Zhang, S. Wu, Y. Jiang, H. Sun, Y. Hu, MicroRNA-145 suppresses mouse granulosa cell proliferation by targeting activin receptor IB., FEBS Lett. 586 (2012) 3263-3270. doi:10.1016/j.febslet.2012.06.048. [DOI] [PubMed]
- 75.Dai A., Sun H., Fang T., Zhang Q., Wu S., Jiang Y., Ding L., Yan G., Hu Y. MicroRNA-133b stimulates ovarian estradiol synthesis by targeting Foxl2. FEBS Lett. 2013;587:2474–2482. doi: 10.1016/j.febslet.2013.06.023. [DOI] [PubMed] [Google Scholar]; A. Dai, H. Sun, T. Fang, Q. Zhang, S. Wu, Y. Jiang, L. Ding, G. Yan, Y. Hu, MicroRNA-133b stimulates ovarian estradiol synthesis by targeting Foxl2., FEBS Lett. 587 (2013) 2474-2482. doi:10.1016/j.febslet.2013.06.023. [DOI] [PubMed]
- 76.Zhang Q., Sun H., Jiang Y., Ding L., Wu S., Fang T., Yan G., Hu Y. MicroRNA-181a suppresses mouse granulosa cell proliferation by targeting activin receptor IIA. PLoS One. 2013;8:e59667. doi: 10.1371/journal.pone.0059667. [DOI] [PMC free article] [PubMed] [Google Scholar]; Q. Zhang, H. Sun, Y. Jiang, L. Ding, S. Wu, T. Fang, G. Yan, Y. Hu, MicroRNA-181a suppresses mouse granulosa cell proliferation by targeting activin receptor IIA., PLoS One. 8 (2013) e59667. doi:10.1371/journal.pone.0059667. [DOI] [PMC free article] [PubMed]
- 77.Carletti M.Z., Fiedler S.D., Christenson L.K. MicroRNA 21 blocks apoptosis in mouse periovulatory granulosa cells. Biol. Reprod. 2010;83:286–295. doi: 10.1095/biolreprod.109.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]; M.Z. Carletti, S.D. Fiedler, L.K. Christenson, MicroRNA 21 blocks apoptosis in mouse periovulatory granulosa cells., Biol. Reprod. 83 (2010) 286-295. doi:10.1095/biolreprod.109.081448. [DOI] [PMC free article] [PubMed]
- 78.Lin F., Li R., Pan Z.X., Zhou B., Yu D.B., Wang X.G., Ma X.S., Han J., Shen M., Liu H.L. miR-26b promotes granulosa cell apoptosis by targeting ATM during follicular atresia in porcine ovary. PLoS One. 2012;7:e38640. doi: 10.1371/journal.pone.0038640. [DOI] [PMC free article] [PubMed] [Google Scholar]; F. Lin, R. Li, Z.X. Pan, B. Zhou, D.B. Yu, X.G. Wang, X.S. Ma, J. Han, M. Shen, H.L. Liu, miR-26b promotes granulosa cell apoptosis by targeting ATM during follicular atresia in porcine ovary., PLoS One. 7 (2012) e38640. doi:10.1371/journal.pone.0038640. [DOI] [PMC free article] [PubMed]
- 79.Yang X., Zhou Y., Peng S., Wu L., Lin H.-Y., Wang S., Wang H. Differentially expressed plasma microRNAs in premature ovarian failure patients and the potential regulatory function of mir-23a in granulosa cell apoptosis. Reproduction. 2012;144:235–244. doi: 10.1530/REP-11-0371. [DOI] [PubMed] [Google Scholar]; X. Yang, Y. Zhou, S. Peng, L. Wu, H.-Y. Lin, S. Wang, H. Wang, Differentially expressed plasma microRNAs in premature ovarian failure patients and the potential regulatory function of mir-23a in granulosa cell apoptosis., Reproduction. 144 (2012) 235-244. doi:10.1530/REP-11-0371. [DOI] [PubMed]
- 80.Kitahara Y., Nakamura K., Kogure K., Minegishi T. Role of microRNA-136-3p on the expression of luteinizing hormone-human chorionic gonadotropin receptor mRNA in rat ovaries. Biol. Reprod. 2013;89:114. doi: 10.1095/biolreprod.113.109207. [DOI] [PubMed] [Google Scholar]; Y. Kitahara, K. Nakamura, K. Kogure, T. Minegishi, Role of microRNA-136-3p on the expression of luteinizing hormone-human chorionic gonadotropin receptor mRNA in rat ovaries., Biol. Reprod. 89 (2013) 114. doi:10.1095/biolreprod.113.109207. [DOI] [PubMed]
- 81.Yin M., Lü M., Yao G., Tian H., Lian J., Liu L., Liang M., Wang Y., Sun F. Transactivation of microRNA-383 by steroidogenic factor-1 promotes estradiol release from mouse ovarian granulosa cells by targeting RBMS1. Mol. Endocrinol. 2012;26:1129–1143. doi: 10.1210/me.2011-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]; M. Yin, M. Lu, G. Yao, H. Tian, J. Lian, L. Liu, M. Liang, Y. Wang, F. Sun, Transactivation of microRNA-383 by steroidogenic factor-1 promotes estradiol release from mouse ovarian granulosa cells by targeting RBMS1., Mol. Endocrinol. 26 (2012) 1129-1143. doi:10.1210/me.2011-1341. [DOI] [PMC free article] [PubMed]
- 82.Xu S., Linher-Melville K., Yang B.B., Wu D., Li J. Micro-RNA378 (miR-378) regulates ovarian estradiol production by targeting aromatase. Endocrinology. 2011;152:3941–3951. doi: 10.1210/en.2011-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]; S. Xu, K. Linher-Melville, B.B. Yang, D. Wu, J. Li, Micro-RNA378 (miR-378) Regulates Ovarian Estradiol Production by Targeting Aromatase, Endocrinology. 152 (2011) 3941-3951. doi:10.1210/en.2011-1147. [DOI] [PMC free article] [PubMed]
- 83.Yao G., Yin M., Lian J., Tian H., Liu L., Li X., Sun F. MicroRNA-224 is involved in transforming growth factor-β-mediated mouse granulosa cell proliferation and granulosa cell function by targeting Smad4. Mol. Endocrinol. 2010;24:540–551. doi: 10.1210/me.2009-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]; G. Yao, M. Yin, J. Lian, H. Tian, L. Liu, X. Li, F. Sun, MicroRNA-224 Is Involved in Transforming Growth Factor-β-Mediated Mouse Granulosa Cell Proliferation and Granulosa Cell Function by Targeting Smad4, Mol. Endocrinol. 24 (2010) 540-551. doi:10.1210/me.2009-0432. [DOI] [PMC free article] [PubMed]
- 84.Dai A., Sun H., Fang T., Zhang Q., Wu S., Jiang Y., Ding L., Yan G., Hu Y. MicroRNA-133b stimulates ovarian estradiol synthesis by targeting Foxl2. FEBS Lett. 2013;587:2474–2482. doi: 10.1016/j.febslet.2013.06.023. [DOI] [PubMed] [Google Scholar]; A. Dai, H. Sun, T. Fang, Q. Zhang, S. Wu, Y. Jiang, L. Ding, G. Yan, Y. Hu, MicroRNA-133b stimulates ovarian estradiol synthesis by targeting Foxl2, FEBS Lett. 587 (2013) 2474-2482. doi:10.1016/j.febslet.2013.06.023. [DOI] [PubMed]
- 85.Yao G., Liang M., Liang N., Yin M., Lü M., Lian J., Wang Y., Sun F. MicroRNA-224 is involved in the regulation of mouse cumulus expansion by targeting Ptx3. Mol. Cell. Endocrinol. 2014;382:244–253. doi: 10.1016/j.mce.2013.10.014. [DOI] [PubMed] [Google Scholar]; G. Yao, M. Liang, N. Liang, M. Yin, M. Lu, J. Lian, Y. Wang, F. Sun, MicroRNA-224 is involved in the regulation of mouse cumulus expansion by targeting Ptx3., Mol. Cell. Endocrinol. 382 (2014) 244-253. doi:10.1016/j.mce.2013.10.014. [DOI] [PubMed]
- 86.Zhang B., Chen L., Feng G., Xiang W., Zhang K., Chu M., Wang P. MicroRNA mediating networks in granulosa cells associated with ovarian follicular development. BioMed Res. Int. 2017;2017:4585213. doi: 10.1155/2017/4585213. [DOI] [PMC free article] [PubMed] [Google Scholar]; B. Zhang, L. Chen, G. Feng, W. Xiang, K. Zhang, M. Chu, P. Wang, MicroRNA Mediating Networks in Granulosa Cells Associated with Ovarian Follicular Development., Biomed Res. Int. 2017 (2017) 4585213. doi:10.1155/2017/4585213. [DOI] [PMC free article] [PubMed]
- 87.Grossman H., Shalgi R. Results Probl. Cell Differ. 2016. A role of MicroRNAs in cell differentiation during gonad development; pp. 309–336. [DOI] [PubMed] [Google Scholar]; H. Grossman, R. Shalgi, A Role of MicroRNAs in Cell Differentiation During Gonad Development, in: Results Probl. Cell Differ., 2016: pp. 309-336. doi:10.1007/978-3-319-31973-5_12. [DOI] [PubMed]
- 88.Ahn H.W., Morin R.D., Zhao H., Harris R.A., Coarfa C., Chen Z.-J., Milosavljevic A., Marra M.A., Rajkovic A. MicroRNA transcriptome in the newborn mouse ovaries determined by massive parallel sequencing. Mol. Hum. Reprod. 2010;16:463–471. doi: 10.1093/molehr/gaq017. [DOI] [PMC free article] [PubMed] [Google Scholar]; H.W. Ahn, R.D. Morin, H. Zhao, R.A. Harris, C. Coarfa, Z.-J. Chen, A. Milosavljevic, M.A. Marra, A. Rajkovic, MicroRNA transcriptome in the newborn mouse ovaries determined by massive parallel sequencing., Mol. Hum. Reprod. 16 (2010) 463-471. doi:10.1093/molehr/gaq017. [DOI] [PMC free article] [PubMed]
- 89.Mishima T., Takizawa T., Luo S.-S., Ishibashi O., Kawahigashi Y., Mizuguchi Y., Ishikawa T., Mori M., Kanda T., Goto T., Takizawa T. MicroRNA (miRNA) cloning analysis reveals sex differences in miRNA expression profiles between adult mouse testis and ovary. Reproduction. 2008;136:811–822. doi: 10.1530/REP-08-0349. [DOI] [PubMed] [Google Scholar]; T. Mishima, T. Takizawa, S.-S. Luo, O. Ishibashi, Y. Kawahigashi, Y. Mizuguchi, T. Ishikawa, M. Mori, T. Kanda, T. Goto, T. Takizawa, MicroRNA (miRNA) cloning analysis reveals sex differences in miRNA expression profiles between adult mouse testis and ovary, REPRODUCTION. 136 (2008) 811-822. doi:10.1530/REP-08-0349. [DOI] [PubMed]
- 90.Di R., He J., Song S., Tian D., Liu Q., Liang X., Ma Q., Sun M., Wang J., Zhao W., Cao G., Wang J., Yang Z., Ge Y., Chu M. Characterization and comparative profiling of ovarian microRNAs during ovine anestrus and the breeding season. BMC Genomics. 2014;15:899. doi: 10.1186/1471-2164-15-899. [DOI] [PMC free article] [PubMed] [Google Scholar]; R. Di, J. He, S. Song, D. Tian, Q. Liu, X. Liang, Q. Ma, M. Sun, J. Wang, W. Zhao, G. Cao, J. Wang, Z. Yang, Y. Ge, M. Chu, Characterization and comparative profiling of ovarian microRNAs during ovine anestrus and the breeding season., BMC Genomics. 15 (2014) 899. doi:10.1186/1471-2164-15-899. [DOI] [PMC free article] [PubMed]
- 91.Tripurani S.K., Xiao C., Salem M., Yao J. Cloning and analysis of fetal ovary microRNAs in cattle. Anim. Reprod. Sci. 2010;120:16–22. doi: 10.1016/j.anireprosci.2010.03.001. [DOI] [PubMed] [Google Scholar]; S.K. Tripurani, C. Xiao, M. Salem, J. Yao, Cloning and analysis of fetal ovary microRNAs in cattle, Anim. Reprod. Sci. 120 (2010) 16-22. doi:10.1016/j.anireprosci.2010.03.001. [DOI] [PubMed]
- 92.Huang J., Ju Z., Li Q., Hou Q., Wang C., Li J., Li R., Wang L., Sun T., Hang S., Gao Y., Hou M., Zhong J. Solexa sequencing of novel and differentially expressed microRNAs in testicular and ovarian tissues in Holstein cattle. Int. J. Biol. Sci. 2011;7:1016–1026. doi: 10.7150/ijbs.7.1016. http://www.ncbi.nlm.nih.gov/pubmed/21912509 [DOI] [PMC free article] [PubMed] [Google Scholar]; J. Huang, Z. Ju, Q. Li, Q. Hou, C. Wang, J. Li, R. Li, L. Wang, T. Sun, S. Hang, Y. Gao, M. Hou, J. Zhong, Solexa sequencing of novel and differentially expressed microRNAs in testicular and ovarian tissues in Holstein cattle., Int. J. Biol. Sci. 7 (2011) 1016-1026. http://www.ncbi.nlm.nih.gov/pubmed/21912509 (accessed November 20, 2018). [DOI] [PMC free article] [PubMed]
- 93.Miles J.R., McDaneld T.G., Wiedmann R.T., Cushman R.A., Echternkamp S.E., Vallet J.L., Smith T.P.L. MicroRNA expression profile in bovine cumulus-oocyte complexes: possible role of let-7 and miR-106a in the development of bovine oocytes. Anim. Reprod. Sci. 2012;130:16–26. doi: 10.1016/j.anireprosci.2011.12.021. [DOI] [PubMed] [Google Scholar]; J.R. Miles, T.G. McDaneld, R.T. Wiedmann, R.A. Cushman, S.E. Echternkamp, J.L. Vallet, T.P.L. Smith, MicroRNA expression profile in bovine cumulus-oocyte complexes: possible role of let-7 and miR-106a in the development of bovine oocytes., Anim. Reprod. Sci. 130 (2012) 16-26. doi:10.1016/j.anireprosci.2011.12.021. [DOI] [PubMed]
- 94.Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]; A.J. Giraldez, Y. Mishima, J. Rihel, R.J. Grocock, S. Van Dongen, K. Inoue, A.J. Enright, A.F. Schier, Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs., Science. 312 (2006) 75-79. doi:10.1126/science.1122689. [DOI] [PubMed]
- 95.Bouchareb A., Le Cam A., Montfort J., Gay S., Nguyen T., Bobe J., Thermes V. Genome-wide identification of novel ovarian-predominant miRNAs: new insights from the medaka (Oryzias latipes) Sci. Rep. 2017;7:40241. doi: 10.1038/srep40241. [DOI] [PMC free article] [PubMed] [Google Scholar]; A. Bouchareb, A. Le Cam, J. Montfort, S. Gay, T. Nguyen, J. Bobe, V. Thermes, Genome-wide identification of novel ovarian-predominant miRNAs: new insights from the medaka (Oryzias latipes), Sci. Rep. 7 (2017) 40241. doi:10.1038/srep40241. [DOI] [PMC free article] [PubMed]
- 96.Juanchich A., Le Cam A., Montfort J., Guiguen Y., Bobe J. Identification of differentially expressed miRNAs and their potential targets during fish ovarian development. Biol. Reprod. 2013;88:128. doi: 10.1095/biolreprod.112.105361. [DOI] [PubMed] [Google Scholar]; A. Juanchich, A. Le Cam, J. Montfort, Y. Guiguen, J. Bobe, Identification of differentially expressed miRNAs and their potential targets during fish ovarian development., Biol. Reprod. 88 (2013) 128. doi:10.1095/biolreprod.112.105361. [DOI] [PubMed]
- 97.Bannister S.C., Smith C.A., Roeszler K.N., Doran T.J., Sinclair A.H., V Tizard M.L. Manipulation of estrogen synthesis alters MIR202* expression in embryonic chicken gonads. Biol. Reprod. 2011;85:22–30. doi: 10.1095/biolreprod.110.088476. [DOI] [PubMed] [Google Scholar]; S.C. Bannister, C.A. Smith, K.N. Roeszler, T.J. Doran, A.H. Sinclair, M.L. V Tizard, Manipulation of estrogen synthesis alters MIR202* expression in embryonic chicken gonads., Biol. Reprod. 85 (2011) 22-30. doi:10.1095/biolreprod.110.088476. [DOI] [PubMed]
- 98.Armisen J., Gilchrist M.J., Wilczynska A., Standart N., Miska E.A. Abundant and dynamically expressed miRNAs, piRNAs, and other small RNAs in the vertebrate Xenopus tropicalis. Genome Res. 2009;19:1766–1775. doi: 10.1101/gr.093054.109. [DOI] [PMC free article] [PubMed] [Google Scholar]; J. Armisen, M.J. Gilchrist, A. Wilczynska, N. Standart, E.A. Miska, Abundant and dynamically expressed miRNAs, piRNAs, and other small RNAs in the vertebrate Xenopus tropicalis., Genome Res. 19 (2009) 1766-1775. doi:10.1101/gr.093054.109. [DOI] [PMC free article] [PubMed]
- 99.Zhao H., Shen J., Medico L., Wang D., Ambrosone C.B., Liu S. A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS One. 2010;5:e13735. doi: 10.1371/journal.pone.0013735. [DOI] [PMC free article] [PubMed] [Google Scholar]; H. Zhao, J. Shen, L. Medico, D. Wang, C.B. Ambrosone, S. Liu, A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer., PLoS One. 5 (2010) e13735. doi:10.1371/journal.pone.0013735. [DOI] [PMC free article] [PubMed]
- 100.Lian J., Zhang X., Tian H., Liang N., Wang Y., Liang C., Li X., Sun F. Altered microRNA expression in patients with non-obstructive azoospermia. Reprod. Biol. Endocrinol. 2009;7:13. doi: 10.1186/1477-7827-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]; J. Lian, X. Zhang, H. Tian, N. Liang, Y. Wang, C. Liang, X. Li, F. Sun, Altered microRNA expression in patients with non-obstructive azoospermia, Reprod. Biol. Endocrinol. 7 (2009) 13. doi:10.1186/1477-7827-7-13. [DOI] [PMC free article] [PubMed]
- 101.Tang D., Huang Z., He X., Wu H., Peng D., Zhang L., Zhang X. Altered miRNA profile in testis of post-cryptorchidopexy patients with non-obstructive azoospermia. Reprod. Biol. Endocrinol. 2018;16:78. doi: 10.1186/s12958-018-0393-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; D. Tang, Z. Huang, X. He, H. Wu, D. Peng, L. Zhang, X. Zhang, Altered miRNA profile in testis of post-cryptorchidopexy patients with non-obstructive azoospermia, Reprod. Biol. Endocrinol. 16 (2018) 78. doi:10.1186/s12958-018-0393-3. [DOI] [PMC free article] [PubMed]
- 102.Wang C., Yang C., Chen X., Yao B., Yang C., Zhu C., Li L., Wang J., Li X., Shao Y., Liu Y., Ji J., Zhang J., Zen K., Zhang C.-Y., Zhang C. Altered profile of seminal plasma MicroRNAs in the molecular diagnosis of male infertility. Clin. Chem. 2011;57:1722–1731. doi: 10.1373/clinchem.2011.169714. [DOI] [PubMed] [Google Scholar]; C. Wang, C. Yang, X. Chen, B. Yao, C. Yang, C. Zhu, L. Li, J. Wang, X. Li, Y. Shao, Y. Liu, J. Ji, J. Zhang, K. Zen, C.-Y. Zhang, C. Zhang, Altered Profile of Seminal Plasma MicroRNAs in the Molecular Diagnosis of Male Infertility, Clin. Chem. 57 (2011) 1722-1731. doi:10.1373/clinchem.2011.169714. [DOI] [PubMed]
- 103.Abu-Halima M., Hammadeh M., Schmitt J., Leidinger P., Keller A., Meese E., Backes C. Altered microRNA expression profiles of human spermatozoa in patients with different spermatogenic impairments. Fertil. Steril. 2013;99:1249–1255. doi: 10.1016/j.fertnstert.2012.11.054. e16. [DOI] [PubMed] [Google Scholar]; M. Abu-Halima, M. Hammadeh, J. Schmitt, P. Leidinger, A. Keller, E. Meese, C. Backes, Altered microRNA expression profiles of human spermatozoa in patients with different spermatogenic impairments., Fertil. Steril. 99 (2013) 1249-1255.e16. doi:10.1016/j.fertnstert.2012.11.054. [DOI] [PubMed]
- 104.Dabaja A.A., Mielnik A., Robinson B.D., Wosnitzer M.S., Schlegel P.N., Paduch D.A. Possible germ cell-Sertoli cell interactions are critical for establishing appropriate expression levels for the Sertoli cell-specific MicroRNA, miR-202-5p, in human testis. Basic Clin. Androl. 2015;25:2. doi: 10.1186/s12610-015-0018-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; A.A. Dabaja, A. Mielnik, B.D. Robinson, M.S. Wosnitzer, P.N. Schlegel, D.A. Paduch, Possible germ cell-Sertoli cell interactions are critical for establishing appropriate expression levels for the Sertoli cell-specific MicroRNA, miR-202-5p, in human testis., Basic Clin. Androl. 25 (2015) 2. doi:10.1186/s12610-015-0018-z. [DOI] [PMC free article] [PubMed]
- 105.Zhao K., Chen Y., Yang R., Bai Y., Li C., Li H., Xiong C. miR-424/322 is downregulated in the semen of patients with severe DNA damage and may regulate sperm DNA damage. Reprod. Fertil. Dev. 2015;28:1598. doi: 10.1071/RD15052. [DOI] [PubMed] [Google Scholar]; K. Zhao, Y. Chen, R. Yang, Y. Bai, C. Li, H. Li, C. Xiong, miR-424/322 is downregulated in the semen of patients with severe DNA damage and may regulate sperm DNA damage., Reprod. Fertil. Dev. 28 (2015) 1598. doi:10.1071/RD15052. [DOI] [PubMed]
- 106.Hanson E.K., Lubenow H., Ballantyne J. Identification of forensically relevant body fluids using a panel of differentially expressed microRNAs. Anal. Biochem. 2009;387:303–314. doi: 10.1016/j.ab.2009.01.037. [DOI] [PubMed] [Google Scholar]; E.K. Hanson, H. Lubenow, J. Ballantyne, Identification of forensically relevant body fluids using a panel of differentially expressed microRNAs., Anal. Biochem. 387 (2009) 303-314. doi:10.1016/j.ab.2009.01.037. [DOI] [PubMed]
- 107.Li Y., Zhou X., John M.A.R. St, Wong D.T.W. RNA profiling of cell-free saliva using microarray technology. J. Dent. Res. 2004;83:199–203. doi: 10.1177/154405910408300303. [DOI] [PubMed] [Google Scholar]; Y. Li, X. Zhou, M.A.R. St. John, D.T.W. Wong, RNA Profiling of Cell-free Saliva Using Microarray Technology, J. Dent. Res. 83 (2004) 199-203. doi:10.1177/154405910408300303. [DOI] [PubMed]
- 108.Sørensen A., Wissing M., Salö S., Englund A., Dalgaard L. MicroRNAs related to polycystic ovary syndrome (PCOS) Genes. 2014;5:684–708. doi: 10.3390/genes5030684. [DOI] [PMC free article] [PubMed] [Google Scholar]; A. Sorensen, M. Wissing, S. Salo, A. Englund, L. Dalgaard, MicroRNAs Related to Polycystic Ovary Syndrome (PCOS), Genes (Basel). 5 (2014) 684-708. doi:10.3390/genes5030684. [DOI] [PMC free article] [PubMed]
- 109.Zhang X., Wei R., Lou J., Zhou J. [Seminal plasma miR-122-3p and miR-141-5p stability and its diagnosis value for idiopathic asthenospermia] Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2016;33:320–323. doi: 10.3760/cma.j.issn.1003-9406.2016.03.009. [DOI] [PubMed] [Google Scholar]; X. Zhang, R. Wei, J. Lou, J. Zhou, [Seminal plasma miR-122-3p and miR-141-5p stability and its diagnosis value for idiopathic asthenospermia]., Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 33 (2016) 320-323. doi:10.3760/cma.j.issn.1003-9406.2016.03.009. [DOI] [PubMed]
- 110.Abu-Halima M., Ludwig N., Hart M., Leidinger P., Backes C., Keller A., Hammadeh M., Meese E. Altered micro-ribonucleic acid expression profiles of extracellular microvesicles in the seminal plasma of patients with oligoasthenozoospermia. Fertil. Steril. 2016;106:1061–1069. doi: 10.1016/j.fertnstert.2016.06.030. e3. [DOI] [PubMed] [Google Scholar]; M. Abu-Halima, N. Ludwig, M. Hart, P. Leidinger, C. Backes, A. Keller, M. Hammadeh, E. Meese, Altered micro-ribonucleic acid expression profiles of extracellular microvesicles in the seminal plasma of patients with oligoasthenozoospermia, Fertil. Steril. 106 (2016) 1061-1069.e3. doi:10.1016/j.fertnstert.2016.06.030. [DOI] [PubMed]
- 111.Wu W., Hu Z., Qin Y., Dong J., Dai J., Lu C., Zhang W., Shen H., Xia Y., Wang X. Seminal plasma microRNAs: potential biomarkers for spermatogenesis status. MHR Basic Sci. Reprod. Med. 2012;18:489–497. doi: 10.1093/molehr/gas022. [DOI] [PubMed] [Google Scholar]; W. Wu, Z. Hu, Y. Qin, J. Dong, J. Dai, C. Lu, W. Zhang, H. Shen, Y. Xia, X. Wang, Seminal plasma microRNAs: potential biomarkers for spermatogenesis status, MHR Basic Sci. Reprod. Med. 18 (2012) 489-497. doi:10.1093/molehr/gas022. [DOI] [PubMed]
- 112.Chen Y., Chen S., Zhang J., Wang Y., Jia Z., Zhang X., Han X., Guo X., Sun X., Shao C., Wang J., Lan T. Expression profile of microRNAs in expressed prostatic secretion of healthy men and patients with IIIA chronic prostatitis/chronic pelvic pain syndrome. Oncotarget. 2018;9:12186–12200. doi: 10.18632/oncotarget.24069. [DOI] [PMC free article] [PubMed] [Google Scholar]; Y. Chen, S. Chen, J. Zhang, Y. Wang, Z. Jia, X. Zhang, X. Han, X. Guo, X. Sun, C. Shao, J. Wang, T. Lan, Expression profile of microRNAs in expressed prostatic secretion of healthy men and patients with IIIA chronic prostatitis/ chronic pelvic pain syndrome, Oncotarget. 9 (2018) 12186-12200. doi:10.18632/oncotarget.24069. [DOI] [PMC free article] [PubMed]
- 113.Guo X., Han T., Hu P., Guo X., Zhu C., Wang Y., Chang S. Five microRNAs in serum as potential biomarkers for prostate cancer risk assessment and therapeutic intervention. Int. Urol. Nephrol. 2018 doi: 10.1007/s11255-018-2009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; X. Guo, T. Han, P. Hu, X. Guo, C. Zhu, Y. Wang, S. Chang, Five microRNAs in serum as potential biomarkers for prostate cancer risk assessment and therapeutic intervention, Int. Urol. Nephrol. (2018). doi:10.1007/s11255-018-2009-4. [DOI] [PMC free article] [PubMed]
- 114.Zhang Y., Jia L., Ji W., Li H. MicroRNA-141 inhibits the proliferation of penile cavernous smooth muscle cells associated with down-regulation of the rhoa/rho kinase signaling pathway. Cell. Physiol. Biochem. 2018;48:348–360. doi: 10.1159/000491741. [DOI] [PubMed] [Google Scholar]; Y. Zhang, L. Jia, W. Ji, H. Li, MicroRNA-141 Inhibits the Proliferation of Penile Cavernous Smooth Muscle Cells Associated with Down-Regulation of the Rhoa/Rho Kinase Signaling Pathway, Cell. Physiol. Biochem. 48 (2018) 348-360. doi:10.1159/000491741. [DOI] [PubMed]
- 115.Hong Y., Wang C., Fu Z., Liang H., Zhang S., Lu M., Sun W., Ye C., Zhang C.-Y., Zen K., Shi L., Zhang C., Chen X. Systematic characterization of seminal plasma piRNAs as molecular biomarkers for male infertility. Sci. Rep. 2016;6:24229. doi: 10.1038/srep24229. [DOI] [PMC free article] [PubMed] [Google Scholar]; Y. Hong, C. Wang, Z. Fu, H. Liang, S. Zhang, M. Lu, W. Sun, C. Ye, C.-Y. Zhang, K. Zen, L. Shi, C. Zhang, X. Chen, Systematic characterization of seminal plasma piRNAs as molecular biomarkers for male infertility, Sci. Rep. 6 (2016) 24229. doi:10.1038/srep24229. [DOI] [PMC free article] [PubMed]
- 116.Zhang L., Liu Z., Li X., Zhang P., Wang J., Zhu D., Chen X., Ye L. Low long non-coding RNA HOTAIR expression is associated with down-regulation of Nrf2 in the spermatozoa of patients with asthenozoospermia or oligoasthenozoospermia. Int. J. Clin. Exp. Pathol. 2015;8:14198–14205. http://www.ncbi.nlm.nih.gov/pubmed/26823733 [PMC free article] [PubMed] [Google Scholar]; L. Zhang, Z. Liu, X. Li, P. Zhang, J. Wang, D. Zhu, X. Chen, L. Ye, Low long non-coding RNA HOTAIR expression is associated with down-regulation of Nrf2 in the spermatozoa of patients with asthenozoospermia or oligoasthenozoospermia., Int. J. Clin. Exp. Pathol. 8 (2015) 14198-14205. http://www.ncbi.nlm.nih.gov/pubmed/26823733 (accessed October 16, 2018). [PMC free article] [PubMed]
- 117.Long W., Zhao C., Ji C., Ding H., Cui Y., Guo X., Shen R., Liu J. Characterization of serum microRNAs profile of PCOS and identification of novel non-invasive biomarkers. Cell. Physiol. Biochem. 2014;33:1304–1315. doi: 10.1159/000358698. [DOI] [PubMed] [Google Scholar]; W. Long, C. Zhao, C. Ji, H. Ding, Y. Cui, X. Guo, R. Shen, J. Liu, Characterization of serum microRNAs profile of PCOS and identification of novel non-invasive biomarkers., Cell. Physiol. Biochem. 33 (2014) 1304-1315. doi:10.1159/000358698. [DOI] [PubMed]
- 118.Sahin C., Mamillapalli R., Yi K.W., Taylor H.S. microRNA Let-7b: a Novel treatment for endometriosis. J. Cell Mol. Med. 2018 doi: 10.1111/jcmm.13807. [DOI] [PMC free article] [PubMed] [Google Scholar]; C. Sahin, R. Mamillapalli, K.W. Yi, H.S. Taylor, microRNA Let-7b: A Novel treatment for endometriosis., J. Cell. Mol. Med. (2018). doi:10.1111/jcmm.13807. [DOI] [PMC free article] [PubMed]
- 119.La Ferlita A., Battaglia R., Andronico F., Caruso S., Cianci A., Purrello M., Di Pietro C. Non-coding RNAs in endometrial physiopathology. Int. J. Mol. Sci. 2018;19:2120. doi: 10.3390/ijms19072120. [DOI] [PMC free article] [PubMed] [Google Scholar]; A. La Ferlita, R. Battaglia, F. Andronico, S. Caruso, A. Cianci, M. Purrello, C. Di Pietro, Non-Coding RNAs in Endometrial Physiopathology, Int. J. Mol. Sci. 19 (2018) 2120. doi:10.3390/ijms19072120. [DOI] [PMC free article] [PubMed]
- 120.Panoutsopoulou K., Avgeris M., Scorilas A. miRNA and long non-coding RNA: molecular function and clinical value in breast and ovarian cancers. Expert Rev. Mol. Diagn. 2018 doi: 10.1080/14737159.2018.1538794. 14737159.2018.1538794. [DOI] [PubMed] [Google Scholar]; K. Panoutsopoulou, M. Avgeris, A. Scorilas, miRNA and long non-coding RNA: molecular function and clinical value in breast and ovarian cancers, Expert Rev. Mol. Diagn. (2018) 14737159.2018.1538794. doi:10.1080/14737159.2018.1538794. [DOI] [PubMed]
- 121.Diez-Fraile A., Lammens T., Tilleman K., Witkowski W., Verhasselt B., De Sutter P., Benoit Y., Espeel M., D'Herde K. Age-associated differential microRNA levels in human follicular fluid reveal pathways potentially determining fertility and success of in vitro fertilization. Hum. Fertil. 2014;17:90–98. doi: 10.3109/14647273.2014.897006. [DOI] [PubMed] [Google Scholar]; A. Diez-Fraile, T. Lammens, K. Tilleman, W. Witkowski, B. Verhasselt, P. De Sutter, Y. Benoit, M. Espeel, K. D’Herde, Age-associated differential microRNA levels in human follicular fluid reveal pathways potentially determining fertility and success of in vitro fertilization., Hum. Fertil. (Camb). 17 (2014) 90-98. doi:10.3109/14647273.2014.897006. [DOI] [PubMed]
- 122.Yan W. Potential roles of noncoding RNAs in environmental epigenetic transgenerational inheritance. Mol. Cell. Endocrinol. 2014;398:24–30. doi: 10.1016/j.mce.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; W. Yan, Potential roles of noncoding RNAs in environmental epigenetic transgenerational inheritance., Mol. Cell. Endocrinol. 398 (2014) 24-30. doi:10.1016/j.mce.2014.09.008. [DOI] [PMC free article] [PubMed]
- 123.Fullston T., Ohlsson Teague E.M.C., Palmer N.O., DeBlasio M.J., Mitchell M., Corbett M., Print C.G., Owens J.A., Lane M. Paternal obesity initiates metabolic disturbances in two generations of mice with incomplete penetrance to the F2 generation and alters the transcriptional profile of testis and sperm microRNA content. FASEB J. 2013;27:4226–4243. doi: 10.1096/fj.12-224048. [DOI] [PubMed] [Google Scholar]; T. Fullston, E.M.C. Ohlsson Teague, N.O. Palmer, M.J. DeBlasio, M. Mitchell, M. Corbett, C.G. Print, J.A. Owens, M. Lane, Paternal obesity initiates metabolic disturbances in two generations of mice with incomplete penetrance to the F2 generation and alters the transcriptional profile of testis and sperm microRNA content., FASEB J. 27 (2013) 4226-4243. doi:10.1096/fj.12-224048. [DOI] [PubMed]
- 124.Gapp K., Jawaid A., Sarkies P., Bohacek J., Pelczar P., Prados J., Farinelli L., Miska E., Mansuy I.M. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice1. Nat. Neurosci. 2014;17:667–669. doi: 10.1038/nn.3695. Gapp K, Jawaid A, Sarkies P, et al. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci. 2014;1. [DOI] [PMC free article] [PubMed] [Google Scholar]; K. Gapp, A. Jawaid, P. Sarkies, J. Bohacek, P. Pelczar, J. Prados, L. Farinelli, E. Miska, I.M. Mansuy, Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice1. Gapp K, Jawaid A, Sarkies P, et al. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci. 2014;1, Nat. Neurosci. 17 (2014) 667-669. doi:10.1038/nn.3695. [DOI] [PMC free article] [PubMed]
- 125.Rodgers A.B., Morgan C.P., Bronson S.L., Revello S., Bale T.L. Paternal stress exposure alters sperm microRNA content and reprograms offspring HPA stress axis regulation. J. Neurosci. 2013;33 doi: 10.1523/JNEUROSCI.0914-13.2013. 9003–12. [DOI] [PMC free article] [PubMed] [Google Scholar]; A.B. Rodgers, C.P. Morgan, S.L. Bronson, S. Revello, T.L. Bale, Paternal stress exposure alters sperm microRNA content and reprograms offspring HPA stress axis regulation., J. Neurosci. 33 (2013) 9003-12. doi:10.1523/JNEUROSCI.0914-13.2013. [DOI] [PMC free article] [PubMed]
- 126.Short A.K., Fennell K.A., Perreau V.M., Fox A., O'Bryan M.K., Kim J.H., Bredy T.W., Pang T.Y., Hannan A.J. Elevated paternal glucocorticoid exposure alters the small noncoding RNA profile in sperm and modifies anxiety and depressive phenotypes in the offspring. Transl. Psychiatry. 2016;6:e837. doi: 10.1038/tp.2016.109. [DOI] [PMC free article] [PubMed] [Google Scholar]; A.K. Short, K.A. Fennell, V.M. Perreau, A. Fox, M.K. O’Bryan, J.H. Kim, T.W. Bredy, T.Y. Pang, A.J. Hannan, Elevated paternal glucocorticoid exposure alters the small noncoding RNA profile in sperm and modifies anxiety and depressive phenotypes in the offspring., Transl. Psychiatry. 6 (2016) e837. doi:10.1038/tp.2016.109. [DOI] [PMC free article] [PubMed]
- 127.Short A.K., Yeshurun S., Powell R., Perreau V.M., Fox A., Kim J.H., Pang T.Y., Hannan A.J. Exercise alters mouse sperm small noncoding RNAs and induces a transgenerational modification of male offspring conditioned fear and anxiety. Transl. Psychiatry. 2017;7:e1114. doi: 10.1038/tp.2017.82. [DOI] [PMC free article] [PubMed] [Google Scholar]; A.K. Short, S. Yeshurun, R. Powell, V.M. Perreau, A. Fox, J.H. Kim, T.Y. Pang, A.J. Hannan, Exercise alters mouse sperm small noncoding RNAs and induces a transgenerational modification of male offspring conditioned fear and anxiety, Transl. Psychiatry. 7 (2017) e1114. doi:10.1038/tp.2017.82. [DOI] [PMC free article] [PubMed]
- 128.Skinner M.K. Epigenetic transgenerational inheritance. Nat. Rev. Endocrinol. 2016;12:68–70. doi: 10.1038/nrendo.2015.206. [DOI] [PMC free article] [PubMed] [Google Scholar]; M.K. Skinner, Epigenetic transgenerational inheritance, Nat. Rev. Endocrinol. 12 (2016) 68-70. doi:10.1038/nrendo.2015.206. [DOI] [PMC free article] [PubMed]
- 129.Metzler-Guillemain C., Victorero G., Lepoivre C., Bergon A., Yammine M., Perrin J., Sari-Minodier I., Boulanger N., Rihet P., Nguyen C. Sperm mRNAs and microRNAs as candidate markers for the impact of toxicants on human spermatogenesis: an application to tobacco smoking. Syst. Biol. Reprod. Med. 2015;61:139–149. doi: 10.3109/19396368.2015.1022835. [DOI] [PubMed] [Google Scholar]; C. Metzler-Guillemain, G. Victorero, C. Lepoivre, A. Bergon, M. Yammine, J. Perrin, I. Sari-Minodier, N. Boulanger, P. Rihet, C. Nguyen, Sperm mRNAs and microRNAs as candidate markers for the impact of toxicants on human spermatogenesis: an application to tobacco smoking, Syst. Biol. Reprod. Med. 61 (2015) 139-149. doi:10.3109/19396368.2015.1022835. [DOI] [PubMed]