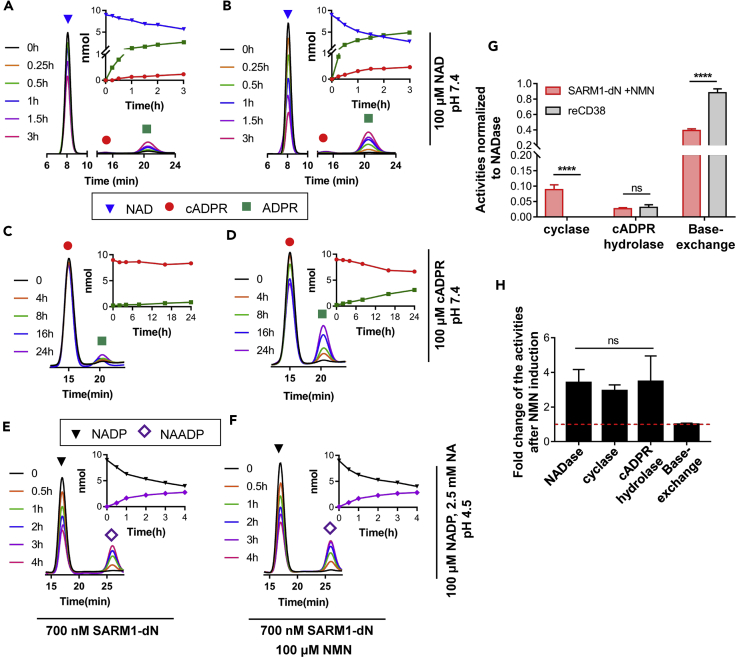
Figure 4.
The Enzymatic Activities of SARM1 with or without NMN Activation
BC2T-tagged SARM1 with the N-terminal location signal truncated (SARM1-dN, c.f. Figure 5A and Methods) was immunoprecipitated by BC2Nb beads and quantified by western blots. Around 700 nM of SARM1-dN (with or without pre-treatment of 100 μM NMN) was used in three reactions.
(A and B) The activities of NAD hydrolase and ADP-ribosyl cyclase. The protein, 700 nM SARM1-dN (A, without NMN; B, with NMN), was incubated with 100 μM NAD in KHM (pH 7.4) for different time periods, and the products were analyzed by HPLC. Insets (above the red dots): enlarged peaks of cADPR in the chromatogram; insets (dot plot): quantification of the products. Blue triangles, NAD; red circles, cADPR; green squares, ADP-ribose.
(C and D) cADPR hydrolase activity. Similar reactions were set and analyzed as in (A and B), except the substrate was replaced by the same amount of cADPR. Insets: Red, cADPR; green, ADP-ribose.
(E and F) Base-exchange reaction. Similar reactions were set and analyzed as in (A and B), except the substrate was replaced by same amount of NADP and 2.5 mM NA in 15 mM acetate buffer (pH 4.5). Insets: black, NADP; purple, NAADP.
(G) The activities of NMN-activated SARM1-dN (in B, D, F) and reCD38 (Graeff et al., 2001) were normalized with their NAD hydrolase activities.
(H) The activities including NADase (ADPR production rate in A and B), ADP-ribosyl cyclase (cADPR production rate in A and B), cADPR hydrolase (ADPR production rate in C and D), and base-exchange activities (NAADP production rate in E and F) were calculated and presented as the fold change after NMN induction. The red dashed line is the activity level of SARM1-dN without NMN. The experiments were repeated at least three times. All the above experiments were repeated at least three times (means ± SDs; n = 3; Student's t test, ****p < 0.0001).