Abstract
Background
Esophageal cancer is the eighth most common cancer globally. Esophageal adenocarcinoma (EA) and esophageal squamous-cell carcinoma (ESCC) are the two major types of esophageal cancer with poor prognosis. The mechanisms of the progression of normal esophagus to Barrett's esophagus (BE) and EA are not fully understood. Mitochondria play a central role in generating energy, apoptosis and cell proliferation. Mutations of mitochondrial DNA (mtDNA) have been identified in many diseases including cancers. Mutations of mtDNA were investigated as a part of carcinogenesis.
Objective
Our objective is to study whether the 5 kb and 7.4 kb mtDNA deletions are important in the progression of normal esophagus to BE and EA.
Method
In this study, the frequency of the 5 kb and 7.4 kb deletions in mtDNA were studied in specimens ranging from normal esophageal tissue to BE and EA and also from ESCC. Seventy six paraffin-embedded tissue samples were studied. Four couple primers were used.
Results
Seventy-six tissue samples were analyzed total. The negative control and the positive control PCR product were detected in all analyzed samples. The fusion PCR products, which represent the presence of the deletions, were not detected in any of the samples.
Conclusion
We can say that, these deletions are not associated with progression of normal esophagus to BE and EA and they do not have an important role in detecting esophagitis, BE, EA, and ESSC.
Keywords: Barrett's esophagus, esophageal cancer, mitochondrial DNA, 4977 bp, 7400 bp
Introduction
Esophageal cancer is the eighth-most common cancer globally with an estimated 16,640 new cases and 14,500 deaths during the year in the United States1. It is very common and aggressive especially for Asian populations2. Esophagus adenocarcinoma (EA) and esophageal squamous-cell carcinoma (ESCC) are the two major types of esophageal cancer. Both of them have a poor prognosis. Symptoms often include difficulty in swallowing, weight loss, pain when swallowing, a hoarse voice, enlarged lymph nodes around the collar bone, a dry cough, and possibly coughing up or vomiting blood3. The most common risk factor for EA is chronic gastroesophageal reflux disease (GERD)4. GERD also causes esophagitis. Barrett's esophagus (BE) is one of the most common premalignant lesions. Esophageal squamous epithelium damaged by GERD may be replaced by Barrett's mucosa, a condition in which the normal squamous epithelium of the esophagus is replaced by a metaplastic, columnar, or glandular epithelium that is predisposed to malignancy5,6. Patients with Barrett's esophagus have 30 to 60 times greater risk of developing EA than the general population7. The progression of BE to EA develops through established histologic changes: intestinal metaplasia (also known as BE) to low grade dysplasia (LGD), high-grade dysplasia (HGD) and EA8,9.
Mitochondria are organelles responsible for generating energy in eukaryotic cells. Mitochondria have their own genome. Each cell contains several hundred to 1000 mitochondria. Each mitochondrion has 2 to 10 copies of mitochondrial DNA (mtDNA), which is transmitted through the maternal lineage10. DNA of mitochondria is circular, double stranded, closed DNA of 16569 bp in size and represents 0.1∼1.0% of the total genomic DNA11. It contains 37 genes encoding 13 peptides for the oxidative phosphorylation apparatus, 7 for subunits of complex I, 1 for subunit of complex III, 3 forsubunits of complex IV, and 2 for subunits of complex V, as well as 22 tRNAs and 2 rRNAs12. It has no introns and replication rate is very high13. Because of the reactive oxygen species, free radicals and lack of a sophisticated DNA repair mechanism it has a very high mutation rate, about 10 times higher than nuclear DNA14.
Mitochondria play a central role in apoptosis and cell proliferation15. Mutations of mtDNA have been associated with seizures, ataxia, cortical blindness, dystonia, exercise intolerance, ophthalmoplegia, optic atrophy, cataracts, diabetes mellitus, short stature, cardiomyopathy and other myopathies, sensorineural hearing loss, and kidney failure (http://www.mitomap.org/). There is also an association between mtDNA and tumors16–18. Many of the mtDNA mutations associated with cancers occur within the non-coding control region (also known as the D-loop) of the mitochondrial genome, which is about 1.1 kb (between bases 16024 and 576) in size19–22. The D-loop was reported to be more susceptible to oxidative damage and sequence variation23.
Common 5 kb or 4977-bp deletion is one of the best-known mutation in mtDNA, between nucleotides 8,470 and 13,447. It is associated with many disorders like myopathies, Alzheimer disease, cancers, aging and used as an indication of mtDNA oxidative damage2,12,24–26. This mutation removes all or part of the genes encoding four complex I subunits, one complex IV subunit, two complex V subunits and five tRNA genes, therefore causing energy production catastrophe and abnormal reactive oxygen species generation27. The 7.4 kb deletion is flanked by direct repeats and it is between the D-loop and the ATPase 6 genes of mtDNA28. It is associated with ageing and cardiac problems28.
In this study the frequency of the 4977 bp and 7400 bp deletions in mtDNA were studied in specimens ranging from normal esophageal tissue to BE and EA and from ESCC. Our objective is to study whether the 5 kb and 7.4 kb mtDNA deletions are important in the progression of normal esophagus to BE and EA.
Methods
Tissue samples
The study was approved by the Ethics Committee of Ataturk University. We used 76 paraffin-embedded tissue samples which were histopathologically confirmed. The tissues were belonging to the patients living in Turkey. Among 33 samples from females, there were 5 normal esophagus, 8 esophagitis, 8 BE, 7 EA and 5 ESCC; and among 43 samples from males there were 5 normal esophagus, 6 esophagitis, 8 BE, 17 EA and 7 ESCC. Five micron thick sections were taken from each patient's tumor in paraffin-embedded tissue blocks.
Mitochondrial DNA extraction
Mitochondrial DNA was extracted from formalin fixed, paraffin-embedded tissue sections using the QIAamp DNA Mini Kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturers' instructions. Paraffin was removed by extraction with xylene and ethanol (96–100%) added to remove residual xylene. Tissue samples were washed with phosphate-buffered saline (PBS) to remove formalin-fixative. Proteinase K was used until the tissue was completely lysed and DNA was extracted. DNA concentration and quality was quantified by absorbance readings taken at 260 and 280 nm using MaestroNano Spectrophotometer (Maestrogen Inc., Hsinchu City, Taiwan).
Polymerase Chain Reactions and gel electrophoresis
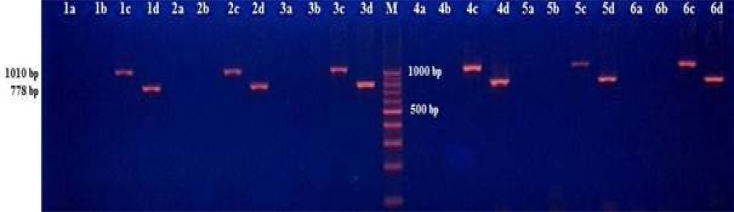
A duplex polymerase chain reaction (PCR) method was used for amplifications of target sequences of mtDNA2. Four couple primers were used: the first (L.9590-H.10368) was to amplify the negative control sequence within the deleted sequence, the second (L.6251-H.7261) was to amplify the positive control sequence out of the deleted sequence, the third (L.7901-H.13631) was to amplify the 4977 bp deletion region and the fourth (L.8531-H.381) was to amplify the 7400 bp deletion region (Table 1). PCR was performed in Sensoquest Labcycler (Sensoquest GmbH, Hannah, Germany). Thermal cycle conditions were as follows: an initial denaturation step of 95°C for 15 min; 32 cycles of 94°C for 60 s, 55°C for 60 s, 72°C for 80 s and a final extension step of 72°C for 15 min. PCR products were resolved together with a marker DNA (DNA 100 bp Ladder) on a 2% agarose gel, stained with ethidium bromide and visualized under UV transilluminator (Figure 1).
Table 1.
The primer sequences and the expected sizes of PCR products of the two fragments in the control regions and two fragments for deletion regions
| Primers | PCR Product Sizes |
Sequences of Primer Pairs |
| Control sequence (negative control) |
778 bp | L(9590) 5′-AGTCCCACTCCTAAACACATCCG-3′ H(10368) 5′-AGGCCAGACTTAGGGCTAGGATGATG-3′ |
| Control sequence (positive control) |
1010 bp | L(6251) 5′-TAT AGT GGA GGC CGG AGC AG-3′ H(7261) 5′-GAA TGA GCC TAC AGA TGA TA-3′ |
| mtDNA4977 deletion region |
753 bp | L(7901) 5′-TGA ACC TAC GAG TAC ACC GA-3′ H(13631) 5′-GGG GAA GCG AGG TTG ACC TG-3′ |
| mtDNA7400 deletion region |
1019 bp | L(8531) 5′-ACG AAA ATC TGT TCG CTT CA-3′ H(381) 5′-AAA TTT GAA ATC TGG TTA GG-3′ |
Figure 1.
The PCR products of esophageal tissues of six patients on 2% agarose gel. “a” represents a 1019 bp PCR product occurring in case of 7400 bp deletion of mtDNA; “b” represents a 753 bp PCR product occurring in case of 4977 bp deletion of mtDNA; “c” represents a 1010 bp PCR product, negative control within deletion region; “d” represents a 778 bp PCR product, positive control out of deletion region; M represented a marker DNA to detect the size of fragments.
Results
In total, seventy-six tissue samples were analyzed. The negative control PCR product of 778 bp and the positive control PCR product of 1010 bp were detected in all analyzed samples. The fusion PCR products of 753 bp which appears if there is the 4977 bp deletion of mtDNA and 1019 bp which appears if there is the 7400 bp deletion of mtDNA, were not seen in any of the samples. Due to the absence of the deletions in any sample, statistical analysis was not performed.
Discussion
Esophageal cancer, the eighth-most common cancer, is aggressive and it has a poor prognosis. GERD causes esophagitis and BE. BE is one of the most common premalignant lesions for EA development7. After the process of carcinogenesis, which is controlled by genetic, epigenetic and environmental factors, the cells gain uncontrollable growth capacity29. Mutations of mtDNA were investigated as a part of this process30–32. Therefore it is important to search for mtDNA alterations in progression of normal esophagus to BE and EA and also for other esophageal malignancies.
The mtDNA4977 deletion is associated with ageing and in many cancers. The mtDNA4977 deletion was less frequent in cancers when compared with the normal tissues; it could be an adaptation mechanism for tumors33–37. Tan et al.38 reported that mtDNA4977 deletion could be used as a biomarker for dysplasia but not for EA38. Abnet et al.2 reported high frequency of mtDNA4977 deletion in esophageal cancer patients from China, but mutation analysis of the whole mitochondrial genome from Northern India did not find this deletion2,17. This maybe because of Chinese population has a high risk for this cancer and it is possible that the pathogenesis of esophageal cancers in this population includes mtDNA mutations.
The 7.4 kb mtDNA deletion has been studied in many diseases, but only found meaningful in ageing and cardiac problems28,39. However the role of the 7.4 kb mtDNA deletion in the progression of normal esophagus to BE, EA and in ESCC is not clear.
In our study the common deletion was considered present if the primers produced a 753 bp fusion product and the 7.4 kb deletion was considered present if the primers produced a 1019 bp fusion product. We used negative control within the deleted sequence, because the common mtDNA region is very susceptible to oxidative damage and deletion of this part could have different hotspots for breakage, and if there is a deletion in this region we would have detected it. However, we could not detect any deletions in these regions and controls.
Conclusion
Our data provides insights for genotype-phenotype correlations of mtDNA mutations with esophageal diseases. Unlike Abnet et al.2, we could say that, the 5 kb and 7.4 kb mtDNA deletions are not associated with progression of normal esophagus to BE and EA and they do not have an important role in detecting and prognosis of esophagitis, BE, EA, and ESSC. Further studies, especially single nucleotide variations and copy number alterations of mtDNA, should be undertaken to investigate the effects of mutations on disease in relation with modifiers.
Ethical publication statement
We confirm that we have read the journal's position on issues involved in ethical publication and we affirm that this report is consistent with those guidelines. The study was approved by the Ethics Committee of Ataturk University (mentioned within “Methods”).
* The study's findings were presented at The Medical Genetics and Clinical Applications Congress (with International Participation) 11–13 February, 2016, Kayseri, Turkey.
Conflict of interest
None.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Abnet CC, Huppi K, Carrera A, Armistead D, McKenney K, Hu N, et al. Control region mutations and the 'common deletion’ are frequent in the mitochondrial DNA of patients with esophageal squamous cell carcinoma. BMC Cancer. 2004;4:30. doi: 10.1186/1471-2407-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsai HW, Chang CC, Sun JT, Liou CB, Lin HC, Lin IH, et al. Clinical features of patients with esophageal and second primary cancers. Asian Pac J Cancer Prev. 2014;15(22):9831–9834. doi: 10.7314/apjcp.2014.15.22.9831. [DOI] [PubMed] [Google Scholar]
- 4.Lagergren J, Bergstrom R, Lindgren A, Nyren O. Symptomatic gastroesophageal reflux as a risk factor for esophageal adenocarcinoma. N Engl J Med. 1999;340(11):825–831. doi: 10.1056/NEJM199903183401101. [DOI] [PubMed] [Google Scholar]
- 5.Sharma P. Clinical practice. Barrett's esophagus. N Engl J Med. 2009;361(26):2548–2556. doi: 10.1056/NEJMcp0902173. [DOI] [PubMed] [Google Scholar]
- 6.Mao WM, Zheng WH, Ling ZQ. Epidemiologic risk factors for esophageal cancer development. Asian Pac J Cancer Prev. 2011;12(10):2461–2466. [PubMed] [Google Scholar]
- 7.Cossentino MJ, Wong RK. Barrett's esophagus and risk of esophageal adenocarcinoma. Semin Gastrointest Dis. 2003;14(3):128–135. [PubMed] [Google Scholar]
- 8.Tharavej C, Hagen JA, Peters JH, Portale G, Lipham J, DeMeester SR, et al. Predictive factors of coexisting cancer in Barrett's high-grade dysplasia. Surg Endosc. 2006;20(3):439–443. doi: 10.1007/s00464-005-0255-x. [DOI] [PubMed] [Google Scholar]
- 9.Shaheen N, Ransohoff DF. Gastroesophageal reflux, barrett esophagus, and esophageal cancer: scientific review. JAMA. 2002;287(15):1972–1981. doi: 10.1001/jama.287.15.1972. [DOI] [PubMed] [Google Scholar]
- 10.Lin CS, Chang SC, Wang LS, Chou TY, Hsu WH, Wu YC, et al. The role of mitochondrial DNA alterations in esophageal squamous cell carcinomas. J Thorac Cardiovasc Surg. 2010;139(1):189–197. doi: 10.1016/j.jtcvs.2009.04.007. e4. [DOI] [PubMed] [Google Scholar]
- 11.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23(2):147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 12.Shen L, Fang H, Chen T, He J, Zhang M, Wei X, et al. Evaluating mitochondrial DNA in cancer occurrence and development. Ann N Y Acad Sci. 2010;1201:26–33. doi: 10.1111/j.1749-6632.2010.05635.x. [DOI] [PubMed] [Google Scholar]
- 13.Fridovich I. Mitochondria: are they the seat of senescence? Aging Cell. 2004;3(1):13–16. doi: 10.1046/j.1474-9728.2003.00075.x. [DOI] [PubMed] [Google Scholar]
- 14.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6(5):389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green DR, Galluzzi L, Kroemer G. Cell biology. Metabolic control of cell death. Science. 2014;345(6203):1250256. doi: 10.1126/science.1250256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berneburg M, Kamenisch Y, Krutmann J. Repair of mitochondrial DNA in aging and carcinogenesis. Photochem Photobiol Sci. 2006;5(2):190–198. doi: 10.1039/b507380d. [DOI] [PubMed] [Google Scholar]
- 17.Gochhait S, Bhatt A, Sharma S, Singh YP, Gupta P, Bamezai RN. Concomitant presence of mutations in mitochondrial genome and p53 in cancer development - a study in north Indian sporadic breast and esophageal cancer patients. Int J Cancer. 2008;123(11):2580–2586. doi: 10.1002/ijc.23817. [DOI] [PubMed] [Google Scholar]
- 18.Xia P, Wang HJ, Geng TT, Xun XJ, Zhou WJ, Jin TB, et al. Mitochondrial DNA levels in blood and tissue samples from breast cancer patients of different stages. Asian Pac J Cancer Prev. 2014;15(3):1339–1344. doi: 10.7314/apjcp.2014.15.3.1339. [DOI] [PubMed] [Google Scholar]
- 19.Liu VW, Shi HH, Cheung AN, Chiu PM, Leung TW, Nagley P, et al. High incidence of somatic mitochondrial DNA mutations in human ovarian carcinomas. Cancer Res. 2001;61(16):5998–6001. [PubMed] [Google Scholar]
- 20.Tan DJ, Bai RK, Wong LJ. Comprehensive scanning of somatic mitochondrial DNA mutations in breast cancer. Cancer Res. 2002;62(4):972–976. [PubMed] [Google Scholar]
- 21.Yacoub HA, Mahmoud WM, El-Baz HA, Eid OM, RI EL, Elhamidy SM, et al. Novel mutations in the displacement loop of mitochondrial DNA are associated with acute lymphoblastic leukemia: a genetic sequencing study. Asian Pac J Cancer Prev. 2014;15(21):9283–9289. doi: 10.7314/apjcp.2014.15.21.9283. [DOI] [PubMed] [Google Scholar]
- 22.Ashtiani ZO, Heidari M, Hasheminasab SM, Ayati M, Rakhshani N. Mitochondrial D-Loop polymorphism and microsatellite instability in prostate cancer and benign hyperplasia patients. Asian Pac J Cancer Prev. 2012;13(8):3863–3868. doi: 10.7314/apjcp.2012.13.8.3863. [DOI] [PubMed] [Google Scholar]
- 23.Mambo E, Gao X, Cohen Y, Guo Z, Talalay P, Sidransky D. Electrophile and oxidant damage of mitochondrial DNA leading to rapid evolution of homoplasmic mutations. Proc Natl Acad Sci U S A. 2003;100(4):1838–1843. doi: 10.1073/pnas.0437910100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai JG, Xiao YB, Min JX, Zhang GQ, Yao K, Zhou RJ. Mitochondrial DNA 4977 BP deletion mutations in lung carcinoma. Indian J Cancer. 2006;43(1):20–25. doi: 10.4103/0019-509x.25771. [DOI] [PubMed] [Google Scholar]
- 25.Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, McKee AC, Beal MF, et al. Marked changes in mitochondrial DNA deletion levels in Alzheimer brains. Genomics. 1994;23(2):471–476. doi: 10.1006/geno.1994.1525. [DOI] [PubMed] [Google Scholar]
- 26.Wu CW, Yin PH, Hung WY, Li AF, Li SH, Chi CW, et al. Mitochondrial DNA mutations and mitochondrial DNA depletion in gastric cancer. Genes Chromosomes Cancer. 2005;44(1):19–28. doi: 10.1002/gcc.20213. [DOI] [PubMed] [Google Scholar]
- 27.Peng TI, Yu PR, Chen JY, Wang HL, Wu HY, Wei YH, et al. Visualizing common deletion of mitochondrial DNA-augmented mitochondrial reactive oxygen species generation and apoptosis upon oxidative stress. Biochim Biophys Acta. 2006;1762(2):241–255. doi: 10.1016/j.bbadis.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 28.Hattori K, Tanaka M, Sugiyama S, Obayashi T, Ito T, Satake T, et al. Age-dependent increase in deleted mitochondrial DNA in the human heart: possible contributory factor to presbycardia. Am Heart J. 1991;121(6 Pt 1):1735–1742. doi: 10.1016/0002-8703(91)90020-i. [DOI] [PubMed] [Google Scholar]
- 29.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 30.Basso D, Navaglia F, Fogar P, Zambon CF, Greco E, Schiavon S, et al. DNA repair pathways and mitochondrial DNA mutations in gastrointestinal carcinogenesis. Clin Chim Acta. 2007;381(1):50–55. doi: 10.1016/j.cca.2007.02.020. [DOI] [PubMed] [Google Scholar]
- 31.Tan DJ, Chang J, Liu LL, Bai RK, Wang YF, Yeh KT, et al. Significance of somatic mutations and content alteration of mitochondrial DNA in esophageal cancer. BMC Cancer. 2006;6:93. doi: 10.1186/1471-2407-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richard SM, Bailliet G, Paez GL, Bianchi MS, Peltomaki P, Bianchi NO. Nuclear and mitochondrial genome instability in human breast cancer. Cancer Res. 2000;60(15):4231–4237. [PubMed] [Google Scholar]
- 33.Bua E, Johnson J, Herbst A, Delong B, McKenzie D, Salamat S, et al. Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers. Am J Hum Genet. 2006;79(3):469–480. doi: 10.1086/507132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohamed SA, Hanke T, Erasmi AW, Bechtel MJ, Scharfschwerdt M, Meissner C, et al. Mitochondrial DNA deletions and the aging heart. Exp Gerontol. 2006;41(5):508–517. doi: 10.1016/j.exger.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 35.Eshaghian A, Vleugels RA, Canter JA, McDonald MA, Stasko T, Sligh JE. Mitochondrial DNA deletions serve as biomarkers of aging in the skin, but are typically absent in nonmelanoma skin cancers. J Invest Dermatol. 2006;126(2):336–344. doi: 10.1038/sj.jid.5700088. [DOI] [PubMed] [Google Scholar]
- 36.Ye C, Shu XO, Wen W, Pierce L, Courtney R, Gao YT, et al. Quantitative analysis of mitochondrial DNA 4977- bp deletion in sporadic breast cancer and benign breast diseases. Breast Cancer Res Treat. 2008;108(3):427–434. doi: 10.1007/s10549-007-9613-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nie H, Shu H, Vartak R, Milstein AC, Mo Y, Hu X, et al. Mitochondrial common deletion, a potential biomarker for cancer occurrence, is selected against in cancer background: a meta-analysis of 38 studies. PLoS One. 2013;8(7):e67953. doi: 10.1371/journal.pone.0067953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan BH, Skipworth RJ, Stephens NA, Wheelhouse NM, Gilmour H, de Beaux AC, et al. Frequency of the mitochondrial DNA 4977bp deletion in oesophageal mucosa during the progression of Barrett's oesophagus. Eur J Cancer. 2009;45(5):736–740. doi: 10.1016/j.ejca.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 39.St John JC, Jokhi RP, Barratt CL. Men with oligoasthenoteratozoospermia harbour higher numbers of multiple mitochondrial DNA deletions in their spermatozoa, but individual deletions are not indicative of overall aetiology. Mol Hum Reprod. 2001;7(1):103–111. doi: 10.1093/molehr/7.1.103. [DOI] [PubMed] [Google Scholar]