Abstract
Background
Avian influence virus H5N1 causes serious public health concern with significant morbidity and mortality from poultry to humans. Interferon-induced transmembrane (IFITM) proteins usually protect cells from many virus infections by viral entry and replication.
Objectives
The purpose of this study was to investigate whether H5N1 viral proteins involved in regulation IFITM1, IFITM2, and IFITM3 following H5N1 infection.
Methods
NS1, M1, NP, PB2, HA and NA genes of H5N1 virus were generated by PCR and cloned into pcDNA3.1/myc-His (+) A vector for genes over-expression experiments. Gene expression levels was performed using Real-time PCR.
Results
Research displayed that NS1, M1, NP, and PB2 proteins of H5N1 virus increased IFITM1, IFITM2, and IFITM3 expression in A549 cells, only IFITM1 was upregulated by M1 in HEK293T cells. However, our study did not find that HA and NA of H5N1 virus affected IFITM genes family or interferon genes expression.
Conclusion
Taken together, our data suggested that IFITM1, IFITM2, and IFITM3 might be directly upregulated via NS1, M1, NP, and PB2 proteins during H5N1 avian influenza virus infection. This study provided new insights into the influence of NS1 and NP proteins on regulation of IFITM1, IFITM2, and IFITM3 expression following H5N1 infection.
Keywords: Influenza Virus, H5N1, NS1 protein, NP protein, IFITMs
Introduction
Avian influenza A virus H5N1 is one of the greatest worldwide pandemic threats to poultry and humans health. H5N1 contains eight negative-sense, single-stranded RNAs that code for eleven proteins (HA, NA, NP, M1, M2, NS1, NEP, PA, PB1, PB1-F2, PB2)1,2. Structural and non-structural proteins of H5N1 are relevant in respect of adaptive and innate immune responses through activating or inhibiting a series of immune-related cytokine. Previous study showed NS1 as the primary influenza virus interferon antagonist with multiple inhibitory effects on host immune pathways3. NS1, as a virulence factor of H5N1 due to mutation at the 42nd residue within the RNA-binding domain (RBD), dramatically changed the degree of pathogenicity of H5N1 in mice4. The IFITM genes belong to a family of interferon-induced transmembrane proteins including IFITM1, IFITM2, IFITM3, IFITM5, and IFITM10. Only IFIM1, IFITM2, and IFITM3 were significantly upregulated by I and II type interferons stimulate5,6. These proteins as innate antiviral cell-intrinsic restriction factors prevent viruses from traversing the lipid bilayer and accessing the cytoplasm, such as influenza A virus (IAV), Japanese encephalitis virus (JEV), and dengue virus (DENV), et al7,8. NP belong to viral nucleoprotein, with viral RNA polymerase together consisting of ribonucleoprotein complexes. Ubiquitination and deubiquitination of NP protein regulated influenza A virus RNA replication9.
To assess whether structural and nonstructural proteins of H5N1 virus involved in regulated IFITM1, IFITM2, and IFITM3 expression, we conducted overpression of NS1, M1, NP, PB2, HA, and NA proteins in order to evaluated the effect of viral proteins on IFITM family. Our study showed that several structural and nonstructural of H5N1 virus distinctly increased IFITM1, IFITM2, and IFITM3 expression, and especially NS1 and NP protein played a stronger role in A549 cells.
Materials and methods
Cell culture
A549 and HEK 293T cells were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM, Gibco) with 10% fetal bovine serum (FCS, Gibco), in a humidified 5% CO2 and 95% air at 37°C.
Gene cloning and plasmids
The open reading frame (ORF) of influenza virus NS1, M1, NP, PB2, HA and NA genes were generated by PCR and cloned into pcDNA3.1/myc-His(+)A vector for genes over-expression experiments. All constructs were verified by sequencing, and primers used in this study are listed in Table 1.
Table 1.
Primer sequences of full-length gene annealing temperature used in construct plasmids
| Gene Symbol |
Sequence(5′–3′) | Annealing(°C) | plamsid |
| NS1 | TACGAATTCATGGATTCCAACACTGTGTC | 54 | 3.1 |
| TACCTCGAGCTTTGGAGAGAGTGGAGGTC | |||
| M1 | TACAAGCTTATGAGCCTTCTAACCGAGGTCGAA | 58 | 3.1 |
| TACCTCGAGCTTGAATCGCTGCATTTGCACTC | |||
| NP | CCCGGATCCATGGCGTCTCAGGGCACCAAACGA | 56 | 3.1 |
| CCCCTCGAGATTGTCATATTCCTCTGCATTGTC | |||
| PB2 | CCCAAGCTTATGGAGAGAATAAAAGAATTAAG | 56 | 3.1 |
| CCCCTCGAGATAGATGGCCATCCGAATCCTTTTG | |||
| HA | TACAAGCTTATGGAGAAAATAGTGCTTCTTCTT | 54 | 3.1 |
| TACCTCGAGAATGCAAATTCTGCATTGTAACG | |||
| NA | TACAAGCTTGCCACCATGAATCCAAATCAGAAGA | ||
| TAATAAC | 54 | 3.1 | |
| TCACTCGAGCTTGTCAATGGTGAATGGCAAC |
Virus Culture
Mouse adapted influenza virus A/environment/Qinghai/1/2008 (H5N1) was propagated in 10-day-old embryonated chicken eggs at 37 °C for 2 days. Allantoic fluid was purified by centrifugation and was stored at −80°C until viral infection was performed when cells were 80% confluent. The serum-free DMEM medium containing allantoic fluid was added at 0.5, 1, 2 multiplicities of infection (MOI) for 1h. Then cells were washed thrice by PBS and cultured with DMEM containing 2% FBS. All experiments with this infectious virus were carried out in a Biosafety Level 3 containment laboratory.
Transfection
Cells were seeded in 12-well plates overnight and transfected with 2 ug plasmids suspended in 3 ul Lipofectamine 3000 (Invitrogen) and 100 ul OPTI-MEM Reduced Serum Medium. After 6 h transfection, the cell medium was replaced by fresh DMEM medium with 10% FBS. A549 and HEK293T cells were transfected for 36 h, then harvested and resolved in TRIzol Reagent for RNA extraction and reverse transcription.
RNA extraction and cDNA synthesis
Total RNA was extracted from A549 and HEK293T cells using the TRIzol Reagent according to the supplier's specifications. Spectrophotometer (Thermo) was used to determine the concentration of RNA at 260/280 nm. cDNA was synthesized using 5ug of total RNA from A549 and HEK293T cells, GoScriptTM Reverse Transcription System (Promega) was used for first-stand synthesis following manufacturer's instructions and was stored at −80°C until used.
Real-time PCR
Real-time PCR was performed in a ABI 7500 fast real-time system (Life Technology) with SYBR green real-time PCR master mix (CWBIO). Real-time PCR conditions were as follows: 95°C for 10 min, then 94°C for 30 s, 60°C for 1 min, 40 cycles, followed by 72°C for 7 min. Reactions were performed in a 20ul volume containing 10 ul SYBR green real-time PCR master mix, 8.5 ul double-distilled water, 1 ul forward and reverse primers, and 0.5 ul cDNA template. The primers are listed in Table 2. β-actin as internal reference in order to normalize the mRNA expression levels of target genes, and generation of specific PCR products was confirmed by melting curve analysis. 2-ΔΔCT method was used to quantify the relative mRNA express levels.
Table 2.
Primer sequences, amplicon size and annealing temperature used in Real-time PCR assays
| Gene Symbol |
Sequence(5′-3′) | Amplicon(bp) | Annealing(°C) |
| β-actin | GCGGGAAATCGTGCGTGACATT | 232 | 60 |
| GATGGAGTTGAAGGTAGTTTCGTG | |||
| IFNA1 | AGAAATACAGCCCTTGTGCC | 150 | 60 |
| TGACCTGGTGTATGAGTCAATAAG | |||
| IFNB1 | CAGCTCTTTC CATGAGCTAC | 330 | 60 |
| CAGCC AGTGC TAGATGAATC | |||
| IFNG | GAGATGACTTCGAAAAGCTGAC | 149 | 60 |
| ACCTCGAAACAGCATCTGAC | |||
| H5N1 | GACCAATCCTGTCACCTCTGA | 251 | 60 |
| GTATATGAGGCCCATRCAACT | |||
| IFITM1 | ATCAACATCCACAGCGAGAC | 253 | 60 |
| CAGAGCCGAATACCAGTAACAG | |||
| IFITM2 | TTCATAGCATTCGCGTACTCC | 296 | 60 |
| GAATACAGGTCAAGGGCAGAG | |||
| IFITM3 | GAGAACCATCCCAGTAACCC | 318 | 60 |
| CAACCATCTTCCTGTCCCTAG |
Statistical analysis
Real-time PCR data of IFITM and interferon family genes are expressed as the means ± s.e.m. (standard error of the mean). Student's t-test was used to determine the significance of differences between two groups, while one-way analysis of variance (ANOVA) was used when there were more than two groups. P-value <0.05 was considered to be statistically significant.
Results
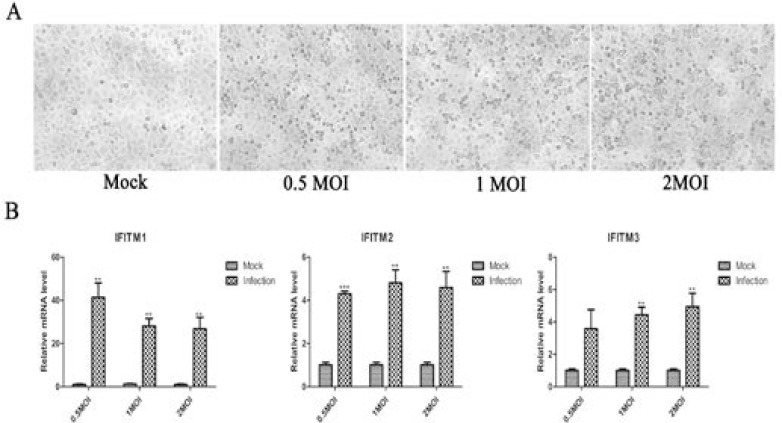
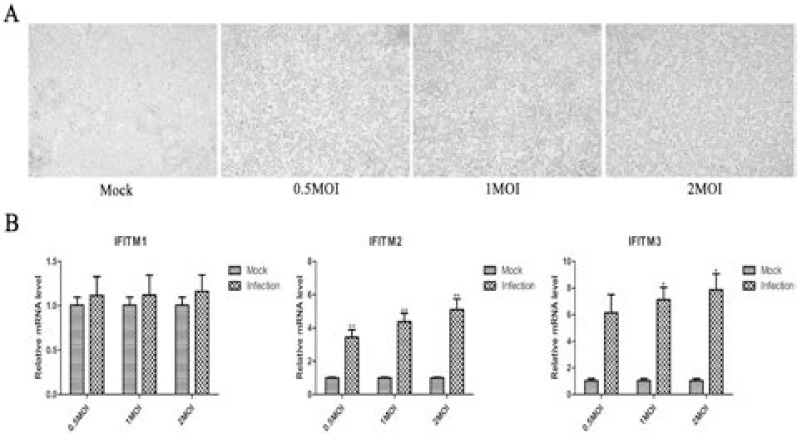
H5N1 promoted expression of IFITM1, 2, and 3 genes in A549 cells
H5N1 virus induced death of A549 and HEK293T cells at 0.5, 1, 2 MOI for 1h, IFITM1, IFITM2, and IFITM3 expression increased in A549 cells, only IFITM2 and IFITM3 were upregulated but IFITM1 had no changes in HEK293T cells. In addition, IFITM1 had higher expression levels than that of IFITM2 and IFITM3 in A549 cells. It was a opposite phenomenon with expression level of IFITM1, IFITM2 and IFITM3 a higher expression than that of IFITM1 in HEK293T cells (Figure 1 and Figure 2).
Figure 1.
Transcription expression pattern of IFITM1, 2, and 3 in A549 cells by H5N1 influenza virus infection.
Figure 2.
Transcription expression pattern of IFITM1, 2, and 3 in HEK293T cells by H5N1 influenza virus infection.
IFNA1, IFNB1, and IFNG were upregulated by H5N1 infection
Our study demonstrated that H5N1 avian influenza virus could upregulate IFNA1, IFNB1, and IFNG in both A549 and HEK293T cells (Figure 3). NS1 and NP proteins were found increased IFNA1 expression, NP could increase IFNB1 expression in A549 cells, and simultaneously NS1 upregulated IFNG in HEK293T cells (Figure 4). HA and NA proteins showed no effect on the interferon genes IFNA1, IFNB1, and IFNG expression (Figure 5).
Figure 3.
Transcription of IFNA1, IFNB1, and IFNG with H5N1 virus infection.
Figure 4.
Influences of influenza virus NS1, M1, NP, and PB2 proteins on the expression of interferons.
Figure 5.
Influences of influenza virus HA and NA proteins on the expression of interferons.
NS1 and NP proteins increased IFITM1, 2, and 3 expression in A549 cells
Experiments displayed NS1, M1, NP, and PB2 could promote IFITM1, IFITM2, and IFITM3 expression in A549 cells, only M1 could upregulate IFITM1 expression in HEK293T cells (Figure 6). Either HA or NA showed no effect on the interferon-related genes IFITM1, IFITM2, and IFITM3 (Figure 7).
Figure 6.
Influences of influenza virus NS1, M1, NP, and PB2 proteins on the gene expression of IFITM family.
Figure 7.
Influences of influenza virus HA and NA proteins on the expression of IFITM family.
Discussion
Our study revealed that H5N1 virus could upregulate IFITM1, IFITM2, and IFITM3, further showing that H5N1 virus also promoted IFNA1, IFNB1, and IFNG overexpression in A549 cell. Above results illustrated H5N1 itself was probably the optimal immunogenic substances, compared to the individual structural or nonstructural proteins of H5N1 virus. To demonstrate which proteins of H5N1 virus involved in immune-related cytokine production, NS1, M1, NP, PB2, HA and NA proteins were overexpressed in A549 and HEK293T cells, respectively. Our study has shown NS1, M1, NP and PB2 of H5N1 virus could increase IFITM1, IFITM2, and IFITM3 expression, and especially NS1 and NP proteins played a stronger role than M1 and PB2 in A549 cells.
The most diverse IAV genes, HA and NA, have been divided into 17 and 10 subtypes, respectively, differences in sequence motifs have been distinguished between subtypes10,11. Previous studies had shown infection with viruses possessing the 1918 virus HA gene caused massive recruitment of polymorphonuclear cells accompanied by intra-alveolar hemorrhage12,13,14. Surprising, HA and NA proteins show no effects on the IFNA1, IFNB1, and IFNG expression, and instead NS1 and NP proteins as viral proteins display important regulatory functions in this study. Billharz R et al. revealed 1918 virus NS1 had potent IFN antagonist activities2. Many evidence had shown that M1 protein played an important role in influenza virus assembly, as it could interact with the viral envelope proteins HA and NA via their cytoplasmic tails and also could interact with the viral RNP (vRNP), which constituted the viral core15,16. NS1 protein of influenza A virus plays important roles in antagonizing the host antiviral response and supporting virus replication, suggesting that the NS1 protein had the ability of suppressing host antiviral defenses at multiple levels17,18. Ubiquitin ligase TRIM25, 2′,5′-oligoadenylate synthetase (OAS), and protein kinase R (PKR) were identified as target by NS1, further induce antiviral effect, which were key regulators of influenza virus transcription/translation processes19,20,21. Our data showed H5N1 virus could promote IFNA1, IFNB1, and IFNG expression, and upregulate IFITM2 and IFITM3, but had no effects on IFITM1 in HEK293T cells. NS1 increased IFNG expression and M1 increased IFITM1 expression in HEK293T cell. IFITM genes can restrict the entry of a wide range of viruses by I and II type interferons, such IFITM3 as an important restrictor against several influenza A viruses22. Moveover, IFN has roles in inhibition of viral replication, stimulation of CTL, increasing MHC I expression, activation of macrophages and neutrophils, and promoting T-cell proliferation23,24. A previous study suggested that the presence of IFN-γ improves the severity of inflammation and lung damage25. Upregulated FAT10 promoted H5N1 viral replication by inhibiting type I IFN26. Furthermore, M1, PB2, HA and NA proteins had no significant effect on IFNA1, IFNB1, and IFNG either in A549 cells or HEK293T cells. Above results demonstrated different cells had differences gene expression profiles, and it also revealed IFNA1, IFNB1, and IFNG specifically performed following distinct infection routes of H5N1.
Conclusion
No articles had reported whether H5N1 virus itself proteins involved in regulation of IFITM1, IFITM2, and IFITM3 with H5N1 influenza virus infection. Our study first found NS1, M1, NP and PB2 of H5N1 virus contributed directly to expression of IFITM1, IFITM2, and IFITM3, revealing a new pathway for understanding activation mechanism of IFITM genes family with influenza virus infection.
Acknowledgments
We thank the Key Scientific Research Projects of Higher Education Institutions in Henan Province under grant number 19A180021, the Young Key Teachers Training Program of Yellow River Conservancy Technical Institute (HYJG[2018]37), the Campus Scientific and Research Fund Project of Yellow River Conservancy Technical Institute under grant number 2017QNKY012, the Research Projects of Employment and Entrepreneurship of Secondary and Higher Education Institutions in Henan Province under grant number JYB2018534, and National Natural Sciences Foundation of China under grant number 31070954.
Conflict of interest
The authors declared that there was no conflict of interests.
References
- 1.Resa-Infante P, Jorba N, Coloma R, Ortin J. The influenza virus RNA synthesis machine: advances in its structure and function. RNA Biol. 2011;8:207–215. doi: 10.4161/rna.8.2.14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rossman JS, Lamb RA. Influenza virus assembly and budding. Virology. 2011;411:229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billharz R, Zeng H, Proll SC, Korth MJ, Lederer S, Albrecht R, et al. The NS1 protein of the 1918 pandemic influenza virus blocks host interferon and lipid metabolism pathways. J Virol. 2009;83:10557–10570. doi: 10.1128/JVI.00330-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kato YS, Fukui K, Suzuki K. Mechanism of a Mutation in Non-Structural Protein 1 Inducing High Pathogenicity of Avian Influenza Virus H5N1. Protein Pept Lett. 2016;23(4):372–378. doi: 10.2174/0929866523666160204124406. [DOI] [PubMed] [Google Scholar]
- 5.Liao TL, Wu CY, Su WC, Jeng KS, Lai MM. Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication. The EMBO Journal. 2010;29:3879–3890. doi: 10.1038/emboj.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nature reviews. Immunology. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey CC, Zhong G, Huang IC, Farzan M. IFITM-Family Proteins: The Cell's First Line of Antiviral Defense. Annual review of virology. 2014;1:261–283. doi: 10.1146/annurev-virology-031413-085537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.John SP, Chin CR, Perreira JM, Feeley EM, Aker AM, Savidis G, et al. The CD225 domain of IFITM3 is required for both IFITM protein association and inhibition of influenza A virus and dengue virus replication. Journal of Virology. 2013;87:7837–7852. doi: 10.1128/JVI.00481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484(7395):519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, et al. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol. 2005;79:2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, et al. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci USA. 2012;109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobasa D, Takada A, Shinya K, Hatta M, Halfmann P, Theriault S, et al. Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature. 2004;431:703–707. doi: 10.1038/nature02951. [DOI] [PubMed] [Google Scholar]
- 13.Kash JC, Basler CF, Garcia-Sastre A, Carter V, Billharz R, Swayne DE, et al. Global host immune response: pathogenesis and transcriptional profiling of type A influenza viruses expressing the hemagglutinin and neuraminidase genes from the 1918 pandemic virus. J Virol. 2004;78:9499–9511. doi: 10.1128/JVI.78.17.9499-9511.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe T, Tisoncik-Go J, Tchitchek N, Watanabe S, Benecke AG, Katze MG, et al. 1918 Influenza Virus Hemagglutinin (HA) and the Viral RNA Polymerase Complex Enhance Viral Pathogenicity, but Only HA Induces Aberrant Host Responses in Mice. Journal of Virology. 2013;87(9):5239–5254. doi: 10.1128/JVI.02753-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitt AP, Lamb RA. Influenza virus assembly and budding at the viral budozone. Adv Virus Res. 2005;64:383–416. doi: 10.1016/S0065-3527(05)64012-2. 26. [DOI] [PubMed] [Google Scholar]
- 16.Wu CY, Jeng KS, Lai MM. The SUMOylation of Matrix Protein M1 Modulates the Assembly and Morphogenesis of Influenza A Virus. Journal of Virology. 2011;85(13):6618–6628. doi: 10.1128/JVI.02401-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hale BG, Randall RE, Ortin J, Jackson D. The multifunctional NS1 protein of influenza A viruses. J Gen Virol. 2008;89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 18.Kochs G, Garcia-Sastre A, Martinez-Sobrido L. Multiple anti-interferon actions of the influenza A virus NS1 protein. J Virol. 2007;81:7011–7021. doi: 10.1128/JVI.02581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, et al. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Min JY, Li S, Sen GC, Krug RM. A site on the influenza A virus NS1protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology. 2007;363:236–243. doi: 10.1016/j.virol.2007.01.038. [DOI] [PubMed] [Google Scholar]
- 21.Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, et al. RIG-I-mediated antiviral responses to singlestranded RNA bearing 5-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 22.Blyth GA, Chan WF, Webster RG, Magor KE. Duck Interferon-Inducible Transmembrane Protein 3 Mediates Restriction of Influenza Viruses. J Virol. 2015;90(1):103–116. doi: 10.1128/JVI.01593-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boehm U, Klamp T, Groot M, Howard JC. Cellular responses to interferon-gamma. Annu Rev Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 24.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma:an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 25.Wiley JA, Cerwenka A, Harkema JR, Dutton RW, Harmsen AG. Production of interferon-gamma by influenza hemagglutinin-specific CD8 effector T cells influences the development of pulmonary immunopathology. Am J Pathol. 2001;158:119–130. doi: 10.1016/s0002-9440(10)63950-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Tang J, Yang N, Liu Q, Zhang Q, Zhang Y, et al. FAT10 Is Critical in Influenza A Virus Replication by Inhibiting Type I IFN. J Immunol. 2016;197(3):824–833. doi: 10.4049/jimmunol.1501563. [DOI] [PubMed] [Google Scholar]