Microcins represent potential alternatives to conventional antibiotics for human and veterinary medicine. For them to be applied in this manner, however, we need to better understand their spectrum of activity, how these proteins interact with susceptible cells, and how producer cells are protected against the antimicrobial properties of the microcins. For microcin PDI (MccPDI), we report that the spectrum of activity likely includes most E. coli strains due to a conserved binding motif found on an outer membrane protein. Shigella has this motif as well and is susceptible to MccPDI killing via damage to the bacterial membrane. Receptor specificity suggests that these proteins could be used without causing large-scale disruptions to a microbiota, but this also increases the likelihood that resistance can evolve via random mutations. As with conventional antibiotics, good stewardship will be needed to preserve the efficacy of microcins should they be deployed for clinical use.
KEYWORDS: microcin PDI, multidrug resistant, UTI, urinary tract infection
ABSTRACT
Microcin PDI (MccPDI), a class IIa microcin that is produced by Escherichia coli strains 25 and 284, is known to inhibit foodborne pathogenic enterohemorrhagic E. coli serotypes O157:H7 and O26. Here we demonstrate that MccPDI can inhibit Shigella strains and E. coli isolates that are multidrug resistant, the latter including strains known to cause urinary tract infections in people and companion animals. Two exceptions out of 17 strains were identified. One of the two resistant E. coli isolates (AR0349) has a mutation in a critical amino acid residue that was identified in previous work as a requisite for the MccPDI precursor protein (McpM) to interact with outer membrane porin F (OmpF) on susceptible cells. The second resistant E. coli strain (MAD 96) had no mutations in ompF, but it was PCR positive for two antimicrobial peptides, of which colicin Ia/Ib likely inhibits the MccPDI-producing strain during coculture. Recombinant McpM was still effective against strain MAD 96. In an assessment of how MccPDI affects susceptible strains, results from both an extracellular ATP assay and a nucleic acid staining assay were consistent with membrane damage, while the addition of 200- to 600-Da polyethylene glycol (PEG) to cocultures protected against MccPDI (>600-Da PEG did not provide protection). Further studies using a paraformaldehyde cross-linking experiment and a bacterial two-hybrid assay demonstrated that MccPDI immunity protein (McpI) forms a multimeric complex with itself and presumably protects the producer strain from within the periplasm through an unknown mechanism.
IMPORTANCE Microcins represent potential alternatives to conventional antibiotics for human and veterinary medicine. For them to be applied in this manner, however, we need to better understand their spectrum of activity, how these proteins interact with susceptible cells, and how producer cells are protected against the antimicrobial properties of the microcins. For microcin PDI (MccPDI), we report that the spectrum of activity likely includes most E. coli strains due to a conserved binding motif found on an outer membrane protein. Shigella has this motif as well and is susceptible to MccPDI killing via damage to the bacterial membrane. Receptor specificity suggests that these proteins could be used without causing large-scale disruptions to a microbiota, but this also increases the likelihood that resistance can evolve via random mutations. As with conventional antibiotics, good stewardship will be needed to preserve the efficacy of microcins should they be deployed for clinical use.
INTRODUCTION
Microcins are antimicrobial peptides (<10 kDa) that are produced by Gram-negative bacteria predominantly from the family Enterobacteriaceae (1–3). The recently described microcin PDI (MccPDI), a class IIa microcin (4) produced by Escherichia coli strains 25 and 284, inhibits growth of foodborne pathogenic E. coli strains, including enterohemorrhagic E. coli serotypes O157:H7 and O26 (5). The name microcin “PDI” was given due to the apparent need for the producing bacteria to be in close proximity to inhibit target bacteria (i.e., proximity-dependent inhibition) (6). The reason why proximity is required is unknown, although only minute quantities of the native protein appear to be secreted, and the proximity requirement may be a consequence of simple concentration dependence. The MccPDI system is plasmid encoded, consisting of five genes: mcpM (precursor protein), mcpI (self-immunity protein), mcpA (putative repressor protein with CaaX protease activity [5]), mcpB (export protein B), and mcpD (export protein D). Export proteins B and D comprise a type I secretion system (T1SS) that functions with TolC (an outer membrane protein) to secrete McpM (5). The excretion of McpM is accompanied by the cleavage of two signal sequences, leaving an 8-kDa mature peptide (Fig. 1), originally described by Zhao et al. (7). E. coli strains are susceptible to MccPDI when outer membrane porin F (OmpF) is present, which was demonstrated in part by heterologous-expression experiments (8). Previous site-directed mutagenesis experiments identified an amino acid motif in extracellular loop 1 of OmpF (K47G48N49) that is required for inhibition of susceptible bacteria (8). This motif is present in multiple reported OmpF sequences from both E. coli and Shigella strains (see Fig. S1 in the supplemental material).
FIG 1.
Full-length amino acid sequence of McpM (GenBank accession number JQ901381). The MccPDI precursor protein, McpM, consists of 120 amino acids. This graphical representation shows McpM and McpM with a 6×His tag (white) and the two signal peptides G17/G18 (dark gray) and G35/A36 (medium gray) where the precursor protein is cleaved during excretion to form the mature peptide (light gray) (7). The excreted protein has an approximate mass of 8.2 kDa and a theoretical pI of 9.58.
Microcins are being investigated as alternatives to medically important antibiotics (3, 9, 10). According to the U.S. Centers for Diseases Control and Prevention (CDC) and the World Health Organization (WHO), the increasing prevalence of multidrug-resistant (MDR) pathogens limits successful clinical outcomes for both people and animals (11–15). Antimicrobial resistance (AMR) is a significant global challenge for both community-acquired (16, 17) and hospital-acquired (18) infections. E. coli strains causing urinary tract infections (UTI) are a particularly challenging problem, with a pandemic of a multilocus sequence type 131 (ST131) strain afflicting people from both high- and lower-income countries (17, 19–21).
Microcins might be a versatile alternative antibiotic for these infections. These low-mass proteins (<10 kDa), of which fewer than 20 have been described (8, 22), appear to be stable and functional under a wide range of pH and ionic conditions (23). Microcins are typically highly specific for conspecific bacteria, making it possible to target specific pathogens causing UTI, pulmonary infections, and septicemia without harming “bystander” bacteria. The present work demonstrates that MccPDI can kill a diversity of bacteria, including multidrug-resistant E. coli strains from urinary infections and Shigella strains. We report two cases whereby UTI strains (Table 1) were resistant to MccPDI when cultured with E. coli strain 25 (Table 1), with the probable mechanisms of resistance involving mutation of a key amino acid residue in the OmpF protein and production of a colicin that likely kills the MccPDI-producing strain before it can affect the susceptible strain.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant genotype or phenotypea | Reference and/or sourceb |
|---|---|---|
| Escherichia coli | ||
| 25 | Wild type; SSuTr PDI+ | 5 |
| 25 ΔmcpM ΔmcpI | SSuTr PDI−; mcpM and mcpI deletion | 7 |
| 25 ΔmcpM ΔmcpI/pMMB207 | SSuTr PDI−; mcpM and mcpI deletion with pMMB207 vector (no-insert control) | This study |
| 25 ΔmcpM ΔmcpI::pMMB207::mcpM | SSuTr Chlr PDI−; mcpM and mcpI deletion complemented with mcpM | This study |
| 25 ΔmcpM::pMMB207::mcpM | SSuTr Chlr PDI−; mcpM deletion complemented with mcpM | 7 |
| 25 ΔmcpM ΔmcpI/pFPV25.1::gfpmut3 | SSuTr Ampr PDI−; mcpM and mcpI deletion complemented with GFPmut3 | This study |
| 25 ΔtraM | SSuTr Kanr PDI+; traM deletion | 5 |
| 25 ΔtraM/pFPV25.1::gfpmut3 | SSuTr Kanr Ampr PDI+; traM deletion with pFPV25.1 complemented with GFPmut3 | This study |
| BW25113 | Nalr; Keio collection wild-type K-12 strain | 65 |
| BW25113 ΔompF | Kanr; Keio collection; ompF deletion | 65 |
| BW25113/pMMB207 | Nalr Chlr; BW25113 with pMMB207 vector (no-insert control) | This study |
| BW25113/pMMB207::mcpI | Nalr Chlr; BW25113 with pMMB207 complemented with mcpI | This study |
| BW25113/pFPV25.1::tdTomato | Nalr Ampr; BW25113 with pFPV25.1 complemented with tdTomato | This study |
| PAP222 | Wild type; Nalr microcin V positive | 66 |
| DH10B | F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ− rpsL nupG | Thermo Fisher |
| DH10B/pCR2.1::pmic-210/0mcpMIADB | Ampr; DH10B with pCR2.1 complemented with the native promoter −210 and PDI operon (4.9 kb) | This study |
| DHM1 | Nalr; F− cya-854 recA1 endA1 gyrA96 thi1 hsdR17 spoT1 rfbD1 glnV44(AS) | Euromedex |
| DHM1/pUT18/pKNT25 | Nalr Ampr Kanr; negative control with vectors pUT18 and pKNT25 | This study |
| DHM1/pUT18::mcpI/pKNT25::mcpI | Nalr Ampr Kanr; complemented with mcpI | This study |
| DHM1/pUT18::mcpI/pKNT25::mcpMΔ36 | Nalr Ampr Kanr; complemented with mcpI and mcpMΔ36 | This study |
| DHM1/pUT18::mcpI/pKNT25::ompF | Nalr Ampr Kanr; complemented with mcpI and ompF | This study |
| DHM1/pUT18C/pKT25 | Nalr Ampr Kanr; negative control with vectors pUT18C and pKT25 | This study |
| DHM1/pUT18C::mcpI/pKT25::mcpI | Nalr Ampr Kanr; complemented with mcpI | This study |
| DHM1/pUT18C::mcpI/pKT25::mcpMΔ36 | Nalr Ampr Kanr; complemented with mcpI and mcpMΔ36 | This study |
| DHM1/pUT18C::mcpI/pKT25::ompF | Nalr Ampr Kanr; complemented with mcpI and ompF | This study |
| DHM1/pUT18::mcpMΔ36/pKNT25::mcpI | Nalr Ampr Kanr; complemented with mcpMΔ36 and mcpI | This study |
| DHM1/pUT18::mcpMΔ36/pKNT25::mcpMΔ36 | Nalr Ampr Kanr; complemented with mcpMΔ36 | This study |
| DHM1/pUT18::mcpMΔ36/pKNT25::ompF | Nalr Ampr Kanr; complemented with mcpMΔ36 and ompF | This study |
| DHM1/pUT18C::mcpMΔ36/pKT25::mcpMΔ36 | Nalr Ampr Kanr; complemented with mcpMΔ36 and mcpI | This study |
| DHM1/pUT18C::mcpMΔ36/pKT25::mcpI | Nalr Ampr Kanr; complemented with mcpMΔ36 and mcpI | This study |
| DHM1/pUT18C::mcpMΔ36/pKT25::ompF | Nalr Ampr Kanr; complemented with mcpMΔ36 and ompF | This study |
| DHM1/pUT18::ompF/pKNT25::mcpI | Nalr Ampr Kanr; complemented with ompF and mcpI | This study |
| DHM1/pUT18::ompF/pKNT25::mcpMΔ36 | Nalr Ampr Kanr; complemented with ompF and mcpMΔ36 | This study |
| DHM1/pUT18::ompF/pKNT25::ompF | Nalr Ampr Kanr; complemented with ompF | This study |
| DHM1/pUT18C::ompF/pKT25::mcpI | Nalr Ampr Kanr; complemented with ompF and mcpI | This study |
| DHM1/pUT18C::ompF/pKT25::mcpMΔ36 | Nalr Ampr Kanr; complemented with ompF and mcpMΔ36 | This study |
| DHM1/pUT18C::ompF/pKT25::ompF | Nalr Ampr Kanr; complemented with ompF | This study |
| DHM1/pUT18C::Zip/pKT25::Zip | Nalr Ampr Kanr; positive control with vectors pUT18C::Zip and pKT25::Zip | Euromedex |
| K-12 | Nalr | 6 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac(F′ proAB lacIqZΔM15 Tn10 [Tetr]) | Agilent |
| XL1-Blue/pUT18::mcpI | Tetr Ampr; complemented with mcpI in vector pUT18 | This study |
| XL1-Blue/pUT18::mcpMΔ36 | Tetr Ampr; complemented with mcpMΔ36 in vector pUT18 | This study |
| XL1-Blue/pUT18::ompF | Tetr Ampr; complemented with ompF in vector pUT18 | This study |
| XL1-Blue/pUT18C | Tetr Ampr; with vector pUT18C | This study |
| XL1-Blue/pUT18C::mcpI | Tetr Ampr; complemented with mcpI in vector pUT18C | This study |
| XL1-Blue/pUT18C::mcpMΔ36 | Tetr Ampr; complemented with mcpMΔ36 in vector pUT18C | This study |
| XL1-Blue/pUT18C::ompF | Tetr Ampr; complemented with ompF in vector pUT18C | This study |
| XL1-Blue/pUT18C::Zip | Tetr Ampr; positive-control vector pUT18C with leucine zipper of GCN4 | This study |
| XL1-Blue/pKT25 | Tetr Kanr; with vector pKT25 | This study |
| XL1-Blue/pKT25::mcpI | Tetr Kanr; complemented with mcpI in vector pKT25 | This study |
| XL1-Blue/pKT25::mcpMΔ36 | Tetr Kanr; complemented with mcpMΔ36 in vector pKT25 | This study |
| XL1-Blue/pKT25::ompF | Tetr Kanr; complemented with ompF in vector pKT25 | This study |
| XL1-Blue/pKT25::Zip | Tetr Kanr; positive-control vector pKT25 with leucine zipper of GCN4 | This study |
| XL1-Blue/pKNT25 | Tetr Kanr; with vector pKNT25 | This study |
| XL1-Blue/pKNT25::mcpI | Tetr Kanr; complemented with mcpI in vector pKNT25 | This study |
| XL1-Blue/pKNT25::mcpMΔ36 | Tetr Kanr; complemented with mcpMΔ36 in vector pKNT25 | This study |
| XL1-Blue/pKNT25::ompF | Tetr Kanr; complemented with ompF in vector pKNT25 | This study |
| Escherichia coli (urinary tract isolates) | ||
| AR0346 | Wild type | CDC |
| AR0349 | Wild type | CDC |
| MAD 92 | Wild type | This study, WADDL |
| MAD 95 | Wild type | This study, WADDL |
| MAD 96 | Wild type | This study, WADDL |
| MAD 98 | Wild type | This study, WADDL |
| MAD 99 | Wild type | This study, WADDL |
| MAD 101 | Wild type | This study, WADDL |
| MAD 102 | Wild type | This study, WADDL |
| MAD 107 | Wild type | This study, WADDL |
| MAD 111 | Wild type | This study, WADDL |
| Shigella | ||
| S. flexneri 24570 | Wild type | BEI Resources |
| S. flexneri 2457T | Wild type | BEI Resources |
| S. sonnei WRAIR I virulent | Wild type | BEI Resources |
| S. dysenteriae Newcastle 1934 | Wild type | BEI Resources |
| Shigella sp. | Wild type | BEI Resources |
Ampr, ampicillin resistant; Chlr, chloramphenicol resistant; Kanr, kanamycin resistant; Nalr, nalidixic acid resistant; SSuTr, resistant to streptomycin, sulfonamide, and tetracycline antibiotics.
E. coli isolates AR0346 and AR0349 were obtained from the CDC and FDA antibiotic resistance isolate bank. WADDL, Washington Animal Disease Diagnostic Lab; BEI Resources, Biodefense and Emerging Infections Research Resources Repository.
Due to technical challenges with obtaining sufficient quantities of recombinant McpM for our experiments, we employed three different methods to evaluate the activity of MccPDI. The inhibition activity of MccPDI was observed by coculturing a wild-type MccPDI producer strain (E. coli 25) in vitro with target bacteria and quantifying changes in the number of target cells over time. This methodology demonstrated that MccPDI affects a wide diversity of E. coli and Shigella strains, and it was used to determine that MccPDI kills susceptible bacteria via membrane damage. Using two different recombinant vectors, we were able to generate sufficient recombinant McpM to verify the bactericidal phenotype of MccPDI using spot assays on agar plates. Finally, we further characterized how the immunity protein (McpI) protects the MccPDI-producing strains. E. coli strain 25 is “immune” to the effects of McpM because of concurrent synthesis of a “self-immunity” protein (McpI) that forms a multimeric structure to protect the PDI producer strain via an unknown mechanism.
RESULTS
Microcin PDI inhibits Shigella strains and UTI E. coli strains.
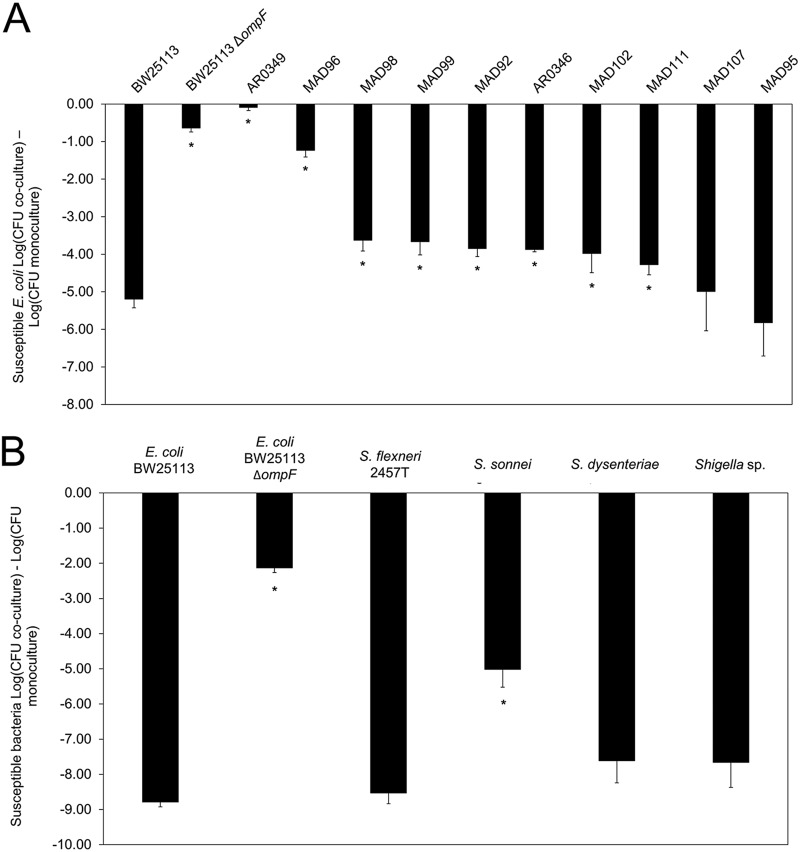
Previously validated coculture competition assays (5–8, 24) were employed to examine the sensitivity of the target bacteria to MccPDI. The coculture competition assays were conducted with E. coli strain 25 (MccPDI producer), six Shigella strains, and nine UTI E. coli strains that were resistant to between one and four classes of antibiotics (Table 2 and Fig. 2). Cell counts (CFU) were then determined by plating on agar with corresponding antibiotic selection. Coculture with strain BW25113 (positive control; MccPDI susceptible) resulted in a 5.2-log CFU reduction relative to monoculture, while coculture with the BW25113 ΔompF strain (negative control; MccPDI nonsusceptible) resulted in a 0.64-log CFU reduction. Eight of the 10 tested UTI strains were inhibited between 3.5 and 8.5 log CFU relative to monoculture results, while Shigella strains were reduced between 5.0 and 8.8 log CFU (Fig. 2A and B; see also Fig. S2A and B in the supplemental material). Two strains, AR0349 and MAD 96, were not inhibited. A reversed coculture competition assay conducted to measure the survival of the MccPDI producer (E. coli 25) against all 12 tested E. coli strains showed that only MAD 96 was capable of inhibiting the producer (Fig. S3). Bacteriocin typing by PCR indicated that E. coli strain MAD 96 harbors genes for two bacteriocins (colicin Ia/Ib [Col Ia/Ib] and microcin V) (Fig. S4) that could inhibit E. coli strain 25, thereby blocking the MccPDI phenotype. We were able to obtain a strain of E. coli that produces only microcin V (strain PAP222) (Table 1) and demonstrated that this microcin does not inhibit E. coli strain 25 (Fig. S5). Presumably, colicin Ia/Ib is responsible for protecting MAD 96 by inhibiting growth of E. coli strain 25.
TABLE 2.
Antibiotic profiles of bacteria used in this studyb
| Strain | Resistance profilea | Sample | Species |
|---|---|---|---|
| Escherichia coli | |||
| MAD 92 | AmcAmpXnl | Urine | Canine |
| MAD 95 | AmcAmpFoxCpdCefEnoGenMarTimSxt | Urine | Canine |
| MAD 96 | AmcAmpCefFoxCpdXnlPenTic | Urine | Canine |
| MAD 98 | AmcAmpLexXnlEno | Urine | Canine |
| MAD 99 | AmcAmpVecFoxCpdXnlCliEryOxaPenTic | Urine | Canine |
| MAD 102 | AmpEno | Urine | Canine |
| MAD 107 | AmcAmpCpdPen | Urine | Canine |
| MAD 111 | AmcAmpVecCpdXnlChlDoxEnoGenMarPenTicTimSxt | Urine | Canine |
| AR0346 | AmkAmpSamAtmCfzFepCtxFoxCipCstGenLvxTetTobSxt | NA | Human |
| AR0349 | AmkAmpSamAtmCfzFepCtxFoxCipCstGenbLvxTetTobSxt | NA | Human |
| 25 | StrSulTet | Fecal | Bovine |
| BW25113 | Nal | NA | NA |
| BW25113 ΔompF | Kan | NA | NA |
| Shigella | |||
| S. flexneri 2457T | Nal | NA | NA |
| S. flexneri 24570 | Nal | NA | NA |
| S. sonnei WRAIR I virulent | Nal | NA | NA |
| S. dysenteriae Newcastle 1934 | Nal | NA | NA |
| Shigella sp. | Nal | NA | NA |
Amc, amoxicillin-clavulanic acid; Amk, amikacin; Amp, ampicillin; Atm, aztreonam; Cef, cephalothin; Cfz, cefazolin; Chl, chloramphenicol; Cip, ciprofloxacin; Cli, clindamycin; Cpd, cefpodoxime; Cst, colistin; Ctx, cefotaxime; Dox, doxycycline; Eno, enrofloxacin; Ery, erythromycin; Fep, cefepime; Fox, cefoxitin; Gen, gentamicin; Kan, kanamycin; Lex, cephalexin; Lvx, levofloxacin; Mar, marbofloxacin; Nal, nalidixic acid; Oxa, oxacillin; Pen, penicillin; Sam, ampicillin-sulbactam; Str, streptomycin; Sul, sulfonamide; Sxt, trimethoprim-sulfamethoxazole; Tet, tetracycline; Tic, ticarcillin; Tim, ticarcillin-clavulanic acid; Tob, tobramycin; Vec, cefovecin; Xnl, ceftiofur. All resistance was determined by the MIC (Sensititre assays) (Washington Animal Disease Diagnostic Laboratory), with the exception of drug resistance of strains MAD 98, MAD 99, MAD 107, and MAD 111, which was determined by a disc diffusion assay. Resistance of E. coli strains AR0346 and AR0349 was determined by the MIC (Sensititre assays) (Centers for Disease Control and Prevention). Strains were considered intermediate at a typical concentration of 8 μg/ml.
NA, information not available.
FIG 2.
MccPDI is effective against UTI E. coli and Shigella strains. (A) Competition between MccPDI-producing strain E. coli 25 and positive and negative controls (BW25113 and BW25113 ΔompF, respectively) as well as 10 other strains in M9 medium for 24 h. (B) Competition between the MccPDI producer E. coli 25 and target Shigella strains (S. flexneri 24570, S. flexneri 2457T, S. sonnei WRAIR I virulent, S. dysenteriae Newcastle 1934, and Shigella sp.) in M9 medium for 24 h. Results are expressed as the difference in mean log CFU during coculture and monoculture of the target strain (n = 3 independent replicates; error bars indicate standard errors of the means [SEM]). *, P < 0.05 compared to wild-type (BW25113) coculture based on one-way ANOVA.
By spotting recombinant McpM (Fig. S6) onto agar plates that were prespread with target bacteria, we were able to confirm the bactericidal activity of McpM (Fig. S7). Interestingly, this was possible even when a key surface outer membrane protein, OmpF, was absent (Fig. S7). Spotted recombinant McpM also inhibited MAD 96 (Fig. S8). Spot assays with E. coli DH10B/pCR2.1::pmic-210/0mcpMIADB (vector construct containing the native mcpM promoter and all five MccPDI genes) confirmed the competition assay results, whereby zones of clearance were clearly evident where this strain was “spotted” onto the lawn of susceptible bacteria (Fig. S8). The zone of clearance in these assays indicates that even the MccPDI-producing strain succumbs to McpM, presumably due to the presence of an abundance of McpM that cannot be fully counteracted by the McpI immunity protein in this assay format. It is also possible that a bacteriocin-producing strain such as MAD 96 could produce a false-positive result with the spot assay by killing DH10B/pCR2.1::pmic-210/0mcpMIADB, thereby leaving a zone of clearance. In this case, it appears that MAD 96 not only kills the MccPDI strain but also grows in the same location, which prevents a false-positive finding (Fig. S9).
OmpF mutations.
Previous work demonstrated that E. coli OmpF is required for MccPDI to inhibit susceptible bacteria (8). An amino acid sequence alignment was used to compare OmpF from E. coli K-12 to OmpF sequences of different Shigella strains and E. coli AR0346, AR0349, and MAD 96 (Table 3). The E. coli AR0349 (GenBank accession no. MH665273) sequence differed from those of E. coli strains K-12 MG1655 and BW25113, Shigella flexneri 2a 2457T, S. sonnei 53G, S. boydii CDC3083-94, and S. dysenteriae Sd197 in multiple locations within extracellular loops 1, 2, 3, 4, 5, and 6 (loop identity is based on the OmpF crystal structure [25]) (Fig. S1), including mutations to a critical OmpF motif in extracellular loop 1 that is necessary for susceptibility to MccPDI (8). Mutations were not detected in loop 7 and loop 8. In contrast to the significant amino acid sequence differences in OmpF from strain AR0349, the amino acid sequences from several other strains, such as Shigella flexneri strain 2a 2457T and S. sonnei 53G, were identical to the conserved E. coli sequence within the extracellular loops.
TABLE 3.
PCR primers used in this study
| Primer | Sequence (5′–3′)a | Purpose | Reference |
|---|---|---|---|
| BACTH construct primers | |||
| M1_36_pUT18_F | catgcatgAAGCTTgCGTAACTCACTGGGTCGAAA | Used to construct mcpMΔ36 | This study |
| M1_36_pUT18_R | catgcatgGAATTCggTCGGTTACATGTTCCGC | ||
| M1_36_pKNT25_F | catgcatgAAGCTTgCGTAACTCACTGGGTCGAAA | Used to construct mcpMΔ36 | This study |
| M1_36_pKNT25_R | catgcatgGAATTCggTCGGTTACATGTTCCGC | ||
| M1_36_pUT18C_F | catgcatgTCTAGAgCGTAACTCACTGGGTCGAAA | Used to construct mcpMΔ36 | This study |
| M1_36_pUT18C_R | catgcatgGAATTCTTATCGGTTACATGTTCCGC | ||
| M1_36_pKT25_F | catgcatgTCTAGAgCGTAACTCACTGGGTCGAAA | Used to construct mcpMΔ36 | This study |
| M1_36_pKT25_R | catgcatgGAATTCTTATCGGTTACATGTTCCGC | ||
| mcpI_pUT18_F | catgcatgAAGCTTgATGGAGGGCGCTACTATGTT | Used to construct mcpI | This study |
| mcpI_pUT18_R | catgcatgGAATTCggTTTCGCGGAGATTGTTCTT | ||
| mcpI_pKNT25_F | catgcatgAAGCTTgATGGAGGGCGCTACTATGTT | Used to construct mcpI | This study |
| mcpI_pKNT25_R | catgcatgGAATTCggTTTCGCGGAGATTGTTCTT | ||
| mcpI_pUT18C_F | catgcatgTCTAGAgATGGAGGGCGCTACTATGTT | Used to construct mcpI | This study |
| mcpI_pUT18C_R | catgcatgGAATTCCTATTTCGCGGAGATTGTTCTT | ||
| mcpI_pKT25_F | catgcatgTCTAGAgATGGAGGGCGCTACTATGTT | Used to construct mcpI | This study |
| mcpI_pKT25_R | catgcatgGAATTCCTATTTCGCGGAGATTGTTCTT | ||
| ompF_pUT18_F | catgcatgAAGCTTgATGATGAAGCGCAATATTCTGG | Used to construct ompF | This study |
| ompF_pUT18_R | catgcatgGAATTCggGAACTGGTAAACGATACCC | ||
| ompF_pKNT25_F | catgcatgAAGCTTgATGATGAAGCGCAATATTCTGG | Used to construct ompF | This study |
| ompF_pKNT25_R | catgcatgGAATTCggGAACTGGTAAACGATACCC | ||
| ompF_pUT18C_F | catgcatgTCTAGAgATGATGAAGCGCAATATTCTGG | Used to construct ompF | This study |
| ompF_pUT18C_R | catgcatgGAATTCTTAGAACTGGTAAACGATACCC | ||
| ompF_pKT25_F | catgcatgTCTAGAgATGATGAAGCGCAATATTCTGG | Used to construct ompF | This study |
| ompF_pKT25_R | catgcatgGAATTCTTAGAACTGGTAAACGATACCC | ||
| M13R49 | GAGCGGATAACAATTTCACACAGG | Used to verify BACTH constructs | This study |
| pUT18C_R | GCATCAGAGCAGATTGTACTGAGAG | ||
| pUT18_R | TTCGCGATCCAGGCCGC | ||
| pKT25_R | CCAGGGTTTTCCCAGTCACG | ||
| pKNT25_R | GCGATTGCTGCATGGTCA | ||
| mcpI construct primers | |||
| mcpI_flag_F | ccgGAATTCATGGAGGGCGCTACTAT | Used to construct pMMB207::mcpI with addition of a Flag tag in the C terminus | This study |
| mcpI_flag_R | acgcGTCGACCTACTTGTCGTCATCGTCTTTGTAGTCTTTCGCGGAGATTGTTCT | ||
| Gene deletion mutant primers | |||
| A1_mcpMI_KO_F | ggaAGATCTATGGCAATGCTGGAAGAG | Used to construct suicide plasmid pDM4ΔmcpM/ΔmcpI(A1+A2) to delete mcpM and mcpI | This study |
| A1_mcpMI_KO_R | AATCTTGCCGATCATATTTCCCCTATCGGT | ||
| A2_mcpMI_KO_F | GAAATATGATCGGCAAGATTCATGGACTA | ||
| A2_mcpMI_KO_R | acgcGTCGACCTTCATAATACGGAACTGTCAG | ||
| mcpM_F | AGATGAGATAACGCTTGTCA | Used to confirm mcpM deletion and complementation | 7 |
| mcpM_R | ACTTCCTCTGTTACCACTTC | ||
| mcpI_F | TATGTGGTTTGTTACTGGGAT | Used to confirm mcpI deletion and complementation | 7 |
| mcpI_R | CGCGGAGATTGTTCTTATTT | ||
| SacI_pMIADB | ggtggtGAGCTCTGGAACGAGATGTACTGAACAGAGGCCGTG | Used to construct pmic-210/0mcpMIADB | This study |
| HindIII_MIADB | ggtggtAAGCTTTCACCTCCTGTTGGGGTGATTATTTATATTATGAGTTCTGGC | ||
| Construct tdTomato primers | |||
| tdtomato_Xbal | tgcTCTAGATTTAAGAAGGAGATATACATATGGTGAGCAAGGGCGAGGA | Construct pFPV-tdTomato for labeling bacteria | This study |
| tdtomato_SphI | catGCATGCCTACTTGTACAGCTCGTCCATGC | ||
| Bacteriocin primers | |||
| mcmM_F | GCATTAGTTGGGGAGCCAGA | Used to detect the gene mcmM (microcin M) | This study |
| mcmM_R | CAACCCCACCAGGAACAGTT | ||
| mchB_F | TGCGAGAAATAACAGAATCACAGT | Used to detect the gene mchB (microcin H47) | This study |
| mchB_R | CCAGCAGAAGAACTGGCACT | ||
| mceA_F | CTTGGCCCGATGAGTACAGG | Used to detect the gene mceA (microcin E492) | This study |
| mceA_R | TTTTGGTGCAGGAGAGACCG | ||
| cvaC_F | TTCAGGGCGTGATATTGCGA | Used to detect the gene cvaC (microcin V) | This study |
| cvaC_R | TCCCGCAGCATAGTTCCATG | ||
| cia_F | TGTAAACCCTCCACGTGTCG | Used to detect the gene cia (colicin 1a) | This study |
| cia_R | GACAACCGGGTGTCCAGAAT | ||
| cib_F | GGTGGTGACAGAGGATGTGG | Used to detect the gene cib (colicin 1b) | This study |
| cib_R | GTCTCCGCCATATGGACCTG | ||
| mcbA_F | TGGAATTAAAAGCGAGTGAATTT | Used to detect the gene mcbA (microcin B17) | This study |
| mcbA_R | CACCGTTTCCACCACTACAA | ||
| ompF sequence primers | |||
| ompF_up | CGCTATCAGGGTAACGGGAG | Used to sequence ompF in E. coli | This study |
| ompF_down | AGCACTTTCACGGTAGCGAA | ||
Restriction enzyme sites are underlined.
Polyethylene glycol protects a susceptible E. coli strain from MccPDI.
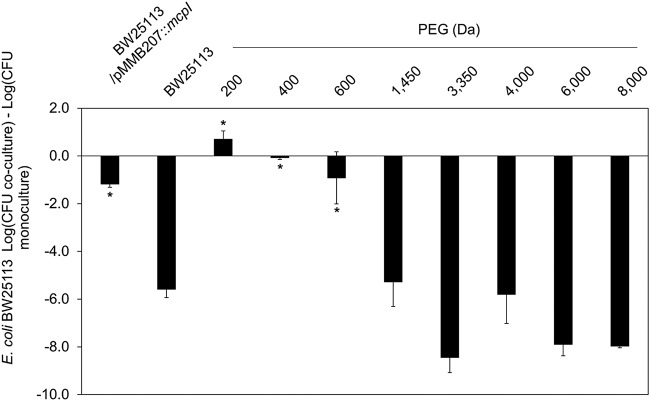
To determine if MccPDI functions by damaging the membrane of susceptible E. coli strains, we first conducted coculture assays in the presence of different mass fractions of polyethylene glycol (PEG) (200 to 8,000 Da). For these experiments, PEG is thought to act as an “osmoprotectant” (26) that blocks the MccPDI phenotype if the effector protein McpM permeabilizes the membrane. When susceptible E. coli strain BW25113 was cocultured with wild-type MccPDI producer E. coli strain 25, there was a typical 5.6-log CFU reduction in the number of BW25113 cells after 24 h, compared with a 1.2-log CFU reduction when E. coli strain BW25113 expressed a recombinant immunity protein (with isopropyl-β-d-1-thiogalactopyranoside [IPTG] induction), McpI (Fig. 3) (5). The addition of 15% (wt/vol) PEG with a molecular mass of between 200 and 600 Da blocked apparent membrane damage, whereas higher mass fractions (1,450 Da and larger) did not affect the MccPDI phenotype.
FIG 3.
The osmoprotectant PEG protects PDI-susceptible bacteria. Results of competition between E. coli 25 and susceptible strain BW25113 with the addition of 15% (wt/vol) PEG of different mass fractions (200 to 8,000 Da). The MccPDI immunity gene (mcpI)-complemented strains of BW25113 served as negative controls for this assay. Results are expressed as the difference in mean log CFU during coculture and monoculture of the target strain (n = 3 independent replicates; error bars indicate SEM). *, P < 0.05 compared to susceptible E. coli strain BW25113 coculture based on one-way ANOVA.
Differential dye testing is consistent with membrane damage.
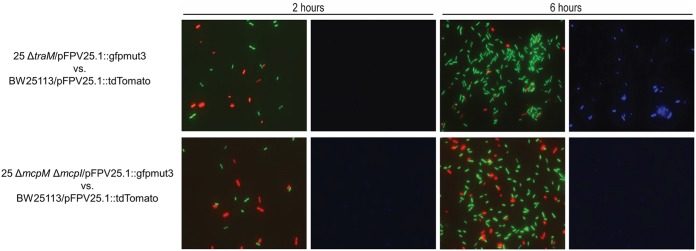
To confirm that MccPDI damages susceptible bacterial membranes, we used a differential staining technique whereby 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) nucleic acid stain was added to cocultures containing an MccPDI producer strain (green fluorescence) and an MccPDI-susceptible strain (red fluorescence). The remaining green/red bacterial cells were quantified after MccPDI exposure. For the concentration of dye used in this experiment, only bacteria with membrane damage will take up the stain for detection by fluorescence (27). The MccPDI-producing strain (E. coli 25 ΔtraM; conjugation negative [5]) and the MccPDI-negative strain (E. coli 25 ΔmcpM ΔmcpI [7]) were transformed with pFPV25.1::gfpmut3 (green). Here, E. coli 25 ΔtraM lacks the ability to be transferred to other cells by conjugation, which limits the likelihood of sharing the MccPDI plasmid and its immunity gene. E. coli 25 ΔmcpM ΔmcpI served as a negative control. MccPDI-susceptible E. coli BW25113 was transformed with pFPV25.1::tdTomato (red). After 2 h of coculture, there were similar proportions of green and red cells and no evidence of DAPI staining between the two groups (E. coli 25 ΔtraM versus BW2511 and E. coli 25 ΔmcpM ΔmcpI versus BW25113) (Fig. 4). After 6 h, however, the MccPDI producer (E. coli 25 ΔtraM) was numerically dominant over susceptible bacteria, and DAPI-stained cells were present (Fig. 4, top row), consistent with death of the susceptible bacteria as a result of membrane damage. In contrast, coculture with MccPDI-negative E. coli 25 (ΔmcpM ΔmcpI) did not result in a difference in the number of cells (green versus red) after 6 h.
FIG 4.
MccPDI induces membrane permeability in susceptible cells. Shown is DAPI staining of the fluorescence-labeled strains in monoculture and coculture for 2 h and 6 h. The green-labeled strains are E. coli 25 ΔtraM and 25 ΔmcpM ΔmcpI, and the red-labeled MccPDI-susceptible strain is BW25113. Blue fluorescence indicates that the cells were stained by DAPI, consistent with increased membrane permeability given exposure to MccPDI.
ATP release assay confirms that MccPDI damages the bacterial cell membrane.
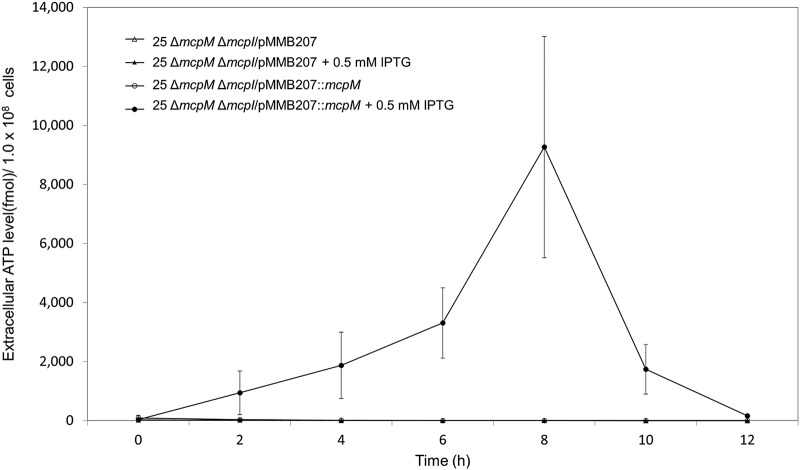
We cultured E. coli 25 ΔmcpM ΔmcpI (microcin and immunity knockout) as a monoculture with or without induction with plasmid pMMB207 containing an IPTG-inducible mcpM sequence. In the presence of 0.5 mM IPTG (Fig. 5), MccPDI was produced, and extracellular levels of ATP increased from an unmeasurable background level to a peak at 8 h before rapidly dropping off. None of the other test conditions resulted in significant changes in the concentration of extracellular ATP. Results were confirmed by a reduction of approximately 4 log CFU at 8 h from the expected population size (Fig. S10). The 8-h peak in the extracellular ATP level and the reduction in CFU also correspond to the expected expression kinetics of ompF (28).
FIG 5.
ATP detected with induced complementation for McpM. E. coli 25 ΔmcpM ΔmcpI with the vector (pMMB207) or with complemented mcpM (pMMB207::mcpM) without IPTG induction showed no detectable ATP in the extracellular environment. Upon induction (0.5 mM IPTG), E. coli 25 ΔmcpM ΔmcpI/pMMB207::mcpM generated elevated levels of ATP (2 to 8 h) compared to the vector-only strain. Results are mean luminescence values for three independent replicates (error bars indicate SEM).
McpI protein-protein interactions.
To characterize how the MccPDI self-immunity protein (McpI) (8 kDa) may function to protect the MccPDI-producing strain, we conducted paraformaldehyde (PFA) cross-linking experiments with a Flag-tagged McpI protein (Fig. 6). Treatment with 2% PFA alone revealed the presence of three protein bands, consistent with protein-protein interactions (Fig. 6, lane 6). The approximate molecular mass of these protein bands was similar to what would be expected if McpI forms multimers. Only a combination of boiling (100°C for 10 min) and the presence of β-mercaptoethanol (βME) eliminated most of the putative dimer and trimer structures (lane 3).
FIG 6.
The McpI immunity protein forms multimers. Shown is detection of the microcin PDI immunity protein (McpI) (with a C-terminal Flag tag) forming a homotrimer structure. Samples consisted of the total bacterial lysate expressing McpI in the presence or absence of the 2% paraformaldehyde (PFA) cross-linking reagent β-mercaptoethanol (βME) and with or without boiling of the sample for 10 min to disrupt cross-linking. Black arrows point toward potential monomer, dimer, and trimer structures upon cross-linking. Crossed-linked and boiled samples show evidence for both dimer and monomer proteins with (lane 1) or without (lane 2) βME. Total reduction of the homotrimer structure into monomers (lane 3) occurred with a combination of βME and boiling. Boiled samples without βME lacked a detectable dimer (lane 4). Heated (60°C) cross-linked samples with (lane 5) or without (lane 6) βME showed evidence of both homotrimer and dimer proteins.
Bacterial two-hybrid system confirms McpI-McpI interaction.
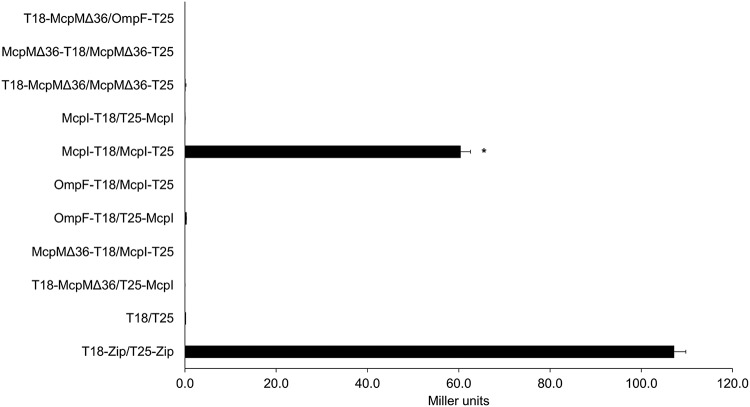
We used a bacterial two-hybrid (BACTH) system (29) to determine if the immunity protein McpI forms a multimer with itself, or with McpM, and to investigate other potential protein-protein interactions with OmpF. This assay works by fusing two proteins of interest with the T18 and T25 subunits of adenylate cyclase from Bordetella pertussis, respectively. If these subunits are located proximally when two proteins interact, this allows adenylate cyclase activity, thereby releasing cAMP into the medium, which can be quantified by measuring β-galactosidase activity (30). McpM is synthesized as a 120-amino-acid (aa) peptide that undergoes two cleavage events (Fig. 1) and is secreted via a type I secretion system (5, 7). We constructed the McpM fusion protein with the putative mature McpM sequence (84 aa long; strain McpMΔ36). Out of nine tested combinations, only the McpI-McpI interaction was positive (McpI-T18/McpI-T25) (Fig. 7). Interestingly, the McpI-T18/T25-McpI interaction did not result in restoration of adenylate cyclase activity, consistent with a nonsymmetric assembly of multimers.
FIG 7.
BACTH McpI-McpI interaction. A beta-galactosidase assay was used to confirm bacterial constructs to quantify the hybrid proteins’ interaction. Compared to the positive control (T18-Zip/T25-Zip) and negative control (vectors only) (T18/T25), the McpI-T18/McpI-T25 interaction was clearly positive given an N-terminal T25 orientation (black bar), but no signal was detected with an McpI-T25 C-terminal orientation (McpI-T18/T25-McpI). No protein interaction was detected with McpMΔ36 or OmpF. All readings were performed in a 96-well plate at an OD420, which are referred to as “Miller units” (n = 3 independent replicates; error bars indicate SEM). *, P < 0.05 in a comparison with the T18-Zip/T25-Zip positive control, based on one-way ANOVA.
DISCUSSION
While MccPDI is demonstrably effective against multiple strains of E. coli (5, 6), this paper reiterates this efficacy in the context of antibiotic-resistant bacteria, including strains recovered from urinary tract infections and several strains of Shigella. Urinary tract infections are of particular interest given that, on a global scale, antibiotic-resistant strains afflict 150 million people each year (31–34). The recent discovery of mcr-1-encoded colistin resistance in combination with extended-spectrum beta-lactamase (ESBL) resistance (e.g., strains AR0346 and AR0349) (Table 2) is emblematic of this growing problem. Even in cases where these strains can still be controlled with traditional antibiotics, the use of these drugs can cause complications for the patient while also providing selective pressure on the normal gastrointestinal tract microflora (34–36). Shigellosis is a problem worldwide (13, 37), particularly in communities suffering from poor sanitation and hygiene. Consequently, there is considerable interest in novel antimicrobials, such as microcins, that have a high degree of specificity and efficacy against E. coli and Shigella infections.
Microcins are a class of antimicrobial peptides that interact with cell surface receptors before translocating into susceptible bacteria (2, 8). Microcin PDI is known to interact with E. coli OmpF, which must be present for MccPDI to kill the target bacteria during coculture (8). The effector protein McpM likely binds to a specific location in OmpF (8), where it probably translocates through the porin, as has been documented for other bacteriocins (e.g., colicin E3 [38]). Interestingly, purified recombinant McpM protein can still inhibit an E. coli strain that does not synthesize OmpF (ΔompF mutant) (see Fig. S7 in the supplemental material). If McpM functions by entering the susceptible cell through OmpF, it is possible that recombinant McpM gains entry via mechanisms (e.g., other outer membrane porins) that may be concentration dependent, although coculture with E. coli 25 and E. coli BW25113 ΔompF showed some evidence that BW25113 was susceptible to “natural concentrations” of MccPDI when OmpF was absent (Fig. 2A and B show reduced CFU for E. coli BW25113 ΔompF, whereas the density of E. coli 25 was elevated when these strains were cocultured [Fig. S3]).
As with any antimicrobial compound, it is inevitable that resistance mechanisms will emerge. A likely “Achilles’ heel” of MccPDI involves mutations in the OmpF protein that is located in the outer membrane of susceptible cells. Zhao et al. (8) determined that a K47G48N49 amino acid motif found in the predicted outer loop 1 of OmpF is required for MccPDI to affect a susceptible cell. Strain AR0349 from the present study has a very divergent OmpF amino acid sequence that includes mutation in this K47G48N49 motif (Fig. S1). We do not know what selection pressures led to the emergence of such mutations, but the K47G48N49-to-K47D48N49 mutation should confer resistance to MccPDI (8). While we surmise that this amino acid motif is required for McpM binding, the glycine-to-aspartic acid substitution exhibited by strain AR0349 would also change the charge characteristics of the OmpF outer vestibule (39), and this might prevent passage of McpM through the porin. Consequently, while the susceptible sequence of OmpF is largely conserved in E. coli strains, and this conservation extends to Shigella species (Fig. S1), potential therapeutic applications of McpM should be judicious to limit widespread selection for resistance mutations.
E. coli strain MAD 96 was also resistant to MccPDI during coculture, although MAD 96 is susceptible to recombinant McpM (Fig. S8), consistent with a susceptible ompF sequence (Fig. S1), or this is an artifact of the high concentration of McpM that was used with the spot assays. Observations in the laboratory (not shown) led us to speculate that MAD 96 produces one or more bacteriocins that might inhibit E. coli strain 25 before McpM can be upregulated sufficiently to impact MAD 96 (Fig. S3 and S4). This would not be unusual given that it is common for uropathogenic E. coli (UPEC) strains to produce bacteriocins (40, 41). MAD 96 was PCR positive for both colicin Ia/Ib and microcin V (Fig. S4). On its own, MccV does not appear to inhibit an MccPDI producer (E. coli 25) in medium (Fig. S5), and thus, it is likely that Col Ia/Ib is primarily responsible for inhibiting the MccPDI-producing strain (Fig. S3). According to Abraham et al. (41), the likelihood of UPEC plasmids carrying both MccV and Col Ia/Ib is as high as 84.5%. This form of “resistance” against MccPDI (and other microcins) would not be relevant if McpM is deployed as a recombinant protein (Fig. S7 and S8) or if it was produced by a non-E. coli heterologous expression host.
Data from multiple assays from the present study are consistent with the conclusion that the MccPDI precursor protein McpM kills susceptible strains via membrane disruption, and experiments here and elsewhere confirm that naturally susceptible strains are widespread (6–8, 24). OmpF is the key ligand for susceptibility (8, 42), but other genes have been identified that are requisite for susceptibility to MccPDI (ompR, atpA, atpF, atpE, atpH, dsbA, and dsbB [8]). OmpR regulates the expression of ompF as part of the EnvZ/OmpR two-component osmoregulatory system (42–44). The roles of ATP synthase (atpA, atpF, atpE, and atpH) and thiol-disulfide interchange protein (dsbA and dsbB) in MccPDI susceptibility remain unknown (8, 45–47). If McpM enters the periplasmic space to affect susceptible cells, we speculate that these other host proteins are needed for McpM maturation before membrane disruption occurs, or they may play an important role in the synthesis and/or function of OmpF. The former would not be unprecedented given that microcin V permeabilizes the bacterial inner membrane, and E492 is a channel-forming bacteriocin (1, 48–50).
Results from the osmoprotectant assay were consistent with low-mass PEG (200 to 600 Da) blocking pores that might be formed by McpM. It is worth noting, however, that PEG molecules of this mass fraction have hydrodynamic radii of 0.56 to 0.8 nm, while the next-highest mass fraction (1,450 Da) did not protect susceptible bacteria, and the expected size of these molecules is 1.2 nm (51). OmpF consists of a trimer complex that forms an “hourglass”-shaped porin, having an external vestibule approximately 1 nm in diameter that reduces in a cone shape to a constricted region of approximately 0.5 nm (39). It is possible that at sufficient concentrations, 200- to 600-Da PEG enters the vestibule and physically blocks McpM from entering the porin or physically blocks access to the binding site. All three of these mechanisms (osmoprotection, physical blockage, and binding blockage) would protect susceptible bacteria from MccPDI.
Like other microcin systems (10, 23), the MccPDI locus encodes a “self-immunity protein” (McpI) that protects the microcin-producing cell from its antibacterial activity (5). Less is known about how microcin immunity functions, although immunity to microcins B17, C, E, and J25 is conferred by concurrently expressed efflux pumps (1, 4, 23, 52). We found no evidence that McpI binds to either McpM or OmpF, but it instead forms a homotrimer structure based on the bacterial two-hybrid (BACTH) system and PFA cross-linking experiments. It is important to note that the BACTH system detects only protein-protein interactions that occur in the cytosolic space (30, 53). Consequently, the McpI-McpI protein-protein interaction revealed by the BACTH system is consistent with the assembly of McpI multimers in the cytosol, although this does not preclude assembly of multimers in the periplasm as well. The function of the multimeric McpI structures is unknown, but analysis of the amino acid sequence using tools such as InterPro and PfamScan suggests the McpI does not function as an enzyme (54, 55). Based on protein structure prediction, McpI consists of two transmembrane domains and was predicted to be a transmembrane protein (Fig. S11) (56–58). It is possible that the multimeric form of McpI enters the periplasm, where it serves as a simple molecular “plug” that blocks the entry of linear McpM peptides into the periplasmic space via OmpF or as a plug to block pores that are formed by McpM from outside the cell membrane. Alternatively, the McpI multimers could remain in the cytosol, where they would block McpM-mediated damage to the inner membrane. Further work is needed to determine how McpM damages the cell membrane and how McpI prevents McpM from killing the producing cell.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Unless otherwise stated, all strains described here (Table 1) were grown in LB Lennox (LB broth) medium (Difco Laboratories, Inc.) or in defined M9 minimal medium (6 g/liter Na2HPO4, 3 g/liter KH2PO4, 0.5 g/liter NaCl, 1 g/liter NH4Cl, 1 mM MgSO4, 0.1 mM CaCl2, and 0.2% glucose) supplemented with thiamine (1 mg/liter) and leucine (100 μg/ml). Bacteria were cultured at 37°C on a shaker table (200 rpm) (24). When used, the following antibiotics were added to media: ampicillin (Amp) at 100 μg/ml, tetracycline (Tet) at 50 μg/ml, kanamycin (Kan) at 50 μg/ml, nalidixic acid (Nal) at 30 μg/ml, and chloramphenicol (Cm) at 32 μg/ml. Unless otherwise noted, basic reagents and antibiotics were purchased from Sigma-Aldrich, Inc., and VWR International, LLC, respectively.
Plasmid/DNA extraction and construction of recombinant protein vectors.
All plasmids were extracted from E. coli using the QIAprep spin miniprep kit (Qiagen), and E. coli genomic DNA was extracted using a DNeasy blood and tissue kit (Qiagen), according to the manufacturer’s instructions. All primer pairs (Eurofins Genomics) and engineered restriction enzyme sequences (New England BioLabs) are listed in Table 3. Platinum PCR supermix (Invitrogen) was used with preparative PCR to engineer plasmids pMMB207, pCR2.1, pFPV25.1gfpmut3, pUT18, pUT18C, pKT25, and pKNT25. PCR was used to verify plasmid constructs (listed in Table 4), and reaction mixtures included DreamTag green PCR master mix (Thermo Scientific). PCR product identity was confirmed by Sanger sequencing (Eurofins Genomics).
TABLE 4.
Plasmids used in this study
| Plasmid | Relevant genotype and/or descriptiona | Source or reference |
|---|---|---|
| pCR2.1-Topo | Ampr; Topo cloning vector | Invitrogen |
| pCR2.1::pmic-210/0mcpMIADB | Ampr; containing mcpM promoter −210/0 and mcpM, mcpI, mcpA, mcpD, and mcpB | This study |
| pMMB207 | Chlr; RSF1010 derivative; IncQ lacIq Tac oriT | 67 |
| pMMB207::mcpM | Chlr; pMMB207 containing the mcpM gene with a 6×His tag at the C terminus | This study |
| pMMB207::mcpI | Chlr; pMMB207 containing the mcpI gene with a Flag tag at the C terminus | This study |
| pFPV25.1::gfpmut3 | Ampr; vector containing GFPmut3 | 62 |
| pFPV25.1::tdTomato | Ampr; vector containing tdTomato | This study |
| pKD4 | Kanr; containing Kanr cassette for PCR amplification | 68 |
| pKD46 | Ampr | 59 |
| pUT18 | Ampr; cloning vector | Euromedex |
| pUT18C | Ampr; cloning vector | Euromedex |
| pUT18C-Zip | Ampr; pUT18 containing the leucine zipper | Euromedex |
| pUT18::mcpMΔ36 | Ampr; pUT18 containing the mcpMΔ36 gene | This study |
| pUT18::mcpI | Ampr; pUT18 containing the mcpI gene | This study |
| pUT18::ompF | Ampr; pUT18 containing the ompF gene | This study |
| pUT18C::mcpMΔ36 | Ampr; pUT18C containing the mcpMΔ36 gene | This study |
| pUT18C::mcpI | Ampr; pUT18C containing the mcpI gene | This study |
| pUT18C::ompF | Ampr; pUT18C containing the ompF gene | This study |
| pKT25 | Kanr; cloning vector | Euromedex |
| pKT25-Zip | Kanr; pKT25 containing the leucine zipper | This study |
| pKT25::mcpMΔ36 | Kanr; pKT25 containing the mcpMΔ36 gene | This study |
| pKT25::mcpI | Kanr; pKT25 containing the mcpI gene | This study |
| pKT25::ompF | Kanr; pKT25 containing the ompF gene | This study |
| pKNT25 | Kanr; cloning vector | Euromedex |
| pKNT25::mcpMΔ36 | Kanr; pKNT25 containing the mcpMΔ36 gene | This study |
| pKNT25::mcpI | Kanr; pKNT25 containing the mcpI gene | This study |
| pKNT25::ompF | Kanr; pKNT25 containing the ompF gene | This study |
Ampr, ampicillin resistant; Chlr, chloramphenicol resistant; Kanr, kanamycin resistant.
Mutant construction.
Methods described previously by Datsenko and Wanner were used to generate gene deletions (59). Briefly, primers (Table 3) were designed to incorporate a 36- to 50-nucleotide DNA sequence that complemented the region flanking the gene of interest. PCR with these primers generated a PCR product that joined the flanking sequences to a kanamycin resistance (Kanr) gene, originally from plasmid pKD4 (Table 4). A QIAquick PCR column purification kit (Qiagen) was used to purify the PCR products, followed by DpnI restriction enzyme digestion (New England Biolabs, Inc.) for 4 h at 37°C to digest the DNA template (pKD4) before repeating the column purification step. The treated PCR product (150 ng) was then electroporated into E. coli strain 25 with a Gene Pulser Xcell instrument (Bio-Rad Laboratories, Inc.), using settings (1.8 kV, 25 μF, 200 Ω, and 1-mm-gap cuvette) described previously (24). In preparation for this procedure, E. coli strain 25 carrying the λ Red plasmid pKD46 (Ampr) was grown to an optical density at 600 nm (OD600) of ∼0.6 in super optimal broth (SOB) medium (Fisher Scientific). Bacterial cells were then washed twice in ice-cold water and once in 10% glycerol before 50 μl of 10% glycerol was added to resuspend the cells for electroporation. Immediately after electroporation, 0.5 ml of SOB with catabolite repression (SOC) recovery medium (Fisher Scientific) was added to cells for recovery (2 h at 30°C with aeration) before plating on LB agar with kanamycin (LB-Kan) and incubation overnight at 30°C in a stationary incubator. PCR was used to verify mcpM and mcpI gene deletions (Table 3).
Coculture competition assay.
Due to difficulties associated with purification of MccPDI, as an indirect method to examine the inhibitory effect of MccPDI, coculture competition assays (in vitro method) were performed using a modified protocol (7, 24, 60). Briefly, bacterial strains were grown individually overnight in LB broth. The following day, cultures grown overnight (50 μl) were transferred into three fresh tubes containing 5 ml M9 medium (E. coli 25 or E. coli BW25113 alone or 50 μl of both strains). M9 cultures were incubated for 24 h at 37°C with shaking. When appropriate, antibiotics and/or 0.5 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG; Fisher Scientific) was added. CFU were enumerated by first preparing serial dilutions (10-fold; 6 replicates each) in a 96-well plate containing sterile 1× phosphate-buffered saline (PBS). After mixing, 5 μl of each dilution was transferred onto an LB agar plate (with or without antibiotics). Plates were incubated at 37°C overnight before CFU were determined according to the methods of Chen et al. (60).
Bacteriocin typing for E. coli strain MAD 96.
E. coli strains 25 and MAD 96 were picked from fresh colonies into 5 ml LB broth and inoculated at 37°C with shaking at 200 rpm. The culture grown overnight (1 μl) was used to PCR amplify different bacteriocin gene candidates using primer sets that were designed using SnapGene software (GSL Biotech) (Table 3).
McpM protein expression and purification.
To generate recombinant McpM (with a C-terminal 6×His tag) for various experiments, 2 liters of a 0.5 mM IPTG (Fisher Scientific)-induced E. coli strain 25 ΔmcpM/pMMB207::mcpM culture grown overnight was pelleted by centrifugation at 4,000 × g at 4°C for 30 min. The pellet was resuspended in 15 ml of PBS (50 mM sodium phosphate, 500 mM NaCl [pH 8.0]). A protease inhibitor cocktail (cOmplete ultra tablets, mini, EDTA free; Roche) was added to the resuspended pellet, and the mixture was sonicated (S-450 Ultrasonics sonifier; Branson) for 6 min with 20-s on/off pulses. Ice was added to the water bath surrounding the tube to avoid sample overheating. After sonication, the tube was centrifuged at 30,000 × g (4°C for 30 min). The supernatant containing soluble McpM was then purified by using high-performance liquid chromatography (HPLC) with nickel-Sepharose Excel resin (GE Healthcare). HPLC conditions were as follows. Two milliliters of resin was packed into a tricorn 5/10 column (GE Healthcare) and attached to an Äkta Avant 25 chromatography system (GE Healthcare). The column was equilibrated at 2 ml/min for 10 column volumes (CV) with PBS (pH 7.6), followed by sample loading using a sample pump at 0.5 ml/min. Washout of the unbound sample was performed at 2 ml/min for 15 CV with PBS (pH 7.6) containing 20 mM imidazole. Sample elution was isocratic for 5 CV at 2 ml/min using PBS (pH 7.6) with 125 mM imidazole. McpM elution was monitored by reading the absorbance at 280 nm, and the peak containing McpM was collected in 500-μl fractions (8°C). Pooled McpM fractions were then desalted into 1× PBS (to remove imidazole) using a HiTrap desalting column (GE Healthcare), according to the manufacturer’s instructions. Desalted McpM was collected by peak fractionation (UV at 280 nm) in 500-μl fractions at 8°C. All data were collected and analyzed using Unicorn chromatography management system version 7.0 (GE Healthcare) (see Fig. S6 in the supplemental material).
High-copy-number McpM expression vector.
A second McpM expression vector was constructed to include the indigenous PDI promoter (−210/0, upstream of the mcpM gene) and the entire MccPDI operon (mcpM, mcpI, mcpA, mcpB, and mcpD), using the primers listed in Table 3. The vector construct (pCR2.1-Topo backbone; Invitrogen) is a high-copy-number plasmid that permits synthesis of more McpM than with pMMB207::mcpM (low-copy-number vector), without the need for induction. The vector construct methods were used as described above to produce pCR2.1::pmic-210/0mcpMIADB, which was subsequently transferred into E. coli strain DH10B by electroporation (see “Spot plating assay,” below).
McpI protein expression.
To generate recombinant McpI, pMMB207::mcpI (with a C-terminal Flag tag) was constructed using the vector construct methods described above. The engineered plasmid was transformed into E. coli BW25113 (Table 4). A fresh BW25113/pMMB207::mcpI colony was inoculated overnight into 1 liter LB broth (0.5 mM IPTG and the appropriate antibiotic) and pelleted by centrifugation at 4,000 × g (4°C for 30 min). Cell pellets were subjected to a cross-linking experiment and Western blot analysis (see below).
Spot plating assay.
Spot plating was developed as a secondary (and more rapid) protocol for detecting MccPDI-susceptible bacteria. Cultures (5 ml) of E. coli and Shigella strains grown overnight were diluted to an OD600 of ∼0.8 with LB broth. Susceptible bacterial strains (e.g., E. coli BW25113) were spread onto an M9 agar or LB agar plate using a sterile cotton swab. Purified McpM protein (5 μl at 40 μg/ml) or a culture (OD600 of ∼0.8) of DH10B/pCR2.1::pmic-210/0mcpMIADB or the microcin PDI producer E. coli 25 (positive control) in PBS or M9 medium (negative control) was spotted onto the agar plate and air dried before being incubated at 37°C overnight (Table 1). Strains that exhibited zones of clearance associated with recombinant McpM or with the MccPDI-producing strain were considered positive for susceptibility to MccPDI.
Western blot analysis.
To detect His-tagged and Flag-tagged recombinant proteins (McpM and McpI, respectively), total bacterial proteins were collected from a 10-ml culture grown overnight (with 0.5 mM IPTG) by centrifugation at 18,000 × g (4°C for 5 min). Cell pellets were resuspended in 1× Laemmli sample buffer (Bio-Rad Laboratories, Inc.) and boiled for 10 min. Any kD Tris-glycine precast gels (Bio-Rad) were used for sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) protein separation. A Trans-Blot turbo transfer starter system (Bio-Rad) was used for protein transfer onto a low-fluorescence polyvinylidene difluoride (PVDF) membrane (Bio-Rad Laboratories, Inc.). Ponceau S staining was used to verify protein transfer prior to the addition of antibodies for specific protein detection. Anti-His tag or anti-Flag tag primary antibody (1:1,000) (Thermo Scientific) was used with secondary goat anti-mouse antibody (1:5,000) (DyLight 650, conjugate). A ChemiDoc MP imaging system (Bio-Rad) was used to detect fluorescent signals.
Osmoprotectant assay.
To determine if pores were present in the bacterial membrane, we added 15% (wt/vol) polyethylene glycols (PEGs) (200 to 8,000 Da) to coculture competition assay mixtures for 24 h (26, 61). Competition assays and enumeration methods were used as described above.
DAPI staining of fluorescently labeled E. coli.
To “tag” bacteria for imaging, we first constructed a reporter plasmid, pFPV25.1::tdTomato, by replacing gfpmut3 in pFPV25.1::gfpmut3 (62) with the tdTomato gene (62) using primers tdtomato_XbaI and tdtomato_SphI (Table 3). Standard cloning procedures were used, and sequencing was used to verify results. E. coli strain 25 ΔtraM and E. coli strain 25 ΔmcpM ΔmcpI (Table 1) were transformed with pFPV25.1::gfpmut3 expressing green fluorescent protein (GFP), while target strain BW25113 was transformed with pFPV251::tdTomato expressing red fluorescent protein (tomato red). E. coli 25 ΔtraM (conjugation negative) was chosen to prevent undesirable conjugation between MccPDI producer and target strains. Competition assays (described above) were conducted with the fluorescently labeled cells, and individual monocultures were run as controls. After 2-h and 6-h incubations, 1-ml samples were taken from each of the cultures, and 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) stain (Thermo Scientific) was added at a final concentration of 0.25 μg/ml for 10 min at room temperature. The stained bacteria were pelleted by centrifugation for 1 min at 12,000 × g and resuspended in the same volume of PBS. Cells were then immobilized onto poly-l-lysine-coated glass slides (Sigma) for 20 min and covered with glass coverslips. Cells were observed and images were captured by using an inverted fluorescence microscope (Evos; Advanced Microscopy Group).
ATP depletion assay.
Extracellular ATP was quantified by using a BacTiter-Glo microbial cell viability assay (Promega). Fresh bacterial colonies of E. coli ΔmcpM ΔmcpI with empty control vector pMMB207 or with complementation with mcpM (pMMB207::mcpM) were transferred to medium and grown at 37°C overnight with appropriate antibiotics (Table 1). Cultures grown overnight (10 μl) were pipetted into fresh M9 medium (90 μl) in a 96-well microplate with appropriate antibiotics and induced with 0.5 mM IPTG as needed (0, 2, 4, 6, 8, 10, and 12 h). According to the manufacturer’s protocol, reagents (BacTiter-Glo buffer and substrate) provided in the kit were then combined and added to each well of inoculant (100 μl reagent to 100 μl culture), and samples were incubated for 5 min at room temperature before luminescence was recorded with an Infiniti M1000 Pro microplate reader (Tecan Systems). A luminescence signal was generated by recombinant luciferase through the detection of ATP. Each assay was repeated for three independent replicates with E. coli ΔmcpM ΔmcpI/pMMB207 and E. coli ΔmcpM ΔmcpI/pMMB207::mcpM (without 0.5 mM IPTG), which served as a negative control. CFU were enumerated at each time point using the method described above.
Paraformaldehyde cross-linking.
We used a cross-linking experiment to determine if the MccPDI self-immunity protein, McpI, forms a multimeric structure. Formaldehyde is the smallest aldehyde (electrophilic molecule) that forms covalent bonds between two different macromolecules (63). Due to its small molecular size, formaldehyde cross-links macromolecules 2.3 to 2.7 Å apart (63, 64), which makes it particularly suitable to study the interactions between proteins in spatial proximity. Furthermore, the ability of PFA to permeate intact cells and cellular membranes easily makes it ideal for this experiment (64). A 5-ml culture containing E. coli BW25113/pMMB207::mcpI was induced overnight with 0.5 mM IPTG (Table 1). A modified protocol of a method previously described by Klockenbusch and Kast (64) was used to cross-link Flag-tagged McpI for analysis by Western blotting. Briefly, a 500-μl culture grown overnight was centrifuged at 18,000 × g for 1 min at room temperature. The resulting cell pellet was washed twice with 100 μl sterile PBS (pH 7.0), and prewarmed (37°C) 2% (wt/vol) PFA was then used to resuspend the pellet. Fixation with PFA proceeded for 30 min at room temperature. PFA-treated cells were recentrifuged at 18,000 × g for 1 min and washed once with 0.5 ml ice-cold 1.25 M glycine in PBS. After another recentrifugation step (18,000 × g for 1 min), the supernatant was discarded. Loading buffer (1× Laemmli buffer; Bio-Rad) was added to the cross-linked samples with or without βME. To reverse cross-linkages, samples were boiled for 20 min. To maintain partial cross-linkage, samples were heated for 10 min at 60°C before loading onto an SDS-PAGE gel for Western blot analysis.
Bacterial two-hybrid assays.
A bacterial two-hybrid (BACTH) system (Euromedex) was used to identify potential protein-protein interactions between different MccPDI proteins (5) and the OmpF ligand from susceptible bacteria (8). Fusion proteins were first constructed for McpI, McpMΔ36 (mature McpM), or OmpF with either the T18 or T25 subunit of adenylate cyclase from Bordetella pertussis (29). According to standard molecular cloning techniques, primers (Table 3) and DNA templates (mcpI, mcpMΔ36, and ompF) were used to prepare PCR products for cloning. Restriction digestion and ligation (T4 ligate; New England Biolabs) were performed to insert PCR amplicons into plasmids pUT18 and pKNT25 (N-terminal T18/T25 adenylate cyclase subunits) or into pUT18C and pKT25 (C-terminal T18/T25). T18 and T25 vector constructs were individually transformed into E. coli XL1-Blue (Agilent) for plasmid propagation, and vector constructs were confirmed by Sanger sequencing (Eurofins Genomics). Plasmid vectors were cotransformed (e.g., a T18 and a T25 construct) into E. coli strain DHM1 (Table 1). The transformed bacteria were inoculated into LB broth with appropriate antibiotics at 30°C for 1 h with aeration before being plated onto LB agar that contained appropriate antibiotics, 50 mM 5-bromo-4-chloro-3-indolyl-d-galactopyranoside (X-gal) (Fisher Scientific), and 0.5 mM IPTG (Fisher Scientific) for blue and white screening. Plates were incubated for 1 to 4 days at 30°C in a stationary incubator.
Beta-galactosidase assays.
We used a beta-galactosidase assay described previously by Karimova et al. (30) to verify potential protein-protein interactions that were identified from the bacterial two-hybrid assay. Briefly, a freshly grown colony was inoculated into 5 ml LB broth with 0.5 mM IPTG and incubated overnight on a shaker table at 37°C with appropriate antibiotics. The inoculant grown overnight (1 ml) was added to 5 ml M63 medium [15.1 mM (NH4)2SO4, 100.0 mM KH2PO4, 1.8 μM FeSO4·7H2O, 3.0 μM vitamin B1 (pH 7.0) with KOH, and 0.4% (wt/vol) maltose], and the resulting OD600 was recorded. The diluted culture (1 ml) was transferred into a new glass 5-ml tube, to which toluene (30 μl) and 30 μl of 0.1% (wt/vol) SDS were added to permeabilize the bacteria, and this mixture was incubated for 30 min at 37°C with vigorous agitation. PM2 assay buffer (0.5 ml) (70 mM Na2HPO4·12H2O, 30 mM NaH2PO4·H2O, 1 mM MgSO4, 0.2 mM MnSO4 at pH 7.0, and 100 mM βME) was added, and the solution was incubated at 28°C for 5 min before the addition of 0.25 ml o-nitrophenol-β-d-galactopyranoside (ONPG). An aliquot of this mixture (100 μl) was transferred to a single well of a 96-well microplate and incubated at 30°C until visible yellow color developed. The reaction was terminated with the addition of 50 μl stop solution (1 M Na2CO3). Data were recorded with a microplate reader at an OD420 (SpectraMax; Molecular Devices, LLC). The enzymatic activity, A (in units per milliliter), was calculated according to the following equation: A = 200 × [(OD420 − OD420 control tube)/total incubation time in minutes] × dilution factor. A level of β-galactosidase activity at least 5-fold higher than that measured for a negative control, DHM1(pKT25/pUT18C) cells, was considered positive for a protein-protein interaction (30).
Statistical analysis.
Where appropriate, experimental results were compared by using one-way analysis of variance (ANOVA) with Dunnett’s one-way multiple-pairwise-comparison test (SigmaPlot version 12.5; Systat Software, Inc., San Jose, CA).
Accession number(s).
Newly determined sequence data for ompF (strain AR0349) were deposited in GenBank under accession number MH665273.
Supplementary Material
ACKNOWLEDGMENTS
L. Knodler provided the pFPV25.1::gfpmut3 vector. We thank M. Laviña for the PAP222 MccV strain. We thank L. Orfe, L. Jones, C. Deobald, and L. Deobald for technical assistance. We kindly thank the CDC for E. coli AR0346 and AR0349 strains and the Washington State University Veterinary Teaching Hospital for clinical canine E. coli isolates.
This project was supported in part by USDA NIFA grant 2010-04487 and by the Agricultural Animal Health Program, Washington Agricultural Research Center, and the Paul G. Allen School for Global Animal Health at Washington State University.
The antibacterial activities of MccPDI are described under U.S. patent no. 9,492,500, for which D.R.C. is an author. The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00371-19.
REFERENCES
- 1.Rebuffat S. 2013. Microcins, p 129–137. In Kastin AJ. (ed), Handbook of biologically active peptides, 2nd ed Elsevier/AP, Amsterdam, Netherlands. [Google Scholar]
- 2.Rebuffat S. 2012. Microcins in action: amazing defence strategies of enterobacteria. Biochem Soc Trans 40:1456–1462. doi: 10.1042/BST20120183. [DOI] [PubMed] [Google Scholar]
- 3.Yang SC, Lin CH, Sung CT, Fang JY. 2014. Antibacterial activities of bacteriocins: application in foods and pharmaceuticals. Front Microbiol 5:241. doi: 10.3389/fmicb.2014.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drider D, Rebuffat S (ed). 2011. Prokaryotic antimicrobial peptides: from genes to applications. Springer-Verlag, New York, NY. [Google Scholar]
- 5.Eberhart LJ, Deringer JR, Brayton KA, Sawant AA, Besser TE, Call DR. 2012. Characterization of a novel microcin that kills enterohemorrhagic Escherichia coli O157:H7 and O26. Appl Environ Microbiol 78:6592–6599. doi: 10.1128/AEM.01067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawant AA, Casavant NC, Call DR, Besser TE. 2011. Proximity-dependent inhibition in Escherichia coli isolates from cattle. Appl Environ Microbiol 77:2345–2351. doi: 10.1128/AEM.03150-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Z, Orfe LH, Liu J, Lu SY, Besser TE, Call DR. 2017. Microcin PDI regulation and proteolytic cleavage are unique among known microcins. Sci Rep 7:42529. doi: 10.1038/srep42529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Z, Eberhart LJ, Orfe LH, Lu SY, Besser TE, Call DR. 2015. Genome-wide screening identifies six genes that are associated with susceptibility to Escherichia coli microcin PDI. Appl Environ Microbiol 81:6953–6963. doi: 10.1128/AEM.01704-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sable S, Pons AM, Gendron-Gaillard S, Cottenceau G. 2000. Antibacterial activity evaluation of microcin J25 against diarrheagenic Escherichia coli. Appl Environ Microbiol 66:4595–4597. doi: 10.1128/AEM.66.10.4595-4597.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillor O, Kirkup BC, Riley MA. 2004. Colicins and microcins: the next generation antimicrobials. Adv Appl Microbiol 54:129–146. doi: 10.1016/S0065-2164(04)54005-4. [DOI] [PubMed] [Google Scholar]
- 11.Tang KL, Caffrey NP, Nobrega DB, Cork SC, Ronksley PE, Barkema HW, Polachek AJ, Ganshorn H, Sharma N, Kellner JD, Ghali WA. 2017. Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: a systematic review and meta-analysis. Lancet Planet Health 1:e316–e327. doi: 10.1016/S2542-5196(17)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 13.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.091101p1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karp BE, Tate H, Plumblee JR, Dessai U, Whichard JM, Thacker EL, Hale KR, Wilson W, Friedman CR, Griffin PM, McDermott PF. 2017. National antimicrobial resistance monitoring system: two decades of advancing public health through integrated surveillance of antimicrobial resistance. Foodborne Pathog Dis 14:545–557. doi: 10.1089/fpd.2017.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woodworth KR, Walters MS, Weiner LM, Edwards J, Brown AC, Huang JY, Malik S, Slayton RB, Paul P, Capers C, Kainer MA, Wilde N, Shugart A, Mahon G, Kallen AJ, Patel J, McDonald LC, Srinivasan A, Craig M, Cardo DM. 2018. Vital signs: containment of novel multidrug-resistant organisms and resistance mechanisms—United States, 2006-2017. MMWR Morb Mortal Wkly Rep 67:396–401. doi: 10.15585/mmwr.mm6713e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wernli D, Jorgensen PS, Harbarth S, Carroll SP, Laxminarayan R, Levrat N, Rottingen JA, Pittet D. 2017. Antimicrobial resistance: the complex challenge of measurement to inform policy and the public. PLoS Med 14:e1002378. doi: 10.1371/journal.pmed.1002378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Phan MD, Gomes Moriel D, Peters KM, Davies M, Rogers BA, Dougan G, Rodriguez-Bano J, Pascual A, Pitout JD, Upton M, Paterson DL, Walsh TR, Schembri MA, Beatson SA. 2014. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A 111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehrad B, Clark NM, Zhanel GG, Lynch JP III.. 2015. Antimicrobial resistance in hospital-acquired Gram-negative bacterial infections. Chest 147:1413–1421. doi: 10.1378/chest.14-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sidjabat HE, Paterson DL. 2015. Multidrug-resistant Escherichia coli in Asia: epidemiology and management. Expert Rev Anti Infect Ther 13:575–591. doi: 10.1586/14787210.2015.1028365. [DOI] [PubMed] [Google Scholar]
- 20.Hasan B, Olsen B, Alam A, Akter L, Melhus A. 2015. Dissemination of the multidrug-resistant extended-spectrum beta-lactamase-producing Escherichia coli O25b-ST131 clone and the role of house crow (Corvus splendens) foraging on hospital waste in Bangladesh. Clin Microbiol Infect 21:1000.e1–1000.e4. doi: 10.1016/j.cmi.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 21.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis 51:286–294. doi: 10.1086/653932. [DOI] [PubMed] [Google Scholar]
- 22.Hammami R, Zouhir A, Le Lay C, Ben Hamida J, Fliss I. 2010. BACTIBASE second release: a database and tool platform for bacteriocin characterization. BMC Microbiol 10:22. doi: 10.1186/1471-2180-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duquesne S, Destoumieux-Garzon D, Peduzzi J, Rebuffat S. 2007. Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat Prod Rep 24:708–734. doi: 10.1039/b516237h. [DOI] [PubMed] [Google Scholar]
- 24.Lu SY, Zhao Z, Avillan JJ, Liu J, Call DR. 2017. Autoinducer-2 quorum sensing contributes to regulation of microcin PDI in Escherichia coli. Front Microbiol 8:2570. doi: 10.3389/fmicb.2017.02570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cowan SW, Garavito RM, Jansonius JN, Jenkins JA, Karlsson R, Konig N, Pai EF, Pauptit RA, Rizkallah PJ, Rosenbusch JP, Rummel G, Schirmer T. 1995. The structure of OmpF porin in a tetragonal crystal form. Structure 3:1041–1050. doi: 10.1016/S0969-2126(01)00240-4. [DOI] [PubMed] [Google Scholar]
- 26.Smith PT, Huang ML, Kirshenbaum K. 2015. Osmoprotective polymer additives attenuate the membrane pore-forming activity of antimicrobial peptoids. Biopolymers 103:227–236. doi: 10.1002/bip.22588. [DOI] [PubMed] [Google Scholar]
- 27.Zink D, Sadoni N, Stelzer E. 2003. Visualizing chromatin and chromosomes in living cells. Methods 29:42–50. doi: 10.1016/S1046-2023(02)00289-X. [DOI] [PubMed] [Google Scholar]
- 28.Pratt LA, Hsing W, Gibson KE, Silhavy TJ. 1996. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol Microbiol 20:911–917. doi: 10.1111/j.1365-2958.1996.tb02532.x. [DOI] [PubMed] [Google Scholar]
- 29.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karimova G, Dautin N, Ladant D. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol 187:2233–2243. doi: 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stamm WE, Norrby SR. 2001. Urinary tract infections: disease panorama and challenges. J Infect Dis 183:S1–S4. doi: 10.1086/318850. [DOI] [PubMed] [Google Scholar]
- 32.Schappert SM, Rechtsteiner EA. 2011. Ambulatory medical care utilization estimates for 2007. Vital Health Stat 13:1–38. [PubMed] [Google Scholar]
- 33.Foxman B. 2014. Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin North Am 28:1–13. doi: 10.1016/j.idc.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 34.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. 2015. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 13:269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Francino MP. 2015. Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front Microbiol 6:1543. doi: 10.3389/fmicb.2015.01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dethlefsen L, Relman DA. 2011. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A 108:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kotloff KL, Riddle MS, Platts-Mills JA, Pavlinac P, Zaidi AKM. 2018. Shigellosis. Lancet 391:801–812. doi: 10.1016/S0140-6736(17)33296-8. [DOI] [PubMed] [Google Scholar]
- 38.Yamashita E, Zhalnina MV, Zakharov SD, Sharma O, Cramer WA. 2008. Crystal structures of the OmpF porin: function in a colicin translocon. EMBO J 27:2171–2180. doi: 10.1038/emboj.2008.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jap BK, Walian PJ. 1996. Structure and functional mechanism of porins. Physiol Rev 76:1073–1088. doi: 10.1152/physrev.1996.76.4.1073. [DOI] [PubMed] [Google Scholar]
- 40.Smajs D, Micenkova L, Smarda J, Vrba M, Sevcikova A, Valisova Z, Woznicova V. 2010. Bacteriocin synthesis in uropathogenic and commensal Escherichia coli: colicin E1 is a potential virulence factor. BMC Microbiol 10:288. doi: 10.1186/1471-2180-10-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abraham S, Chapman TA, Zhang R, Chin J, Mabbett AN, Totsika M, Schembri MA. 2012. Molecular characterization of Escherichia coli strains that cause symptomatic and asymptomatic urinary tract infections. J Clin Microbiol 50:1027–1030. doi: 10.1128/JCM.06671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai SJ, Inouye M. 2002. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J Biol Chem 277:24155–24161. doi: 10.1074/jbc.M110715200. [DOI] [PubMed] [Google Scholar]
- 43.Qin L, Yoshida T, Inouye M. 2001. The critical role of DNA in the equilibrium between OmpR and phosphorylated OmpR mediated by EnvZ in Escherichia coli. Proc Natl Acad Sci U S A 98:908–913. doi: 10.1073/pnas.031383098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forst S, Delgado J, Inouye M. 1989. Phosphorylation of OmpR by the osmosensor EnvZ modulates expression of the ompF and ompC genes in Escherichia coli. Proc Natl Acad Sci U S A 86:6052–6056. doi: 10.1073/pnas.86.16.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inaba K, Ito K. 2008. Structure and mechanisms of the DsbB-DsbA disulfide bond generation machine. Biochim Biophys Acta 1783:520–529. doi: 10.1016/j.bbamcr.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 46.Cingolani G, Duncan TM. 2011. Structure of the ATP synthase catalytic complex (F(1)) from Escherichia coli in an autoinhibited conformation. Nat Struct Mol Biol 18:701–707. doi: 10.1038/nsmb.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Capaldi RA, Schulenberg B, Murray J, Aggeler R. 2000. Cross-linking and electron microscopy studies of the structure and functioning of the Escherichia coli ATP synthase. J Exp Biol 203:29–33. [DOI] [PubMed] [Google Scholar]
- 48.Bieler S, Silva F, Soto C, Belin D. 2006. Bactericidal activity of both secreted and nonsecreted microcin E492 requires the mannose permease. J Bacteriol 188:7049–7061. doi: 10.1128/JB.00688-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lagos R, Tello M, Mercado G, Garcia V, Monasterio O. 2009. Antibacterial and antitumorigenic properties of microcin E492, a pore-forming bacteriocin. Curr Pharm Biotechnol 10:74–85. doi: 10.2174/138920109787048643. [DOI] [PubMed] [Google Scholar]
- 50.Gerard F, Pradel N, Wu LF. 2005. Bactericidal activity of colicin V is mediated by an inner membrane protein, SdaC, of Escherichia coli. J Bacteriol 187:1945–1950. doi: 10.1128/JB.187.6.1945-1950.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scherrer R, Gerhardt P. 1971. Molecular sieving by the Bacillus megaterium cell wall and protoplast. J Bacteriol 107:718–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Severinov K, Semenova E, Kazakov A, Kazakov T, Gelfand MS. 2007. Low-molecular-weight post-translationally modified microcins. Mol Microbiol 65:1380–1394. doi: 10.1111/j.1365-2958.2007.05874.x. [DOI] [PubMed] [Google Scholar]
- 53.Bobrov AG, Kirillina O, Forman S, Mack D, Perry RD. 2008. Insights into Yersinia pestis biofilm development: topology and co-interaction of Hms inner membrane proteins involved in exopolysaccharide production. Environ Microbiol 10:1419–1432. doi: 10.1111/j.1462-2920.2007.01554.x. [DOI] [PubMed] [Google Scholar]
- 54.Mistry J, Bateman A, Finn RD. 2007. Predicting active site residue annotations in the Pfam database. BMC Bioinformatics 8:298. doi: 10.1186/1471-2105-8-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kallberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, Xu J. 2012. Template-based protein structure modeling using the RaptorX Web server. Nat Protoc 7:1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 58.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 Web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen CY, Nace GW, Irwin PL. 2003. A 6 × 6 drop plate method for simultaneous colony counting and MPN enumeration of Campylobacter jejuni, Listeria monocytogenes, and Escherichia coli. J Microbiol Methods 55:475–479. doi: 10.1016/S0167-7012(03)00194-5. [DOI] [PubMed] [Google Scholar]
- 61.Cox CS. 1966. Bacterial survival in suspension in polyethylene glycol solutions. J Gen Microbiol 45:275–281. doi: 10.1099/00221287-45-2-275. [DOI] [PubMed] [Google Scholar]
- 62.Valdivia RH, Falkow S. 1996. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol Microbiol 22:367–378. doi: 10.1046/j.1365-2958.1996.00120.x. [DOI] [PubMed] [Google Scholar]
- 63.Hoffman EA, Frey BL, Smith LM, Auble DT. 2015. Formaldehyde crosslinking: a tool for the study of chromatin complexes. J Biol Chem 290:26404–26411. doi: 10.1074/jbc.R115.651679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klockenbusch C, Kast J. 2010. Optimization of formaldehyde cross-linking for protein interaction analysis of non-tagged integrin beta1. J Biomed Biotechnol 2010:927585. doi: 10.1155/2010/927585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pugsley AP. 1985. Escherichia coli K12 strains for use in the identification and characterization of colicins. J Gen Microbiol 131:369–376. doi: 10.1099/00221287-131-2-369. [DOI] [PubMed] [Google Scholar]
- 67.Morales VM, Backman A, Bagdasarian M. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39–47. doi: 10.1016/0378-1119(91)90007-X. [DOI] [PubMed] [Google Scholar]
- 68.Milton DL, O’Toole R, Horstedt P, Wolf-Watz H. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol 178:1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.