Acinetobacter baumannii is a high-priority pathogen for which research on mechanisms of resistance and virulence is a critical need. Commonly used antibiotic selection markers are not suitable for use in MDR and XDR isolates of A. baumannii due to the high antibiotic resistance of these isolates, which poses a barrier to the study of this pathogen. This study demonstrates the practical potential of using apramycin and zeocin mini-Tn7- and Flp recombinase-encoded constructs to carry out genomic manipulations in clinical isolates of A. baumannii displaying MDR and XDR phenotypes.
KEYWORDS: single copy, apramycin, cloning, gene expression, zeocin
ABSTRACT
The purpose of this study was to create single-copy gene expression systems for use in genomic manipulations of multidrug-resistant (MDR) and extensively drug-resistant (XDR) clinical isolates of Acinetobacter baumannii. In this study, mini-Tn7 vectors with zeocin and apramycin selection markers were created by cloning the ble and aac(3)-IV genes, respectively, enabling either inducible gene expression (pUC18T-mini-Tn7T-Zeo-LAC and pUC18T-mini-Tn7T-Apr-LAC) or expression from native or constitutive promoters (pUC18T-mini-Tn7T-Zeo and pUC18T-mini-Tn7T-Apr). The selection markers of these plasmids are contained within a Flp recombinase target (FRT) cassette, which can be used to obtain unmarked mini-Tn7 insertions upon introduction of a source of Flp recombinase. To this end, site-specific excision vectors pFLP2A and pFLP2Z (containing apramycin and zeocin selection markers, respectively) were created in this study as an accessory to the mini-Tn7 vectors described above. Combinations of these novel mini-Tn7 plasmids and their compatible pFLP2Z or pFLP2A accessory plasmid were used to generate unmarked insertions in MDR clinical isolates of A. baumannii. In addition, several fluorescent markers were cloned and inserted into MDR and XDR clinical isolates of A. baumannii via these apramycin and zeocin mini-Tn7 constructs to demonstrate their application.
IMPORTANCE Acinetobacter baumannii is a high-priority pathogen for which research on mechanisms of resistance and virulence is a critical need. Commonly used antibiotic selection markers are not suitable for use in MDR and XDR isolates of A. baumannii due to the high antibiotic resistance of these isolates, which poses a barrier to the study of this pathogen. This study demonstrates the practical potential of using apramycin and zeocin mini-Tn7- and Flp recombinase-encoded constructs to carry out genomic manipulations in clinical isolates of A. baumannii displaying MDR and XDR phenotypes.
INTRODUCTION
Acinetobacter baumannii is an opportunistic, Gram-negative coccobacillus that has become a serious health concern due to an increasing incidence of multidrug-resistant (MDR) hospital-acquired infections (1). Recently, the WHO listed A. baumannii as a global priority pathogen for which research and development of antibiotics is critically needed (http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/). In order to study the resistance and virulence mechanisms of A. baumannii, availability of effective molecular tools for genetic manipulation in this species is necessary. However, one of the major challenges in studying A. baumannii in the laboratory setting is the scarcity of molecular tools available for genetic manipulation in this important pathogen (2).
Clinical isolates of Acinetobacter present an even greater challenge to researchers as far as genetic manipulations are concerned, as these strains often display decreased susceptibility to most commonly used antibiotics. This makes such antibiotic markers impractical in genetic manipulation procedures. A promising set of molecular tools for use in MDR strains of Acinetobacter are mini-Tn7 vectors, a collection of Tn7 transposon-based plasmids that were first used in other Gram-negative bacteria such as Pseudomonas spp. and Burkholderia spp. (3–6).
Mini-Tn7 vectors can be used for a range of applications, including single-copy gene complementation, molecular tagging, and promoter analysis (7). The major advantages of these vectors include the ability to offer site-specific, single-copy chromosomal insertion at a neutral location within the genome, thus bypassing the need for continued antibiotic selection, and their broad-host-range capabilities (6, 7). With rare exceptions (8), chromosomal insertion of mini-Tn7 occurs downstream of the glmS gene, which encodes glutamine-fructose-6-phosphate aminotransferase, a highly conserved essential enzyme of bacteria (9). Bacteria with multiple glmS genes contain multiple Tn7 insertion (attTn7) sites (3, 5). Unmarked insertions can be obtained by introduction of a site-specific excision vector that encodes a source of Flp recombinase, an enzyme that recognizes the Flp recombinase target (FRT) sites flanking the selectable marker in mini-Tn7 and allows for its excision (7, 10).
A limitation of mini-Tn7 vectors currently available is the lack of antibiotic resistance markers for use in MDR bacteria. Versions of mini-Tn7 vectors shown for use thus far in A. baumannii utilized gentamicin (Gm), tetracycline, and trimethoprim resistance markers for selection (6, 7). Using these selection markers in genetic manipulations of A. baumannii clinical isolates is often impractical, as clinical strains can be highly resistant to these antibiotics (11, 12).
However, antibiotics/antimicrobials that are not widely used clinically can be utilized for selection purposes, since incidences of resistance against them are likely to be very low due to the lack of selection pressure. Zeocin (Zeo) resistance markers have recently been shown to be useful for selection purposes in MDR clinical isolates of A. baumannii (13, 14). Similarly, apramycin (Apr) has also been shown to have promising activity against MDR A. baumannii strains (15). Although apramycin is used in veterinary medicine, both apramycin and zeocin are antibiotics that are not used in human clinical settings (14, 16), and therefore they are less prone to already-acquired resistance due to lack of selective pressure.
The objective of this study was to create a set of mini-Tn7 insertion vectors and Flp recombinase excision vectors containing apramycin resistance (Aprr) and zeocin resistance (Zeor) markers that can be used to carry out genomic manipulations in clinical isolates of A. baumannii displaying MDR and extensively drug-resistant (XDR) phenotypes. The tools developed in this study include mini-Tn7 vectors containing apramycin or zeocin selection markers that allow for both inducible and constitutive expression of target gene, as well as pFLP2A (Aprr) and pFLP2Z (Zeor) site-specific excision vectors that can be used to obtain unmarked insertions. Finally, we also created mini-Tn7 vectors with fluorescent markers to be used in MDR A. baumannii isolates.
RESULTS AND DISCUSSION
Genetic manipulations in bacteria require the use of selection markers. Frequently, selection markers confer resistance to antibiotics that are or have been used in clinical settings. This presents a challenge for work with organisms that display reduced susceptibility to these antibiotics. A. baumannii is one such organism. Clinical isolates of A. baumannii display resistance to a majority of antibiotics, making treatment of infections a challenge. Thus, there is an urgent need to understand the molecular mechanisms of antibiotic resistance of A. baumannii. These efforts frequently require genetic manipulations in clinical isolates of A. baumannii, but paradoxically, it is the antibiotic resistance of clinical isolates that hinders the molecular characterization of antibiotic resistance mechanisms in A. baumannii. One solution to this problem is the use of selection markers that confer resistance to antibiotics that are not used clinically. In this work, we constructed mini-Tn7-based vectors containing Zeo or Apr resistance markers and demonstrated their application in multi- and pan-drug-resistant isolates of A. baumannii. Zeocin is a glycopeptide antibiotic that causes cell death by intercalating into DNA and causing its cleavage (17). Apramycin is an aminoglycoside that inhibits translation by binding the 30S rRNA (16). Due to its widespread toxicity, zeocin is not used therapeutically in humans (17). Apramycin, on the other hand, has been used widely in animals but not as an antibiotic in humans (16).
In this study, three different clinical isolates of A. baumannii were used: AB030, AB031, and LAC-4. Both LAC-4 and AB030 are resistant to gentamicin, while AB031 is susceptible (Table 1). High resistance of LAC-4 (MIC, 64 μg/ml) and AB030 (MIC, ≥1,024 μg/ml) to gentamicin means that the Gmr marker is not ideal in these strains. Conversely, all three strains had relatively low MIC values for Apr and Zeo (Table 1). Therefore, both of these resistance markers are suitable for genetic manipulations in multidrug-resistant isolates of A. baumannii. Using Apr and Zeo resistance markers, we created site-specific mini-Tn7-based gene insertion systems. Further, plasmids that express yeast Flp recombinase were also modified by cloning Apr or Zeo resistance genes, allowing for the creation of unmarked insertions.
TABLE 1.
MICs for A. baumannii strains used in this study
| Strain | MIC (µg/ml)a
|
||
|---|---|---|---|
| Gentamicinb | Apramycin | Zeocin | |
| ATCC 17978 | 1 (S) | 2–4 | 16 |
| AB030 | >1,024 (R) | 16–32 | 128–256 |
| AB031 | 2 (S) | 8 | 16 |
| LAC-4 | 64 (R) | 8 | 4–8 |
R, resistant; S, susceptible.
Gentamicin susceptibility results were interpreted using CSLI guidelines (22).
The utility of single-copy gene expression systems using the mini-Tn7-based plasmids has been recognized in various bacterial species (4–7, 18). We recently showed the utility of mini-Tn7 vectors with an inducible promoter (Ptac) where the expression was controlled by addition of IPTG (isopropyl-β-d-thiogalactopyranoside) (19). The Tn7 tnsABCD genes encode the site-specific transposition pathway and mediate insertion into the chromosome at attTn7 sites (9). The chromosomal sequence recognized by TnsD resides in the 3′-terminal portion of the bacterial glmS gene; however, insertion occurs at the attTn7 insertion site usually located in an intergenic region downstream of glmS (9). The glmS genes appear to be highly conserved in a diversity of bacterial genomes, conferring a broad host range to the Tn7 transposition pathway of mini-Tn7 vectors (4). Although A. baumannii has two glmS genes, only one (glmS2) contains the attTn7-associated sequence (7). Sequence alignment of the attTn7-associated sequence and the Tn7 insertion site of A. baumannii type strain ATCC 17978 and clinical isolates AB030, AB031, and LAC-4 showed 100% conserved sequence homology between the strains (Fig. 1). This represents the versatility of mini-Tn7 vectors for genomic manipulation in A. baumannii clinical isolates.
FIG 1.
Sequence alignment of the attTn7-associated sequence and the Tn7 insertion site of Acinetobacter baumannii ATCC 17978 and clinical isolates used in this study, showing the highly conserved sequence homology between the strains. The insertion site is located 24 bp downstream of the glmS2 gene in a neutral location (as shown by the arrow). The stop codon of the glmS2 gene is outlined in the red box. The consensus sequence is shown at the bottom of the alignment and displays 100% identity.
Once the feasibility of using mini-Tn7 plasmids with Apr or Zeo resistance markers was confirmed, mini-Tn7 plasmids were constructed for use in multidrug-resistant A. baumannii isolates. First, we created the pUC18T-mini-Tn7T-Apr-LAC and pUC18T-mini-Tn7T-Zeo-LAC plasmids, which contain the IPTG-inducible Escherichia coli trp-lac hybrid promoter Ptac and the lacIq gene (Fig. 2). These new mini-Tn7 plasmids share the following relevant characteristics: (i) an origin of transfer for conjugation (oriT), (ii) a high-copy-number ColE1 origin of replication for maintenance in E. coli but not in A. baumannii, (iii) an ampicillin resistance gene for E. coli selection (bla), and (iv) the following elements within the Tn7 transposon left and right ends (Tn7L and Tn7R): a multiple-cloning site (MCS) for cloning of sequences under the control of the tac promoter, a lac operator, the lac repressor (lacI) gene under the control of the lacIq promoter, and an FRT cassette flanking Apr or Zeo resistance markers. The utility of mini-Tn7 vectors containing inducible promoters has previously been shown in various organisms (5, 20). Such plasmids are particularly useful for genetic complementation studies, as they allow regulated expression of genes in single copy and thus avoidance of undesirable “multicopy effects” that may result from plasmid-based gene expression.
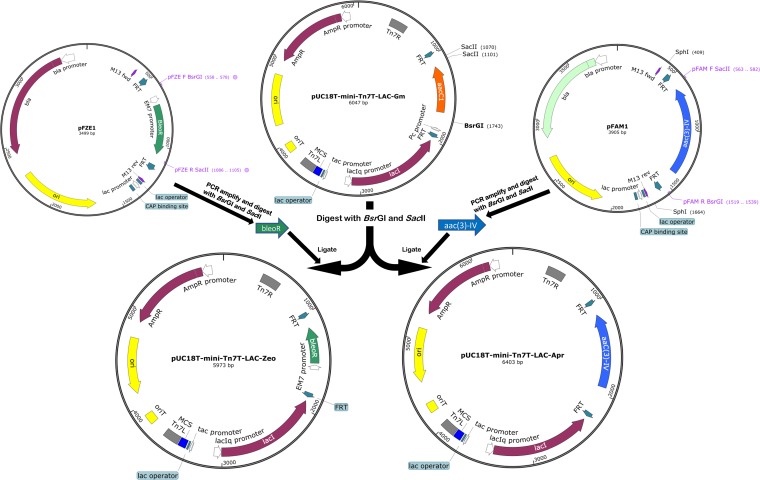
FIG 2.
Construction of the pUC18T-mini-Tn7T-Apr-LAC and pUC18T-mini-Tn7T-Zeo-LAC plasmids. The plasmids were created by replacing the aacC1 gentamicin resistance gene of pUC18T-mini-Tn7T-Gm-LAC with apramycin and zeocin resistance genes, respectively. Plasmid sequences were confirmed using Illumina Hi-Seq next-generation sequencing (MGH CCIB DNA Core Facility, Cambridge, MA, USA). Maps were constructed using SnapGene software (GSL Biotech).
Two other versions of mini-Tn7 plasmids that lack the inducible Ptac promoters were created. These two plasmids, pUC18T-mini-Tn7T-Apr and pUC18T-mini-Tn7T-Zeo, allow for the cloning of the genes with their native promoters. Alternatively, these can also be used for tagging of A. baumannii isolates using fluorescent markers, as we show below.
One of the salient features of mini-Tn7-based systems is the unmarked insertion of the plasmid in the chromosome (6). Unmarked insertions are often desired to avoid complicating the study of mutant strains with an antibiotic resistance marker, and they can be obtained by introducing a source of Flp recombinase into the mini-Tn7 insertion mutant. This allows for excision of the resistance marker through FRT site-specific recognition. The site-specific excision vector pFLP2 created for such a purpose in manipulations of Pseudomonas aeruginosa (10) was modified for A. baumannii by insertion of the origin of replication from the plasmid pWH1266 (21). The pFLP2A and pFLP2Z plasmids created in this study were made using this modified plasmid, pFLP2ab. Both pFLP2A and pFLP2Z, therefore, share the following relevant characteristics: an Flp recombinase encoding gene, the pWH1266 origin of replication, and a sacB counterselectable marker for curing of pFLP. These plasmids differ with respect to the selection marker; pFLP2Z contains a Zeor marker, and pFLP2A contains an Aprr marker. These two plasmids were created with the intention of providing a compatible source of Flp recombinase to obtain markerless insertions when using either the Apr or the Zeo mini-Tn7 construct created in this study. For example, in an MDR or XDR clinical isolate where the MICs for Zeo and Apr are of practical use for selection purposes, the mini-Tn7 Apr construct may be used for insertion and the pFLP2Z excision vector can be used subsequently to obtain the markerless insert. Some clinical isolates may not be susceptible to Apr or Zeo, which will hinder achieving markerless insertions. For example, AB030 is resistant to Zeo, and as a result we were not able to create markerless insertions in the strain using plasmid pFLP2Z. Nevertheless, we were able to achieve marked insertions of the fluorescent proteins in AB030 (Fig. 3). Thus, the insertion system can still have wide applications in clinical isolates, as long as they are susceptible to at least one of the two antibiotics, i.e., Apr or Zeo. Insertion of these plasmids did not have an effect on the fitness of any of the tested strains in the rich growth medium (see Fig. S3 in the supplemental material).
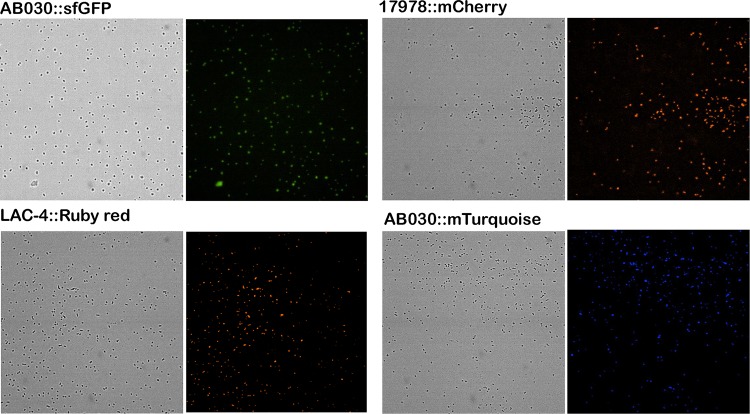
FIG 3.
Visualization of the inserted fluorescent-protein genes in different A. baumannii strains using the ImageXpress Micro 4 high-content imaging system with a 40× objective. Images for LAC-4::Ruby Red and 17978::mCherry was acquired using a Texas Red filter, while those for AB030::sfGFP and AB030::mTurquoise were acquired using an FITC filter and a DAPI filter, respectively. The images acquired using the filters were compared to the bright-field images of the same to observe the presence and distribution of cells. Emission and excitation wavelengths are provided in Materials and Methods.
To test the utility of the plasmids constructed, we cloned fluorescent markers in pUC18T-mini-Tn7T-Apr. Four different plasmids containing different fluorescent markers were created, as listed in Table 2. This was followed by inserting pUC18T-mini-Tn7T-Apr-sfGFP in AB030, pUC18T-mini-Tn7T-Apr-mRuby in LAC-4, pUC18T-mini-Tn7T-Apr-mCherry in ATCC 17978, and pUC18T-mini-Tn7T-Apr-mTurquoise in AB030. The cells were visualized using a fluorescence imager. As seen in Fig. 3, single-copy expression of fluorescent genes can be effectively used to visualize A. baumannii strains. There are a number of applications of such a system; for example, fluorescently labeled cells can be used in virulence studies (such as in infection models), monitoring of biofilm growth, fitness competition assays, etc.
TABLE 2.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant characteristicsa | Source or reference/GenBank accession number |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α(λpir+) | F– φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK– mK+) phoA supE44 λ– thi-1 gyrA96 relA1 | Kumar lab collection |
| HB101 | F– mcrB mrr hsdS20(rB− mB−) recA13 leuB6 ara-14 proA2 lacY1 galK2 xyl-5 mtl-1 rpsL20 (Smr) glnV44 λ– | Kumar lab collection |
| SM10(λpir+) | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Km λpir+ | Kumar lab collection |
| A. baumannii | ||
| ATCC 17978 | Type strain | ATCC |
| AB030 | XDR clinical isolate from Winnipeg, MB | 27 |
| AB031 | Clinical isolate from Toronto, ON | 27 |
| LAC-4 | MDR clinical isolate from Los Angeles County, CA | 26 |
| AB234 | ATCC 17978::mini-Tn7T-Apr-LAC | This study |
| AB235 | ATCC 17978::mini-Tn7T-Zeo-LAC | This study |
| AB240 | LAC-4::mini-Tn7T-Zeo-LAC | This study |
| AB241 | AB031::mini-Tn7T-Zeo-LAC | This study |
| AB242 | AB030::mini-Tn7T-Apr-LAC | This study |
| AB243 | AB031::mini-Tn7T-Apr-LAC | This study |
| AB244 | LAC-4::mini-Tn7T-Apr-LAC | This study |
| AB271 | ATCC 17978::mini-Tn7T-Apr-mCherry | This study |
| AB272 | LAC-4::mini-Tn7T-Apr-RubyRed | This study |
| AB273 | AB030::mini-Tn7T-Apr-sfGFP | This study |
| AB274 | AB030::mini-Tn7T-Apr-mTurquoise | This study |
| Plasmids | ||
| pUC18T-mini-Tn7T-Gm-LAC | Gmr Ampr | 4 |
| pUC18T-miniTn7T-Gm | Gmr Ampr | 4 |
| pUC18T-miniTn7T-Apr-LAC | Aprr Ampr | This study/MH290812.1 |
| pUC18T-miniTn7T-Apr | Aprr Ampr | This study/MK356268 |
| pUC18T-miniTn7T-Zeo-LAC | Zeor Ampr | This study/MH290813.1 |
| pUC18T-miniTn7T-Zeo | Zeor Ampr | This study/MK356269 |
| pFLP2ab | pFLP2 containing the origin of replication from pWH1266, Ampr | This study |
| pFLP2A | Aprr | This study/MK356266 |
| pFLP2Z | Zeor | This study/MK356267 |
| pFAM1 | Aprr Ampr | Schweizer Lab collection |
| pFZE1 | Zeor Ampr | 5 |
| pTNS2 | Ampr | 4 |
| pRK2013 | Kmr | 28 |
| pUC18T-mini-Tn7T-Apr-sfGFP | Aprr | This study/MH976505 |
| pUC18T-mini-Tn7T-Apr- mCherry | Aprr | This study/MH976504 |
| pUC18T-mini-Tn7T-Apr-mTurquoise | Aprr | This study/MK213339 |
| pUC18T-mini-Tn7T-Apr-RubyRed | Aprr | This study/MK213338 |
| pFPV-mCherry/2 | Ampr | 29 |
| pFPV-mTurquoise2 | Ampr | Chao lab collection. |
| pFPV-mRuby2 | Ampr | Chao lab collection |
| pUC57-sfGFP | Ampr | This study |
Abbreviations: Amp, ampicillin; Apr, apramycin; Gm, gentamicin; Km, kanamycin; Sm, streptomycin; Zeo, zeocin.
In conclusion, availability of diverse molecular tools for effective genetic manipulation is necessary to study pathogens, such as A. baumannii, that demonstrate high degrees of genetic variability and resistance. The single-copy gene expression systems created in this study aid in genetic manipulations in MDR and XDR isolates of A. baumannii in a highly efficient and reproducible manner.
MATERIALS AND METHODS
Plasmids, bacterial strains, and growth conditions.
All plasmids and bacterial strains used in this study are listed in Table 2. Bacterial strains were grown in lysogeny broth (LB) (Lennox) (Becton, Dickinson and Company, Cockeysville, MD, USA) medium at 37°C unless otherwise indicated. Apramycin (Apr) (Gold Biotechnology, St. Louis, MO, USA) concentrations for selection were 30 µg/ml for E. coli and 100 µg/ml for the A. baumannii type strain and clinical isolates. Zeocin (Zeo) (InvivoGen, San Diego, CA, USA) concentrations for selection were 25 µg/ml for E. coli and 100 µg/ml for the A. baumannii type strain and clinical isolates, unless otherwise stated. Gentamicin (Gm) (BioBasic, Markham, ON, Canada) concentrations for selection were 15 µg/ml for E. coli and 50 µg/ml for A. baumannii strains.
Antibiotic susceptibility assays.
Antibiotic susceptibility assays were carried out using a broth microdilution method according to CSLI guidelines (22). Gentamicin, zeocin, and apramycin susceptibilities were tested for A. baumannii type strain ATCC 17978 and A. baumannii clinical isolates AB030, AB031, and LAC-4. Briefly, antibiotics were serially diluted 2-fold in cation-adjusted Mueller-Hinton broth (MHB) (Oxoid, Nepean, ON, Canada) from a starting concentration of 1,024 µg/ml. Log-phase bacterial cultures were diluted in 0.85% saline to the 0.5 McFarland turbidity standard. Standardized cultures were further diluted in MHB (1:50, vol/vol) before being aliquoted into the antibiotic dilutions in a 96-well plate. Cells were incubated overnight at 37°C without shaking for 18 h before the MICs were recorded.
Determination of attTn7 site in A. baumannii clinical isolates.
The genome sequences of A. baumannii ATCC 17978 (23), AB030 (24), AB031 (25), and LAC-4 (26) were used to determine the presence of attTn7-associated sequences as described previously (7). Alignment was carried out using Clustal Omega multiple-sequence alignment tool (https://www.ebi.ac.uk/Tools/msa/clustalo/). The aligned sequences were visualized using the GeneDoc multiple-sequence alignment editor and shading utility, v 2.7.000 (K. B. Nicholas and H. B. Nicholas, Jr., GeneDoc: a tool for editing and annotating multiple sequence alignments, 1997 [distributed by the authors]).
Construction of mini-Tn7 vectors with Apr and Zeo resistance markers.
The strategy for the construction of mini-Tn7 plasmids is summarized in Fig. 2. The Gmr marker from pUC18T-mini-Tn7T-LAC-Gm was excised using BsrGI and SacII (New England Biolabs, ON, Canada), leaving the FRT sites in the plasmid backbone intact. The resulting plasmid backbone (5,374-bp fragment) was purified from the agarose gel using the E.Z.N.A. gel extraction kit (Omega Bio-Tek, GA, USA) according to the manufacturer’s protocol. The 998-bp aac(3)-IV (Aprr)- and 569-bp ble (Zeor)-containing fragments were PCR amplified from the pFAM1 and pFZE1 plasmids (primer sequences are provided in Table 3), respectively, and purified from the agarose gel using the E.Z.N.A. gel extraction kit (Omega Bio-Tek). PCR products were digested with BsrGI and SacII (New England Biolabs) and the fragments ligated into the mini-Tn7 plasmid backbone digested with the same enzymes mentioned above. The ligated plasmids were transformed into E. coli DH5α, and colonies were selected on LB plus 30 μg/ml Apr (Apr30) for pUC18T-mini-Tn7T-LAC-Apr transformants or on LB plus 25 μg/ml Zeo (Zeo25) for pUC18T-mini-Tn7T-LAC-Zeo transformants. Construction of pUC18T-mini-Tn7T-LAC-Apr and pUC18T-mini-Tn7T-LAC-Zeo was initially confirmed by colony PCR as well as restriction digestion. Further confirmation was carried out by sequencing both plasmids by Illumina Hi-Seq next-generation sequencing (MGH CCIB DNA Core Facility, Cambridge, MA, USA).
TABLE 3.
Primers used in this study
| Name | Target | Sequencea | Application |
|---|---|---|---|
| ABglmS_F_New | glmS2 | 5′TATGGAAGAAGTTCAGGCTC | Screening for mini-Tn7 genomic insertion |
| Tn7R | Tn7 right end | 5′CACAGCATAACTGGACTGATTTC | |
| pFAM F SacII | Aprr gene | 5′TGCATCCGCGGTAGATAATTCGGCTTCTCC | Cloning of Aprr fragments, screening for Aprr gene |
| pFAM R BsrGI | Aprr gene | 5′CAGTGTGTACAATTCCCTTTGTCAACAGCAAT | |
| pFZE F BsrGI | Zeor gene | 5′CGCTGTGTACAAATTCTAGATATTCGGCTTCG | Cloning of Zeor fragments, screening for Zeor gene |
| pFZE R SacII | Zeor gene | 5′TAGTACCGCGGTCTCTGATGTTACATTGCAC |
Restriction sites are underlined.
For construction of pUC18T-mini-Tn7T plasmids with Aprr and Zeor resistance markers, the Aprr and Zeor genes [aac(3)-IV and ble, respectively] were obtained, using the restriction enzymes BsrGI and SacII, from pUC18T-mini-Tn7T-LAC-Apr and pUC18T-mini-Tn7T-LAC-Zeo, respectively. The resistance genes were extracted from the agarose gel using the E.Z.N.A. gel extraction kit (Omega Bio-Tek) and ligated to pUC18T-mini-Tn7-Gm digested with BsrGI and SacII (to remove aacC1 [Gmr marker]). The ligated plasmids were transformed into E. coli DH5α, and colonies were selected on LB plus Apr30 for pUC18T-mini-Tn7T-Apr transformants or on LB plus Zeo25 for pUC18T-mini-Tn7T-Zeo transformants. Both pUC18T-mini-Tn7T-Apr and pUC18T-mini-Tn7T-Zeo were confirmed using restriction digestion.
All plasmid maps were created using SnapGene software (GSL Biotech).
Construction of pFLP2A and pFLP2Z.
Plasmid pFLP2ab was created by cloning the Acinetobacter-compatible origin of replication from plasmid pWH1266 (21) into plasmid pFLP2 (4). The pFLP2A and pFLPZ plasmids were created by cloning the aac(3)-IV (Aprr) and ble (Zeor) genes into pFLP2ab. Briefly, pFLP2ab was extracted from E. coli DH5α using the EZ-10 spin column kit (BioBasic), and the bla gene was excised using AhdI and XmnI (New England Biolabs). The aac(3)-IV (Aprr) and ble (Zeor) fragments were PCR amplified with Q5 DNA polymerase (New England Biolabs) using pFAM1 and pFZE1 as templates, respectively. The PCR products were cloned into the pFLP2ab plasmid backbone (10,041 bp) using T4 DNA ligase (Thermo Scientific). Transformants were selected on LB plus Apr30 or LB plus Zeo25, respectively. The resulting pFLP2A and pFLP2Z plasmids were confirmed by restriction digestion (maps are provided in Fig. S1 in the supplemental material).
Optimizing GFP for expression in A. baumannii.
The nucleotide sequence for the codon-optimized superfolder green fluorescent protein gene (sfGFP) for use in Acinetobacter spp. was obtained from Marek Basler (Center for Molecular Life Sciences, University of Basel). This sequence was used to construct constitutively expressed synthetic sfGFP. Briefly, the nucleotide sequence of the tac promoter (Ptac) along with a 47-bp downstream region from the pUC18T-mini-Tn7T-Gm-LAC vector was fused upstream of sfGFP to create the synthetic gene. Restriction enzyme recognition sites for KpnI, NotI, and HindIII were introduced immediately upstream of Ptac, immediately upstream of the sfGFP start codon, and immediately downstream of the sfGFP stop codon, respectively. The synthetic construct was cloned in plasmid pUC57 using the commercial gene synthesis service offered by BioBasic Inc. The synthetic sfGFP was excised from pUC57 by restriction digestion using KpnI and HindIII. The gene was gel purified and ligated into the pUC18T-mini-Tn7T-Apr vector digested with the same enzymes, to be used in further experiments.
Construction of pUC18T-mini-Tn7T-Apr vectors containing genes for fluorescent proteins sfGFP, mRuby, mCherry, and mTurquoise.
The genes for the fluorescent proteins along with their native promoters were excised from their parent plasmids (Table 2) using HindIII and KpnI (New England Biolabs) and ligated to pUC18T-mini-Tn7T-Apr digested with the same enzymes before transformation into E. coli DH5α. Colonies were selected on LB plus Apr30. The constructs were confirmed via restriction digestion, and maps of these plasmids are provided in Fig. S2 in the supplemental material.
Insertion of mini-Tn7 vectors into A. baumannii strains.
Delivery of mini-Tn7 vectors into A. baumannii was carried out via either electroporation (6) or four-parental mating (7) methods. For electroporation, cells were incubated in 4 ml LB broth culture overnight at 37°C, aliquoted into microcentrifuge tubes, and centrifuged at 13,000 × g for 2 min. The cell pellet was washed three times with sterile ice-cold water. The washed pellets were pooled in 100 µl cold distilled water and incubated on ice for 15 min with 100 ng each of mini-Tn7 and pTNS2 helper plasmids. The pTNS2 plasmid (4) provides TnsABCD in trans in order to facilitate the high-frequency integration of the mini-Tn7 vector into attTn7. The plasmid is nonreplicative in A. baumannii and is spontaneously lost by cells. Following incubation on ice, cells were electroporated using the Eppendorf Electroporator 2510 at the following settings: 25 μF, 200 Ω, and 2.0 kV. One milliliter of SOC broth (BioBasic) was immediately added to the cell suspension. Cells were recovered for 1 h at 37°C with shaking (250 rpm). Recovered cells were plated on LB-plus-Apr100 or LB-plus-Zeo100 plates, and the resulting colonies were screened for the mini-Tn7 insert by colony PCR as described below.
Occasionally depending on the plasmid/strain combinations, a four-parental conjugation method, as described previously (7), was used with minor modifications. Briefly, 100 µl each of overnight (37°C) cultures of the E. coli DH5α/mini-Tn7 donor strain, the respective A. baumannii recipient strain, and the E. coli DH5α/pTNS2 and E. coli HB101/pRK2013 helper strains was added to 600 µl LB broth and centrifuged at 13,000 × g for 2 min. The cells were washed three times using 1 ml LB broth by centrifuging at 13,000 × g for 2 min. The pellet was resuspended in 10 µl LB broth and spotted onto a 2- by 2-cm sterile nitrocellulose filter paper placed on a prewarmed (37°C) LB agar plate. Following incubation at 30°C for 24 h, the cells were washed off the nitrocellulose filter paper with 0.85% sterile saline and plated on Simmons citrate agar (Sigma-Aldrich, St. Louis, MO, USA) supplemented with Apr100 or Zeo100. Colonies from the Simmons citrate plates were patched onto LB agar supplemented with Apr100 or Zeo100 and screened for the mini-Tn7 insert by colony PCR as described below.
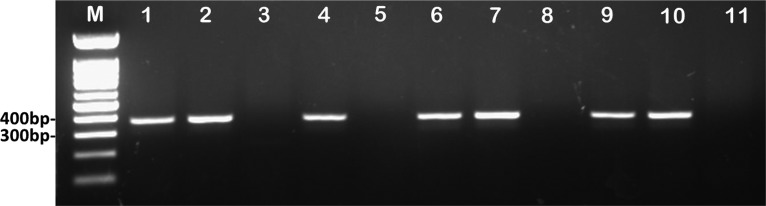
Colonies from LB-plus-Apr100 or LB-plus-Zeo100 plates were resuspended in 20 µl sterile water and incubated for 10 min at 100°C to lyse cells. One microliter of cell lysate was used as the template in each PCR. PCR was set up using Taq DNA polymerase (Froggabio), the ABglmS_F_N and Tn7R primers (Table 3), and the following cycle conditions: denaturing at 94°C for 30 s, annealing at 48°C for 30 s, and extension at 72°C for 30 s. Successful insertion produced a PCR product of 368 bp (Fig. 4).
FIG 4.
PCR confirmation of mini-Tn7T-Apr-LAC and mini-Tn7T-Zeo-LAC genomic insertions in the A. baumannii strains used in this study. The mini-Tn7T-Apr-LAC and mini-Tn7T-Zeo-LAC genomic insertions in A. baumannii strains were confirmed using the ABglmS_F_New and Tn7R primers, which give the expected 368-bp band. Lane M, 100-bp molecular size marker (New England Biolabs); lane 1, A. baumannii ATCC 17978::mini-Tn7T-Apr-LAC; lane 2, A. baumannii ATCC 17978::mini-Tn7T-Zeo-LAC; lane 3, negative control for A. baumannii ATCC 17978; lane 4, A. baumannii AB030::mini-Tn7T-Apr-LAC; lane 5, negative control for A. baumannii AB030; lane 6, A. baumannii AB031::mini-Tn7T-Apr-LAC; lane 7, A. baumannii AB031::mini-Tn7T-Zeo-LAC; lane 8, negative control for A. baumannii AB031; lane 9, A. baumannii LAC-4::mini-Tn7T-Apr-LAC; lane 10, A. baumannii LAC-4::mini-Tn7T-Zeo-LAC; lane 11, negative control for A. baumannii LAC-4.
Removal of Aprr/Zeor markers from mini-Tn7 insertions in A. baumannii.
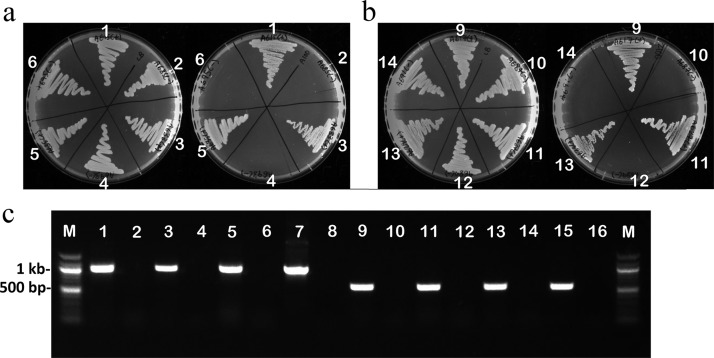
The removal of aac(3)-IV (Aprr marker) from miniTn7T-Apr-LAC or of ble (Zeor marker) from mini-Tn7T-Zeo-LAC genomic insertions in A. baumannii strains was achieved using pFLP2Z or pFLP2A excision vectors, respectively. Briefly, for the removal of the resistance marker, 100 ng of pFLP2Z or pFLP2A plasmid DNA was used for electroporation. After recovery following electroporation, cells were plated on LB agar supplemented with either Apr100 (for pFLP2A) or Zeo100 (for pFLP2Z) and incubated overnight at 37°C. Colonies from these plates were cross patched onto LB-plus-Zeo100 and LB-plus-Apr100 plates. Colonies that were apramycin susceptible and zeocin resistant (indicating loss of the Aprr marker from the genome) or Zeo susceptible and Apr resistant (indicating loss of the Zeor marker from the genome) were confirmed by colony PCR for the loss of respective resistance gene. The pFLP2Z or pFLP2A plasmids were cured by streaking cells on LB agar supplemented with 10% sucrose and incubating overnight at 37°C. Loss of pFLP2Z or pFLP2A was confirmed by cross patching onto LB agar and LB agar supplemented with the appropriate antibiotic (Zeo or Apr), followed by PCR using the primers for Zeor or Aprr resistance genes (Table 3) to confirm the loss of the resistance gene (Fig. 5a, b, and c).
FIG 5.
Deletion of apramycin or zeocin resistance markers from A. baumannii using pFLP2A. (a) Photographs of the LB agar plate (left) and the LB agar plate supplemented with 100 µg/ml of apramycin (right) to screen for the loss of the Aprr resistance gene by using pFLP2Z, encoding Flp recombinase. The absence of growth on LB plus Apr confirms the loss of aac(3)-IV (Aprr marker). (b) Photographs of the LB agar plate (left) and the LB agar plate supplemented with 100 µg/ml of zeocin (right) to screen for the loss of Zeor resistance gene. The absence of growth on LB plus Zeo confirms the loss of ble (Zeor marker). (c) The loss of the relevant resistance markers was further confirmed with PCR. Lanes 1, 3, and 5 show the 998-bp band corresponding to the aac(3)-IV gene in ATCC 17978, LAC-4, and AB031, respectively. Lanes 2, 4, and 6 show the loss of the 998-bp band in the pFLP2Z-treated ATCC 17978, LAC-4, and AB031 strains, respectively. Lanes 7 and 8, positive and negative controls for the 998-bp aac(3)-IV band, respectively. Lanes 9, 11, and 13 show the 569-bp band corresponding to the ble gene in ATCC 17978, LAC-4, and AB031, respectively. Lanes 10, 12, and 14 show the loss of the 569-bp band in the pFLP2A-treated ATCC 17978, LAC-4, and AB031 strains, respectively. Lanes 15 and 16, positive and negative controls for the 569-bp ble band, respectively. Lanes M, 100-bp molecular size marker.
Fluorescent-image acquisition.
Image acquisition was conducted using the ImageXpress Micro 4 high-content imaging system from Molecular Devices. Bacterial cultures for image acquisition were grown in LB broth overnight, and 10 µl of the overnight culture was placed on a clean glass slide and spread using a coverslip. The sample was air dried and heat fixed, after which the coverslip was placed on the slide before imaging. Images were taken using a 40× Super Plan Fluor ELWD objective and sCMOS camera. Images of fluorescence-tagged proteins were with mTurquoise, enhanced GFP (eGFP), mCherry, and Ruby Red. The following filter cubes were used to visualize the cells: TL10 (transmitted light), DAPI (4′,6′-diamidino-2-phenylindole) (excitation at 377/50 nm and emission at 447/60 nm, for mTurquoise), fluorescein isothiocyanate (FITC) (excitation at 424/27 nm and emission at 520/35 nm, for eGFP), and Texas Red (excitation at 562/40 nm and emission at 624/40 nm, for mCherry and Ruby Red). The software used to analyze images was MetaXpress version 6.2.3.733 from Molecular Devices (San Jose, CA, USA).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada to A.K. (RGPIN 05550-2015). K.D.-M. was supported by an NSERC summer fellowship. P.M.D.S. is supported by fellowships from the University of Manitoba and Faculty of Science.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00066-19.
REFERENCES
- 1.Howard A, O'Donoghue M, Feeney A, Sleator RD. 2012. Acinetobacter baumannii: an emerging opportunistic pathogen. Virulence 3:243–250. doi: 10.4161/viru.19700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yildirim S, Thompson MG, Jacobs AC, Zurawski DV, Kirkup BC. 2016. Evaluation of parameters for high efficiency transformation of Acinetobacter baumannii. Sci Rep 6:22110. doi: 10.1038/srep22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi KH, DeShazer D, Schweizer HP. 2006. Mini-Tn7 insertion in bacteria with multiple glmS-linked attTn7 sites: example Burkholderia mallei ATCC 23344. Nat Protoc 1:162–169. doi: 10.1038/nprot.2006.25. [DOI] [PubMed] [Google Scholar]
- 4.Choi KH, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods 2:443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- 5.Choi KH, Mima T, Casart Y, Rholl D, Kumar A, Beacham IR, Schweizer HP. 2008. Genetic tools for select-agent-compliant manipulation of Burkholderia pseudomallei. Appl Environ Microbiol 74:1064–1075. doi: 10.1128/AEM.02430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi KH, Schweizer HP. 2006. Mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc 1:153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- 7.Kumar A, Dalton C, Cortez-Cordova J, Schweizer HP. 2010. Mini-Tn7 vectors as genetic tools for single copy gene cloning in Acinetobacter baumannii. J Microbiol Methods 82:296–300. doi: 10.1016/j.mimet.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Choi KH, Schweizer HP. 2006. Mini-Tn7 insertion in bacteria with secondary, non-glmS-linked attTn7 sites: example Proteus mirabilis HI4320. Nat Protoc 1:170–178. doi: 10.1038/nprot.2006.26. [DOI] [PubMed] [Google Scholar]
- 9.Peters JE, Craig NL. 2001. Tn7: smarter than we thought. Nat Rev Mol Cell Biol 2:806–814. doi: 10.1038/35099006. [DOI] [PubMed] [Google Scholar]
- 10.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 11.Liu F, Zhu Y, Yi Y, Lu N, Zhu B, Hu Y. 2014. Comparative genomic analysis of Acinetobacter baumannii clinical isolates reveals extensive genomic variation and diverse antibiotic resistance determinants. BMC Genomics 15:1163. doi: 10.1186/1471-2164-15-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maraki S, Mantadakis E, Mavromanolaki VE, Kofteridis DP, Samonis G. 2016. A 5-year surveillance study on antimicrobial resistance of Acinetobacter baumannii clinical isolates from a tertiary Greek hospital. Infect Chemother 48:190–198. doi: 10.3947/ic.2016.48.3.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lucidi M, Runci F, Rampioni G, Frangipani E, Leoni L, Visca P. 2018. New shuttle vectors for gene cloning and expression in multidrug-resistant Acinetobacter species. Antimicrob Agents Chemother 62:e02480-17. doi: 10.1128/AAC.02480-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luna BM, Ulhaq A, Yan J, Pantapalangkoor P, Nielsen TB, Davies BW, Actis LA, Spellberg B. 2017. Selectable markers for use in genetic manipulation of extensively drug-resistant (XDR) Acinetobacter baumannii HUMC1. mSphere 2:e00140-17. doi: 10.1128/mSphere.00140-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang AD, Smith KP, Eliopoulos GM, Berg AH, McCoy C, Kirby JE. 2017. In vitro apramycin activity against multidrug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa. Diagn Microbiol Infect Dis 88:188–191. doi: 10.1016/j.diagmicrobio.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Paget E, Davies J. 1996. Apramycin resistance as a selective marker for gene transfer in mycobacteria. J Bacteriol 178:6357–6360. doi: 10.1128/jb.178.21.6357-6360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Utashima Y, Yamashita S, Arima T-H, Masaki K. 2017. Codon optimization enables the Zeocin resistance marker’s use in the ascomycete yeast Debaryomyces occidentalis. J Gen Appl Microbiol 63:254–257. doi: 10.2323/jgam.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Crepin S, Harel J, Dozois CM. 2012. Chromosomal complementation using Tn7 transposon vectors in Enterobacteriaceae. Appl Environ Microbiol 78:6001–6008. doi: 10.1128/AEM.00986-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ducas-Mowchun K, De Silva PM, Patidar R, Schweizer HP, Kumar A. 2019. Tn7-based single-copy insertion vectors for Acinetobacter baumannii. Methods Mol Biol 1946:135–150. doi: 10.1007/978-1-4939-9118-1_13. [DOI] [PubMed] [Google Scholar]
- 20.Kumar A, Chua KL, Schweizer HP. 2006. Method for regulated expression of single-copy efflux pump genes in a surrogate Pseudomonas aeruginosa strain: identification of the BpeEF-OprC chloramphenicol and trimethoprim efflux pump of Burkholderia pseudomallei 1026b. Antimicrob Agents Chemother 50:3460–3463. doi: 10.1128/AAC.00440-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunger M, Schmucker R, Kishan V, Hillen W. 1990. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87:45–51. doi: 10.1016/0378-1119(90)90494-C. [DOI] [PubMed] [Google Scholar]
- 22.Clinical and Laboratory Standards Institute. 2017. M100-27. Performance standards for antimicrobial susceptibility testing, 27th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 23.Smith MG, Gianoulis TA, Pukatzki S, Mekalanos JJ, Ornston LN, Gerstein M, Snyder M. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev 21:601–614. doi: 10.1101/gad.1510307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loewen PC, Alsaadi Y, Fernando D, Kumar A. 2014. Genome sequence of an extremely drug-resistant clinical isolate of Acinetobacter baumannii strain AB030. Genome Announc 2:e01035-14. doi: 10.1128/genomeA.01035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loewen PC, Alsaadi Y, Fernando D, Kumar A. 2014. Genome sequence of a tigecycline-resistant clinical isolate of Acinetobacter baumannii strain AB031 obtained from a bloodstream infection. Genome Announc 2:e01036-14. doi: 10.1128/genomeA.01036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ou H-Y, Kuang SN, He X, Molgora BM, Ewing PJ, Deng Z, Osby M, Chen W, Xu HH. 2015. Complete genome sequence of hypervirulent and outbreak-associated Acinetobacter baumannii strain LAC-4: epidemiology, resistance genetic determinants and potential virulence factors. Sci Rep 5:8643. doi: 10.1038/srep08643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernando D, Zhanel GG, Kumar A. 2013. Antibiotic resistance and expression of resistance-nodulation-division pumps and outer membrane porins in Acinetobacter species isolated from Canadian hospitals. Can J Infect Dis Med Microbiol 24:17–21. doi: 10.1155/2013/696043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drecktrah D, Levine-Wilkinson S, Dam T, Winfree S, Knodler LA, Schroer TA, Steele-Mortimer O. 2008. Dynamic behavior of Salmonella-induced membrane tubules in epithelial cells. Traffic 9:2117–2129. doi: 10.1111/j.1600-0854.2008.00830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.