Fresh produce is an important vehicle for STEC transmission, and experimental evidence shows that STEC can colonize plants as secondary hosts, but differences in the capacity to colonize occur between different plant species and tissues. Therefore, an understanding of the impact that these plant factors have on the ability of STEC to grow and establish is required for food safety considerations and risk assessment. Here, we determined whether growth and the ability of STEC to form biofilms in plant extracts could be related to specific plant metabolites or could predict the ability of the bacteria to colonize living plants. Growth rates for sprouted seeds (alfalfa and fenugreek) but not those for leafy vegetables (lettuce and spinach) exhibited a positive relationship between plant extracts and living plants. Therefore, the detailed variations at the level of the bacterial isolate, plant species, and tissue type all need to be considered in risk assessment.
KEYWORDS: Escherichia coli O157:H7, EHEC, alfalfa, fenugreek, lettuce, spinach
ABSTRACT
Contamination of fresh produce with pathogenic Escherichia coli, including Shiga-toxigenic E. coli (STEC), represents a serious risk to human health. Colonization is governed by multiple bacterial and plant factors that can impact the probability and suitability of bacterial growth. Thus, we aimed to determine whether the growth potential of STEC for plants associated with foodborne outbreaks (two leafy vegetables and two sprouted seed species) is predictive of the colonization of living plants, as assessed from growth kinetics and biofilm formation in plant extracts. The fitness of STEC isolates was compared to that of environmental E. coli isolates at temperatures relevant to plant growth. Growth kinetics in plant extracts varied in a plant-dependent and isolate-dependent manner for all isolates, with spinach leaf lysates supporting the highest rates of growth. Spinach extracts also supported the highest levels of biofilm formation. Saccharides were identified to be the major driver of bacterial growth, although no single metabolite could be correlated with growth kinetics. The highest level of in planta colonization occurred on alfalfa sprouts, though internalization was 10 times more prevalent in the leafy vegetables than in sprouted seeds. Marked differences in in planta growth meant that the growth potential of STEC could be inferred only for sprouted seeds. In contrast, biofilm formation in extracts related to spinach colonization. Overall, the capacity of E. coli to colonize, grow, and be internalized within plants or plant-derived matrices was influenced by the isolate type, plant species, plant tissue type, and temperature, complicating any straightforward relationship between in vitro and in planta behaviors.
IMPORTANCE Fresh produce is an important vehicle for STEC transmission, and experimental evidence shows that STEC can colonize plants as secondary hosts, but differences in the capacity to colonize occur between different plant species and tissues. Therefore, an understanding of the impact that these plant factors have on the ability of STEC to grow and establish is required for food safety considerations and risk assessment. Here, we determined whether growth and the ability of STEC to form biofilms in plant extracts could be related to specific plant metabolites or could predict the ability of the bacteria to colonize living plants. Growth rates for sprouted seeds (alfalfa and fenugreek) but not those for leafy vegetables (lettuce and spinach) exhibited a positive relationship between plant extracts and living plants. Therefore, the detailed variations at the level of the bacterial isolate, plant species, and tissue type all need to be considered in risk assessment.
INTRODUCTION
Contamination of fresh produce from Shiga-toxigenic Escherichia coli (STEC) presents a serious hazard as a cause of foodborne illnesses, diarrhea, and enterohemorrhagic disease. Fresh produce is a major vehicle of transmission of STEC, with foods of plant origin accounting for the majority of E. coli and Shigella outbreaks in the United States (1). Fresh produce is often eaten raw or minimally processed, and contamination of the produce can occur at any point along the food chain from farm to fork, with major outbreaks from, e.g., spinach (2) and sprouted seeds (3) being documented. STEC has been shown to interact with plants and can use them as secondary hosts (4, 5), which has implications for preharvest contamination, as well as persistence on postharvest produce (6–8).
Colonization of host plants by E. coli is governed by a range of environmental, bacterial, and plant factors. Initial contact with and the attachment of bacteria to plant tissue are defined by motility, adherence factors, and plant cell wall components (9, 10), while establishment is influenced by a range of plant biotic (11, 12) and abiotic (13, 14) factors. The ability of bacteria to grow in the presence of plant material is a key factor in assessing risk, and although proliferation is well-known to be influenced by physiochemical factors (15, 16), risk assessments for STEC on fresh produce tend to consider plants as a homogeneous whole (17–19).
STEC preferentially colonizes the roots and rhizosphere of fresh produce plants over leafy tissue and has been shown to be internalized by plant tissue, where it can persist in the apoplastic space as an endophyte (20, 21). The apoplast contains metabolites, such as solutes, sugars, proteins, and cell wall components (22), and as such provides a rich environment for many bacterial species, including both commensal bacteria and human pathogens (23, 24). The rate of STEC internalization is dependent on multiple factors, including the plant species and tissue (25) and how plants are propagated (26, 27). Specificity in the response of STEC to different plant species and tissue types has been demonstrated at the transcriptional level (28, 29). Therefore, there is a need to take into account the specificity of the STEC-plant interactions that could impact risk.
Determination of the growth potential of a bacterial population takes into account the probability of growth together with the suitability of the growing population for a particular environment (30). It is used as a measure in risk assessment, e.g., for the growth of STEC in water (31). In plant hosts, bacterial growth potential is governed by several factors, including bacterial growth rates, initial adherence, and colony establishment, which is often in biofilms, as well as plant-dependent factors, including metabolite availability and plant defense responses (32). Therefore, the aim here was to determine whether the in vitro growth kinetics of and biofilm formation by STEC in plant extracts, together with plant metabolite analysis, could be related to the colonization of plants that are associated with foodborne outbreaks and, hence, inform the growth potential of STEC in planta. The use of genetically distinct E. coli isolates (two STEC isolates, two environmental isolates, and one laboratory isolate) enabled comparison of bacterial phenotypic variation within plants and plant-derived matrices. Growth kinetics and biofilm formation were quantified in different tissue extracts of two leafy vegetables, lettuce and spinach, and two sprouted seeds, fenugreek and alfalfa sprouts. Growth kinetics were related to the metabolomics of the extracts. Quantification of in planta colonization and internalization allowed a correlation analysis to be performed for the two STEC isolates.
RESULTS
E. coli growth rates in plant extracts.
To relate growth potential to the colonization of fresh produce plants by STEC, in vitro growth rates were first measured in plant extracts. Primary modeling of in vitro growth data in plant extracts successfully fitted 86.7% (117 of 135) of the growth curves with a nonlinear Baranyi model (see Methods SM1 in the supplemental material). Misfits were improved by manually truncating the growth curves to the time before the observed decrease in cell density that occurred in stationary phase, resulting in an adjusted R2 (R2adj) value of 0.996 (see Fig. S1 and Table S1a in the supplemental material). Comparison of the maximum growth rates (μ) showed that the highest growth rates were in spinach extracts, with the fastest growth occurring in leaf lysates at 18°C or the apoplast at 25°C (Fig. 1A), while in lettuce the fastest growth occurred in the apoplastic extract at all temperatures tested (Fig. 1B). All isolates grew consistently faster in fenugreek sprout extracts than in alfalfa, and either sprout extract supported faster growth than defined medium [rich defined 3-(N-morpholino)propanesulfonic acid (MOPS) glycerol (RDMG)] (Fig. 1C). The E. coli O157:H7 isolates showed differential responses in the different extracts, and their growth rates were as high as or higher than those of the environmental isolates in almost all extracts. The lowest growth rates occurred for the laboratory-adapted isolate MG1655. The plant extract tissue type as well as the bacterial isolate significantly impacted μ, as determined from a two-way analysis of variance (ANOVA), at 18°C [F (4, 7,363) = 76.3 (P < 0.0001) and F (8, 7,363) = 436.4 (P < 0.0001) for bacterial isolate and extract type, respectively] and at 20°C [F (4, 8,387) = 160.3 (P < 0.0001) and F (8, 8,387) = 416.1 (P < 0.0001) for bacterial isolate and extract type, respectively].
FIG 1.
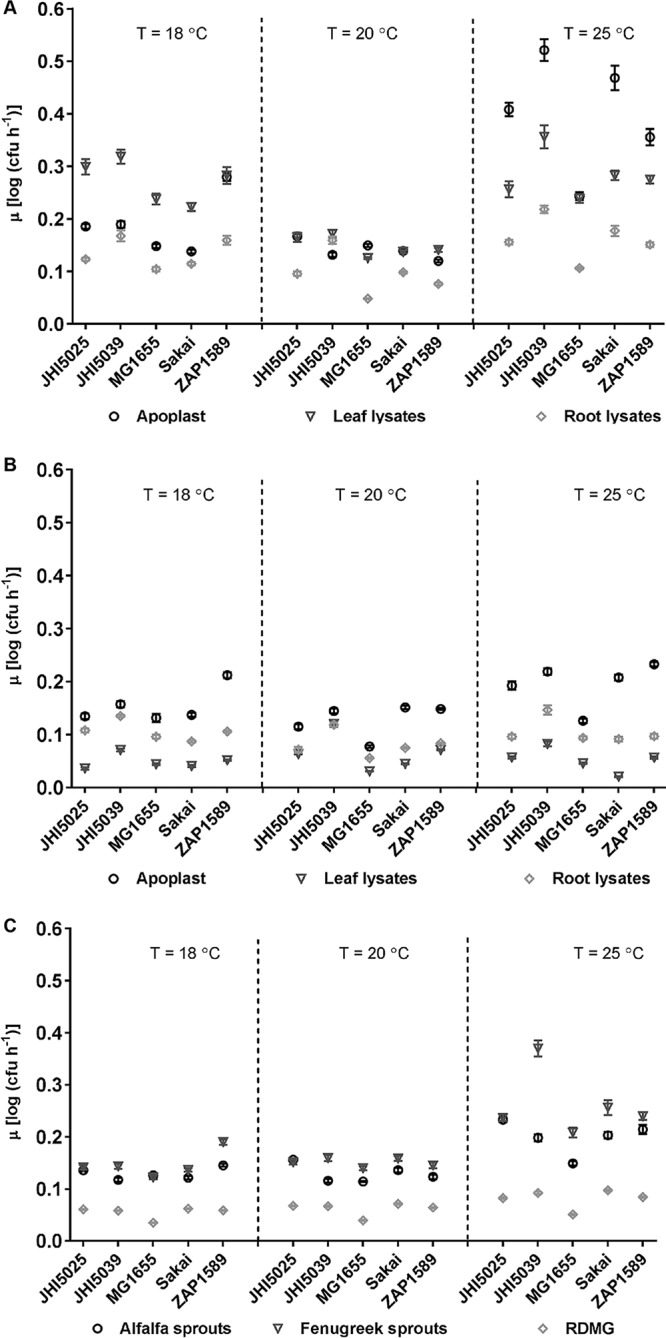
Maximum growth rates (μ) of reference E. coli isolates in plant extracts. μ were calculated using the Baranyi model for the reference E. coli isolates in spinach (A) or lettuce (B) apoplast (circles), leaf lysate (triangles), and root lysate (diamonds) extracts or in alfalfa (circles) or fenugreek (triangles) sprout lysate extracts (C), with RDMG (diamonds) as a no-plant-extract control, at a temperature (T) of 18, 20, or 25°C. Each point is the average rate (n = 12), with standard errors indicated by bars. P value summaries from multiple-comparison analysis by isolate versus MG1655 or by extract type versus RDMG are provided in Tables S1b and S1c in the supplemental material, respectively.
Growth was almost always the highest at 25°C, although there were exceptions, e.g., for E. coli O157:H7 isolate ZAP1589 in lettuce extracts. Growth characteristics were similar at both 18 and 20°C, but μ were, in general, lower at 20°C than at 18°C. This counterintuitive result was reproducible and occurred in all growth experiments. It meant that secondary modeling for temperature was not possible. It was possible, however, for the effects of temperature on growth in the defined medium without plant extracts, which produced a linear distribution for temperature for all five E. coli isolates (R2 = 0.996 to 1) (Methods SM2), indicating that the effect was due to the plant extracts and was not a systemic error.
Metabolite analysis of fresh produce plant extracts.
To establish the impacts of different plant components on the growth of the E. coli isolates, metabolite analysis was performed for the extracts. Determination of the absolute levels of mono- and disaccharides (sucrose, fructose, glucose, arabinose) showed that the highest abundance was in fenugreek sprout extracts, followed by lettuce apoplast and lettuce leaf lysates (Table 1). Sucrose was the most abundant sugar in all species and cultivars, except for alfalfa, which had high levels of fructose and glucose. Arabinose was detected only in the apoplastic fluid of spinach and lettuce, accounting for 0.36% and 0.23% of all sugars, respectively. A two-way ANOVA showed significant differences for tissue types [F (7, 60) = 16.5, P < 0.0001].
TABLE 1.
Quantification of saccharides from plant extracts
| Plant extract | Concn (μg mg−1)a
|
|||
|---|---|---|---|---|
| Glucose | Fructose | Sucrose | Arabinose | |
| Fenugreek | 24.5 ± 3.1 | 24.9 ± 3.7 | 75.6 ± 6.3 | ND |
| Alfalfa | 35.4 ± 0.8 | 35.8 ± 18.6 | 3.5 ± 0.3 | ND |
| Lettuce apoplast | 19.4 ± 1.8 | 23.4 ± 2.8 | 53.4 ± 20.7 | 0.226 ± 0.001 |
| Lettuce leaf lysates | 10.7 ± 0.3 | 14.6 ± 0.4 | 50.1 ± 3.1 | ND |
| Lettuce root lysates | 9.9 ± 0.1 | 20.0 ± 0.9 | 22.5 ± 0.4 | ND |
| Spinach apoplast | 11.8 ± 2.0 | 8.0 ± 1.7 | 38.3 ± 7.0 | 0.211 ± 0.049 |
| Spinach leaf | 21.9 ± 2.9 | 6.1 ± 0.8 | 32.8 ± 2.6 | ND |
| Spinach root | 17.4 ± 1.2 | 9.00 ± 0.9 | 29.4 ± 1.5 | ND |
The concentrations of the mono- and disaccharides were determined by HPLC. ND, not detected.
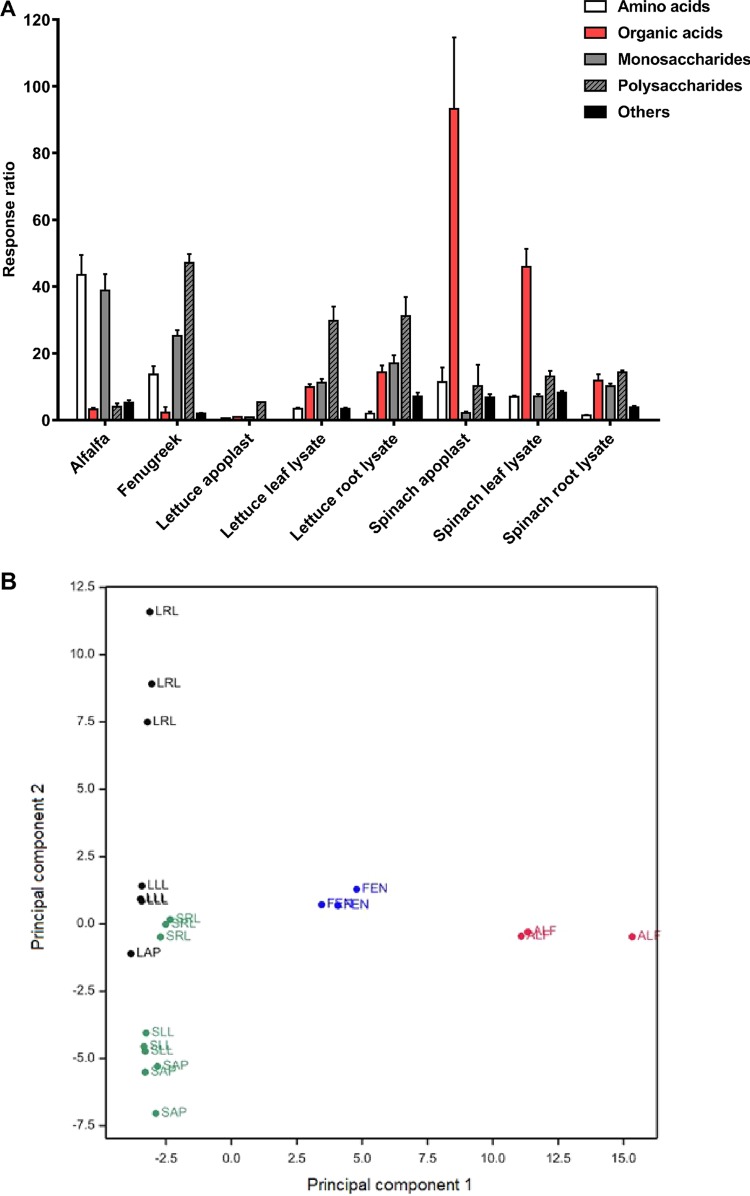
The levels of amino acids and other metabolites were determined from the identification of 116 polar metabolites, of which 60 were assigned and mapped onto a simplified polar metabolite pathway for plants to visualize the metabolite availability for the bacteria (Fig. S2). The abundance ratio of each compound against the internal standard, ribitol, generated a response ratio (RR) to allow a semiquantitative comparison (Table S2). Differences between species and tissue types occurred in a pattern similar to that for the mono- and disaccharides (Table 1), and for 12 metabolites, including fructose, glucose, and sucrose, there were significantly different RR [two-way ANOVA and the Tukey multiple-comparison test, F (7, 854) = 37.2, P < 0.0001]. Small amounts of arabinose could be found in all tissues, with no significant differences being found between host species or tissue types. Grouping of metabolites by structure (Fig. 2A) for monosaccharides, polysaccharides, amino acids, organic acids, and other metabolites showed that the largest amounts of total saccharides were present in fenugreek sprouts, while alfalfa was higher in monosaccharides and amino acids. The organic acids in the spinach apoplast consisted mainly of oxalic acid, and the amount was almost double the amount in spinach leaf lysates. The percent composition showed that the majority of metabolites in all lettuce extracts were polysaccharides, whereas they were mainly organic acids in all spinach extracts.
FIG 2.
Plant extract metabolomics and grouping. (A) The 60 assigned metabolites from all species and tissues are separated into amino acids, organic acids, mono- and polysaccharides, and others by their mean total response ratio (with the SD indicated by bars). (B) Score plot of principal component 1 (31% variance) and principal component 2 (19%) for all 116 polar metabolites for alfalfa (ALF) in red, fenugreek (FEN) in blue, spinach (spinach apoplast [SAP], spinach leaf [SLL], spinach roots [SRL]) in green, and lettuce (lettuce apoplast [LAP], lettuce leaf [LLL], lettuce roots [LRL]) in black.
A significant variation in the metabolite content occurred between plant tissues, as well as for individual metabolites [two-way ANOVA, assuming a parametric distribution, F (420, 854) = 43.15, P < 0.001]. A principal-component analysis (PCA) showed that the first five components accounted for ∼85% of the variance, and 50% of the variance for all detectable polar metabolites (n = 116) was attributed to principal component 1 (PC1) and PC2 (Fig. 2B). This was supported by a significant positive correlation for leaf lysates and apoplast extracts of lettuce and spinach (R2 > 0.97), a weak correlation for the root lysates based on species (R2 = 0.542 to 0.757), and no significant correlation between any species for the tissues.
Influence of plant extract metabolites on E. coli growth.
To relate any specific plant metabolites to bacterial growth, a correlation analysis was carried out between the plant extract growth rates for two E. coli O157:H7 isolates (Sakai and ZAP1589) and the assigned metabolites. Several organic acids were positively associated with maximal growth rates (μ), although there was a temperature-dependent effect. The metabolites associated with growth at 18°C for isolate Sakai were galactosyl glycerol, threonic acid, and oxoproline (P = ∼0.04); those associated with growth at 20°C were malic acid, fumaric acid, and quinic acid (P = 0.014 to 0.048); and those associated with growth at 25°C were oxalic acid (P = 0.009), aspartic acid (P = 0.038), glutamic acid (P = 0.046), coumaric acid (P = 0.011), and uridine (P = 0.011). Chlorogenic acid (trans-5-O-caffeoyl-d-quinate) was consistently associated with growth at all temperatures (P = 0.04 at 18°C, P = 0.004 at 20°C, and P = 0.04 at 25°C). E. coli isolate ZAP1589 gave similar results, although there was also a bacterial isolate effect, as there were no significant associations at 20°C. Therefore, no single metabolite was identified to be a major factor influencing the E. coli growth rate with a significant impact from the growth temperature.
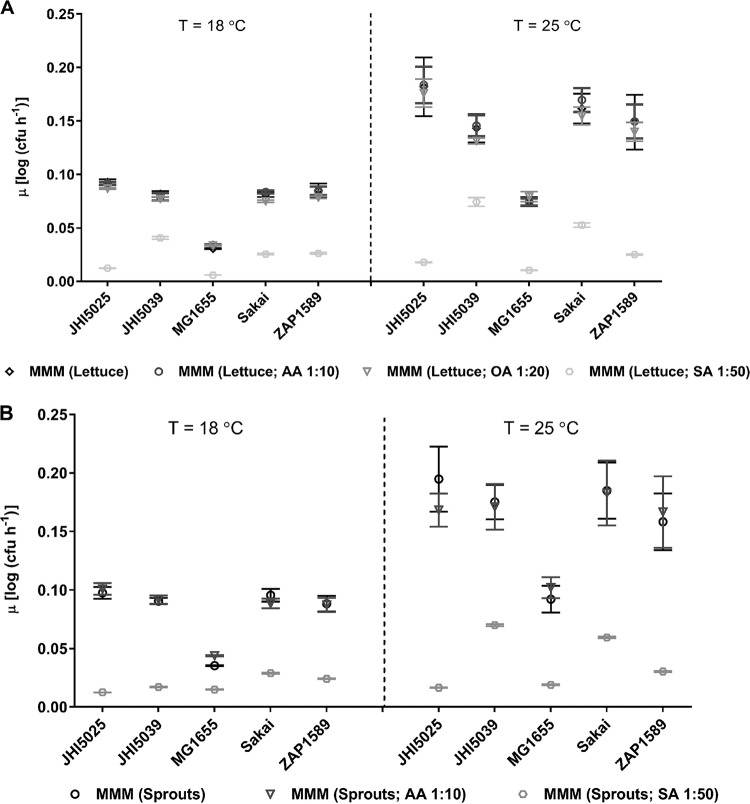
We then investigated the main metabolite groups to determine if they could influence bacterial growth by generating defined artificial growth media comprising the main plant extract metabolites. The six most abundant metabolites were selected from lettuce apoplast or sprout extracts to represent contrasting metabolite profiles (Table 2). Each of the major groups of saccharides (SAs), organic acids (OA), or amino acids (AAs) was assessed independently by dilution, to restrict their effect, and at temperatures relevant to lettuce (18°C) and sprouts (25°C). Maximal growth rates were similar in the sprout and lettuce extract artificial media (Fig. 3), although they were reduced compared to those in the complete, natural extracts (Fig. 1). Growth rates were significantly reduced when the concentration of the saccharide (SA) group was reduced for both artificial media (P < 0.0049 for all comparisons), while restriction of the amino acids (AA) or organic acids (OA) had no impact (Fig. 3). The SA-dependent effect occurred for all E. coli isolates, although there were also significant isolate dependencies [two-way ANOVA, F (16, 28,637) = 39.5 (P < 0.0001) at 25°C; two-way ANOVA, F (4, 9,544) = 401.3 (P < 0.0001) at 18°C].
TABLE 2.
Composition of defined artificial medium supplements
| Metabolite | Concn (μg ml−1)a
|
|
|---|---|---|
| Sprouts | Lettuce apoplast | |
| Saccharides (SA) | ||
| Sucrose | 3,021.4 | 2,116.2 |
| Fructose | 1,443.4 | 926.5 |
| Glucose | 1,425.0 | 769.8 |
| Amino acids (AA) | ||
| Asparagine | 814.3 | NA |
| Alanine | 766.1 | NA |
| Serine | 327.4 | NA |
| Oxoproline | NA | 63.4 |
| Organic acids (OA) | ||
| Malic acid | NA | 194.0 |
| 2,3-Dihydroxy-propanoic acid | NA | 143.5 |
The concentrations of the major six components in sprout extracts (alfalfa and fenugreek combined) and lettuce apoplast used to generate defined artificial media were determined by HPLC and GC-MS. NA, not applicable.
FIG 3.
Maximum growth rates (μ) in artificial media mimicking plant extracts. μ were calculated using the Baranyi model for the E. coli isolates at 18°C and 25°C in medium mimicking lettuce apoplast (A) or sprout lysates (a mixture of alfalfa and fenugreek sprout metabolites) (B) with specified dilutions. The base minimal MOPS medium (MMM) was supplemented with saccharides (SA), organic acids (OA), or amino acids (AA) at the dilution specified. Each point is the average rate, with standard errors indicated by bars.
Influence of plant extracts on E. coli biofilm formation.
On host tissue in planta, bacterial colonies are more likely to be present in biofilms than as single cells. Therefore, the influence of the plant extracts of the leafy vegetables on E. coli biofilm formation ability was tested in isolation, i.e., on polystyrene surfaces. Spinach leaf lysates and root lysates were the only extracts that induced biofilms for all isolates, albeit it was minimal for isolate MG1655 (P < 0.0011 compared to isolate MG1655) (Table 3). The remaining extracts were not as conducive for biofilm formation, with the exception of the biofilm formation by one of the environmental isolates (JHI5025). This was not explained by different growth rates, since this isolate did not exhibit the highest growth rates in the extracts compared to the rates for the other isolates (Fig. 1), and presumably therefore reflects increased adherence in the presence of the plant extracts. A qualitative risk ranking was determined for implementation of biofilm formation as a risk factor for the E. coli O157:H7 isolates (Sakai and ZAP1589) and identified spinach roots to be at highest risk. The ranking was as follows (from highest to lowest): spinach roots > spinach leaves > lettuce roots > lettuce leaves > spinach apoplast > lettuce apoplast.
TABLE 3.
Biofilm formation by reference E. coli isolates in plant tissue extractsa
| Treatment | Avg ± variance density of crystal violet at OD590b
|
||||
|---|---|---|---|---|---|
| Sakai | ZAP1589 | JHI5025 | JHI5039 | MG1655 | |
| Sp. apoplast | 0.002 ± 0.001 (NS) | 0.011 ± 0.001 (NS) | 0.372 ± 0.007**** | 0.013 ± 0.000 (NS) | 0.001 ± 0.002 (NS) |
| Sp. leaf | 0.071 ± 0.000*** | 0.128 ± 0.001**** | 0.218 ± 0.034**** | 0.113 ± 0.001**** | 0.000 ± 0.000 (NS) |
| Sp. root | 0.173 ± 0.000**** | 0.148 ± 0.017**** | 0.179 ± 0.015**** | 0.126 ± 0.000**** | 0.013 ± 0.000 (NS) |
| Lt. apoplast | 0.000 ± 0.002 (NS) | 0.005 ± 0.000 (NS) | 0.125 ± 0.005**** | 0.001 ± 0.000 (NS) | 0.000 ± 0.000 (NS) |
| Lt. leaf | 0.000 ± 0.000 (NS) | 0.018 ± 0.001 (NS) | 0.151 ± 0.002**** | 0.007 ± 0.000 (NS) | 0.001 ± 0.000 (NS) |
| Lt. root | 0.008 ± 0.000 (NS) | 0.029 ± 0.001 (NS) | 0.066 ± 0.001 (NS) | 0.025 ± 0.000 (NS) | 0.000 ± 0.000 (NS) |
| RDMG | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.013 ± 0.000 | 0.000 ± 0.000 | 0.000 ± 0.000 |
Biofilms were formed on polystyrene multiwall plates following incubation in spinach (Sp.) and lettuce (Lt.) extracts (apoplast, leaf, root) and rich defined MOPS medium with glycerol (RDMG) at 18°C for 48 h under static conditions.
P value summaries are provided per isolate for each extract type versus RDMG (NS, not significant [P > 0.05]; ***, P ≤ 0.001; ****, P ≤ 0.0001).
E. coli O157:H7 colonization and internalization in planta.
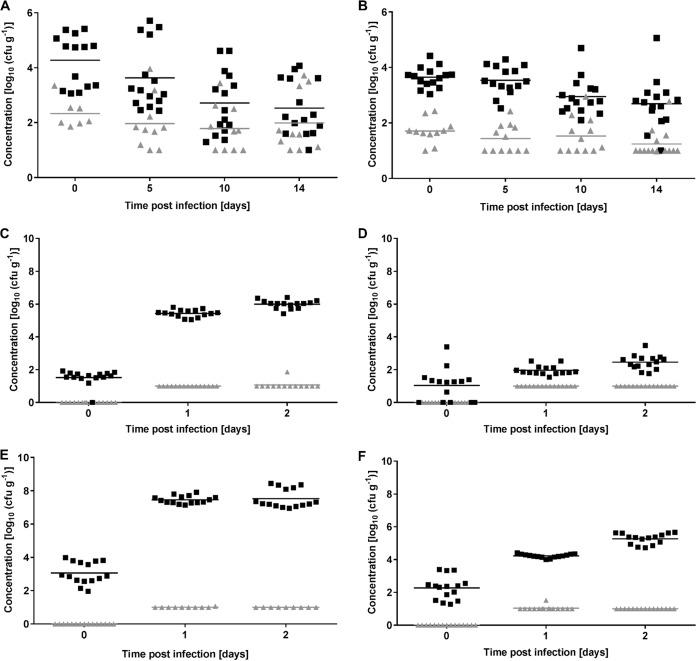
E. coli O157:H7 colonization of leafy vegetables and sprouts was quantified to determine whether growth kinetics and biofilm formation in the extracts were predictive of in planta colonization. Colonization by one E. coli O157:H7 isolate (ZAP1589) on spinach and lettuce and by both O157:H7 isolates (ZAP1589 and Sakai) on sprouted seeds was quantified. Our previous in planta data for lettuce and spinach plants showed that the highest levels of E. coli isolate Sakai occurred on spinach roots (25). Inoculation of spinach and lettuce with the high dose (107 CFU ml−1) of E. coli isolate ZAP1589 also resulted in higher levels of bacteria on the roots than on the leaves, with similar levels being found on spinach and lettuce roots, e.g., 2.53 ± 0.97 and 2.69 ± 0.88 log(CFU g−1) at day 14, respectively (Fig. 4A and B). In planta colonization of sprouted seeds by the two E. coli O157:H7 reference isolates was quantified for plants grown under conditions that mimic industry settings (hydroponically at 25°C for 3 days) (Fig. 4C to F). A low inoculation dose of 103 CFU ml−1 was used, and total viable counts on day 0 were estimated by the most probable number (MPN) method since they fell below the direct plating detection threshold. The total counts of isolate Sakai increased by 4.5 log(CFU g−1) on alfalfa sprouts and 3 log(CFU g−1) on fenugreek sprouts at between 0 and 2 days postinfection (dpi). Viable counts for isolate ZAP1589 were generally lower than those for isolate Sakai on both sprouted seeds, but they still reached 6.00 ± 0.253 log(CFU g−1) on alfalfa at 2 dpi.
FIG 4.
Total and internalized counts for E. coli O157:H7 in planta. (A and B) The number of E. coli isolate ZAP1589 bacteria recovered from inoculation (107 CFU ml−1) of spinach (var. Amazon) (A) or lettuce (var. All Year Round) (B) roots at 0, 5, 10, and 14 dpi. (C to F) The number of E. coli isolate ZAP1589 bacteria recovered from alfalfa (C) or fenugreek (D) and E. coli isolate Sakai bacteria recovered from alfalfa (E) or fenugreek sprouts (F) after inoculation at 103 CFU ml−1. Samples were recovered at 0, 1, and 2 dpi. Averages (lines) and individual sample counts are shown for the total population (black) or the internalized population (gray) (n = 15; ∼1.5 g per sample for sprouts, individual plants for spinach and lettuce). Sprout day 0 data were assessed by the MPN method (level of detection = 0); otherwise, minimum counts were manually leveled to the direct plating detection limit of 10 CFU g−1 on day 1.
Internalization was also assessed since endophytic behavior is a feature of E. coli O157:H7 colonization of fresh produce plants and growth potential could be reflected by growth in the apoplast washings. Internalization of isolate ZAP1589 occurred at higher levels in spinach roots than in lettuce roots (Fig. 4A and B), although the prevalence was similar in both plant species (60% and 58.3% of spinach and lettuce plants, respectively, contained endophytic bacteria). In contrast, internalization in sprouts occurred on only three occasions in all the experiments: for isolate Sakai in alfalfa [1.07 log(CFU g−1)] and fenugreek [1.53 log(CFU g−1)] on day 1 and for isolate ZAP1589 in alfalfa [1.87 log(CFU g−1)] on day 2. The prevalence was 7.1% (1/14 samples were positive), although the viable counts were close to the limit of detection by direct plating. Therefore, internalization of E. coli O157:H7 isolates Sakai and ZAP1589 appeared to be a rare event for sprouted seeds, although they colonized the external sprout tissue to higher levels than they did external lettuce or spinach tissue.
Correlating in planta colonization with plant extract growth rate kinetics.
To relate growth kinetics in extracts to in planta growth, growth rates were estimated for in planta growth. This was possible for sprouted seeds since the colonization levels increased over time (Fig. 4). Alfalfa plants supported significantly higher growth rates for both E. coli O157:H7 isolates than fenugreek did: 2.23 ± 0.213 log(CFU g−1) per day (R2 = 0.720) and 1.50 ± 0.0913 log(CFU g−1) per day (R2 = 0.863) for Sakai on alfalfa and fenugreek sprouts, respectively, and 2.24 ± 0.159 log(CFU g−1) per day (R2 = 0.822) and 0.710 ± 0.116 log(CFU g−1) per day (R2 = 0.464) for isolate ZAP1589 on alfalfa and fenugreek sprouts, respectively. The difference in the growth rates between the isolates on fenugreek sprouts was significant (P < 0.0001). Although the in planta growth rates of E. coli isolate Sakai for spinach tissues (on leaves, on roots, or internalized in leaf apoplast) or lettuce (on leaves and roots) were estimated from a low inoculation dose (103 CFU ml−1) (25), these were nonsignificant since growth over the 10-day period was minimal or completely constrained, with a high degree of plant-to-plant variation being found. Growth rate estimates were not made when a high starting inoculum was used since the colonization levels decreased over time (Fig. 4).
Comparisons of the in planta and extract growth rate estimates were made for E. coli O157:H7 isolates both on sprouted seeds (at 25°C) or in spinach and lettuce (at 18°C) (Fig. 5). A positive correlation occurred for growth rate estimates in the sprouted seeds (R2 = 0.516), although this was not significant. Since in planta growth in spinach or lettuce tissues was minimal, there was no correlation with the growth rates in the corresponding extracts. Therefore, the restrictions on bacterial growth that occurred with living plants meant that the growth rates in extracts could not be extrapolated to the in planta growth potential for leafy vegetables but did bear a positive relationship for sprouted seeds.
FIG 5.
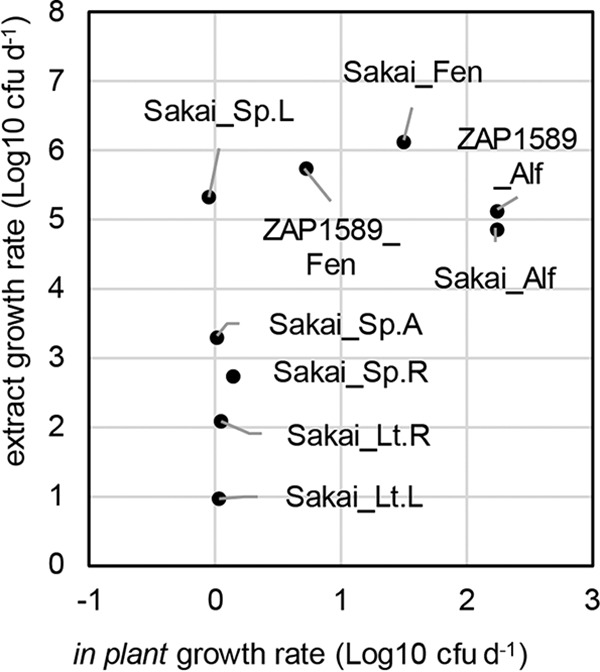
Comparison of in planta and extract growth rates for E. coli isolates Sakai and ZAP1589. Growth rates for in planta estimates were plotted against estimates for plant extracts on a log10(number of CFU day−1 [d−1]) basis for E. coli isolates Sakai and ZAP1589 and were normalized per gram (fresh weight) for plant tissues or per milliliter for plant extracts. Estimates were obtained for growth at 25°C for sprouted seeds (alfalfa [Alf], fenugreek [Fen]) and at 18°C for spinach (Sp.) or lettuce (Lt.) tissues (A, apoplast; L, leaves; R, roots).
DISCUSSION
The potential for foodborne bacteria to grow in fresh produce food commodities is a key consideration in quantitative risk assessment. Factors that influence bacterial growth are the plant species and tissue, the bacterial species or isolate, and the surrounding environment. The growth potential of a bacterial population consists of the proportion of the growing subpopulation and the suitability of the environment for growth, and it provides a quantitative description of the probability of growth (30). Therefore, the factors that influence the growth potential of STEC in edible plants include plant-dependent and physiochemical factors, as well as bacterial isolate-specific responses. Metabolically active components of plants can be extrapolated from plant extracts for bacterial growth dynamic measurements coupled with metabolite analysis. They also represent a bacterial growth substrate in their own right that could arise during the postharvest production process, e.g., from cut surfaces. A number of studies have shown the growth of foodborne bacteria on plant extracts during the production process (33–35), and the growth potential for E. coli O157:H7 has been evaluated in water (31). Here, maximum growth rates in plant extracts were strongly influenced by the plant tissue type and species, as well as the E. coli isolate tested, and were overlaid by temperature-dependent effects. In planta growth rates, however, were markedly different between the sprouted seeds and leafy vegetables, with a growth restriction being evident in the leafy vegetables. The plant-dependent factors that could account for this difference include plant age, defense response, growth conditions, and associated microbiomes. As such, the growth rates in the extracts could not be used to infer the in planta growth potential for spinach or lettuce. In contrast, proliferation on sprouted seeds did bear a positive relationship to the growth rates in extracts, although it was also dependent on the plant species and on the bacterial isolate tested.
Saccharides were shown to be the major driving force for E. coli growth, which is unsurprising, given their role in central metabolism (36). Although the levels of the most abundant sugars, glucose, fructose, and sucrose (the disaccharide of glucose and fructose), could explain the high growth rates in sprout extracts, similarly rapid growth did not occur in lettuce leaf lysate extract, despite the presence of an abundance of sugars, indicating that plant species-specific inhibitory compounds exist. This is supported by the occurrence of more rapid growth rates in spinach leaf extracts than in lettuce. Plant-dependent factors that could influence bacterial growth potential include the innate defense response (37) and the antimicrobial activity of plant secondary metabolites (38). Plant development stage is an important factor, since sprouted seeds, which were abundant in glucose and fructose, are at a developmental stage distinct from that of mature plants, and young plants of a variety of species can serve as preferential secondary plant hosts for STEC (39).
Bacterial growth rates were not significantly impacted by manipulation of the major amino or organic acids from the extracts, although the phenolic acid chlorogenate (trans-5-O-caffeoyl-d-quinate) was positively associated with growth. This is in contrast to reports of its ability to inhibit fatty acid synthesis in E. coli isolate MG1655 (40) and prevent E. coli growth (41) but may be explained by differences in concentration between the extracts and exogenous application. Oxalate levels were relatively high in spinach, in keeping with previous reports that show an average level as high as ∼1,000 mg/100 g (fresh weight) (42), and correlated with growth for isolate Sakai at 25°C. Amino acids levels were substantially higher in sprouted seed extracts than in the leafy vegetables, which is likely a reflection of the different developmental stages of the plants (43). It was notable that the artificial media did not support growth rates equivalent to those of the complete, natural extract medium, indicating that other, minor nutrients in the extracts were utilized for maximal bacterial growth and also need to be accounted for in growth dynamics.
Bacteria, including STEC, tend to form biofilms in association with plant tissue (25, 39, 44). Here, a risk ranking could be inferred from biofilm formation in the extracts, with spinach roots being ranked the highest. Curli is an important biofilm component for STEC associated with plants (45), but other biofilm components are likely to be responsible for the biofilm formation in extracts, since isolate Sakai did not form biofilms in the spinach apoplast extract in vitro, although it does produce curli during endophytic colonization and biofilm formation in leaves (25). This indicates that specific in planta cues induce different biofilm components. Alternative biofilm components that may be involved include type 1 fimbriae, which were shown to be expressed by the environmental isolates JHI5025 and JIH5039 at 20°C and which promoted binding to spinach roots (46).
Internalization of STEC into apoplastic spaces in plants presents a hazard, as pathogens cannot be removed from apoplastic spaces by conventional sanitation methods. However, the growth potential for internalized E. coli O157:H7 could not be inferred from growth in apoplast extracts since endophytic proliferation in the apoplast was prevented or reduced (25). As the apoplast is a habitat for plant-associated endophytes (47) and phytopathogens (48), it appears that for E. coli additional factors, such as the plant defense response, need to be considered. The increased likelihood of internalization into the tissues of leafy vegetables than into sprouted seeds for the E. coli O157:H7 isolates could be due to multiple factors, including plant age, the competing microbiota, and access to nutrients. Plant-dependent factors have also been shown to impact the colonization of lettuce cultivars by STEC (49).
The in planta colonization by E. coli O157:H7 isolate Sakai was significantly higher than that by isolate ZAP1589 in both leafy tissue types and on both sprouted seed species (25). In contrast, growth rates in the plant extracts and in the artificial media overlapped, albeit with specific extract-specific differences. Since isolate ZAP1589 was found to be flagellate but nonmotile, this may reflect a role for flagella in plant colonization (10). The growth rates for ZAP1589 on sprouted seeds were similar to the rates reported for other E. coli O157:H7 isolates on 2-day-old alfalfa sprouts (50). The growth rates of both E. coli O157:H7 isolates in the extracts were, in general, as high as those of the environmental isolates, indicating similarities in the fitness levels of STEC and environmental E. coli isolates in the plant environment. As anticipated, almost all growth rates were lowest for the laboratory-adapted K-12 isolate, and biofilm formation was essentially absent.
The ability of E. coli isolates to metabolize different carbon sources varies and could contribute to the isolate-dependent variations in growth rates. Although less than 50% of E. coli isolates can metabolize sucrose (36), the E. coli O157:H7 isolate Sakai genome encodes the sucrose transport genes (51), and sucrose degradation genes were expressed by this isolate on exposure to spinach extracts (28). The sucrose translocator from Salmonella enterica serovar Typhimurium was expressed by a related epiphyte in planta (52). In contrast, fructose and glucose are sufficient sole carbon source metabolites for E. coli, and their role in bacterial metabolism is well characterized (36). An E. coli fructose metabolism gene has also been expressed in a related epiphyte in planta (53).
Growth rates normally positively correlate with temperature (54), as was observed for growth rates in the defined medium without plant extracts, which exhibited a linear distribution from 18°C to 25°C. However, maximal growth rates in the extracts were influenced in a nonlinear manner by temperature. Similarly, a nonlinear effect was reported in a meta-study of the growth of STEC on lettuce (55). Since E. coli Sakai exhibits distinct metabolic responses to different plant tissues (28), it is possible that a temperature-dependent effect on metabolite content similarly impacted bacterial metabolism and the resultant growth. This may explain the different organic acid-growth correlations that occurred at 20°C versus 18°C. The implications are that a linear approximation, e.g., such as a Ratkowsky model, is not sufficient to describe E. coli growth in plant extracts, although it has been used to model growth on plants (54, 56).
In conclusion, growth potential in planta was described, in part, by growth rates in plant extracts, but only for sprouted seeds. On the other hand, biofilm formation in plant extracts showed some relation to in planta colonization in leafy vegetables. The plant species- and tissue-type dependent differences in metabolites meant that no single metabolite could be correlated with growth, and the only positive association was with the combined group of saccharides. The marked differences in in planta colonization between the sprouted seeds and leafy vegetables reinforce the higher risk associated with very young plants grown under conditions conducive for bacterial growth (39). Therefore, although these data can inform hazard identification and risk analyses, it is evident that important specificities within each plant-microbe system need to be considered, and it is not possible to take a generalized view of STEC-plant colonization.
MATERIALS AND METHODS
Bacteria and media.
The bacterial isolates panel comprised five isolates: two E. coli O157:H7 isolates, two environmental E. coli isolates, and an E. coli K-12 isolate (Table 4). E. coli ZAP1589 is an Stx-negative derivative generated from isolate H110320350. The regions flanking the stx genes were amplified using primers specific for stx1 and stx2: primers No-stx1 (5ʹ-ttgctggtctcggtacccgggAGTGCTGTGACGATGATGCGATG), Ni-stx1 (5ʹ-cgctcttgcggccgcttggaacggATTACACAATACTCCTTGAGCAC), Co-stx1 (5ʹ-tcccattcgccaccggtcgacGCGGGTCCGGACGGTCATATGTC), Ci-stx1 (5ʹ-ccgttccaagcggccgcaagagcgCAGAATAGCTCAGTGAAAATAGC), No-stx2 (5ʹ-ttgctggtctcggtacccgggCCAAGCACGCCATTGCATCTTAC), Ni-stx2 (5ʹ-cgctcttgcggccgcttggaacggATACAAGGTGTTCCTTTTGGCTG), Co-stx2 (5ʹ-tcccattcgccaccggtcgacAACCTCTCCTGCCGCCAGCAAAG), and Ci-stx2 (5ʹ-ccgttccaagcggccgcaagagcgGGCATAACCTGATTCGTGGTATG) (chromosomal target sequences are in uppercase and cloning sequences are in lowercase). The PCR fragments were cloned into pTOF25 and verified by sequencing. The kanamycin resistance gene from pTOF2 (57) was cloned into the stx1 deletion construct, and the tetracycline resistance gene from pTOF1-TcR (58) was cloned into the stx2 deletion construct. The plasmids were transformed into isolate H110320350 for allelic exchange to delete stx1 and stx2 sequentially, and these were confirmed to be absent by PCR using primers 5ʹ-ATAAATCGCCATTCGTTGACTAC and 5ʹ-AGAACGCCCACTGAGATCATC for stx1 and 5ʹ-GGCACTGTCTGAAACTGCTCC and 5ʹ-TCGCCAGTTATCTGACATTCTG for stx2. The motility of isolate ZAP1589 and isolate H110320350 was tested on motility agar (0.7%), and the presence of the H7 flagellum was confirmed by agglutination with the monoclonal H7 antibody.
TABLE 4.
Bacterial isolates used in this studya
| Isolate name | Serotype | ST | Stx | Source | Reference | Accession no.b |
|---|---|---|---|---|---|---|
| MG1655 | OR:H48 | 98 | NA | Fecal/lab | 65 | NC_000913.3 |
| JHI5025 | ND | 2055 | NA | Soil | 66 | ERS1939526 |
| JHI5039 | ND | 2303 | NA | Root | 66 | ERS1939531 |
| Sakai | O157:H7 | 11 | Negative | Sprout/clinical | 67 | NC_002695.2 |
| ZAP1589 | O157:H7 | 11 | Negative | Leek/clinical | 49c | PRJNA248042 |
ST, sequence type; Stx, Shiga toxin presence; ND, not determined; NA, not applicable. The Sakai isolate used here is the stx-inactivated derivative (67).
GenBank, ENA or BioProject accession numbers are provided for whole genomes.
Isolate ZAP1589, derived from strain H110320350 (68), has both stx-encoding regions removed and is H7 positive but nonmotile.
Bacteria were cultured overnight in lysogeny broth medium (LB) at 37°C (59) with shaking at 200 rpm. Prior to experimentation, an aliquot of the overnight culture was inoculated 1:100 in rich defined 3-(N-morpholino)propanesulfonic acid (MOPS) medium (60) with 0.2% glycerol and essential and nonessential amino acids, termed rich defined MOPS glycerol (RDMG), for 24 h at 18°C and 200 rpm. Bacteria were collected by centrifugation, washed in phosphate-buffered saline (PBS), and adjusted to the required starting optical density at 600 nm (OD600). The medium was supplemented with 30 μg ml−1 kanamycin, if required. Defined artificial lettuce apoplast or sprout extract medium was generated by adding each group of constituents (Table 2) to a base minimal MOPS medium (MMM) lacking a carbon source and amino acids. Each component group was added at the defined concentration to represent the concentrations and composition present in lettuce apoplast or sprout extracts and by dilution of one major group at a time at 1:50 saccharides (SA), 1:10 amino acids (AA), or 1:20 organic acids (OA). The other groups were present at 1:1. The pH of the sprout defined medium was 7.2, and that of the lettuce apoplast defined medium was 7.05. Viable counts were determined from 10-fold dilutions plated on MacConkey (MAC) agar, which were incubated overnight at 37°C and counted manually the next day. All experiments were conducted in triplicate. Viable counts and the OD600 were plotted in Excel 2010 software.
Plant extracts and metabolite analysis.
Lettuce (Lactuca sativa) var. All Year Round and spinach (Spinacia oleracea) var. Amazon were grown individually in 9-cm3 pots in compost for microbiological assays or in vermiculite for metabolite analysis in a greenhouse for 3 weeks. Fenugreek (Trigonella foenum-graecum) and alfalfa (Medicago sativa) seeds were soaked in sterile distilled water (SDW) for 3 h at room temperature (RT), surface sterilized with 3% calcium hypochlorite (20,000 ppm ml−1 active chlorite) for 15 min, washed five times with SDW, and soaked for 2 h in SDW at RT. Sprouts were transferred aseptically on distilled water agar (DWA; 0.5% agar) and sprouted for 2 (alfalfa) or 5 (fenugreek) days at 25°C in the darkness. Leaf apoplastic washings were collected as described previously (see Methods SM3 in the supplemental material), optimized for spinach and lettuce to minimize cytoplasmic contamination (61). All tissue extracts were made as described previously (28). In brief, vermiculite was gently washed off the roots with tap water and rinsed with SDW. Leaves and roots were separated with a sterile scalpel, macerated in liquid nitrogen with a pestle in a mortar, and stored at −20°C until use. The macerated material was preprocessed for sample clarification by mixing 1 g with 20 ml SDW, and the mixture was soaked on a shaker for 4 h and centrifuged at 5,000 relative centrifugal force (rcf) for 15 min. The supernatant was then heated to 50°C for 30 min. The extract was centrifuged at 5,000 rcf for 15 min and filter sterilized through a 0.45-μm-pore-size filter for root tissue or a 0.1-μm-pore-size filter for leaf tissue. Sprouts were macerated in liquid nitrogen, processed as described above without a washing step to remove vermiculite, and filter sterilized through a 0.22-μm-pore-size filter. Apoplast extracts were filtered sterilized through a 0.1-μm-pore-size filter (Durapore; Merck, Germany). Extracts were made from ∼5 plants per sample for leaves and roots and up to 24 plants for apoplastic washings or for sprouts. Ten milliliters of plant extract samples was used for gas chromatography-mass spectrometry (GC-MS) analysis as described in Methods SM4 in the supplemental material. Lysates were prepared for high-performance liquid chromatography (HPLC) as described previously (62).
Growth rate parameterization.
Representative edible species associated with foodborne outbreaks were used: two leafy greens (lettuce, spinach) and two sprouted seeds (fenugreek, alfalfa). The plant tissues used represented edible, nonedible, and internalized tissues of the leafy greens from total lysates of leaves or roots and apoplastic washings recovered from the leaves, respectively, while total sprout lysates were used to represent edible sprouts. A panel of five E. coli isolates was assessed (Table 4) to compare the relative fitness of two STEC O157:H7 Stx-negative isolates to that of two environmental isolates from plant roots and soil. A feces-derived and laboratory-adapted K-12 isolate was included for reference. Growth was assessed at three temperatures (18, 20, and 25°C), to represent the relevant growth temperatures of field-grown leafy greens in northern temperate zones and sprouted seeds grown under controlled conditions. Growth kinetics were measured from optical densities derived from a plate reader (as described by others [30]).
Bacterial growth rates.
Bacterial growth rates were determined using a prewarmed plate reader (Bioscreen C plate reader; Oy Growth Curves Ab Ltd., Finland), set to different temperatures. The E. coli isolates were grown as described above, adjusted to an OD600 of 0.05 in PBS (∼2.1 × 107 CFU ml−1), and inoculated at a 1:10 dilution in plant extracts (at 1:20 [wt/vol] in distilled H2O) or defined media (Table 2) in a 200-μl total volume in multiwell plates. The growth of the E. coli isolates was measured at 18, 20, and 25°C in 100-well microwell plates (Honeycomb; Thermo Fisher, USA). The contents of the wells were randomized in duplicate on the plate, with negative controls being included. All growth curves for extracts were repeated three times with four replicates on the plates. Measurements were recorded every 15 min for 48 h, and the multiwell plates were shaken for 60 s pre- and postmeasurement. The results were exported from plate reader proprietary software as tab-delimited files. For model fitting, the results for 12 replicates of each isolate and medium type were averaged and converted to viable counts [log(number of CFU hour−1)] (Methods SM5 in the supplemental material). A conversion factor of 4.2 × 108 CFU ml−1 was applied so that all growth curves could be modeled using DM-Fit (Methods SM1 in the supplemental material). Secondary modeling was applied for the different temperatures as described in Methods SM2 in the supplemental material. A two-way ANOVA was carried out for multiple comparisons (isolate/extract type) in Prism (v6) software (GraphPad Software Inc., USA).
Biofilms.
Bacterial biofilms were measured as described previously (63). Bacteria were grown aerobically in LB at 37°C for 12 h, subcultured (1:1,000 [vol/vol]) in RDMG for 18 h at 18°C, diluted in PBS to an OD600 of 0.05, inoculated into plant extracts as described for the growth rate determination in a 96-well polystyrene plate, and incubated statically for 48 h at 18°C. The washed wells were stained with 0.1% crystal violet solution and solubilized with 95% ethanol. The solution was transferred into a fresh plate, and the absorbance was measured at 590 nm with a plate reader (Multiskan Go; Thermo Scientific, USA). Results were exported with the software SkanIt (Thermo Scientific, USA) to Microsoft Excel 2010 software for analysis. A two-way ANOVA was carried out for multiple comparisons (isolate/extract type) in Prism (v6) software (GraphPad Software Inc., USA).
Plant colonization assay.
Lettuce and spinach plants (∼3 weeks old) were transferred to a growth chamber (Snijders) at 21°C with 75% humidity and a 16-h light and 8-h dark cycle (400 μE/m2 · s [30,000 lx]) 3 days prior to inoculation and were not watered for ∼18 h prior to inoculation. The roots were inoculated by placing pots in a plastic box containing a 1-liter suspension of E. coli Sakai or ZAP1589 diluted to an OD600 of 0.02 (equivalent to 107 CFU ml−1) in SDW, which partially submerged the pots. After 1 h of inoculation, the pots were transferred to the growth chamber until sampling. Sprouts were inoculated with 103 CFU ml−1 bacteria in 0.5 liter SDW for 1 h, rinsed with 0.5× Murashige and Skoog (MS) basal medium (no sucrose), transferred to petri dishes containing distilled water agar (DWA; 0.8% agar), and incubated for up to 3 days at 25°C. Negative controls were incubated with SDW without bacteria.
Lettuce and spinach roots were sampled at 0, 5, 10, and 14 days postinfection (dpi) and aseptically removed from aerial tissue with a sterile scalpel, the compost was removed by washing with SDW, the roots were transferred into 50-ml tubes and washed with PBS, and the fresh weight was determined. The sprouts were sampled at 0, 1, and 2 dpi, where half of the samples were used to enumerate the total viable counts of E. coli and stored in PBS until further use (∼ 30 min) and surface-associated bacteria were removed from the other half of the samples by surface sterilization with 200 ppm Ca(ClO)2 for lettuce/spinach roots or 20,000 ppm Ca(ClO)2 for sprouts for 15 min. Surface decontamination of sprout tissue required at least 15,000 ppm of Ca(ClO)2 to eradicate the external E. coli bacteria, but endophytes appeared to be protected from the active chlorite since endemic internalized bacteria occurred on the recovery medium after surface decontamination with 20,000 ppm Ca(ClO)2. The root/sprouts were washed five times with PBS to ensure removal of all loosely adherent bacterial cells and residual chlorine. Surface sterilization was validated as described previously (25). Any samples containing surface-associated bacterial colonies were removed from subsequent analysis. Roots/sprouts were macerated using a mortar and pestle in 2 ml PBS and ∼50 mg sterile sand. The supernatant was diluted with PBS once for spinach and lettuce (1:1), three times for fenugreek (1:3), or four times for alfalfa (1:4), and 100 μl was plated on MAC plates using a spiral plater (WASP; Don Whitley Scientific, UK) and incubated for 24 h at 37°C. The colonies on the plates were counted using a counting grid (WASP; Don Whitley Scientific, UK), multiplied by the dilution factor, and converted to the number of CFU milliliter−1. The experiment was repeated three times with five replicate samples per time point, and sprout samples comprised multiple (>15) sprouts. The limit of detection from direct plating was 20 CFU ml−1, below which values were manually leveled to <1 log(CFU ml−1) for lettuce and spinach root data. Since the level of inoculation of sprouts for day 0 was below the detection limit, the numbers were semiquantified by the most probable number (MPN) method for the 3-tube assay, as described by Oblinger and Koburger (64). Samples were diluted 6-fold in buffered peptone water (BPW) and incubated overnight at 37°C, and the results for positive samples were confirmed by plating triplicate 100-μl samples on MAC agar and incubating overnight at 37°C.
Supplementary Material
ACKNOWLEDGMENTS
N.J.H. and S.M. were supported by an FSA grant (FS101056); B.M. was supported by a Ph.D. award to N.J.H., N.J.C.S., F.B., and K.J.F.; and N.J.H. was partly funded by the Rural & Environment Science & Analytical Services Division of the Scottish government.
We are grateful to Susan Verrall and Raymond Campbell (Hutton Institute) for assistance with GC-MS and HPLC and David Gally (University of Edinburgh) for the use of CL3 facilities.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00123-19.
REFERENCES
- 1.Painter JA, Hoekstra RM, Ayers T, Tauxe RV, Braden CR, Angulo FJ, Griffin PM. 2013. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998-2008. Emerg Infect Dis 19:407–415. doi: 10.3201/eid1903.111866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jay MT, Cooley MB, Carychao D, Wiscomb GW, Sweitzer RA, Crawford-Miksza L, Farrar JA, Lau DK, O’Connell J, Millington A, Asmundson RV, Atwill ER, Mandrell RE. 2007. Escherichia coli O157:H7 in feral swine near spinach fields and cattle, central California coast. Emerg Infect Dis 13:1908–1911. doi: 10.3201/eid1312.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchholz U, Bernard H, Werber D, Böhmer MM, Remschmidt C, Wilking H, Deleré Y, An der Heiden M, Adlhoch C, Dreesman J, Ehlers J, Ethelberg S, Faber M, Frank C, Fricke G, Greiner M, Höhle M, Ivarsson S, Jark U, Kirchner M, Koch J, Krause G, Luber P, Rosner B, Stark K, Kühne M. 2011. German outbreak of Escherichia coli O104:H4 associated with sprouts. N Engl J Med 365:1763–1770. doi: 10.1056/NEJMoa1106482. [DOI] [PubMed] [Google Scholar]
- 4.Erickson MC, Liao J, Payton AS, Webb CC, Ma L, Zhang GD, Flitcroft I, Doyle MP, Beuchat LR. 2013. Fate of Escherichia coli O157:H7 and Salmonella in soil and lettuce roots as affected by potential home gardening practices. J Sci Food Agric 93:3841–3849. doi: 10.1002/jsfa.6321. [DOI] [PubMed] [Google Scholar]
- 5.Holden N, Jackson RW, Schikora A. 2015. Plants as alternative hosts for human and animal pathogens. Front Microbiol 6:397. doi: 10.3389/fmicb.2015.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang L. 2012. Mathematical modeling and numerical analysis of the growth of non-O157 Shiga toxin-producing Escherichia coli in spinach leaves. Int J Food Microbiol 160:32–41. doi: 10.1016/j.ijfoodmicro.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 7.Jensen DA, Friedrich LM, Harris LJ, Danyluk MD, Schaffner DW. 2015. Cross contamination of Escherichia coli O157:H7 between lettuce and wash water during home-scale washing. Food Microbiol 46:428–433. doi: 10.1016/j.fm.2014.08.025. [DOI] [PubMed] [Google Scholar]
- 8.Koseki S, Isobe S. 2005. Prediction of pathogen growth on iceberg lettuce under real temperature history during distribution from farm to table. Int J Food Microbiol 104:239–248. doi: 10.1016/j.ijfoodmicro.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 9.Rossez Y, Holmes A, Lodberg-Pedersen H, Birse L, Marshall J, Willats WGT, Toth IK, Holden NJ. 2014. Escherichia coli common pilus (ECP) targets arabinosyl residues in plant cell walls to mediate adhesion to fresh produce plants. J Biol Chem 289:34349–34365. doi: 10.1074/jbc.M114.587717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossez Y, Holmes A, Wolfson EB, Gally DL, Mahajan A, Pedersen HL, Willats WGT, Toth IK, Holden NJ. 2014. Flagella interact with ionic plant lipids to mediate adherence of pathogenic Escherichia coli to fresh produce plants. Environ Microbiol 16:2181–2195. doi: 10.1111/1462-2920.12315. [DOI] [PubMed] [Google Scholar]
- 11.Melotto M, Panchal S, Roy D. 2014. Plant innate immunity against human bacterial pathogens. Front Microbiol 5:411. doi: 10.3389/fmicb.2014.00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seo S, Matthews KR. 2012. Influence of the plant defense response to Escherichia coli O157:H7 cell surface structures on survival of that enteric pathogen on plant surfaces. Appl Environ Microbiol 78:5882–5889. doi: 10.1128/AEM.01095-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elhadidy M, Álvarez-Ordóñez A. 2016. Diversity of survival patterns among Escherichia coli O157:H7 genotypes subjected to food-related stress conditions. Front Microbiol 7:322. doi: 10.3389/fmicb.2016.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Record MT Jr, Courtenay ES, Cayley DS, Guttman HJ. 1998. Responses of E. coli to osmotic stress: large changes in amounts of cytoplasmic solutes and water. Trends Biochem Sci 23:143–148. doi: 10.1016/S0968-0004(98)01196-7. [DOI] [PubMed] [Google Scholar]
- 15.Buchanan RL, Klawitter LA. 1992. The effect of incubation temperature, initial pH, and sodium chloride on the growth kinetics of Escherichia coli O157:H7. Food Microbiol 9:185–196. doi: 10.1016/0740-0020(92)80046-7. [DOI] [Google Scholar]
- 16.Presser KA, Ross T, Ratkowsky DA. 1998. Modelling the growth limits (growth/no growth interface) of Escherichia coli as a function of temperature, pH, lactic acid concentration, and water activity. Appl Environ Microbiol 64:1773–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danyluk MD, Schaffner DW. 2011. Quantitative assessment of the microbial risk of leafy greens from farm to consumption: preliminary framework, data, and risk estimates. J Food Prot 74:700–708. doi: 10.4315/0362-028X.JFP-10-373. [DOI] [PubMed] [Google Scholar]
- 18.Franz E, Tromp SO, Rijgersberg H, van der Fels-Klerx HJ. 2010. Quantitative microbial risk assessment for Escherichia coli O157:H7, Salmonella, and Listeria monocytogenes in leafy green vegetables consumed at salad bars. J Food Prot 73:274–285. doi: 10.4315/0362-028X-73.2.274. [DOI] [PubMed] [Google Scholar]
- 19.Pang H, Lambertini E, Buchanan RL, Schaffner DW, Pradhan AK. 2017. Quantitative microbial risk assessment for Escherichia coli O157:H7 in fresh-cut lettuce. J Food Prot 80:302–311. doi: 10.4315/0362-028X.JFP-16-246. [DOI] [PubMed] [Google Scholar]
- 20.Deering AJ, Mauer LJ, Pruitt RE. 2012. Internalization of E. coli O157:H7 and Salmonella spp. in plants: a review. Food Res Int 45:567–575. doi: 10.1016/j.foodres.2011.06.058. [DOI] [Google Scholar]
- 21.Wright KM, Chapman S, McGeachy K, Humphris S, Campbell E, Toth IK, Holden NJ. 2013. The endophytic lifestyle of Escherichia coli O157:H7: quantification and internal localization in roots. Phytopathology 103:333–340. doi: 10.1094/PHYTO-08-12-0209-FI. [DOI] [PubMed] [Google Scholar]
- 22.Pignocchi C, Foyer CH. 2003. Apoplastic ascorbate metabolism and its role in the regulation of cell signalling. Curr Opin Plant Biol 6:379–389. doi: 10.1016/S1369-5266(03)00069-4. [DOI] [PubMed] [Google Scholar]
- 23.Erlacher A, Cardinale M, Grube M, Berg G. 2015. Biotic stress shifted structure and abundance of Enterobacteriaceae in the lettuce microbiome. PLoS One 10:e0118068. doi: 10.1371/journal.pone.0118068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou Z, Fink RC, Radtke C, Sadowsky MJ, Diez-Gonzalez F. 2013. Incidence of naturally internalized bacteria in lettuce leaves. Int J Food Microbiol 162:260–265. doi: 10.1016/j.ijfoodmicro.2013.01.027. [DOI] [PubMed] [Google Scholar]
- 25.Wright KM, Crozier L, Marshall J, Merget B, Holmes A, Holden NJ. 2017. Differences in internalization and growth of Escherichia coli O157:H7 within the apoplast of edible plants, spinach and lettuce, compared with the model species Nicotiana benthamiana. Microb Biotechnol 10:555–569. doi: 10.1111/1751-7915.12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erickson MC, Webb CC, Davey LE, Payton AS, Flitcroft ID, Doyle MP. 2014. Biotic and abiotic variables affecting internalization and fate of Escherichia coli O157:H7 isolates in leafy green roots. J Food Prot 77:872–879. doi: 10.4315/0362-028X.JFP-13-432. [DOI] [PubMed] [Google Scholar]
- 27.Erickson MC, Webb CC, Diaz-Perez JC, Phatak SC, Silvoy JJ, Davey L, Payton AS, Liao J, Ma L, Doyle MP. 2010. Infrequent internalization of Escherichia coli O157:H7 into field-grown leafy greens. J Food Prot 73:500–506. doi: 10.4315/0362-028X-73.3.500. [DOI] [PubMed] [Google Scholar]
- 28.Crozier L, Hedley P, Morris J, Wagstaff C, Andrews SC, Toth I, Jackson RW, Holden N. 2016. Whole-transcriptome analysis of verocytotoxigenic Escherichia coli O157:H7 (Sakai) suggests plant-species-specific metabolic responses on exposure to spinach and lettuce extracts. Front Microbiol 7:1088. doi: 10.3389/fmicb.2016.01088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van der Linden I, Cottyn B, Uyttendaele M, Vlaemynck G, Heyndrickx M, Maes M, Holden N. 2016. Microarray-based screening of differentially expressed genes of E. coli O157:H7 Sakai during preharvest survival on butterhead lettuce. Agriculture 6:6. doi: 10.3390/agriculture6010006. [DOI] [Google Scholar]
- 30.George SM, Métris A, Baranyi J. 2015. Integrated kinetic and probabilistic modeling of the growth potential of bacterial populations. Appl Environ Microbiol 81:3228–3234. doi: 10.1128/AEM.04018-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vital M, Stucki D, Egli T, Hammes F. 2010. Evaluating the growth potential of pathogenic bacteria in water. Appl Environ Microbiol 76:6477–6484. doi: 10.1128/AEM.00794-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holden N, Pritchard L, Toth I. 2009. Colonization outwith the colon: plants as an alternative environmental reservoir for human pathogenic enterobacteria. FEMS Microbiol Rev 33:689–703. doi: 10.1111/j.1574-6976.2008.00153.x. [DOI] [PubMed] [Google Scholar]
- 33.Koukkidis G, Haigh R, Allcock N, Jordan S, Freestone P. 2017. Salad leaf juices enhance Salmonella growth, colonization of fresh produce, and virulence. Appl Environ Microbiol 83:e02416-16. doi: 10.1128/AEM.02416-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Posada-Izquierdo G, Del Rosal S, Valero A, Zurera G, Sant'Ana AS, Alvarenga VO, Pérez-Rodríguez F. 2016. Assessing the growth of Escherichia coli O157:H7 and Salmonella in spinach, lettuce, parsley and chard extracts at different storage temperatures. J Appl Microbiol 120:1701–1710. doi: 10.1111/jam.13122. [DOI] [PubMed] [Google Scholar]
- 35.Posada-Izquierdo GD, Pérez-Rodríguez F, López-Gálvez F, Allende A, Gil MI, Zurera G. 2014. Modeling growth of Escherichia coli O157:H7 in fresh-cut lettuce treated with neutral electrolyzed water and under modified atmosphere packaging. Int J Food Microbiol 177:1–8. doi: 10.1016/j.ijfoodmicro.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 36.Mayer C, Boos W. 2005, posting date Hexose/pentose and hexitol/pentitol metabolism. EcoSal Plus 2005. doi: 10.1128/ecosalplus.3.4.1. [DOI] [PubMed] [Google Scholar]
- 37.Klerks MM, Franz E, van Gent-Pelzer M, Zijlstra C, van Bruggen AH. 2007. Differential interaction of Salmonella enterica serovars with lettuce cultivars and plant-microbe factors influencing the colonization efficiency. ISME J 1:620–631. doi: 10.1038/ismej.2007.82. [DOI] [PubMed] [Google Scholar]
- 38.Wallace RJ. 2004. Antimicrobial properties of plant secondary metabolites. Proc Nutr Soc 63:621–629. doi: 10.1079/PNS2004393. [DOI] [PubMed] [Google Scholar]
- 39.Wright KM, Holden NJ. 2018. Quantification and colonisation dynamics of Escherichia coli O157:H7 inoculation of microgreens species and plant growth substrates. Int J Food Microbiol 273:1–10. doi: 10.1016/j.ijfoodmicro.2018.02.025. [DOI] [PubMed] [Google Scholar]
- 40.Li B-H, Ma X-F, Wu X-D, Tian W-X. 2006. Inhibitory activity of chlorogenic acid on enzymes involved in the fatty acid synthesis in animals and bacteria. IUBMB Life 58:39–46. doi: 10.1080/15216540500507408. [DOI] [PubMed] [Google Scholar]
- 41.Zheng Y, Liu J, Cao ML, Deng JM, Kou J. 2016. Extrication process of chlorogenic acid in Crofton weed and antibacterial mechanism of chlorogenic acid on Escherichia coli. J Environ Biol 37(5 Spec No):1049–1055. [PubMed] [Google Scholar]
- 42.Mou B. 2008. Evaluation of oxalate concentration in the U.S. spinach germplasm collection. HortScience 43:1690–1693. doi: 10.21273/HORTSCI.43.6.1690. [DOI] [Google Scholar]
- 43.Bewley JD, Black M. 1978. Physiology and biochemistry of seeds in relation to germination: 1 development, germination, and growth. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 44.Danhorn T, Fuqua C. 2007. Biofilm formation by plant-associated bacteria. Annu Rev Microbiol 61:401–422. doi: 10.1146/annurev.micro.61.080706.093316. [DOI] [PubMed] [Google Scholar]
- 45.Carter MQ, Louie JW, Feng D, Zhong W, Brandl MT. 2016. Curli fimbriae are conditionally required in Escherichia coli O157:H7 for initial attachment and biofilm formation. Food Microbiol 57:81–89. doi: 10.1016/j.fm.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 46.Marshall J, Rossez Y, Mainda G, Gally DL, Daniell T, Holden N. 2016. Alternate thermoregulation and functional binding of Escherichia coli type 1 fimbriae in environmental and animal isolates. FEMS Microbiol Lett 363:fnw251. doi: 10.1093/femsle/fnw251. [DOI] [PubMed] [Google Scholar]
- 47.Sattelmacher B. 2001. The apoplast and its significance for plant mineral nutrition. New Phytol 149:167–192. doi: 10.1046/j.1469-8137.2001.00034.x. [DOI] [PubMed] [Google Scholar]
- 48.Rico A, Preston GM. 2008. Pseudomonas syringae pv. tomato DC3000 uses constitutive and apoplast-induced nutrient assimilation pathways to catabolize nutrients that are abundant in the tomato apoplast. Mol Plant Microbe Interact 21:269–282. doi: 10.1094/MPMI-21-2-0269. [DOI] [PubMed] [Google Scholar]
- 49.Quilliam RS, Williams AP, Jones DL. 2012. Lettuce cultivar mediates both phyllosphere and rhizosphere activity of Escherichia coli O157:H7. PLoS One 7:e33842. doi: 10.1371/journal.pone.0033842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Charkowski AO, Barak JD, Sarreal CZ, Mandrell RE. 2002. Differences in growth of Salmonella enterica and Escherichia coli O157:H7 on alfalfa sprouts. Appl Environ Microbiol 68:3114–3120. doi: 10.1128/AEM.68.6.3114-3120.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baumler DJ, Peplinski RG, Reed JL, Glasner JD, Perna NT. 2011. The evolution of metabolic networks of E. coli. BMC Syst Biol 5:21. doi: 10.1186/1752-0509-5-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller WG, Brandl MT, Quiñones B, Lindow SE. 2001. Biological sensor for sucrose availability: relative sensitivities of various reporter genes. Appl Environ Microbiol 67:1308–1317. doi: 10.1128/AEM.67.3.1308-1317.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leveau JHJ, Lindow SE. 2001. Appetite of an epiphyte: quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc Natl Acad Sci U S A 98:3446–3453. doi: 10.1073/pnas.061629598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ratkowsky DA, Lowry RK, McMeekin TA, Stokes AN, Chandler RE. 1983. Model for bacterial culture growth rate throughout the entire biokinetic temperature range. J Bacteriol 154:1222–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McKellar RC, Delaquis P. 2011. Development of a dynamic growth-death model for Escherichia coli O157:H7 in minimally processed leafy green vegetables. Int J Food Microbiol 151:7–14. doi: 10.1016/j.ijfoodmicro.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 56.McKellar RC, Lu X. 2004. Modeling microbial responses in food. CRC Press LLC, Boca Raton, FL. [Google Scholar]
- 57.Merlin C, McAteer S, Masters M. 2002. Tools for characterization of Escherichia coli genes of unknown function. J Bacteriol 184:4573–4581. doi: 10.1128/JB.184.16.4573-4581.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tree JJ, Granneman S, McAteer SP, Tollervey D, Gally DL. 2014. Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli. Mol Cell 55:199–213. doi: 10.1016/j.molcel.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bertani G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J Bacteriol 186:595–600. doi: 10.1128/JB.186.3.595-600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J Bacteriol 119:736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lohaus G, Pennewiss K, Sattelmacher B, Hussmann M, Hermann Muehling K. 2001. Is the infiltration-centrifugation technique appropriate for the isolation of apoplastic fluid? A critical evaluation with different plant species. Physiol Plant 111:457–465. doi: 10.1034/j.1399-3054.2001.1110405.x. [DOI] [PubMed] [Google Scholar]
- 62.Shepherd LV, McNicol JW, Razzo R, Taylor MA, Davies HV. 2006. Assessing the potential for unintended effects in genetically modified potatoes perturbed in metabolic and developmental processes. Targeted analysis of key nutrients and anti-nutrients. Transgenic Res 15:409–425. doi: 10.1007/s11248-006-0012-5. [DOI] [PubMed] [Google Scholar]
- 63.Merritt JH, Kadouri DE, O’Toole GA. 2005. Growing and analyzing static biofilms. Curr Protoc Microbiol Chapter 1:Unit 1B.1. doi: 10.1002/9780471729259.mc01b01s00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oblinger JL, Koburger JA. 1975. Understanding and teaching the most probable number technique. J Milk Food Technol 38:540–545. doi: 10.4315/0022-2747-38.9.540. [DOI] [Google Scholar]
- 65.Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, Horiuchi T. 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol Syst Biol 2:2006.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holden NJ, Wright F, MacKenzie K, Marshall J, Mitchell S, Mahajan A, Wheatley R, Daniell TJ. 2014. Prevalence and diversity of Escherichia coli isolated from a barley trial supplemented with bulky organic soil amendments: green compost and bovine slurry. Lett Appl Microbiol 58:205–212. doi: 10.1111/lam.12180. [DOI] [PubMed] [Google Scholar]
- 67.Dahan S, Knutton S, Shaw RK, Crepin VF, Dougan G, Frankel G. 2004. Transcriptome of enterohemorrhagic Escherichia coli O157 adhering to eukaryotic plasma membranes. Infect Immun 72:5452–5459. doi: 10.1128/IAI.72.9.5452-5459.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perry N, Cheasty T, Dallman T, Launders N, Willshaw G. 2013. Application of multilocus variable number tandem repeat analysis to monitor Verocytotoxin-producing Escherichia coli O157 phage type 8 in England and Wales: emergence of a profile associated with a national outbreak. J Appl Microbiol 115:1052–1058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.