Abstract
BRMS1 is a 246-residue-long protein belonging to the family of metastasis suppressors. It is a predominantly nuclear protein, although it can also function in the cytoplasm. At its C terminus, it has a region that is predicted to be a nuclear localization sequence (NLS); this region, NLS2, is necessary for metastasis suppression. We have studied in vitro and in silico the conformational preferences in aqueous solution of a peptide (NLS2-pep) that comprises the NLS2 of BRMS1, to test whether it has a preferred conformation that could be responsible for its function. Our spectroscopic (far-UV circular dichroism, DOSY-NMR and 2D-NMR) and computational (all-atom molecular dynamics) results indicate that NLS2-pep was disordered in aqueous solution. Furthermore, it did not acquire a structure even when experiments were performed in a more hydrophobic environment, such as the one provided by 2,2,2-trifluoroethanol (TFE). The hydrodynamic radius of the peptide in water was identical to that of a random-coil sequence, in agreement with both our molecular simulations and other theoretical predictions. Thus, we suggest that NLS2 is a disordered region, with non pre-formed structure, that participates in metastasis suppression.
Keywords: Nuclear localization sequence, Nuclear magnetic resonance, Circular dichroism, Molecular dynamics
1. Introduction
Many types of cancer are relatively curable in the case they are diagnosed at an early stage. However, if neoplastic cells are spread toward organs, where they establish colonies, the prognosis is often poor. Thus, it is widely acknowledged that understanding the mechanism of metastasis, and the molecules that modulate it, will lead to improvements to cancer treatment. Several genetic steps are necessary during metastasis, including the reduced expression of different genes in aggressive human tumours. Restoration of normal levels for some of these genes, in metastatic tumour cell lines, yields a significant reduction of the metastasis in vivo without effects on primary tumour growth [1]. The molecular events triggered by this family of genes, known as metastasis suppressors, are not fully known, although their role seems to be key at the final stages of the metastatic cascade [2].
Breast cancer metastasis suppressor 1 (BRMS1) is a 246-residue-long member of the family of metastasis suppressors, which was identified by differential display [3]. It significantly decreases metastasis without blocking breast, ovarian carcinoma or melanoma primary tumour growth [3,4]. BRMS1 transcriptional repression reduces metastasis and appears to be epigenetically regulated [4,5]. Furthermore, BRMS1 acts at the levels of cell-cell communication and abrogation of phosphoinositide signalling [4,6], probably forming complexes with proteins involved in the regulation of transcription [4,7–9], specifically with chromatin remodelling complexes [4,10]. Despite its wide biomedical interest, molecular characterization of BRMS1, including structural studies, remains largely elusive, and only recently the X-ray structure of the region comprising its nuclear export signal has been solved [11,12]. In solution, this region appears to be disordered, whereas in the crystal, it forms an oligomeric coiled-coil.
BRMS1 is a predominantly nuclear protein that inhibits melanoma cell invasion [13], although recent studies have shown its localization also in the cytoplasm, where it inhibits tumour progression. BRMS1 has two predicted nuclear localization sequences (NLSs) located at its C terminus: NLS1, comprising residues 198–205, and NLS2, including residues 238–244 [11]. NLSs are specific polypeptide patches, which act as signals and mediate the active transport of proteins through the nuclear envelope, a double membrane that is penetrated by the nuclear pore complexes, allowing macromolecular exchange between the nucleus and the cytoplasm [14]. We have previously shown that NLS2 was not necessary for nuclear localization of BRMS1 (because of its small size), but it was important for metastasis suppression [15]. Therefore, it is interesting to find out whether isolated NLS2 has any structural propensity under physiological conditions.
In this work, we have investigated the behaviour in solution of an NLS2-mimicking cognate peptide (hereafter named NLS2-pep), which closely reproduces the sequence of the parent NLS2 peptide. In particular, by means of NMR and far-UV circular dichroism (CD), we examined whether NLS2-pep had any tendency to adopt a well defined conformation in aqueous solution. Furthermore, we carried out molecular dynamics (MD) simulations to obtain atomic details on its molecular structure. Our in vitro and in silico results show that isolated NLS2-pep was disordered in solution and that it did not have any tendency to adopt a folded conformation, even in the presence of a more hydrophobic environment, like the one provided by TFE. Therefore, the involvement of NLS2 in metastasis suppression did not implicate the presence of any pre-formed type of secondary structure.
2. Materials and methods
2.1. Materials
Deuterium oxide, ampicillin, GdmCl and isopropyl-β-D-1-tiogalactopyranoside (IPTG) were obtained from Apollo Scientific (Stockport, UK). Sodium trimethylsilyl [2,2,3,3-2H4] propionate (TSP), imidazole, Trizma base, urea, TFE, and deuterated acetic acid and its sodium salt, were from Sigma-Aldrich (Madrid, Spain). GST-resin was from GE Healthcare (Barcelona, Spain). Dialysis tubing, with a molecular weight cut-off of 3500 Da, was from Spectrapor (Spectrum Laboratories, Shiga, Japan). Water was deionized and purified on a Millipore system.
2.2. NLS2-pep design
Since NLS2 is necessary for metastasis suppression, we attempted to express such region in a pET41a vector, with a GST-tag at its N terminus and a TEV cleavage site (Fig. S1). However, although expression was satisfactory in BL21 (DE3) and C41 E. coli strains for the construct after IPTG induction, the expressed protein was accumulated as inclusion bodies (IBs) within the cell. Rescue of these IBs was carried out in the presence of either 8M urea or 7M GdmCl. Attempts to remove the denaturant by using dialysis led to precipitation; furthermore, attempts to remove the denaturant during the purification in the GST-resin led to precipitation over the resin. The same happened when we tried to express the whole NLS1-linker-NLS2 region (residues 198 to 244 of BRMS1) (Fig. S1). Therefore, we decided to reproduce only NLS2 by using peptide synthesis. The sequence of the designed peptide, NLS2-pep, was YK231ARAAVSPQKRKSDGP246 (here and throughout the whole text, the numbering of residues refers to human BRMS1). The Y at the N terminus, which is not present in the sequence of BRMS1, was added to allow a direct measurement of peptide concentration by calculating the molar extinction coefficient of the isolated Tyr [16]. This residue was also acetylated, to remove the N-terminal charge which is not present in NLS2 along the backbone of BRMS1. Furthermore, the C-terminal Pro was amidated for three distinct reasons: to improve the stability and increase the half-life of the peptide in solution; to mimic a longer protein main chain due to the potential presence of other C-terminal amino acid residues, as it happens in at least one of BRMS1 isoforms; and to remove any negative charge in an otherwise highly positively charged peptide.
2.3. CD
The far-UV CD spectra were collected on a Jasco J815 spectropolarimeter (Jasco, USA) fitted with a thermostated cell holder, and interfaced with a Peltier unit. The instrument was periodically calibrated with (+)-10-camphorsulphonic acid. Data in aqueous solution or in the presence of co-solvent (TFE) were acquired with the same experimental set-up already reported [17]. Typically, steady-state spectra were performed at a scan speed of 20 nm/min with a response time of 2 s and averaged over 6 scans with a bandwidth of 1 nm. Spectra were corrected by subtracting the baseline in all cases. A concentration of 60 μM of peptide was used in buffer phosphate (50 mM, pH 7.0) at a temperature of 5 °C. Molar ellipticity was calculated as previously described [17].
TFE titrations were carried out at the same pH and temperature as those in aqueous solution, with a range of co-solvent concentrations from 0 to 80%. Peptide concentration was 30 μM in the TFE titrations.
2.4. NMR spectroscopy
The NMR data were acquired on a Bruker Avance DRX-500 (Bruker GmbH, Germany) spectrometer equipped with a triple-resonance probe and z-gradients; on a Bruker AMX 600 (Bruker GmbH, Germany) spectrometer equipped with a triple-resonance probe and z-gradients; and on a Bruker AV 800 (Bruker GmbH, Germany) spectrometer equipped with an inverse triple-resonance TCI cryoprobe and z-gradients. All experiments were carried out at 5 °C in the three spectrometers in deuterated acetate buffer (50 mM, pH 4.5, not corrected by isotope effects). The NLS2-pep concentration was in all cases ~1.8 mM.
-
1D 1H-NMR spectra- Homonuclear 1D1H NMR experiments of NLS2-pep in aqueous solution were performed in the 500 and 800 MHz spectrometers, with 16 K points, 128 scans and a spectral width of 5000 Hz (10 ppm). Signal from water was suppressed with the WATERGATE sequence [18]. Baseline correction and zero-filling were applied before data processing. TSP was used as the internal chemical shift reference, taking into account the pH-dependence of its resonance [19]. Experiments were also carried out in 100% D2O in the 500 MHz spectrometer, at the same pH (not corrected by isotope effects), by dissolving the corresponding amount of peptide in D2O and acquiring the spectrum immediately thereafter.
Experiments with the peptide in 80% TFE at pH 4.5 were carried out in the 600 MHz spectrometer, by using the same experimental set-up described above.
- Translational diffusion measurements- DOSY experiments were carried out in the 500 MHz spectrometer with the pulse-field gradient (PFG) spin-echo sequence, as described previously [17] and in the presence of 2% of dioxane, with sixteen gradient strengths ranging linearly from 2 to 95% of the total power of the gradient unit. Samples were prepared by dissolving the corresponding amount of peptide in D2O buffer. The methyl region was used for intensity measurements, which were fitted to:
where I is the peak intensity of methyl groups; I0 is the maximum peak intensity of the same resonance(s) at the smallest gradient strength; δ is the duration of the gradient (2.5 ms); G is the gradient strength (in T cm−1); Δ is the time between the gradients (250 ms); γH is the gyromagnetic constant of the proton; and, τ is the recovery delay between the bipolar gradients (100 μs).(1) -
2D 1H-NMR spectroscopy- Two-dimensional experiments of the peptide in aqueous solution with a spectral width of 5000 Hz in each dimension were performed in the 500 MHz spectrometer in phase-sensitive mode by using the time-proportional-phase incrementation technique (TPPI) [20]. Standard TOCSY (60 and 80 ms, with the MLEV-17 spin-lock sequence [21]), ROESY (300 ms) and NOESY (200 and 300 ms) experiments were performed. All data were acquired with a matrix size of 4K × 512, and 1 s of relaxation time. Typically, 80 scans were acquired per t1 increment in the TOCSY experiment, and the residual water signal was removed by using the WATERGATE sequence [18]. NOESY and ROESY spectra [22,23] were typically collected with 128 scans per t1 increment, with the WATERGATE sequence. In all the experiments, the data were zero-filled, resolution-enhanced with square sine-bell window functions optimized in each spectrum, baseline-corrected and processed with the Bruker Topspin 2.1 software. 1H NMR resonances were assigned by standard sequential assignment processes [24]. The random-coil chemical shift values of Hα protons were obtained from tabulated data [24], corrected by neighbouring residue effects [25].
The same 2D NMR experiments were acquired in the 600 MHz spectrometer for the sample in 80% TFE at the same pH. All data were collected with a matrix size of 2K × 1K, and 1 s of relaxation time. Typically, 16 to 32 scans were acquired per t1 increment in each of the experiments, and the residual water signal was also removed by using the WATERGATE sequence [18].
A 2D 1H-15N HSQC spectrum [26] was acquired in the 800 MHz spectrometer with 2K in the F2 dimension (with a spectral width of 12 ppm) and 64 in the F1 one (with a spectral width of 40 ppm). The number of scans was 512. Signal from water was suppressed with the WATERGATE sequence; the 1H carrier was located at 4.73 ppm, and the 15N was at 116.5 ppm. The resulting matrix was zero-filled to double the number of original points in all dimensions and shifted squared sine-bell apodization functions were applied in all dimensions prior to Fourier transformation, using the Bruker Topspin 2.1 software. The 15N dimension was calibrated externally as described [19].
2.5. Molecular modelling
All-atom MD was performed with the GROMACS simulation suite [27], following a protocol we have already used for other IDPs and unfolded protein fragments [28,29]. The starting conformation of NLS2-pep was extended along the dihedral main chain angles (φ=−180°, ψ=180°). The force field adopted was AMBER [30], combined with three different water models to test the variability of the simulated conformational ensemble of the peptide. In particular, we used the popular 3-site TIP3P model [31], the more complex 4-site OPC model [32], parameterized to accurately reproduce the electrostatics around a water molecule, and the other 4-site variant TIP4P-D [33], which optimizes the description of London dispersion interactions and therefore improves simulations of the key features of the unfolded protein states [34].
The peptide was centered in a box with the shape of a rhombic dodecahedron and with a minimum distance from the edges of 1 nm, and Cl− counterions were added to neutralize its charge. Structures were sampled for 200 ns in the isothermal-isobaric (NPT) ensemble, after brief equilibration with positional restraints for 50 ps under isothermal-isochoric (NVT) conditions. Details of the reference values and coupling times used for the thermostat/barostat, and other conditions (including the treatment of electrostatic and van der Waals interactions, and long-range dispersion corrections) have been described elsewhere [35,36]. Data analysis was performed with the help of the VMD software [37]; the default distance value (3.2 Å) was used to as cut-off to assess the formation of a salt bridge, and the STRIDE algorithm [38] was employed for determining the peptide secondary structure.
3. Results
3.1. Isolated NLS2-pep was disordered under different solution conditions
To characterize the conformational features of NLS2 in solution, we used far-UV CD and NMR. Since the cognate peptide NLS2-pep has a single Tyr residue at the N terminus, fluorescence does not give any useful information about the conformation of the parent NLS2, and it was only used to determine peptide concentration. In contrast, far-UV CD provides reliable information about the amount of secondary structures. Moreover, NMR gives further insight into the presence of secondary and tertiary structures of the peptide, and the DOSY-NMR measurements provide the translation diffusion coefficient, D, which gives an estimate of the size of the molecule.
-
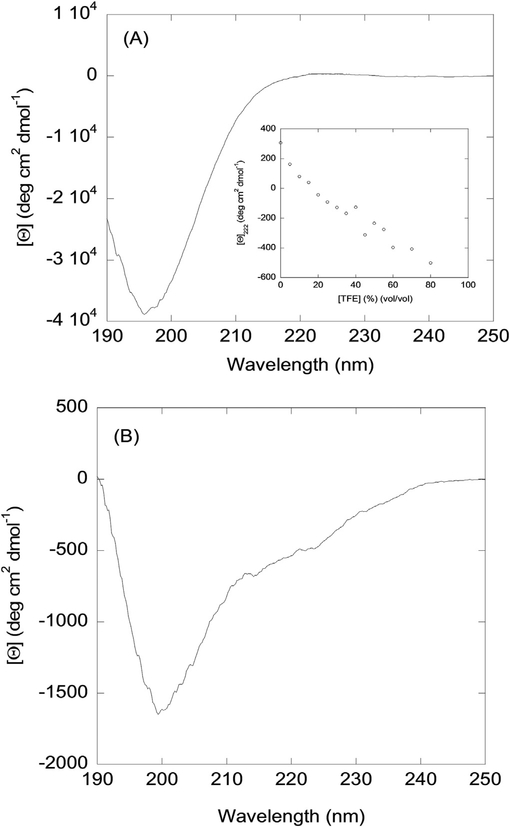
Far-UV CD: The far-UV CD spectrum of NLS2-pep had a minimum at 196 nm, and a small maximum at ~222 nm, suggesting that the peptide was predominantly disordered (Fig. 1 A). The maximum observed at 222 nm could be due to the presence of a little amount of structure resembling β-turns or polyproline type II [39,40]. The deconvolution of the far-UV CD spectrum, using the programs CDSSTR, CONTIN, SELCON3 and k2D from the Dichro-Web online server [41,42], indicated a possible presence of 0–5% of α-helix, 0–25% β-sheet; 20–30% β-turns; and 40–100% random-coil. Therefore, NLS2-pep, from CD measurements, appeared to be mostly disordered.
We also carried out TFE titrations of NLS2-pep followed by far-UV CD. The goal of these titrations is to determine whether the presence of a hydrophobic environment makes NLS2-pep acquire a folded conformation. We observed that as the concentration of the co-solvent was raised, there was a linear increase (in absolute value) of the molar ellipticity at 222 nm; thus, we did not observe a sigmoidal curve (Fig. 1 A inset). The spectrum of NLS2-pep at 80% TFE (vol/vol) had a minimum at 200 nm and a wide shoulder at 222 nm (Fig. 1 B). Therefore, NLS2-pep did not acquire any preferred structure conformation, even in a more hydrophobic environment.
-
NMR: We first carried out DOSY-NMR measurements (Fig. 2 A) to find out the size and compactness of NLS2-pep at pH 4.5. The translational diffusion coefficient, D, of NLS2-pep was (1.08 ± 0.06) × 10−6 cm2 s−1. By taking into account the hydrodynamic radius, RS, of dioxane (2.12 Å), and considering its D ((4.92 ± 0.07) × 10−6 cm2 s−1), the estimated RS for NLS2-pep, assuming an approximately globular shape (Stokes-Einstein equation), was 9 ± 1 Å. This value is an underestimation of the RS of NLS2-pep when compared with the one calculated from a theoretical equation obtained from hydrodynamic measurements in other disordered peptides [43], which gave an RS value of 11 ± 2 Å; in fact, the latter is compatible but larger. In either case, from the size determined we can conclude that NLS2-pep was monomeric and disordered.
The 1D 1H NMR spectrum of NLS2-pep showed all the amide protons clustered in a narrow region (Fig. 2 B top); furthermore, there was no dispersion in the methyl region (Fig. 2 B bottom). In addition, experiments in the presence of D2O did not show any protected amide proton, indicating the absence of stable hydrogen-bonds.
Fig. 1. Far-UV CD characterization of NLS2-pep:
(A) Far-UV CD spectrum of NLS2-pep at 5 °C and pH 7.0 in aqueous solution. Inset: The molar ellipticity at 222 nm, [Θ]222, as the concentration of TFE was increased. (B) Far-UV CD spectrum of NLS2-pep at 5 °C and pH 7.0 in 80% TFE (vol/vol).
Fig. 2. NMR characterization of NLS2-pep:
(A) Intensity decay (arbitrary units) of the methyl signals as the pulse field gradient strength was increased (x-axis) in the DOSY experiments. The line is the fitting of Eq. (1) to the data. (B) 1D 1H- NMR spectrum of NLS2-pep in aqueous solution, showing the amide (top) and the methyl (bottom) regions. (C) Summary of NMR data for NLS2-pep in aqueous solution. NOEs are classified into strong, medium or weak, and represented with a different height of the bar underneath the sequence; signal intensity was judged by visual inspection from the ROESY. The Hα NOE with the following Hδ of a proline residue is indicated by an open bar in the row corresponding to the αN sequential contacts. The dashed line in the first line indicate NOE contacts that could not be unambigously assaiged due to signal overlap.
The 2D 1H-15N HSQC spectrum (Fig. S2), as expected from the 1D 1 H NMR spectrum, also showed a lack of dispersion of the signals, with all the amide protons clustered between 8.7 and 8.3 ppm. We also carried out homonuclear 2D 1H NMR experiments in aqueous solution to assign the spectrum of NLS2-pep. The peptide was mainly disordered in solution, as suggested by two lines of evidence. First, the sequencecorrected conformational shifts [24,25] (Δδ) of Hα protons were within the commonly accepted range for random-coil peptides (that is, |Δδ|≤ 0.1 ppm) (Table S1, values in parenthesis in the column of Hα); only that of Ser237 was larger than + 0.1, but that high value was due to the presence of the adjacent Pro238 residue. And second, no long- or medium-range NOEs were detected (Fig. 2C), but rather, only sequential NOEs were observed. The presence of an NOE between the Hα of Ser237 and the Hδ of Pro238, and the absence of other minor NOE signals, both suggest that the Ser237-Pro238 peptide bond adopts mainly a trans conformation [24]. We could not obtain information on the conformation of the other proline bond (Gly245-Pro246) due to the impossibility to assign the Hδ of Pro246 (Table S1). The assignment of the peptide in aqueous solution has been deposited in the BRMB with the accession number 27759.
As a control to test whether more helical structures were formed in the presence of the co-solvent, as indicated by the far-UV CD spectrum (Fig. 1 B), we also carried out the assignment of NSL2pep in the presence of 80% TFE (the highest concentration of co-solvent used). The assignments are reported in Table S2. All the signals had lower dispersion than in aqueous solution, and we could only observe NN(i,i+1) NOEs between the following pairs of residues: Ala232/Arg233; Ala235/Val236; Gln239/Lys240; Lys242/Ser243; and Asp244/Gly245. However, we did not observe the presence of long- or medium-range NOEs. Furthermore, only residues Gln239 and Lys240 showed up-field shifted chemical shifts in the Hα protons (Table S2). Therefore, it seems that the peptide in the presence of those high co-solvent concentrations populated turn-like conformations, whose presence would explain the shoulder at 222 nm in the far-UV CD spectrum under the same conditions (Fig. 1 B). Furthermore, those turn-like conformations are similar to those observed in the peptide in the MD simulations carried out in aqueous solution (see below).
3.2. NLS2-pep ensemble populated a few transient intramolecular structures
The experimental techniques used to characterize the conformation of NLS2-pep in aqueous solution agree in describing this peptide as essentially unstructured, but could provide little details on either the absence or presence of intramolecular residue contacts. To further study the conformation of NLS2-pep at atomic detail, we performed MD simulations in explicit solvent, using three distinct water models: the widely used TIP3P [31], the all-purpose OPC [32], and the TIP4P-D model [33] specifically developed for IDPs. As shown in Fig. 3, an ensemble of coiled peptide conformations was obtained in all cases. The most evident difference was observed in the gyration radius, RG: the values in the three simulations were 10.8 ± 2.0 Å, 13.2 ± 1.7 Å and 13.4 ± 1.4 Å, respectively.
Fig. 3. Simulated structural ensemble of NLS2-pep:
(Top) Conformations obtained by using the water models TIP3P, OPC and TIP4P-D. Sampling is every 10 ns, and the peptide structures are shown in tube representation at increasing simulation time (colour scale: blue → white → red). Superposition was obtained with a least-squares fit on Cα atoms with respect to the starting structure. (Bottom) Distributions of the radius of gyration, RG, in the three corresponding cases. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The value of RG obtained with TIP3P water was apparently in better agreement with our NMR results, but we observed that such a smaller size compared to that obtained in the other two MD runs was due to a tendency towards over-compaction of the NLS2-pep structure. This artefact is commonly observed for simulation of IDPs when the solvent model is not appropriate [33], and was visible as a peak in the distribution of the values of RG in the range 8–9Å (Fig. 3, bottom). In contrast, the structures obtained in the runs performed with the OPC model showed no sign of over-compaction, and only a moderate asymmetry in the size distribution. This effect could be due (at least in part) to the presence of the Pro238 residue at the centre of the NLS2-pep sequence, which confers a boomerang shape to the peptide conformation, and restrains its possibility to extend – whereas there is no molecular restrain that precludes a collapsed conformation. This shape, which is markedly different from a globular one and entails that the centre of mass is outside the volume occupied by the peptide, could explain the discrepancy with the RG value estimated in our DOSY-NMR experiments. The simulation ensemble obtained with TIP4P-D water was similar to the one obtained with the OPC model regarding the peptide shape and both the average and standard deviation of its size, whereas the distribution of RG values was slightly more symmetrical compared with the one obtained with that model.
In summary, simulations with OPC and TIP4P-D water led to an equilibrated ensemble with comparable statistical properties, and are therefore in good reciprocal agreement. The overall findings indicate an inherent propensity of NLS2-pep to maintain an unfolded structure with a curved but relatively extended conformation, possibly due to the electrostatic repulsion because of the abundance of residues with the same (positive) charge in the sequence.
A detailed investigation of the conformational propensity of NLS2-pep was successively carried out on the simulated peptide ensembles obtained with both OPC and TIP4P-D water. The results revealed that the most specific intramolecular interaction consists in the possibility for the negatively charged side-chain of Asp244 to form transient salt-bridges with the three consecutive and positively charged residues Lys240, Arg241 and Lys242. As shown in Fig. 4, simulations with both water models agree that there is a small preference for the formation of the Asp244-Arg241 bond over the other two electrostatic interactions, but the data essentially indicate that it consists of a flickering structure when considered on a time scale ≥1 ns. Similarly, no indication was found of persistent (≥10 ns) secondary structure for NLS2-pep, except for the formation of labile turns in the peptide main chain (with the MD run in OPC water showing a slightly larger propensity to structure formation than the one with TIP4P-D water, Fig. S3).
Fig. 4. Formation of salt bridges in the simulated structures of NLS2-pep:
Bond formation between the negatively charged side chain of Asp244 and the three positively charged and consecutive residues Lys240, Arg241 and Lys242 in simulation with (A) OPC water model and (B) TIP4P-D solvent, as a function of time.
To sum up, the overall simulation results confirm the conclusion obtained by spectroscopic techniques, which described NLS2-pep as essentially unstructured in solution, and contribute to suggest that NLS2 is a totally disordered region within BRMS1.
4. Discussion and conclusions
As a member of the family of metastasis suppressors, BRMS1 plays an active role in blocking tumour progression. This protein has several regions or domains [8,11], and for some of which their structures have been solved [11,12]. In this work we have focused on characterizing the conformation of the second NLS, at the C-terminal region of BMRS1, which is necessary for metastasis suppression [15]. To this aim, we have studied the conformational preference of the cognate peptide NLS2-pep by using a combination of far-UV CD, DOSY-NMR and 2D-NMR, and MD simulations.
Our main finding is that NLS2-pep is a highly-charged region that has not any structure when isolated in aqueous solution. This conclusion follows from a general agreement among the results obtained with all the techniques employed, both in vitro and in silico. In particular, the simulated structure of NLS2-pep only showed the existence of transient salt-bridges and turn-like conformations in a few regions involving the peptide side-chains and backbone, respectively. Furthermore, experiments in the presence of TFE indicate that the peptide did not have any tendency to fold even in the presence of an apolar solvent, which mimics a hydrophobic environment that may more closely resemble the crowded milieu within cells. Only in the presence of a high concentration of solvent (80%), the peptide acquired turn-like conformations, resembling those observed in the in silico experiments in aqueous solution.
We have previously shown that NLS2 was not necessary for nuclear localization of BRMS1 (because of its small size), but it was important for metastasis suppression [15]. However, we do not know at the moment whether BRMS1 can use the active transport system of proteins through the nuclear envelope under stress conditions, and therefore whether in such cases NLS2 would play an active biological role. Therefore, we believe it is instructive to compare the conformational propensities we found for NLS2 with those of other NLSs that act as proper localization sequences. Other NLSs are also disordered within their parental protein [44–48]; for instance, the NLSs of several antigens remain disordered when bound to karyopherin α (importin α) [44,46–48], their natural partner. In other cases, NLSs could show more chameleonic structural properties, as for example the isolated NLS of RelA, the canonical nuclear factor-κB family member. In fact, that specific NLS is disordered in the free state (with evidence of helical propensity in some residues); however, the NLS of RelA folds upon binding to IκBα, adopting a helical conformation [49], whereas it remains disordered when NF-κB, a transcription factor, is bound to DNA [50].
Based on our results in TFE, since NLS2 of BRMS1 does not undergo a disorder-to-order transition in the presence of a higher hydrophobic environment, we speculate that this specific NLS could not acquire a structure in the presence of any canonical binding partner of BRMS1. The unfolded nature of NLS2 may facilitate acquisition of versatile conformations to bind the multiple targets of BRMS1 (even those proteins not included in the nuclear envelope); such an intrinsically disordered state was also observed in solution for the isolated nuclear export signal of BRMS1 [11,12], although this region shows an inherent tendency to self-associate.
To conclude, the evidence we presented that NLS2 is a disordered structure within BRMS1 adds such important sequence involved in metastasis suppression to the increasing list of unfolded regions that are found to perform their biological function without relying on a well-defined conformation. Although difficult to study both from a theoretical point of view and in experiments, these biological systems are a challenge to extend our understanding of the molecular basis of signalling in cells. To this aim, further analysis will be necessary to elucidate the full functional properties of NLS2. Our study represents a step towards this direction, and will provide a contribution in the identification of the basic elements that allow BRMS1 to participate to the metastasis pathway in vivo.
Supplementary Material
Acknowledgements
We thank the two anonymous reviewers for suggestions and discussion. We thank Prof. Paul Fitzpatrick for handling the manuscript. This work was supported by Spanish Ministry of Economy and Competitiveness and European Funds [CTQ2015–64445-R (to JLN)]. Partial support was received from Susan G. Komen for the Cure [SAC11037], the National Foundation for Cancer Research and the American Cancer Society [PF-16–227-O1-CSM]. DP-U acknowledges the use of the “Manuel Rico” NMR laboratory, LMR (CSIC), a Spanish largescale national NMR facility ICTS R-LRB. BR acknowledges kind hospitality and use of computational resources in the European Magnetic Resonance Center (CERM), Sesto Fiorentino (Florence), Italy.
Abbreviations
- BRMS1
breast cancer metastasis suppressor 1
- CD
circular dichroism
- DOSY
diffusion ordered spectroscopy
- GdmCl
guanidine hydrochloride
- GST
glutathione S-transferase
- IB
inclusion body
- IPTG
isopropyl-β-D-1-tiogalactopyranoside
- MD
molecular dynamics
- NLS
nuclear localization sequence
- NLS2-pep
NLS2-mimicking cognate peptide
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhuaser effect
- NOESY
nuclear Overhauser effect spectroscopy
- NPT
isothermal-isobaric ensemble
- NVT
isothermal-isochoric ensemble
- PFG
pulse-field gradient
- ROESY
rotating frame NOE spectroscopy
- TOCSY
total correlation spectroscopy
- TFE
2,2,2-trifluoroethanol
- TSP
Sodium trimethylsilyl [2,2,3,3-2H4] propionate
- UV
ultraviolet
Footnotes
The authors declare no competing interests.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.abb.2019.01.035.
References
- [1].Rinker-Schaeffer CW, O’Keefe JP, Welch DR, Theodorescu D, Metastasis suppressor proteins: discovery, molecular mechanisms, and clinical application, Clin. Canc. Res 12 (2006) 3882–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Steeg PS, Ouatas T, Halverson D, Palmieri D, Salerno M, Metastasis suppressor genes: basic biology and potential clinical use, Clin. Breast Canc 4 (2003) 51–62. [DOI] [PubMed] [Google Scholar]
- [3].Seraj MJ, Samant RS, Verderame MF, Welch DR, Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13, Cancer Res. 60 (2000) 2764–2769. [PubMed] [Google Scholar]
- [4].Welch DR, Manton CA, Hurst DR, Breast cancer metastasis suppressor 1 (BRMS1): robust biological and pathological data but still enigmatic mechanism of action, Adv. Cancer Res 132 (2016) 111–137. [DOI] [PubMed] [Google Scholar]
- [5].Metge BJ, Frost AR, King JA, Dyess DL, Welch DR, Samant RS, Shevde LA, Epigenetic silencing contributes to the loss of BRMS1 expression in breast cancer, Clin. Exp. Metastasis 25 (2008) 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Saunders MM, Seraj MJ, Li Z, Zhou Z, Winter CR, Welch DR, Donahue HJ, Breast cancer metastatic potential correlates with a break-down in homospecific and heterospecific gap junctional intercellular communication, Cancer Res. 61 (2001) 1765–1767. [PubMed] [Google Scholar]
- [7].Cicek M, Fukuyama R, Cicek MS, Sizemore S, Welch DR, Sizemore N, Casey G, BRMS1 contributes to the negative regulation of uPA gene expression through recruitment of HDAC1 to the NF-kappaB binding site of the uPA promoter, Clin. Exp. Metastasis 26 (2009) 229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rivera J, Megías D, Bravo J, Sorting nexin 6 interacts with breast cancer metastasis suppressor-1 and promotes transcriptional repression, J. Cell. Biochem 111 (2010) 1464–1472. [DOI] [PubMed] [Google Scholar]
- [9].Hurst DR, Mehta A, Moore BP, Phadke PA, Meehan WJ, Accavitti MA, Shevde LA, Hopper JE, Xie Y, Welch DR, Samant RS, Breast cancer metastasis suppressor 1 (BRMS1) is stabilized by the Hsp90 chaperone, Biochem. Biophys. Res. Commun 348 (2006) 1429–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hurst DR, Welch RR, Unravelling the enigmatic complexities of of BRMS1-mediated metastasis suppression, FEBS Lett. 585 (2011) 3185–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Spínola-Amilibia M, Rivera J, Ortiz-Lombardía M, Romero A, Neira JL, Bravo J, The structure of the BRMS1 nuclear export signal and SNX6 interacting region reveals an hexamer formed by antiparallel coiled coils, J. Mol. Biol 411 (2011) 1114–1127. [DOI] [PubMed] [Google Scholar]
- [12].Spínola-Amilibia M, Rivera J, Ortiz-Lombardía M, Romero A, Neira JL, Bravo J, BRMS151–98 and BRMS151–84 are crystal oligomeric coiled coils with different oligomerization states, which behave as disordered protein fragments in solution, J. Mol. Biol 425 (2013) 2147–2163. [DOI] [PubMed] [Google Scholar]
- [13].Slipicevic A, Holm R, Emilsen E, Ree-Roseness AK, Welch DR, Mælandsmo GR, Flørenes VA, Cytoplasmic BRMS1 expression in malignant melanoma is associated with increased disease-free survival, BMC Canc. 12 (2012) 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Görlich D, Kutay U, Transport between the cell nucleus and the cytoplasm, Annu. Rev. Cell Dev. Biol 15 (1999) 607–660. [DOI] [PubMed] [Google Scholar]
- [15].Hurst DR, Xie Y, Thomas J:W, Liu J, Edmons MD, Stewart MD, Welch DR, The C-terminal putative nuclear localization sequence of Breast Cancer Metastasis Suppressor 1, BRMS1, is necessary for metastasis suppression, PLoS One 8 (2013) e55966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gill SC, von Hippel PH, Calculation of protein extinction coefficients from amino acid sequence data, Anal. Biochem 182 (1989) 319–326. [DOI] [PubMed] [Google Scholar]
- [17].Neira JL, Hornos F, Bacarizo J, Cámara-Artigas A, Gómez J, The monomeric species of the regulatory domain of Tyrosine Hydroxylase has a low conformational stability, Biochemistry 55 (2016) 6209–6220. [DOI] [PubMed] [Google Scholar]
- [18].Piotto M, Saudek V, Sklenar V, Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions, J. Biomol. NMR 2 (1993) 661–665. [DOI] [PubMed] [Google Scholar]
- [19].Cavanagh JF, Wayne J, Palmer AG III, Skelton NJ, Protein NMR Spectroscopy: Principles and Practice, first ed., Academic Press, San Diego, 1996. [Google Scholar]
- [20].Marion D, Wüthrich K, Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins, Biochem. Biophys. Res. Commun 11 (1983) 967–974. [DOI] [PubMed] [Google Scholar]
- [21].Bax A, Davis DG, MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy, J. Magn. Reson 65 (1985) 355–360. [Google Scholar]
- [22].Kumar A, Ernst RR, Wüthrich K, A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules, Biochem. Biophys. Res. Commun. 95 (1980) 1–6. [DOI] [PubMed] [Google Scholar]
- [23].Bax A, Davis DG, Practical aspects of two-dimensional Transverse NOE spectroscopy, J. Magn. Reson 63 (1985) 207–213. [Google Scholar]
- [24].Wüthrich K, NMR of Proteins and Nucleic Acids, John Wiley and Sons, New York, 1986. [Google Scholar]
- [25].Schwarzinger S, Kroon GK, Foss TR, Chung J, Wright PE, Dyson HJ, Sequence-dependent correction of random coil NMR chemical shifts, J. Am. Chem. Soc 123 (2001) 2970–2978. [DOI] [PubMed] [Google Scholar]
- [26].Bodenhausen G, Ruben DJ, Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy, Chem. Phys. Lett 69 (1980) 185–189. [Google Scholar]
- [27].Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl J, GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers, Software 1–2 (2015) 19–25. [Google Scholar]
- [28].Neira JL, Rizzuti B, Iovanna JL, Determinants of the pKa values of ionizable residues in an intrinsically disordered protein, Arch. Biochem. Biophys 598 (2016) 18–27. [DOI] [PubMed] [Google Scholar]
- [29].Santofimia-Castaño P, Rizzuti B, Abián O, Velázquez-Campoy A, Iovanna JL, Neira JL, Amphipathic helical peptides hamper protein-protein interactions of the intrinsically disordered chromatin nuclear protein 1 (NUPR1), BBA-Gen. Subjects 1862 (2018) 1283–1295. [DOI] [PubMed] [Google Scholar]
- [30].Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE, Improved side-chain torsion potentials for the Amber ff99SB protein force field, Proteins 78 (2010) 1950–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML, Comparison of simple potential functions for simulating liquid water, J. Chem. Phys 79 (1983) 926–935. [Google Scholar]
- [32].Izadi S, Anandakrishnan R, Onufriev AV, Building water models: a different approach, J. Phys. Chem. Lett 5 (2014) 3863–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Piana S, Donchev AG, Robustelli P, Shaw DE, Water dispersion interactions strongly influence simulated structural properties of disordered protein states, J. Phys. Chem. B 119 (2015) 5113–5123. [DOI] [PubMed] [Google Scholar]
- [34].Rizzuti B, Daggett V, Using simulations to provide the framework for experimental protein folding studies, Arch. Biochem. Biophys 531 (2013) 128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rizzuti B, Bartucci R, Sportelli L, Guzzi R, Fatty acid binding into the highest affinity site of human serum albumin observed in molecular dynamics simulation, Arch. Biochem. Biophys 579 (2015) 18–25. [DOI] [PubMed] [Google Scholar]
- [36].Evoli S, Mobley DL, Guzzi R, Rizzuti B, Multiple binding modes of ibuprofen in human serum albumin identified by absolute binding free energy calculations, Phys. Chem. Chem. Phys 18 (2016) 32358–32368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Humphrey W, Dalke A, Schulten K, VMD: visual molecular dynamics, J. Mol. Graph. Model 14 (1996) 33–38. [DOI] [PubMed] [Google Scholar]
- [38].Frishman D, Argos P, Knowledge-based protein secondary structure assignment, Proteins 23 (1995) 566–579. [DOI] [PubMed] [Google Scholar]
- [39].Shi Z, Olson CA, Rose GD, Baldwin RL, Kallenbach NR, Polyproline II structure in a sequence of seven alanine residues, Proc. Natl. Acad. Sci. U.S.A 99 (2002) 9190–9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chemes LB, Alonso LG, Noval MG, de Prat-Gay G, Circular dichroism techniques for the analysis of intrinsically disordered proteins and domains, Methods Mol. Biol 895 (2012) 387–404. [DOI] [PubMed] [Google Scholar]
- [41].Whitmore L, Wallace BA, Protein secondary structure analysis from circular dichroism spectroscopy: methods and reference databases, Biopolymers 89 (2008) 392–400. [DOI] [PubMed] [Google Scholar]
- [42].Whitmore L, Wallace BA, DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data, Nucleic Acids Res. 32 (2004) W668–W673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Danielsson J, Jarvet J, Damberg P, Gräslund A, Translational diffusion measured by PFG-NMR on full length and fragments of the Alzheimer Aβ(1–40) peptide. Determination of hydrodynamic radii of random coil peptides of varying length, Magn. Reson. Chem 40 (2002) S89–S97 2002. [Google Scholar]
- [44].Süel KE, Cansiglou AE, Chook YM, Atomic resolution structures in nuclear transport, Methods 39 (2006) 342–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yamagishi R, Okuyama T, Oba S, Shimada J, Chaen S, Kaneko H, Comprehensive analysis of the dynamic structure of nuclear localization signals, Biochem. Biophys. Reports 4 (2015) 392–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Conti E, Kuriyan J, Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by kariopherin α, Structure 8 (2000) 329–338. [DOI] [PubMed] [Google Scholar]
- [47].Lee BJ, Cansizoglu AE, Süel K:E, Louis TH, Zhang Z, Chook YM, Rules for nuclear localization sequence recognition by kariopherin β2, Cell 126 (2006) 543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Fontes MRM, The T, Toth G, John A, Pavo I, Jans DA, Kobe B, Role for flanking sequences and phosphorylation in the recognition of the simian-virus-40 large T-antigen nuclear localization sequences by importin-α, Biochem. J 375 (2003) 339–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Cervantes CF, Bergqvist S, Kjaergaard M, Kroon G, Sue S-C, Dyson HJ, Komives EA, The RelA nuclear localization signal folds upon binding to IκBα, J. Mol. Biol 405 (2011) 754–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Jacobs MD, Harrison SC, Structure of an Iκbα/NF-κB complex, Cell 95 (1998) 749–758. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.