Viruses utilize viral glycoproteins to efficiently enter target cells during infection. How viruses acquire viral glycoproteins has been studied to understand the pathogenesis of viruses and develop safer and more efficient viral vectors for gene therapies. The CTs of viral glycoproteins have been shown to regulate various stages of glycoprotein biogenesis, but a gap still remains in understanding the molecular mechanism of glycoprotein acquisition and functionality regarding the CT. Here, we studied the mechanism of how specific mutations in the CT of a gammaretroviral envelope glycoprotein distinctly affect infectivity of two different viruses. Different mutations caused failure of glycoproteins to function in a virus-specific manner due to distinct fusion defects, suggesting that there are virus-specific characteristics affecting glycoprotein functionality.
KEYWORDS: Env incorporation, fusogenicity, HIV-1, MLV, R peptide, retrovirus, assembly, cytoplasmic tail, glycoprotein
ABSTRACT
Viruses can incorporate foreign glycoproteins to form infectious particles through a process known as pseudotyping. However, not all glycoproteins are compatible with all viruses. Despite the fact that viral pseudotyping is widely used, what makes a virus/glycoprotein pair compatible is poorly understood. To study this, we chose to analyze a gammaretroviral glycoprotein (Env) whose compatibility with different viruses could be modulated through small changes in its cytoplasmic tail (CT). One form of this glycoprotein is compatible with murine leukemia virus (MLV) particles but incompatible with human immunodeficiency virus type 1 (HIV-1) particles, while the second is compatible with HIV-1 particles but not with MLV particles. To decipher the factors affecting virus-specific Env incompatibility, we characterized Env incorporation, maturation, cell-to-cell fusogenicity, and virus-to-cell fusogenicity of each Env. The HIV-1 particle incompatibility correlated with less efficient cleavage of the R peptide by HIV-1 protease. However, the MLV particle incompatibility was more nuanced. MLV incompatibility appeared to be caused by lack of incorporation into particles, yet incorporation could be restored by further truncating the CT or by using a chimeric MLV Gag protein containing the HIV-1 MA without fully restoring infectivity. The MLV particle incompatibility appeared to be caused in part by fusogenic repression in MLV particles through an unknown mechanism. This study demonstrates that the Env CT can dictate functionality of Env within particles in a virus-specific manner.
IMPORTANCE Viruses utilize viral glycoproteins to efficiently enter target cells during infection. How viruses acquire viral glycoproteins has been studied to understand the pathogenesis of viruses and develop safer and more efficient viral vectors for gene therapies. The CTs of viral glycoproteins have been shown to regulate various stages of glycoprotein biogenesis, but a gap still remains in understanding the molecular mechanism of glycoprotein acquisition and functionality regarding the CT. Here, we studied the mechanism of how specific mutations in the CT of a gammaretroviral envelope glycoprotein distinctly affect infectivity of two different viruses. Different mutations caused failure of glycoproteins to function in a virus-specific manner due to distinct fusion defects, suggesting that there are virus-specific characteristics affecting glycoprotein functionality.
INTRODUCTION
An essential protein encoded by all retroviruses is Env, a transmembrane glycoprotein that mediates viral entry into target cells. Env is translated in the endoplasmic reticulum (ER), transported to the Golgi compartment, and glycosylated and cleaved by a furin-type cellular protease into two subunits, SU (surface) and TM (transmembrane). The two subunits remain associated after cleavage as they traffic to the viral assembly site and are incorporated into viral particles. During entry of virus into the target cell, the SU subunit of Env first binds to the host cell receptor, triggering a conformational change, and the TM subunit subsequently facilitates fusion between the viral membrane and the host cell (1–3).
Some retroviral Env proteins go through a priming step before becoming fusogenically active. For gammaretroviruses, the activated viral protease in the viral particle cleaves off a short segment of the Env cytoplasmic tail (CT), called the R peptide, allowing Env to become fusogenic (4–6). This cleavage affects trimeric interactions of the TM subunit and results in the splaying of the three TM legs, which is essential for receptor triggering (6). Although this type of Env activation has been most extensively studied in gammaretroviruses, the Env proteins from the deltaretrovirus Mason-Pfizer monkey virus (MPMV) and the lentivirus equine infectious anemia virus (EIAV) have also been reported to have R peptides (7–11). Viral maturation can also be involved in priming Env in other retroviruses; in the case of HIV-1, even though a specific R peptide is not present, maturation affects Env conformation and fusogenicity in a CT-dependent manner (12–14).
In addition to the direct regulation of fusion, the Env CT has also been reported to be involved in multiple roles in the viral life cycle, including signaling, trafficking, incorporation, and interaction with host factors (15–19). Importantly, the Env CTs of several proteins have been reported to affect their incorporation into foreign viral particles (8, 20–23). We reported previously that the CT of MLV Env is dispensable for recruitment to HIV assembly sites. However, we also found that MLV Env is selectively packaged into MLV particles over HIV particles if both are present in the same cell and that this selective incorporation into MLV particles was CT dependent, suggesting that specific interactions exist between the CT and the viral particle (24).
In this study, we characterized two distinct Env glycoproteins with changes in their CTs that cause virus-specific incompatibility with either lentiviral (HIV-1) or gammaretroviral (MLV) particles. Our results demonstrate that mutations in the CT can have distinct effects on two different retroviruses, suggesting that the distinct environment within different types of retroviral particles can affect the functionality of Env.
RESULTS
Mutations in the GaLV Env CT dictate virus-specific compatibility.
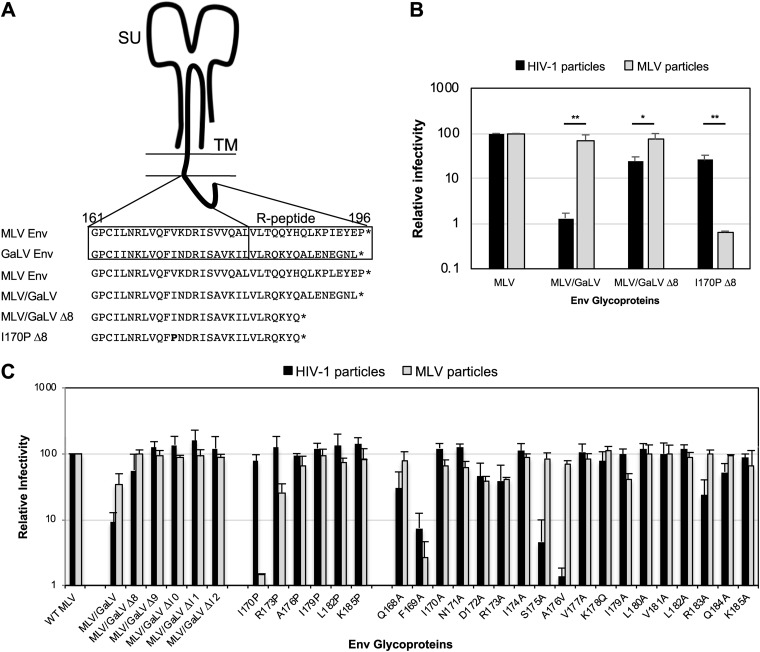
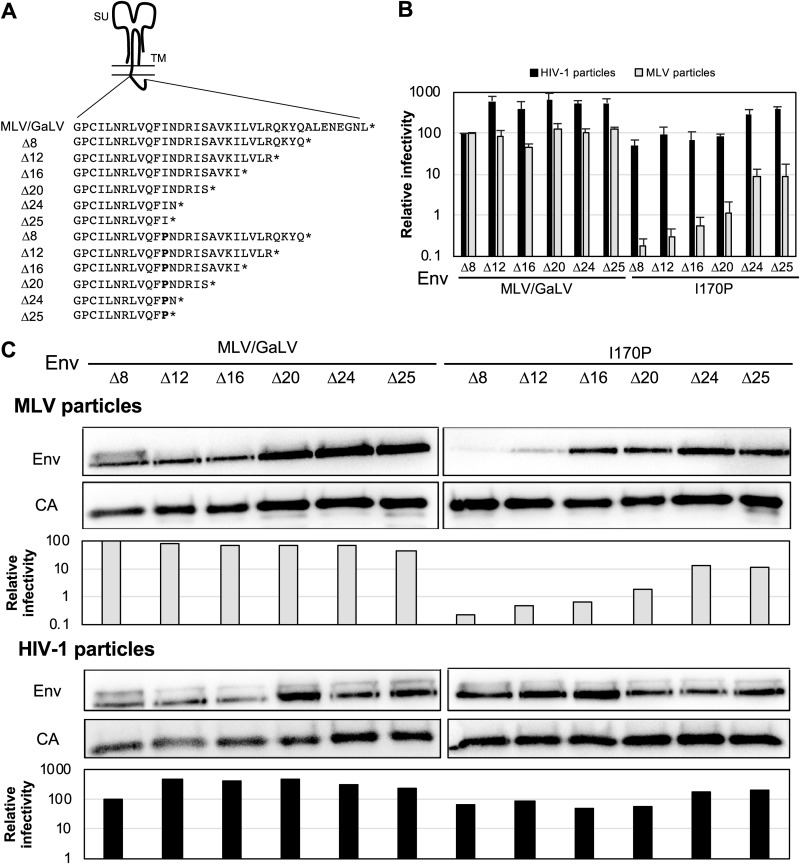
We along with others have previously demonstrated that gibbon ape leukemia virus (GaLV) Env is incompatible with HIV-1 and that this incompatibility is dictated by the CT. This incompatibility can be imparted to MLV Env by replacing the MLV Env CT with the GaLV Env CT (Fig. 1A, MLV/GaLV Env). The incompatibility of the GaLV Env CT was found to be largely dependent on the HIV-1 accessory protein Vpu (25, 26). However, even in the absence of Vpu, HIV-1 infectivity with MLV/GaLV Env is reduced by more than 10-fold compared to that with MLV Env (Fig. 1B). This incompatibility of MLV/GaLV Env with HIV-1 is relieved by deletion of 8 amino acids from the C terminus of the CT (denoted MLV/GaLV Δ8).
FIG 1.
Mutations in the Env cytoplasmic tail cause virus-specific incompatibility. (A) Schematic of Env cytoplasmic tail variants. Numbering follows from the N terminus of MLV Env TM. (B) Relative percent infectivity with different Env variants normalized to that of MLV Env. Averages and standard deviations from three independent experiments are shown. (C) Relative percent infectivity with various mutated Env proteins normalized to that of MLV Env. Numbering follows from the N terminus of MLV Env TM. Averages and standard deviations from three independent experiments are shown. **, P < 0.01; *, P < 0.05 (by unpaired and two-tailed Student's t test for each Env).
We showed previously that the CT of MLV Env can dictate selective packaging of MLV Env into MLV particles (24). While exploring the sequences in the CT that dictate selective packaging into MLV particles, we serendipitously found that an additional point mutation in MLV/GaLV Δ8, I170P (referred to as I26P in the previous report; nomenclature defined by the 170th residue of the MLV TM subunit), resulted in a glycoprotein that was compatible with HIV-1 particles but remarkably less compatible (∼100-fold) with MLV particles (Fig. 1B and C) (27). Henceforth, this construct will be referred to as I170P Δ8. Because these few differences in MLV/GaLV Env and I170P Δ8 present opposite phenotypes in virus-specific compatibility, we sought to identify the factors dictating these incompatibilities.
Vpu-independent HIV-1 particle incompatibility motifs in the GaLV Env CT reside on 171N, 178K, and 179I.
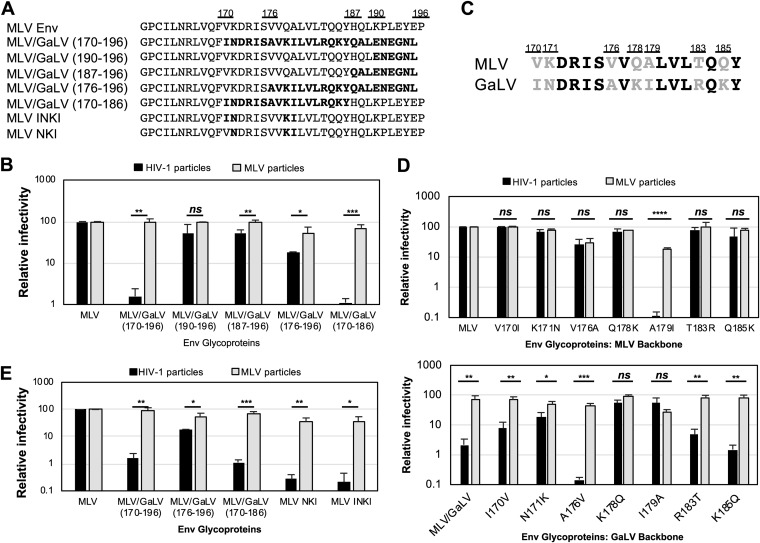
MLV/GaLV Env consisting of residues 170 to 196 [MLV/GaLV(170–196) Env] consistently shows a 10-fold or greater drop in infectivity with HIV-1 particles than that with the parent MLV Env. To narrow down the precise sequence responsible for the incompatibility, several additional chimeras of Env were generated with various amounts of GaLV Env CT sequence (Fig. 2A). Interestingly, when most of the MLV Env CT was exchanged with GaLV sequence (residues 176 to 196, which include the protease cleavage site), the chimeric Env remained more compatible than the MLV/GaLV(170–196) Env with HIV-1 particles (Fig. 2B). However, when the chimeric MLV/GaLV(170–186) Env was generated, the infectivity dropped to a level similar to that of MLV/GaLV(170–196) Env, suggesting that the region between residues 170 and 186 dictates the incompatibility. Among the 17 amino acids in this region, seven positions differ between the MLV and GaLV Env CTs (Fig. 2C). We created point mutations between these residues to determine the precise residues in the GaLV Env CT that dictate HIV-1 particle incompatibility (Fig. 2D). Starting from the MLV Env CT backbone, which is compatible with HIV-1 particles, only one of the substitutions in GaLV Env sequence, A179I, caused a great reduction in infectivity with HIV-1 particles although this mutation also reduced infectivity with MLV particles. This suggests that more than one amino acid change to MLV Env would be required to enforce virus-specific incompatibility. Starting from the GaLV Env CT, which is not compatible with HIV-1 particles, three point mutations, N171K, K178Q, and I179A, alleviated the incompatibility with HIV-1 particles (Fig. 2D). I170V in the GaLV Env backbone showed somewhat moderate recovery of HIV-1 particle infectivity although this mutation still showed a statistically significant difference (P < 0.01). Introduction of the GaLV Env amino acids N171, K178, and I179 or of amino acids I170, N171, K178, and I179 (subsequently referred to as MLV Env NKI and INKI, respectively) into the MLV Env CT was sufficient to make MLV Env incompatible with HIV-1 particles (Fig. 2E). These chimeras retain compatibility with MLV particles.
FIG 2.
GaLV incompatibility with HIV-1 particles is modulated by residues 171N, 178K, and 179I. (A) Amino acid sequences of MLV and GaLV Env cytoplasmic tails. Chimeric mutations are indicated in boldface. (B) Relative percent infectivity with different chimeric Env proteins. (C) Comparison of amino acid sequences between MLV and GaLV in the region of residues 170 to 186. Variable amino acids are indicated in gray font. (D) Relative percent infectivity of Env point mutants. (E) Relative percent infectivity of MLV Env combination mutants. MLV Env proteins with K171N, Q178K, and A179I mutations (MLV NKI) and the V170I additional mutation (MLV INKI) are indicated. Infectivity is normalized to that of MLV Env. Averages and standard deviations from three independent experiments are shown. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant (by unpaired and two-tailed Student's t test for each Env).
MLV Env NKI incompatibility with HIV-1 particles is related to Env maturation while I170P Env incompatibility with MLV particles is related to Env incorporation.
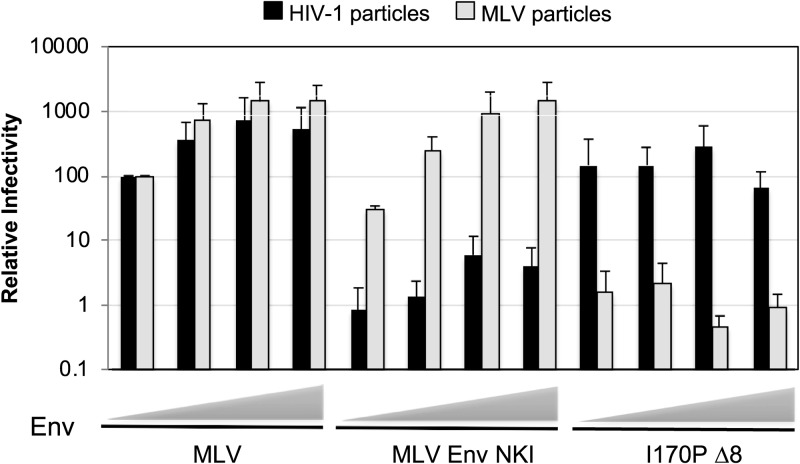
Next, we sought to investigate the mechanism behind virus-specific Env incompatibility of MLV Env NKI and I170P Δ8. First, the effects of increasing amounts of Env in transfections were tested to determine whether the incompatibility is due to the Env expression level (Fig. 3). Viral infectivity with an incompatible Env was not restored with either Env at higher concentrations, suggesting that the incompatibility is not related to Env expression levels.
FIG 3.
Env incompatibility is expression level independent. Relative percent infectivity with increasing amounts of Env (20 ng, 80 ng, 320 ng, and 1,280 ng), normalized to that with 20 ng of MLV Env is shown. Averages and standard deviations from three independent experiments are shown.
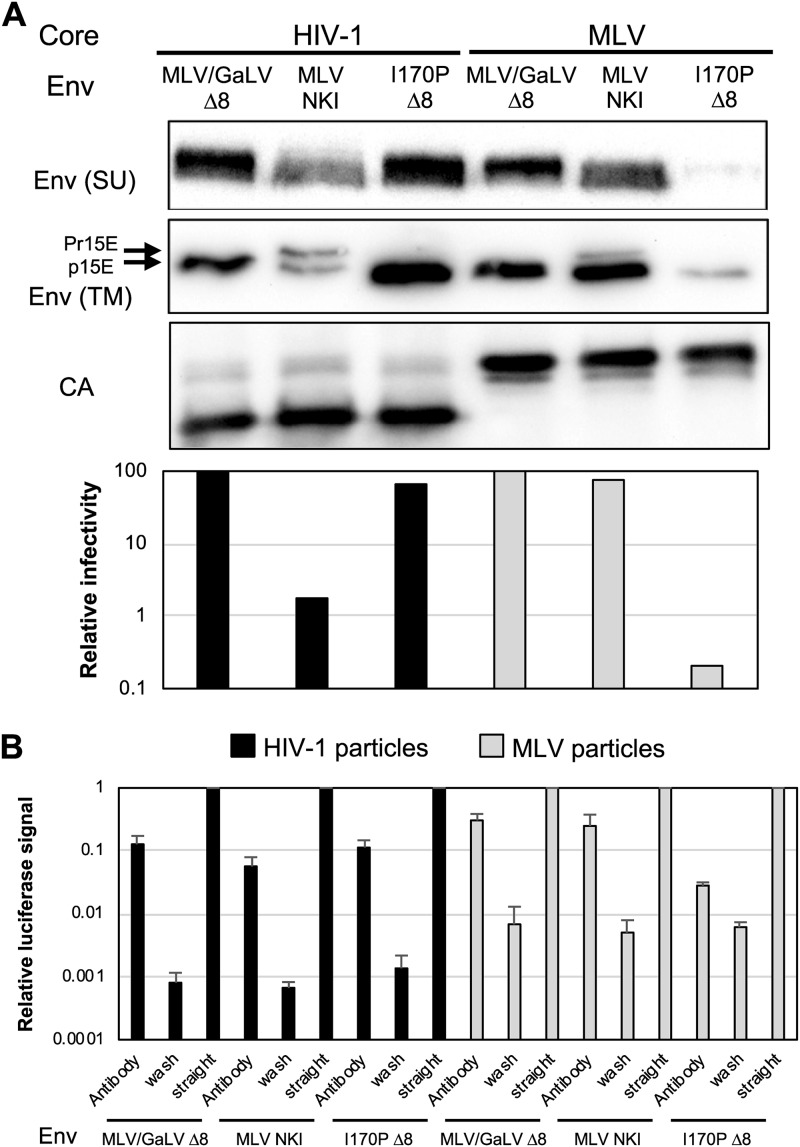
We also examined whether the Env incompatibility was due to loss of Env incorporation. Viruses with each Env were produced and concentrated to conduct Western blotting (Fig. 4A). Incorporation of I170P Δ8 into MLV particles was significantly lower than that in HIV-1 particles, suggesting that the incompatibility is at least in part due to an incorporation block. MLV Env NKI was incorporated into HIV-1 particles slightly less efficiently than into MLV particles though this reduction did not seem to correlate with the large loss of infectivity.
FIG 4.
I170P Env is incorporation defective in MLV particles, but MLV Env NKI is maturation defective in HIV particles. (A) Western blotting was performed with purified virus on SU (HA), p15E TM, and CA. Relative percent infectivity normalized to that of MLV/GaLV Δ8 is shown from the same viral medium used for Western blotting. Shown is a representative example of three independent experiments. (B) Virus capture assay with HIV-1 and MLV iGluc vectors and GFP-tagged Envs. Viruses in the collected supernatant were captured with GFP-antibody coated on ELISA plates, washed, and infected with 293FT cells with cocaptured VSV-G, and Gaussia luciferase activity was measured. Env incorporation was represented by signal from the infected cells with captured virus. Each sample was normalized to level of the positive control (straight infection without wash). Averages and standard deviations from three independent experiments are shown.
Next, we sought to examine whether the incompatibility of incorporated MLV Env NKI is due to a defect in Env activation during virus maturation. To test if the incompatibility is due to the lack of R-peptide cleavage, the same viruses from the experiment described above were also blotted for the Env TM subunit (Fig. 4A). As shown, MLV/GaLV Δ8 was cleaved efficiently in both HIV-1 and MLV particles, while MLV Env NKI was only partially cleaved in HIV-1 particles. Moreover, at least half of the TM domain remained in the noncleaved Pr15E form, suggesting that the MLV Env NKI incompatibility with HIV-1 particles is due to inefficient cleavage, hence, a fusogenic defect. I170P Δ8 was found to be properly cleaved in HIV-1 and MLV particles though very little was present in MLV particles.
Another approach, viral capture, was also used to verify loss of I170P Δ8 incorporation. Previously, we found that different Env glycoproteins can be copackaged into the same viral particles (28). By taking advantage of this copackaging, virus can be captured with an antibody to one glycoprotein, and then the infectivity, driven by the second glycoprotein, can be measured. Thus, the infectivity can be used as a readout for incorporation of the captured glycoprotein into particles. The advantage of this assay is that the signal can only come from incorporation into actual viral particles and not any other forms of glycoprotein shedding. Viral particles were produced with green fluorescent protein (GFP)-tagged Envs and vesicular stomatitis virus G protein (VSV-G). The particles were captured by GFP antibody, and the infectivity was measured by copackaged VSV-G (Fig. 4B). In this experiment, MLV/GaLV Env and I170P Δ8 were incorporated equally into HIV-1 particles, but I170P Δ8 was incorporated at least 10 times less than MLV/GaLV Env into MLV particles.
The I170P Δ8 incorporation defect in MLV particles is alleviated by CT truncations or by replacing MLV MA.
Since the I170P residue is located on the N-terminal portion of the CT, we sought to investigate whether the C terminus of the Env CT contributes to the incompatibility. Additional C-terminal truncations of MLV/GaLV Env and I170P Env were generated (Fig. 5A). As shown in Fig. 5B, these truncations partially restored the infectivity of Envs with the I170P mutation with MLV particles, but even the truncation with the highest infectivity (I170P Δ25, with a deletion of 25 residues in the CT) was still down over 10-fold compared to the level with MLV/GaLV Δ25. To investigate whether the Env incorporation correlated with infectivity, Western blotting was conducted (Fig. 5C). Despite the poor recovery in infectious particle production, these truncations led to efficient Env incorporation into MLV particles. For example, the 16-amino-acid truncation restored incorporation to nearly the wild-type level even though infectivity remained down almost 100-fold. This phenotype was recapitulated with the virus capture assay (data not shown). Each of the truncations was also tested with HIV-1 particles. Incorporation into HIV-1 particles was relatively constant among all of the Envs. Surprisingly, smaller truncations to MLV/GaLV Env (Δ12, Δ16, and Δ20) enhanced the infectivity 3- to 7-fold with HIV-1 particles, but the same truncations in Env proteins with the I170P mutation did not enhance infectivity (Fig. 5B and C).
FIG 5.
I170P Env incompatibility is partially alleviated by truncation of the Env CT. (A) Schematic diagram with amino acid sequences with Env truncations. (B) Relative percent infectivity normalized to that of MLV/GaLV Δ8. The average and standard deviation from three independent experiments for each are shown. (C) Western blot of purified HIV-1 and MLV particles with the indicated Env shows Env incorporation and capsid release. Relative percent infectivity of the medium used in Western blotting, normalized to that of MLV/GaLV Δ8, is shown. Shown is a representative example of two independent experiments.
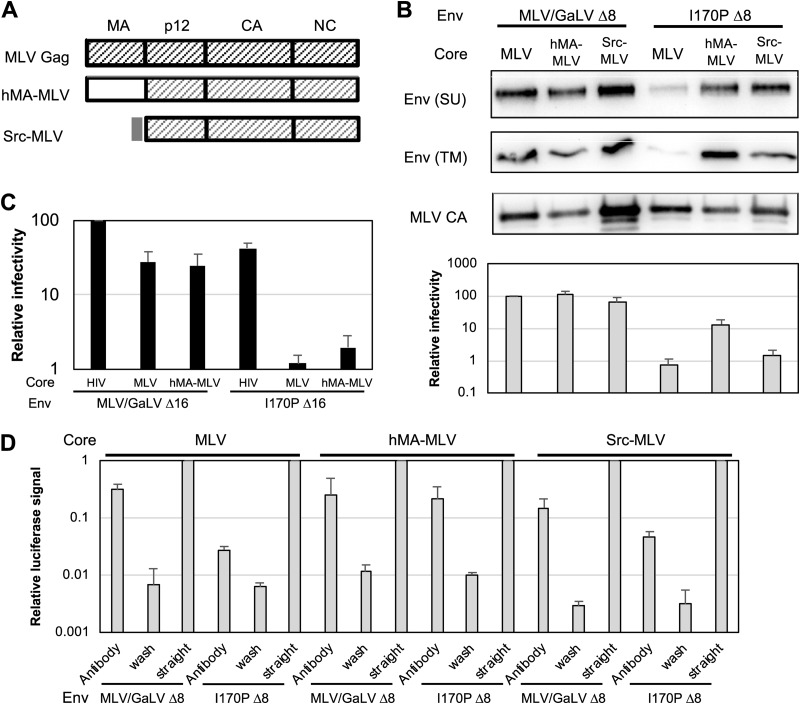
If direct interactions exist between Gag and the Env CT, these interactions would most likely involve the membrane-binding MA domain of Gag. Indeed, several mutations in HIV MA have been shown to affect Env incorporation, and in one case a mutation in HIV MA was even shown to affect R-peptide cleavage of MLV Env (29–37). To test if MA contributes to the I170P incompatibility, the MA domain of MLV Gag was replaced with HIV-1 MA (hMA-MLV) or the short membrane binding domain from Src (Src-MLV) (Fig. 6A). Both of the chimeric proteins were able to incorporate MLV/GaLV Env and form infectious particles (Fig. 6B and D). With I170P Δ8, incorporation was enhanced with both chimeras relative to the level with wild-type MLV Gag, but the infectivity with the Gag chimeras was only modestly increased with I170P Δ8 and remained down ∼8-fold or more relative to that with MLV/GaLV Δ8. To determine if inefficient R-peptide cleavage was occurring, we probed the TM domain and found that it was efficiently cleaved with all three constructs. As further evidence that the I170P defect is not related to maturation, truncation of the R peptide (I170P Δ16) did not restore the infectivity of hMA-MLV (Fig. 6C). These data suggest that the MA domain plays a role in Env incorporation but that there are additional factors that contribute to the virus-specific functionality of the Env.
FIG 6.
The I170P Env incompatibility is partially alleviated by replacing MLV MA. (A) Schematic diagram of chimeric MLV Gag constructs used. (B) Western blot of purified HIV-1 and MLV particles with the indicated Env shows Env incorporation and capsid release. Averages and standard deviations from six independent experiments including infectivity from medium used for Western blotting are shown. The Western blot image is a representative example of two independent experiments. (C) Relative percent infectivity normalized to that of HIV-1 with MLV/GaLV Δ16. Averages and standard deviations from three independent experiments are shown. (D) Virus capture assay with MLV, hMA-MLV, and Src-MLV Gag with iGluc vectors and GFP-tagged Envs. Env incorporation was represented by signal from the infected cells with captured virus. Each sample was normalized to the level of positive control (straight infection without wash). Averages and standard deviations from three independent experiments are shown.
I170P Env retains fusogenicity out of virions but not within the MLV particles.
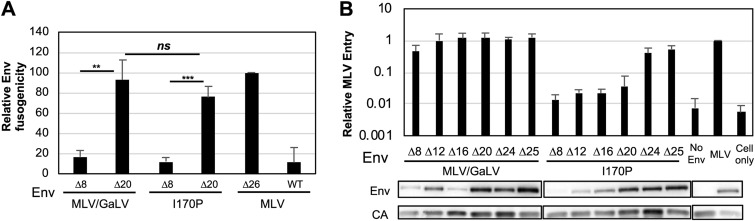
Because the various Env proteins display different levels of functionality depending on the type of viral particles they are incorporated into, we next sought to test the functionality of the Env proteins when expressed alone. To do this, Env constructs were transfected along with the transcriptional activator tTA (tetracycline [Tet]-off) in 293FT cells, and the transfected cells were cocultured with cells stably expressing the MLV Env receptor mCAT-1 (murine cationic amino acid transporter-1) and a Tet-responsive element (TRE)-driven Gaussia luciferase (TRE-Gluc). When the Env on the transfected cell surface fuses with the mCAT-1 receptor on the neighboring cell, tTA is transmitted to the reporter cells and turns on the TRE-induced Gaussia luciferase gene. Luciferase activity was measured and normalized to the level of constitutively active MLV Env Δ26 (Fig. 7A). Both MLV/GaLV Δ8 and I170P Δ8 showed low signals, as expected, since both Envs contain a portion of the R peptide. MLV/GaLV Δ20 and I170P Δ20 were chosen to test the fusogenicity since both Envs lacked an R peptide and were incorporated at close to wild-type levels, yet I170P Δ20 displayed more than 100-fold lower infectivity with MLV particles than MLV/GaLV Δ20 (Fig. 5). As expected MLV/GaLV Δ20 displayed greatly enhanced fusogenicity compared to that of MLV/GaLV Δ8. Fusogenicity of I170P Δ20 was slightly lower than that of MLV/GaLV Δ20 though the difference was not statistically significant.
FIG 7.
I170P Env is fusogenic out of virions but not within MLV particles. (A) Cell-to-cell fusogenic activity of indicated Env proteins relative to that of MLV Δ26 Env. (B) Virus-to-cell fusogenic activity of the indicated Env proteins relative to that of wild-type MLV Env. Env incorporation in MLV Gag-Cre virus was determined by Western blotting. A representative example of two Western blots is shown. Fusion assays represent the averages and standard deviations from at least three independent experiments. **, P < 0.01; ***, P < 0.001; ns, not significant (by unpaired and two-tailed Student's t test).
Next, we proceeded to investigate the fusogenicity of I170P Env proteins within MLV particles. To test this, we used a system developed by Ahi et al. (38) which utilized MLV Gag fused with Cre recombinase and an HT1080/mCAT-1 Cre reporter cell line. When the Env binds the mCAT-1 receptor and mediates fusion, cleaved Cre recombinase enters the cell and induces recombination that turns on the luciferase gene. Luciferase signal was measured and normalized to that of MLV Env. As shown, I170P Δ8, Δ12, Δ16, and Δ20 Envs all displayed at least 25-fold less fusogenicity from within MLV particles than their MLV/GaLV Env counterparts (Fig. 7B). Further truncation of the I170P Env CT (Δ24 and Δ25) increased fusogenicity within particles up to about half that of the MLV/GaLV Env counterparts. As before, truncation of I170P Env enhanced incorporation into viral particles, but incorporation did not correlate with fusogenicity. Most notably, I170P Δ20 had essentially a wild-type level of incorporation, but its fusogenicity was still reduced 25-fold.
Hydrophobicity is important at the 170th residue of the GaLV Env CT.
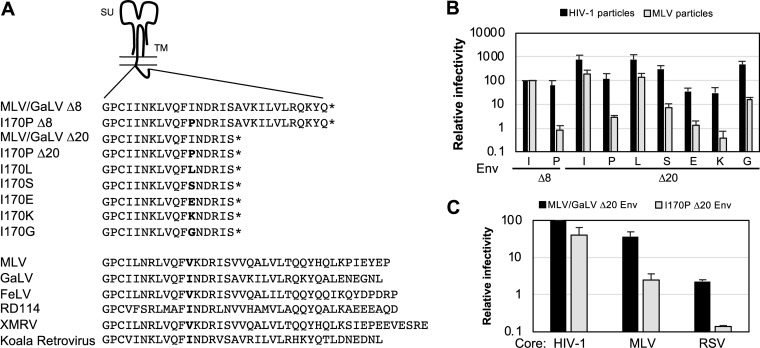
Several reports suggest that retroviral Env CTs contain amphipathic helixes, including the lytic peptides in the long CT of lentiviral Envs (17, 18). Gammaretroviral Env CTs also have conserved amphipathic regions with a proposed alpha-helical structure (27, 39–43). Among the amphipathic sequences present on the gammaretroviral Env CT, hydrophobic residues appear every three or four amino acids, supporting the possibility of the helix having a hydrophobic face. This helix has been proposed to affect membrane curvature during fusion (42). Because the 170th residues on MLV and GaLV Envs would be predicted to be part of this hydrophobic face, we hypothesized that the hydrophobicity of the I170 residue is critical in Env fusogenicity in MLV particles and introduced mutations to that residue to test this idea (Fig. 8A). I170P Δ20 was chosen for introducing point mutations because it retains MLV incompatibility, is constitutively active, and is incorporated into MLV particles at nearly wild-type levels. Among the Envs tested with a mutation at the 170th residue, a mutation leading to an alternative hydrophobic residue, leucine, exhibited wild-type infectivity with both HIV-1 and MLV particles (Fig. 8B). Mutations that introduced charged residues, glutamic acid and lysine, showed low infectivity, with a 25-fold reduction in infectious HIV-1 particle production and a 100-fold or greater reduction in infectious MLV particle production, which is an even greater effect than that observed with the proline mutation. Introduction of glycine and serine residues produced intermediate phenotypes. With these mutations, infectivity dropped about 2-fold in HIV-1 particles while the infectivity was down about 15- to 30-fold in MLV particles. These results suggest that hydrophobicity is functionally important at this position, supporting the importance of the proposed alpha-helical structure of the CT. The original proline mutation (I170P) exhibits about a 75-fold reduction in MLV infectivity, which could be due to both the reduced hydrophobicity and the disruption of the helix structure in the CT. The hydrophobic 170th residue is very well conserved in other gammaretroviruses, further suggesting the importance of this residue (Fig. 8A). For all the mutations tested, HIV-1 particles were shown to be generally more tolerant to changes than MLV particles.
FIG 8.
Hydrophobicity is important at the 170th residue of TM. (A) Schematic diagram of Env CT with mutations. Below is the comparison of the CTs from different gammaretroviruses. (B) Relative percent infectivity with mutant Env normalized to that of MLV/GaLV Δ8. (C) Relative percent infectivity of HIV-1, MLV, and RSV normalized to that of MLV/GaLV Δ20 with HIV-1. The average and standard deviation from three independent experiments for each are shown. FeLV, feline leukemia virus; RD114, a feline endogenous retrovirus; XMRV, xenotropic murine leukemia virus-related virus.
To test if the incompatibility is specific to MLV particles, next we investigated whether the I170P Env retains incompatibility in a different retrovirus family, Rous sarcoma virus (RSV; strain RCAS) (Fig. 8C). RSV particle infectivity with I170P Δ20 was also greatly reduced compared to that with MLV/GaLV Δ20, and the reduction was similar to that seen with MLV particles.
The 170th residue in GaLV Env CT is critical for MLV infectivity.
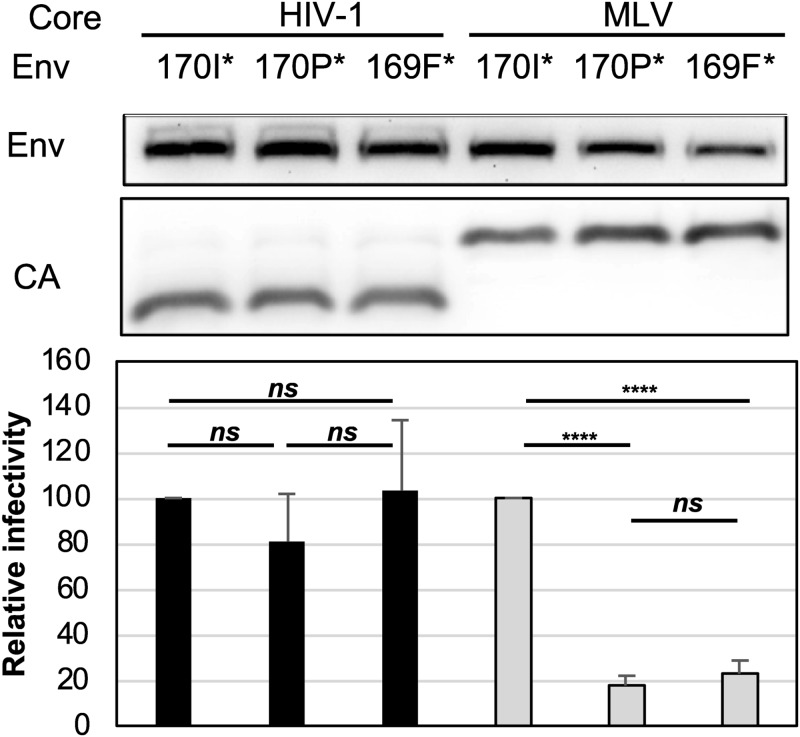
Truncation analysis (Fig. 5) showed that the mutation I170P affected MLV particle compatibility even if the protein was truncated immediately after that residue. Next, we analyzed the effects of truncating Env at position 169 (169F*), eliminating the position all together. This truncated Env was efficiently incorporated into MLV and HIV-1 particles, but the functionality was considerably lower in MLV particles than in HIV-1 particles (Fig. 9). Adding the 170th residue enhanced functionality with MLV particles by over 4-fold if the 170th residue was an isoleucine (170I*) but did not enhance it at all if it was a proline (170P*). All three of the glycoproteins were similarly functional with HIV-1 particles. These data suggest that the I170 residue of Env is critical for functionality with MLV particles but not with HIV-1 particles. In a previous study we showed that large truncations in MLV Env affected functionality with MLV particles more than with HIV-1 particles (24), but we were surprised to find here that the loss of functionality was not due to reduced incorporation. These data suggest that Env functionality is modulated in a virus-specific manner through an unknown mechanism.
FIG 9.
The 170th residue in MLV/GaLV Env CT is critical for MLV infectivity. Representative Env incorporation with the indicated Env from three independent experiments is shown. Average relative infectivity normalized to that of MLV/GaLV Δ26 (170I*) and standard deviations from three independent experiments are shown. ****, P < 0.0001; ns, not significant (by unpaired and two-tailed Student's t test).
DISCUSSION
Viral Env glycoprotein acquisition has been studied not only to understand the biological mechanisms of viral assembly and pathogenesis but also to enhance current knowledge on more efficient and safer application of gene therapy. Even though various studies address Env incompatibility in retroviral vectors, a gap remains in understanding the mechanism of Env recruitment and incorporation especially regarding the CT.
In this study, we investigated the mechanism behind virus-specific incompatibility dependent on the Env CT. The GaLV Env CT incompatibility with HIV-1 has been extensively studied, and we along with others reported that the incompatibility was partly dependent on Vpu (8, 20, 25, 26, 44, 45). Here, we addressed the GaLV Env CT incompatibility remaining in the absence of Vpu. Interestingly, from the exchanged segments of the CT between MLV and GaLV, we found that residues 170 to 186 of GaLV Env recapitulated the incompatibility with HIV-1 particles, but residues 176 to 196 of GaLV Env did not (Fig. 2). This finding initially led us to believe that the incompatibility was not due to a cleavage defect because the R-peptide cleavage site-containing regions (residues 176 to 186) were identical in the two chimeras. This was surprising because a previous study had suggested that the GaLV Env R peptide is poorly cleaved in HIV-1 particles (26). However, because the MLV Env with NKI residues from the region of amino acids 170 to 186 of GaLV displayed a cleavage defect, it became clear that amino acids upstream of the cleavage site also influenced cleavage efficiency. Among the three residues, two were at the cleavage site (Q178K and A179I), and one was just upstream of the cleavage site (K171N). This trio of changes modestly reduced Env incorporation but noticeably reduced R-peptide cleavage and infectivity with HIV-1 particles.
For the MLV particle-specific incompatibility, MLV/GaLV I170P Δ8 Env was shown to be functional with HIV particles but not with MLV particles. This particular mutant gained our attention because the defect was both striking and virus specific. Introduction of the I170P mutation to MLV/GaLV Δ8 decreased the infectivity of MLV particles by approximately 100-fold but had essentially no effect on the infectivity of HIV-1 particles. Our analyses have revealed that this stark apparent difference in virus-specific incompatibility levels was in fact due to three different factors.
The first factor causing incompatibility of Env proteins containing the I170P mutation with MLV particles is lack of incorporation. While incorporation of Env proteins into HIV-1 particles is not affected by the I170P mutation, the incorporation into MLV particles is severely reduced. We postulate that the loss of incorporation is because the I170P mutation changes the overall structure of the CT, making it unable to fit into the lattice formed by the MLV MA domain. Similarly, the MA domain of HIV-1 Gag has been shown to affect HIV-1 Env incorporation, and compensatory mutations near the MA trimer interface rescue Env mutants with incorporation defects (34, 35, 37). Consistent with our postulate, we found that the incorporation defect of I170P Env in MLV particles was alleviated by replacing MLV MA with HIV-1 MA or the Src membrane-binding domain (Fig. 6). In addition, truncation of the Env CT progressively restored incorporation into MLV particles (Fig. 5). However, the infectivity of MLV particles in both of these cases was only partially restored, suggesting that other factors contribute to the I170P defect in MLV particles.
The second factor contributing to the apparent virus-specific defect was the observation that the I170P mutation appears to have opposing positive and negative effects on infectivity with HIV-1 particles. Because the introduction of I170P into MLV/GaLV Δ8 Env had no effect on HIV-1 infectivity, we first interpreted this to mean that the mutation did not alter the functionality of the protein. However, further truncations of wild-type MLV/GaLV Env (Δ12, Δ16, and Δ20 constructs) were found to noticeably enhance the infectivity of HIV-1 particles (3- to 7-fold), but these same truncations had no effect on HIV-1 particle infectivity in the presence of the I170P mutation. We interpret this to mean that there is something in the R-peptide region that suppresses HIV-1 infectivity but is alleviated by the I170P mutation. The most likely explanation is that there is a difference in the efficiency of R-peptide cleavage as was observed with other full-length GaLV CT constructs (Fig. 4). Although we did not observe a difference in R-peptide cleavage in HIV-1 particles, it is possible that such a difference was too small to be apparent or that other factors such as the kinetics of R-peptide cleavage contributed to the effect. Nonetheless, the introduction of I170P into MLV/GaLV Env did have a negative impact on HIV-1 particle infectivity, but this impact was only obvious if the Env had most or all of its R peptide deleted (Δ12, Δ16, and Δ20 constructs) (Fig. 5). However, this 5- to 8-fold reduction in infectivity with HIV-1 particles was still noticeably less than the 50-fold or greater reduction seen with MLV particles, suggesting that the incompatibility is still partially virus specific.
The third factor contributing to the apparent virus-specific defect was the virus-specific compatibility of I170P Env proteins in particles. Even when incorporation levels were equivalent and the R peptide was removed, MLV/GaLV Env proteins containing I170P were less functional in MLV particles than in HIV-1 particles (Fig. 5). We postulate that this difference in the two types of particles is due to differences in the lipid membrane that interacts with the Env CT. The CTs of MLV and GaLV Env are thought to form amphipathic helixes (27, 39, 40), and a series of hydrophobic amino acids including I170 are highly conserved and would form one face of that helix. The amphipathic membrane-proximal region in the cytoplasmic tail of MLV Env was also previously reported to be critical for Env fusogenicity and was hypothesized to affect membrane curvature during the fusion (42, 43). In our study, in the context of MLV/GaLV Δ20, mutation of the 170th position to any nonhydrophobic amino acid greatly reduced infectivity although the effect was always more pronounced in MLV particles than in HIV-1 particles (Fig. 8). We reported previously that large truncations in the CT of MLV Env had a greater effect on MLV infectivity than on HIV-1 infectivity (24), but we were surprised to find here that the effect was not due to differences in Env incorporation. An Env protein truncated to one residue before I170 (169F*) was over 4-fold less functional in MLV particles than an Env truncated immediately after I170 (170I*); yet the two Env proteins were equally infectious in HIV-1 particles, and the two Env proteins were equally incorporated into both HIV-1 and MLV particles. We propose that this amino acid interacts with the viral membrane to aid in fusion activity and that minor differences between the HIV-1 and MLV membranes make the position more critical in MLV particles. If this model is correct, it is surprising that replacing MLV MA with HIV-1 MA does not alleviate the necessity of this residue in MLV particles. However, because MLV Gag contains the p12 domain between MA and CA, the MA domain of HIV-1 Gag would not necessarily be in the same configuration in MLV particles as in HIV-1 particles and thus not necessarily interact with the membrane in the same way.
Overall, we show two different mutations in the CT of Env that lead to virus-specific incompatibility in distinct ways. While GaLV Env incompatibility with HIV-1 is largely related to insufficient R-peptide cleavage, I170P Env incompatibility with MLV is due to a mixture of factors. The 170th residue in the Env TM subunit is critical in MLV particles but not in HIV-1 particles. How these mutations in the GaLV cytoplasmic tail affect virus-specific fusion remains a matter of speculation.
MATERIALS AND METHODS
Antibody generation.
A humanized version of the rat IgM monoclonal antibody 42/114 was generated for detection of the MLV TM (p15E) protein. To accomplish this, RNA from the 42/114 hybridoma (46) was extracted, cDNA was generated, and the sequence for the variable regions of μ heavy chain (forward primer, GAGGTGCAGCTTCAGGAGTCAGGACCTGGCCTT; reverse primer, ATCAGACAGGGGGCTCTCGCAGGAGA) and κ light chain (forward primer, TTAGGGCTGCTGCTGCTTTGG; reverse primer, CGTTTCAGTTCCAGCTTGGT) were PCR amplified. The extracted variable regions were subcloned using an In-Fusion cloning system (Clontech) into variable regions of pVITRO1-Trastizimab-IgG1/K, a gift from Andrew Beavil (plasmid 61883; Addgene) (47). Complete γ heavy chain and κ light chain from the vector were then PCR amplified and inserted into retroviral vectors pQCXIP and pQCXIH, respectively (Clontech) and used to generate a stable 293FT cell line secreting human IgG. Collected antibodies secreted in medium were further purified with a HiTrap Protein G column (GE Health Science), dialyzed, and concentrated by filter centrifugation for 45 to 90 min at 4,000 rpm (Amicon Ultra-15 filter unit).
Cell culture.
The 293FT cell line was obtained from Invitrogen. The 293T mCAT-1 cell line expressing the ecotropic Friend MLV (F-MLV) Env receptor was kindly provided by Walter Mothes (Yale University). The HT1080/mCAT-1 Cre reporter cell line was kindly provided by Alan Rein (38). 293T mCAT-1 cells stably expressing Gaussia luciferase downstream of a Tet-responsive element (TRE) promoter was previously described (48). All cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 1 mM sodium pyruvate, 10 mM nonessential amino acids, and 1% minimal essential medium (MEM) vitamins.
Plasmids.
NL4-3 derived HIV-CMV-GFP (where CMV is cytomegalovirus) was kindly provided by Vineet Kewal-Ramani (National Cancer Institute [NCI]-Frederick). This proviral vector lacks Vif, Vpr, Vpu, Nef, and Env and contains CMV-GFP in place of Nef. CMV-MLV-GagPol and the ecotropic Friend MLV Env expression vector were kindly provided by Walter Mothes (Yale University). The MLV reporter constructs pQCXIP-TdTomato and pQCXIP-GFP were previously described (25, 49). The plasmid encoding Cre recombinase fused with MLV Gag (MLV Gag-Cre) was kindly provided by Alan Rein (38). For Src-MLV GagPol, the Src variant (MGSNKSKPKDASSQR) was cloned in the place of MA in CMV-MLV-GagPol. A CMV-driven tTA (Tet-off) protein expression vector was previously described (48). The HIV-1 vector with CMV-driven, reverse-intron-interrupted Gaussia luciferase (HIV-iGluc) was adapted from a vector kindly provided by David Derse and was described previously (28, 48, 50, 51). The MLV reporter vector with a CMV-driven, reverse-intron-interrupted Gaussia luciferase gene (MLV-iGluc) was constructed by subcloning the CMV-driven reverse-intron-interrupted Gaussia luciferase gene from HIV-iGluc into pQCXIP (Clontech). The MLV GagPol construct with the HIV-1 MA domain (hMA-MLV) was previously described (52). The MLV Env with GaLV Env CT was previously described (25). The MLV/GaLV Env Δ8 construct and MLV/GaLV Env I170P Δ8 (I26P in the previous report) construct were previously described (27). Point mutations and truncations of MLV and MLV/GaLV Env were created by oligonucleotide-mediated mutagenesis. Some Env constructs used in Western blotting contained a hemagglutinin (HA) tag (YPYDVPDYA) in a proline-rich region in the SU subunit (48). For virus capture assay, codon-optimized MLV/GaLV Δ8 Env with an internal HA tag in the proline-rich region in SU (Addgene) was inserted in pQCXIP (Clontech), and the HA tag was replaced with GFP. The VSV-G expression construct was obtained from the NIH AIDS Reagent Program (53). For the RSV proviral construct, CMV was inserted upstream of the GFP gene in the previously described RCAS.GFP-Tomato vector (54). pUC19 was obtained from Invitrogen.
Infectivity assay.
Infectivity assays using HIV-CMV-GFP or CMV-MLV-GagPol and pQCXIP-GFP with various Env constructs were performed by transfection in 293FT cells with polyethylenimine (PEI) (55). Medium was changed about 24 h posttransfection and collected after another 24 h. Supernatant containing the virus was frozen at −80°C for at least 4 h, thawed, and spun at 3,200 × g for 5 min, and the same volume of medium was added to target cells with 20 μg of hexadimethrine bromide per ml (H9268; Sigma). Infected cells were collected at about 48 h postinfection, fixed with 4% paraformaldehyde, washed with phosphate-buffered saline (PBS), and analyzed on an Accuri C6 flow cytometer.
Except where noted, transfections for the infectivity assay were performed in six-well plates with a total of 2 μg of plasmids. For HIV-1 infectivity, 1,800 ng of HIV-CMV-GFP and 200 ng of Env constructs were cotransfected. For MLV infectivity, 900 ng of CMV-MLV-GagPol and 900 ng of either pQCXIP-GFP or pQCXIP-TdTomato were transfected along with 200 ng of Env. For the Env titration assay, 400 ng of HIV-CMV-GFP or 400 ng of CMV-MLV GagPol and 400 ng of pQCXIP-TdTomato were transfected with various amounts of Env (20 ng, 80 ng, 320 ng, and 1,280 ng), and a filler plasmid, pUC19, was used to normalize total DNA quantity in the transfection. For Western blotting and infectivity in the experiments shown in Fig. 6C, 8C, and 9, a total of 1 μg of plasmid was transfected at a 9-to-1 ratio of virus to Env. For Western blotting and the infectivity experiments in shown in Fig. 4 and 5C, 1,500 ng of virus and 500 ng of Env were transfected.
Virus capture assay.
The virus capture assay was performed using a modified version of our published protocol (28, 48). Briefly, 293FT cells in 12-well plates were transfected with HIV-iGluc (400 ng) or MLV-iGluc (200 ng) and MLV-CMV-GagPol (200 ng) along with various GFP-tagged Env expression plasmids (40 ng for HIV-1 particles and 50 ng for MLV particles) and the VSV-G expression plasmid (10 ng for HIV-1 and 25 ng for MLV). These amounts were derived empirically from titration curves to yield similar infectivity levels between the two different virus constructs. Medium was changed after 24 h and collected after another 24 h. Twenty microliters (or 100 μl for Env truncations) of PBS with or without previously described polyclonal GFP antibodies (1:10,000) (48) was added to the wells of an enzyme-linked immunosorbent assay (ELISA) plate and incubated overnight at 4°C. The antibody or PBS was then removed and replaced with blocking buffer (PBS with 1% bovine serum albumin [BSA], 5% sucrose, 0.05% sodium azide) for 1 h. After blocking buffer was removed, 10 μl of viral supernatant and 10 μl of PBS were added to the each well and incubated for 3 h at 37°C. After the incubation, supernatant was removed, and all wells were washed twice with 100 μl of PBS except for the positive-control wells, where the viral supernatant was not removed. Twenty microliters of PBS was then added to washed wells to normalize the volume, and 293FT cells were added on top for infection. Two days later, 20 μl of supernatant from each well was transferred to a black 96-well plate for measuring Gaussia luciferase activity with 50 μl of 10 μM coelenterazine in 0.1 M Tris (pH 7.4) and 0.3 M sodium ascorbate (NanoLight Technology). Luminescence, representing infectivity, was measured from the supernatant using a PerkinElmer Enspire 2300 Multilabel Reader. Each sample was normalized to the level of the straight infection (no wash) as a positive control.
Viral entry assay.
The viral entry assay was performed as previously described (38), with a few modifications. Briefly, 293FT cells in 12-well plates were transfected with 50 ng of each Env expression plasmid, 225 ng of CMV-MLV-GagPol, 225 ng of pQXIP-GFP, and 45 ng of MLV Gag-Cre fusion plasmid. In some instances, these transfections were performed in six-well plates with appropriate scaling of plasmids. After 24 h of transfection, medium was changed, and the supernatant was harvested after another 24 h. Medium was frozen at −80°C for at least 4 h and thawed, and Cre reporter cells were transduced with equal volumes of the virus samples. After 48 h, firefly luciferase activity was measured using Bright-Glo reagent (Promega).
Env fusion assay.
293FT cells were transfected with 100 ng of Env and 100 ng of tTA expression plasmid in a six-well plate, or half this amount was transfected in 12 wells. The next day, medium was changed to remove residual transfection reagent, and 293 mCAT-1 TRE-Gluc cells were plated on top of the transfected 293FT cells. After an additional 24 h, 20 μl of supernatant from cocultured cells was assayed for luminescence in triplicate, as previously mentioned.
Western blotting.
Virus samples were concentrated by centrifugation at 30,000 × g for 2 h through a 20% sucrose cushion. One milliliter of medium was used for probing SU, and 1.5 ml of medium was used to probe for p15E. For probing the SU subunits of the 169F*, 170I*, and 170P* constructs, 0.6 ml of medium was used. Viral pellets were resuspended and denatured by heating in sample buffer (50 mM Tris, 2% sodium dodecyl sulfate [SDS], 20% glycerol, 5% β-mercaptoethanol) before loading. For probing SU, a 10% PAGE gel and 0.45-μm-pore-size polyvinylidene difluoride (PVDF) membrane for transfer were used. For probing p15E, a 15% PAGE gel and 0.22-μm-pore-size PVDF membrane for transfer were used. In both experiments, membranes were blocked with 2% nonfat dry milk prior to antibody incubation. For detecting SU, goat anti-MLV Env gp70 (kindly provided by Alan Rein, NCI-Frederick) diluted 1:10,000 was used. HA-tagged SU Env was detected using anti-HA antibody diluted 1:1,000 (H3663; Sigma). TM p15E antibody was generated in human IgG form, and 6 μg of purified antibody was used per 1 ml of PBS with Tween 20 (PBS-T) without milk. Anti-HIV p24 hybridoma medium was used at 1:500 (HIV-1 p24 hybridoma [183-H12-5C], obtained from NIH AIDS Reagent Program) from Bruce Chesebro (56), and anti-MLV capsid (CA) medium from hybridoma (R187; ATCC) (57) was used at 1:250. After blots were washed with PBS, horseradish peroxidase (HRP)-conjugated secondary antibody was used at 1:10,000 for all blots. Horseradish peroxidase-linked anti-mouse (A5278), anti-goat (A8919), anti-rat (A5795), and anti-human (A0293) were obtained from Sigma. Luminata Classico Western HRP substrate (Millipore) was used for visualization of the membranes with a chemiluminescence image analyzer (Fuji Film LAS-3000 or UVP BioSpectrum 815 Imaging System).
ACKNOWLEDGMENTS
We thank Alan Rein for providing the materials for the MLV virus-to-cell fusion assay. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: pHEF-VSVG from Lung-Ji Chang (catalog no. 4693) and anti-HIV-1 p24 hybridoma (183-H12-5C) (catalog no. 1513) from Bruce Chesebro.
This work was supported by the National Institute of General Medicine under award R01GM110776.
REFERENCES
- 1.Johnson MC. 2011. Mechanisms for Env glycoprotein acquisition by retroviruses. AIDS Res Hum Retroviruses 27:239–247. doi: 10.1089/AID.2010.0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murakami T. 2012. Retroviral Env glycoprotein trafficking and incorporation into virions. Mol Biol Int 2012:682850. doi: 10.1155/2012/682850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison SC. 2015. Viral membrane fusion. Virology 479-480:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ragheb JA, Anderson WF. 1994. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J Virol 68:3220–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rein A, Mirro J, Haynes JG, Ernst SM, Nagashima K. 1994. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J Virol 68:1773–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loving R, Wu SR, Sjoberg M, Lindqvist B, Garoff H. 2012. Maturation cleavage of the murine leukemia virus Env precursor separates the transmembrane subunits to prime it for receptor triggering. Proc Natl Acad Sci U S A 109:7735–7740. doi: 10.1073/pnas.1118125109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brody BA, Rhee SS, Sommerfelt MA, Hunter E. 1992. A viral protease-mediated cleavage of the transmembrane glycoprotein of Mason-Pfizer monkey virus can be suppressed by mutations within the matrix protein. Proc Natl Acad Sci U S A 89:3443–3447. doi: 10.1073/pnas.89.8.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christodoulopoulos I, Cannon PM. 2001. Sequences in the cytoplasmic tail of the gibbon ape leukemia virus envelope protein that prevent its incorporation into lentivirus vectors. J Virol 75:4129–4138. doi: 10.1128/JVI.75.9.4129-4138.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bobkova M, Stitz J, Engelstadter M, Cichutek K, Buchholz CJ. 2002. Identification of R-peptides in envelope proteins of C-type retroviruses. J Gen Virol 83:2241–2246. doi: 10.1099/0022-1317-83-9-2241. [DOI] [PubMed] [Google Scholar]
- 10.Rice NR, Henderson LE, Sowder RC, Copeland TD, Oroszlan S, Edwards JF. 1990. Synthesis and processing of the transmembrane envelope protein of equine infectious anemia virus. J Virol 64:3770–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karshin WL, Arcement LJ, Naso RB, Arlinghaus RB. 1977. Common precursor for Rauscher leukemia virus gp69/71, p15(E), and p12(E). J Virol 23:787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang J, Aiken C. 2007. Maturation-dependent human immunodeficiency virus type 1 particle fusion requires a carboxyl-terminal region of the gp41 cytoplasmic tail. J Virol 81:9999–10008. doi: 10.1128/JVI.00592-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joyner AS, Willis JR, Crowe JE Jr, Aiken C. 2011. Maturation-induced cloaking of neutralization epitopes on HIV-1 particles. PLoS Pathog 7:e1002234. doi: 10.1371/journal.ppat.1002234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wyma DJ, Jiang J, Shi J, Zhou J, Lineberger JE, Miller MD, Aiken C. 2004. Coupling of Human Immunodeficiency virus type 1 fusion to virion maturation: a novel role of the gp41 cytoplasmic tail. J Virol 78:3429–3435. doi: 10.1128/JVI.78.7.3429-3435.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santos da Silva E, Mulinge M, Perez Bercoff D. 2013. The frantic play of the concealed HIV envelope cytoplasmic tail. Retrovirology 10:54. doi: 10.1186/1742-4690-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Postler TS, Desrosiers RC. 2013. The tale of the long tail: the cytoplasmic domain of HIV-1 gp41. J Virol 87:2–15. doi: 10.1128/JVI.02053-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steckbeck JD, Kuhlmann AS, Montelaro RC. 2014. Structural and functional comparisons of retroviral envelope protein C-terminal domains: still much to learn. Viruses 6:284–300. doi: 10.3390/v6010284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tedbury PR, Freed EO. 2015. The cytoplasmic tail of retroviral envelope glycoproteins. Prog Mol Biol Transl Sci 129:253–284. doi: 10.1016/bs.pmbts.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spearman P. 2018. Viral interactions with host cell Rab GTPases. Small GTPases 9:192–201. doi: 10.1080/21541248.2017.1346552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stitz J, Buchholz CJ, Engelstadter M, Uckert W, Bloemer U, Schmitt I, Cichutek K. 2000. Lentiviral vectors pseudotyped with envelope glycoproteins derived from gibbon ape leukemia virus and murine leukemia virus 10A1. Virology 273:16–20. doi: 10.1006/viro.2000.0394. [DOI] [PubMed] [Google Scholar]
- 21.Sandrin V, Boson B, Salmon P, Gay W, Negre D, Le Grand R, Trono D, Cosset FL. 2002. Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34+ cells derived from human and nonhuman primates. Blood 100:823–832. doi: 10.1182/blood-2001-11-0042. [DOI] [PubMed] [Google Scholar]
- 22.Sandrin V, Muriaux D, Darlix JL, Cosset FL. 2004. Intracellular trafficking of Gag and Env proteins and their interactions modulate pseudotyping of retroviruses. J Virol 78:7153–7164. doi: 10.1128/JVI.78.13.7153-7164.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandrin V, Cosset FL. 2006. Intracellular versus cell surface assembly of retroviral pseudotypes is determined by the cellular localization of the viral glycoprotein, its capacity to interact with Gag, and the expression of the Nef protein. J Biol Chem 281:528–542. doi: 10.1074/jbc.M506070200. [DOI] [PubMed] [Google Scholar]
- 24.Lucas TM, Lyddon TD, Grosse SA, Johnson MC. 2010. Two distinct mechanisms regulate recruitment of murine leukemia virus envelope protein to retroviral assembly sites. Virology 405:548–555. doi: 10.1016/j.virol.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lucas TM, Lyddon TD, Cannon PM, Johnson MC. 2010. Pseudotyping incompatibility between HIV-1 and gibbon ape leukemia virus Env is modulated by Vpu. J Virol 84:2666–2674. doi: 10.1128/JVI.01562-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christodoulopoulos I, Droniou-Bonzom ME, Oldenburg JE, Cannon PM. 2010. Vpu-dependent block to incorporation of GaLV Env into lentiviral vectors. Retrovirology 7:4. doi: 10.1186/1742-4690-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janaka SK, Lucas TM, Johnson MC. 2011. Sequences in gibbon ape leukemia virus envelope that confer sensitivity to HIV-1 accessory protein Vpu. J Virol 85:11945–11954. doi: 10.1128/JVI.05171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gregory DA, Olinger GY, Lucas TM, Johnson MC. 2014. Diverse viral glycoproteins as well as CD4 co-package into the same human immunodeficiency virus (HIV-1) particles. Retrovirology 11:28. doi: 10.1186/1742-4690-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freed EO, Martin MA. 1995. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J Virol 69:1984–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mammano F, Kondo E, Sodroski J, Bukovsky A, Göttlinger HG. 1995. Rescue of human immunodeficiency virus type 1 matrix protein mutants by envelope glycoproteins with short cytoplasmic domains. J Virol 69:3824–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freed EO, Martin MA. 1996. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol 70:341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ono A, Huang M, Freed EO. 1997. Characterization of human immunodeficiency virus type 1 matrix revertants: effects on virus assembly, Gag processing, and Env incorporation into virions. J Virol 71:4409–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brandano L, Stevenson M. 2012. A highly conserved residue in the C-terminal helix of HIV-1 matrix is required for envelope incorporation into virus particles. J Virol 86:2347–2359. doi: 10.1128/JVI.06047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tedbury PR, Ablan SD, Freed EO. 2013. Global rescue of defects in HIV-1 envelope glycoprotein incorporation: implications for matrix structure. PLoS Pathog 9:e1003739. doi: 10.1371/journal.ppat.1003739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tedbury PR, Mercredi PY, Gaines CR, Summers MF, Freed EO. 2015. Elucidating the mechanism by which compensatory mutations rescue an HIV-1 matrix mutant defective for Gag membrane targeting and envelope glycoprotein incorporation. J Mol Biol 427:1413–1427. doi: 10.1016/j.jmb.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiernan RE, Freed EO. 1998. Cleavage of the murine leukemia virus transmembrane Env protein by human immunodeficiency virus type 1 protease: transdominant inhibition by matrix mutations. J Virol 72:9621–9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tedbury PR, Novikova M, Ablan SD, Freed EO. 2016. Biochemical evidence of a role for matrix trimerization in HIV-1 envelope glycoprotein incorporation. Proc Natl Acad Sci U S A 113:E182–E190. doi: 10.1073/pnas.1516618113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahi YS, Zhang S, Thappeta Y, Denman A, Feizpour A, Gummuluru S, Reinhard B, Muriaux D, Fivash MJ, Rein A. 2016. Functional interplay between murine leukemia virus glycogag, serinc5, and surface glycoprotein governs virus entry, with opposite effects on gammaretroviral and ebolavirus glycoproteins. mBio 7:e01985-16. doi: 10.1128/mBio.01985-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang C, Compans RW. 1996. Analysis of the cell fusion activities of chimeric simian immunodeficiency virus-murine leukemia virus envelope proteins: inhibitory effects of the R peptide. J Virol 70:248–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor GM, Sanders DA. 2003. Structural criteria for regulation of membrane fusion and virion incorporation by the murine leukemia virus TM cytoplasmic domain. Virology 312:295–305. doi: 10.1016/S0042-6822(03)00297-6. [DOI] [PubMed] [Google Scholar]
- 41.Yang C, Compans RW. 1997. Analysis of the murine leukemia virus R peptide: delineation of the molecular determinants which are important for its fusion inhibition activity. J Virol 71:8490–8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Epand RF, Zhang YL, Mirzabekov T, Kagan B, Silberstein A, Hubbell WL, Epand RM, Chakraborti S, Dimitrov DS, Anderson WF, Rozenberg-Adler Y. 2008. Membrane activity of an amphiphilic alpha-helical membrane-proximal cytoplasmic domain of the MoMuLV envelope glycoprotein. Exp Mol Pathol 84:9–17. doi: 10.1016/j.yexmp.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 43.Rozenberg-Adler Y, Conner J, Aguilar-Carreno H, Chakraborti S, Dimitrov DS, Anderson WF. 2008. Membrane-proximal cytoplasmic domain of Moloney murine leukemia virus envelope tail facilitates fusion. Exp Mol Pathol 84:18–30. doi: 10.1016/j.yexmp.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 44.Janaka SK, Faurot J, Johnson MC. 2013. Functional complementation of a model target to study Vpu sensitivity. PLoS One 8:e68507. doi: 10.1371/journal.pone.0068507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song YE, Cyburt D, Lucas TM, Gregory DA, Lyddon TD, Johnson MC. 2018. βTrCP is required for HIV-1 Vpu modulation of CD4, GaLV Env, and BST-2/tetherin. Viruses 10:E573. doi: 10.3390/v10100573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pinter A, Honnen WJ, Tung JS, O'Donnell PV, Hämmerling U. 1982. Structural domains of endogenous murine leukemia virus gp70s containing specific antigenic determinants defined by monoclonal antibodies. Virology 116:499–516. doi: 10.1016/0042-6822(82)90143-X. [DOI] [PubMed] [Google Scholar]
- 47.Dodev TS, Karagiannis P, Gilbert AE, Josephs DH, Bowen H, James LK, Bax HJ, Beavil R, Pang MO, Gould HJ, Karagiannis SN, Beavil AJ. 2014. A tool kit for rapid cloning and expression of recombinant antibodies. Sci Rep 4:5885. doi: 10.1038/srep05885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Janaka SK, Gregory DA, Johnson MC. 2013. Retrovirus glycoprotein functionality requires proper alignment of the ectodomain and the membrane-proximal cytoplasmic tail. J Virol 87:12805–12813. doi: 10.1128/JVI.01847-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salamango DJ, Johnson MC. 2015. Characterizing the murine leukemia virus envelope glycoprotein membrane-spanning domain for its roles in interface alignment and fusogenicity. J Virol 89:12492–12500. doi: 10.1128/JVI.01901-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mazurov D, Ilinskaya A, Heidecker G, Lloyd P, Derse D. 2010. Quantitative comparison of HTLV-1 and HIV-1 cell-to-cell infection with new replication dependent vectors. PLoS Pathog 6:e1000788. doi: 10.1371/journal.ppat.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aloia AL, Duffy L, Pak V, Lee KE, Sanchez-Martinez S, Derse D, Heidecker G, Cornetta K, Rein A. 2013. A reporter system for replication-competent gammaretroviruses: the inGluc-MLV-DERSE assay. Gene Ther 20:169–176. doi: 10.1038/gt.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gregory DA, Lyddon TD, Johnson MC. 2013. Multiple Gag domains contribute to selective recruitment of murine leukemia virus (MLV) Env to MLV virions. J Virol 87:1518–1527. doi: 10.1128/JVI.02604-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang LJ, Urlacher V, Iwakuma T, Cui Y, Zucali J. 1999. Efficacy and safety analyses of a recombinant Human Immunodeficiency Virus type 1 derived vector system. Gene Ther 6:715–728. doi: 10.1038/sj.gt.3300895. [DOI] [PubMed] [Google Scholar]
- 54.Jorgenson RL, Vogt VM, Johnson MC. 2009. Foreign glycoproteins can be actively recruited to virus assembly sites during pseudotyping. J Virol 83:4060–4067. doi: 10.1128/JVI.02425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. 1995. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A 92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chesebro B, Wehrly K, Nishio J, Perryman S. 1992. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol 66:6547–6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chesebro B, Britt W, Evans L, Wehrly K, Nishio J, Cloyd M. 1983. Characterization of monoclonal antibodies reactive with murine leukemia viruses: use in analysis of strains of friend MCF and Friend ecotropic murine leukemia virus. Virology 127:134–148. doi: 10.1016/0042-6822(83)90378-1. [DOI] [PubMed] [Google Scholar]