HIV-1 replication requires interaction of the viral Rev protein with a cis-acting regulatory RNA, the Rev response element (RRE), whose sequence changes over time during infection within a single host. In this study, we show that the RRE is subject to selection pressure and that RREs from later time points in infection tend to have higher functional activity. Differences in RRE functional activity are attributable to specific changes in RNA structure. Our results suggest that RRE evolution during infection may be important for HIV pathogenesis and that efforts to develop therapies acting on this viral pathway should take this into account.
KEYWORDS: HIV, HIV RRE, HIV Rev, RNA export, RNA structure, viral sequence evolution, viral sequence variation
ABSTRACT
The HIV-1 Rev response element (RRE) is a cis-acting RNA element characterized by multiple stem-loops. Binding and multimerization of the HIV Rev protein on the RRE promote the nucleocytoplasmic export of incompletely spliced mRNAs, an essential step in HIV replication. Most of our understanding of the Rev-RRE regulatory axis comes from studies of lab-adapted HIV clones. However, in human infection, HIV evolves rapidly, and mechanistic studies of naturally occurring Rev and RRE sequences are essential to understanding this system. We previously described the functional activity of two RREs found in circulating viruses in a patient followed during the course of HIV infection. The early RRE was less functionally active than the late RRE, despite differing in sequence by only 4 nucleotides. In this study, we describe the sequence, function, and structural evolution of circulating RREs in this patient using plasma samples collected over 6 years of untreated infection. RRE sequence diversity varied over the course of infection, with evidence of selection pressure that led to sequence convergence as disease progressed being found. An increase in RRE functional activity was observed over time, and a key mutation was identified that correlates with a major conformational change in the RRE and increased functional activity. Additional mutations were found that may have contributed to increased activity as a result of greater Shannon entropy in RRE stem-loop II, which is key to primary Rev binding.
IMPORTANCE HIV-1 replication requires interaction of the viral Rev protein with a cis-acting regulatory RNA, the Rev response element (RRE), whose sequence changes over time during infection within a single host. In this study, we show that the RRE is subject to selection pressure and that RREs from later time points in infection tend to have higher functional activity. Differences in RRE functional activity are attributable to specific changes in RNA structure. Our results suggest that RRE evolution during infection may be important for HIV pathogenesis and that efforts to develop therapies acting on this viral pathway should take this into account.
INTRODUCTION
All retroviruses produce mRNAs that retain introns, and these mRNAs must be exported to the cytoplasm for packaging into viral particles and translation of essential viral proteins. Eukaryotic cells have RNA surveillance mechanisms that would normally restrict nucleocytoplasmic export and translation of these mRNA species (1). Thus, retroviruses have evolved specific mechanisms to overcome this restriction (2). In HIV, the Rev protein, in conjunction with its RNA-binding partner, the Rev response element (RRE), mediates this important function (3–6).
HIV Rev contains several well-characterized functional domains that facilitate shuttling between the nucleus/nucleolus and the cytoplasm by accessing cellular pathways for nuclear import and export. One of these domains is a basic, arginine-rich motif that functions as a nuclear/nucleolar localization signal (7–10). This domain also serves as an RNA-binding domain that binds specifically to the RRE (11, 12). It is flanked on both sides by oligomerization domains that are required for Rev multimerization on the RRE (13, 14). Toward the carboxy terminus is a leucine-rich domain which docks on the Crm1-RanGTP complex (15) and functions as the nuclear export signal (NES) (12, 16, 17).
The RRE is a cis-acting RNA element located in a highly conserved and functionally important region of env at the N terminus of the gp41 fusion protein (3–6). At this position, it is present in all HIV RNAs that retain introns. The minimal functional RRE, or “short” RRE, has been mapped in infectious laboratory clones to a 234-nucleotide (nt) region (see Fig. 3). It forms a highly branched structure where the 5′ and 3′ ends pair to form stem-loop I (SL-I) (18). SL-I opens into a central loop, from which several additional stem-loops branch out. Stem-loop II contains stem IIA, which branches out of the central loop and opens into a three-way junction. The junction opens into stem-loops IIB and IIC (19, 20). SL-III is a small stem-loop that comes off the central loop, while SL-IV and SL-V can adopt alternative topologies (21). The maximal functional activity of the RRE requires a somewhat larger structure, often referred to as the “long” (351-nt) RRE, characterized by an extended SL-I (18, 22, 23).
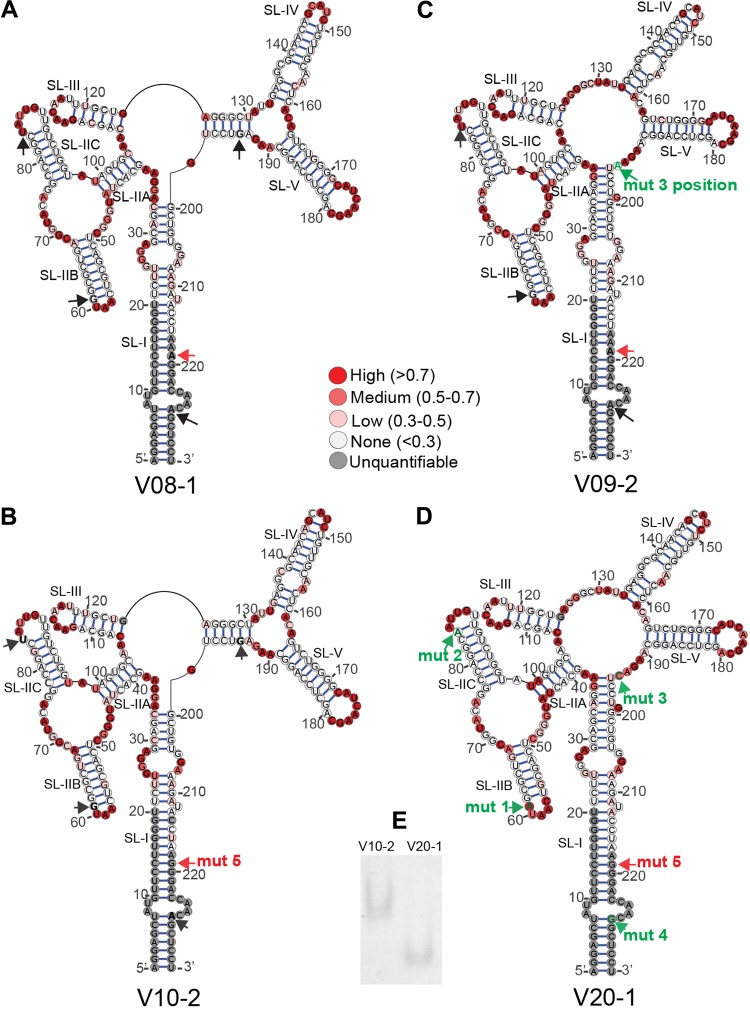
FIG 3.
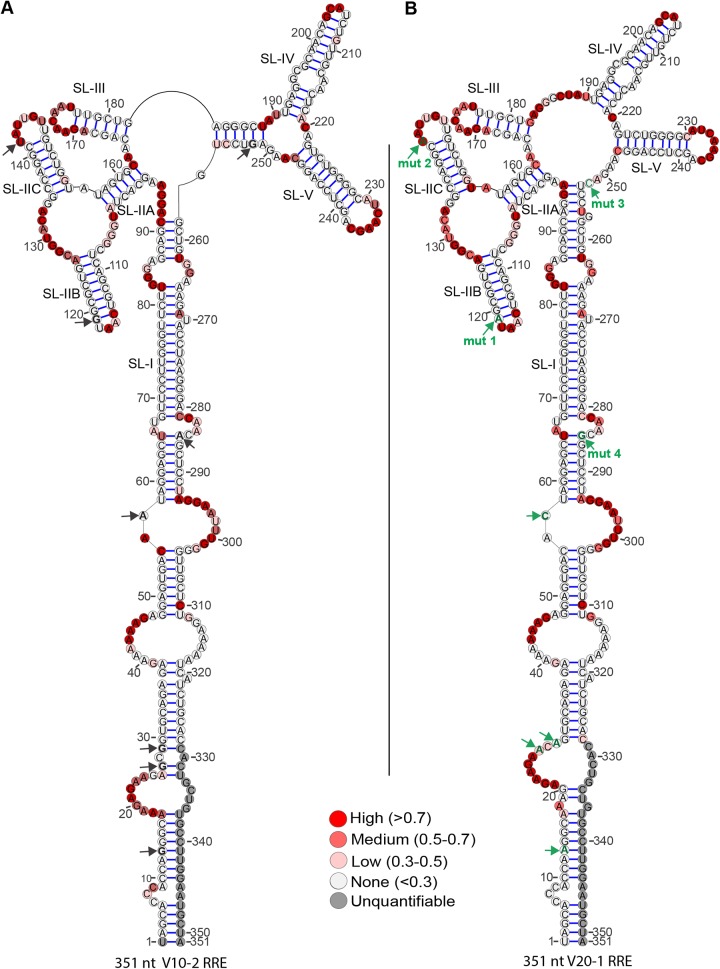
Structures of selected SC3 RREs. Secondary structures of V08-1 (A), V10-2 (B), V09-2 (C), and V20-1 (D) 234-nt RREs were determined by SHAPE-MaP. The 1M7 reactivities of the RRE nucleotides are color coded and superimposed on the structures. The positions of the mut 1, mut 2, mut 3, and mut 4 single-nucleotide changes are represented by black arrows. Green labeled arrows show where the nucleotide at this position varies from V08-1, the presumptive founder sequence, and V10-2. In V09-2, the mut 3 position shows a G-to-A change, while the mut 3 position in V20-1 shows a G-to-C change. The mut 5 position is shown with a red arrow and is additionally labeled with “mut 5” in structures where the nucleotide varies from V08-1. (E) The migration of V10-2 and V20-1 RRE on a native PAGE gel was visualized after 22 h by UV shadowing.
Primary RRE binding initiates cooperative assembly of additional Rev molecules (about 6 to 8) in a process that requires electrostatic Rev-RRE interactions and hydrophobic Rev-Rev interactions (13, 18, 24–32). Rev oligomerization on the RRE increases its binding affinity ∼500-fold (30). Oligomerization also arranges the NES domains for binding to the RanGTP-bound Crm1 dimer (15), forming an export-competent ribonucleoprotein complex (33, 34). The complex is targeted to the nuclear pore, where it interacts with the nucleoporins, resulting in its translocation to the cytoplasmic side. Once in the cytoplasm, RRE-containing mRNAs are translated into Gag, Gag-Pol, Env, and accessory viral proteins.
Molecular details of the Rev-RRE pathway have been delineated mostly from studies of lab-adapted HIV clones. However, there is mounting evidence that subtle variations in Rev and RRE sequences among primary isolates may contribute to pathogenesis. Differences in Rev-RRE functional activity up to 24-fold were observed for naturally occurring viruses in different patients (35). In addition, a study performed with a Thai cohort demonstrated RRE changes over the course of infection, and higher RRE activity was associated with a more rapid CD4 count decline (36). Low Rev activity has also been associated with slower disease progression (37, 38) and reduced susceptibility of infected cells to T-cell killing (39). In equine infectious anemia virus, a related lentivirus that also utilizes the Rev/RRE axis, the functional evolution of Rev has been observed during infection, and Rev activity has been found to correlate with the disease state in ponies (40–42). Further studies of the structural and functional evolution of the Rev-RRE system in natural infection are necessary to understand the role that this regulatory axis plays in the adaptation of HIV to diverse immune environments, and this may benefit the development of Rev-RRE-targeted therapeutics.
We previously investigated the activity of Rev-RRE cognate pairs from HIV isolates recovered from five different patients at two time points during their course of infection (43). The sequences were obtained by single-genome sequencing of viruses from blood plasma samples, and their functional activity was determined using a subgenomic reporter assay. We observed significant activity differences between Rev-RRE cognate pairs from different patients and from different time points in the same patient. The evolution of Rev and the RRE observed in patient SC3 was particularly striking. In this patient, the RRE converged on one predominant sequence at the later time point (the M57A RRE, here designated V20-1), suggesting that it was subject to strong selective pressures. Furthermore, this RRE had only 4 nucleotide changes (mutation 1 [mut 1] to mut 4) relative to the early-time-point RRE (M0-A RRE, here designated V10-2) but was 2- to 3-fold more functionally active. Gel mobility shift assays revealed that the V20-1 RRE promoted Rev multimerization at a lower concentration of Rev protein than the V10-2 RRE. It was also notable that the predominant Rev sequence present at the early time point persisted at the late time point, suggesting that the limited nucleotide changes in the RRE were the major driver of differential Rev-RRE activity.
In the current study, we conducted a detailed examination of RRE evolution in the blood plasma of patient SC3 using samples collected at 6-month intervals, from the time that viral RNA was first detected through year 6 of infection. DNA deep sequencing, selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) chemical probing, and a Rev-RRE functional assay were used to determine sequence evolution, RRE secondary structure, and Rev-RRE functional activity over time and to explore the mechanism underlying the observed activity differences. This study highlights, for the first time, the structure-function relationship of longitudinal RRE sequence evolution in a single patient and the underlying molecular mechanisms.
(This article was submitted to an online preprint archive [44].)
RESULTS
RRE sequence evolution in an HIV-infected individual followed over many years.
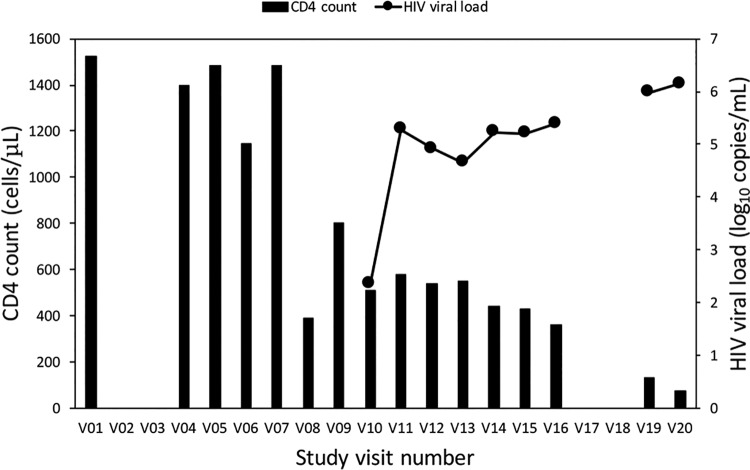
Plasma samples from patient SC3 were collected over a period of 10 years during standardized visits spaced about 6 months apart as part of the Women’s Interagency HIV Study (WIHS) Consortium in the Bronx, NY. Patient SC3 was enrolled in the WIHS cohort prior to HIV seroconversion, was followed through visit 20 (V20), and never received antiretroviral therapy (ART). Samples for this patient were available for all time points, with the exception of four missed visits (visits 2, 3, 17, and 18). Plasma samples from each WIHS visit were tested for p24 antibody and CD4 count (Fig. 1). The viral load was measured once the patient tested positive for p24 antibodies. In this report, each visit is denominated VXX, with XX representing the visit number. Although p24 antibodies were not detected under the original WIHS protocol until V10, a significant fall in her CD4 count was noted at V08, and we were able to readily amplify HIV from plasma taken at this time point, suggesting infection between V07 and V08. After a short-lived rebound at V09, the CD4 count continued to fall and reached a nadir of 73 cells/μl at V20. Seroconversion and viremia were noted in the data obtained from the WIHS protocol at V10, and the viral load continued to rise through V20, reaching a peak of 1.4 × 106 copies/ml. As expected, without ART treatment, we observed a general increase in the viral load concomitant with a decline in the CD4 count as disease progressed.
FIG 1.
Patient SC3 HIV viral load and CD4 count at each visit during participation in the WIHS cohort. Sequential study visits occurred about 6 months apart, and blood samples were obtained at each visit. V01 refers to the first visit, V04 to the fourth, etc. Data are missing for unattended visits. HIV sequences could first be amplified from plasma samples obtained at V08 and onwards. However, HIV seroconversion was first appreciated under the WIHS protocol at V10, and viral loads were performed only from this visit on. The patient died after V20.
We investigated RRE sequence evolution in viruses isolated from V08 and onwards. A total of 12 plasma samples representing visits 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 19, and 20 were received from the WIHS Consortium. Viral RNA isolation was attempted from each sample, and, if successful, RRE sequences were determined by next-generation sequencing. As stated above, the first visit to yield HIV sequences that could be amplified by PCR was V08. The prevalence of each RRE sequence at a given visit was calculated as a percentage of the total number of sequences present at that time point (see Fig. S1 in the supplemental material). Sequences present with a frequency of <5% were then excluded from consideration, as it was not possible to distinguish true rare RREs from artifacts due to PCR errors, and the prevalence of significant variants was recalculated after exclusion of these minority sequences (Fig. 2 and S2). RRE sequences were assigned a code in the form VXX-Y, where XX refers to the visit number and Y refers to the prevalence rank order of that sequence.
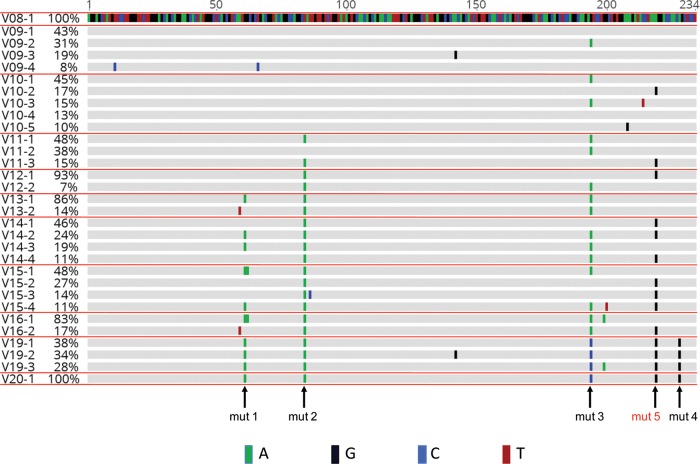
FIG 2.
Evolutionary alignment of SC3 RREs. Deep sequencing of viral RNA was performed from plasma collected at each study visit. This alignment includes RRE contigs that were present in at least 5% of the total sequences, as explained in Materials and Methods. For some visits, the total percentage shown is 101%, due to rounding errors from these calculations. Sequence labels are in the form of VXX-Y, where XX refers to the visit number and Y refers to the rank order of the contig within that visit plasma sample. Nucleotide changes relative to the presumptive founder sequence, V08-1, are highlighted. Mutation 1 (mut 1) to mut 5 refer to single-nucleotide changes that occurred in V20-1 relative to V08-1. Mut 5 is highlighted in red, as this change was also found in V10-2 and was not recognized in our previous study of the early and late RREs from patient SC3 (43). (See also Fig. S1 in the supplemental material for the alignment, including minority variants, and Fig. S2 for the complete nucleotide sequences included here.)
As shown in Fig. 1, data obtained from the WIHS indicated that patient SC3 seroconverted to HIV-positive status at V10. Thus, in our previous study, V10 was believed to represent a time point within 6 months of infection and was designated month 0 (M0). An RRE was identified at this visit by single-genome sequencing. It was believed to be the major species present at that time and designated an early RRE. Our deep DNA sequencing results showed that this early RRE sequence represented 17% of the sequences present at that visit, and it is therefore labeled V10-2 in the present study. This sequence was very similar to the single founder RRE sequence found at V08 (V08-1), differing only by a single nucleotide near the 3′ end and mapping to the right-hand side of SL-1 (here designated mut 5). Our previous study also identified a dominant RRE from V20 which we designated a late RRE. This RRE is identical to the V20-1 sequence generated by deep sequencing.
Since the V10-2 RRE previously described as the early RRE has been studied intensively by us, we decided to compare its properties to those of the founder V08-1 RRE. Both RREs were transcribed in vitro, and their secondary structures were chemically probed by 1-methyl-7-nitroisatoic anhydride (1M7), using selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) (45–51). In this procedure, RNAs are first modified with chemical reagents that selectively acylate unpaired ribonucleotides at their 2′-hydroxyl positions and then reverse transcribed under conditions that introduce mutations in cDNA opposite the sites of modification (48, 50). The position and frequencies of these mutations are used to create reactivity profiles indicating which RNA nucleotides are likely to be single or double stranded (data available upon request). This information is in turn used to guide the RNAstructure software (52) to generate lowest-Gibbs-free-energy secondary structure models.
SHAPE-MaP studies revealed that V08-1 and V10-2 formed similar structures with only minor local differences (Fig. 3A and B). These structures are similar to the previously reported ARV2/SF2 RRE structure (22), where a part of the central loop (nt 125 to 130) between SL-III and SL-IV paired with nucleotides from the upper stem of SL-I (nt 193 to 198), forming a stem that bridges the central loop and SL-IV and SL-V.
To determine the activity of each of these RREs, as well as that of V20-1, each RNA was cloned into a two-color fluorescence-based proviral reporter vector (53) which carried inactivating mutations in rev. RRE activity was measured by cotransfecting a separate plasmid that expressed the previously described M0-B/M57-A-SC3 Rev into 293T/17 cells. This Rev protein has been shown by single-genome sequencing to pair with the V10-2 RRE (43). It was also the unique founder Rev sequence present at V08. Unexpectedly, this sequence still persisted at V20 and was found in single genomes with the V20-1 RRE (43). The reporter construct expressed enhanced green fluorescent protein (eGFP) from the intron-containing Rev-dependent gag mRNA and the TagBFP blue fluorescent protein (BFP) from the intron-less Rev-independent nef-like transcript. Thus, in this assay, the ratio of the fluorescent signals (eGFP/TagBFP) is a measure of Rev-RRE functional activity. Control experiments showed that the assay is highly sensitive to the addition of Rev. For example, when the amount of Rev plasmid in the assay system is increased from 0 ng to 100 ng, there is a 268-fold increase in GFP expression, with only a slight decrease in BFP expression (2.6-fold) (53).
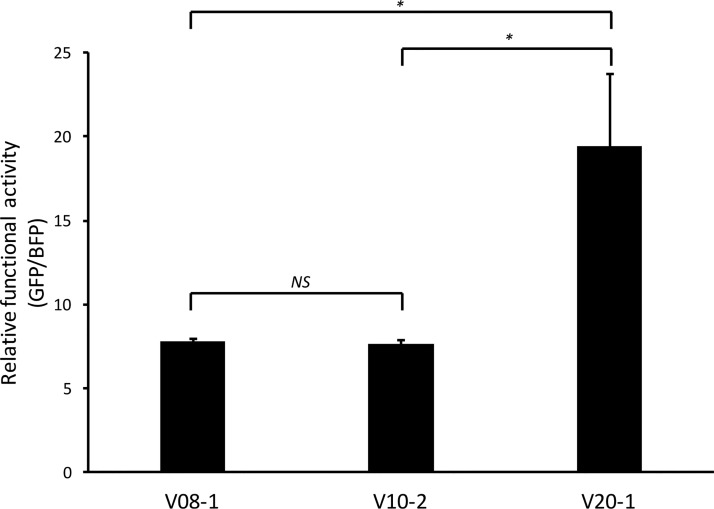
All three RREs displayed clear Rev responsiveness, as the fluorescent signal ratio in the non-Rev-containing transfections was at least 500-fold less than that in Rev-containing transfections (data not shown). Despite their local structural differences, both the V08-1 and V10-2 RREs showed similar levels of RRE activity (P = 0.999) (Fig. 4). Therefore, although mutation 5 arose after infection and increased in prevalence through V20, it did not increase RRE functional activity or significantly change the RRE structure. Additionally, the activities of V08-1 and V10-2 RREs were substantially less than that of V20-1 (Fig. 4) (P = 0.022 and P = 0.021, respectively), consistent with our previous finding that the V20-1 RRE is more active (43). Notably, we also replicated the finding that V20-1 is significantly more active than V10-2, using a lentiviral vector system, where packaging of genomic RNA and the titer are dependent on Rev-RRE function (data available upon request) (35, 54).
FIG 4.
Functional activity of SC3 founder (V08-1), early (V10-2), and late (V20-1) RREs. The functional activity of the three RREs was determined by transfecting reporter constructs containing the different RREs into 293T/17 cells along with 50 ng of SC3 Rev and measuring the ratio of the mean fluorescent intensity of GFP to that of BFP. The SEM is represented by error bars (n = 4). *, P < 0.05; NS, nonsignificant difference.
V10-2 and V20-1 RREs form distinctly different secondary structures.
We previously reported that there are only four single-nucleotide changes (mut 1 to 4) between the V10-2 and V20-1 RREs (43). A fifth mutation (mut 5) is present in both V10-2 and V20-1 and therefore did not score as a difference between the two RREs identified as early and late in our previous study. On a canonical 234-nt five-stem-loop (5SL) RRE structure (21), these changes map near the apical loop of SL-IIB (nt 61, mutation 1 [mut 1]) and SL-IIC (nt 84, mutation 2 [mut 2]), in the central loop region between SL-V and SL-I (nt 194, mutation 3 [mut 3]), and at the base of stem of SL-I (nt 228, mutation 4 [mut 4]) (Fig. 3B and D). We next explored whether the differences in activity between the two RREs could be attributed to structural differences caused by any individual nucleotide change.
In vitro-transcribed and folded V10-2 and V20-1 short RREs migrated at a significantly different rate when analyzed by native polyacrylamide gel electrophoresis (PAGE) (Fig. 3E), suggesting major structural differences. Consistent with the gel migration data, SHAPE-MaP of the two 234-nt RREs revealed that the V10-2 and V20-1 RREs adopt distinctly different conformations (Fig. 3B and D). Unlike V10-2, which had part of the central loop region base paired, V20-1 formed the canonical 5SL structure, where five distinct stem-loop structures (SL-I to SL-V) radiate out directly from the single-stranded central loop.
Closer analysis of the location of each of the four nucleotide changes suggested that the major structural shift in the two RREs might have been caused by mut 3. The nucleotide at the mut 3 position (G) is base paired in the bridging stem in V10-2. In V20-1, it mutates to C, disrupting base pairing at this position, thereby destabilizing the stem formed by nt 125 to 130 and nt 193 to 198. Consequently, nt 34 to 36 were significantly more reactive in V10-2 than in V20-1, while nt 125, 127, and 130 were significantly less reactive in V10-2 than in V20-1. Mut 1 and mut 2 produced more limited local structural changes, i.e., opening and formation of the base pair near the apical loop of SL-IIB and SL-IIC in V20-1 relative to V10-2. Since mut 4 was located at the primer hybridization site (see the PCR1 step of SHAPE-MaP in Materials and Methods) for the 234-nt RREs, we were unable to assess its effect on the secondary structure.
A 3-dimensional structural analysis of a 351-nt RRE by small-angle X-ray scattering has shown that the extended SL-I folds back on regions in and around SL-II to expose a cryptic Rev-binding site (22). Data obtained in connection with our previous study (43) showed that the extended SL-I of the 351-nt V10-2 and V20-1 RREs has an additional four nucleotide changes at nucleotide positions 13, 27, 29, and 57 (using the 351-nt RRE numbering system). We therefore investigated if these nucleotide changes in the extended SL-I affect RRE structure. Additionally, probing of these extended RREs provided an opportunity to determine the effect of mut 4 on their structure, as this position no longer fell on the primer hybridization site during SHAPE analysis of the 351-nt RREs. The long RREs were investigated by capillary electrophoresis-based SHAPE (Fig. 5) (CE-SHAPE), using N-methylisatoic anhydride (NMIA) as the electrophile (55, 56). CE-SHAPE differs from SHAPE-MaP in that the chemically modified nucleotides are identified as stops during primer extension. CE-SHAPE analysis of the 351-nt V10-2 and V20-1 revealed that mut 4 did not produce any structural changes between the V10-2 and V20-1 RREs, as it changed a U-A base pair to U-G. Additionally, the nucleotide differences in the extended SL-I region of these RREs produced only highly localized structural changes, if any. Changes at nt 13 and nt 57 produced no structural changes at all, whereas nucleotide differences at nt 27 and nt 29 induced opening of base pairs at these positions in V20-1. As these mutations in SL-1 did not alter the structure of the comparable region of the 234-nt RRE, subsequent studies investigated their effects only in the short RRE forms.
FIG 5.
Secondary structures of full-length (351-nt) V10-2 and V20-1 SC3 RREs. The secondary structures of V10-2 (A) and V20-1 (B) SC3 RREs were determined by CE-SHAPE. NMIA nucleotide reactivities are color coded and superimposed on the structures. The positions of the 8 nucleotides differing between the two RREs are represented by arrows. Mutations within the short 234-nt RRE are designated mut 1, mut 2, mut 3, and mut 4 and shown in green on the V20-1 RRE. Four additional mutations within stem SL-I are shown as green arrows without labels on the V20-1 RRE. The position of each of these changes is shown by black arrows on the V10-2 RRE.
Structural and functional analysis of patient SC3 RREs containing individual single-nucleotide mutations.
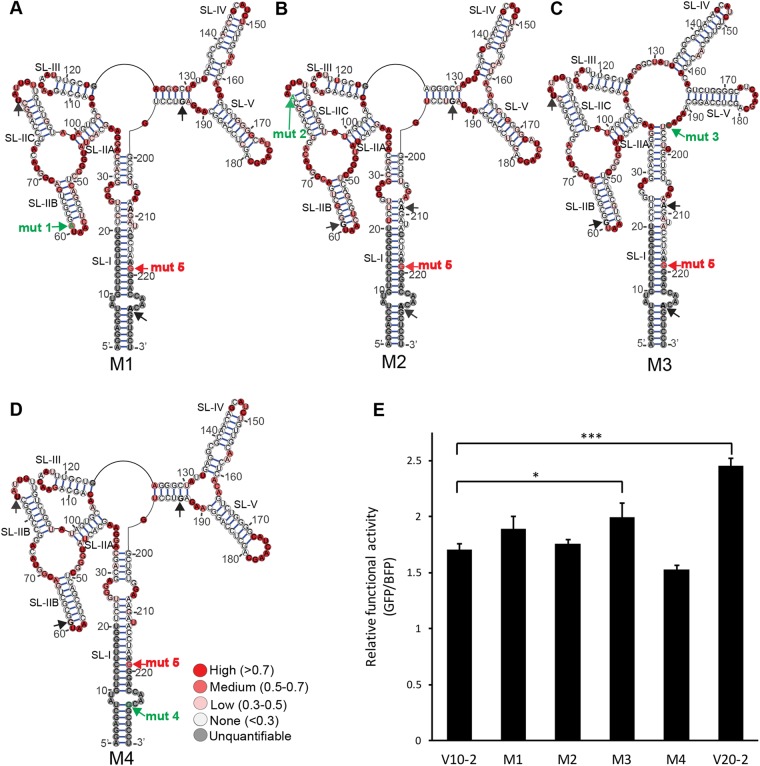
We next investigated the contribution of each of the 4 nucleotides which differed between the V10-2 and the V20-1 RREs to alterations in structure and functional activity. To this end, we created four new synthetic 234-nt RRE sequences, each containing one of the single-nucleotide changes in a V10-2 RRE background. The new RREs containing mut 1, 2, 3, and 4 were termed, respectively, M1, M2, M3, and M4. The secondary structure of each RRE was probed by SHAPE-MaP (Fig. 6A to D). As predicted, the M1, M2, and M4 RREs formed V10-2-like structures, whereas the M3 RRE formed a V20-1-like structure. The M1 RRE resembled the V10-2 RRE, with the exception that the G-to-A mutation eliminated a base pair adjacent to the SL-II apical loop. Similarly, the M2 RRE differed from the V10-2 RRE only in the formation of an extra base pair next to the apical loop of SL-IIC. The M4 RRE formed a structure identical to that of the V10-2 RRE, since the mut 4 A-to-G change preserved noncanonical base pairing at this position by replacing A-U with a G-U base pair. The major structural shift involving the central loop, SL-IV, SL-V, and SL-I between the V10-2 and V20-1 RREs was reproduced by the single G-to-C mutation in the M3 RRE. Therefore, we reasoned that mut 3 might be the major driver of the functional difference between the V10-2 and V20-1 RREs.
FIG 6.
Secondary structures and function of V10-2 RRE single mutants. Secondary structures of the M1 (A), M2 (B), M3 (C), and M4 (D) SC3 RREs were determined by SHAPE-MaP. 1M7 reactivities are color coded and superimposed on the structures. The positions of mut 1 to mut 5 are represented by black and green arrows. Mut 1, mut 2, mut 3, and mut 4 are indicated in green on the RREs, where the nucleotide at that position differs from that in V10-2, while mut 5 is represented in red and is present on every structure. (E) Rev-RRE functional activity of the M1 to M4 RREs compared with that of V10-2 and V20-1 in the presence of 100 ng SC3 Rev. SEM are represented by error bars (n = 2). *, P = 0.072; ***, P = 0.001.
We next measured the contribution of each mutation to the functional activity difference between the V10-2 and V20-1 RREs in 293T/17 cells (Fig. 6E). The activity of RREs M1 to M4 was determined using the fluorescence-based transient proviral reporter with the same cognate SC3 Rev used in the assay whose results are presented in Fig. 4. As before, in the presence of SC3 Rev, the functional activity of the V20-1 RRE was significantly higher than that of the V10-2 RRE (P = 0.001). Among all single SC3 mutants, only M3 displayed activity that was statistically significantly higher than that of V10-2 (P = 0.072), even though it did not quite reach the activity level of V20-1. The M1 and M2 RREs were only slightly more active than V10-2, but these differences did not reach statistical significance. The activity of the M4 RRE was also not statistically distinguishable from that of V10-2. However, the M4 RRE had a tendency toward lower, rather than higher, functional activity. Thus, the difference in functional activity between V10-2 and V20-1 could not be explained by any single nucleotide mutation.
Structural and functional analysis of selected RRE sequences derived from patient SC3 at different time points.
To further understand how specific sequence changes between the V10-2 and V20-1 RREs affected functional activity, we studied intermediate RRE sequences from viruses that arose during disease progression in patient SC3. SC3 RREs that allowed us to test combinations of these mutations in the order in which they appeared in the natural isolates were selected from this evolutionary data set (Fig. 2 and S2). The secondary structure and functional activity of these RREs were then determined by SHAPE-MaP and the fluorescence-based reporter assay, respectively.
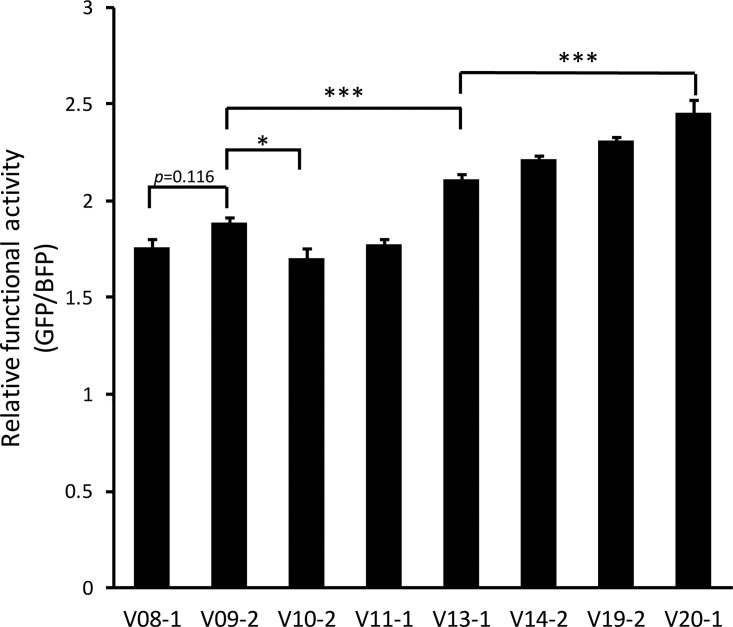
The position designated mut 3 starts as a G in V08-1, remains a G in V10-2, and changes to an A in many sequences in V09 through V16 and then to a C in V19 and V20. As this is the position that causes the major structural shift between V10-2 and V20-1, we sought to determine if the A at the mut 3 position would result in the same structure as its replacement by C. To test this, we determined the secondary structure of the V09-2 RRE by SHAPE-MaP and compared it to the structure of V08-1 and V10-2 (Fig. 3A to C). The V09-2 RRE sequence is identical to V08-1 except at the mut 3 position, where the G in V08-1 is replaced with A. We hypothesized that if disruption of the G-C base pair at this position induced the structural shift observed between V10-2 and V20-1, then such a shift should also be reflected between V08-1 and V09-2. Indeed, the secondary structure of V09-2 resembled that of the V20-1 RRE, confirming our hypothesis. The V09-2 RRE was also slightly more active than the V08-1 RRE (though this trend did not reach statistical significance [P = 0.116]) and the V10-2 RRE (P = 0.027) (Fig. 7). Consistent with our previous observation with the M3 RRE, the structural shift invoked in V20-1 and V09-2 relative to V08-1 and V10-2 showed a trend toward increased SC3 RRE activity.
FIG 7.
Rev-RRE activity of SC3 RRE haplotypes. The activity of selected RREs was determined in the presence of 100 ng Rev using the fluorescent assay system. SEM are represented by the error bars (n = 2 for all RREs, except V9-2 and V13-1, where n = 4). *, P < 0.05; ***, P < 0.001.
Mut 2 was first noted in the sample from V11 and was present in all RRE sequences from subsequent visits. We tested the V11-1 RRE, as it provided the opportunity of investigating the combination of mut 2 together with A in the position of mut 3. Although it was predicted to have higher activity than M3 and the V09-2 RRE, the V11-1 RRE had a similar level of RRE activity as the founder RRE, V08-1, suggesting that while A at the position of mut 3 increases functional activity alone, it does not do so in the context of the additional mut 2 (P = 1.000).
We first detected mut 1 in plasma samples from visit 13. This mutation was frequently observed on subsequent visits and was present in the majority of RREs from visit 15 onwards. The V13-1 RRE contained not only mut 1 but also mut 2 and A in the mut 3 position. This RRE had significantly higher activity than both the V08-1 and V09-2 variants (P < 0.001 for both comparisons). Therefore, mut 1 not only contributes to RRE activity in itself but also rescues the activity of the combination of mut 2 and an A in the mut 3 position.
We also tested the activity of V14-2, which allowed us to test the combined phenotype of mut 1, mut 2, A in the mut 3 position, and mut 5. The activity of this RRE did not significantly differ from that of V13-1, suggesting that acquisition of mut 5 does not contribute to RRE activity (P = 0.332). However, the activity of the V20-1 RRE, which harbors C in the position of mut 3 rather than the A of V13-1, as well as the other mutations, was significantly higher than that of V13-1 (P < 0.001). This suggests that C in the mut 3 position is important for imparting higher RRE activity when all other mutations are present. It is also possible that although mut 4 does not contribute to RRE activity individually, it can contribute to activity when combined with the other additional changes.
This set of SC3 RREs permitted examination of changes in functional activity over time. The activities of V08-1, V10-2, and V11-1 were very similar, demonstrating that the combinations of mutations observed in these sequences are not sufficient in themselves to confer higher functional activity. V13-1 was the first tested RRE to arise, other than V09-2, that showed significantly higher activity than V08-1, and it did so with the combination of mut 1, mut 2, and A in the mut 3 position. The accumulation of additional changes in V14-2 (addition of mut 5), V19-2 (addition of mut 4, the A-to-C change at mut 3, and an additional A-to-G change at nt 142), and V20-1 (reversion of nt 142 to A) corresponded to a trend toward increasing functional activity at each step. This steady trend suggests functional evolution of the RRE in the later phase of the disease course of patient SC3, as new combinations of mutations arose and those conferring greater functional activity showed preferential selection.
Shannon entropy profile of SC3 RRE variants and single mutants.
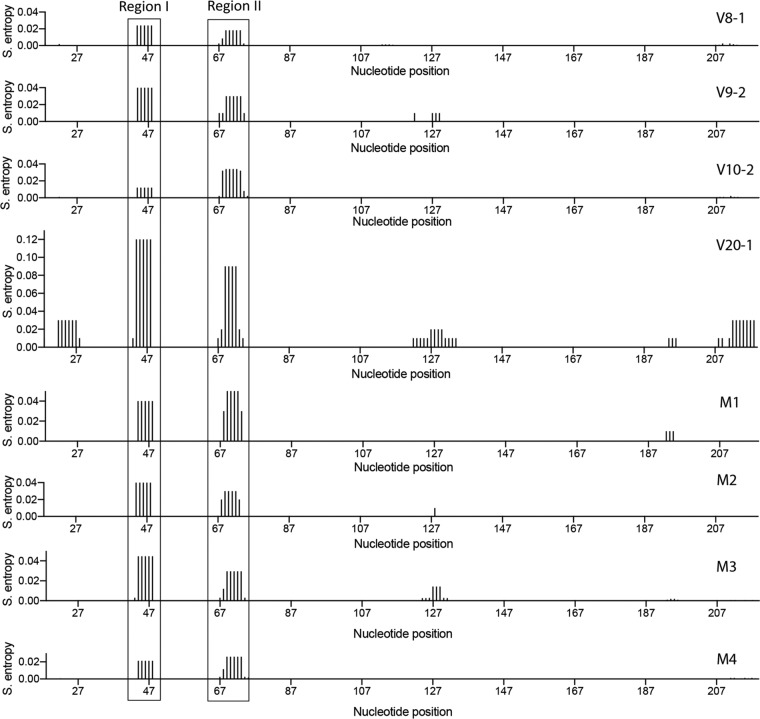
Since the energetically most favorable secondary structures of SC3 RRE variants and single mutants were insufficient to explain the functional activity differences observed between the V08-1 and V20-1 RREs, we next assessed the ability of the RREs to adopt alternative conformers by evaluating their SHAPE-guided Shannon entropies and base pairing probability at a single-nucleotide resolution. Shannon entropies were calculated based on the probability for each base pair appearing across all possible structures predicted for the RNA. Regions with highly stable well-defined RNA structures are characterized by lower Shannon entropies. Conversely, regions with high Shannon entropy are likely to form alternative conformers.
We compared the Shannon entropy profiles of the V08-1, V09-2, V10-2, V20-1, M1, M2, M3, and M4 SC3 RREs (Fig. 8). Most regions of these RREs exhibited low Shannon entropy, suggesting that their overall secondary structures are highly stable. This result is consistent with the observation that these RRE RNAs migrate as a single discrete band on a native agarose gel, suggesting a high degree of structural homogeneity (data available upon request).
FIG 8.
Shannon entropy (S. entropy) profiles of SC3 RRE variants and single mutants. The Shannon entropy values of the SHAPE-MaP-generated RRE structures, smoothed over centered 11-nt sliding windows, are plotted as a function of the nucleotide position. Higher Shannon entropy suggests regions that are structurally dynamic. The boxed regions correspond to the loop between SL-IIA and SL-IIB (region I) and the loop between SL-IIB and SL-IIC (region II). These regions display higher Shannon entropy in the higher-activity V20-1 structure and lower entropy in the lower-activity V08-1 structure.
However, we observed differences in Shannon entropy values at two different regions. Specifically, values of the loop regions between SL-IIA and SL-IIB (region I) and between SL-IIB and SL-IIC (region II) varied between RREs. The values for the V20-1 RRE were, respectively, 5- to 6-fold and 4-fold higher than those for the V08-1 and V10-2 RREs, suggesting that the nucleotides in these two regions of the V20-1 RRE are not always single stranded, as depicted in the secondary structures generated by SHAPE-MaP. This finding is further corroborated by the SHAPE-MaP-guided base pairing probability calculated for each of these RREs (data available upon request), which shows that the two higher-entropy regions of the V20-1 RRE can base pair with each other with a probability of 10% to 80%. Both the Shannon entropy and the pairing probability data suggest that these two regions of the V20-1 RRE are structurally dynamic, a feature that confers accessibility to protein interaction with the RNA by serving as landing pads for protein cofactors (45). This accessibility may have implications for Rev binding, possibly contributing to the higher activity of V20-1 RRE.
Furthermore, both the V09-2 and M3 RREs have 2-fold higher Shannon entropy for the loop region between SL-IIA and SL-IIB (region I) than the V08-1 and V10-2 RREs. Their entropy values for the nucleotides between SL-IIB and SL-IIC (region II), however, did not change relative to those of V08-1 and V10-2. While both V09-2 and M3 undergo the structural shift characteristic of V20-1 and are modestly more active than V08-1 and V10-2, the inability of the V09-1 and M3 RREs to reach the degree of structural flexibility of the V20-1 RRE in the internal loop of SL-II might explain why these RREs are not as functionally active as V20-1. The Shannon entropy profile of the M2 RRE was similar to that of the M3 and V09-1 RREs, whereas the M1 RRE had a 2-fold higher Shannon entropy than V08-1 and V10-2 in both regions of SL-II. The modestly higher entropy in the M1, M2, and M3 RREs relative to V08-1 and V10-2 might help to explain their trend toward higher functional activity (Fig. 6). The M4 RRE, on the other hand, did not show any increase in entropy over V08-1 and V10-2 and also displayed similar or lower functional activity.
Taken together, our analysis suggests that both the major structural shift observed between the V08-1 and V20-1 RREs and possibly also the difference in structural dynamism in the internal loop of SL-II contribute to the significant increase in functional activity over time.
DISCUSSION
In this study, we describe a complex relationship between the RRE structure and functional activity using naturally occurring sequences obtained from a single patient. This study is the first detailed examination of the longitudinal evolution of the HIV-1 RRE over about 6 years of naturally occurring infection and demonstrates clear selection pressures acting on the RRE sequence, with a tendency toward increased functional activity over time. Increased activity can be explained by large-scale conformational changes within the RRE and a decrease in base-pairing stability at the initial Rev-binding site in SL-II.
The evolution of the RRE sequence is strongly suggestive of selection pressures operating on the RRE itself. The single founder RRE sequence found at visit 8 diversified initially and by visit 20 converged on only a single circulating RRE sequence differing from V08-1 by 5 single nucleotides. The early appearance and persistence of mutations at only a few locations, as well as the reduced viral diversity by visit 20, argue that the phenotype resulting from these changes was subject to strong selection pressures. Functional activity analysis of the SC3 RREs suggests that the RREs with higher activity were selected as disease progressed, with the early-time-point RREs (V08-1 and V10-2) displaying the lowest functional activity and the late-time-point RRE (V20-1) displaying the highest functional activity. The tendency toward increased functional activity corresponded with gradual accumulation, in different combinations, of five mutations that characterize V20-1 relative to V08-1.
There was no direct correspondence between specific mutations and functional activity phenotypes. Changes at the mutation 3 position, predicted to disrupt base pairing, appeared to result in an activity increase. However, the V09-2 RRE containing this change alone still had significantly lower activity than V20-1, showing that the other mutations also contribute to a change in RRE activity in combination. While all of mutations 1 to 5 were necessary to achieve the highest activity, their contributions were not merely additive. For example, V11-1 resembled V09-2 with the addition of mut 2, but it did not have increased activity relative to the founder. Additionally, mutations which had no impact on activity in isolation could increase activity in combination. For example, mut 4 alone did not change activity but was responsible for the increase in activity between V14-2 and V20-1 in the context of the other four mutations.
Two types of structural changes that can explain differences in activity were uncovered in this study: major conformational changes and regions of increased base-pairing entropy. Previously, we have shown that alternative conformations of the laboratory strain NL4-3 RRE display significantly different activities, with a four-stem-loop conformation supporting lower replicative capacity than a five-stem-loop structure (21). In this study, we found that the higher-activity RRE (V20-1) formed a five-stem-loop conformation resembling that observed for NL4-3, while the lower-activity RREs (V08-1 and V10-2) adopted a different conformation with a more collapsed central loop. As expected from functional analysis of the M3 and V09-2 RREs, both the G-to-A (V09-2) and G-to-C (M3) changes at the mutation 3 position induced a five-stem-loop structure associated with increased activity. The additional mutations were not individually associated with large-scale conformational changes in SHAPE-MaP modeling.
The combination of mutations 1 to 5, though not each mutation in isolation, also corresponded to increased Shannon entropy in stem-loop II. This is the region where the first and second dimers of Rev bind (57). The disordered base pairing in this region, implied by greater entropy, may facilitate Rev-RNA binding at the primary Rev-binding site and subsequent multimerization, leading to enhanced activity. This is consistent with our previous in vitro gel-shift observations (43) that demonstrated the ability of the V20-1 RRE to promote Rev multimerization more efficiently than the V10-2 RRE. In the absence of major conformational changes attributable to mutations 1, 2, 4, and 5, this mechanism may account for the difference in functional activity between the RREs containing unpaired nucleotides at the mutation 3 position (e.g., V09-2 and M3) that lack these additional mutations and the V20-1 RRE. It remains unclear how the presence of these mutations in combination results in increased stem-loop II entropy.
This study also provides further support for the hypothesis that the Rev-RRE regulatory axis plays an important role in HIV pathogenesis. An important consideration in the interpretation of these findings is that all five mutations in V20-1 relative to V08-1 are nonsynonymous in the overlapping gp41-coding sequence, raising the possibility that changes in Env could contribute to the selective pressure. We searched for known cytotoxic T-lymphocyte (CTL) epitopes in the Env open reading frame overlapping the V08-1 RRE sequence using the Los Alamos HIV database Epitope Location Finder tool (www.hiv.lanl.gov). CTL epitopes have been described in the V08-1 regions containing three of the five mutations (mutations 3, 4, and 5). Thus, selection of these mutations could plausibly reflect immune evasion, though it is notable that neither mutation 3 nor mutation 5 was consistently found in all RRE sequences at all time points after mutations 3 and 5 were first observed at visits 9 and 10.
While the selection evidenced by sequence convergence at visit 20 might be due to pressure on Env in terms of immune evasion, replication efficiency, or other factors, it is likely that functional differences in Rev-RRE activity also play a significant role in overall viral fitness. Nucleotide changes at the mutation 3 position highlight this likelihood. The nonsynonymous mutations from G to A at visit 9 and A to C at visit 19 are both consistent with CTL evasion. From the standpoint of the RRE, however, the G-to-A change shifts the RRE structure to a higher-activity conformation, while the A-to-C change is structurally and functionally neutral. Thus, the A-to-C mutation preserves high RRE activity while also modifying the Env amino acid sequence. The convergence of circulating RRE sequences by visit 20 is particularly striking, in light of the very low CD4 count at this time point, as viral fitness would likely have been more influenced by Rev-RRE activity than by immune surveillance.
We were unable to assess the functional evolution of the Rev protein over time in this study due to the sequencing strategy used, which created reads too short to allow linkage of both of the Rev-coding exons and the RRE. However, previous work using single-genome sequencing with samples from this patient (43) demonstrated that a single Rev amino acid sequence was most prevalent at both visits V10 and V20. This is the Rev sequence that was used in this study. Its persistence throughout the infection period suggests that it likely occurred in conjunction with most or all of the RREs tested here. It is also notable that m6A methylation of RRE RNA has recently been proposed to be an additional mechanism of modulating HIV replication capacity (58). This was not assessed in the present study but could additionally contribute to differences in Rev-RRE activity.
The generalizability of the findings described here awaits further longitudinal studies, analyzing both Rev and RRE sequences, in patients with various courses of disease. A clear understanding of RRE evolution during natural infection will help to further our understanding of the role that this regulatory axis may play in key aspects of the viral life cycle, including transmission and the establishment of latency. Furthermore, since the Rev-RRE interaction is an essential step in viral replication, it is also a promising target for drug development. This study demonstrates that RRE structures can vary over the course of natural infection and, thus, that rational drug design must account for the spectrum of potential Rev-RRE interaction conformations.
MATERIALS AND METHODS
Clinical samples and ethics statement.
All studies involving human subjects were approved by the Institutional Review Board at Montefiore Hospital, Bronx, NY, as part of the Women’s Interagency HIV Study (WIHS). Written informed consent was provided by all study subjects. Plasma samples from one patient (SC3), collected over a period of 7 years, were obtained from the WIHS Consortium, Bronx, NY (Kathryn Anastos, principal investigator), as previously described (43). A total of 11 samples representing different time points were received for analysis.
Viral RNA extraction and cDNA synthesis.
Viral RNA was extracted from 1 ml of plasma using a guanidinium extraction protocol (59). Viral cDNA synthesis was performed immediately after viral RNA was extracted as described previously (43). Briefly, each viral RNA pellet was resuspended in 40 μl of 5 mM Tris-HCl, pH 8.0, prior to cDNA synthesis with 50 μM oligo(dT) and 100 U of SuperScript III reverse transcriptase per reaction mixture (Invitrogen, Carlsbad, CA). The reverse transcription (RT) reaction mixture was incubated at 50°C for 1 h, then 55°C for 1 h, and then 70°C for 15 min, after which 4 units of RNase H (Invitrogen) was added and samples were incubated at 37°C for 20 min. Samples were stored at −80°C prior to amplicon generation.
PCR amplification and purification.
Amplicons (∼3 kb) spanning both HIV Rev exons and the RRE were produced for each time point. The first-round PCR was performed by adding 1 μl of cDNA template to a 20-μl reaction mixture containing 0.005 U of Platinum Taq Hi-Fidelity polymerase (Invitrogen) as previously described (60). Using 1 μl of the first-round PCR product as a template, nested PCR resulted in amplicons ∼3 kb in length. The primers for the first-round PCR were 2302 (5′-AAGCCACCTTTGCCTAGTG-3′) and 2278 (5′-TTGCTACTTGTGATTGCTCCATGT-3′), while those for the nested PCR were 2277 (5′-TAGAGCCCTGGAAGCATCCAGGAAG-3′) and 2280 (5′-GTCTCGAGATACTGCTCCCACCC-3′) (43). Amplicon size was verified by agarose gel electrophoresis.
Each 20-μl PCR mixture was purified with 36 μl of AMPure XP beads (Agencourt). After addition of beads, samples were pipetted up and down 10 times and incubated at room temperature for 10 min prior to being placed on a magnetic stand. After a 2-min incubation, the supernatant was removed and the beads were washed twice with 80% ethanol. After the beads were air dried on a magnetic stand for 10 min, they were removed from the magnet, resuspended in 52.5 μl of elution buffer, and incubated at room temperature for 2 min. The beads were then placed on the magnetic stand for 2 min or until the supernatant was cleared. The cleared supernatant containing the purified PCR amplicons was removed and placed in a new 96-well plate and then stored at −20°C.
DNA library preparation and sequencing.
Prior to preparing the DNA library, purified amplicon products were quantified with a double-stranded DNA (dsDNA) high-sensitivity (HS) assay kit (Invitrogen) using a Qubit (v2.0) fluorometer (Invitrogen). The DNA library was prepared using the standard Nextera XT DNA library preparation protocol (Illumina). A total of 1 ng of input DNA for each sample was added to the reaction buffer, which fragments the input DNA and adds adapter sequences to the ends to allow for PCRs downstream in the library preparation process. A brief PCR cycle followed the fragmentation protocol to add indexes used to identify each individual sample and additional sequences for proper cluster formation. PCR samples were purified and size selected for 300- to 500-bp amplicons using 25 μl of AMPure XP beads. Amplicons were pooled together in equimolar concentrations and then sequenced with an Illumina MiSeq reagent kit (v3; 600 cycles).
RRE sequence analysis.
Paired-end reads for each time point were generated using an Illumina MiSeq kit. The two overlapping paired reads were merged using FLASH (61), a plug-in in the Geneious tool (Biomatters, Auckland, New Zealand), to produce a single read for each pair. These reads were then aligned to the NL4-3 genome, and reads overlapping the RRE were extracted. Extracted reads were filtered for individual reads that spanned the full length of the short RRE. These reads were assembled into contigs using the Geneious de novo assembly tool, set for a minimum overlap identity of 100% and a minimum overlap of the entire 234-nt minimal RRE. The frequency of each contig was calculated based on the number of reads assigned to each contig. Contigs comprising less than 5% of the total reads were then removed from each time point (to eliminate minor HIV species and/or potential PCR errors), and the frequencies of the remaining contigs were recalculated based on the total number of reads remaining. The sequence of each contig was then aligned to the sequence from the major V08 contig to generate the alignment shown in Fig. 2 (see also Fig. S1 and S2 in the supplemental material). RREs were named using the format VXX-Y, where XX corresponds to the WIHS cohort visit number at which the plasma sample generating the sequence was obtained and Y corresponds to the frequency rank order of the sequence. Thus, V10-2 refers to the second most prevalent RRE from the sequencing of the plasma sample obtained at WIHS visit 10.
In vitro transcription of RRE RNAs.
RRE RNAs were in vitro transcribed by T7 RNA polymerase using a MEGAshort-script kit (Life Technologies) per the manufacturer’s guidelines. Templates for in vitro transcription were generated by PCR amplification of RREs from plasmids containing synthetic sequences (Integrated DNA Technologies) using oligonucleotides designed to introduce both a T7 promoter sequence at the 5′ end of the 234-nt and 351-nt RREs and a structure cassette at the 3′ end of the 351-nt RREs (Table S1). Plasmids used for generating a transcription DNA template for the 234-nt V08-1, V09-2, V10-2, V20-1, M1, M2, M3, and M4 RREs were designated pHR 5334, pHR 5766, pHR 4784, pHR 4788, pHR 5326, pHR 5328, pHR 5330, and pHR 5332, respectively. Those used to generate the 351-nt V10-2 and V20-1 RREs were designated pHR 5321 and pHR 5322, respectively. In vitro transcription reactions were treated with Turbo DNase I (Life Technologies) for 1 h at 37°C to digest the DNA template and heated to 85°C for 2 min, and RNA was fractionated on a denaturing gel (5% polyacrylamide [19:1], 1× Tris-borate-EDTA [TBE], 7 M urea) at a constant temperature (45°C, 30 W maximum). RRE bands were located by UV shadowing, excised, electroeluted at 200 V for 2 h at 4°C, ethanol precipitated, and stored at 4°C in TE buffer (10 mM Tris, pH 7.6, 0.1 mM EDTA).
Folding and SHAPE-MaP profiling of RRE RNA.
For each RNA, reaction mixtures with 1M7 [1M7 (+)], reaction mixtures without 1M7 [1M7 (−)], and denaturation control reaction mixtures were generated. Approximately 5 pmol of RRE RNA was incubated with renaturation buffer (1×; 10 mM Tris, pH 8.0, 100 mM KCl, 0.1 mM EDTA) in a total volume of 5 μl, heated to 85°C, and renatured by slow cooling (0.1°C/s) to 25°C. Renatured RNA was incubated with RNA folding buffer (1×; 35 mM Tris, pH 8.0, 90 mM KCl, 0.15 mM EDTA, 5 mM MgCl2, 2.5% glycerol) in a total volume of 9 μl at 37°C for 30 min. RNA was modified by addition of 1 μl of 25 mM 1M7. The negative-control reaction contained 1 μl of dimethyl sulfoxide (DMSO) instead of 1M7. The denaturation control was produced by incubating 5 pmol of RNA with denaturation buffer (1×; 50% formamide, 50 mM HEPES, 4 mM EDTA) in a volume of 9 μl at 95°C for 1 min. One microliter of 20 mM 1M7 was added, and the mixture was incubated at 95°C for another 1 min. RNA was recovered from the 1M7 (+), 1M7 (−), and denatured control reaction mixtures by ethanol precipitation, resuspended in TE buffer (10 mM Tris, pH 7.6; 0.1 mM EDTA), and stored at −20°C.
Mutational profiling of modified RNA.
RNA from the 1M7 (+), 1M7 (−), and denatured control reaction mixtures was reverse transcribed using the corresponding oligonucleotides to generate a cDNA library (Table S1). Reverse transcription was performed by first annealing 2 μM RT oligonucleotide to the RNA in a reaction volume of 11 μl by incubation at 65°C for 5 min, followed by cooling on ice. cDNA synthesis was initiated by incubating the annealing mixture with 8 μl of 2.5× RT reaction mixture (2.5×; 125 mM Tris [pH 8.0], 187.5 mM KCl, 15 mM MnCl2, 25 mM dithiothreitol, 1.25 mM deoxynucleoside triphosphates [dNTPs]) and 1 μl of SuperScript II reverse transcriptase (200 U/μl; Thermo Fisher) for 42°C for 3 h. The RNA template was then hydrolyzed by adding 1 μl 2 N NaOH, followed by neutralization by addition of 1 μl 2 N HCl. The cDNA library was purified using G50 spin columns (GE Healthcare).
For mutational profiling, cDNA was converted to dsDNA with Illumina adapters for high-throughput sequencing on an Illumina platform. This was achieved in two consecutive PCRs, namely, PCR1 and PCR2. The first reaction (PCR1) appended partial Illumina adapters to the ends of the amplicons. The entire cDNA library was used as template in a 100-μl PCR1 reaction mixture (1.1 μl each of 50 pmol of forward and reverse oligonucleotides, 2 μl of 10 mM dNTPs, 20 μl of 5× Q5 reaction buffer, 1 μl of Hot Start High-Fidelity DNA polymerase [NEB]) using cycling conditions of 98°C for 30 s; 15 cycles of 98°C for 10 s, 50°C for 30 s, and 72°C for 30 s; and 72°C for 2 min. The PCR product was gel purified, and 10% was used as the template DNA in PCR2. PCR2 completed the Illumina adapters while adding appropriate barcoding indices. PCR2 and the cycling conditions were the same as those described above for PCR1. The resulting amplicon library was fractionated through a 2% agarose gel, and amplicons were purified from the gel slices by electroelution at room temperature for 2 h, followed by ethanol precipitation. Each amplicon library was quantified by real-time PCR using a KAPA universal library quantification kit (Illumina) per the manufacturer’s protocol. Sequencing libraries were pooled and mixed with 20% PhiX and sequenced on an Illumina MiSeq sequencer to generate 2 × 150 paired-end reads. Sequence files were fed into ShapeMapper (v1.2) software (50) to generate SHAPE reactivity profiles by aligning the reads to 234-nt RRE RNA sequence using the software with default settings. Reactivity values obtained from ShapeMapper were input to RNAstructure software (52) to generate the minimum-free-energy RNA secondary structure model.
RRE gel migration assay.
RRE structural homogeneity was assessed by comparing the migration rate of the folded RREs on native agarose and polyacrylamide gels. Approximately 20 pmol of RNA was suspended in 5 μl renaturation buffer (1×; 10 mM Tris, pH 8.0, 100 mM KCl, 0.1 mM EDTA), heated to 85°C, and renatured by slow cooling (0.1°C/s) to 25°C. Renatured RNA was incubated with 5 μl of 2× RNA folding buffer (1×; 70 mM Tris, pH 8.0, 180 mM KCl, 0.3 mM EDTA, 10 mM MgCl2, 5% glycerol) at 37°C for 30 min and fractionated through a native 8% polyacrylamide gel (29:1) run for 22 h at 4°C or on a 2% native agarose. RRE bands were visualized by UV shadowing and ethidium bromide staining, respectively.
Plasmid constructs for functional assays of Rev-RRE activity.
The functional activity of the Rev-RRE pairs was determined by means of a fluorescence-based transient-transfection assay. To test RRE sequences, a plasmid containing a full-length NL4-3 HIV-1 sequence (62) was modified to express blue fluorescent protein (BFP) in a Rev-independent fashion and green fluorescent protein (GFP) in a Rev-dependent fashion. The native 351-nt RRE within env was flanked by XmaI and XbaI sites to permit exchange of this sequence with other RREs of interest. Additional modifications were made to ensure that the construct was not replication competent and could not express Rev. To create additional RRE constructs, the native RRE was removed by digesting the plasmid with XmaI and XbaI. Commercially synthesized 234-nt RRE sequences of interest with appropriate flanking sequences (Integrated DNA Technologies) were then cloned into the opened env region using Gibson assembly.
Rev was provided in trans using one of two constructs. The predominant Rev previously identified at V10 and V20 by single-genome sequencing had identical amino acid sequences. This Rev was designated M0-B/M57-A-SC3 and used for functional assays, except as otherwise noted (43).
For the analysis whose results are described in Fig. 4, the M0-B/M57-A-SC3 rev sequence was cloned into a cytomegalovirus (CMV) expression plasmid. For the remaining functional assays, a separate Rev-expressing construct was created by modifying the murine stem cell virus (MSCV) construct pMSCV-IRES-mCherry FP (Addgene plasmid 52114) to express both the M0-B/M57-A-SC3 Rev and the mCherry fluorescent protein by means of a bicistronic transcript including an internal ribosome entry site (IRES) (63). The MSCV vector was a gift from Dario Vignali (unpublished). The constructs utilized in the functional assays are listed in Table S2.
Selection of RREs for functional analysis.
Both naturally occurring RREs sequenced from patient SC3 and synthetic RREs containing individual mutations of interest were tested for functional activity. All RREs tested were 234 nt in length. The naturally occurring RREs assessed were V08-1, V09-2, V10-2, V11-1, V13-1, V14-2, V19-2, and V20-1. Additionally, synthetic RREs were created based on single-nucleotide differences between the sequences of V10-2 and V20-1. The mutation 1 (M1) RRE is identical to V10-2, with the exception of a G-to-A change at position 61, reflecting the G-to-A change seen at that position in V20-1. The mutation 2 (M2) RRE is identical to V10-2, with the exception of a T-to-A change at position 84. Of note, this RRE sequence was also observed to occur naturally as sequence V12-1. The mutation 3 (M3) RRE is identical to V10-2, with the exception of a G-to-C change at position 194. The mutation 4 (M4) RRE is identical to V10-2, with the exception of an A-to-G change at position 228.
Rev-RRE functional activity assays.
RRE-containing constructs were designed such that GFP expression occurs in a Rev-dependent fashion, while BFP expression occurs in a Rev-independent fashion. In cells transfected with both the RRE-containing and Rev-containing constructs, the degree of GFP expression relative to BFP expression is proportional to the functional activity of the tested Rev-RRE pair (53).
Except as noted, transfections were performed using the polyethylenimine method and 8 × 105 293T/17 cells in each well of a 12-well plate. Cells were maintained in 1 ml Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 5% bovine calf serum. Each RRE-containing construct was tested individually with the SC3 Rev-containing construct. Additionally, each RRE-containing construct was transfected into cells without the addition of Rev to ensure that GFP expression in this system was truly Rev dependent. In transfections performed without the Rev-containing construct, an empty CMV construct was used to compensate such that each transfection was performed with a constant mass of plasmid. For each transfection, 1,000 ng of an RRE-containing construct and 100 ng of either the MSCV-Rev construct or the empty vector were used. Simultaneously, different cultures of 293T/17 cells were transfected with constructs expressing GFP, BFP, or mCherry alone as single-color controls to permit color compensation during flow cytometry. Each set of transfections was performed in duplicate for all RREs with the exception of V09-2 and V13-1, for which four replicates were performed. After transfection, cells were incubated for 48 h and then suspended in phosphate-buffered saline. Flow cytometry was performed on the resulting suspension using an Attune NxT flow cytometer with an autosampler attachment (Thermo Fischer Scientific). Data acquisition was performed on the Attune NxT software package using the channels listed in Table 1.
TABLE 1.
Channel settings for the Attune flow cytometer
| Channel | Laser line (nm) | Emission filter (nm) | Fluorescent protein |
|---|---|---|---|
| 1 | 488 | 530/30 | eGFP |
| 2 | 405 | 440/50 | TagBFP |
| 3 | 561 | 620/15 | mCherry |
Postacquisition color compensation and data analysis were performed using FlowJo (v10) software (FlowJo, LLC). For each analysis, gating was performed on single 293T/17 cells. Next, a daughter population of cells that expressed the RRE-containing transcript was identified by gating on positivity for GFP or BFP. Using this population, the ratio of the arithmetic mean fluorescent intensity (MFI) of eGFP to that of TagBFP for successfully transfected cells was calculated in both the presence and the absence of Rev. A low ratio in the absence of Rev and a high ratio in its presence indicated that GFP expression was Rev dependent.
After determining that the RRE-containing constructs expressed GFP in a Rev-dependent fashion, an additional analysis was performed to determine the functional activity of specific Rev-RRE pairs. In this further analysis, only those transfections including both an RRE-containing construct and the Rev-containing construct were considered. An additional daughter population was created from that described above by gating on mCherry positivity. This final population consisted only of single 293T/17 cells expressing GFP and/or BFP and also mCherry. This pattern of expression ensured that selected cells were successfully cotransfected with both plasmid constructs. Once this population was defined, the ratio of the GFP MFI to the BFP MFI was recalculated. This final GFP/BFP ratio was used to determine the relative functional activity of different RREs assessed in the presence of SC3 Rev. Differences in functional activity values between RREs were evaluated using SPSS Statistics (v25) software (IBM). P values were calculated using a two-tailed one-way analysis of variance (ANOVA) with Tukey’s honestly significant difference test for the assays whose results are shown in Fig. 4 and 7 and with a one-tailed one-way ANOVA with Dunnett’s test for the assay whose results are shown in Fig. 6E.
Analysis of the V08-1, V10-2, and V20-1 RREs, described in Fig. 4, was performed with minor modifications. For this analysis, Rev was provided in trans by the CMV-SC3 Rev construct. Each well of a 12-well plate was seeded with 4 × 105 293T/17 cells maintained in 2 ml IMDM supplemented with 10% bovine calf serum. One day after plating, medium was removed from each well and replaced with 1 ml IMDM supplemented with 5% bovine calf serum. The cells in each well were then transfected with 1,000 ng of an RRE-containing construct and 50 ng of the CMV-Rev construct using the polyethylenimine method. Four transfection replicates were performed. Simultaneously, different cultures of 293T/17 cells were transfected with constructs expressing GFP or BFP alone as single-color controls to permit color compensation during flow cytometry. Cells were harvested, and flow cytometry was performed 24 h after transfection. Gating was performed as described above, identifying cells successfully transduced with the RRE-containing construct and expressing GFP or BFP, and the ratio of GFP/BFP was calculated. Additional gating on mCherry-positive cells was not performed.
CE-SHAPE.
RNA was folded as described above for the gel migration assay, except that the final volume of the folded RNA mix was 150 μl and glycerol was excluded from the folding buffer. Folded RNAs were probed using 3 mM NMIA. For this, RNAs were divided into experimental (NMIA-positive) and control (NMIA-negative) aliquots (72 μl each), to which 8 μl 30 mM NMIA in anhydrous DMSO or DMSO alone was added, respectively. Modification reaction mixtures were incubated at 37°C for 50 min, ethanol precipitated, and resuspended in 13 μl nuclease-free water. Reverse transcription of modified RNAs, cDNA processing/fractionation, and SHAPE data analysis were conducted as previously described (64).
Creating probability pairing and Shannon entropy profiles.
Pairing probability (data available upon request) and Shannon entropy values were calculated by feeding SuperFold software (50) with the 1M7 reactivities of the RREs. Shannon entropy measurements were calculated over a centered 11-nt sliding window and plotted against the nucleotide position.
Data availability.
The nucleotide sequences corresponding to RREs from viral quasispecies found in samples from patient SC3, which were generated through the procedure described above, were deposited in GenBank under accession numbers MK190736 through MK190867. The SC3 Rev sequence used in the functional assays was previously deposited in GenBank under accession number KF559146. The RRE sequences listed here as V10-2 and V20-1 were previously deposited in GenBank under accession numbers KF559160 and KF559162, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants GM 110009, AI134208, AI087505, and AI068501 as well as Women’s Interagency HIV Study (WIHS) grant UO1-AI-35004 from the National Institutes of Health (NIH). The patient samples used in this study were collected by the Women’s Interagency HIV Study (WIHS). The Bronx WHIS repository provided the initial patient materials, and the WIHS central repository provided the materials from the intermediate time points (Bronx WIHS [Kathryn Anastos and Anjali Sharma], U01-AI-035004). S.F.J.L.G. and C.S. were supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health, U.S. Department of Health and Human Services. P.E.H.J. was supported by grant K08AI136671 from the National Institutes of Health. Salary support for M.-L.H. and D.R. was provided by the Charles H. Ross, Jr., and Myles H. Thaler Endowments at the University of Virginia.
Jason W. Rausch (NCI) is acknowledged for his contribution in setting up the SHAPE-MaP analysis pipeline, and we thank Jing Huang for outstanding technical support.
The contents of this publication are solely the responsibility of the authors and do not represent the official views of NIH or the WIHS. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.02102-18.
REFERENCES
- 1.Rekosh D, Hammarskjold ML. 2018. Intron retention in viruses and cellular genes: detention, border controls and passports. Wiley Interdiscip Rev RNA 9:e1470. doi: 10.1002/wrna.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leblanc J, Weil J, Beemon K. 2013. Posttranscriptional regulation of retroviral gene expression: primary RNA transcripts play three roles as pre-mRNA, mRNA, and genomic RNA. Wiley Interdiscip Rev RNA 4:567–580. doi: 10.1002/wrna.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandes JD, Booth DS, Frankel AD. 2016. A structurally plastic ribonucleoprotein complex mediates post-transcriptional gene regulation in HIV-1. Wiley Interdiscip Rev RNA 7:470–486. doi: 10.1002/wrna.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammarskjold ML, Heimer J, Hammarskjold B, Sangwan I, Albert L, Rekosh D. 1989. Regulation of human immunodeficiency virus env expression by the rev gene product. J Virol 63:1959–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hadzopoulou-Cladaras M, Felber BK, Cladaras C, Athanassopoulos A, Tse A, Pavlakis GN. 1989. The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region. J Virol 63:1265–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 7.Berger J, Aepinus C, Dobrovnik M, Fleckenstein B, Hauber J, Bohnlein E. 1991. Mutational analysis of functional domains in the HIV-1 Rev trans-regulatory protein. Virology 183:630–635. doi: 10.1016/0042-6822(91)90992-K. [DOI] [PubMed] [Google Scholar]
- 8.Cochrane AW, Perkins A, Rosen CA. 1990. Identification of sequences important in the nucleolar localization of human immunodeficiency virus Rev: relevance of nucleolar localization to function. J Virol 64:881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubota S, Nosaka T, Cullen BR, Maki M, Hatanaka M. 1991. Effects of chimeric mutants of human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex on nucleolar targeting signals. J Virol 65:2452–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dundr M, Leno GH, Hammarskjold ML, Rekosh D, Helga-Maria C, Olson MO. 1995. The roles of nucleolar structure and function in the subcellular location of the HIV-1 Rev protein. J Cell Sci 108:2811–2823. [DOI] [PubMed] [Google Scholar]
- 11.Kjems J, Frankel AD, Sharp PA. 1991. Specific regulation of mRNA splicing in vitro by a peptide from HIV-1 Rev. Cell 67:169–178. doi: 10.1016/0092-8674(91)90580-R. [DOI] [PubMed] [Google Scholar]
- 12.Battiste JL, Mao H, Rao NS, Tan R, Muhandiram DR, Kay LE, Frankel AD, Williamson JR. 1996. Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex. Science 273:1547–1551. doi: 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- 13.Daugherty MD, Liu B, Frankel AD. 2010. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat Struct Mol Biol 17:1337–1342. doi: 10.1038/nsmb.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas SL, Oft M, Jaksche H, Casari G, Heger P, Dobrovnik M, Bevec D, Hauber J. 1998. Functional analysis of the human immunodeficiency virus type 1 Rev protein oligomerization interface. J Virol 72:2935–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Booth DS, Cheng Y, Frankel AD. 2014. The export receptor Crm1 forms a dimer to promote nuclear export of HIV RNA. Elife 3:e04121. doi: 10.7554/eLife.04121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wen W, Meinkoth JL, Tsien RY, Taylor SS. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 17.Fischer U, Huber J, Boelens WC, Mattaj IW, Luhrmann R. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 18.Mann DA, Mikaelian I, Zemmel RW, Green SM, Lowe AD, Kimura T, Singh M, Butler PJ, Gait MJ, Karn J. 1994. A molecular rheostat. Co-operative rev binding to stem I of the rev-response element modulates human immunodeficiency virus type-1 late gene expression. J Mol Biol 241:193–207. doi: 10.1006/jmbi.1994.1488. [DOI] [PubMed] [Google Scholar]
- 19.Dayton ET, Powell DM, Dayton AI. 1989. Functional analysis of CAR, the target sequence for the Rev protein of HIV-1. Science 246:1625–1629. doi: 10.1126/science.2688093. [DOI] [PubMed] [Google Scholar]
- 20.Kjems J, Brown M, Chang DD, Sharp PA. 1991. Structural analysis of the interaction between the human immunodeficiency virus Rev protein and the Rev response element. Proc Natl Acad Sci U S A 88:683–687. doi: 10.1073/pnas.88.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherpa C, Rausch JW, Le Grice SF, Hammarskjold ML, Rekosh D. 2015. The HIV-1 Rev response element (RRE) adopts alternative conformations that promote different rates of virus replication. Nucleic Acids Res 43:4676–4686. doi: 10.1093/nar/gkv313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bai Y, Tambe A, Zhou K, Doudna JA. 2014. RNA-guided assembly of Rev-RRE nuclear export complexes. Elife 3:e03656. doi: 10.7554/eLife.03656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW Jr, Swanstrom R, Burch CL, Weeks KM. 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cook KS, Fisk GJ, Hauber J, Usman N, Daly TJ, Rusche JR. 1991. Characterization of HIV-1 REV protein: binding stoichiometry and minimal RNA substrate. Nucleic Acids Res 19:1577–1583. doi: 10.1093/nar/19.7.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daly TJ, Rennert P, Lynch P, Barry JK, Dundas M, Rusche JR, Doten RC, Auer M, Farrington GK. 1993. Perturbation of the carboxy terminus of HIV-1 Rev affects multimerization on the Rev responsive element. Biochemistry 32:8945–8954. doi: 10.1021/bi00085a028. [DOI] [PubMed] [Google Scholar]
- 26.Malim MH, McCarn DF, Tiley LS, Cullen BR. 1991. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J Virol 65:4248–4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heaphy S, Finch JT, Gait MJ, Karn J, Singh M. 1991. Human immunodeficiency virus type 1 regulator of virion expression, rev, forms nucleoprotein filaments after binding to a purine-rich “bubble” located within the rev-responsive region of viral mRNAs. Proc Natl Acad Sci U S A 88:7366–7370. doi: 10.1073/pnas.88.16.7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zapp ML, Hope TJ, Parslow TG, Green MR. 1991. Oligomerization and RNA binding domains of the type 1 human immunodeficiency virus Rev protein: a dual function for an arginine- rich binding motif. Proc Natl Acad Sci U S A 88:7734–7748. doi: 10.1073/pnas.88.17.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zemmel RW, Kelley AC, Karn J, Butler PJ. 1996. Flexible regions of RNA structure facilitate co-operative Rev assembly on the Rev-response element. J Mol Biol 258:763–777. doi: 10.1006/jmbi.1996.0285. [DOI] [PubMed] [Google Scholar]
- 30.Daugherty MD, D'Orso I, Frankel AD. 2008. A solution to limited genomic capacity: using adaptable binding surfaces to assemble the functional HIV Rev oligomer on RNA. Mol Cell 31:824–834. doi: 10.1016/j.molcel.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DiMattia MA, Watts NR, Stahl SJ, Rader C, Wingfield PT, Stuart DI, Steven AC, Grimes JM. 2010. Implications of the HIV-1 Rev dimer structure at 3.2 Å resolution for multimeric binding to the Rev response element. Proc Natl Acad Sci U S A 107:5810–5814. doi: 10.1073/pnas.0914946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jain C, Belasco JG. 2001. Structural model for the cooperative assembly of HIV-1 Rev multimers on the RRE as deduced from analysis of assembly-defective mutants. Mol Cell 7:603–614. doi: 10.1016/S1097-2765(01)00207-6. [DOI] [PubMed] [Google Scholar]
- 33.Fornerod M, Ohno M, Yoshida M, Mattaj IW. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051–1060. doi: 10.1016/S0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 34.Otero GC, Harris ME, Donello JE, Hope TJ. 1998. Leptomycin B inhibits equine infectious anemia virus Rev and feline immunodeficiency virus Rev function but not the function of the hepatitis B virus posttranscriptional regulatory element. J Virol 72:7593–7597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson PE, Tebit DM, Rekosh D, Hammarskjold ML. 2016. Rev-RRE functional activity differs substantially among primary HIV-1 isolates. AIDS Res Hum Retroviruses 32:923–934. doi: 10.1089/AID.2016.0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phuphuakrat A, Paris RM, Nittayaphan S, Louisirirotchanakul S, Auewarakul P. 2005. Functional variation of HIV-1 Rev response element in a longitudinally studied cohort. J Med Virol 75:367–373. doi: 10.1002/jmv.20279. [DOI] [PubMed] [Google Scholar]
- 37.Iversen AK, Shpaer EG, Rodrigo AG, Hirsch MS, Walker BD, Sheppard HW, Merigan TC, Mullins JI. 1995. Persistence of attenuated rev genes in a human immunodeficiency virus type 1-infected asymptomatic individual. J Virol 69:5743–5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Churchill MJ, Chiavaroli L, Wesselingh SL, Gorry PR. 2007. Persistence of attenuated HIV-1 rev alleles in an epidemiologically linked cohort of long-term survivors infected with nef-deleted virus. Retrovirology 4:43. doi: 10.1186/1742-4690-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bobbitt KR, Addo MM, Altfeld M, Filzen T, Onafuwa AA, Walker BD, Collins KL. 2003. Rev activity determines sensitivity of HIV-1-infected primary T cells to CTL killing. Immunity 18:289–299. doi: 10.1016/S1074-7613(03)00031-1. [DOI] [PubMed] [Google Scholar]
- 40.Baccam P, Thompson RJ, Li Y, Sparks WO, Belshan M, Dorman KS, Wannemuehler Y, Oaks JL, Cornette JL, Carpenter S. 2003. Subpopulations of equine infectious anemia virus Rev coexist in vivo and differ in phenotype. J Virol 77:12122–12131. doi: 10.1128/JVI.77.22.12122-12131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belshan M, Baccam P, Oaks JL, Sponseller BA, Murphy SC, Cornette J, Carpenter S. 2001. Genetic and biological variation in equine infectious anemia virus Rev correlates with variable stages of clinical disease in an experimentally infected pony. Virology 279:185–200. doi: 10.1006/viro.2000.0696. [DOI] [PubMed] [Google Scholar]
- 42.Belshan M, Harris ME, Shoemaker AE, Hope TJ, Carpenter S. 1998. Biological characterization of Rev variation in equine infectious anemia virus. J Virol 72:4421–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sloan EA, Kearney MF, Gray LR, Anastos K, Daar ES, Margolick J, Maldarelli F, Hammarskjold ML, Rekosh D. 2013. Limited nucleotide changes in the Rev response element (RRE) during HIV-1 infection alter overall Rev-RRE activity and Rev multimerization. J Virol 87:11173–11186. doi: 10.1128/JVI.01392-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sherpa C, Jackson P, Gray LR, Anastos K, Le Grice SFJ, Hammarskjold M-L, Rekosh D. 2019. Evolution of the HIV-1 RRE during natural infection reveals nucleotide changes that correlate with altered structure and increased activity over time. bioRxiv 10.1101/483511. [DOI] [PMC free article] [PubMed]
- 45.Smola MJ, Christy TW, Inoue K, Nicholson CO, Friedersdorf M, Keene JD, Lee DM, Calabrese JM, Weeks KM. 2016. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc Natl Acad Sci U S A 113:10322–10327. doi: 10.1073/pnas.1600008113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sztuba-Solinska J, Rausch JW, Smith R, Miller JT, Whitby D, Le Grice S. 2017. Kaposi's sarcoma-associated herpesvirus polyadenylated nuclear RNA: a structural scaffold for nuclear, cytoplasmic and viral proteins. Nucleic Acids Res 45:6805–6821. doi: 10.1093/nar/gkx241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu YF, Mauger DM, Goldstein DB, Urban TJ, Weeks KM, Bradrick SS. 2015. IFNL3 mRNA structure is remodeled by a functional non-coding polymorphism associated with hepatitis C virus clearance. Sci Rep 5:16037. doi: 10.1038/srep16037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM. 2014. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat Methods 11:959–965. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smola MJ, Calabrese JM, Weeks KM. 2015. Detection of RNA-protein interactions in living cells with SHAPE. Biochemistry 54:6867–6875. doi: 10.1021/acs.biochem.5b00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smola MJ, Rice GM, Busan S, Siegfried NA, Weeks KM. 2015. Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat Protoc 10:1643–1669. doi: 10.1038/nprot.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sherpa C, Rausch JW, Le Grice S. 2018. Structural characterization of maternally expressed gene 3 RNA reveals conserved motifs and potential sites of interaction with polycomb repressive complex 2. Nucleic Acids Res 46:10432–10447. doi: 10.1093/nar/gky722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bellaousov S, Reuter JS, Seetin MG, Mathews DH. 2013. RNAstructure: web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res 41:W471–W474. doi: 10.1093/nar/gkt290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jackson PEH, Huang J, Sharma M, Rasmussen SK, Hammarskjold M-L, Rekosh D. 2019. A novel retroviral vector system to analyze expression from mRNA with retained introns using fluorescent proteins and flow cytometry. bioRxiv 551846. 10.1101/551846. [DOI] [PMC free article] [PubMed]
- 54.Srinivasakumar N, Chazal N, Helga-Maria C, Prasad S, Hammarskjöld ML, Rekosh D. 1997. The effect of viral regulatory protein expression on gene delivery by human immunodeficiency virus type 1 vectors produced in stable packaging cell lines. J Virol 71:5841–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deigan KE, Li TW, Mathews DH, Weeks KM. 2009. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci U S A 106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McGinnis JL, Dunkle JA, Cate JH, Weeks KM. 2012. The mechanisms of RNA SHAPE chemistry. J Am Chem Soc 134:6617–6624. doi: 10.1021/ja2104075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tiley LS, Malim MH, Tewary HK, Stockley PG, Cullen B. 1992. Identification of a high-affinity RNA-binding site for the human immunodeficiency virus type 1 Rev protein. Proc Natl Acad Sci U S A 89:758–762. doi: 10.1073/pnas.89.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, Mason CE, Rana T. 2016. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol 1:16011. doi: 10.1038/nmicrobiol.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palmer S, Wiegand AP, Maldarelli F, Bazmi H, Mican JM, Polis M, Dewar RL, Planta A, Liu S, Metcalf JA, Mellors JW, Coffin JM. 2003. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol 41:4531–4536. doi: 10.1128/JCM.41.10.4531-4536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kearney M, Maldarelli F, Shao W, Margolick JB, Daar ES, Mellors JW, Rao V, Coffin JM, Palmer S. 2009. Human immunodeficiency virus type 1 population genetics and adaptation in newly infected individuals. J Virol 83:2715–2727. doi: 10.1128/JVI.01960-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Magoc T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 59:284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 64.Lusvarghi S, Sztuba-Solinska J, Purzycka KJ, Pauly GT, Rausch JW, Grice SF. 2013. The HIV-2 Rev-response element: determining secondary structure and defining folding intermediates. Nucleic Acids Res 41:6637–6649. doi: 10.1093/nar/gkt353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The nucleotide sequences corresponding to RREs from viral quasispecies found in samples from patient SC3, which were generated through the procedure described above, were deposited in GenBank under accession numbers MK190736 through MK190867. The SC3 Rev sequence used in the functional assays was previously deposited in GenBank under accession number KF559146. The RRE sequences listed here as V10-2 and V20-1 were previously deposited in GenBank under accession numbers KF559160 and KF559162, respectively.