REPLY
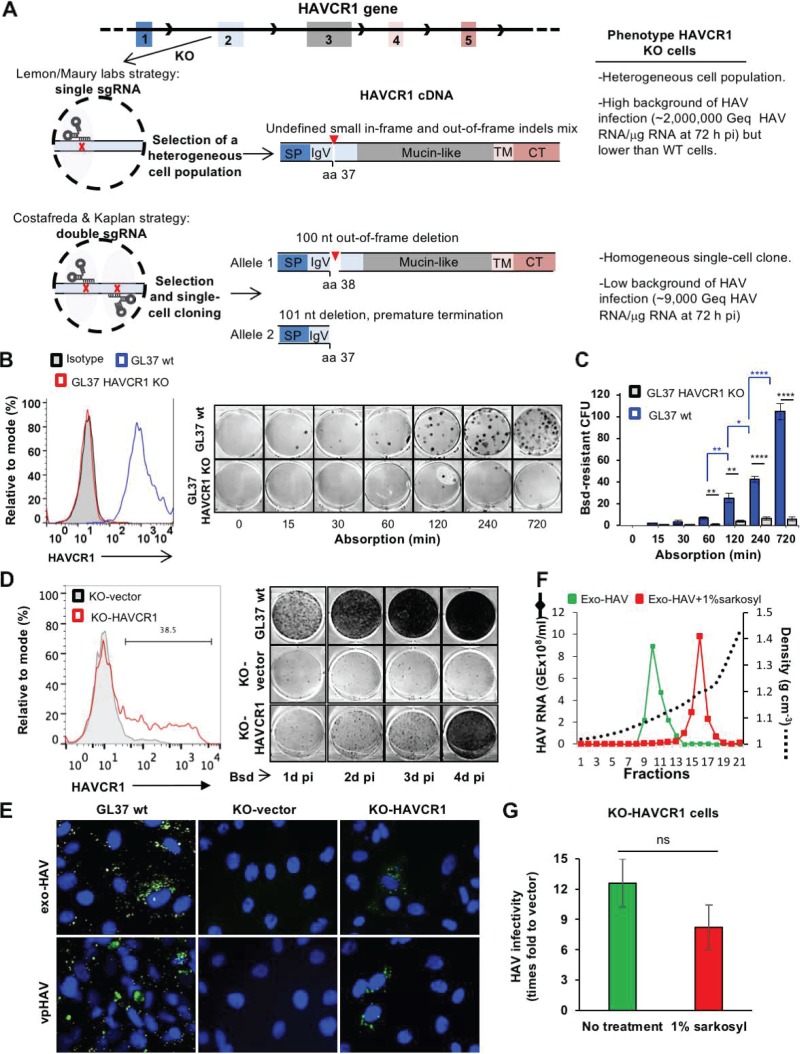
We have recently shown that HAVCR1 is a functional hepatitis A virus (HAV) cellular receptor in clone GL37 of African green monkey kidney (GL37) cells that mediates cell entry of viral particles (vpHAV) and exosomes produced in HAV-infected cells (exo-HAV or eHAV) (1). We also showed that HAVCR1 is the predominant HAV receptor in GL37 cells, whereas alternative, yet-unidentified HAV receptors are coexpressed with HAVCR1 in other primate cells, such as Vero E6 and Huh7 cells (1). The knockout (KO) of HAVCR1 using CRISPR/Cas9 technology significantly reduced the susceptibility of GL37 cells to HAV infection, which was regained upon transfection of HAVCR1 cDNA (1), supporting the notion that resistance to HAV infection in these GL37 HAVCR1 KO cells is due to the lack of HAV receptors rather than other factors. Taken together with our previous work showing the direct interaction of HAV with HAVCR1 in cellular and cell-free systems (2–10), our data clearly indicated that HAVCR1 is indeed a functional HAV receptor of vpHAV and exo-HAV. For the letter to the editor of Das et al. (11), the Lemon lab and Maury lab (Lemon/Maury labs) tried to reproduce our results using GL37 cells and one of the sgRNAs that we described in our paper (1) but used a flawed experimental design that led them to conclude incorrectly that HAVCR1 was not a functional vpHAV receptor. The Lemon/Maury lab data clearly show that the susceptibility of vpHAV is reduced significantly in their GL37 HAVCR1 KO cells compared to that in wild-type GL37 (GL37 wt) cells (see Fig. 1D and E in reference 11). However, the HAV background level in their GL37 HAVCR1 KO cells was ∼200 times higher than in ours (1), which is likely due to a combination of factors, including the CRISPR/Cas9 strategy used to generate their HAVCR1 KO cells (Fig. 1A), the short vpHAV adsorption times, and the high multiplicities of infection used in their experiments. The Lemon/Maury labs produced their GL37 HAVCR1 KO cells using a single sgRNA cloned into a lentivirus vector to mediate a single cut in exon 2 of the HAVCR1 gene, selected the pool of puromycin-resistant transduced cells, verified that heterogeneous small insertions and deletions (indels) were introduced in exon 2, and used the uncloned heterogeneous cell population to analyze the function of HAVCR1 as an HAV receptor. This simple and fast procedure resulted in the selection of a pool of heterogeneous puromycin-resistant cells consisting mainly of HAVCR1 KO cells but also most likely containing cells with various degrees of susceptibility to vpHAV infection due to (i) small in-frame indels that did not prevent the HAV receptor function of HAVCR1, (ii) off-target events that conferred puromycin resistance but did not knock out HAVCR1, and (iii) expression of different levels of alternative HAV receptors that are not predominant in AGMK GL37 cells compared to in Vero and Huh7 cells (1), which would significantly increase the background level of HAV infection in their heterogenous pool of puromycin-resistant cells. To circumvent the problems of using a heterogeneous pool of cells to evaluate HAVCR1 as a functional HAV receptor, we (i) utilized double sgRNAs to produce large and well-defined out-of-frame deletions of 100 and 101 nucleotides in the HAVCR1 gene and (ii) single-cell cloned transfectants to generate a homogeneous HAVCR1 KO cell clone highly resistant to vpHAV and exo-HAV infection (1). The Lemon/Maury labs also argued that vpHAV did not bind to HAVCR1 at the cell surface by comparing levels of binding to GL37 wt and GL37 HAVCR1 KO cells but used a suboptimal adsorption time of 2 h, which limited binding of vpHAV to HAVCR1 (Fig. 1B and C), in contrast to the 12 h used in our experiments, which significantly increases binding of vpHAV to GL37 wt but not GL37 HAVCR1 KO cells. To rebut their additional suggestions, we show in Fig. 1D and E that the low background levels of HAV produced in HAVCR1 KO cells did not spread to the remaining HAVCR1 KO cells in the monolayer, as expected from the knockout of a functional viral receptor but not an accessory attachment factor. In our recent paper (1), we showed that treatment of purified naked vpHAV with 1% Sarkosyl, an ionic detergent, did not affect their infectivity in our GL37 HAVCR1 KO cells transfected with HAVCR1 cDNA, indicating that lipids are not required for the infection of naked vpHAV mediated by HAVCR1. Here, we extracted lipids from exo-HAV using 1% Sarkosyl and purified the naked HAV particles in iodixanol gradients (Fig. 1F). The infectivity of these naked HAV particles in GL37 HAVCR1 KO cells transfected with HAVCR1 cDNA was similar to that of the untreated exo-HAV cells (Fig. 1G), further showing that lipids are not required for the HAVCR1-mediated infection of naked HAV particles. In summary, we showed that HAVCR1 plays an essential role as an HAV functional receptor of vpHAV and exo-HAV in GL37 cells by using a double sgRNA CRISPR/Cas9 knockout strategy and single-cell cloning of the KO cells, which resulted in the isolation of a GL37 HAVCR1 KO clone resistant to HAV infection with a low background of susceptibility to HAV infection. In contrast, the Lemon/Maury labs used a single sgRNA strategy to select a heterogeneous pool of cells with a high background of susceptibility to HAV infection as well as comparatively short HAV incubation times that obscured their data analysis.
FIG 1.
(A) Schematic representation of the CRISPR/Cas9 strategies used to knock out the HAVCR1 gene at exon 2 in GL37 cells. The strategy used by Das et al. (11) (Lemon/Maury labs) was based on a single sgRNA to introduce a single cut in exon 2 and generate small and undefined indels. A heterogeneous population of cells containing indels was used to assess the role of HAVCR1 as a functional HAV receptor. The strategy adopted by Costafreda and Kaplan (1) was based on two sgRNAs that cut exon 2 at two different sites, resulting in defined out-of-frame deletions of 100 nucleotides in one allele and 101 nucleotides in the other allele of the selected single-cell clone used to assess the role of HAVCR1 as a functional HAV receptor. The HAVCR1 cDNA codes for a type I integral membrane glycoprotein containing a signal peptide (SP), a variable immunoglobulin-like domain (IgV), a mucin-like domain, a transmembrane domain (TM), and a cytoplasmic tail (CT). aa, amino acid; Geq, genome equivalents; pi, postinfection; nt, nucleotide. (B) Adsorption kinetics of vpHAV in GL37 wt and GL37 HAVCR1 KO cells. (Left) Analysis of the expression of HAVCR1 at the cell surface of GL37 wt (blue line) and GL37 HAVCR1 KO (red line) cells by flow cytometry analysis using anti-HAVCR1 MAb 1D12. GL37 wt cells were also stained with an isotype control (filled gray curve). (Right) Blasticidin (Bsd)-resistant CFU assay (1) using purified vpHAV containing a Bsd-selectable marker (vpHAV-Bsd) adsorbed to GL37 wt or GL37 HAVCR1 KO cells for 0 to 12 h. Briefly, monolayers were washed, incubated at 37°C for 24 h, trypsinized, and selected with Bsd. At 12 days postinfection, cell colonies were fixed and stained with crystal violet (dark spots), and 6-well plates were imaged using a flatbed scanner. (C) Quantitation of Bsd-resistant CFU in GL37 wt and GL37 HAVCR1 KO cells from panel B. Data are means ± SEM of colony numbers from four independent experiments. Differences between GL37 wt and GL37 HAVCR1 KO cells at each adsorption time point (black lines and asterisks) and between GL37 wt at different time points (blue lines and asterisks) were analyzed by Student’s unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (D) Spread of HAV infection at different times postinfection in GL37 HAVCR1 KO cells transfected with HAVCR1 cDNA (KO-HAVCR1) or a vector (KO-vector). (Left) Expression of HAVCR1 at the cell surface of the transfectants by flow cytometry using anti-HAVCR1 MAb 1D12. The bar indicates HAVCR1-positive cells shown as a percentage. Transfectants were infected with HAV-Bsd at a multiplicity of infection (MOI) of 0.1 50% tissue culture infectious dose (TCID50)/cell, incubated at 37°C for 24 to 96 h, trypsinized, and plated in 6-well plates for a Bsd-resistant CFU assay (1). Results are representative of two independent experiments, each with two replicates. The number of Bsd-resistant CFU increased in GL37 wt cells and KO-HAVCR1 transfectants but not in KO-vector cells, indicating that the exo-HAV and vpHAV produced in infected cells cannot spread to other GL37 HAVCR1 KO cells in the absence of HAVCR1 expression. (E) Immunofluorescence analysis of HAV infection in HAVCR1 KO cells. GL37 wt cells and GL37 HAVCR1 KO cells transfected with a vector (KO-vector) or HAVR1 cDNA (KO-HAVCR1) were infected at a low MOI (0.1 TCID50/ml) of exo-HAV or vpHAV for 4 days, fixed, permeabilized, stained with anti-HAV neutralizing MAbs (green fluorescence), and counterstained with DAPI (4′,6-diamidino-2-phenylindole) nuclear dye (blue fluorescence). Micrographs of representative fields were taken at a magnification of ×400, showing that all GL37 wt cells are infected but that only ∼30% of HAVCR1 KO cells transfected with HAVCR1 cDNA were infected, which correlated with the number of transfected cells that express HAVCR1 at the cell surface (Fig. 1D, left) and indicated that exo-HAV and vpHAV cannot infect HAVCR1 KO cells in the absence of HAVCR1. (F) Gradient-purified exo-HAV not treated (green line) or treated with 1% Sarkosyl (red line) were repurified through iodixanol gradients. The HAV RNA in gradient fractions collected after isopycnic ultracentrifugation was quantified by reverse transcription-quantitative PCR (RT-qPCR), and the density of each fraction was determined by refractometry. Fractions 10 and 11, containing the peak of exo-HAV, were collected and used for further experimentation. Fraction 16, containing the peak of vpHAV extracted from exo-HAV by detergent treatment, was also collected. GE, genome equivalents. (G) Comparison of the infectivities in GL37 HAVCR1 KO cell transfectants of purified exo-HAV and naked HAV particles extracted from exo-HAV by detergent treatment. GL37 HAVCR1 KO cells transfected with HAVCR1 (KO-HAVCR1) or a vector (KO-vector) were infected with untreated exo-HAV (green bar), fractions 10 and 11 from panel F, or naked HAV particles extracted with detergent from exo-HAV (red bar), fraction 16 from panel F. Virus growth was determined by RT-qPCR at 72 h postinfection. Data are means ± SEM from three independent experiments. ns, nonsignificant.
Finally, we agree with the Lemon/Maury labs’ definition of what constitutes an HAV functional receptor. We have previously shown that monoclonal antibodies against the IgV domain of HAVCR1 block vpHAV infection (2–4), that vpHAV binds to IgV of HAVCR1 and deletion of the IgV domain prevents binding of vpHAV to HAVCR1 (5), that soluble forms of HAVCR1 neutralize vpHAV infectivity (6–8) and alter vpHAV to release the viral genome (7), that the binding site of vpHAV maps to the CC’FG epitope of the HAVCR1 IgV domain (9), and that mutations in the FG loop of HAVCR1 prevent binding and neutralization of vpHAV (10). Taken together with our recent data showing that transfection of HAVCR1 cDNA rescued the infectivity of vpHAV in AGMK GL37 HAVCR1 KO (1), our results clearly show that HAVCR1 is indeed an essential functional HAV receptor required for infection of GL37 cells. In other primate cells lines, such as Vero E6 and Huh7 cells, which coexpress HAVCR1 and additional, yet-unidentified HAV receptors, HAVCR1 also mediates HAV infection, as shown in our blocking studies with anti-HAVCR1 monoclonal antibodies (1). Therefore, HAVCR1 is a functional HAV receptor that mediates binding, cell entry, and uncoating of viral particles as well as binding and cell entry of exosomes, including exo-HAV, and not a mere attachment factor that only enhances HAV infection.
ACKNOWLEDGMENTS
This work was supported in part by Food and Drug Administration (FDA) intramural funding to G.K. and a program project grant from the National Institute of Allergy and Infectious Diseases (2P01-AI054456-06A1 to G.K. as a collaborator). This project was supported in part by an appointment to the Research Fellowship Program at the Office of Blood Research and Review, Center for Biologics Evaluation and Research, Food and Drug Administration, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the FDA (to M.I.C.).
Our contributions are an informal communication and represent our own best judgment. These comments do not bind or obligate the FDA.
Footnotes
This is a response to a letter by Das et al. (https://doi.org/10.1128/JVI.01793-18).
REFERENCES
- 1.Costafreda MI, Kaplan G. 2018. HAVCR1 (CD365) and its mouse ortholog are functional hepatitis A virus (HAV) cellular receptors that mediate HAV infection. J Virol 92:e02065-17. doi: 10.1128/JVI.02065-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaplan G, Totsuka A, Thompson P, Akatsuka T, Moritsugu Y, Feinstone SM. 1996. Identification of a surface glycoprotein on African green monkey kidney cells as a receptor for hepatitis A virus. EMBO J 15:4282–4296. doi: 10.1002/j.1460-2075.1996.tb00803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feigelstock D, Thompson P, Mattoo P, Kaplan GG. 1998. Polymorphisms of the hepatitis A virus cellular receptor 1 in African green monkey kidney cells result in antigenic variants that do not react with protective monoclonal antibody 190/4. J Virol 72:6218–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feigelstock D, Thompson P, Mattoo P, Zhang Y, Kaplan GG. 1998. The human homolog of HAVcr-1 codes for a hepatitis A virus cellular receptor. J Virol 72:6621–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson P, Lu J, Kaplan GG. 1998. The Cys-rich region of hepatitis A virus cellular receptor 1 is required for binding of hepatitis A virus and protective monoclonal antibody 190/4. J Virol 72:3751–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silberstein E, Dveksler G, Kaplan GG. 2001. Neutralization of hepatitis A virus (HAV) by an immunoadhesin containing the cysteine-rich region of HAV cellular receptor-1. J Virol 75:717–725. doi: 10.1128/JVI.75.2.717-725.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silberstein E, Xing L, van de Beek W, Lu J, Cheng H, Kaplan GG. 2003. Alteration of hepatitis A virus (HAV) particles by a soluble form of HAV cellular receptor 1 containing the immunoglobin- and mucin-like regions. J Virol 77:8765–8774. doi: 10.1128/JVI.77.16.8765-8774.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silberstein E, Konduru K, Kaplan GG. 2009. The interaction of hepatitis A virus (HAV) with soluble forms of its cellular receptor 1 (HAVCR1) share the physiological requirements of infectivity in cell culture. Virol J 6:175. doi: 10.1186/1743-422X-6-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santiago C, Ballesteros A, Tami C, Martinez-Munoz L, Kaplan GG, Casasnovas JM. 2007. Structures of T cell immunoglobulin mucin receptors 1 and 2 reveal mechanisms for regulation of immune responses by the TIM receptor family. Immunity 26:299–310. doi: 10.1016/j.immuni.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manangeeswaran M, Jacques J, Tami C, Konduru K, Amharref N, Perrella O, Casasnovas JM, Umetsu DT, Dekruyff RH, Freeman GJ, Perrella A, Kaplan GG. 2012. Binding of hepatitis A virus to its cellular receptor 1 inhibits T-regulatory cell functions in humans. Gastroenterology 142:1516–1525.e3. doi: 10.1053/j.gastro.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das A, Maury W, Lemon SM. 2019. TIM1 (HAVCR1): an essential “receptor” or an “accessory attachment factor” for hepatitis A virus? J Virol 93:e01793-18. doi: 10.1128/JVI.01793-18. [DOI] [PMC free article] [PubMed] [Google Scholar]