HCMV UL138 curtails the activity of the MRP1 drug transporter by reducing its steady-state levels, leaving cells susceptible to killing by cytotoxic agents normally exported by MRP1. It has been suggested in the literature that capitalizing on this UL138-induced vulnerability could be a potential antiviral strategy against virally infected cells, particularly those harboring a latent infection during which UL138 is one of the few viral proteins expressed. Therefore, identifying the regions of UL138 required for MRP1 downregulation and predicting genetic variants that may be selected upon UL138-targeted chemotherapy are important ventures. Here we present the first structure-function examination of UL138 activity and determine that its transmembrane domain and acidic cluster dileucine Golgi sorting motif are required for functional MRP1 downregulation.
KEYWORDS: HCMV, MRP1, trafficking, UL138
ABSTRACT
The human cytomegalovirus (HCMV) UL138 protein downregulates the cell surface expression of the multidrug resistance-associated protein 1 (MRP1) transporter. We examined the genetic requirements within UL138 for MRP1 downregulation. We determined that the acidic cluster dileucine motif is essential for UL138-mediated downregulation of MRP1 steady-state levels and inhibition of MRP1 efflux activity. We also discovered that the naturally occurring UL138 protein isoforms, the full-length long isoform of UL138 and a short isoform missing the N-terminal membrane-spanning domain, have different abilities to inhibit MRP1 function. Cells expressing the long isoform of UL138 show decreased MRP1 steady-state levels and fail to efflux an MRP1 substrate. Cells expressing the short isoform of UL138 also show decreased MRP1 levels, but the magnitude of the decrease is not the same, and they continue to efficiently efflux an MRP1 substrate. Thus, the membrane-spanning domain, while dispensable for a UL138-mediated decrease in MRP1 protein levels, is necessary for a functional inhibition of MRP1 activity. Our work defines the genetic requirements for UL138-mediated MRP1 downregulation and anticipates the possible evolution of viral escape mutants during the use of therapies targeting this function of UL138.
IMPORTANCE HCMV UL138 curtails the activity of the MRP1 drug transporter by reducing its steady-state levels, leaving cells susceptible to killing by cytotoxic agents normally exported by MRP1. It has been suggested in the literature that capitalizing on this UL138-induced vulnerability could be a potential antiviral strategy against virally infected cells, particularly those harboring a latent infection during which UL138 is one of the few viral proteins expressed. Therefore, identifying the regions of UL138 required for MRP1 downregulation and predicting genetic variants that may be selected upon UL138-targeted chemotherapy are important ventures. Here we present the first structure-function examination of UL138 activity and determine that its transmembrane domain and acidic cluster dileucine Golgi sorting motif are required for functional MRP1 downregulation.
INTRODUCTION
Human cytomegalovirus (HCMV) is a major human pathogen infecting between 60% and 90% of the human population (1). Though generally asymptomatic in healthy adults, infection can have severe consequences in immunosuppressed individuals, making HCMV a major concern in transplant medicine (2–4). HCMV infection is also a leading infectious cause of birth defects, resulting in hearing loss, developmental delays, and microcephaly (5, 6). The virus productively replicates in many cell types, including fibroblasts and smooth muscle, epithelial, endothelial, and glial cells (1). Current treatments for HCMV target only productive replication, carry significant toxicities (7), and select for drug-resistant mutants (8).
HCMV establishes a latent infection in CD34+ hematopoietic progenitor cells, granulocyte-macrophage progenitors, and monocytes (9–17). Reactivation from latency occurs in response to inflammatory signals as infected cells differentiate through the myeloid lineage (15, 18–21). There are currently no available clinical treatments that target the latent virus, but an increasing understanding of the molecular events that establish and maintain HCMV latency has led to novel preclinical antiviral interventions (22–24).
The UL138 gene is the target of one of the investigatory antiviral approaches that focus on the ability of UL138 to downregulate the cell surface expression of multidrug resistance-associated protein 1 (MRP1) (22). MRP1 is an ATP-binding cassette (ABC) multipass transmembrane protein initially discovered as a drug resistance factor in cancer cell lines and subsequently found to be upregulated in drug-resistant tumors (25–27). Vincristine, a cytotoxic agent exported from cells by MRP1, was shown to preferentially kill HCMV-infected cells. This increased susceptibility to vincristine was attributed to UL138-mediated MRP1 downregulation, impairing the ability of the infected cells to export the drug (22).
MRP1 functions as an efflux pump that upon synthesis traffics from the endoplasmic reticulum (ER) through the Golgi apparatus into the vesicular transport system and eventually to its final destination in the plasma membrane. During HCMV infection, MRP1 is degraded by lysosomal proteases in a UL138-dependent manner in multiple cell types, including fibroblasts, THP-1 monocytes, HeLa cells, and primary CD34+ cells (22). The amino acid sequence of UL138 reveals some potential mechanisms for MRP1 degradation. UL138 possesses a transmembrane domain and four Golgi sorting motifs (28). These domains may allow UL138 to be incorporated as an integral-membrane or membrane-associated component into Golgi-derived vesicles on the endosomal route to the lysosome. Through UL138’s interaction with MRP1 (22), this could provide a route for MRP1 lysosomal degradation.
The putative amino-terminal transmembrane sequences of UL138 are largely absent in a naturally occurring isomer that initiates translation from the internal methionine-16 codon (29), expressing an N-terminally truncated short isoform. It is not known which isoform, i.e., the (full-length) long isoform or the short isoform, is more important during clinical infection. In vitro, the long isoform (expressed from methionine-1) is the major species; however, the short isoform is expressed and appears to play a role in the establishment and maintenance of latency (29). Latency establishment and maintenance in vitro are most efficient when the two isoforms of UL138 are naturally simultaneously expressed from the wild-type (WT) gene (29). The primary subcellular location of both UL138 isoforms is the Golgi apparatus (28, 29).
Potential roles for the Golgi sorting motifs in UL138 for Golgi localization or MRP1 downregulation have not been previously examined. Golgi sorting motifs are linear sequences that direct cargo into vesicles destined for the endosomal pathway and, in some cases, to the lysosome for destruction. UL138 possesses three consensus tyrosine Golgi sorting motifs (YXXØ [where X is any amino acid and ø is a bulky hydrophobic amino acid]) and one acidic cluster dileucine Golgi sorting motif (DXXLL [where X is any amino acid]) (28). Golgi sorting motifs are recognized by adaptor proteins (APs) that recruit clathrin and other accessory factors to sort cargo to specific destinations throughout the endocytic network (30). Tyrosine sorting motifs bind the AP complexes AP-1, AP-2, AP-3, and AP-4 (31), while acidic cluster dileucine motifs bind the Golgi-localized gamma-eared Arf binding family of proteins (GGAs) (32, 33).
Because UL138 downregulates MRP1 through a lysosome-dependent pathway (22), any or all of the Golgi sorting motifs of UL138 could be required for MRP1 downregulation, and the transmembrane domain may be required to help tether UL138 to the membranes of the Golgi apparatus or endocytic vesicles. Therefore, we examined the ability of UL138 proteins lacking Golgi sorting motifs or the transmembrane domain to downregulate MRP1. Here we show that the acidic cluster dileucine motif and the putative transmembrane domain are required for UL138-mediated MRP1 inactivation.
RESULTS
The transmembrane domain and Golgi sorting motifs must be disrupted to displace UL138 from the Golgi apparatus.
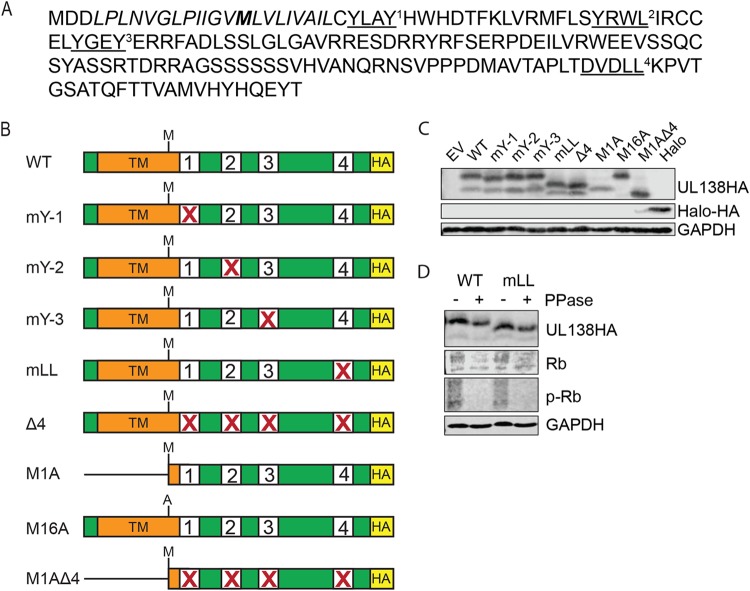
Potential roles for the Golgi sorting motifs of UL138 (Fig. 1A) have not been examined. Each Golgi sorting motif was individually and collectively disrupted in the otherwise WT C-terminally hemagglutinin (HA)-tagged allele of UL138 that simultaneously generates the full-length UL138 long isoform and a short isoform originating from an internal in-frame methionine (Fig. 1B; see also Table 1). To express only one UL138 isoform at a time, we created the following two mutants: M1A, expressing only the short isoform of UL138, and M16A, expressing only the long isoform (Fig. 1B; see also Table 1). The sorting motifs were also collectively disrupted in the M1A mutant allele (Fig. 1B; see also Table 1). Each protein was expressed during transient transfection of primary normal human dermal fibroblasts (NHDFs) (Fig. 1C). The short isoform, whether expressed from a WT or M1A allele, accumulated to lower steady-state levels than the long isoform (Fig. 1C), similarly to previously published results (29).
FIG 1.
HCMV UL138 has four putative Golgi sorting motifs. (A) UL138 protein sequence from HCMV. Tyrosine sorting motifs (YXXØ) and the acidic cluster dileucine motif (DXXLL) are underlined and denoted with a superscript number. The transmembrane domain is italicized, and the M16 alternate start site for the short isoform is indicated in bold. (B) Schematic of UL138 wild-type (WT) and mutant alleles with C-terminal HA tag and disruptions (X) in the indicated Golgi sorting motifs. Numbers correspond to the superscript numbers in the coding sequence. (C) Western blotting of cell lysates from NHDF cells transfected with the indicated expression plasmid. GAPDH served as a loading control. EV, empty vector. Images representative of results from three independent biological replicates are shown. (D) Western blotting of NHDF cell lysates treated (+) or not treated (-) with λ phosphatase. Cells were transfected with the WT or with mLL mutant UL138 and harvested 48 h later. The cellular retinoblastoma (Rb) protein was used as a control for phosphatase treatment. Images representative of results from three independent biological replicates are shown.
TABLE 1.
Mutant UL138 alleles used in this study
| Mutant name | Mutant explanation | Specific substitutions |
Corresponding mutant virus |
Reference(s) or source |
|---|---|---|---|---|
| WT | No mutations in UL138 | TB138-HA or TB138-Flag | 29, 64 | |
| mY-1 | Mutation of first tyrosine motif | Y26LAY to A26AAY | This study | |
| mY-2 | Mutation of second tyrosine motif | Y44RWL to A44AAA | This study | |
| mY-3 | Mutation of third tyrosine motif | Y54GEY to A54GEY | This study | |
| mLL | Mutation of acidic cluster dileucine motif | D142VDLL to A142AAAA | TB138 mLL-HA | This study |
| Δ4 | Mutation of four Golgi motifs | The mutations listed above combined in one allele | TB138 Δ4-HA | This study |
| M1A | Transmembrane mutant; expresses only the short isoform of UL138 from M16 | M1 to A1 | TB138 M1A-Flag | 29 |
| M16A | Expresses only the full-length long isoform of UL138 from M1 | M16 to A16 | TB138 M16A-Flag | 29 |
| M1AΔ4 | Mutation of four Golgi motifs in the context of the transmembrane mutant; M1A | M1 to A1 combined with the Δ4 mutations | This study |
All mutants in which the acidic cluster dileucine sorting motif was disrupted displayed an increase in electrophoretic mobility that manifested as a downward shift of ∼2 kDa (Fig. 1C). Although phosphorylation events have been reported in conjunction with acidic cluster dileucine sorting motifs (30, 34) and although phosphorylation is sometimes responsible for electrophoretic shifts in proteins (35), we were unable to shift the migration of the wild-type protein by treatment with lambda phosphatase in the lysates where the enzyme was capable of removing retinoblastoma (Rb) protein phosphorylation (Fig. 1D). The size of the shift appears to eliminate possible ubiquitination or SUMOylation. A previous report (36) indicated failure of attempts to shift UL138 electrophoretic mobility with endoglycosidase H, appearing to eliminate possible N-linked glycosylation. We have not explored other potential posttranslational modifications or intrinsic properties of the substituted amino acids as explanations for the differential migration.
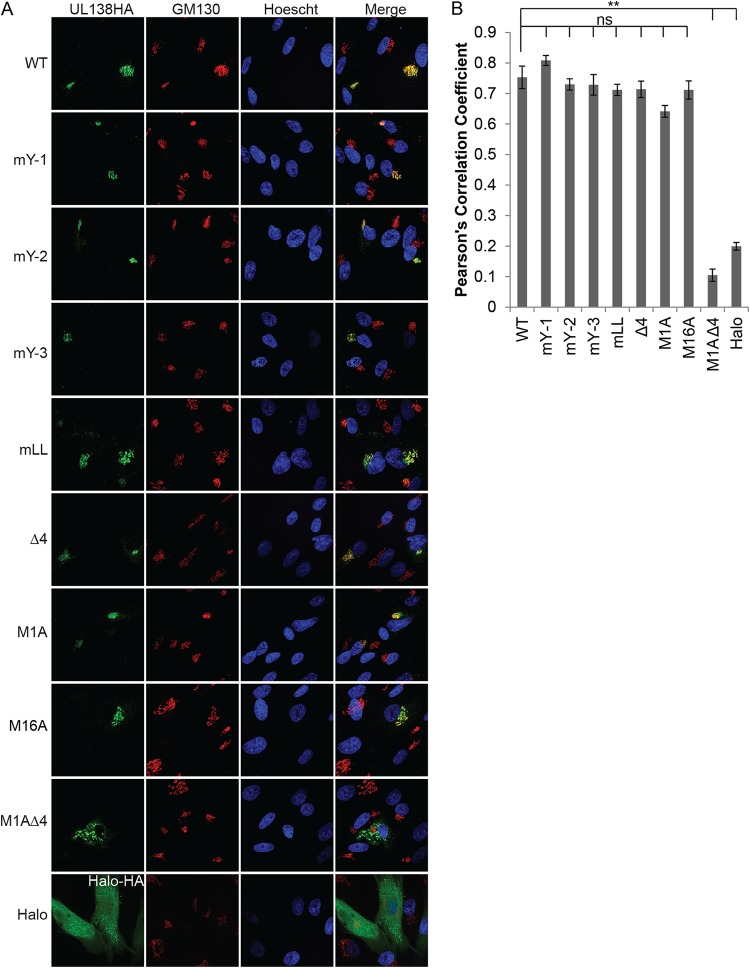
Similarly to the WT protein, all of the singly substituted mutants colocalized with Golgi marker GM130 in transfected NHDFs in qualitative (Fig. 2A) and quantitative (Fig. 2B) assays. In fact, disruption of all four Golgi sorting motifs was unable to prevent UL138 from localizing to the Golgi apparatus. As previously described (29), the M1A mutant expressing only the short isoform of UL138 that lacked the majority of the transmembrane domain also localized to the Golgi apparatus in qualitative (Fig. 2A) and quantitative (Fig. 2B) assays. It was only when all four Golgi sorting motifs were disrupted in the context of the M1A mutant (M1AΔ4) that UL138 protein failed to localize to the Golgi apparatus (Fig. 2). The lack of M1AΔ4 colocalization with the Golgi was similar to the results seen with the HaloTag (Halo) peptide, a 297-residue epitope tag used in protein purification and subcellular localization studies (37) that we used as a non-Golgi-localized control (Fig. 2). While Halo has a diffuse pan-cytoplasmic staining pattern, the M1AΔ4 mutant had a punctate pan-cytoplasmic staining pattern. We conclude that disruption of the transmembrane domain and of all four Golgi sorting motifs is sufficient to displace UL138 from the Golgi apparatus.
FIG 2.
The Golgi sorting motifs and the transmembrane domain work in concert to localize UL138 to the Golgi apparatus. (A) Indirect immunofluorescence of NHDF cells transfected with plasmids expressing UL138 WT or the indicated mutants. Cells were harvested, fixed, and stained 48 h posttransfection. GM130 is a Golgi marker. Nuclei were counterstained with Hoechst stain. Halo served as a non-Golgi localized control. Images representative of results from three independent biological replicates are shown. (B) A total of 10 cells per condition per experiment were analyzed for colocalization, and values representing Pearson’s correlation coefficients were determined. Data represent means ± standard errors of the means (SEM) of results from three independent biological replicates. **, P < 0.01; ns, not significant (P > 0.05) (Student's t test).
Efficient downregulation of MRP1 expression requires the UL138 transmembrane domain and acidic cluster dileucine motif.
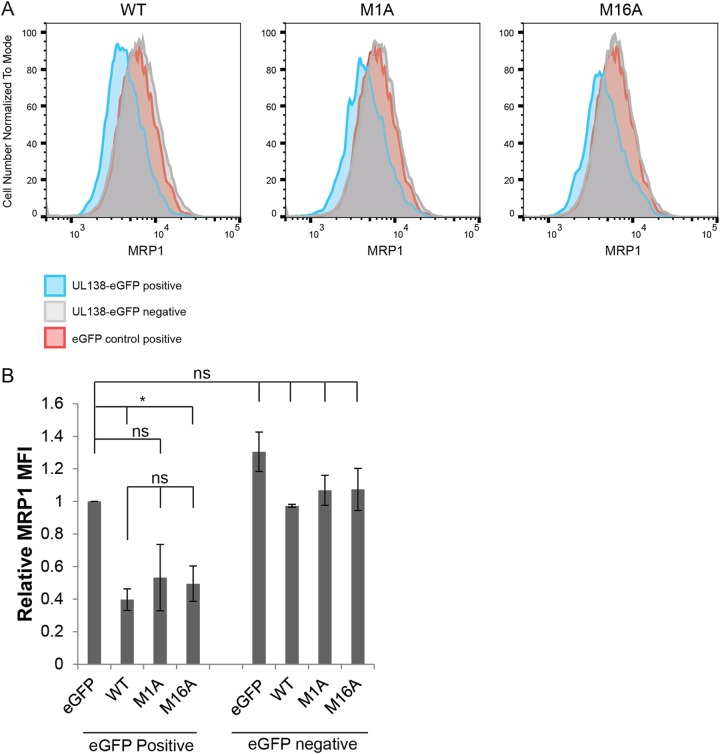
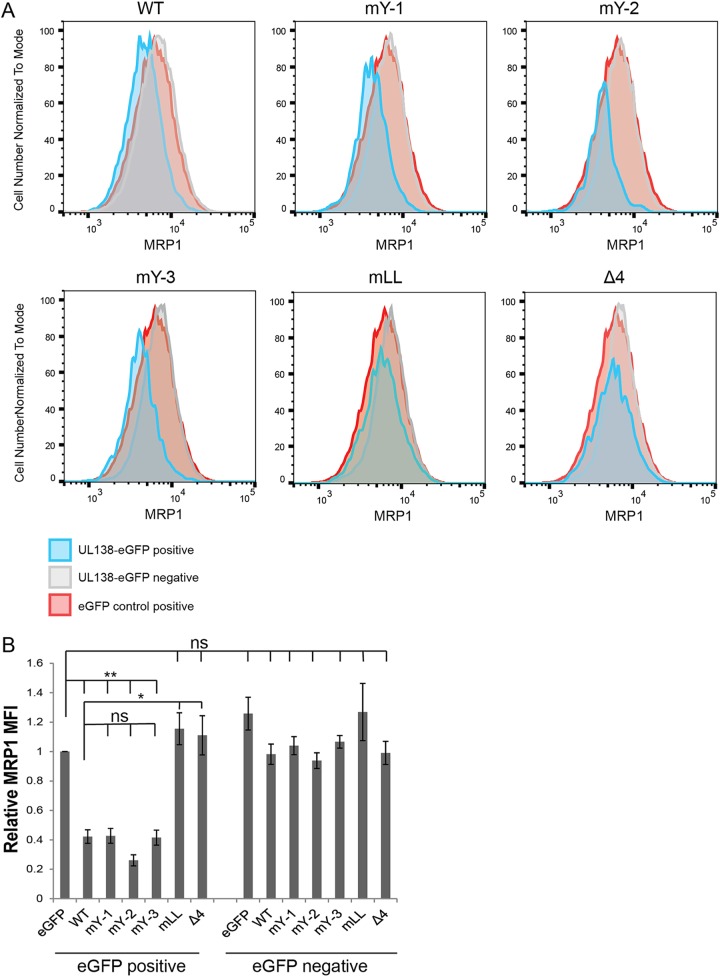
The presence of UL138 is both necessary (during HCMV infection) and sufficient (during transient transfection) to decrease the cell surface expression of MRP1 and to direct the protein to the lysosome for degradation (22). To screen our UL138 alleles for defects in MRP1 downregulation, we used a transient-transfection–flow cytometry assay in NHDF cells expressing endogenous MRP1. Expression plasmids encoding the UL138 WT, single-isoform mutants, or Golgi sorting motif mutant alleles fused to enhanced green fluorescent protein (eGFP), or an eGFP-alone control, were transfected into NHDFs. We then analyzed MRP1 expression by flow cytometry. Blue histograms (Fig. 3A; see also Fig. 4A) represent either the UL138-eGFP WT or mutant allele transgene-expressing cells (UL138-eGFP positive). Gray histograms (Fig. 3A; see also Fig. 4A) represent cells that did not express the eGFP transgene in the UL138-eGFP transfections (UL138-eGFP negative). Red histograms (Fig. 3A; see also Fig. 4A) represent eGFP-alone-control-expressing cells (eGFP control positive). Comparing eGFP-alone-control-expressing cells to the WT UL138-eGFP-expressing cells, we saw a downregulation in MRP1 (Fig. 3; see also Fig. 4), replicating previous results (22). Comparing eGFP-alone-control-expressing cells to UL138-eGFP-negative cells, we observed no differences in MRP1 levels, indicating that eGFP expression alone had no effect on MRP1 expression (Fig. 3; see also Fig. 4).
FIG 3.
The transmembrane domain of UL138 increases the magnitude of MRP1 downregulation. (A) NHDF cells were transfected with expression plasmids for eGFP (eGFP control positive) or WT UL138 or the indicated UL138 mutant alleles fused to eGFP (UL138-eGFP positive), harvested 48 h later, and analyzed by flow cytometry for MRP1. Representative histograms of results from three independent biological replicates are shown. Blue histograms (UL138-eGFP positive cells) and gray histograms (UL138-eGFP-negative cells) represent cells from the same transfection. Red histograms represent control eGFP-alone-positive cells (eGFP control positive). (B) Quantitation of mean fluorescence intensity (MFI) of MRP1 staining in the eGFP-positive or eGFP-negative population of the indicated UL138 or control transfections relative to the eGFP-control-positive cells. Data represent means ± SEM of results from three independent biological replicates. *, P < 0.05; ns, not significant (P > 0.05) (Student's t test).
FIG 4.
The acidic cluster dileucine motif of UL138 is required for MRP1 downregulation. (A) NHDF cells were transfected with expression plasmids for eGFP (eGFP control positive), WT UL138, or the indicated UL138 Golgi sorting mutants (mY-1, mY-2, mY-3, mLL, or Δ4) fused to eGFP (UL138-eGFP positive), harvested 48 h later, and analyzed by flow cytometry for MRP1 expression. Representative histograms of results from three independent biological replicates are shown. Blue histograms (UL138-eGFP-positive cells) and gray histograms (UL138-eGFP-negative cells) represent cells from the same transfection. Red histograms represent control eGFP-alone-positive cells (eGFP control positive). (B) Quantification of the MFI of MRP1 staining in the eGFP-positive or eGFP-negative cells of the indicated UL138 or control transfections relative to the eGFP control-positive cells. Data represent means ± SEM of results from three independent biological replicates. *, P < 0.05; **, P < 0.01; ns, not significant (P > 0.05) (Student's t test).
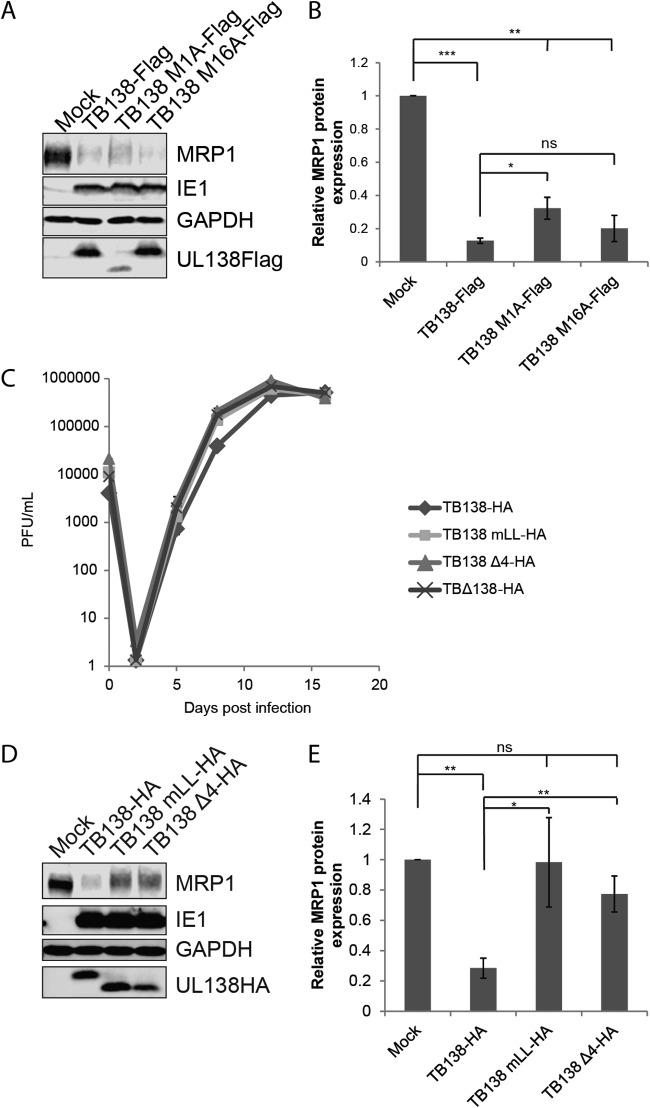
Cells expressing the M16A allele (only the long isoform) displayed decreased MRP1 levels compared to the eGFP-alone control (Fig. 3). The reductions in MRP1 steady-state levels mediated by the M16A allele were similar to those seen with WT UL138 (Fig. 3). Cells expressing the M1A allele (only the short isoform) showed reduced MRP1 levels compared to the eGFP-alone control in three independent transfections. The reductions in the levels of the M1A transfections were of decreased magnitude and of greater variability than those observed in cells expressing the WT UL138 allele (Fig. 3) and did not reach statistical significance. We conclude that the transmembrane domain, though not absolutely required, certainly enhances MRP1 downregulation. For the Golgi sorting motifs, cells expressing alleles with mutations in any of the three tyrosine sorting motifs (mY-1, mY-2, or mY-3) displayed decreased MRP1 levels compared to the eGFP-alone control that were similar to the levels seen with cells expressing the WT UL138 allele (Fig. 4). Cells expressing alleles with mutations in the acidic cluster dileucine motif (mLL or Δ4) did not show decreased MRP1 levels compared to the eGFP-alone control (Fig. 4), indicating that they failed to downregulate MRP1. The UL138-eGFP-negative populations showed no differences in MRP1 expression from the eGFP-alone-control-expressing cells (Fig. 4). We conclude that the acidic cluster dileucine motif is required for the ability of UL138 to downregulate MRP1.
To determine if the transmembrane domain of UL138 was required to downregulate MRP1 during infection, we utilized recombinant TB40/E-based UL138-Flag-tagged viruses that synthesize only the long (TB138 M16A-Flag) or only the short (TB138 M1A-Flag) isoform of UL138 (29). Cells infected with the TB138 M16A-Flag virus showed reductions in MRP1 levels similar to those seen with cells infected with the WT virus, TB138-Flag (Fig. 5A and B). Cells infected with the TB138 M1A-Flag virus also showed reduced MRP1 levels that were statistically different from those seen with mock-infected cells, but the reductions did not reach the level achieved during WT virus infection (Fig. 5A and B). We conclude that, similarly to the results of the transfection experiments (Fig. 3), the transmembrane domain enhances the ability of UL138 to downregulate MRP1 but is not absolutely required for the downregulation.
FIG 5.
Both the transmembrane-spanning peptide and acidic cluster dileucine motif of UL138 participate in MRP1 downregulation during HCMV infection. (A) Lysates from NHDFs infected with the indicated viruses at an MOI of 3 for 48 h were analyzed by Western blotting with the indicated antibodies. Images representative of results from three independent biological replicates are shown. (B) MRP1 levels normalized to GAPDH and compared to a mock-infected control were determined by densitometry using data from the experiments whose results are presented in panel A. Data represent means ± standard deviations of results from three independent biological replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant (P > 0.05) (Student's t test). (C) NHDFs were infected with TB40/E-based viruses encoding HA-tagged WT UL138 (TB138-HA) or the indicated Golgi sorting motif mutants (TB138 mLL-HA; TB138 Δ4-HA) or a UL138 null virus (TBΔ138-HA) at an MOI of 0.1. Infectious progeny were isolated on the indicated day postinfection, and virus titers were determined by plaque assay. Data represent means ± SEM of results from three independent biological replicates. (D) Lysates from NHDFs infected with the indicated viruses at an MOI of 3 for 48 h were analyzed by Western blotting with the indicated antibodies. Images representative of results from three independent biological replicates are shown. (E) MRP1 levels normalized to GAPDH and compared to a mock-infected control were determined by densitometry using data from the experiments whose results are presented in panel D. Data represent means ± SEM of results from three independent biological replicates. *, P < 0.05; **, P < 0.01; ns, not significant (P > 0.05) (Student's t test).
To determine if the acidic cluster dileucine motif of UL138 was required for downregulation of MRP1 during infection, we generated new recombinant TB40/E-based UL138-HA-tagged viruses with alanine substitutions in the acidic cluster dileucine motif (TB138 mLL-HA) or in all four Golgi sorting motifs (TB138 Δ4-HA) (Table 1). All viruses grew to titers similar to those measured for the WT parental control, TB138-HA, during productive replication in NHDFs (Fig. 5C). Cells infected with the TB138 mLL-HA and TB138 Δ4-HA viruses showed no significant reduction in MRP1 levels compared to a mock-infected control (Fig. 5D and E). We conclude that, consistent with the results from the transfection experiments (Fig. 4), the acidic cluster dileucine sorting motif is required for the ability of UL138 to downregulate MRP1 during infection.
The transmembrane domain and acidic cluster dileucine motif are required for UL138-mediated MRP1 inactivation.
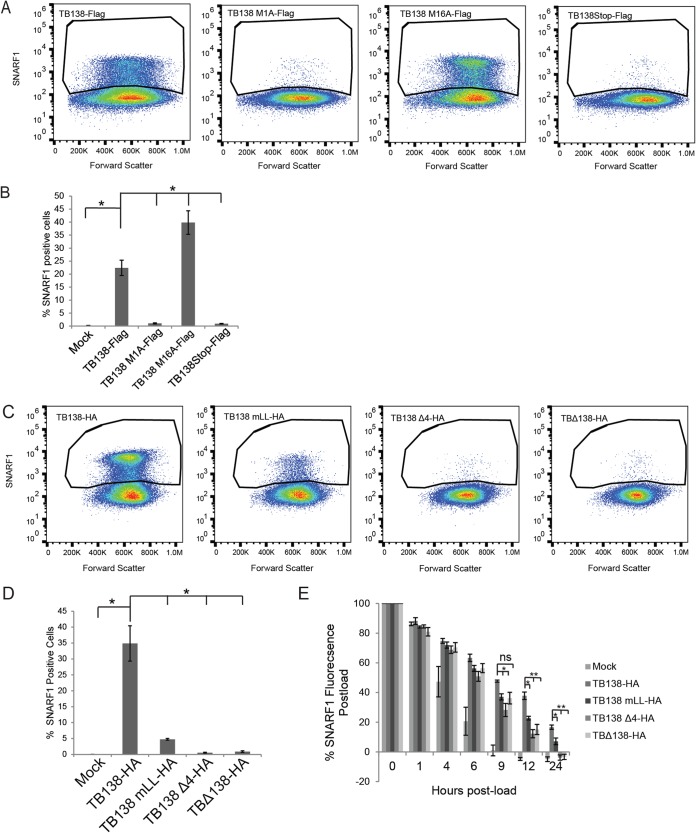
Downregulation of MRP1 levels by UL138 impairs the ability of cells to export one of its known substrates (22), 5-carboxyseminaptharhodafluor (SNARF1) (38). At 24 h after loading cells with SNARF1 and 48 h postinfection, <1% of mock-infected and UL138-null HCMV (TB138Stop-Flag)-infected cells remained SNARF positive, while ∼20% to 25% of the WT TB40/E-infected (TB138-Flag) cells remained SNARF1 positive (Fig. 6A and B). These data indicate that UL138-mediated downregulation of MRP1 levels leads to functional inactivation of MRP1, as previously reported (22). Cells infected with the TB138 M16A-Flag virus (expressing only the long isoform) showed percentages of SNARF1-positive cells even higher than those seen with cells infected with the WT virus (Fig. 6A and B). Cells infected with the TB138 M1A-Flag virus (expressing only the short isoform) showed percentages of SNARF1-positive cells similar to the levels seen with mock-infected or TB138Stop-Flag-infected negative controls, leading us to conclude that the transmembrane-spanning peptide is required to functionally inhibit MRP1.
FIG 6.
The acidic cluster dileucine motif and transmembrane domain of UL138 are required to functionally inhibit MRP1 during HCMV infection. (A) NHDFs infected for 24 h at an MOI of 3 with the indicated virus were loaded with SNARF1. At 24 h later, SNARF1-positive cells were quantitated by flow cytometry. Images representative of results from three independent biological replicates are shown. (B) Quantitation of data from the experiments whose results are presented in panel A. Data represent means ± SEM of results from three independent biological replicates *, P < 0.05; **, P < 0.01; ns, not significant (P > 0.05) (Student's t test). (C) NHDFs infected for 24 h at an MOI of 3 with the indicated virus were loaded with SNARF1. At 24 h later, SNARF1-positive cells were quantitated by flow cytometry. Images representative of results from three independent biological replicates are shown. (D) Quantitation of data from the experiments whose results are presented in panel C. Data represent means ± SEM of results from three independent biological replicates. *, P < 0.05; **, P < 0.01; ns, not significant (P > 0.05) (Student's t test). (E) The MFI of SNARF1-loaded NHDFs infected with the indicated virus at an MOI of 3 for 24 h was measured on a fluorescent plate reader at the indicated time post-SNARF1 load. Background fluorescence was subtracted, and MFI data were normalized to the MFI measured immediately postload (time zero). Data represent means ± SEM of results from three independent biological replicates. *, P < 0.05; **, P < 0.01; ns, not significant (P > 0.05) (Student's t test).
Cells infected with the TB138 mLL-HA and TB138 Δ4-HA viruses also showed percentages of SNARF1-positive cells similar to those seen with mock-infected and TBΔ138-HA (the matched UL138-null virus)-infected negative controls (Fig. 6C and D). Again, the cells infected with the matched WT control (TB138-HA) had significantly more SNARF1-positive cells than either of the Golgi mutants, the UL138 null (TBΔ138-HA) or mock-infected control. When we monitored SNARF1 export over time, we noted that the cells infected with TB138 mLL-HA or TB138 Δ4-HA exported SNARF1 in a time frame similar to that seen with a UL138 null (TBΔ138-HA) infection and faster than cells infected with a virus expressing WT UL138 (TB138-HA) (Fig. 6E). Taking the results together, we conclude that the transmembrane domain and the acidic cluster dileucine motif are required for the ability of UL138 to inhibit MRP1 function during HCMV infection.
DISCUSSION
Here we show that the Golgi localization of UL138 is lost when the transmembrane domain and all four Golgi sorting motifs are missing from the protein. We identify the UL138 acidic cluster dileucine motif as required for MRP1 downregulation and inactivation of MRP1 activity. We also show that the UL138 short isoform lacking the putative transmembrane-spanning peptide, while still able to partially downregulate MRP1, is unable to inactivate its function.
As integral membrane proteins (28), newly synthesized copies of the long isoform (29) of UL138 likely traverse the ER and Golgi stacks to set up residence in the trans-Golgi network (TGN). Resident Golgi proteins are retained in part by selective blocking of incorporation into transport vesicles (39). Golgi retention mechanisms include protein aggregation, specific (generally short) lengths of transmembrane domains, and the presence of an (F/L)-(L/V)-(S/T) motif (40). Aggregation of UL138 has not been reported, and the length of the UL138 transmembrane domain has not been experimentally determined. UL138 does, however, contain a consensus Golgi retention motif (the FLS motif; amino acids 41 to 43) (see Fig. 1).
Resident Golgi proteins also maintain Golgi localization by iterative rounds of anterograde transport (into transport vesicles) and retrograde transport (back to the Golgi apparatus). The AP-1 cargo adaptor protein can mediate retrograde transport from endosomes back to the Golgi apparatus by interacting with tyrosine or dileucine sorting motifs (41), but a UL138 mutant lacking all four Golgi sorting motifs still resides in the Golgi apparatus (Fig. 2), indicating that these motifs are not required for UL138 retrograde transport. Proteins are also transported from endosomes back to the Golgi apparatus through retrograde transport mediated by the sorting nexins (SNXs) in cooperation with one of two multiprotein retrieval complexes (called “retromer” and “retriever”) (42, 43). These heterogeneous protein complexes mediate the retrieval and sorting of a diverse set of cargos from the endosomal compartment. The retromer complex is the canonical retrograde retrieval complex and binds cargo either directly or through interactions with SNX proteins (43–45). The retriever complex works in association with CCDC22, CCDC93, and COMMD (CCC complex) and with the Wiskott-Aldrich syndrome protein and SCAR homologue (WASH complex) (42). The CCC complex and retriever complex recycle cell surface proteins from the endosome, preventing their degradation by the lysosome, independently of the retromer complex (46). Both the retromer complex and the retriever complex interact with the WASH complex to create actin-rich endosomal domains from which cargo is retrieved (47). UL138 (Fig. 1) contains multiple potential direct retromer-binding motifs and SNX binding PDZ-binding motifs [Ø-X-(L/M) and S/T-X-Ø, respectively] and interacts in vitro with SNX 21 (48). While UL138 encodes potential sites to interact with the endosomal retrieval machinery of the cell, roles for the retromer, retriever, or SNX proteins in UL138 localization or function have not yet been examined, but such roles could explain why the Golgi sorting motif mutant, allele Δ4, still retained Golgi localization (Fig. 2).
The short isoform of UL138 localizes to the Golgi apparatus (29) but is missing the predicted membrane targeting signal sequence and most of the predicted transmembrane domain. Therefore, it is unlikely that the short isoform is an integral membrane protein, and it is unlikely to be incorporated into transport vesicles. However, the short isoform of UL138 may associate with the surface of transport vesicles. The UL138 M1AΔ4 protein lacking the putative transmembrane peptide and all of the Golgi sorting motifs failed to localize to the Golgi apparatus (Fig. 2). This result indicates that at least one of the Golgi motifs, perhaps acting through membrane-independent soluble protein-protein interactions with AP and/or GGA proteins or with other Golgi-adjacent proteins, has a Golgi localization function for the UL138 short isoform. In both transfections and infections, the UL138 short isoform accumulates to levels substantially lower than those seen with the long isoform (Fig. 1C; see also Fig. 5A) (29) and fails to fully downregulate MRP1 (Fig. 3; see also Fig. 5A and B). We cannot determine whether the inability of the short isoform to fully downregulate the levels of MRP1 or to inhibit its function (Fig. 6A and B) is due to a critical function of the missing transmembrane domain or due to an inability to achieve a high enough steady-state level. Without noticeably increased UL138 protein levels, the virus that expresses only the long isoform of UL138 (TB138 M16A-Flag) appears to be more effective at inhibiting MRP1-dependent efflux than the WT virus, which is capable of expressing both isoforms (TB138-Flag) (Fig. 6A and B). Perhaps both isoform-specific functional efficiencies and steady-state levels allow the virus to fine-tune UL138-dependent outcomes.
The requirement of the acidic cluster dileucine motif of UL138 for MRP1 downregulation (Fig. 4; see also Fig. 5 and Fig. 6) implicates GGA proteins in mediating the delivery of newly synthesized MRP1 from the Golgi apparatus or endocytosed MRP1 from the plasma membrane into the endosomes destined for the lysosome in a UL138-dependent fashion. We hypothesize that the interaction between UL138 and MRP1 (22) allows UL138- and GGA-dependent entry of MRP1 into the endosomal pathway. From there, MRP1 may be internalized into multivesicular bodies by the endosomal sorting complex required for transport (ESCRT) (49), which then fuse to the lysosomes for degradation (Fig. 7). ESCRT is required to remodel endosomal membranes to expose internalized membrane proteins to the lumen of the multivesicular body and, eventually, the lysosome for degradation (49, 50). While AP proteins (specifically, AP-3) can also deliver proteins directly to the lysosome, they are more often associated with simple dileucine motifs rather than with acidic cluster dileucine motifs like those found in UL138 (Fig. 1) (30, 51–53) and are therefore unlikely to play a role in UL138 function.
FIG 7.
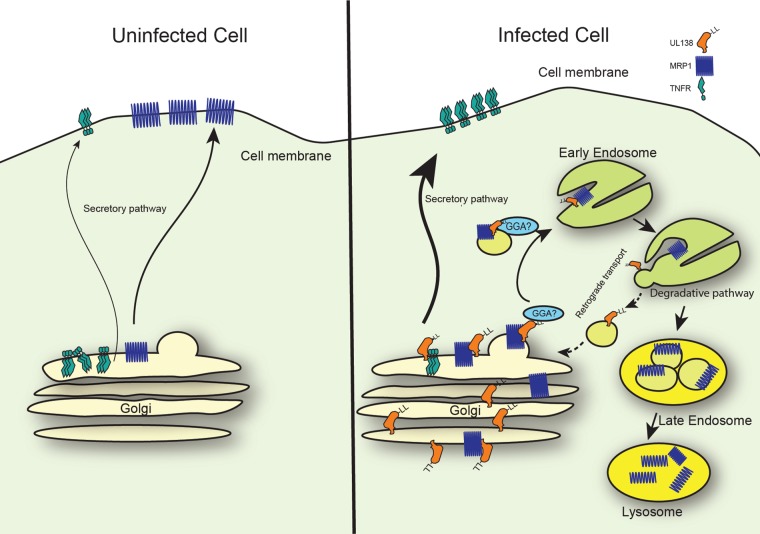
Model of UL138-mediated trafficking. In uninfected cells, TNFR resides predominantly in the Golgi apparatus and MRP1 at the plasma membrane. During HCMV infection, UL138 promotes TNFR1 cell surface expression by an unknown mechanism. UL138 downregulates the levels of MRP1 through a process that requires the acidic cluster dileucine motif and that is made more efficient by the presence of the full transmembrane domain. In our speculative model, UL138 interacts with MRP1 in the Golgi apparatus. UL138 binds through its acidic cluster dileucine motif to cellular GGA proteins and directs MRP1 to endosomes. Within the endosomes, UL138 and MRP1 dissociate, resulting in MRP1 incorporation into intraluminal vesicles for transport to lysosomes and UL138 recycling to the Golgi apparatus in a process potentially requiring retromer and/or SNX proteins. We suspect that the coexpressed short isoform of UL138 does not play a major role in UL138-mediated trafficking events, as it is expressed at low levels and fails to efficiently inhibit MRP1 function.
In addition to downregulating the cell surface expression of MRP1, UL138 upregulates the cell surface expression of tumor necrosis factor receptor 1 (TNFR1) (36, 54). Cell surface TNFR1 is continuously internalized and degraded by lysosomes without being recycled, a process accelerated by the presence of bound ligand (55). However, the majority of TNFR1 is sequestered in the Golgi apparatus (56) at the TGN (57) by the C-terminal 23 amino acids of the protein (58), with only a small fraction of molecules reaching the cell surface. The mechanisms and consequences of UL138-mediated upregulation of TNFR1 have not been examined. It is becoming clear that HCMV infection drastically alters the endosomal sorting compartment and selectively redirects cargo proteins (59). UL138 could be acting as a linker between these cargo proteins and the cellular sorting machinery, selectively sorting cargo to alternate fates such as it does for MRP1 and TNFR1.
We provide a speculative but testable model for UL138 and its functions in regulating MRP1 and TNFR1 cell surface expression (Fig. 7). We hypothesize that UL138 disrupts the ill-defined tethering interactions that keep TNFR1 at the Golgi apparatus, allowing the incorporation of TNFR1 into transport vesicles and thereby upregulating its cell surface expression. Mutations that upregulate TNFR1 at the cell surface have been theorized to mask a Golgi retention or retrieval signal (60). Perhaps UL138 binding to TNFR occludes the same signal. Because the Golgi sorting motifs analyzed here direct traffic to endosomes and not the plasma membrane, we suspect that they are not required for UL138-mediated TNFR1 upregulation. The acidic cluster dileucine motif is required for UL138-mediated MRP1 inactivation (Fig. 4; see also Fig. 5 and Fig. 6). Thus, we hypothesize that UL138 interacts with MRP1 in the Golgi apparatus and delivers it to transport vesicles through interactions with GGA proteins mediated by the acidic cluster dileucine motif. Within the acidifying endosomes, UL138 and MRP1 dissociate. MRP1 is incorporated into intraluminal vesicles, perhaps in an ESCRT-dependent manner, for delivery to lysosomes for degradation (50, 61, 62), and UL138 is recycled back to the Golgi apparatus by retrograde transport (Fig. 7).
HCMV remains a significant clinical pathogen, with current treatments plagued by resistance, toxicities, and an inability to target latently infected cells (7, 8). A novel proposal to target infected cells capitalizes on the UL138-mediated downregulation of MRP1 for the ex vivo selection of HCMV-negative subpopulations or the killing of HCMV-positive subpopulations within hematopoietic stem cell (bone marrow) transplantations (22). Should such strategies become a clinical reality, an understanding of the genetics and mechanism of UL138-mediated MRP1 downregulation as determined (in part) here may help predict or manage the inevitable emergence of resistant viral mutants. If the cytotoxic agents that rely on MRP1 export were to be used clinically, our data suggest that selective pressure would be placed on the acidic cluster dileucine motif and transmembrane-spanning peptide of UL138.
MATERIALS AND METHODS
Cells and infections.
Normal human dermal fibroblasts (NHDFs; Clonetics) were maintained as described previously (63). TB138-Flag, TB138 M1A-Flag, TB138 M16A-Flag, and TB138Stop-Flag were described previously (29). The UL138-HA tagged TB40/E virus (TB138-HA) and UL138-HA deletion virus (TBΔ138-HA) were described previously (64). Novel UL138 Golgi sorting motif mutant viruses were derived from TB138-HA bacterial artificial chromosomes (BAC) (64) and generated using a two-step red recombination procedure (65) and the primers listed in Table 2. Virus titers were determined by plaque assay on NHDFs. Cells were infected in a minimal volume of media at the indicated multiplicity of infection (MOI) for 1 h. Viral inoculum was then aspirated, and cells were returned to normal culture conditions for the indicated time periods.
TABLE 2.
Primers used in this study
| Use | Primer namea | Primer sequence |
|---|---|---|
| UL138 SDM primers | UL138 mY-1 SDM Forward | 5′-GATCGTGGCCATTCTCTGCGCGGCCGCTTACCATTGGCACGACAC |
| UL138 mY-1 SDM Reverse | 5′-GTGTCGTGCCAATGGTAAGCGGCCGCGCAGAGAATGGCCACGATC | |
| UL138 mY-2 SDM Forward | 5′-GTGCGCATGTTTTTGAGCGCCGCGGCGGCGATCCGCTGTTGCGAGCTGTAC | |
| UL138 mY-2 SDM Reverse | 5′-GTACAGCTCGCAACAGCGGATCGCCGCCGCGGCGCTCAAAAACATGCGCAC | |
| UL138 mY-3 SDM Forward | 5′-CGCTGTTGCGAGCTGGCCGGCGAATACGAGCGCCGGTTC | |
| UL138 mY-3 SDM Reverse | 5′-GAACCGGCGCTCGTATTCGCCGGCCAGCTCGCAACAGCG | |
| UL138 mLL SDM Forward | 5′-GGTGACGGCGCCGCTGACCGCGGCGGCGGCGGCGAAACCCGTGACGGGATCC | |
| UL138 mLL SDM Reverse | 5′-GGATCCCGTCACGGGTTTCGCCGCCGCCGCCGCGGTCAGCGGCGCCGTCACC | |
| UL138-eGFP fusion cloning | UL138 eGFP fusion Forward | 5′-TATGACGTGCCTGACTATGCCGTGAGCAAGGGCGAGGAGCTGTTC |
| UL138 eGFP fusion Reverse | 5′-AGCTCCTCGCCCTTGCTCACGGCATAGTCAGGCACGTCATAAG | |
| UL138 pSG5 infusion Forward | 5′-ACTATAGGGCGAATTCGATTCCGAAATTCATGGACGATC | |
| eGFP pSG5 infusion Reverse | 5′-ATCTGGATCCGAATTCTTACTTGTACAGCTCGTCCATGCCGAG | |
| HaloTag cloning | Halo pSG5 infusion Forward | 5′-ACTATAGGGCGAATTCATGGCAGAAATCGGTACTGGCTTTC |
| Halo pSG5 infusion Reverse | 5′-ATCTGGATCCGAATTCTCAGGCATAGTCAGGCACGTCATAAGGATAGCCGGAAATCTCGAGCGTCGACAG | |
| BAC mutagenesis primers | UL138 ins KanR-BglII F | 5′-TTGAGTAGATCTTGGTCCGTTGGGAGGAAGTGTCTTCCCAGTGCAGCTACGCGTCGTCGCGGAACCCACTGCTTACTGGCTTATCG |
| UL138 ins KanR-BglII R | 5′-TTTCGTAGATCTGCCAGTGTTACAACCAATTAACC | |
| TB40E-BAC ins UL138 F | 5′-TGTACAAAAGAGAGAGACTGGGACGTAGATCCGGACAGAGGACGGTCACCATGGACGATCTGCCGCTGAAC | |
| TB40E-BAC ins UL138 R | 5′-GTCAAAACGACATTACCGCGATCCGCTCCCCTCTTTTTTCTTTTTCTCATTCAGGCATAGTCAGGCACGTC | |
F, forward; R, reverse.
Plasmids, transfections, and enzymes.
UL138 mutants were generated in pSG5 from a WT clone (64). The specific substitutions are listed in Table 1. Briefly, the mutagenesis primers listed in Table 2 were used to amplify UL138HA-pSG5 (64) using CloneAmp HiFi polymerase (Clontech; 639298). PCR products were digested with DpnI enzyme (NEB; R0176S) and transformed into Stellar competent cells (Clontech; 636766). Positive clones were recovered and verified via Sanger sequencing. The HaloTag peptide (Halo) was cloned into pSG5 from pFC14a (Promega; G9651) using an In-fusion cloning kit (Clontech; 638920) and primers listed in Table 2. Cells were transfected using an Amaxa Nucleofector II and a Lonza Amaxa NHDF Nucleofector kit (Lonza; VPD-1001) according to the manufacturer’s instructions. λ protein phosphatase treatment was performed according to the instructions of the manufacturer (New England Biolabs; P0753S).
Antibodies and Western blotting.
The antibody against IE1 (1B12) has been described previously (66). The following antibodies were obtained from the indicated commercial sources: GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Invitrogen; 6C5), HA (BioLegend; HA.11), MRP1 (Enzo; MRPr1), Rb-pS608 (Cell Signaling Technologies; 2181), Rb (Cell Signaling Technologies; 9309). For Western blotting, equal numbers of cells were lysed in 2% SDS containing 4% β-mercaptoethanol or radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 8], 5 mM EDTA, 150 mM NaCl, 10% glycerol, 0.1% SDS, 1% NP-40, 1% Triton X-100) and separated by SDS-PAGE. Gels were blotted onto nitrocellulose membranes (GE Amersham; Protran). Membranes were blocked with 5% bovine serum albumin (Research Products International Corp.)–TBST (10 mM Tris [pH 8], 150 mM NaCl, 0.05% Tween 20), stained with the indicated primary antibody, and washed in TBST. Membranes were stained with Li-Cor IRDye 680 and 800 secondary antibodies (Li-Cor) and imaged on a Li-Cor Odyssey FC system.
SNARF1 release assay.
The SNARF1 release assay was modified from one previously described (22). Briefly, NHDFs were infected for 24 h at an MOI of 3. Cells were loaded with 10 μM 5-carboxyseminaptharhodafluor acetoxymethyl ester (SNARF1) (Thermo Fisher; C1272) for 45 min, after which cells were washed with phosphate-buffered saline (PBS) and fluorescence was read on a Citation 5 plate reader (Biotek) (excitation, 485 nm; emission, 595 nm) to obtain postload fluorescence intensity (time zero). Cells were returned to complete media and maintained under standard culture conditions. At the indicated time points, the cells were washed with PBS and read on the plate reader as described above. After 24 h, the cells were collected by the use of trypsin, fixed, and analyzed using an Attune flow cytometer (Thermo Fisher) or a Fortessa flow cytometer (BD).
Flow cytometry.
Cells were harvested by the use of trypsin and washed with 2% fetal bovine serum (FBS)–PBS. They were then stained with Live/Dead Violet (Invitrogen; L34955) for 30 min, washed, fixed with 1% formaldehyde for 30 min, and washed again. For analyses using internal antigens, cells were permeabilized with 0.1% saponin (Sigma)–PBS for 15 min. Cells were stained with MRP1-PE (MRP1-phycoerythrin) (Miltenyi; REA481) antibody, washed, and analyzed on a LSRII system (BD).
Immunofluorescence.
Transfected cells were plated on coverslips in a 6-well dish. After 48 h, coverslips were harvested and fixed with 1% formaldehyde. Cells were then permeabilized with 0.1% Triton X-100 (Sigma) and 0.05% Tween 20 (Sigma) and stained with the HA (BioLegend; HA.11) and GM130 (Cell Signaling Technology; D6B1) antibodies. Cells were washed and stained with secondary antibodies Alexa 488 and Alexa 594 (Molecular Probes; A11029 and A11012). Cells were counterstained with Hoechst stain. Coverslips were mounted on slides using Fluoromount-G (Southern Biotech; 0100-01). Slides were imaged on a Prairie laser scanning confocal microscope (Prairie Technologies). Images were analyzed using Fiji (67). Colocalization data were quantified using the FIJI plugin Colocalization Threshold.
ACKNOWLEDGMENTS
We thank Dick van den Boomen for advice on detecting MRP1 by Western blotting, Jon Audhya for discussions about the secretory pathway, and members of our laboratory for helpful comments and suggestions.
This work was supported by grants from the NIH to R.F.K. (AI130089 and AI139180), and to C.B.G. (AI131449-01A1). Flow cytometry support was provided by the University of Wisconsin (UW) Carbone Cancer Center (support grant P30CA014520) and the UW Flow Fortessa (instrumentation grant S100OD018202-01).
C.B.G. and R.F.K. designed the experiments, analyzed the data, and wrote the paper. C.B.G. performed all the experiments.
REFERENCES
- 1.Mocarski ES, Shenk T, Pass RF. 2007. Cytomegaloviruses, p 2701–2772. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Koval CE. 2018. Prevention and treatment of cytomegalovirus infections in solid organ transplant recipients. Infect Dis Clin North Am 32:581–597. doi: 10.1016/j.idc.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Ljungman P, Brand R, Hoek J, de la Camara R, Cordonnier C, Einsele H, Styczynski J, Ward KN, Cesaro S. 2014. Donor cytomegalovirus status influences the outcome of allogeneic stem cell transplant: a study by the European Group for Blood and Marrow Transplantation. Clin Infect Dis 59:473–481. doi: 10.1093/cid/ciu364. [DOI] [PubMed] [Google Scholar]
- 4.Snydman DR, Torres-Madriz G, Boucher HW. 2008. Perspectives in the treatment and prophylaxis of cytomegalovirus disease in solid-organ transplant recipients. Clin Infect Dis 47:702–711. doi: 10.1086/590934. [DOI] [PubMed] [Google Scholar]
- 5.Britt W. 2015. Controversies in the natural history of congenital human cytomegalovirus infection: the paradox of infection and disease in offspring of women with immunity prior to pregnancy. Med Microbiol Immunol 204:263–271. doi: 10.1007/s00430-015-0399-9. [DOI] [PubMed] [Google Scholar]
- 6.Kenneson A, Cannon MJ. 2007. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev Med Virol 17:253–276. doi: 10.1002/rmv.535. [DOI] [PubMed] [Google Scholar]
- 7.Britt WJ, Prichard MN. 2018. New therapies for human cytomegalovirus infections. Antiviral Res 159:153–174. doi: 10.1016/j.antiviral.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Fisher CE, Knudsen JL, Lease ED, Jerome KR, Rakita RM, Boeckh M, Limaye AP. 2017. Risk factors and outcomes of ganciclovir-resistant cytomegalovirus infection in solid organ transplant recipients. Clin Infect Dis 65:57–63. doi: 10.1093/cid/cix259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maciejewski JP, Bruening EE, Donahue RE, Mocarski ES, Young NS, Jeor SS. 1992. Infection of hematopoietic progenitor cells by human cytomegalovirus. Blood 80:170–178. [PubMed] [Google Scholar]
- 10.Söderberg-Nauclér C, Fish KN, Nelson JA. 1997. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell 91:119–126. doi: 10.1016/S0092-8674(01)80014-3. [DOI] [PubMed] [Google Scholar]
- 11.Sindre H, Tjøonnfjord GE, Rollag H, Ranneberg-Nilsen T, Veiby OP, Beck S, Degré M, Hestdal K. 1996. Human cytomegalovirus suppression of and latency in early hematopoietic progenitor cells. Blood 88:4526–4533. [PubMed] [Google Scholar]
- 12.Mendelson M, Monard S, Sissons P, Sinclair J. 1996. Detection of endogenous human cytomegalovirus in CD34+ bone marrow progenitors. J Gen Virol 77:3099–3102. doi: 10.1099/0022-1317-77-12-3099. [DOI] [PubMed] [Google Scholar]
- 13.Goodrum FD, Jordan CT, High K, Shenk T. 2002. Human cytomegalovirus gene expression during infection of primary hematopoietic progenitor cells: a model for latency. Proc Natl Acad Sci U S A 99:16255–16260. doi: 10.1073/pnas.252630899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodrum F, Jordan CT, Terhune SS, High K, Shenk T. 2004. Differential outcomes of human cytomegalovirus infection in primitive hematopoietic cell subpopulations. Blood 104:687–695. doi: 10.1182/blood-2003-12-4344. [DOI] [PubMed] [Google Scholar]
- 15.Taylor-Wiedeman J, Sissons P, Sinclair J. 1994. Induction of endogenous human cytomegalovirus gene expression after differentiation of monocytes from healthy carriers. J Virol 68:1597–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hargett D, Shenk TE. 2010. Experimental human cytomegalovirus latency in CD14+ monocytes. Proc Natl Acad Sci U S A 107:20039–20044. doi: 10.1073/pnas.1014509107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondo K, Kaneshima H, Mocarski ES. 1994. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc Natl Acad Sci U S A 91:11879–11883. doi: 10.1073/pnas.91.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reeves MB, MacAry PA, Lehner PJ, Sissons JGP, Sinclair JH. 2005. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc Natl Acad Sci U S A 102:4140–4145. doi: 10.1073/pnas.0408994102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinclair J. 2008. Human cytomegalovirus: latency and reactivation in the myeloid lineage. J Clin Virol 41:180–185. doi: 10.1016/j.jcv.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 20.Reeves MB, Sinclair JH. 2013. Circulating dendritic cells isolated from healthy seropositive donors are sites of human cytomegalovirus reactivation in vivo. J Virol 87:10660–10667. doi: 10.1128/JVI.01539-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sinclair J, Reeves M. 2014. The intimate relationship between human cytomegalovirus and the dendritic cell lineage. Front Microbiol 5:389. doi: 10.3389/fmicb.2014.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weekes MP, Tan SYL, Poole E, Talbot S, Antrobus R, Smith DL, Montag C, Gygi SP, Sinclair JH, Lehner PJ. 2013. Latency-associated degradation of the MRP1 drug transporter during latent human cytomegalovirus infection. Science 340:199–202. doi: 10.1126/science.1235047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishna BA, Spiess K, Poole EL, Lau B, Voigt S, Kledal TN, Rosenkilde MM, Sinclair JH. 2017. Targeting the latent cytomegalovirus reservoir with an antiviral fusion toxin protein. Nat Commun 8:14321. doi: 10.1038/ncomms14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spiess K, Jeppesen MG, Malmgaard-Clausen M, Krzywkowski K, Dulal K, Cheng T, Hjortø GM, Larsen O, Burg JS, Jarvis MA, Garcia KC, Zhu H, Kledal TN, Rosenkilde MM. 2015. Rationally designed chemokine-based toxin targeting the viral G protein-coupled receptor US28 potently inhibits cytomegalovirus infection in vivo. Proc Natl Acad Sci U S A 112:8427–8432. doi: 10.1073/pnas.1509392112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cole SP, Bhardwaj G, Gerlach JH, Mackie JE, Grant CE, Almquist KC, Stewart AJ, Kurz EU, Duncan AM, Deeley RG. 1992. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science 258:1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- 26.Zaman GJ, Flens MJ, van Leusden MR, de Haas M, Mülder HS, Lankelma J, Pinedo HM, Scheper RJ, Baas F, Broxterman HJ. 1994. The human multidrug resistance-associated protein MRP is a plasma membrane drug-efflux pump. Proc Natl Acad Sci U S A 91:8822–8826. doi: 10.1073/pnas.91.19.8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nooter K, Westerman AM, Flens MJ, Zaman GJ, Scheper RJ, van Wingerden KE, Burger H, Oostrum R, Boersma T, Sonneveld P. 1995. Expression of the multidrug resistance-associated protein (MRP) gene in human cancers. Clin Cancer Res 1:1301–1310. [PubMed] [Google Scholar]
- 28.Petrucelli A, Rak M, Grainger L, Goodrum F. 2009. Characterization of a novel Golgi apparatus-localized latency determinant encoded by human cytomegalovirus. J Virol 83:5615–5629. doi: 10.1128/JVI.01989-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SH, Caviness K, Albright ER, Lee J-H, Gelbmann CB, Rak M, Goodrum F, Kalejta RF. 2016. Long and short isoforms of the human cytomegalovirus UL138 protein silence IE transcription and promote latency. J Virol 90:9483–9494. doi: 10.1128/JVI.01547-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonifacino JS, Traub LM. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem 72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 31.Mardones GA, Burgos PV, Lin Y, Kloer DP, Magadan JG, Hurley JH, Bonifacino JS. 12 February 2013. Structural basis for the recognition of tyrosine-based sorting signals by the Mu3A subunit of the AP-3 adaptor complex. J Biol Chem doi: 10.1074/jbc.M113.450775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Misra S, Puertollano R, Kato Y, Bonifacino JS, Hurley JH. 2002. Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains. Nature 415:933–937. doi: 10.1038/415933a. [DOI] [PubMed] [Google Scholar]
- 33.Shiba T, Takatsu H, Nogi T, Matsugaki N, Kawasaki M, Igarashi N, Suzuki M, Kato R, Earnest T, Nakayama K, Wakatsuki S. 2002. Structural basis for recognition of acidic-cluster dileucine sequence by GGA1. Nature 415:937–941. doi: 10.1038/415937a. [DOI] [PubMed] [Google Scholar]
- 34.Méresse S, Hoflack B. 1993. Phosphorylation of the cation-independent mannose 6-phosphate receptor is closely associated with its exit from the trans-Golgi network. J Cell Biol 120:67–75. doi: 10.1083/jcb.120.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hume AJ, Finkel JS, Kamil JP, Coen DM, Culbertson MR, Kalejta RF. 2008. Phosphorylation of retinoblastoma protein by viral protein with cyclin-dependent kinase function. Science 320:797–799. doi: 10.1126/science.1152095. [DOI] [PubMed] [Google Scholar]
- 36.Le VTK, Trilling M, Hengel H. 2011. The cytomegaloviral protein pUL138 acts as potentiator of tumor necrosis factor (TNF) receptor 1 surface density to enhance ULb′-encoded modulation of TNF-α signaling. J Virol 85:13260–13270. doi: 10.1128/JVI.06005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV. 2008. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol 3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 38.Jin J, Jones AT. 2009. The pH sensitive probe 5-(and-6)-carboxyl seminaphthorhodafluor is a substrate for the multidrug resistance-related protein MRP1. Int J Cancer 124:233–238. doi: 10.1002/ijc.23892. [DOI] [PubMed] [Google Scholar]
- 39.Banfield DK. 2011. Mechanisms of protein retention in the Golgi. Cold Spring Harb Perspect Biol 3:a005264. doi: 10.1101/cshperspect.a005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao C, Cai Y, Wang Y, Kang B-H, Aniento F, Robinson DG, Jiang L. 2014. Retention mechanisms for ER and Golgi membrane proteins. Trends Plant Sci 19:508–515. doi: 10.1016/j.tplants.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 41.Progida C, Bakke O. 2016. Bidirectional traffic between the Golgi and the endosomes – machineries and regulation. J Cell Sci 129:3971–3982. doi: 10.1242/jcs.185702. [DOI] [PubMed] [Google Scholar]
- 42.McNally KE, Cullen PJ. 2018. Endosomal retrieval of cargo: retromer is not alone. Trends Cell Biol 28:807–822. doi: 10.1016/j.tcb.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 43.Burd C, Cullen PJ. 2014. Retromer: a master conductor of endosome sorting. Cold Spring Harb Perspect Biol 6:a016774. doi: 10.1101/cshperspect.a016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lucas M, Gershlick DC, Vidaurrazaga A, Rojas AL, Bonifacino JS, Hierro A. 2016. Structural mechanism for cargo recognition by the retromer complex. Cell 167:1623–1635.e14. doi: 10.1016/j.cell.2016.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gallon M, Clairfeuille T, Steinberg F, Mas C, Ghai R, Sessions RB, Teasdale RD, Collins BM, Cullen PJ. 2014. A unique PDZ domain and arrestin-like fold interaction reveals mechanistic details of endocytic recycling by SNX27-retromer. Proc Natl Acad Sci U S A 111:E3604-13. doi: 10.1073/pnas.1410552111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McNally KE, Faulkner R, Steinberg F, Gallon M, Ghai R, Pim D, Langton P, Pearson N, Danson CM, Nägele H, Morris LL, Singla A, Overlee BL, Heesom KJ, Sessions R, Banks L, Collins BM, Berger I, Billadeau DD, Burstein E, Cullen PJ. 2017. Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling. Nat Cell Biol 19:1214–1225. doi: 10.1038/ncb3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puthenveedu MA, Lauffer B, Temkin P, Vistein R, Carlton P, Thorn K, Taunton J, Weiner OD, Parton RG, von Zastrow M. 2010. Sequence-dependent sorting of recycling proteins by actin-stabilized endosomal microdomains. Cell 143:761–773. doi: 10.1016/j.cell.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen W, Lin K, Zhang L, Guo G, Sun X, Chen J, Ye L, Ye S, Mao C, Xu J, Zhang L, Jiang L, Shen X, Xue X. 2015. The cytomegalovirus protein UL138 induces apoptosis of gastric cancer cells by binding to heat shock protein 70. Oncotarget 7:5630–5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hurley JH, Emr SD. 2006. The Escrt complexes: structure and mechanism of a membrane-trafficking network. Annu Rev Biophys Biomol Struct 35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katzmann DJ, Babst M, Emr SD. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145–155. doi: 10.1016/S0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 51.Boehm M, Bonifacino JS. 2002. Genetic analyses of adaptin function from yeast to mammals. Gene 286:175–186. doi: 10.1016/S0378-1119(02)00422-5. [DOI] [PubMed] [Google Scholar]
- 52.Höning S, Sandoval IV, von Figura K. 1998. A di-leucine-based motif in the cytoplasmic tail of LIMP-II and tyrosinase mediates selective binding of AP-3. EMBO J 17:1304–1314. doi: 10.1093/emboj/17.5.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bresnahan PA, Yonemoto W, Ferrell S, Williams-Herman D, Geleziunas R, Greene WC. 1998. A dileucine motif in HIV-1 Nef acts as an internalization signal for CD4 downregulation and binds the AP-1 clathrin adaptor. Curr Biol 8:1235–1238. doi: 10.1016/S0960-9822(07)00517-9. [DOI] [PubMed] [Google Scholar]
- 54.Montag C, Wagner JA, Gruska I, Vetter B, Wiebusch L, Hagemeier C. 2011. The latency-associated UL138 gene product of human cytomegalovirus sensitizes cells to tumor necrosis factor alpha (TNF-alpha) signaling by upregulating TNF-alpha receptor 1 cell surface expression. J Virol 85:11409–11421. doi: 10.1128/JVI.05028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe N, Kuriyama H, Sone H, Neda H, Yamauchi N, Maeda M, Niitsu Y. 1988. Continuous internalization of tumor necrosis factor receptors in a human myosarcoma cell line. J Biol Chem 263:10262–10266. [PubMed] [Google Scholar]
- 56.Bradley JR, Thiru S, Pober JS. 1995. Disparate localization of 55-kd and 75-kd tumor necrosis factor receptors in human endothelial cells. Am J Pathol 146:27–32. [PMC free article] [PubMed] [Google Scholar]
- 57.Jones SJ, Ledgerwood EC, Prins JB, Galbraith J, Johnson DR, Pober JS, Bradley JR. 1999. TNF recruits TRADD to the plasma membrane but not the trans-Golgi network, the principal subcellular location of TNF-R1. J Immunol 162:1042–1048. [PubMed] [Google Scholar]
- 58.Storey H, Stewart A, Vandenabeele P, Luzio JP. 2002. The p55 tumour necrosis factor receptor TNFR1 contains a trans-Golgi network localization signal in the C-terminal region of its cytoplasmic tail. Biochem J 366:15–22. doi: 10.1042/bj20020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeltzer S, Zeltzer CA, Igarashi S, Wilson J, Donaldson JG, Goodrum F. 2018. Virus control of trafficking from sorting endosomes. mBio 9:e00683-18. doi: 10.1128/mBio.00683-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nedjai B, Hitman GA, Yousaf N, Chernajovsky Y, Stjernberg-Salmela S, Pettersson T, Ranki A, Hawkins PN, Arkwright PD, McDermott MF, Turner MD. 2008. Abnormal tumor necrosis factor receptor I cell surface expression and NF-κB activation in tumor necrosis factor receptor-associated periodic syndrome. Arthritis Rheum 58:273–283. doi: 10.1002/art.23123. [DOI] [PubMed] [Google Scholar]
- 61.Raiborg C, Stenmark H. 26 March 2009. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 62.Christ L, Raiborg C, Wenzel EM, Campsteijn C, Stenmark H. 2017. Cellular functions and molecular mechanisms of the ESCRT membrane-scission machinery. Trends Biochem Sci 42:42–56. doi: 10.1016/j.tibs.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 63.VanDeusen HR, Kalejta RF. 2015. The retinoblastoma tumor suppressor promotes efficient human cytomegalovirus lytic replication. J Virol 89:5012–5021. doi: 10.1128/JVI.00175-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee SH, Albright ER, Lee J-H, Jacobs D, Kalejta RF. 2015. Cellular defense against latent colonization foiled by human cytomegalovirus UL138 protein. Sci Adv 1:e1501164. doi: 10.1126/sciadv.1501164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol Biol 634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 66.Zhu H, Shen Y, Shenk T. 1995. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol 69:7960–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]