Influenza virus defective interfering particles (DIPs) that harbor internal deletions within their genomes occur naturally during infection in humans and during cell culture. They have been hypothesized to influence the pathogenicity of the virus; however, their specific function remains elusive. The accurate detection of DIP-associated deletion junctions is crucial for understanding DIP biology but is complicated by an array of technical issues that can bias or confound results. Here, we demonstrate a combined experimental and computational framework for detecting DIP-associated deletion junctions using next-generation sequencing (NGS). We detail how to validate pipeline performance and provide the bioinformatics pipeline for groups interested in using it. Using this optimized pipeline, we detect hundreds of distinct deletion junctions generated during infection with a diverse panel of influenza viruses and use these data to test a long-standing hypothesis concerning the molecular details of DIP formation.
KEYWORDS: defective interfering particles, influenza, next-generation sequencing
ABSTRACT
The mechanisms and consequences of defective interfering particle (DIP) formation during influenza virus infection remain poorly understood. The development of next-generation sequencing (NGS) technologies has made it possible to identify large numbers of DIP-associated sequences, providing a powerful tool to better understand their biological relevance. However, NGS approaches pose numerous technical challenges, including the precise identification and mapping of deletion junctions in the presence of frequent mutation and base-calling errors, and the potential for numerous experimental and computational artifacts. Here, we detail an Illumina-based sequencing framework and bioinformatics pipeline capable of generating highly accurate and reproducible profiles of DIP-associated junction sequences. We use a combination of simulated and experimental control data sets to optimize pipeline performance and demonstrate the absence of significant artifacts. Finally, we use this optimized pipeline to reveal how the patterns of DIP-associated junction formation differ between different strains and subtypes of influenza A and B viruses and to demonstrate how these data can provide insight into mechanisms of DIP formation. Overall, this work provides a detailed roadmap for high-resolution profiling and analysis of DIP-associated sequences within influenza virus populations.
IMPORTANCE Influenza virus defective interfering particles (DIPs) that harbor internal deletions within their genomes occur naturally during infection in humans and during cell culture. They have been hypothesized to influence the pathogenicity of the virus; however, their specific function remains elusive. The accurate detection of DIP-associated deletion junctions is crucial for understanding DIP biology but is complicated by an array of technical issues that can bias or confound results. Here, we demonstrate a combined experimental and computational framework for detecting DIP-associated deletion junctions using next-generation sequencing (NGS). We detail how to validate pipeline performance and provide the bioinformatics pipeline for groups interested in using it. Using this optimized pipeline, we detect hundreds of distinct deletion junctions generated during infection with a diverse panel of influenza viruses and use these data to test a long-standing hypothesis concerning the molecular details of DIP formation.
INTRODUCTION
Influenza virus defective interfering particles (DIPs) were first described over 60 years ago and are classically defined by their ability to interfere with the production of wild-type virus (1, 2). This ability has been linked to the ability of defective interfering (DI) RNAs to both outcompete wild-type (WT) genomic RNAs for resources and packaging into virions and to more potently stimulate the induction of antiviral immunity through cytosolic RNA sensors (3–6). DIPs have also been implicated in influencing the outcome of influenza virus infection in humans (7). The specific mechanisms and broader functional consequences of DIP formation during influenza infection remain poorly understood.
Influenza DIPs are characterized by the presence of large internal deletions in one or more genome segments that disrupt essential open reading frames while retaining the sequences required for replication and packaging (5). As such, the mapping of DIP-associated deletions has helped to define the minimum sequences required for genome replication and packaging (8, 9). These deletions are believed to result from a poorly defined process by which the viral RNA-dependent RNA polymerase (RdRp) ceases RNA polymerization at one site of the viral RNA template (donor site), only to resume at another site downstream (acceptor site), resulting in a failure to copy an internal stretch of the WT template (10). Until recently, the ability to characterize these DIP-associated deletion junction sites (breakpoints) has been limited based on the need to clone and Sanger sequence individual DIP-associated RNAs. As a result, the number of individual DIP-associated RNA sequences that have been analyzed has been relatively small, hindering efforts to define the factors that govern DIP deletion formation.
The advent of next-generation sequencing (NGS) has increased the number of individual recombinant sequences that can be identified within a given sample by orders of magnitude. However, the identification and analysis of DIP-associated RNAs by NGS poses new challenges, including the successful alignment of junction-containing (or junction-spanning) reads to the viral reference sequence, the precise definition and localization of DIP-associated deletion breakpoints, and the differentiation of true DIP deletion sequences from the artifactual recombinants that can form during reverse transcription, PCR, and/or sequencing. Without careful optimization and validation, these issues can easily compromise efforts to define the genetic profile of DIP populations.
Here, we describe the development and validation of an Illumina-based sequencing framework for the identification and analysis of influenza virus DIP-associated deletion junctions. The bioinformatics pipeline combines the Bowtie 2 alignment algorithm with the Viral Recombination Mapper (ViReMa) algorithm developed by Andrew Routh and a collection of additional scripts for data processing and analysis (11, 12).
We used simulated NGS data sets and a panel of experimental control samples to optimize and quantify the sensitivity, precision, and reproducibility of our pipeline. Subsequently, we used the optimized pipeline to fine-tune the experimental protocol from sample preparation to sequencing to better detect and map DIP-associated deletions generated during experimental infection with a panel of influenza A (IAV) and influenza B viruses (IBV). This work highlights the computational and experimental controls needed for Illumina-based NGS studies of viral recombination and provides an optimized, user-friendly sequencing and bioinformatics pipeline for the identification and analysis of DIP-associated sequences. Higher-resolution analysis of these deletion sequences can shed light on the specific molecular mechanisms of DIP formation, as well as on how DIPs may affect the overall behavior of viral populations.
(This article was submitted to an online preprint archive [13].)
RESULTS
Overview of the pipeline.
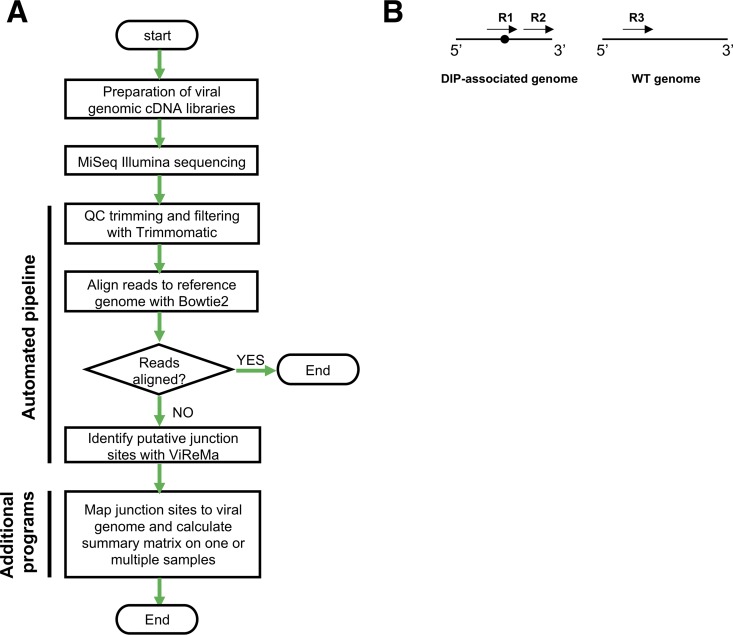
The sequencing framework we describe here encompasses sample preparation, sequencing, and data analysis (Fig. 1A). In brief, we generate 8-segment, full-length amplicons from viral samples and sequence them using the Illumina MiSeq platform. Datasets are quality filtered and aligned to the viral reference genome using Bowtie 2 in a conservative manner that disallows soft clipping. Thus, reads containing deletion junctions fail to align and are fed into the ViReMa algorithm to detect DIP-associated deletion junctions. To exclude short indels, we ignored all deletions of 20 nucleotides (nt) or shorter. Finally, the identified junctions are mapped to the viral genome and output as a matrix containing the segment name, junction start and end sites, and NGS read support that can easily be analyzed using additional software tools. Below, we outline the approaches we have taken to optimize and validate the various steps in the process.
FIG 1.
Overview of sequencing/bioinformatics framework. (A) Flowchart outlining the steps of the sequencing pipeline. QC, quality control. (B) Simple depiction of the possible types of NGS reads in relation to a deletion junction within a sample. Arrows represent individual NGS reads, and the black circle denotes the location of a deletion junction. R1, reads derived from a deletion-containing sequence that span the deletion junction; R2, reads derived from a deletion-containing sequence that do not span the deletion junction; R3, reads derived from sequences that do not contain deletions.
Optimization of analysis pipeline using simulated data.
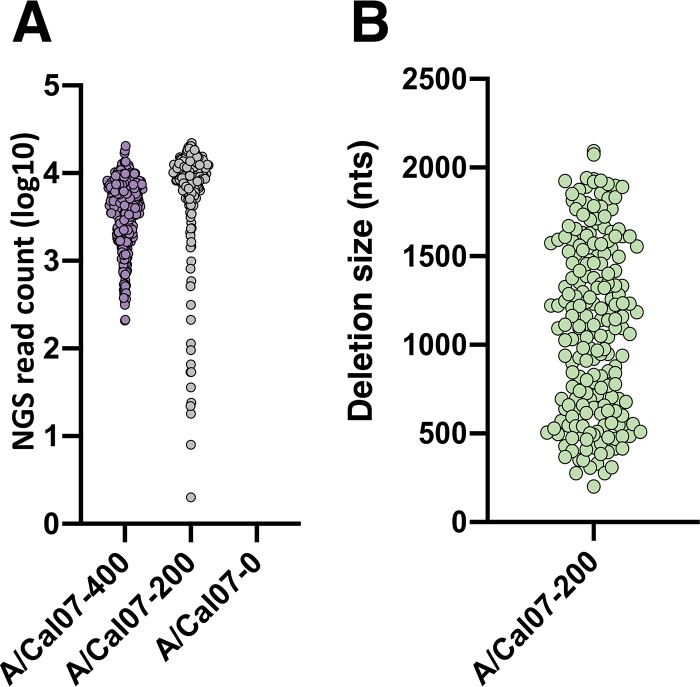
All bioinformatics pipelines have the potential to introduce artifacts and biases during data analysis. Therefore, we first aimed to optimize the sensitivity and precision of our bioinformatics pipeline using simulated NGS data sets where we absolutely know the identities and frequencies of all DIP-associated deletion sequences present. IAV DIP-associated deletions can be found in nearly all (if not all) genome segments at a wide range of frequencies (10, 14). To mimic this natural variation, we used MetaSim to generate a panel of Illumina MiSeq-based NGS simulated data sets that contain DIP-associated deletions in all genome segments at various frequencies, locations, and deletion sizes (see Table 1, Fig. 2). We used a simple Perl script to randomly generate deletion junctions within the terminal ∼600 nt of A/California/07/09 (A/Cal07), since these regions have been shown to be hotspots for DIP-associated deletions (9, 10, 15). We also generated a negative-control data set that lacks deletions to quantify the occurrence of false positives generated by the pipeline. Critically, we introduced a nucleotide substitution frequency of ∼1% into these data sets, based on the published Illumina MiSeq empirical error model (16, 17). Each data set comprised ∼1million 2 × 250-nt paired-end reads, mirroring the read depth that we expect per sample on a typical sequencing run.
TABLE 1.
Description of the simulated data sets used in this study
| Data set namea | Total junction count (n) | Total no. of junction NGS reads | Total no. of WT NGS individual reads | Junction count (n) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PB2 | PB1 | PA | HA | NP | NA | M | NS | ||||
| A/Cal07-400 | 400 | 1,838,898 | 116,548 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 |
| A/Cal07-200 | 200 | 177,4920 | 225,080 | 25 | 25 | 25 | 25 | 25 | 25 | 25 | 25 |
| A/Cal07-0 | 0 | 0 | 2,000,000 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
The total number of 2 × 250-bp paired-end reads for each data set was ∼1 million, with a total of 2 million individual reads.
FIG 2.
Simulated data set features. (A) Read support numbers for the individual deletion junctions in the indicated data sets. (B) The deletion sizes of all junctions in the A/Cal07-200 data set. In A and B, each dot represents a unique junction.
Optimization of alignment.
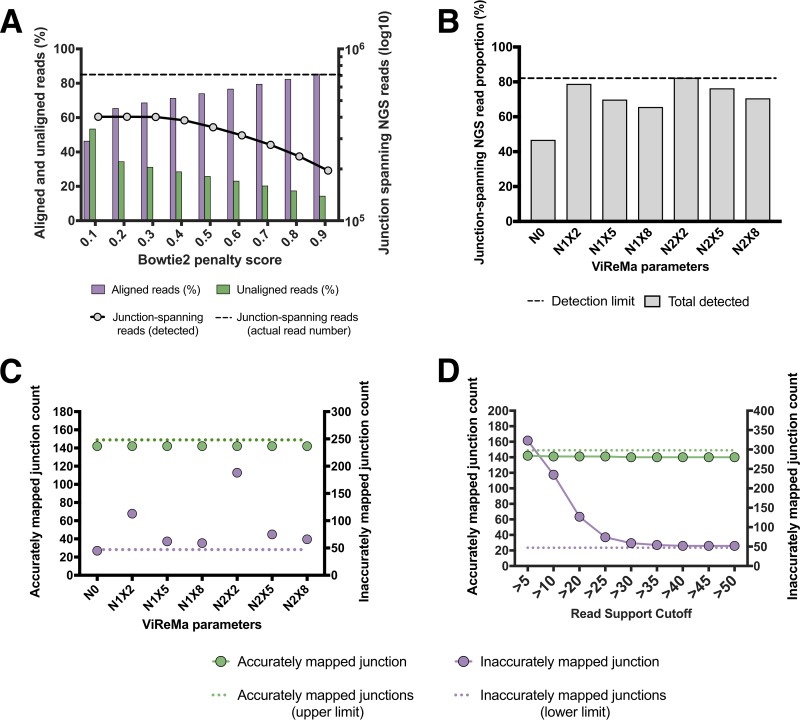
We first optimized the filtering of reads that contain deletion junctions (Fig. 1B, R1) from those that do not include junctions (Fig. 1B, R2 and R3). To do this, we aligned all reads to the WT reference genome using Bowtie 2. Reads that successfully align should not contain deletion junctions and are saved for further analysis, while reads that fail to align are fed into the ViReMa algorithm. The performance of this alignment step is highly dependent upon the mismatch penalty score that is used during alignment. The penalty score setting determines the number of mismatches and/or gaps that are tolerated within an alignment. If penalties are too stringent, reads with random mutations or base-calling errors will fail to align and be sent to ViReMa, increasing both the chances of false positives and the total computational time per sample; if they are too lenient, true junction-spanning reads will successfully align and be excluded from downstream analysis.
We used a junction-rich simulated data set (A/Cal07-400) to test the effects of various alignment penalty scores on the output of ViReMa (Fig. 3A). We observed that a penalty score of 0.3 minimized the number of unaligned reads (and thus the potential for false positives) without diminishing the number of junction-spanning reads detected. This value was used for all subsequent analysis.
FIG 3.
Optimization of bioinformatics pipeline using simulated data. (A) Quantification of the effects of various Bowtie 2 penalty scores on the number of junction spanning reads detected by ViReMa in the A/Cal07-400 simulated data set (dashed line represents the actual number of junction-spanning reads present in the data set). The percentages of reads that aligned to reference genome (purple) and failed to align (green) are also shown for each penalty score. (B) The effects of ViReMa –X and –N parameters on the percentage of junction-spanning reads present in the simulated data set that were successfully detected. The dashed line shows the maximum theoretical sensitivity (∼81.5%) based on the ViReMa seed length of 25 nt. (C) The effects of ViReMa –X and –N parameters on the number of accurately (green) and inaccurately mapped (purple) deletion junctions reported by the pipeline using the A/Cal07-200 simulated data set. The maximum possible number of accurate junctions and the minimum number of inaccurate junctions (resulting from junctions adjacent to direct repeat sequences) are shown for comparison. (D) Effects of various minimum read support cutoffs (RSCs) on junction detection. Analysis performed on the A/Cal07-200 simulated data set using N1X8 ViReMa values.
Optimization of ViReMa operation.
We next optimized the sensitivity and precision with which the pipeline detects deletion junctions. The ability of ViReMa to accurately map true junction-containing reads is affected by three factors. The first is the method the algorithm uses to identify breakpoints. ViReMa extracts and aligns a seed sequence of 20 to 30 nt (the default value of 25 was used in this study) from the beginning of each read and begins aligning the downstream nucleotides. If at any point the downstream alignment fails (as would be the case for a deletion breakpoint), ViReMa generates a new seed sequence starting from that location for realignment. Thus, breakpoints cannot be detected if they occur within the terminal 25 nt of a read.
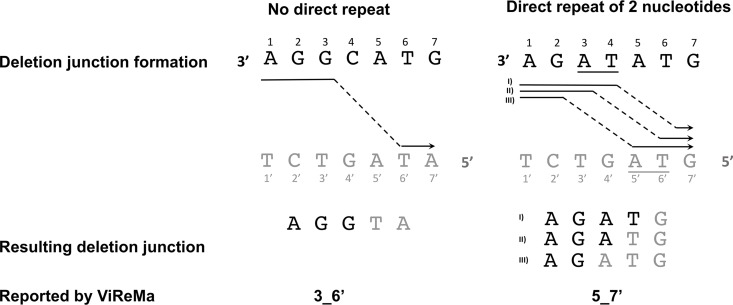
The second factor is the presence of short direct repeats adjacent to the junction site. These repeats result in a situation where multiple potential breakpoints can give rise to the same final sequence, making precise definition of the true breakpoints impossible (Fig. 4). ViReMa deals with these “fuzzy” regions through the parameter “Defuzz,” which can be set to report the junction either to the 5′ end, 3′ end or to the middle of the ambiguous region. For the sake of consistency, we pushed all fuzzy junctions toward the 3′ end of the ambiguous region (not the 3′ end of the entire read). The effects of direct repeats on breakpoint mapping are impossible to avoid and vary somewhat between influenza genome segments. Importantly, while this effect reduces the precision of breakpoint mapping, it does not affect the ability of the pipeline to determine the actual sequences of DIP-associated RNAs.
FIG 4.
Illustration of a “fuzzy” junction caused by an adjacent direct repeat. The black letters represent the sequence where the polymerase detaches (donor site), and the gray letters represent the sequence where polymerase reinitiates (acceptor site). The arrows represent the actual path of the polymerase, and the underlined letters denote the direct repeat sequence. The resulting DIP-associated sequences are represented by the black and gray letters to highlight the donor and acceptor sites, respectively. The junction sites reported by ViReMa are at the bottom and use the following nomenclature: DonorSite_AcceptorSite. Note that ViReMa was adjusted to push the junction site toward the 3′ site of the direct repeat. The left side illustrates a situation where no direct repeats are present and, as a result, ViReMa reports the correct junction. On the right, a situation where a direct repeat of 2 nucleotides is present, resulting in ambiguity. In this case, ViReMa pushes the junction toward the 3′ end of the repeat and incorrectly reports the junction as 5 to 7′ (5_7′). Shown are three possible paths for the polymerase that yield different junction locations with the same sequence. In all cases, the junction will be reported as 5_7′.
The third factor is the potential for base-calling errors or mutations to result in erroneous junction mapping. Even though reported junctions in this category are derived from real junctions, they can be viewed as false positives in that they are reported as distinct junctions that do not actually exist in the viral population.
Altogether, these three factors set a ceiling on the maximum number of deletion junctions that can be accurately detected and mapped. Using our simulated data sets, we knew how many deletion junctions actually existed, exactly where they were located, and whether or not they were adjacent to direct repeats (see Materials and Methods) that could result in incorrect mapping. This allowed us to systematically optimize the sensitivity and precision of the software pipeline.
We tested how various ViReMa operating parameters affected both junction-spanning read detection and actual junction reporting. We used the A/Cal07-200 data set to challenge ViReMa across a range of –N parameter (number of mismatches allowed) and –X parameter (mismatch distance from the putative junction location) values. We first asked how various –N and –X parameters influenced the total number of junction-spanning reads detected (Fig. 3B). We found that using N = 0 (–X is irrelevant at this condition) significantly decreased the number of junction-spanning reads detected compared with nonzero –N and –X values. We next asked how increasing the –N and –X values affected the number of accurately and inaccurately mapped junctions reported (Fig. 3C). We observed a clear correlation between the –X parameter and junction-mapping precision, as increasing the –X value decreased the number of inaccurately mapped junctions. Overall, we found that using N = 1 and X = 8 reduced inaccurate junction mapping to the minimum amount possible, given the occurrence of direct repeats adjacent to 23.5% (47 of 200) of junctions in the data set.
We next asked whether setting a minimum read support cutoff (RSC) to report a junction affected the numbers of both accurate and inaccurate junctions that the pipeline identified. Requiring that a given junction be represented within a minimum number of reads can decrease the number of erroneously mapped junctions arising from base-calling errors but could also result in some true junctions being lost due to insufficient read coverage. We aligned our simulated A/Cal07-200 data set with Bowtie 2 and used the resulting unaligned reads to challenge ViReMa using different RSC values (Fig. 3D). We found that the number of true junctions reported by the pipeline was very close to the theoretical maximum, with minimal dropoff across the range of RSCs tested. In contrast, we observed that the number of inaccurately reported junctions was highly sensitive to the RSC value used. An RSC of >30 was needed to lower the number of inaccurately reported junctions to the minimal limit (determined by the number of “fuzzy” junctions with adjacent direct repeats in the data set). Together, these data highlight the importance of optimizing RSC values and the ViReMa –N and –X parameters for maximizing the sensitivity of junction detection while minimizing the number of false positives. We set our default values at an RSC of >30, an –N of 1, and an –X of 8 for the subsequent analysis.
Validation of sequencing pipeline.
After optimizing the bioinformatics component of our pipeline using simulated data sets, we examined the ability of the pipeline to detect DIP-associated deletions within complex viral populations from experimental samples. Our overall strategy was based on the universal, 8-segment reverse transcription-PCR (RT-PCR) approach pioneered by Zhou et al. (18). Critically, there are a number of steps among the library preparation and sequencing steps that have the potential to introduce artifacts that can compromise junction detection and analysis. In particular, we were concerned about the potential for recombination during reverse transcription, PCR, and/or sequencing to generate junctions that will be called by the pipeline (19, 20). To address this, we prepared several control sample libraries, sequenced them on the MiSeq platform, and ran the results through our optimized pipeline.
To quantify generation of false positives during the PCR and/or sequencing steps, we constructed libraries without using actual viral RNA or reverse transcriptase. To do this, we generated an equimolar mixture of full-length PCR amplicons from each of the eight IAV genome segments, using reverse genetics plasmids encoding the gene segments from A/Puerto Rico/8/1934 (A/PR8) as the templates. These amplicons were gel purified to ensure correct, full-length size, and then used as the templates for the universal amplification PCR and subsequent library preparation. Our analysis pipeline detected no deletion junctions in this control, indicating that none of the steps in our pipeline from PCR onwards were significant sources of false-positive signals.
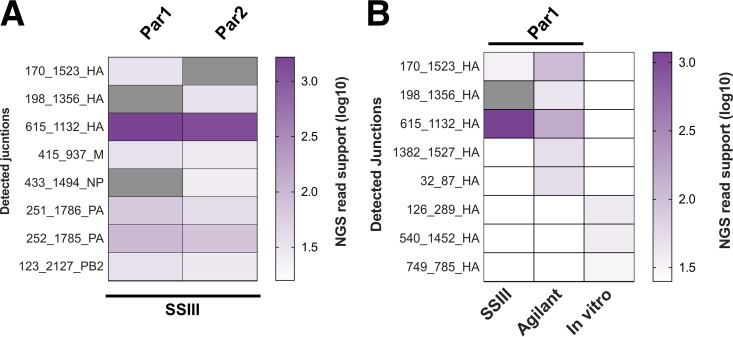
We next sequenced a recombinant A/Cal07 stock that was grown under low-multiplicity-of-infection (MOI) conditions to minimize the frequency of DIPs (21). We performed two independent RNA extractions and reverse transcription reactions on this stock to serve as technical replicates (named Par1 and Par2). ViReMa detected 6 and 7 DIP-associated deletion junctions from Par1 and Par2, respectively, with junction-spanning reads representing ∼0.1 to 0.2% of the total reads (Fig. 5A). The majority of these reads were derived from a single shared deletion junction in hemagglutinin (HA) (615_1132_HA, where the nomenclature is [5′ junction position]_[3′ junction position]_[gene segment]). On the other hand, 4 other DIP-associated junctions were shared between replicates, each with low NGS read depth (ranging between 19 and 94). Two unshared junctions in Par1 and one in Par2 were actually reported in both replicates but failed to reach the level of detection in one replicate.
FIG 5.
Reverse transcription is not a significant source of deletions. We performed two independent RNA extractions, RT reactions, PCR amplifications, and library preparations from a single recombinant A/Cal07 working stock grown at a low MOI (Par1 and Par2). Gray blocks represent deletions with nonzero read support that fell below our read support cutoff (RSC), and white blocks denote deletions that were not detected. (A) Comparison of deletion junctions detected in Par1 and Par2 samples. (B) Comparison of HA segment junctions detected in two libraries generated from independent RT reactions using two different RT enzymes, along with a library generated from in vitro T7-transcribed viral RNA.
The significant overlap between the specific junctions that were reported from the 2 replicates suggested that these junctions were produced by the viral polymerase (and were thus bona fide DIP-associated sequences) rather than by the reverse transcriptase. However, the generation of the same junction in independent reverse transcriptase (RT) reactions could also indicate the existence of strong hotspots for RT recombination. To more directly address the potential contribution of RT-derived recombinants, we performed two independent experiments. First, we compared the junctions detected in HA segment libraries generated from Par 1 using two different RT enzymes, Invitrogen Superscript III and Agilent AccuScript. Second, we performed in vitro transcription of a plasmid-derived A/Cal07 HA segment using T7 RNA polymerase, which then was used as a template for RT-PCR to produce the amplicon library for sequencing. The IAV polymerase was not involved in this control; thus, any deletions detected would have been generated by T7 polymerase or the RT enzyme.
The junctions reported from libraries generated by the two RT enzymes had significant overlap and were both dominated by 615_1132_HA (Fig. 5B). In contrast, we detected none of the Par1-derived junctions in the library generated from T7-transcribed HA (Fig. 5B). Although the read depth coverage was comparable to Par1, 615_1132_HA was completely absent, and the 3 junctions that were detected had minimal read support and were not seen in virus-derived libraries. Together, these results suggest that the formation of deletion junctions during the reverse transcription reaction is rare and that the Par1-derived junctions we observed are most likely derived from true DIPs present within our viral stock, despite the stock having been prepared at a low MOI. This highlights the difficulty in producing a completely DIP-free virus preparation.
Generation of DIP-enriched populations through high-MOI passage.
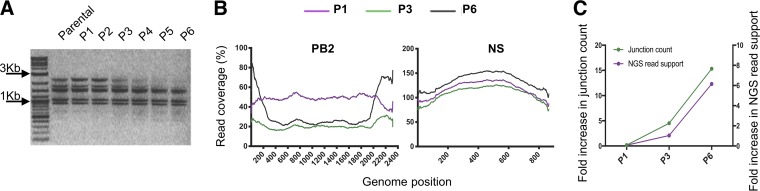
To test the ability of the pipeline to detect real DIP-associated RNAs, we enriched for DIPs through serial undiluted passage of A/Cal07 in Madin-Darby canine kidney (MDCK) cells. We confirmed the presence of DIPs by amplifying full-length genomic cDNA at each passage and examining the size distribution of PCR products by gel electrophoresis (Fig. 6A), as previously described (21). The gradual disappearance of the polymerase segments, which are thought to form the majority of DIPs, and the appearance of a smear below the shortest IAV segment (NS, ∼0.9 kb) were consistent with the accumulation of DIPs over successive passages. Based on these results, we picked passage 1 (P1), P3, and P6 as representative samples for sequencing.
FIG 6.
Generation of DIP-rich populations through high-MOI passage. A/Cal07 was serially passaged 6 times (P1 to P6) in MDCK cells at a sustained high MOI. (A) PCR products from the indicated A/Cal07 populations following 8-segment whole-genome amplification, visualized on a 1% agarose gel. The accumulation of deletion junctions is reflected by the disappearance of the polymerase segments (∼2.3 kb) and the appearance of a smear below the NS segment (∼0.9 kb) ranging from ∼0.3 to 0.8 kb. (B) Coverage depth of aligned reads from the indicated passages for PB2 and NS genome segments. The coverage was normalized against the read coverage of the parental sample (Par1). (C) Fold increase in the number of deletion junctions (left y axis) and total read support for those junctions (right y axis) over the parental sample (Par1).
We further confirmed the presence of DIPs by plotting the read coverage of the aligned reads from passages 1, 3, and 6 (Fig. 6B). These coverage plots clearly reveal the characteristic pattern of DIP-rich populations, with a much lower depth of read alignment in the middle portion of the segment than those in the termini. As expected, the number of DIP-associated deletion junctions detected by the pipeline also increased across passages, reaching the highest level at passage 6 (Fig. 6C). To confirm that these junction-containing sequences were derived from virion rather than cellular RNA, we measured the number of reads that aligned to the host (canine) genome in our samples. We found very few reads derived from the canine genome in all the passages, compared with about 40% of the reads from RNA extracted from infected cells (data not shown).
Optimization of minimum read support cutoffs.
Our experiments using simulated data sets revealed the importance of setting minimal RSCs for maximizing the accuracy of pipeline performance and suggested that the optimal RSC may differ between data sets. We next attempted to optimize RSC values for our experimental data set, for which we did not actually know the precise location and number of junctions present in the population (as we did with our simulated data sets). To quantify precision in junction detection for our experimental data set, we assumed that base-calling errors and mutations that result in inaccurate junction reporting would be stochastic and thus read support for these inaccurate junctions would be highly variable between technical replicates. In contrast, read support for real junctions should be consistent between replicates.
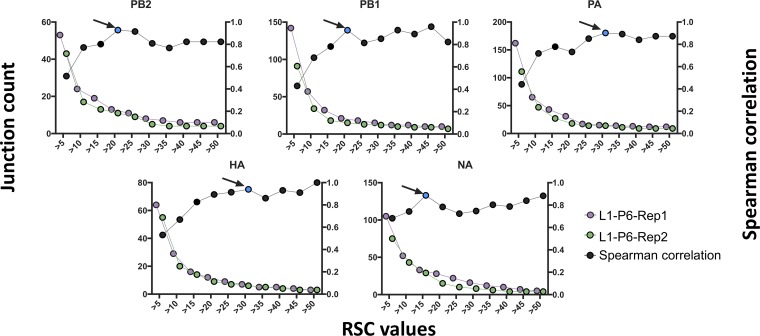
We assessed the effects of various RSC values on the degree of correlation between junctions identified in two technical replicates generated from the same passage 6 population (hereinafter, L1-P6-Rep1 and L1-P6-Rep2, where L refers to lineage, P to passage number, and Rep to technical replicate). We varied the RSC values from 1 to 50 for each individual genome segment and examined the effect on the number of reported junctions (Fig. 7). We observed a similar pattern to that observed for our simulated data, where raising the RSC to 10 or higher resulted in a large dropoff in the number of reported junctions. We next determined the RSC value that yielded the highest degree of correlation between the two replicates. We identified the following distinct optimal RSC cutoff values for each segment: 20, 20, 30, 30, and 15 for PB2, PB1, PA, HA, and neuraminidase (NA), respectively. The average of these values was used as an RSC for the remaining segments, for which not enough junctions were detected to perform the correlation test (see below).
FIG 7.
Determination of optimal read support cutoffs for experimental data. Plots showing the numbers of deletion junctions (left y axis) reported in the indicated genome segments for two technical replicates (L1-P6-Rep1 and L1-P6-Rep2) across various RSC values. Black dots represent the results of Spearman correlation tests between the replicates at each RSC condition (right y axis). Blue dots indicate the point with the highest degree of correlation and minimum decrease in junction count for each genome segment.
We do not expect these values to be universal, as they are likely influenced by a number of factors that will vary between individual sequencing runs. Also, for different applications, it may be beneficial to lower the RSC to improve detection sensitivity at the cost of precision. Thus, we suggest running two technical replicates with each NGS run to establish optimal per-segment RSC values for that run. It is also worth mentioning that we removed reads with identical sequences (duplicates) using the “dedup” option in ViReMa because there is no possible way to define the source of duplication (e.g., PCR duplicates versus identical reads that occur by chance), and we wanted to minimize the influence of PCR biasing. The presence or absence of PCR duplicates will likely change the optimal RSC values. Finally, read depth was highly consistent between samples within our individual sequencing runs. In situations where this is not the case, RSCs may need to be normalized to per-sample sequencing depth.
Effect of various template inputs on pipeline performance.
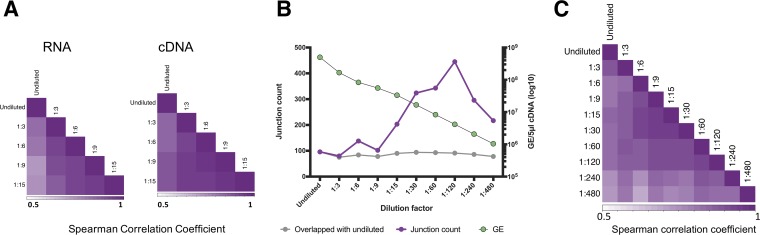
We next asked whether the amount of cDNA template that goes into the library preparation affects the sensitivity and/or stochasticity of junction detection by the pipeline. We serially diluted both the amount of viral RNA template used in the RT reaction and the amount of cDNA template used in the PCR and compared pipeline outputs from the DIP-rich L1-P6-Rep1 population. We first tested the correlation between detected DIP-associated junctions at a limited number of dilutions ranging from 1:3 to 1:15. We observed that the correlation of read support values between specific junctions across dilutions was more consistent when cDNA was diluted, rather than RNA, suggesting that RNA dilution may increase the stochasticity of downstream PCR amplification (Fig. 8A).
FIG 8.
Effects of viral template input on the detection of DIP-associated junctions. We serially diluted RNA or cDNA generated from the L1-P6-Rep1 sample and compared sequencing results between libraries generated with these dilutions as the templates. (A) Serial dilutions (1:3 to 1:15) was carried out on either the RNA or cDNA of the L1-P6-Rep1 sample, and a correlation test between the detected DIP-associated junctions was performed. (B) For each dilution, the total number of detected junctions (purple) is shown, along with the number of specific junctions that were also detected in the undiluted sample (gray). The copy number of viral cDNA molecules included in downstream PCR and library preparation for each dilution was determined by RT-qPCR (green; right y axis). GE, gene equivalent. (C) Read support values for all deletion junctions common across the diluted (at the cDNA level) and undiluted samples were normalized to the total number of deletion junction-spanning reads for each sample and used to perform a Spearman correlation between all pairs of samples using the R cor function.
Based on this, we performed whole-genome PCR using a dilution series of L1-P6-Rep1-derived cDNAs (spanning roughly 4 × 108 to 4 × 106 nucleoprotein [NP] genome equivalents per PCR) as the template (Fig. 8B). We observed that there is an optimal amount of input cDNA template for maximizing junction detection. Diluting the input cDNA 1:120 (corresponding to ∼4 × 106 NP genome equivalents) increased the number of detected junctions by more than 4-fold compared with that with undiluted input. Further dilution of input template beyond 1:120 resulted in a decrease in sensitivity. Importantly, dilution across the range tested did not result in a failure to detect any of the junctions reported in the undiluted sample. We also observed that the correlation of read support values between specific junctions across dilutions tracked closely with the sensitivity (Fig. 8C). Together, these observations indicate that optimization of the cDNA template input amount can significantly improve the sensitivity of DIP-associated junction detection.
Reproducibility of pipeline performance.
Multiple steps in the combined experimental/computational pipeline could introduce stochasticity into the pipeline performance, thus diminishing overall consistency and reproducibility of output. To examine the reproducibility of our pipeline’s performance, we sequenced two separate extractions of a single P6 population (L1-P6-Rep1 and L1-P6-Rep2) and compared the pipeline outputs between the two replicates. We found that the normalized read support values of individual junctions were highly correlated between the two replicate samples regardless of whether the replicates were sequenced on the same MiSeq flow cell (Spearman’s R = 0.92) or separate ones (Spearman’s R = 0.91). Thus, the combined steps from RNA extraction to sequence analysis introduce minimal noise into the pipeline output, and pipeline performance is highly reproducible between experiments.
Commonalities and differences in DIP formation between IAV and IBV subtypes and strains.
After optimizing the pipeline, we used it to examine the patterns of DIP formation across diverse strains of IAV and IBV. We serially passaged A/Perth/16/2009 (A/Perth09; seasonal H3N2), A/California/07/2009 (A/Cal07; seasonal H1N1), A/New Caledonia/1999 (A/NC99; seasonal H1N1), and B/Lee/1940 (B/Lee; lab-adapted IBV) at a high MOI in duplicate independent lineages (for the sake of consistency, we performed a new A/Cal07 passage series in parallel with the other viruses rather than using samples from the passage series shown in Fig. 4). For each virus, we assessed the presence of DIPs across passages by 8-segment PCR and gel electrophoresis and sequenced 2 DIP-enriched passages from both passage lineages after normalizing virus input amounts across all samples based on Fig. 8. To empirically determine optimal RSCs (as in Fig. 7), we sequenced two technical replicates of a single population for each virus.
For each virus strain, we detected hundreds of distinct deletion junctions spread across the individual genome segments. When we compared the repertoires of distinct junctions between the two independent passage lineages for each strain, we observed very little overlap, suggesting that most of the junctions we observed were generated de novo during passage and that there is a substantial degree of stochasticity in the deletion generation process (Fig. 9A). The total number of unique junctions decreased between early and late passages for all viruses except B/Lee, consistent with competition between DIPs during passage (data not shown).
FIG 9.
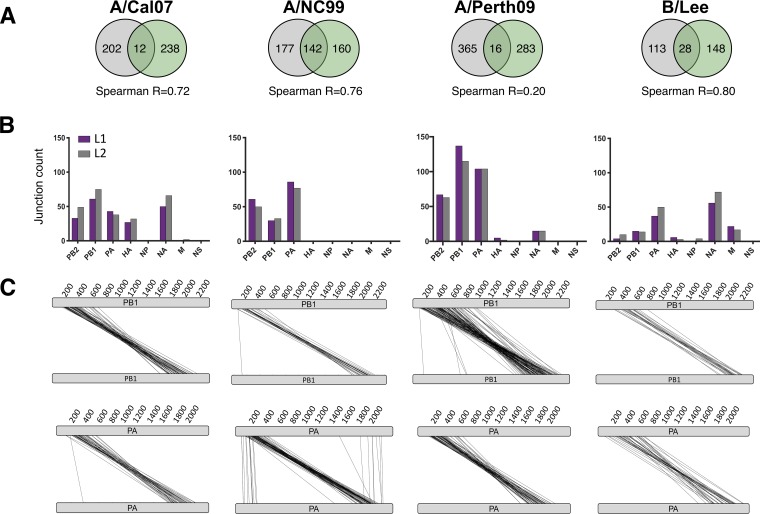
Identification of DIP-associated junctions in different influenza types, subtypes, and strains following high-MOI passage. Each virus was serially passaged in MDCK cells at a high MOI in two independent, parallel lineages (L1 and L2). The earliest passages that showed high accumulation of DIPs (based on gel analysis shown in Fig. S6A) were picked for NGS. (A) Venn diagrams showing the numbers of shared versus unique DIP-associated junctions between the two passage lineages for each virus. (B) The numbers of distinct junctions detected within each genome segment for both passage lineages of the indicated viruses. (C) Parallel coordinate diagrams showing the specific locations of deletion junctions found in the PB1 and PA segments of lineage 1 of the indicated viruses. Each individual junction is represented by a black line that connects the donor and acceptor sites of the breakpoint. Data in panels A to C are all placed below the relevant virus strain labels at the top of the figure.
Despite variation in the specific locations of individual junctions that arose in replicate lineages, the overall distribution of unique junctions across genome segments was highly reproducible between lineages (Fig. 9B). Comparison between strains revealed both common and strain-dependent patterns. For all IAV strains tested, the majority of DIP-associated deletions occurred in the three polymerase genes, mirroring previous studies, while the NP, matrix (M), and NS segments were almost entirely deletion-free. In contrast, the occurrence of deletions within the HA and NA segments varied significantly between strains: A/NC99 had no detectable junctions in these segments, while A/Cal07 had dozens of deletions in both HA and NA. B/Lee differed significantly from the IAV strains in that (i) there were relatively fewer unique junctions in the polymerase segments compared with the NA segment, and (ii) there were numerous deletion junctions detected in the M segment. Together, these results indicate clear virus strain and type differences in patterns of DIP formation within individual genome segments.
While the precise locations of deletion junctions varied significantly, plotting individual junction locations within the genome segments revealed that they were confined within clear hotspots toward the termini of the segments (with a few exceptions) (Fig. 9C). This suggests that while the specific nucleotide locations of deletion breakpoints may be highly stochastic, the overall contours of where they form in the context of the genome segment are highly defined and nonrandom.
Lack of association between direct repeats and junction formation.
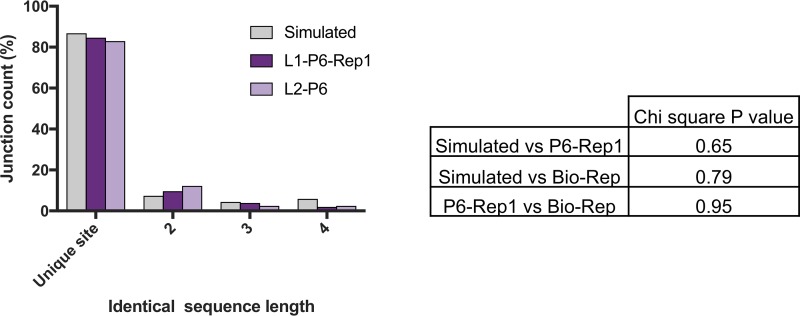
Direct repeat sequences (detailed in Fig. 4) are common across the IAV genome and have previously been hypothesized to contribute to DIP-associated deletion formation by promoting viral polymerase slippage (10, 15). We leveraged the large number of DIP-associated deletion junctions that we identified in this study to test this hypothesis. We asked whether the deletion junctions in the DIP-enriched sample L1-P6-Rep1 were more frequently found adjacent to direct repeats than would be expected if the junctions were located randomly in the viral genome. We compared the frequency of deletion junctions associated with direct repeats between the L1-P6-Rep1 and L2-P6 populations, i.e., biological replicates derived from two independent lineages (where all deletions are formed by the viral RdRp) and the A/Cal07-200 simulated data set, where all deletions are randomly localized (Fig. 10). The frequency of direct repeats of various lengths at junction sites in the real viral populations was not significantly different than that seen in the simulated data, indicating that direct repeat sequences are not enriched at DIP-associated junctions and arguing against a significant role for direct repeats in DIP formation.
FIG 10.
Direct repeat sequences are not overrepresented at DIP-associated deletion junctions. The percentages of deletion junctions within the polymerase segments that occurred at unique sites or at sites with direct nucleotide repeats 2 to 4 nt in length were compared between L1-P6-Rep1, L2-P6 (passage 6 from two independent lineages), and the A/Cal07-200 simulated data set. The number of junctions was plotted and compared by chi square. The table shows the chi-square P values between every possible pair of the samples.
DISCUSSION
Sensitive and accurate detection of DIP-associated sequences within viral populations is critical for defining the causes and consequences of DIP formation during influenza virus infection. Here, we outline a pipeline to detect DIP-associated junctions within viral populations using Illumina-based short read sequencing, validate its performance using a combination of simulated and experimental control data sets, and use this pipeline to reveal variation in patterns of DIP formation across viral strains and types.
Our primary goal was to develop and optimize a reasonably simple and straightforward sequencing framework that accounts for the potential artifacts that can potentially confound NGS-based DIP detection efforts. We chose the Illumina sequencing platform because it is widely available, easy to use, and conducive to sample multiplexing, and because it has a relatively low rate of base-calling errors. One concern that we had initially was that recombination during reverse transcription, PCR, or sequencing might make identification of bona fide DIP-associated junctions a challenge. Two recently developed technologies, CirSeq and ClickSeq, largely eliminate this issue, but they also significantly increase the amount of labor involved in library preparation (22, 23). We observed that the occurrence of nonviral recombination that occurs during our library preparation and sequencing procedures was vanishingly small and can effectively be ignored. Thus, while both CirSeq and ClickSeq are enormously useful in certain circumstances, our data indicate that such methods are not required to generate highly accurate and sensitive profiles of influenza virus DIPs.
We chose ViReMa to identify deletion breakpoints because it is the only published algorithm specifically designed to identify viral recombination junction locations while explicitly accounting for the high error rates inherent in RNA virus replication and NGS. Previously published studies have used different splicing aligners, such as TopHat (7), or alternative packages, like CLC bio Genomics Workbench (24) or MUMmer (15), to identify junctions. Although these methods have generated useful data that has contributed significantly to efforts to dissect DIP biology, each has significant limitations that can bias (due to a preference for splice donor/acceptor sequences) or reduce the sensitivity of junction detection (due to high read support requirements for deletion calling or the need to map specific breakpoints by hand).
A significant shortcoming of the method we detail here is that the measured read support for individual deletion junctions does not necessarily reflect the actual frequencies of these deletions within the viral population. This is due to biasing of PCR amplification toward shorter products, as well as to the uneven distribution of read coverage across the viral genome. For situations where the accurate measurement of individual DIP genotype frequencies is critical, we recommend pairing a cDNA barcoding method, such as primer identification (ID) (25, 26), with a platform capable of long-read sequencing, such as PacBio or Oxford Nanopore (27). Alternatively, direct sequencing of viral RNA using the Oxford Nanopore platform may also prove to be useful for accurate measurement of junction frequencies (28). Note that these long-read sequencing methods are not compatible with components of our pipeline (such as Bowtie 2) and would thus require additional pipeline development.
When we used our pipeline to examine DIP-enriched viral populations generated through serial passage, we detected hundreds of distinct DIP-associated deletion junctions, revealing a high degree of diversity within the DIP population. Although the majority of these DIP-associated junctions were derived from the polymerase segments as expected, we also detected a substantial proportion of deletions within the HA and NA segments, while deletions within the NP, M, and NS segments were rare or nonexistent. This nonrandom distribution of junctions across the genome segments mirrors what has been reported elsewhere and highlights how little we know about the specific molecular mechanisms that regulate DIP formation. The variation in DIP formation patterns that we observed between viral strains may aid in defining the molecular determinants that govern deletion formation within individual genome segments. Furthermore, it raises the question, given the potential role of DIPs in influencing infection outcome (7), of whether these strain differences in DIP formation have phenotypic consequences during infection.
We hope that the approach detailed here, and the associated bioinformatics pipeline, proves useful to other groups interested in defective interfering particle biology. Our approach is optimized for influenza virus sequences; however, the approaches and controls detailed here can easily be adapted to other RNA virus systems.
MATERIALS AND METHODS
Viruses and cells.
Madin-Darby canine kidney cells (MDCK; obtained from Jonathan Yewdell) and human embryonic kidney 293 cells (293T; obtained from Joanna Shisler) were grown in minimal essential medium (MEM) plus GlutaMax (Gibco), supplemented with 8.3% fetal bovine serum (Seradigm), at 37°C and 5% CO2. Recombinant A/California/07/09 (Cal07) virus was rescued via the standard 8-plasmid reverse genetics approach. Briefly, 60 to 90% confluent 293T cells were transfected with 500 ng of various plasmids (pDZ::PB2, pDZ::PB1, pDZ::PA, pDZ::HA, pDZ::NP, pDZ::NA, pDZ::M, and pDZ::NS) by using JetPrime (Polyplus) according to the manufacturer’s instructions. A/Cal07 reverse genetics plasmids were originally obtained as A/California/04/2009-encoding plasmids from Jonathan Yewdell. We introduced A660G and A335G substitutions into the HA and NP plasmids, respectively, to convert them to match the amino acid sequences of A/California/07/2009 HA and NP (NCBI accession numbers CY121680 and CY121683). Recombinant A/Perth/16/2009 was generated via the same method from plasmids kindly provided by Seema Lakdawala. Seed stocks of virus were prepared by amplifying plaque isolates from the rescue supernatants. Virus working stocks were generated by infecting MDCK cells with seed stock at an MOI of 0.001 50% tissue culture infectious dose (TCID50)/cell and collecting and clarifying supernatants at 48 hours postinfection (hpi). A/New Caledonia/1999 and B/Lee/1940 virus stocks were both provided by Jonathan Yewdell.
Generation of DIP stocks through high MOI passage.
Confluent MDCK cells in 6-well plates were infected in duplicate with undiluted stocks of A/Cal07, A/Perth09, or A/NC99 at 37°C or with B/Lee at 35°C. Supernatants (2 ml total per well) were harvested at 24 hpi (passage 1). An aliquot of 500 μl of this supernatant was used to infect a 6-well plate of fresh MDCK cells to generate the next passage. This process was repeated up to 9 times for both independent lineages of all viruses examined. Note that the MOIs across different passages were not consistent between viruses due to differences in starting titer and replication rates in MDCK cells.
IAV genome amplification.
Viral RNA was extracted from 140 μl of cell culture supernatant using the QIAamp viral RNA kit (Qiagen) and eluted in 60 μl distilled H2O (dH2O). For cDNA reactions, 3 μl of RNA was mixed with 1 μl (2 μM) MBTUni-12 primer (5′-ACGCGTGATCAGCAAAAGCAGG-3′) + 1 μl (10 μM) deoxynucleoside triphosphates (dNTPs) + 8 μl dH2O. The mixture was incubated for 5 min at 65°C and then placed on ice for 2 min. Subsequently, the mixture was removed from ice and the following were added: 1 μl SuperScript III (SSIII) RT (Invitrogen), 4 μl of × first-strand buffer (comes with SSIII kit), 1 μl of dithiothreitol (DTT), and 1 μl RNase-in (Invitrogen). The reaction mixture was incubated at 45°C for 50 min, followed by a 15 min incubation at 70°C for inactivation. cDNA product (5 μl) was mixed with the following for PCR amplification: 2.5 μl (10 μM) MBTUni-12 primer, 2.5 μl (10 μM) MBTUni-13 primer (5′-ACGCGTGATCAGTAGAAACAAGG-3′), 0.5 μl Phusion polymerase (NEB), 10 μl 5× high-fidelity (HF) buffer, 1 μl (10 mM dNTP mix), and 28.5 μl dH2O. The PCR conditions used were 98°C (30 s) followed by 25 cycles of 98°C (10 s), 57°C (30 s), and 72°C (90 s); a terminal extension of 72°C (5 min); and a final 10°C hold. PCR products were purified using the PureLink PCR purification kit (Invitrogen) with the <300-nt cutoff option and eluted in 30 μl dH2O. There was no difference in deletion junction detection when we purified the PCR products with the lower-cutoff option (data not shown); note that this may not be the case for all DIP-enriched samples. For B/Lee, we used the universal amplification method published by Zhou et al (29).
NGS library preparation.
We started with ∼20 ng of the PCR products in a volume of 50 μl. The Covaris M220 sonicator (Covaris) was used to fragment the DNA. The following three conditions were used to generate average fragment lengths of 300, 500, and 700 bp: (i) 300 bp = Peak Power 50, Duty Factor 20 and Cycles/Burst 200 for 2:40 min, (ii) 500 bp = Peak Power 50, Duty Factor 10 and Cycles/Burst 200 for 1:30 min, and (iii) ∼600 bp = fragment length Peak Power 50, Duty Factor 10 and Cycles/Burst 200 for 1 min. In our hands, the fragmentation length did not have any effect on our sequencing results (data not shown). For the sake of consistency, we used the 300-bp fragmentation length. To confirm the PCR products, we visualized the amplicons on a Fragment Analyzer (AATI) with the DNF-486 high-sensitivity NGS kit before and after fragmentation. Next, we used a Kapa Hyper prep kit (Roche) according to the manual to construct the libraries. To eliminate the possibility of index hopping (or index switching), we used the TruSeq unique dual indexes (UDI) from Illumina. The Adapter ligation step was carried out with 5 μl of TruSeq UDIs diluted 1:10 with 10 nM Tris. For maximum efficiency, we increased the ligation time to 30 min. We then performed 3 cycles of PCR with the Kapa library amplification primers diluted 1:5 in water, followed by a cleanup step with 40 μl of AxyPrep Mag PCR beads (Thermo Fisher). We then mixed the libraries at an equimolar ratio and carried out a quantitative PCR (qPCR) to accurately quantitate the library pool and maximize the number of clusters in the sequencing flow cell. A size selection step was not needed. Finally, the pooled libraries were sequenced with paired-end 2 × 250-nt reads on an Illumina MiSeq instrument using V2 chemistry. The fastq files were generated and demultiplexed with the bcl2fastq Conversion Software v2.20 (Illumina).
Simulated data sets.
All of the simulated data sets used in this study were generated by MetaSim (v0.9.1) (30), a genomics and metagenomics simulator. Several reference library sequences composed of WT reference sequences of IAV Cal07 (see Table 2 for NCBI accession numbers), mixed with a defined DIP sequence population—generated randomly within the first and last 600 nt of all the segments—were used in MetaSim for data simulation. The configurations were fixed across all data sets to maintain the preferable conditions. The reference sequences were fragmented into 350-nt fragment lengths with a standard deviation of ±50 and were simulated into ∼1 million 2 × 250-nt paired-end reads per sample, with a total substitution rate of ∼1% based on the published Illumina empirical error model. One data set was simulated with no DIP sequences as a control sample for any computational artifacts. MetaSim generated two FASTA files of 1 million reads per file per sample (∼2 million single-end reads = 1 million paired-end reads), which were subsequently used for the optimization process.
TABLE 2.
NCBI accession numbers for influenza viruses used in this study
| Gene segment | NCBI accession no. |
||||
|---|---|---|---|---|---|
| A/PR8 | A/Cal07 | A/NC/99 | A/Perth/09 | B/Lee/40 | |
| PB2 | AF389115.1 | CY121687 | CY147325 | KJ609203.1 | CY115118.1 |
| PB1 | AF389116.1 | CY121686 | CY147324 | KJ609204.1 | CY115117.1 |
| PA | AF389117.1 | CY121685 | CY147323 | KJ609205.1 | CY115116.1 |
| HA | AF389118.1 | CY121680 | CY147318 | KJ609206.1 | CY115111.1 |
| NP | AF389119.1 | CY121683 | CY147321 | KJ609207.1 | CY115114.1 |
| NA | AF389120.1 | CY121682 | CY147320 | KJ609208.1 | CY115113.1 |
| M | AF389121.1 | CY121681 | CY147319 | KJ609209.1 | CY115112.1 |
| NS | AF389122.1 | CY121684 | CY147322 | KJ609210.1 | CY115115.1 |
Sequencing analysis of DIP-associated junctions.
The raw sequencing reads were quality-filtered by Trimmomatic (v0.36) (parameters: ILLUMINACLIP:TruSeq3-PE-2.fa:2:15:10 SLIDINGWINDOW:3:20 LEADING:28 TRAILING:28) (31), and any reads shorter than 75-nt were removed from the data sets. The paired reads were concatenated into one file and treated as single-end when aligned end-to-end to the WT reference sequences (Table 2) using Bowtie 2 (v2.3.1) (parameters: –score-min L,0, −0.3). Bowtie 2 passes these values into a linear function to calculate the total penalty score [f(x) = −0.3 × x], where x is the read length. This option allows Bowtie 2 to align based on the read length, i.e., the penalty score of a 250-nt read is relatively the same as that of a 200-nt read.
Subsequently, the algorithm ViReMa (v0.10) was used to analyze the remaining unaligned reads (putative junction-spanning reads) (parameters: -DeDup –MicroInDel_Length 20 –Defuzz 3 –N 1 –X 8). Next, the DIP-associated deletion junctions and their read support were extracted from ViReMa output files and sorted per segment, using an in-house Perl script, for data analysis and visualization. To detect any MDCK genome leakage, the data sets were aligned against the dog genome (assembly CanFam3.1). All scripts are available at https://github.com/BROOKELAB/Influenza-virus-DI-identification-pipeline.
Quantification of sensitivity and precision.
To calculate the actual number of junction-spanning reads in Fig. 2A, reads that derived from DIP-associated sequences were counted by their FASTA headers (produced by MetaSim), which contain the source of each read. To calculate the maximum theoretical sensitivity of ViReMa (Fig. 3B) based on a seed length of 25-nt and two allowed mutations (–N = 2), the number of mutations was subtracted from the seed length, which in turn was multiplied by 2 to account for both termini [(25 − 2) × 2 = 46]. Subsequently, this number was subtracted from the possible cutting site of a 250-nt read and divided by the total number of cutting sites and multiplied by 100 ([(249 − 46)/249] × 100 = 81.5%). To calculate the number of accurately and inaccurately mapped junctions in Fig. 3C and D, the original sequences generated to simulate the data set A/Cal07-200 were used in ViReMa with –N set to 0, and the remaining parameters were kept the same (the junctions that occurred within the first or last 25 nt were removed). Subsequently, the junctions that accurately mapped were counted, resulting in 149 accurately mapped versus 47 inaccurately mapped junctions.
Correlation analysis.
For the correlation tests, the NGS read support count for each DIP-associated junction was normalized against the total detected junction-spanning reads of every sample. Next, the correlation was calculated based on Spearman’s rank correlation using R (cor function).
Data availability.
All NGS data sets generated in this study may be found under BioProject accession number PRJNA527853.
ACKNOWLEDGMENTS
We are grateful to other members of the lab for helpful comments and critical readings of the manuscript, as well as to Alvaro Hernandez and the staff at the high-through sequencing and genotyping unit within the Roy J. Carver Biotechnology Center for excellent advice and technical assistance.
This work was generously funded by the Defense Advanced Research Projects Agency under contract DARPA-16-35-INTERCEPT-FP-018.
REFERENCES
- 1.Von Magnus P. 1954. Incomplete forms of influenza virus. Adv Virus Res 2:59–79. doi: 10.1016/S0065-3527(08)60529-1. [DOI] [PubMed] [Google Scholar]
- 2.von Magnus P. 1951. Propagation of the PR8 strain of influenza A virus in chick embryos. II. The formation of incomplete virus following inoculation of large doses of seed virus. Acta Pathol Microbiol Scand 28:278–293. [DOI] [PubMed] [Google Scholar]
- 3.Rezelj VV, Levi LI, Vignuzzi M. 2018. The defective component of viral populations. Curr Opin Virol 33:74–80. doi: 10.1016/j.coviro.2018.07.014. [DOI] [PubMed] [Google Scholar]
- 4.Baum A, Sachidanandam R, García-Sastre A. 2010. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci U S A 107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nayak DP, Chambers TM, Akkina RK. 1985. Defective-interfering (DI) RNAs of influenza viruses: origin, structure, expression, and interference. Curr Top Microbiol Immunol 114:103–151. [DOI] [PubMed] [Google Scholar]
- 6.Brooke CB. 2017. Population diversity and collective interactions during influenza virus infection. J Virol 91:e01164-17. doi: 10.1128/JVI.01164-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasilijevic J, Zamarreño N, Oliveros JC, Rodriguez-Frandsen A, Gómez G, Rodriguez G, Pérez-Ruiz M, Rey S, Barba I, Pozo F, Casas I, Nieto A, Falcón A. 2017. Reduced accumulation of defective viral genomes contributes to severe outcome in influenza virus infected patients. PLoS Pathog 13:e1006650. doi: 10.1371/journal.ppat.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sherry L, Punovuori K, Wallace LE, Prangley E, DeFries S, Jackson D. 2016. Identification of cis-acting packaging signals in the coding regions of the influenza B virus HA gene segment. J Gen Virol 97:306–315. doi: 10.1099/jgv.0.000358. [DOI] [PubMed] [Google Scholar]
- 9.Hutchinson EC, von Kirchbach JC, Gog JR, Digard P. 2010. Genome packaging in influenza A virus. J Gen Virol 91:313–328. doi: 10.1099/vir.0.017608-0. [DOI] [PubMed] [Google Scholar]
- 10.Jennings PA, Finch JT, Winter G, Robertson JS. 1983. Does the higher order structure of the influenza virus ribonucleoprotein guide sequence rearrangements in influenza viral RNA? Cell 34:619–627. doi: 10.1016/0092-8674(83)90394-X. [DOI] [PubMed] [Google Scholar]
- 11.Routh A, Johnson JE. 2014. Discovery of functional genomic motifs in viruses with ViReMa-a Virus Recombination Mapper-for analysis of next-generation sequencing data. Nucleic Acids Res 42:e11. doi: 10.1093/nar/gkt916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alnaji FG, Holmes JR, Rendon G, Vera JC, Fields CJ, Martin BE, Brooke CB. 2018. Illumina-based sequencing framework for accurate detection and mapping of influenza virus defective interfering particle-associated RNAs. bioRXiv 10.1101/440651. [DOI] [PMC free article] [PubMed]
- 14.Janda JM, Davis AR, Nayak DP, De BK. 1979. Diversity and generation of defective interfering influenza virus particles. Virology 95:48–58. doi: 10.1016/0042-6822(79)90400-8. [DOI] [PubMed] [Google Scholar]
- 15.Saira K, Lin X, DePasse JV, Halpin R, Twaddle A, Stockwell T, Angus B, Cozzi-Lepri A, Delfino M, Dugan V, Dwyer DE, Freiberg M, Horban A, Losso M, Lynfield R, Wentworth DN, Holmes EC, Davey R, Wentworth DE, Ghedin E, INSIGHT FLU002 Study Group, INSIGHT FLU003 Study Group. 2013. Sequence analysis of in vivo defective interfering-like RNA of influenza A H1N1 pandemic virus. J Virol 87:8064–8074. doi: 10.1128/JVI.00240-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ. 2012. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 17.Schirmer M, Ijaz UZ, D’Amore R, Hall N, Sloan WT, Quince C. 2015. Insight into biases and sequencing errors for amplicon sequencing with the Illumina MiSeq platform. Nucleic Acids Res 43:e37. doi: 10.1093/nar/gku1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, Kawaoka Y, Wentworth DE. 2009. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses. J Virol 83:10309–10313. doi: 10.1128/JVI.01109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Görzer I, Guelly C, Trajanoski S, Puchhammer-Stöckl E. 2010. The impact of PCR-generated recombination on diversity estimation of mixed viral populations by deep sequencing. J Virol Methods 169:248–252. doi: 10.1016/j.jviromet.2010.07.040. [DOI] [PubMed] [Google Scholar]
- 20.Lahr DJG, Katz LA. 2009. Reducing the impact of PCR-mediated recombination in molecular evolution and environmental studies using a new-generation high-fidelity DNA polymerase. Biotechniques 47:857–866. doi: 10.2144/000113219. [DOI] [PubMed] [Google Scholar]
- 21.Xue J, Chambers BS, Hensley SE, López CB. 2016. Propagation and characterization of influenza virus stocks that lack high levels of defective viral genomes and hemagglutinin mutations. Front Microbiol 7:326. doi: 10.3389/fmicb.2016.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Routh A, Head SR, Ordoukhanian P, Johnson JE. 2015. ClickSeq: fragmentation-free next-generation sequencing via click ligation of adaptors to stochastically terminated 3′-azido cDNAs. J Mol Biol 427:2610–2616. doi: 10.1016/j.jmb.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Acevedo A, Andino R. 2014. Library preparation for highly accurate population sequencing of RNA viruses. Nat Protoc 9:1760–1769. doi: 10.1038/nprot.2014.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Timm C, Akpinar F, Yin J. 2014. Quantitative characterization of defective virus emergence by deep sequencing. J Virol 88:2623–2632. doi: 10.1128/JVI.02675-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kosik I, Ince WL, Gentles LE, Oler AJ, Kosikova M, Angel M, Magadán JG, Xie H, Brooke CB, Yewdell JW. 2018. Influenza A virus hemagglutinin glycosylation compensates for antibody escape fitness costs. PLoS Pathog 14:e1006796. doi: 10.1371/journal.ppat.1006796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom R. 2011. Accurate sampling and deep sequencing of the HIV-1 protease gene using a primer ID. Proc Natl Acad Sci U S A 108:20166–20171. doi: 10.1073/pnas.1110064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaworski E, Routh A. 2017. Parallel ClickSeq and Nanopore sequencing elucidates the rapid evolution of defective-interfering RNAs in Flock House virus. PLoS Pathog 13:e1006365. doi: 10.1371/journal.ppat.1006365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garalde DR, Snell EA, Jachimowicz D, Sipos B, Lloyd JH, Bruce M, Pantic N, Admassu T, James P, Warland A, Jordan M, Ciccone J, Serra S, Keenan J, Martin S, McNeill L, Wallace EJ, Jayasinghe L, Wright C, Blasco J, Young S, Brocklebank D, Juul S, Clarke J, Heron AJ, Turner DJ. 2018. Highly parallel direct RNA sequencing on an array of nanopores. Nat Methods 15:201–206. doi: 10.1038/nmeth.4577. [DOI] [PubMed] [Google Scholar]
- 29.Zhou B, Lin X, Wang W, Halpin RA, Bera J, Stockwell TB, Barr IG, Wentworth DE. 2014. Universal influenza B virus genomic amplification facilitates sequencing, diagnostics, and reverse genetics. J Clin Microbiol 52:1330–1337. doi: 10.1128/JCM.03265-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richter DC, Ott F, Auch AF, Schmid R, Huson DH. 2008. MetaSim—a sequencing simulator for genomics and metagenomics. PLoS ONE 3:e3373. doi: 10.1371/journal.pone.0003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All NGS data sets generated in this study may be found under BioProject accession number PRJNA527853.