HIV-1 has evolved to acquire several strategies to limit the exposure of its envelope glycoproteins (Env) on the surface of infected cells. In this study, we show that antibody-induced Env internalization is conformation specific and reduces the susceptibility of infected cells to antibody-dependent cellular cytotoxicity (ADCC). Thus, a better understanding of this mechanism might help develop antibodies with improved capacities to mediate ADCC.
KEYWORDS: ADCC, dynamin, Env, Env conformation, HIV-1, Internalization, bNAbs, nonneutralizing antibodies
ABSTRACT
To minimize immune responses against infected cells, HIV-1 limits the surface expression of its envelope glycoprotein (Env). Here, we demonstrate that this mechanism is specific for the Env conformation and affects the efficiency of antibody-dependent cellular cytotoxicity (ADCC). Using flow cytometry and confocal microscopy, we show that broadly neutralizing antibodies (bNAbs) targeting the “closed” conformation of Env induce its internalization from the surface. In contrast, non-neutralizing antibodies (nNAbs) are displayed on the cell surface for prolonged period of times. The bNAb-induced Env internalization can be decreased by blocking dynamin function, which translates into higher susceptibilities of infected cells to ADCC. Our results suggest that antibody-mediated Env internalization is a mechanism used by HIV-1 to evade immune responses against the “closed” conformation of Env expressed on HIV-1-infected cells.
IMPORTANCE HIV-1 has evolved to acquire several strategies to limit the exposure of its envelope glycoproteins (Env) on the surface of infected cells. In this study, we show that antibody-induced Env internalization is conformation specific and reduces the susceptibility of infected cells to antibody-dependent cellular cytotoxicity (ADCC). Thus, a better understanding of this mechanism might help develop antibodies with improved capacities to mediate ADCC.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) envelope glycoproteins (Env) are exposed on the surface of viral particles and infected cells. Antibodies (Abs) targeting Env can therefore either neutralize viral particles or mediate different immune responses, including antibody-dependent cellular cytotoxicity (ADCC) against infected cells. ADCC has been associated with decreased HIV-1 transmission and disease progression (1–5). Antibodies with ADCC activity, in the presence of low plasma IgA Env-specific Abs, inversely correlated with HIV-1 acquisition in the partially successful Thai RV144 vaccine trial (6).
HIV-1 has evolved several mechanisms to avoid the elimination of infected cells via ADCC. Env in its “open” CD4-bound conformation is particularly vulnerable to ADCC activity (7–9). HIV-1 keeps Env in its “closed” (state 1) conformation on the surfaces of infected cells by preventing the accumulation of the CD4 receptor via Nef- and Vpu-mediated downregulation (9–11). The virus also evades ADCC by antagonizing BST-2 (9, 12–14) through its Vpu accessory protein. In addition to these evasion mechanisms, the virus minimizes Env accumulation at the cell surface by internalizing Env (15). HIV-1 Env has a long C-terminal cytoplasmic domain, which is involved in the regulation of Env trafficking and contains several endocytic signals, including membrane-proximal tyrosine-based sorting motifs and dileucine motifs (16–18). Mutations of these motifs have been shown to result in increased cell surface expression of Env, which correlates with increased ADCC responses (15, 19, 20). Internalizing envelope glycoproteins from the cell surface to avoid humoral immune responses does not appear to be restricted to HIV-1; equine herpesvirus 1 (21) and pseudorabies virus (22) also downregulate their envelope glycoproteins in order to avoid antibody-dependent complement-mediated lysis. Thus, internalization of viral envelope glycoproteins from the surface of infected host cells appears to be a mechanism employed by many viruses to minimize recognition by the immune system (23–26).
The ability of antigen-Ab complexes to remain on the surfaces of infected cells might be another factor modulating humoral responses against different viruses. It has been reported previously that the interaction of some Abs with Env glycoproteins expressed on the surfaces of respiratory syncytial virus (RSV) (27)- and feline coronavirus (28)-infected cells accelerates their internalization and decreases their exposure to the immune system. We evaluated here the fate of HIV-1 Env expression at the surfaces of cells upon antibody binding and made the surprising observation that the binding of broadly neutralizing antibodies (bNAbs) but not that of nonneutralizing antibodies (nNAbs) induced Env-Ab complex internalization and reduced ADCC responses mediated by bNAbs. These data indicate that HIV-1 specifically evades immune responses against the closed conformation of Env.
RESULTS
bNAbs induce Env internalization from the cell surface.
To determine whether Env-Abs complexes can remain stable over time at the surface of infected cells, we selected nine anti-Env Abs that recognize different conformations and epitopes of Env. These Abs target different sites on Env, such as the gp120-gp41 interface (PGT151) (29), glycans on the gp120 outer domain (2G12) (30), the V1/V2 apex (PG9) (31), and V3 glycans (PGT121, PGT126) (32), as well as CD4-induced (CD4i) targeting epitopes, including the coreceptor binding site (CoRBS; 17b and N12-i2) and the cluster A region in the gp120 inner domain (A32, N5-i5) (33). This panel of antibodies was selected because it can distinguish “closed” versus “open” trimers. Indeed, PGT151, PG9, PGT121, and PGT126 preferentially recognize the “closed” trimer (34–37), whereas CD4i Abs 17b, N12-i2, A32, and N5-i5 bind epitopes only exposed in the “open” trimer. 2G12 is an antibody that can recognize both forms of trimers (9, 37) but has a preference for the “closed” form (36).
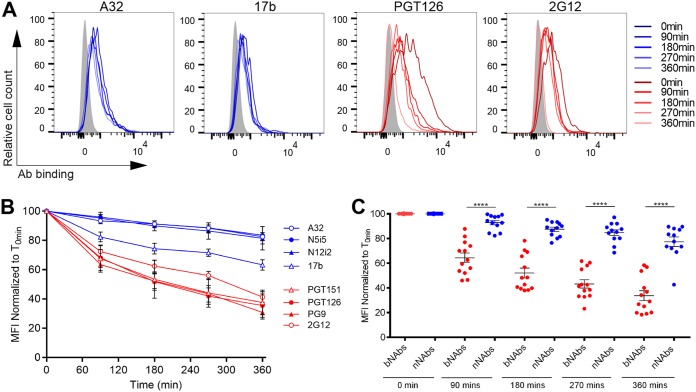
Primary CD4+ T cells were infected with a previously reported wild-type (wt) HIV-1 NL4.3 strain encoding a gfp reporter gene and an R5-tropic (ADA) envelope (9). Infected cells were identified based on GFP expression (GFP+). Cells were first incubated with monoclonal Abs (MAbs) at 4°C for 30 min, washed to remove excess antibody, and then incubated at 37°C for internalization to occur. After incubation at 37°C for different time intervals, Env-Ab complexes remaining at the cell surface were visualized with a fluorescent secondary anti-human antibody by flow cytometry. Under these experimental settings we observed that the binding of bNAbs (PG126, PGT151, PG9, and 2G12) significantly declined over time indicating Env internalization (Fig. 1). In contrast, binding of the nNAbs (17b, N12-i2, A32, and N5-i5) was fairly steady. The reduction in the cell surface levels of Env was ∼60% after 6 h of incubation with PGT126, PG9, PGT151, and 2G12 at 37°C. Conversely, binding to surface Env was only reduced by ∼20% upon incubation for the same time period at 37°C with N12-i2, N5-i5, A32, and 17b (Fig. 1B and C).
FIG 1.
Broadly neutralizing antibodies but not nonneutralizing antibodies induce Env internalization. A panel of bNAbs (PGT121, PGT126, PG9, and 2G12) and nNAbs (A32, N5-i5, 17b, and N12-i2) were used to stain the surface of primary CD4+ T cells infected with the NL4.3 GFP ADA virus. (A) Histograms depicting representative staining of infected cells (GFP+) with A32, 17b, PGT126, and 2G12 MAbs over time or with mock-infected cells (gray). (B and C) Quantification of remaining antibody-Env complexes on the cell-surface over different time points is expressed as percentage of the MFI relative to the 0-min time control. Error bars indicate means ± the standard errors of the mean (SEM). Statistical significance was tested using an unpaired t test or a Mann-Whitney U test based on statistical normality (****, P < 0.0001).
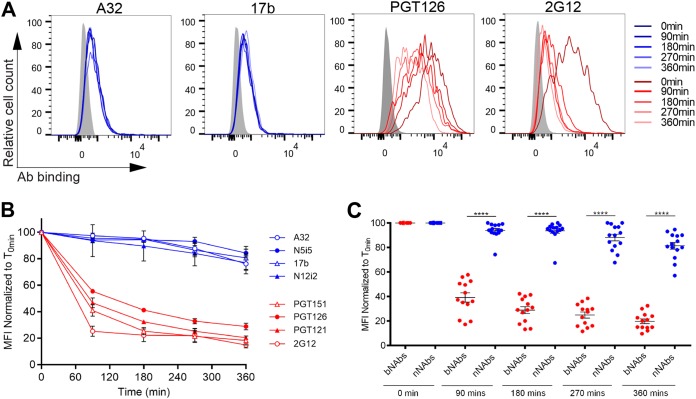
To evaluate whether this phenotype was restricted to the infectious molecular clone used, we infected primary CD4+ T cells with the transmitted/founder (T/F) virus CH58 (CH58 T/F) and performed the same experiments described above with the exception that infected cells were identified by intracellular Gag staining. Similar to what we obtained with NL4.3 ADA-infected cells (Fig. 1), bNAbs reduced the cell surface levels of CH58T/F Env by 80% after 6 h of incubation at 37°C, whereas incubation with nNAbs only reduced Env by ∼20% during the same time period (Fig. 2).
FIG 2.
Binding of bNAbs but not nNAbs decreases Env expression from the surfaces of T/F virus-infected cells. A panel of bNAbs (PGT121, PGT126, PG9, and 2G12) and nNAbs (A32, N5-i5, 17b, and N12-i2) were used to stain the surfaces of primary CD4+ T cells infected with CH58 T/F virus. (A) Histograms depicting representative staining of infected cells (Gag+) with A32, 17b, PGT126, and 2G12 MAbs over time or with mock-infected cells. (B and C) Quantification of remaining antibody-Env complexes on the cell surface over different time points is expressed as the percentage of the MFI relative to the 0-min time control. Error bars indicate means ± the SEM. Statistical significance was tested using an unpaired t test or a Mann-Whitney U test based on statistical normality (****, P < 0.0001).
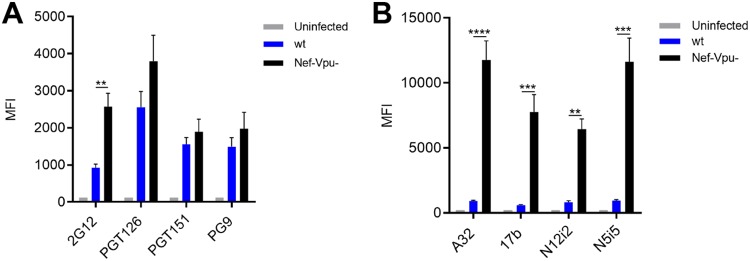
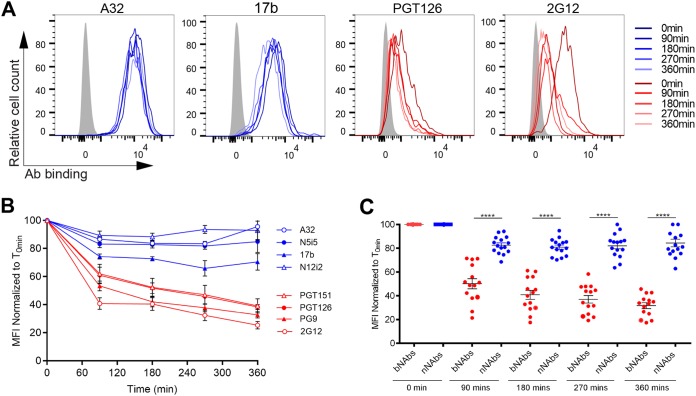
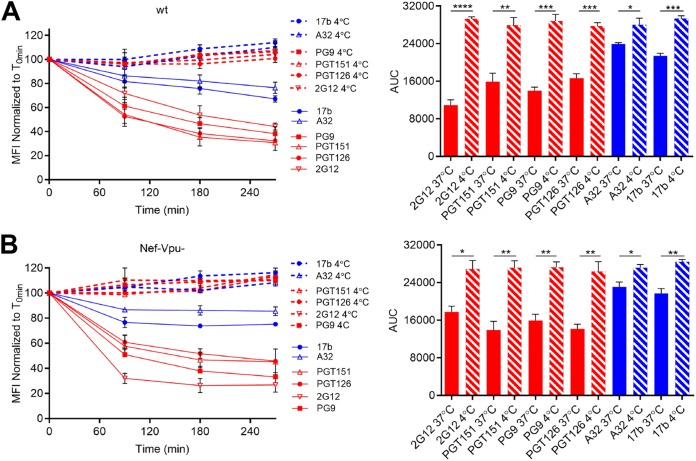
It is now well established that the conformation of Env at the cell surface influences antibody binding and ADCC responses (38–40). Nonneutralizing antibodies and bNAbs target different Env conformations, with most bNAbs preferentially recognizing Env in its “closed” conformation (state 1) (36), whereas the nNAbs used here target epitopes that are only exposed upon Env interaction with CD4 (9, 33, 37, 42). To verify whether the differences in cell surface reduction of Env between bNAbs and nNAbs was due to their differential recognition of infected cells, we infected primary CD4+ T cells with NL4.3 ADA defective for Nef and Vpu accessory proteins (Nef-Vpu-) that fails to downregulate CD4. This virus was used to present Env in its CD4-bound “open” conformation at the surfaces of infected cells and enhance the recognition of infected cells by the nNAbs used in this study (8, 9) (Fig. 3). As expected, deletion of Vpu enhances the overall levels of Env at the cell surface as measured by 2G12, known to bind to both “open” and “closed” conformations of Env (7, 9, 43) (Fig. 3A). This phenomenon has been well established and is due to the accumulation of BST-2-trapped viral particles at the cell surface, which results in Env accumulation (9, 12, 14). In agreement with their CD4-induced nature, the recognition of infected cells by the nNAbs tested (A32, 17b, N12-i2, and N5-i5) was dramatically enhanced by deletion of Nef and Vpu. Deletion of these accessory genes impair the ability of HIV-1 to downregulate CD4 from the cell surface, thus resulting in increased Env-CD4 interactions and the subsequent exposure of CD4i epitopes (Fig. 3B) (9). Despite their improved capacity to recognize Env at the surfaces of Nef-Vpu-infected cells, nNAbs only reduced Env from the cell surface by ∼20% after 6 h, while bNAbs reduced Env levels by ∼60% within the same time period (Fig. 4). Altogether, these results indicate that the conformation specificity of antibody-mediated internalization of Env is not affected by changes in surface expression and that Nef and Vpu accessory proteins are not involved in the bNAb-mediated reduction of Env from the cell surface.
FIG 3.
Nef and Vpu protect infected cells from recognition by CD4i antibodies. Primary CD4+ T cells infected with NL4.3 GFP ADA-based viruses either wt or defective for Nef and Vpu expression (Nef-Vpu-) were stained with bNAbs (2G12, PGT126, PGT151, and PG9) (A) or anti-cluster A (A32 and N5-i5) and CoRBS (N12-i2 and 17b) antibodies (B). Error bars indicate means ± the SEM. Statistical significance was tested using an unpaired t test or a Mann-Whitney U test based on statistical normality (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
FIG 4.
Nef and Vpu are dispensable for bNAbs-mediated Env internalization. Cell-surface staining of primary CD4+ T cells infected with NL4.3 GFP ADA-based virus defective for Nef and Vpu expression using a panel of bNAbs (PGT126, PGT151, PG9, and 2G12) and nNAbs (A32, N5-i5, 17b, and N12-i2). (A) Histograms depicting representative staining of infected cells (GFP+) with A32, 17b, PGT126, and 2G12 MAbs over time or with mock-infected cells (gray). (B and C) Quantification of remaining antibody-Env complexes on the cell surface over different time points is expressed as percentage of the MFI relative to the 0-min time control. Error bars indicate means ± the SEM. Statistical significance was tested using an unpaired t test or a Mann-Whitney U test based on statistical normality (****, P < 0.0001).
Antibody-induced internalization of cell surface-expressed HIV-1 Env.
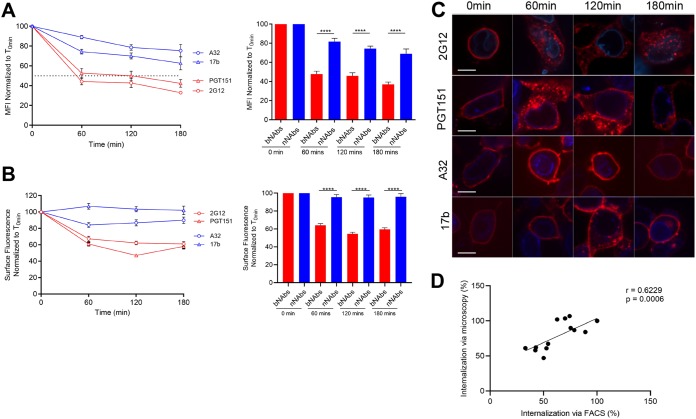
To confirm that Env-bNAb complexes were internalized upon incubation at 37°C, we performed confocal microscopy studies in parallel to flow cytometry. Since primary CD4+ T cells are small and poorly adherent, we used HEK 293T cells for these studies. 293T cells were transfected with a primary tier 2 Env (JRFL) and incubated 48 h posttransfection with Alexa Fluor-conjugated bNAbs (2G12 and PGT151) or nNAbs (17b and A32) for various time points at 37°C before fixing and analysis by flow cytometry and confocal imaging. Since A32 and 17b do not recognize the unliganded EnvJRFL, Env was cotransfected, together with the CD4 receptor, to trigger the exposure of the CD4i epitopes recognized by these nNAbs (9, 43). Flow cytometry confirmed that the bNAbs 2G12 and PGT151 were internalized much faster than nNAbs 17b and A32, thus phenocopying the results from T cells (Fig. 5A). Parallel confocal imaging demonstrated that, upon the addition of bNAbs PGT151 and 2G12, Env moved from the plasma membrane into intracellular compartments after 60 min of incubation at 37°C. In contrast, the binding of A32 and 17b did not change the cell surface localization of Env (Fig. 5B and C). These results were consistent with those obtained using infected primary CD4+ T cells (Fig. 1, 2, and 4). Interestingly, the quantification of internalization measured by microscopy correlated well with the quantification of cell surface Env measured by flow cytometry (Fig. 5D), suggesting that both assays measure Env internalization. Further supporting this possibility, we observed that bNAb-induced Env reduction from the cell surface was blocked if cells were incubated at 4°C, a temperature known to block molecular trafficking events, for the entirety of the internalization assay (Fig. 6).
FIG 5.
Antibody-induced internalization of Env from the surface of transfected cells. 293T cells were transfected with plasmids encoding HIV-1JRFL Env alone or together with human CD4 receptor to expose CD4i epitopes. Env expressed in the absence of CD4 were visualized with 2G12 and PGT151. Env coexpressed with CD4 was visualized with 17b and A32. (A) Flow cytometric analysis of Env internalization from the surface of 293T-transfected cells. The level of the remaining surface-expressed Env after internalization is expressed as percentage of the MFI relative to the 0-min time control. (B) Confocal microscopy analysis of antibody-induced internalization. The remaining antibody-Env complexes over different time points are expressed as percentages of the surface fluorescence relative to the 0-min control. (C) For confocal microscopy, 293T cells were also transfected with the lamin B receptor-CFP plasmid (a nuclear envelope marker used to identify transfected cells). Images show the localization of antibody-Env complexes at different time points (0, 60, 120, and 180 min). Images represent a single confocal z-section through the middle of the cell; 20 cells were imaged per condition, and representative images are shown. Scale bar, 10 μm. (D) Correlation of the quantification of Env internalization by confocal microscopy with internalization by flow cytometry using a Pearson correlation test. Error bars indicate means ± the SEM. Statistical significance was tested using an unpaired t test or a Mann-Whitney U test based on statistical normality (***, P < 0.001; ****, P < 0.0001).
FIG 6.
bNAb-triggered Env internalization is temperature-dependent manner. Cell surface staining of primary CD4+ T cells infected with NL4.3 GFP ADA-based viruses either wt (A) or defective for Nef and Vpu (Nef-Vpu-) expression (B) with A32, 17b, PGT126, PGT151, PG9, and 2G12 MAbs. (Left) Quantification of remaining antibody-Env complexes on the cell surface over different time points (90, 180, and 270 min) at 4°C (dotted lines) or 37°C (solid lines) is expressed as the percentage of MFI relative to the 0-min time control. (Right) Areas under the curve (AUC) were calculated based on MFI data sets using GraphPad Prism software. Error bars indicate means ± the SEM. Statistical significance was tested using an unpaired t test or a Mann-Whitney U test based on statistical normality (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
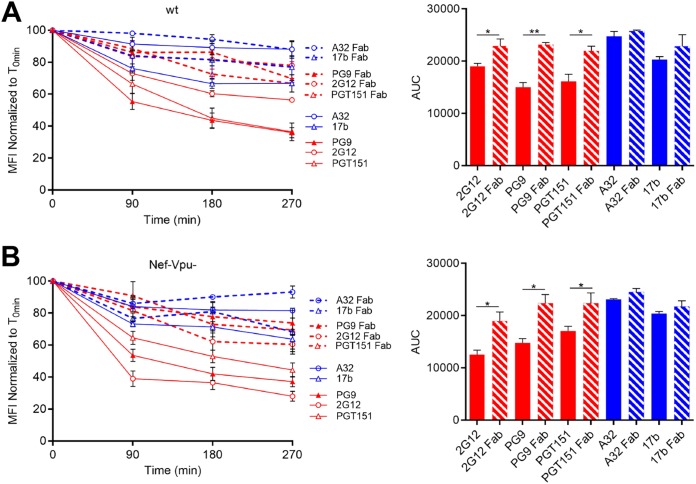
It has been previously suggested that Ab-mediated cross-linking facilitates internalization of RSV fusion proteins. In this study, RSV Env endocytosis was significantly reduced when Fab fragments were used instead of their full MAb counterpart (27). To verify whether this was the case for HIV-1 Env, we performed side-by-side comparisons on the ability of full-length Abs versus their Fab fragments to reduce Env levels at the cell surface upon incubation at 37°C, as measured by flow cytometry. A32 and 17b Fab fragments behaved similar to the full-length Abs. However, PG9, PGT151 and 2G12 Fab fragments were significantly less efficient than their IgG counterparts at inducing internalization of Env (Fig. 7). These data suggest that the cross-linking of Env trimers at the cell surface stimulates bNAb-mediated internalization.
FIG 7.
Fab fragments fail to induce Env internalization. Cell surface staining of primary CD4+ T cells infected with NL4.3 GFP ADA (A) or defective for Nef and Vpu (Nef-Vpu-) expression (B) was performed with A32, 17b, PGT151, PG9, and 2G12 MAbs (solid lines) or their Fab fragments (dotted lines). (Left) Quantification of remaining antibody-Env complexes on the cell surface over different time points (90, 180, and 270 min) is expressed as the percentage of MFI relative to the 0-min time control. (Right) Areas under the curve (AUC) were calculated based on MFI data sets using GraphPad Prism software. Error bars indicate means ± the SEM. Statistical significance was tested using an unpaired t test or a Mann-Whitney U test based on statistical normality (*, P < 0.05; **, P < 0.01).
A dynamin inhibitor attenuates bNAb-induced Env internalization and increases the susceptibility of infected cells to ADCC.
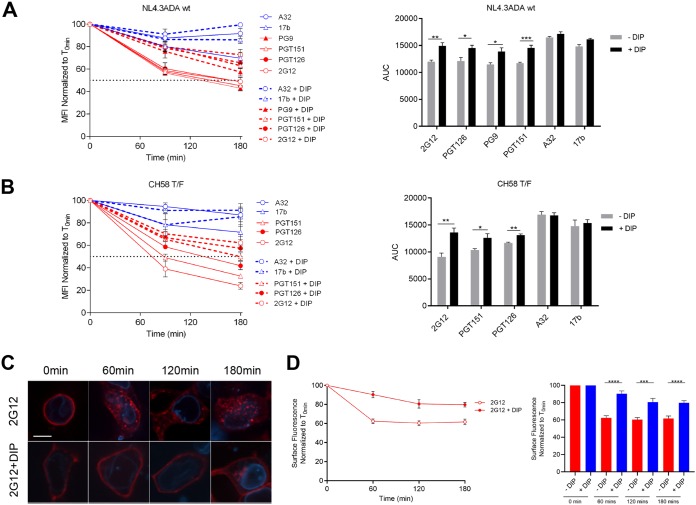
To further confirm that internalization was involved in the observed bNAb-induced reduction of Env from the cell surface, we decided to block the function of dynamin, a GTPase implicated in endocytic membrane fission events (44). Primary CD4+ T cells were infected with NL4.3 ADA green fluorescent protein (GFP) or CH58 T/F and dynamin was inhibited with the dynamin inhibitory peptide (DIP). This peptide blocks the binding of dynamin to amphiphysin and has been shown to reduce endocytic events (27, 45, 46). The addition of DIP significantly reduced bNAb-induced Env internalization in infected cells as measured by flow cytometry (Fig. 8A and B) and confocal microscopy using EnvJRFL-transfected 293T cells (Fig. 8C and D). No statistical differences in the internalization rates of Env bound by nNAbs, with or without added DIP, were observed. Thus, this reiterates the observation that the binding of these Abs does not promote Env internalization.
FIG 8.
bNAb-mediated Env internalization is dynamin dependent. Cell surface staining of primary CD4+ T cells infected with either NL4.3 GFP ADA-based virus (A) or CH58 T/F virus (B) was performed with A32, 17b, PGT151, PGT126, PG9, and 2G12 MAbs in the presence or absence of 50 μM DIP. (Left) Quantification of remaining antibody-Env complexes on the cell surface over different time points (90 and 180 min) is expressed as a percentage of MFI relative to the 0-min time control. (Right) AUC values were calculated based on MFI data sets using GraphPad Prism software. Cell surface staining of 293T cells transfected with HIV-1JRFL Env, along with the lamin B receptor-CFP plasmid, was performed to locate the nuclear envelope with Alexa Fluor 594-conjugated 2G12 MAb in the presence or absence of 40 μM DIP. (C) Images show the localization of antibody-Env complexes at different time points (0, 60, 120, and 180 min). Images represent a single confocal z-section through the middle of the cell; 20 cells were imaged per condition, and representative images are shown. Scale bar, 10 μm. (D) Quantification of remaining antibody-Env complexes over different time points is expressed as the percentage of surface fluorescence relative to the 0-min control. Error bars indicate means ± the SEM. Statistical significance was tested using an unpaired t test or a Mann-Whitney U test based on statistical normality (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Since DIP treatment resulted in an accumulation of bNAb/Env complexes at the surface of HIV-1-infected cells, we evaluated whether it had any impact on the ability of bNAbs to mediate ADCC. Primary CD4+ T cells were infected with the NL4.3 ADA GFP wt virus, and their susceptibility to ADCC was measured using a previously described fluorescence-activated cell sorting (FACS)-based assay (9, 47). Interestingly, we observed a significant increase in ADCC responses mediated by PGT126, PGT151, PG9, and 2G12 upon DIP treatment (Fig. 9) but not of direct killing (i.e., in the absence of Abs [not shown]). Thus, this suggests that under normal conditions bNAb-mediated Env internalization reduces their capacity to trigger ADCC.
FIG 9.
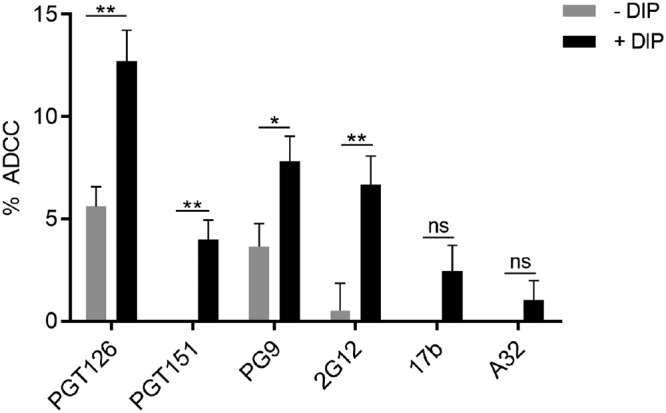
A dynamin inhibitor enhances the ADCC activity of bNAbs. Primary CD4 T cells isolated from at least three different healthy donors were infected with NL4.3 GFP ADA virus and used as target cells. Autologous peripheral blood mononuclear cells were used as effector cells in a FACS-based ADCC assay (9, 47). The percentages of ADCC-mediated killing obtained with A32, 17b, PGT126, PGT151, PG9, and 2G12 MAbs in the presence of 50 μM DIP or the vehicle control are shown. Error bars indicate means ± the SEM. Statistical significance was tested using an unpaired t test or a Mann-Whitney U test based on statistical normality (*, P < 0.05; **, P < 0.01).
DISCUSSION
Here we report that antibody-bound Env proteins are internalized from the surface of HIV-1-infected cells in a conformation-specific manner. bNAbs that preferentially recognize “closed” Env conformations trigger rapid Env internalization. When this process was decreased using a dynamin inhibitor, the susceptibility of infected cells to ADCC mediated by bNAbs was enhanced, suggesting that bNAb-triggered Env internalization impairs their ability to mediate robust ADCC responses. In contrast, nNAb specific for “open” Env conformation remain exposed on the surfaces of infected cells for prolonged periods of time. As such, differential internalization of Env-antibody complexes is likely an immune evasion mechanism that HIV-1 evolved to limit the surface exposure of Env in “closed” conformations while distracting the immune system with the display of Env proteins in “open” conformations that ultimately result in nonneutralizing immune responses.
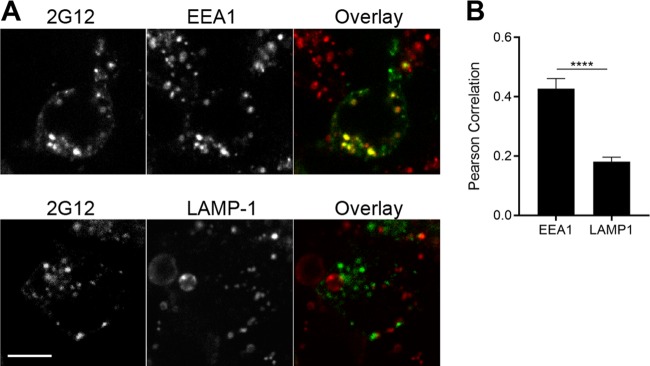
Specifically, the native Env trimer mainly exists in an untriggered “closed” conformation (state 1). The interaction with the CD4 receptor lowers the energy barrier to reach the “open” states 2/3 (34, 36). Natural HIV-1 infection elicits mainly nNAbs, which poorly recognize the “closed” Env and are only able to recognize highly conserved epitopes exposed upon the “opening” of Env (8, 48). Despite poor neutralizing activity, these nNAbs have been shown to exert a constant selection pressure and alter the course of HIV-1-infection in vivo (49–51) and can mediate potent ADCC activity against cells presenting Env in its “open” CD4-bound conformation (7, 9, 38, 52). It is interesting to note that the nNAbs used in this study can form a stable complex with Env on the cell surface for a prolonged amount of time. Conversely, the bNAbs tested induced faster Env internalization rates. Since these bNAbs recognize the “closed” conformation of Env, these results suggest that Env sampling its State 1 conformation might be located in discrete membrane microdomains that could be prone to antibody-mediated internalization. Additional studies are required to explore this interesting possibility. Of note, bNAb-Env complexes do not appear to follow the degradative pathway since the intracellular compartments where they accumulate over time are negative for the lysosomal marker Lamp1 (Fig. 10). Rather, Env accumulated in endosomes positive for early endosome marker EEA1 (Fig. 10). It is intriguing to speculate that this endocytic pathway could be related to the observed role for recycling endosomes in the incorporation of Env into budding virions (53–55).
FIG 10.
bNAb-Env complexes accumulate in an early endosome compartment. 293T cells were transfected with a codon-optimized JR-FL Env plasmid. At 48 h posttransfection, the cells were incubated with Alexa Fluor 488-conjugated 2G12 for 180 min. (A) Cells were then fixed, permeabilized, and stained for endogenous EEA1 or LAMP1 proteins, followed by Alexa Fluor 568-conjugated secondary antibodies. Representative images are shown. (B) Colocalization was quantified for 20 cells per condition using the Pearson correlation. Values shown represent means ± the SEM. Scale bar, 10 μm. Statistical significance was tested using an unpaired t test (****, P < 0.001).
Previous reports have shown that humoral immune responses, such as antibody-dependent complement-mediated lysis, are decreased by internalization of surface-expressed viral glycoproteins upon the binding of antibodies (22, 23). We show here that this also applies to ADCC. The surprising differences observed between bNAbs and nNAbs studied here suggest that different Env populations sampling different conformations coexist at the surfaces of infected cells. Envs sampling the “closed” conformation could potentially facilitate the elicitation of bNAbs. Therefore, it is tempting to speculate that the virus minimizes the exposure of Env sampling the “closed” conformation while tolerating the exposure of limited amounts of Env in the “open” conformation. The “open” Env could act as a decoy, since it is well established that this conformation fails to elicit bNAbs. The nNAbs that are elicited instead fail to neutralize viral particles or mediate ADCC against wild type-infected cells (9–11, 43, 56, 57).
It is becoming increasingly clear that several factors contribute to ADCC responses against HIV-1-infected cells, i.e., Env conformation (38), CD4 (8, 9, 11) and BST-2 (12–14) downregulation, gp120 shedding (37, 59), and the stability of Env-Ab complexes at the cell surface (60), which are driven by the affinity of Abs for Env (61), Env internalization (15), cell surface expression of stress ligands (56, 62), and now antibody-induced Env internalization. Additional work will be required to tease apart the differential contribution of each of these factors in ADCC. Further dissecting the mechanisms underlying antibody-induced Env internalization might help in the development of new generations of bNAbs that are able to efficiently eliminate HIV-1-infected cells.
MATERIALS AND METHODS
Plasmids and cell lines.
293T human embryonic kidney cells (American Type Culture Collection) were maintained at 37°C under 5% CO2 in Dulbecco modified Eagle medium (Invitrogen) containing 5% fetal bovine serum (Sigma) and 100 μg/ml of penicillin-streptomycin (Mediatech). The E168K mutation was introduced into the previously described pcDNA3.1 expressing codon-optimized HIV-1 JRFL envelope glycoproteins (34) using a QuikChange II XL site-directed mutagenesis protocol (Stratagene). Other plasmids used to transfect 293T cells include pcDNA3.1 human CD4 expressor and pIRES-GFP vector (9, 43). The pCAGGS codon-optimized JR-FL gp160 plasmid was kindly provided by Joseph Sodroski (63). The pECFP lamin B receptor plasmid was kindly provided by Melissa Rolls and Tom Rapoport (64).
Isolation of primary cells, viral production, and infections.
CD4+ T lymphocytes were purified from resting PBMCs by negative selection and activated as previously described (8, 9). Briefly, PBMC were obtained by leukapheresis from at least five different healthy HIV-uninfected individuals. CD4+ T lymphocytes were purified by negative selection using immunomagnetic beads as per the manufacturer’s instructions (StemCell Technologies). CD4+ T lymphocytes were activated with phytohemagglutinin-L (10 μg/ml) for 48 h and then maintained in RPMI 1640 complete medium supplemented with rIL-2 (100 U/ml).
To ensure similar levels of infection between viruses, vesicular stomatitis virus G-pseudotyped viruses were produced and titrated as described previously (8). Viruses were used to infect activated primary CD4 T cells from healthy HIV-1-negative donors by spin infection at 800 × g for 1 h in 96-well plates at 25°C.
Antibodies.
Anti-HIV-1 gp120 MAbs recognizing CD4-induced epitopes (A32 and 17b; obtained from the NIH AIDS Reagent Program), the outer domain (2G12; obtained from the NIH AIDS Reagent Program), and the gp120-gp41 interface (PGT151; obtained from International AIDS Vaccine Initiative [IAVI]) were conjugated with Alexa Fluor 488 or Alexa Fluor 594 (Thermo Fisher Scientific) according to the manufacturer’s protocol and used for confocal microscopy analyses of cell surface staining of 293T-transfected cells. In addition, the following anti-Env MAbs were also used for cell surface staining: PGT126, PGT121 (IAVI), PG9 (Polymun), N5-i5, and N12-i2. Fab fragments were generated by papain digest of the corresponding antibody. Briefly, purified IgGs were incubated at 37°C overnight with immobilized papain (G Biosciences) and then filtered to remove the papain. Fabs were separated from undigested IgG and Fc by passage over a HiTrap protein A column (GE Healthcare) equilibrated with phosphate-buffered saline (PBS; pH 7.2). Flowthrough fractions were concentrated and further purified by gel filtration chromatography on a Superdex 200 gel filtration column (GE Healthcare) equilibrated in 25 mM Tris-HCl (pH 8.5) and 150 mM sodium chloride. Fab fractions with elution times roughly correlating to 50 kDa were combined and concentrated prior to use. Goat anti-human Alexa Fluor 647-conjugated secondary Ab (Thermo Fisher Scientific) was used to determine overall antibody binding and AquaVivid (Thermo Fisher Scientific) as a viability dye. Mouse monoclonal EEA1 (14/EEA1) and LAMP1 (H4A3) antibodies were obtained from BD Transduction Laboratories. Alexa Fluor 568-conjugated goat anti-mouse was obtained from Thermo Fisher Scientific.
Flow cytometry analysis of cell surface staining.
Cell surface staining was performed as previously described (8, 9). Cells infected with HIV-1 primary isolates were identified by intracellular staining of HIV-1 Gag using the Cytofix/Cytoperm fixation/permeabilization kit (BD Biosciences) and the PE-conjugated anti-Gag MAb, clone KC57 (Beckman Coulter). The percentage of infected cells (Gag+ or GFP+ cells) was determined by gating the living cell population based on viability dye staining with AquaVivid (Thermo Fisher Scientific). For the cell surface staining of transfected 293T cells, 3 × 105 293T cells were transfected by the calcium phosphate method with the Env-expressing plasmids, along with a pIRES-GFP vector, at a ratio of 2 μg of pcDNA3.1 or JRFL Env to 0.5 μg of GFP. At 16 h posttransfection, cells were washed with fresh medium, and cell surface staining was carried out 24 h later. Samples were analyzed on an LSRII cytometer (BD Biosciences), and data analysis was performed using FlowJo vX.0.7 (Tree Star).
Antibody-induced internalization assay.
48 h postinfection or posttransfection, HIV-1-infected primary CD4+ T cells or Env-transfected 293T cells, respectively, were incubated with 5 μg/ml anti-Env antibodies for 30 min at 4°C. Excess antibodies were washed three times with cold PBS. This was followed by incubation at 37°C in complete media to start the internalization process. After different time points, cells were fixed with 2% paraformaldehyde. For flow cytometry analyses, to visualize remaining antigen-antibody complexes on the cell surface, cells were stained with a goat anti-human conjugated with Alexa Fluor-647 secondary Ab (Thermo Fisher Scientific). As a control, cells were fixed after 4°C incubation (time point, 0 min). Dead cells were excluded by staining the cells with live/dead fixable AquaVivid stain (Thermo Fisher Scientific). The reduction in surface expression for a given time point was normalized by using the following equation: [(mean fluorescence intensity at x min)/(mean fluorescence intensity at 0 min)] × 100.
For confocal microscopy analyses, 293T cells were plated in 10-mm MatTek dishes with #0 coverslip bottoms. 293T cells were transfected with 1 μg of pCAGGS JR-FL Env (codon-optimized) plasmid and 100 ng of lamin B receptor-CFP plasmid (to locate the nuclear envelope) with or without 1 μg of human CD4 plasmid using Fugene 6 reagent according to the manufacturer’s instructions. At 48 h posttransfection, the cells were incubated with prelabeled antibodies diluted 1:250 in fresh medium for the indicated times, washed once with PBS plus 0.5% bovine serum albumin (BSA), and fixed with PBS plus 4% paraformaldehyde for 10 min. Cells were then placed in PBS plus 0.01% sodium azide prior to imaging. For the 0-min control, cells were fixed prior to antibody staining for 1 h in PBS plus 0.5% BSA. For endosomal staining, cells were first incubated with prelabeled 2G12 antibody for 180 min, followed by fixation, permeabilization (5 min with 0.1% Triton X-100), and staining for endogenous EEA1 or LAMP1.
Internalization inhibition assay.
To inhibit antibody-induced internalization, a dynamin inhibitor was used before and during the internalization assay. Myristoylated dynamin inhibitory peptide (DIP; Tocris Bioscience) was diluted in water prior to dilution in serum-free cell culture medium. After 1 h of pretreatment of infected or transfected cells with 50 and 40 μM DIP, respectively, at 37°C, fresh inhibitor was added to the cells, together with antibodies, to allow for their attachment and then during incubations at 37°C to induce their internalization. As a control, 20 μg/ml of biotinylated transferrin was used to confirm the effectiveness of DIP, and this was visualized with streptavidin-Alexa Fluor 647 (data not shown).
Antibody-dependent cellular cytotoxicity assay.
Measurement of ADCC using the FACS-based assay was performed at 48 h postinfection as previously described (9, 47). Briefly, infected primary CD4+ T cells were stained with AquaVivid viability dye and cell proliferation dye (eFluor670; eBioscience) and used as target cells. Next, the target cells were treated with and without 50 μM DIP for 1 h at 37°C. Autologous PBMC effector cells, stained with another cellular marker (cell proliferation dye eFluor450; eBioscience), were added at an effector/target ratio of 10:1 in 96-well V-bottom plates (Corning). DIP was washed out before incubating the infected cells with effector cells to guarantee that the drug affected only the target cells and not the effector cells. Then, 5 μg/ml concentrations of MAbs were added to appropriate wells, and cells were incubated for 15 min at room temperature. Subsequently the plates were centrifuged for 1 min at 300 × g and incubated at 37°C and 5% CO2 for 5 h before being fixed in a 2% PBS-formaldehyde solution. Samples were acquired on an LSRII cytometer (BD Biosciences), and data analysis was performed using FlowJo vX.0.7 (Tree Star). The percentage of cytotoxicity was calculated using the following formula: (% of GFP+ cells in targets plus effectors) − (% of GFP+ cells in targets plus effectors plus MAbs)/(% of GFP+ cells in targets) by gating on infected live target cells.
Confocal microscopy.
Confocal imaging was performed using a Nikon Eclipse TE2000-E microscope equipped with 444-, 488-, and 561-nm lasers, a Yokogawa CSU 10 spinning disc confocal laser scanning unit, and an Andor Zyla sCMOS camera. Images were analyzed manually using ImageJ (65). Briefly, regions of interest were drawn around entire cells and cytoplasmic regions. Total fluorescence for regions of interest were calculated as the area × (mean – minimum). Surface fluorescence was calculated as the total fluorescence minus the cytoplasmic fluorescence. Values are represented as surface/total and cytoplasmic/total ratios, normalized to the 0-min time point. The average intensity and area were measured and used to calculate the total fluorescence and the cytoplasmic fluorescence. The minimum intensity was subtracted from the mean intensity to correct for cytoplasmic background fluorescence. Colocalization between Env and endosomal markers was quantified with the Pearson correlation function using the JACoP plugin (66) for ImageJ.
Statistical analyses.
Statistics were analyzed using Prism v6.01 (GraphPad, San Diego, CA). Every data set was tested for statistical normality, and this information was used to apply the appropriate (parametric or nonparametric) statistical test. P values of <0.05 were considered significant; significance values are indicated by asterisks in the figures (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
ACKNOWLEDGMENTS
This study was supported by a CIHR foundation grant 352417 to A.F., by NIH grant R01 AI129769 to M.P. and A.F., by NIH grants R01 GM116654 and P01-GM56550 to W.M., by NIH grant R01 AI116274 to M.P., and by NIH grant R01 AI121135 to D.T.E. A.F. is the recipient of a Canada Research Chair on Retroviral Entry. J.P. is the recipient of a CIHR doctoral fellowship. J.R. is the recipient of an amfAR Mathilde Krim Fellowship in Basic Biomedical Research, and S.D. is a recipient of an FRQS postdoctoral fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors have no conflicts of interest to report.
The views expressed in this presentation are those of the authors and do not reflect the official policy or position of the Uniformed Services University, U.S. Army, the Department of Defense, or the U.S. Government.
REFERENCES
- 1.Baum LL, Cassutt KJ, Knigge K, Khattri R, Margolick J, Rinaldo C, Kleeberger CA, Nishanian P, Henrard DR, Phair J. 1996. HIV-1 gp120-specific antibody-dependent cell-mediated cytotoxicity correlates with rate of disease progression. J Immunol 157:2168–2173. [PubMed] [Google Scholar]
- 2.Chung AW, Isitman G, Navis M, Kramski M, Center RJ, Kent SJ, Stratov I. 2011. Immune escape from HIV-specific antibody-dependent cellular cytotoxicity (ADCC) pressure. Proc Natl Acad Sci U S A 108:7505–7510. doi: 10.1073/pnas.1016048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forthal DN, Landucci G, Haubrich R, Keenan B, Kuppermann BD, Tilles JG, Kaplan J. 1999. Antibody-dependent cellular cytotoxicity independently predicts survival in severely immunocompromised human immunodeficiency virus-infected patients. J Infect Dis 180:1338–1341. doi: 10.1086/314988. [DOI] [PubMed] [Google Scholar]
- 4.Mabuka J, Nduati R, Odem-Davis K, Peterson D, Overbaugh J. 2012. HIV-specific antibodies capable of ADCC are common in breastmilk and are associated with reduced risk of transmission in women with high viral loads. PLoS Pathog 8:e1002739. doi: 10.1371/journal.ppat.1002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun Y, Asmal M, Lane S, Permar SR, Schmidt SD, Mascola JR, Letvin NL. 2011. Antibody-dependent cell-mediated cytotoxicity in simian immunodeficiency virus-infected rhesus monkeys. J Virol 85:6906–6912. doi: 10.1128/JVI.00326-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prevost J, Richard J, Ding S, Pacheco B, Charlebois R, Hahn BH, Kaufmann DE, Finzi A. 2018. Envelope glycoproteins sampling states 2/3 are susceptible to ADCC by sera from HIV-1-infected individuals. Virology 515:38–45. doi: 10.1016/j.virol.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Veillette M, Coutu M, Richard J, Batraville LA, Dagher O, Bernard N, Tremblay C, Kaufmann DE, Roger M, Finzi A. 2015. The HIV-1 gp120 CD4-bound conformation is preferentially targeted by antibody-dependent cellular cytotoxicity-mediating antibodies in sera from HIV-1-infected individuals. J Virol 89:545–551. doi: 10.1128/JVI.02868-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veillette M, Desormeaux A, Medjahed H, Gharsallah NE, Coutu M, Baalwa J, Guan Y, Lewis G, Ferrari G, Hahn BH, Haynes BF, Robinson JE, Kaufmann DE, Bonsignori M, Sodroski J, Finzi A. 2014. Interaction with cellular CD4 exposes HIV-1 envelope epitopes targeted by antibody-dependent cell-mediated cytotoxicity. J Virol 88:2633–2644. doi: 10.1128/JVI.03230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alsahafi N, Ding S, Richard J, Markle T, Brassard N, Walker B, Lewis GK, Kaufmann DE, Brockman MA, Finzi A. 2016. Nef proteins from HIV-1 elite controllers are inefficient at preventing antibody-dependent cellular cytotoxicity. J Virol 90:2993–3002. doi: 10.1128/JVI.02973-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prevost J, Richard J, Medjahed H, Alexander A, Jones J, Kappes JC, Ochsenbauer C, Finzi A. 2018. Incomplete downregulation of CD4 expression affects HIV-1 Env conformation and antibody-dependent cellular cytotoxicity responses. J Virol 92:e00484-18. doi: 10.1128/JVI.00484-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arias JF, Heyer LN, von Bredow B, Weisgrau KL, Moldt B, Burton DR, Rakasz EG, Evans DT. 2014. Tetherin antagonism by Vpu protects HIV-infected cells from antibody-dependent cell-mediated cytotoxicity. Proc Natl Acad Sci U S A 111:6425–6430. doi: 10.1073/pnas.1321507111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alvarez RA, Hamlin RE, Monroe A, Moldt B, Hotta MT, Rodriguez Caprio G, Fierer DS, Simon V, Chen BK. 2014. HIV-1 Vpu antagonism of tetherin inhibits antibody-dependent cellular cytotoxic responses by natural killer cells. J Virol 88:6031–6046. doi: 10.1128/JVI.00449-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richard J, Prevost J, von Bredow B, Ding S, Brassard N, Medjahed H, Coutu M, Melillo B, Bibollet-Ruche F, Hahn BH, Kaufmann DE, Smith AB III, Sodroski J, Sauter D, Kirchhoff F, Gee K, Neil SJ, Evans DT, Finzi A. 2017. BST-2 expression modulates small CD4-mimetic sensitization of HIV-1-infected cells to antibody-dependent cellular cytotoxicity. J Virol 91. doi: 10.1128/JVI.00219-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Bredow B, Arias JF, Heyer LN, Gardner MR, Farzan M, Rakasz EG, Evans DT. 2015. Envelope glycoprotein internalization protects human and simian immunodeficiency virus infected cells from antibody-dependent cell-mediated cytotoxicity. J Virol 89:10648–10655. doi: 10.1128/JVI.01911-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boge M, Wyss S, Bonifacino JS, Thali M. 1998. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J Biol Chem 273:15773–15778. doi: 10.1074/jbc.273.25.15773. [DOI] [PubMed] [Google Scholar]
- 17.Byland R, Vance PJ, Hoxie JA, Marsh M. 2007. A conserved dileucine motif mediates clathrin and AP-2-dependent endocytosis of the HIV-1 envelope protein. Mol Biol Cell 18:414–425. doi: 10.1091/mbc.e06-06-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blot G, Janvier K, Le Panse S, Benarous R, Berlioz-Torrent C. 2003. Targeting of the human immunodeficiency virus type 1 envelope to the trans-Golgi network through binding to TIP47 is required for env incorporation into virions and infectivity. J Virol 77:6931–6945. doi: 10.1128/JVI.77.12.6931-6945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bu Z, Ye L, Vzorov A, Taylor D, Compans RW, Yang C. 2004. Enhancement of immunogenicity of an HIV Env DNA vaccine by mutation of the Tyr-based endocytosis motif in the cytoplasmic domain. Virology 328:62–73. doi: 10.1016/j.virol.2004.06.041. [DOI] [PubMed] [Google Scholar]
- 20.Fultz PN, Vance PJ, Endres MJ, Tao B, Dvorin JD, Davis IC, Lifson JD, Montefiori DC, Marsh M, Malim MH, Hoxie JA. 2001. In vivo attenuation of simian immunodeficiency virus by disruption of a tyrosine-dependent sorting signal in the envelope glycoprotein cytoplasmic tail. J Virol 75:278–291. doi: 10.1128/JVI.75.1.278-291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Meulen KM, Nauwynck HJ, Pensaert MB. 2003. Absence of viral antigens on the surface of equine herpesvirus-1-infected peripheral blood mononuclear cells: a strategy to avoid complement-mediated lysis. J Gen Virol 84:93–97. doi: 10.1099/vir.0.18864-0. [DOI] [PubMed] [Google Scholar]
- 22.Van de Walle GR, Favoreel HW, Nauwynck HJ, Pensaert MB. 2003. Antibody-induced internalization of viral glycoproteins and gE-gI Fc receptor activity protect pseudorabies virus-infected monocytes from efficient complement-mediated lysis. J Gen Virol 84:939–948. doi: 10.1099/vir.0.18663-0. [DOI] [PubMed] [Google Scholar]
- 23.Favoreel HW, Van Minnebruggen G, Van de Walle GR, Ficinska J, Nauwynck HJ. 2006. Herpesvirus interference with virus-specific antibodies: bridging antibodies, internalizing antibodies, and hiding from antibodies. Vet Microbiol 113:257–263. doi: 10.1016/j.vetmic.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Vogt C, Eickmann M, Diederich S, Moll M, Maisner A. 2005. Endocytosis of the Nipah virus glycoproteins. J Virol 79:3865–3872. doi: 10.1128/JVI.79.6.3865-3872.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popa A, Carter JR, Smith SE, Hellman L, Fried MG, Dutch RE. 2012. Residues in the Hendra virus fusion protein transmembrane domain are critical for endocytic recycling. J Virol 86:3014–3026. doi: 10.1128/JVI.05826-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moll M, Klenk HD, Herrler G, Maisner A. 2001. A single amino acid change in the cytoplasmic domains of measles virus glycoproteins H and F alters targeting, endocytosis, and cell fusion in polarized Madin-Darby canine kidney cells. J Biol Chem 276:17887–17894. doi: 10.1074/jbc.M010183200. [DOI] [PubMed] [Google Scholar]
- 27.Leemans A, De Schryver M, Van der Gucht W, Heykers A, Pintelon I, Hotard AL, Moore ML, Melero JA, McLellan JS, Graham BS, Broadbent L, Power UF, Caljon G, Cos P, Maes L, Delputte P. 2017. Antibody-induced internalization of the human respiratory syncytial virus fusion protein. J Virol 91:e00184-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dewerchin HL, Cornelissen E, Nauwynck HJ. 2006. Feline infectious peritonitis virus-infected monocytes internalize viral membrane-bound proteins upon antibody addition. J Gen Virol 87:1685–1690. doi: 10.1099/vir.0.81692-0. [DOI] [PubMed] [Google Scholar]
- 29.Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang CH, McBride R, von Bredow B, Shivatare SS, Wu CY, Chan-Hui PY, Liu Y, Feizi T, Zwick MB, Koff WC, Seaman MS, Swiderek K, Moore JP, Evans D, Paulson JC, Wong CH, Ward AB, Wilson IA, Sanders RW, Poignard P, Burton DR. 2014. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity 40:657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, Wilson IA. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 31.Julien JP, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ, Sanders RW, Moore JP, Wilson IA, Ward AB. 2013. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci U S A 110:4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anand SP, Prevost J, Baril S, Richard J, Medjahed H, Chapleau JP, Tolbert WD, Kirk S, Smith AB III, Wines BD, Kent SJ, Hogarth PM, Parsons MS, Pazgier M, Finzi A. 2019. Two families of Env antibodies efficiently engage Fc-gamma receptors and eliminate HIV-1-infected cells. J Virol 93:e01823-18. doi: 10.1128/JVI.01823-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herschhorn A, Ma X, Gu C, Ventura JD, Castillo-Menendez L, Melillo B, Terry DS, Smith AB III, Blanchard SC, Munro JB, Mothes W, Finzi A, Sodroski J. 2016. Release of gp120 restraints leads to an entry-competent intermediate state of the HIV-1 envelope glycoproteins. mBio 7:e01598-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma X, Lu M, Gorman J, Terry DS, Hong X, Zhou Z, Zhao H, Altman RB, Arthos J, Blanchard SC, Kwong PD, Munro JB, Mothes W. 2018. HIV-1 Env trimer opens through an asymmetric intermediate in which individual protomers adopt distinct conformations. Elife 7:e34271. doi: 10.7554/eLife.34271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB III, Kwong PD, Blanchard SC, Mothes W. 2014. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richard J, Prevost J, Baxter AE, von Bredow B, Ding S, Medjahed H, Delgado GG, Brassard N, Sturzel CM, Kirchhoff F, Hahn BH, Parsons MS, Kaufmann DE, Evans DT, Finzi A. 2018. Uninfected bystander cells impact the measurement of HIV-specific antibody-dependent cellular cytotoxicity responses. mBio 9:e00358-18. doi: 10.1128/mBio.00358-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richard J, Prevost J, Alsahafi N, Ding S, Finzi A. 2017. Impact of HIV-1 envelope conformation on ADCC responses. Trends Microbiol 26:253–265. doi: 10.1016/j.tim.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 39.Veillette M, Richard J, Pazgier M, Lewis GK, Parsons MS, Finzi A. 2016. Role of HIV-1 envelope glycoproteins conformation and accessory proteins on ADCC responses. Curr HIV Res 14:9–23. [DOI] [PubMed] [Google Scholar]
- 40.Forthal DN, Finzi A. 2018. Antibody-dependent cellular cytotoxicity in HIV infection. AIDS 32:2439–2451. doi: 10.1097/QAD.0000000000002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reference deleted.
- 42.Richard J, Pacheco B, Gohain N, Veillette M, Ding S, Alsahafi N, Tolbert WD, Prevost J, Chapleau JP, Coutu M, Jia M, Brassard N, Park J, Courter JR, Melillo B, Martin L, Tremblay C, Hahn BH, Kaufmann DE, Wu X, Smith AB III, Sodroski J, Pazgier M, Finzi A. 2016. Co-receptor binding site antibodies enable CD4-mimetics to expose conserved anti-cluster A ADCC epitopes on HIV-1 envelope glycoproteins. EbioMedicine 12:208–218. doi: 10.1016/j.ebiom.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veillette M, Coutu M, Richard J, Batraville LA, Desormeaux A, Roger M, Finzi A. 2014. Conformational evaluation of HIV-1 trimeric envelope glycoproteins using a cell-based ELISA assay. J Vis Exp 2014:51995. doi: 10.3791/51995:51995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferguson SM, De Camilli P. 2012. Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol 13:75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grabs D, Slepnev VI, Songyang Z, David C, Lynch M, Cantley LC, De Camilli P. 1997. The SH3 domain of amphiphysin binds the proline-rich domain of dynamin at a single site that defines a new SH3 binding consensus sequence. J Biol Chem 272:13419–13425. doi: 10.1074/jbc.272.20.13419. [DOI] [PubMed] [Google Scholar]
- 46.Falgairolle M, O’Donovan MJ. 2015. Pharmacological investigation of Fluoro-Gold entry into spinal neurons. PLoS One 10:e0131430. doi: 10.1371/journal.pone.0131430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richard J, Veillette M, Batraville LA, Coutu M, Chapleau JP, Bonsignori M, Bernard N, Tremblay C, Roger M, Kaufmann DE, Finzi A. 2014. Flow cytometry-based assay to study HIV-1 gp120 specific antibody-dependent cellular cytotoxicity responses. J Virol Methods 208:107–114. doi: 10.1016/j.jviromet.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 48.Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med 201:1407–1419. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwong PD, Mascola JR. 2012. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity 37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horwitz JA, Bar-On Y, Lu CL, Fera D, Lockhart AAK, Lorenzi JCC, Nogueira L, Golijanin J, Scheid JF, Seaman MS, Gazumyan A, Zolla-Pazner S, Nussenzweig MC. 2017. Non-neutralizing antibodies alter the course of HIV-1 infection in vivo. Cell 170:637–648. doi: 10.1016/j.cell.2017.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moody MA, Gao F, Gurley TC, Amos JD, Kumar A, Hora B, Marshall DJ, Whitesides JF, Xia SM, Parks R, Lloyd KE, Hwang KK, Lu X, Bonsignori M, Finzi A, Vandergrift NA, Alam SM, Ferrari G, Shen X, Tomaras GD, Kamanga G, Cohen MS, Sam NE, Kapiga S, Gray ES, Tumba NL, Morris L, Zolla-Pazner S, Gorny MK, Mascola JR, Hahn BH, Shaw GM, Sodroski JG, Liao HX, Montefiori DC, Hraber PT, Korber BT, Haynes BF. 2015. Strain-specific V3 and CD4 binding site autologous HIV-1 neutralizing antibodies select neutralization-resistant viruses. Cell Host Microbe 18:354–362. doi: 10.1016/j.chom.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ding S, Tolbert WD, Prevost J, Pacheco B, Coutu M, Debbeche O, Xiang SH, Pazgier M, Finzi A. 2016. A highly conserved gp120 inner domain residue modulates env conformation and trimer stability. J Virol 90:8395–8409. doi: 10.1128/JVI.01068-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kirschman J, Qi M, Ding L, Hammonds J, Dienger-Stambaugh K, Wang JJ, Lapierre LA, Goldenring JR, Spearman P. 2018. HIV-1 envelope glycoprotein trafficking through the endosomal recycling compartment is required for particle incorporation. J Virol 92:01893-17. doi: 10.1128/JVI.01893-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qi M, Chu H, Chen X, Choi J, Wen X, Hammonds J, Ding L, Hunter E, Spearman P. 2015. A tyrosine-based motif in the HIV-1 envelope glycoprotein tail mediates cell-type- and Rab11-FIP1C-dependent incorporation into virions. Proc Natl Acad Sci U S A 112:7575–7580. doi: 10.1073/pnas.1504174112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qi M, Williams JA, Chu H, Chen X, Wang JJ, Ding L, Akhirome E, Wen X, Lapierre LA, Goldenring JR, Spearman P. 2013. Rab11-FIP1C and Rab14 direct plasma membrane sorting and particle incorporation of the HIV-1 envelope glycoprotein complex. PLoS Pathog 9:e1003278. doi: 10.1371/journal.ppat.1003278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alsahafi N, Richard J, Prevost J, Coutu M, Brassard N, Parsons MS, Kaufmann DE, Brockman M, Finzi A. 2017. Impaired downregulation of NKG2D ligands by Nef protein from elite controllers sensitizes HIV-1-infected cells to ADCC. J Virol 91:e00109-17. doi: 10.1128/JVI.00109-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding S, Veillette M, Coutu M, Prevost J, Scharf L, Bjorkman PJ, Ferrari G, Robinson JE, Sturzel C, Hahn BH, Sauter D, Kirchhoff F, Lewis GK, Pazgier M, Finzi A. 2016. A highly conserved residue of the HIV-1 gp120 inner domain is important for antibody-dependent cellular cytotoxicity responses mediated by anti-cluster A antibodies. J Virol 90:2127–2134. doi: 10.1128/JVI.02779-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reference deleted.
- 59.Richard J, Veillette M, Ding S, Zoubchenok D, Alsahafi N, Coutu M, Brassard N, Park J, Courter JR, Melillo B, Smith AB III, Shaw GM, Hahn BH, Sodroski J, Kaufmann DE, Finzi A. 2016. Small CD4 mimetics prevent HIV-1 uninfected bystander CD4+ T cell killing mediated by antibody-dependent cell-mediated cytotoxicity. EBioMedicine 3:122–134. doi: 10.1016/j.ebiom.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bruel T, Guivel-Benhassine F, Amraoui S, Malbec M, Richard L, Bourdic K, Donahue DA, Lorin V, Casartelli N, Noel N, Lambotte O, Mouquet H, Schwartz O. 2016. Elimination of HIV-1-infected cells by broadly neutralizing antibodies. Nat Commun 7:10844. doi: 10.1038/ncomms10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ivan B, Sun Z, Subbaraman H, Friedrich N, Trkola A. 2019. CD4 occupancy triggers sequential pre-fusion conformational states of the HIV-1 envelope trimer with relevance for broadly neutralizing antibody activity. PLoS Biol 17:e3000114. doi: 10.1371/journal.pbio.3000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parsons MS, Richard J, Lee WS, Vanderven H, Grant MD, Finzi A, Kent SJ. 2016. NKG2D acts as a coreceptor for natural killer cell-mediated anti-HIV-1 antibody-dependent cellular cytotoxicity. AIDS Res Hum Retroviruses 32:1089–1086. doi: 10.1089/AID.2016.0099. [DOI] [PubMed] [Google Scholar]
- 63.Mao Y, Wang L, Gu C, Herschhorn A, Desormeaux A, Finzi A, Xiang SH, Sodroski JG. 2013. Molecular architecture of the uncleaved HIV-1 envelope glycoprotein trimer. Proc Natl Acad Sci U S A 110:12438–12443. doi: 10.1073/pnas.1307382110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rolls MM, Stein PA, Taylor SS, Ha E, McKeon F, Rapoport TA. 1999. A visual screen of a GFP-fusion library identifies a new type of nuclear envelope membrane protein. J Cell Biol 146:29–44. doi: 10.1083/jcb.146.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bolte S, Cordelieres FP. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]