Abstract
Background
Mammographic breast density, adjusted for age and body mass index, and a polygenic risk score (PRS), comprised of common genetic variation, are both strong risk factors for breast cancer and increase discrimination of risk models. Understanding their joint contribution will be important to more accurately predict risk.
Methods
Using 3628 breast cancer cases and 5126 controls of European ancestry from eight case-control studies, we evaluated joint associations of a 77-single nucleotide polymorphism (SNP) PRS and quantitative mammographic density measures with breast cancer. Mammographic percent density and absolute dense area were evaluated using thresholding software and examined as residuals after adjusting for age, 1/BMI, and study. PRS and adjusted density phenotypes were modeled both continuously (per 1 standard deviation, SD) and categorically. We fit logistic regression models and tested the null hypothesis of multiplicative joint associations for PRS and adjusted density measures using likelihood ratio and global and tail-based goodness of fit tests within the subset of six cohort or population-based studies.
Results
Adjusted percent density (odds ratio (OR) = 1.45 per SD, 95% CI 1.38–1.52), adjusted absolute dense area (OR = 1.34 per SD, 95% CI 1.28–1.41), and the 77-SNP PRS (OR = 1.52 per SD, 95% CI 1.45–1.59) were associated with breast cancer risk. There was no evidence of interaction of the PRS with adjusted percent density or dense area on risk of breast cancer by either the likelihood ratio (P > 0.21) or goodness of fit tests (P > 0.09), whether assessed continuously or categorically. The joint association (OR) was 2.60 in the highest categories of adjusted PD and PRS and 0.34 in the lowest categories, relative to women in the second density quartile and middle PRS quintile.
Conclusions
The combined associations of the 77-SNP PRS and adjusted density measures are generally well described by multiplicative models, and both risk factors provide independent information on breast cancer risk.
Electronic supplementary material
The online version of this article (10.1186/s13058-019-1138-8) contains supplementary material, which is available to authorized users.
Keywords: Breast density, Breast cancer risk, Polygenic risk score, Genetic variation, Risk models
Introduction
Large consortia have identified multiple common genetic susceptibility markers associated with risk of breast cancer [1–4]. Although each single nucleotide polymorphism (SNP) is associated with a small increase in risk, their combined effects are more substantial [5, 6]. Effects of multiple SNPs have been shown to combine multiplicatively, such that the combined effects can be efficiently summarized as polygenic risk scores (PRS) [2, 6]. Based on a 77-SNP PRS, women in the top 10% of the risk distribution have been estimated to have a two-fold risk of breast cancer, relative to those of median PRS, and this risk increases to three-fold for women in the top 1% [6]. In addition, several studies have shown that the PRS is a strong risk factor for young women [7], those with family history [8], BRCA1 and BRCA2 mutation carriers [7, 9–11], and for women with contralateral breast cancer [12]. Use of the PRS has also been shown to increase the discrimination of risk models [4, 13–16]. The PRS therefore has the potential to add information to the established risk factors for breast cancer and improve individualized risk prediction [17].
Understanding joint associations of the PRS with other risk factors is important for accurate risk prediction [6, 17–19]. In the most comprehensive study to examine the joint association of a breast cancer PRS and environmental factors (defined as reproductive, anthropometric, lifestyle factors and exogenous hormones) on risk, based on up to 28,241 and 30,445 controls, most associations were consistent with independent (i.e., multiplicative) associations [17]. This implies that the higher a woman’s genetic risk, the greater the absolute risk associated with environmental risk factors [17, 20].
Mammographic density adjusted for age and BMI is one of the strongest breast cancer risk factors [21, 22] but few studies have examined the joint relationship of mammographic density measures and PRS on breast cancer risk. We previously estimated the contribution of the American College of Radiology (ACR) Breast Imaging Reporting and Data System (BI-RADS) four category density measure and a 76-SNP PRS with breast cancer risk using three studies [19]. We found the PRS and BI-RADS density were independent breast cancer risk factors (with no evidence that their joint association deviated from multiplicative) and that the PRS improved discrimination of the Breast Cancer Surveillance Consortium (BCSC) risk model [11, 19]. However, these prior studies lacked the precision of a quantitative mammographic density measure, did not examine absolute dense area, and had limited power for evaluating interactions. A continuous measure could provide better risk discrimination than a categorical measure such as BI-RADS (which has only four categories) [22] and particularly in the tails of the distributions, where clinical implications will be the greatest; the highest risk women could be offered more intensive screening or interventions while women with lowest risk could have reduced or less frequent screening [18].
Here, we evaluate the joint associations on breast cancer risk of a 77-SNP PRS for breast cancer and quantitative mammographic density measures, including percent density and absolute dense area, adjusted for age and BMI, using data from eight studies in the Breast Cancer Association Consortium (BCAC) [1, 23–27].
Methods
Subjects
The study sample consisted of 3628 cases and 5126 controls of European ancestry from eight studies in BCAC; of these, six studies were population-based, contributing 2439 cases and 3895 controls, and the others were clinic based. Each study had available genotyping information on the 77 SNPs included in the PRS, mammographic density and other breast cancer risk factors. Each study obtained informed consent and had relevant ethics and institutional approvals. A summary of study design, sample sizes, and mammographic and genotyping characteristics is given in Additional file 1: Table S1.
Mammographic density measures
All mammographic density measurements were performed on digitized analogue films using either the Cumulus [28] or Madena [29] programs (Additional file 1: Table S1) which apply a thresholding technique to measure total area of the breast and absolute dense area, from which percent dense area and absolute non-dense area are derived. Absolute dense area values were converted to square centimeters according to the pixel size used in the digitization. Measurements were conducted by observers blind to genotype, case status, and breast cancer risk factor data. For cases, mammograms prior to diagnosis or, when this was not possible, those from the contralateral breast taken at the time of diagnosis were used (Additional file 1: Table S1). The mammographic density measurements were made for both craniocaudal (CC) and mediolateral oblique (MLO) views, which have consistently been shown to be highly correlated (range 0.87–0.90) [30]. All studies have previously contributed to genetic analyses of mammographic density, and similar associations were found across studies [31–34].
Genotyping
The 77 SNPs used to compute the PRS [6] were genotyped for the eight studies either as part of a GWAS (Illumina, Human Hap550) [34] or on a custom Illumina iSelect genotyping array comprising 211,155 SNPs (iCOGS, described in [1]). Quality control was conducted at the study level, as previously described [1, 35]; call rates were > 95% for all SNPs. Thus, 77 SNPs associated with breast cancer and their published odds ratios were used to form the PRS.
Statistical methods
Mammographic density measures were first square root transformed and adjusted for age, 1/BMI and study, as described previously [36, 37], and residuals were used for analyses.
The 77 SNP PRS was calculated as previously described [6, 19]. Briefly, the PRS was derived for each study subject using the formula:
where xk is the number of minor alleles (0, 1 or 2) for SNP k, βk are weights, and n = 77 was the total number of SNPs. Under the assumption of no non-multiplicative interactions, the optimum weights βk are the estimated per-allele log-odds ratios, and we used these weights to derive the 77 SNP PRS as previously described [6]. For missing genotypes or those excluded based on Hardy–Weinberg equilibrium P values < 0.001 (1.1% of the 77 genotypes), we used simple MCMC imputation to assign a probable dosage value based on the other available genotypes and risk factors [38, 39].
Pearson correlation coefficients between the continuously distributed PRS and adjusted mammographic density measures were estimated for controls separately. ORs and 95% confidence intervals (CIs) for breast cancer risk were estimated using logistic regression models and presented as the change in odds per each standard deviation of the adjusted measures (based on using controls [37]). Likelihood ratio statistics were computed to measure the strength of association of density measures, PRS, and their combinations with breast cancer risk; the baseline model for comparison was comprised of age, 1/BMI, and study. Parity, menopausal status, family history, and HT (in postmenopausal women) were also evaluated as confounders of the associations of PRS and adjusted density measures with breast cancer risk.
We estimated interactions between the adjusted mammographic density measures and the PRS and tested their significance using the likelihood ratio test (LRT). To assess the goodness of fit of a model that assumes PRS and mammographic density act multiplicatively on breast cancer risk, we performed a global Hosmer-Lemeshow goodness of fit test using deciles [40] as well as a tail-based goodness of fit test [41] to assess deviations, especially at the extremes of the risk distribution. Although primary analyses used continuous measures of density and PRS (per 1 SD), we also evaluated quintiles of PRS and quartiles of adjusted density measures to be consistent with prior studies [6, 29, 42] as well as allow for ease of interpretation, in particular for those in the lowest quartile of density. Tests of interaction and goodness of fit were performed on the subset of six population-based studies, as done in [17], given the potential for biased estimates of main effects when analyzing non-population-based studies.
Heterogeneity of association across studies was tested by including an interaction term between density measures or PRS and study, using the LRT. Statistical analyses were conducted using SAS 9.4 and R (version 3.3.1). All tests were two-sided and P ≤ 0.05 was considered statistically significant.
Results
The characteristics of the 3628 cases and 5126 controls are described in Table 1 (Additional file 1: Table S2). Cases were more likely to be postmenopausal and to have a family history of breast cancer. Among postmenopausal women, cases were also more likely to have used hormone therapy (HT) (Table 1).
Table 1.
Summary characteristics at time of mammogram and by breast case status for the eight participating studies
| Characteristic | Category | Cases N = 3628 |
Controls N = 5126 |
||
|---|---|---|---|---|---|
| N | % | N | % | ||
| Study type | Population-based or cohort | 2439 | 67.2 | 3895 | 76.0 |
| Hospital-based | 1189 | 32.8 | 1231 | 24.0 | |
| Age (years) | < 50 | 432 | 11.9 | 553 | 10.8 |
| 50–59 | 1150 | 31.7 | 1462 | 28.5 | |
| ≥ 60 | 2046 | 56.4 | 3111 | 60.7 | |
| Parity | Nulliparous | 431 | 12.1 | 582 | 11.5 |
| Parous | 3141 | 87.9 | 4459 | 88.5 | |
| Unknown | 56 | 85 | |||
| Menopausal status | Pre-menopausal | 540 | 15.0 | 878 | 17.4 |
| Post-menopausal | 3058 | 85.0 | 4194 | 82.6 | |
| Unknown | 30 | 54 | |||
| Postmenopausal HT use in post-menopausal women | Ever | 1737 | 59.6 | 2116 | 57.4 |
| Never | 1179 | 40.4 | 1568 | 42.6 | |
| Unknown | 142 | 510 | |||
| BMI (kg/m2) | < 25 | 1542 | 42.9 | 2095 | 41.3 |
| ≥ 25 | 2049 | 57.1 | 2982 | 58.7 | |
| Unknown | 37 | 49 | |||
| Family history breast cancer in first degree relatives | No | 2808 | 81.5 | 3944 | 85.1 |
| Yes | 637 | 18.5 | 688 | 14.9 | |
| Unknown | 183 | 494 | |||
HT hormone therapy
Adjusted percent density (PD) and dense area (DA) measures were positively associated with breast cancer across all studies (Additional file 1: Table S3). For adjusted PD, there was a 1.45-fold increased risk (95% CI, 1.38–1.52) per SD of the adjusted PD (Table 2; χ2 = 156, P < 0.001 compared to baseline model). Further, compared to women with density in the second quartile PD, women in the top quartile had a 64% greater risk and women in the lowest quartile had a 40% lower risk of breast cancer. The associations for adjusted DA were slightly weaker than for PD, but still significant (e.g., OR 1.34 (1.28–1.41) per SD adjusted DA) (Table 2) (Additional file 1: Table S3). Associations were similar but attenuated when using population-based studies alone (Table 2) and did not materially change after adjustment for parity, menopausal status, family history, and HT (in postmenopausal women) [data not shown]. Among the population-based studies, there was some evidence for study heterogeneity (PD phet = 0.08; DA phet = 0.04), largely due to MMHS which had stronger associations compared to the other studies. Removal of MMHS resulted in similar associations of adjusted density measures with breast cancer (data not shown) but reduced heterogeneity (PD phet = 0.42; DA phet = 0.25).
Table 2.
Associations (odds ratios, OR) for adjusted percent density (PD) and dense area (DA) measures with breast cancer risk, with or without polygenic risk score (PRS). All eight studies and restricted to cohort/population-based studies only
| Adjusted density measure* | Full sample | Cohort/population-based studies only | ||||
|---|---|---|---|---|---|---|
| N case/N control | OR (95% CI) | Adj for PRS OR (95% CI) |
N case/N control | OR (95% CI) | Adj for PRS OR (95% CI) |
|
| PD (per 1 SD) | 3628/5126 | 1.45 (1.38, 1.52) | 1.42 (1.36, 1.50) | 2439/3895 | 1.42 (1.34, 1.50) | 1.40 (1.32, 1.48) |
| PD quartiles | ||||||
| 1 | 531/1282 | 0.60 (0.52, 0.70) | 0.61 (0.52, 0.70) | 450/1064 | 0.61 (0.51, 0.72) | 0.62 (0.52, 0.73) |
| 2 (Ref) | 723/1281 | Ref | Ref | 519/984 | Ref | Ref |
| 3 | 946/1282 | 1.25 (1.09, 1.42) | 1.22 (1.06, 1.39) | 615/939 | 1.20 (1.03, 1.41) | 1.18 (1.00, 1.38) |
| 4 | 1428/1281 | 1.64 (1.44, 1.87) | 1.60 (1.40, 1.82) | 855/908 | 1.51 (1.29, 1.76) | 1.48 (1.27, 1.74) |
| DA (per 1 SD) | 3628/5126 | 1.34 (1.28, 1.41) | 1.32 (1.26, 1.39) | 2439/3895 | 1.36 (1.29, 1.44) | 1.35 (1.28, 1.44) |
| DA quartiles | ||||||
| 1 | 530/1283 | 0.56 (0.49, 0.65) | 0.57 (0.50, 0.66) | 427/1040 | 0.58 (0.40, 0.69) | 0.59 (0.50, 0.70) |
| 2 (Ref) | 764/1281 | Ref | Ref | 535/1003 | Ref | Ref |
| 3 | 964/1281 | 1.15 (1.01, 1.31) | 1.15 (1.00, 1.31) | 644/969 | 1.14 (0.98, 1.33) | 1.13 (0.97, 1.33) |
| 4 | 1370/1281 | 1.41 (1.23, 1.61) | 1.38 (1.21, 1.59) | 833/883 | 1.45 (1.24, 1.69) | 1.44 (1.22, 1.69) |
*Residuals from models adjusted for age, 1/BMI, and study
Heterogeneity of density association across population-based studies: PD phet = 0.08; DA phet = 0.04 (Exclusion of MMHS results in PD phet = 0.42; DA phet = 0.25)
SD standard deviation, CI confidence interval, Ref reference group, Adj adjusted
PRS was associated with breast cancer risk both when modeled continuously (OR = 1.52 (1.45–1.59) per SD, Table 3; χ2 = 255, P < 0.001 compared to baseline model), or in quintiles (Table 3). Estimates were similar when adjusted for parity, menopausal status, family history, and HT [data not shown] but slightly stronger when only including population-based studies (Table 3). There was no evidence for heterogeneity by study.
Table 3.
Association of polygenic risk score (PRS) with breast cancer risk and evaluation of confounding due to family history and density measures (adjusted PD and adjusted DA). All eight studies combined and subset to cohort/population-based studies. All models adjusted for age, 1/BMI, and study
| Model | N case/N control | PRS OR (95% CI) |
+Family history OR (95% CI) |
+adjusted PD OR (95% CI) |
+adjusted DA OR (95% CI) |
|---|---|---|---|---|---|
| Overall sample | |||||
| Overall PRS (per 1 SD) | 3628/5126 | 1.52 (1.45, 1.59) | 1.52 (1.44, 1.59) | 1.50 (1.42, 1.57) | 1.50 (1.43, 1.58) |
| PRS quintile | |||||
| 1 | 349/1033 | 0.51 (0.44, 0.60) | 0.51 (0.44, 0.61) | 0.53 (0.45, 0.63) | 0.53 (0.45, 0.63) |
| 2 | 535/1008 | 0.80 (0.69, 0.93) | 0.78 (0.67, 0.92) | 0.80 (0.69, 0.94) | 0.80 (0.69, 0.94) |
| 3 (Ref) | 687/1024 | 1.00 (Ref) | 1.00 (Ref) | 1.00 (Ref) | 1.00 (Ref) |
| 4 | 887/1032 | 1.31 (1.14, 1.50) | 1.31 (1.14, 1.51) | 1.32 (1.14, 1.53) | 1.33 (1.15, 1.53) |
| 5 | 1170/1029 | 1.66 (1.45, 1.90) | 1.65 (1.44, 1.90) | 1.64 (1.43, 1.89) | 1.65 (1.44, 1.91) |
| Cohort/population-based studies only | |||||
| Overall PRS (per 1 SD) | 2439/3895 | 1.56 (1.48, 1.66) | 1.55 (1.47, 1.65) | 1.54 (1.45, 1.63) | 1.55 (1.46, 1.64) |
| PRS quintile | |||||
| 1 | 232/786 | 0.51 (0.42, 0.62) | 0.51 (0.42, 0.62) | 0.53 (0.43, 0.65) | 0.53 (0.43, 0.64) |
| 2 | 361/776 | 0.80 (0.66, 0.95) | 0.79 (0.66, 0.94) | 0.80 (0.67, 0.96) | 0.80 (0.67, 0.96) |
| 3 (Ref) | 464/789 | 1.00 (Ref) | 1.00 (Ref) | 1.00 (Ref) | 1.00 (Ref) |
| 4 | 589/793 | 1.30 (1.10, 1.53) | 1.28 (1.08, 1.52) | 1.29 (1.09, 1.54) | 1.30 (1.10, 1.54) |
| 5 | 793/751 | 1.79 (1.52, 2.11) | 1.76 (1.50, 2.08) | 1.76 (1.49, 2.08) | 1.77 (1.50, 2.09) |
Heterogeneity of PRS association by study: P = 0.84 for population based studies
PRS quintiles: quintile 1, − 1.411 to − 0.014; quintile 2, − 0.015 to 0.280; quintile 3, 0.281 to 0.542; quintile 4, 0.543 to 0.885; quintile 5, 0.886 to 2.583
SD standard deviation, CI confidence interval, Ref reference group, Adj adjusted
PRS and adjusted density measures were only weakly correlated (Pearson correlation 0.06, P < 0.001 for adjusted PD and 0.05, P < 0.001 for adjusted DA using controls). Adjusting for PRS made little change to the association between adjusted density measures and breast cancer risk (e.g., OR per 1 SD for adjusted PD, 1.42, 95% CI 1.36–1.50; Table 2). Similarly, adjustment for density measures had very little impact on the association between PRS and risk (Table 3).
Interactions between adjusted density measures and PRS on breast cancer risk
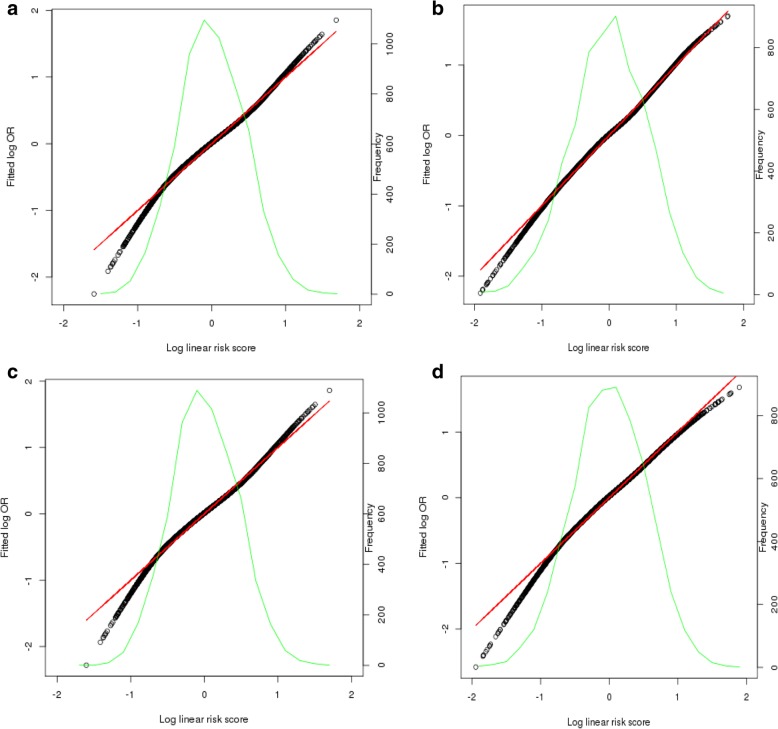
Among the population-based studies, there was no evidence of an interaction between PRS and adjusted PD, whether assessed as continuous (per 1 SD) or categorical (quartiles PD/quintiles PRS) variables; this included evaluation by likelihood ratio tests [[ORint (95% CI) = 0.96 (0.91,1.02), χ2LRT = 1.6, PLRT = 0.21 for continuous and P = 0.42 for categorical] (Fig. 1) and global (P > 0.09) or tail-based (P > 0.23) goodness of fit tests (Fig. 2; Additional file 1: Table S4). Findings were generally similar for adjusted DA and PRS on breast cancer (Additional file 1: Table S4) (Figs. 1 and 2). Results were unchanged when excluding MMHS.
Fig. 1.
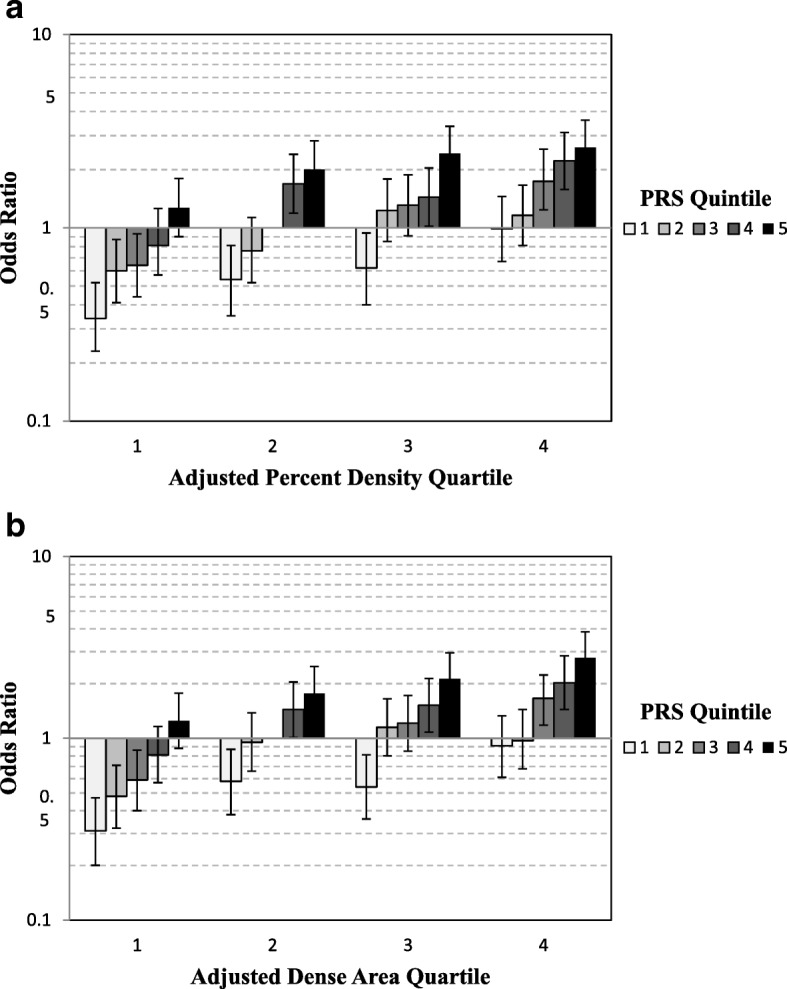
Joint association of quartiles of adjusted density phenotypes and quintiles PRS with breast cancer risk, adjusted for age, 1/BMI, and study. Quartiles adjusted percent density and PRS quintile with breast cancer risk (a). Quartiles of adjusted dense area and PRS quintile with breast cancer risk (b). PRS quintiles: quintile 1, − 1.411 to − 0.014; quintile 2, − 0.015 to 0.280; quintile 3, 0.281 to 0.542; quintile 4, 0.543 to 0.885; quintile 5, 0.886 to 2.583. Reference category is PRS quintile 3 and density quartile 2
Fig. 2.
Tail-based test results from models with continuous adjusted density measures and PRS on breast cancer risk. Population-based studies. Models of adjusted percent density and PRS without interaction (a) and with multiplicative interaction included (b). Models with adjusted dense area and PRS without interaction (c) and with multiplicative interaction included (d)
The multiplicative association can be seen in the similarities of the risk estimates from the joint association of PRS and density measures to the risk expected from the individual main effect models (Tables 2 and 3; Fig. 1). The joint association (OR) was 2.60 in the highest categories of adjusted PD and PRS and 0.34 in the lowest categories relative to the reference category (Fig. 1). These estimates are very similar to the predicted relative risk estimates resulting from multiplying the individual OR associated with each category of PRS and adjusted PD from the main effect models [i.e., OR(fourth quartile PD) × OR(fifth quintile PRS) = 2.70; OR(first quartile PD) × OR(first quintile PRS) = 0.31].
Discussion
This is the first study to have examined the joint association of a 77-SNP PRS and continuous mammographic density measures on breast cancer risk. We found that the combined associations of the PRS and adjusted density measures on breast cancer are well described by a multiplicative model. These results imply that either adjusted percent density or absolute dense area measures can be incorporated as continuous measures into risk models with PRS in a straightforward manner, without interaction terms. Moreover, the two measures are close to uncorrelated; as a result, the risk discrimination afforded by using both measures is much greater than using either alone.
Our findings are consistent with those from the few prior studies evaluating PRS and density measures. Two studies evaluated the contribution of the PRS to the Breast Cancer Surveillance Consortium or BCSC model, which includes the BI-RADS four category density measure [11, 19]. Both studies found significant improvement in the discrimination with the PRS. One of these [19] evaluated an interaction between the BI-RADS density measure and the PRS, finding no departure from a multiplicative model and little correlation between the BI-RADS density and PRS measures. van Veen et al. [43] recently examined an 18-SNP PRS, IBIS 10-year risk, and a visual measure of mammographic density (adjusted for BMI, age) with breast cancer risk among women in a mammography screening practice. Assuming independence between the PRS, IBIS model, and mammographic density, they found that the PRS added substantial information to a model with IBIS risk and mammographic density. They also found only a weak correlation between the PRS, adjusted density, and the IBIS risk model estimate, consistent with our results and those of Vachon et al. [19] Similarly, using data from the Nurses’ Health Studies, Zhang et al. [16] found significant improvement in discrimination when a 67-SNP PRS was added to either the BCRAT model or the Rosner-Colditz model. Addition of a continuous measure of mammographic density also significantly improved the discrimination of both models. However, this paper did not specifically evaluate any interaction between mammographic density and the PRS. Our paper is the first to examine the joint association of continuous density measures with a PRS.
Our findings are consistent with joint effects of PRS with other breast cancer risk factors. Two studies have examined the joint association of a PRS with reproductive variables (age at menarche, parity, age at first birth), alcohol intake, postmenopausal hormone therapy, and BMI on breast cancer. The first involved a 77-SNP PRS examined in up to 28,241 cases and 30,445 controls in BCAC, finding that most associations were consistent with a multiplicative association [17]. An earlier study examined a 24-SNP PRS with the same risk factors, using 17,171 cases and 19,862 controls from the Breast and Prostate Cancer Cohort Consortium (BPC3) and also did not find deviation from the multiplicative model [18].
Our study used quantitative density measures assessed from digitized film mammograms, primarily using the Cumulus software. Although this measure allowed us to evaluate a more precise density measure than the four BI-RADS categories evaluated previously, it is not used in clinical practice as it is not fully automated. Some centers now use commercially available automated density measures such as Volpara which provide a continuous volumetric density in addition to a BI-RADS-like categorical estimate. We and others have shown that these automated measures have a similar ability to predict risk as the BI-RADS clinical density measure and the area-based measures used in the current report [42, 44]. It is likely that our results on the combined effect of the SNP and thresholding density measures presented here will translate to the automated volumetric measures, but this still needs to be evaluated directly.
Since the 77-SNP PRS was established and validated, additional common genetic variants have been identified for breast cancer risk, and these will allow a more informative PRS to be developed [2–4, 35]. The 77 SNPs contributing to the PRS used in this report are estimated to explain 14% of the familial risk, and additional SNPs identified are estimated to explain another 4% [2]. Of the 77 SNPs, nine have been shown to be associated with mammographic density phenotypes but together they explain less than 1.5% of the between-woman variation in these adjusted density traits [31]. Despite this overlap, the PRS is almost uncorrelated with the density measures, and adjustment for PRS resulted in minimal changes in the association between adjusted density measures and breast cancer and vice versa.
Some SNPs have been identified to be more strongly associated with ER-positive or ER-negative disease [2, 4, 45, 46]. The majority of the 77 SNPs in the current PRS are associated with ER-positive breast cancer, with only 27 associated at P < 10− 4 with ER-negative breast cancer [1, 2, 6]. Mammographic density has been shown to be a risk factor for both ER-positive and ER-negative breast cancer and all 9 of the SNPs associated with density are associated with both subtypes [1, 2, 47–49]. Future analyses which consider the joint associations of the subtype-specific PRS and mammographic density with breast cancer risk will be worthwhile.
Important strengths of this analysis include the largest dataset to date to examine the combined associations of PRS and mammographic density on breast cancer risk, the use of quantitative density measures that were standardized across studies, and a common genotyping platform with standard quality control procedures for the majority of studies. We recognize, however, that our results are strictly generalizable to women of European ancestry only. We also note some overlap between our studies with those used to identify the associated SNPs and develop the PRS. However, while this could have led to some overfitting and hence overestimation of the risk gradient for the PRS, this would not have affected the primary conclusion that associations of the PRS and density are almost completely independent and not confounded. Also, two of the largest cohort studies in our sample (MMHS, NHS) consisting of over 1300 cases were not included in the studies used for development of the 77-SNP PRS and estimation of the PRS risk gradients (1.7 and 1.4 per unit SD for PD) were similar to those estimated from the remaining studies. Finally, as noted above, additional work will be necessary to confirm our findings with an updated PRS and novel mammographic density measures [50, 51] as they become available.
Conclusion
In summary, we confirm continuous mammographic density measures and PRS are two of the strongest risk factors for breast cancer and can be included in risk models without interaction terms. Absolute risk associated with higher density will be larger for women at high SNP-based inherited risk.
Additional file
Table S1. Study design and characteristics for eight participating studies. Table S2. Characteristics of cases and non-cases by study (% of non-missing). Table S3. Associations (OR, 95% CI) of adjusted percent density (PD), adjusted dense area (DA) and polygenic risk score (PRS) with breast cancer by study. Adjusted for age and BMI. Table S4. Goodness of fit test P values for joint effect of adjusted* density measures with 77-SNP polygenic risk score (PRS) on breast cancer, based on six population-based studies. PRS and density measures are continuous (per 1 standard deviation, SD). (DOCX 34 kb)
Acknowledgements
We would like to thank the participants and staff of the NHS and NHSII for their valuable contributions as well as the state cancer registries of the following states for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, HI, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, and WY.
Funding
This work was supported by the National Cancer Institute (R01 CA128931, R01 CA122340, R01 CA128978, R01 CA97396, P50 CA116201, R01 CA240386, K24 CA169004, R21 CA179442, P01 CA154292, P01CA87969, R01 CA085265, UM1CA176726, UM1CA186107, and U01 CA164973) and the Breast Cancer Research Foundation. BBCC was supported in part by the ELAN program of the Medical Faculty, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg. Funding for the genotyping of BBCC and MCBCS as well as the iCOGS Illumina array is provided by grants from the EU FP7 programme (COGS) and from Cancer Research UK. Collaborative Oncological Gene-environment Study (COGS) enabled the genotyping for this study. Funding for the BCAC component is provided by grants from the EU FP7 programme (COGS) and from Cancer Research UK.
BCAC is funded by Cancer Research UK [C1287/A16563] and by the European Community’s Seventh Framework Programme under grant agreement n° 223175 (HEALTH-F2-2009-223175) (COGS) and by the European Union’s Horizon 2020 Research and Innovation Programme under grant agreements 633784 (B-CAST) and 634935 (BRIDGES).
The SASBAC study was supported by the Märit and Hans Rausing’s Initiative Against Breast Cancer, the National Institutes of Health (RO1 CA58427), the Agency for Science, Technology and Research (A*STAR; Singapore), and the Swedish Research Council. Jingmei Li is a recipient of a National Research Foundation Singapore Fellowship (NRF-NRFF2017–02).
The Melbourne Collaborative Cohort Study (MCCS) cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian National Health and Medical Research Council grants 209057 and 396414 and by infrastructure provided by Cancer Council Victoria. Cases and their vital status were ascertained through the Victorian Cancer Registry and the Australian Institute of Health and Welfare, including the National Death Index and the Australian Cancer Database.
Availability of data and materials
The datasets generated and/or analyzed during the current study are part of the Breast Cancer Association Consortium and would be available with the appropriate permissions, including an application process and appropriate data transfer agreements.
Abbreviations
- BCAC
Breast Cancer Association Consortium
- BCRAT
Breast Cancer Risk Assessment Tool
- BCSC
Breast Cancer Surveillance Consortium
- BI-RADS
Breast Imaging Reporting and Data System
- BMI
Body mass index
- BPC3
Breast and Prostate Cancer Cohort Consortium
- CC
Craniocaudal
- CI
Confidence interval
- DA
Dense area
- GWAS
Genome-wide association study
- HT
Hormone therapy
- IBIS
International Breast Cancer Intervention Study
- LR
Likelihood-ratio
- MCMC
Markov Chain Monte Carlo
- MLO
Mediolateral oblique
- OR
Odds ratio
- PD
Percent density
- PRS
Polygenic risk score
- SD
Standard deviation
- SNP
Single nucleotide polymorphism
Authors’ contributions
CMV, CS, DFE, NC, and MGC contributed to the conception of design. CMV, CS, RMT, DJT, PAF, JS, MCS, JL, RLM, LB, JL, KC, MKB, QW, JEO, AN, JD, DFE, GM, LLM, CAH, JLH, GGG, RJM, FJC, PH, RJM, and NW contributed to the acquisition of data. CS, SW, DJT, SL, NC, MKB, JLH, JD, DFE, and RL contributed to the design and analysis. CMV, CS, RMT, DJT, PAF, JS, MCS, SW, SL, RLM, LB, JL, KC, QW, ME, NC, JLH, JEO, AN, DFE, GM, LLM, GGG, LH, FJC, and PH interpreted the data: CMV is accountable for all aspects of work. All authors approved the final manuscript.
Ethics approval and consent to participate
Obtained on all studies and statement is included in methods.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Celine M. Vachon, Phone: 507-284-9977, Email: vachon.celine@mayo.edu
Christopher G. Scott, Email: scott.christopher@mayo.edu
Rulla M. Tamimi, Email: nhrmt@channing.harvard.edu
Deborah J. Thompson, Email: djt25@medschl.cam.ac.uk
Peter A. Fasching, Email: Peter.Fasching@uk-erlangen.de
Jennifer Stone, Email: jennifer.stone@uwa.edu.au.
Melissa C. Southey, Email: msouthey@unimelb.edu.au
Stacey Winham, Email: winham.stacey@mayo.edu.
Sara Lindström, Email: saralind@uw.edu.
Jenna Lilyquist, Email: lilyquist.jenna@gmail.com.
Graham G. Giles, Email: graham.giles@cancervic.org.au
Roger L. Milne, Email: roger.milne@cancervic.org.au
Robert J. MacInnis, Email: robert.macinnis@cancervic.org.au
Laura Baglietto, Email: lba@unimelb.edu.au.
Jingmei Li, Email: lijm1@gis.a-star.edu.sg.
Kamila Czene, Email: Kamila.Czene@ki.se.
Manjeet K. Bolla, Email: mkh39@medschl.cam.ac.uk
Qin Wang, Email: qw232@medschl.cam.ac.uk.
Joe Dennis, Email: jgd29@cam.ac.uk.
Lothar Haeberle, Email: lothar.haeberle@uk-erlangen.de.
Mikael Eriksson, Email: Mikael.Eriksson@ki.se.
Peter Kraft, Email: pkraft@hsph.harvard.edu.
Robert Luben, Email: robert.luben@phpc.cam.ac.uk.
Nick Wareham, Email: nick.wareham@mrc-epid.cam.ac.uk.
Janet E. Olson, Email: olsonj@mayo.edu
Aaron Norman, Email: norman.aaron@mayo.edu.
Eric C. Polley, Email: polley.eric@mayo.edu
Gertraud Maskarinec, Email: gertraud@cc.hawaii.edu.
Loic Le Marchand, Email: loic@cc.hawaii.edu.
Christopher A. Haiman, Email: haiman@usc.edu
John L. Hopper, Email: j.hopper@unimelb.edu.au
Fergus J. Couch, Email: couch.fergus@mayo.edu
Douglas F. Easton, Email: dfe20@medschl.cam.ac.uk
Per Hall, Email: per.hall@ki.se.
Nilanjan Chatterjee, Email: nchatte2@jhu.edu.
Montse Garcia-Closas, Email: montserrat.garcia-closas@nih.gov.
References
- 1.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45(4):353–361. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michailidou K, Lindstrom S, Dennis J, Beesley J, Hui S, Kar S, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551(7678):92–94. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milne RL, Kuchenbaecker KB, Michailidou K, Beesley J, Kar S, Lindstrom S, et al. Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat Genet. 2017;49(12):1767–1778. doi: 10.1038/ng.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lilyquist J, Ruddy KJ, Vachon CM, Couch FJ. Common Genetic Variation and Breast Cancer Risk-Past, Present, and Future. Cancer Epidemiol Biomarkers Prev. 2018;27(4):380–94. [DOI] [PMC free article] [PubMed]
- 5.Chatterjee N, Shi J, Garcia-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat Rev Genet. 2016;17(7):392–406. doi: 10.1038/nrg.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mavaddat N, Pharoah PD, Michailidou K, Tyrer J, Brook MN, Bolla MK, et al. Prediction of breast cancer risk based on profiling with common genetic variants. J Natl Cancer Inst. 2015;107(5). [DOI] [PMC free article] [PubMed]
- 7.Dite GS, MacInnis RJ, Bickerstaffe A, Dowty JG, Allman R, Apicella C, et al. Breast cancer risk prediction using clinical models and 77 independent risk-associated SNPs for women aged under 50 years: Australian Breast Cancer Family Registry. Cancer Epidemiol Biomark Prev. 2016;25(2):359–365. doi: 10.1158/1055-9965.EPI-15-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Feng B, Miron A, Chen X, Beesley J, Bimeh E, et al. Breast cancer risk prediction using a polygenic risk score in the familial setting: a prospective study from the Breast Cancer Family Registry and kConFab. Genet Med. 2017;19(1):30–35. doi: 10.1038/gim.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuchenbaecker KB, McGuffog L, Barrowdale D, Lee A, Soucy P, Dennis J, et al. Evaluation of polygenic risk scores for breast and ovarian cancer risk prediction in BRCA1 and BRCA2 mutation carriers. J Natl Cancer Inst. 2017;109(7). [DOI] [PMC free article] [PubMed]
- 10.Muranen TA, Greco D, Blomqvist C, Aittomaki K, Khan S, Hogervorst F, et al. Genetic modifiers of CHEK2*1100delC-associated breast cancer risk. Genet Med. 2017;19(5):599–603. doi: 10.1038/gim.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shieh Y, Hu D, Ma L, Huntsman S, Gard CC, Leung JWT, et al. Joint relative risks for estrogen receptor-positive breast cancer from a clinical model, polygenic risk score, and sex hormones. Breast Cancer Res Treat. 2017;166(2):603–612. doi: 10.1007/s10549-017-4430-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robson ME, Reiner AS, Brooks JD, Concannon PJ, John EM, Mellemkjaer L, et al. Association of common genetic variants with contralateral breast cancer risk in the WECARE study. J Natl Cancer Inst. 2017;109(10). [DOI] [PMC free article] [PubMed]
- 13.Gail MH. Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk. J Natl Cancer Inst. 2008;100(14):1037–1041. doi: 10.1093/jnci/djn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Closas M, Gunsoy NB, Chatterjee N. Combined associations of genetic and environmental risk factors: implications for prevention of breast cancer. J Natl Cancer Inst. 2014;106(11). [DOI] [PMC free article] [PubMed]
- 15.Maas P, Barrdahl M, Joshi AD, Auer PL, Gaudet MM, Milne RL, et al. Breast cancer risk from modifiable and nonmodifiable risk factors among white women in the United States. JAMA Oncol. 2016;2(10):1295–1302. doi: 10.1001/jamaoncol.2016.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Rice M, Tworoger SS, Rosner BA, Eliassen AH, Tamimi RM, et al. Addition of a polygenic risk score, mammographic density, and endogenous hormones to existing breast cancer risk prediction models: a nested case-control study. PLoS Med. 2018;15(9):e1002644. doi: 10.1371/journal.pmed.1002644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudolph A, Song M, Brook MN, Milne RL, Mavaddat N, Michailidou K, et al. Joint associations of a polygenic risk score and environmental risk factors for breast cancer in the Breast Cancer Association Consortium. Int J Epidemiol. 2018;47(2):526–536. doi: 10.1093/ije/dyx242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joshi AD, Lindstrom S, Husing A, Barrdahl M, VanderWeele TJ, Campa D, et al. Additive interactions between susceptibility single-nucleotide polymorphisms identified in genome-wide association studies and breast cancer risk factors in the Breast and Prostate Cancer Cohort Consortium. Am J Epidemiol. 2014;180(10):1018–1027. doi: 10.1093/aje/kwu214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vachon CM, Pankratz VS, Scott CG, Haeberle L, Ziv E, Jensen MR, et al. The contributions of breast density and common genetic variation to breast cancer risk. J Natl Cancer Inst. 2015;107(5). [DOI] [PMC free article] [PubMed]
- 20.Hopper JL, Dite GS, MacInnis RJ, Liao Y, Zeinomar N, Knight JA, et al. Age-specific breast cancer risk by body mass index and familial risk: prospective family study cohort (ProF-SC) Breast Cancer Res. 2018;20(1):132. doi: 10.1186/s13058-018-1056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pettersson A, Graff RE, Ursin G, Santos Silva ID, McCormack V, Baglietto L, et al. Mammographic density phenotypes and risk of breast cancer: a meta-analysis. J Natl Cancer Inst. 2014;106(5). [DOI] [PMC free article] [PubMed]
- 22.Bond-Smith D, Stone J. Methodological Challenges and Updated Findings from a Meta-analysis of the Association between Mammographic Density and Breast Cancer. Cancer Epidemiol Biomarkers Prev. 2019;28(1):22–31. [DOI] [PubMed]
- 23.Bojesen SE, Pooley KA, Johnatty SE, Beesley J, Michailidou K, Tyrer JP, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nature Genet. 2013;45(4):371–384. doi: 10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nature Genet. 2013;45(4):392–398. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, Cox DG, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41(5):579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cox A, Dunning AM, Garcia-Closas M, Balasubramanian S, Reed MW, Pooley KA, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39(3):352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 27.Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39(7):865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 28.Boyd NF, Guo H, Martin LJ, Sun L, Stone J, Fishell E, et al. Mammographic density and the risk and detection of breast cancer. N Engl J Med. 2007;356(3):227–236. doi: 10.1056/NEJMoa062790. [DOI] [PubMed] [Google Scholar]
- 29.Vachon CM, Brandt KR, Ghosh K, Scott CG, Maloney SD, Carston MJ, et al. Mammographic breast density as a general marker of breast cancer risk. Cancer Epidemiol Biomark Prev. 2007;16:43–49. doi: 10.1158/1055-9965.EPI-06-0738. [DOI] [PubMed] [Google Scholar]
- 30.Maskarinec G, Meng L. A case-control study of mammographic densities in Hawaii. Breast Cancer Res Treat. 2000;63(2):153–161. doi: 10.1023/A:1006486319848. [DOI] [PubMed] [Google Scholar]
- 31.Stone J, Thompson DJ, Dos Santos Silva I, Scott C, Tamimi RM, Lindstrom S, et al. Novel associations between common breast cancer susceptibility variants and risk-predicting mammographic density measures. Cancer Res. 2015;75(12):2457–2467. doi: 10.1158/0008-5472.CAN-14-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vachon CM, Scott CG, Fasching PA, Hall P, Tamimi RM, Li J, et al. Common breast cancer susceptibility variants in LSP1 and RAD51L1 are associated with mammographic density measures that predict breast cancer risk. Cancer Epidemiol Biomark Prev. 2012;21(7):1156–1166. doi: 10.1158/1055-9965.EPI-12-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varghese JS, Thompson DJ, Michailidou K, Lindstrom S, Turnbull C, Brown J, et al. Mammographic breast density and breast cancer: evidence of a shared genetic basis. Cancer Res. 2012;72(6):1478–1484. doi: 10.1158/0008-5472.CAN-11-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindstrom S, Vachon CM, Li J, Varghese J, Thompson D, Warren R, et al. Common variants in ZNF365 are associated with both mammographic density and breast cancer risk. Nat Genet. 2011;43(3):185–187. doi: 10.1038/ng.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mavaddat N, Michailidou K, Dennis J, Lush M, Fachal 4, Lee A, et al. Polygenic Risk Scores for Prediction of Breast Cancer and Breast Cancer Subtypes. Am J Hum Genet. 2019;104(1):21-34. [DOI] [PMC free article] [PubMed]
- 36.Krishnan K, Baglietto L, Stone J, McLean C, Southey MC, English DR, et al. Mammographic density and risk of breast cancer by tumor characteristics: a case-control study. BMC Cancer. 2017;17(1):859. doi: 10.1186/s12885-017-3871-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hopper JL. Odds per adjusted standard deviation: comparing strengths of associations for risk factors measured on different scales and across diseases and populations. Am J Epidemiol. 2015;182(10):863–867. doi: 10.1093/aje/kwv193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li KH. Imputation using markov chains. J Stat Comput Simul. 1988;30(1):57–79. doi: 10.1080/00949658808811085. [DOI] [Google Scholar]
- 39.Rubin DB. Multiple imputation for nonresponse in surveys. New York: Wiley; 1987.
- 40.Hosmer DW, Lemeshow S, Sturdivant RX. Applied logistic regression. 3. New York: Wiley; 2013. [Google Scholar]
- 41.Song M, Kraft P, Joshi AD, Barrdahl M, Chatterjee N. Testing calibration of risk models at extremes of disease risk. Biostatistics. 2015;16:143–54. doi: 10.1093/biostatistics/kxu034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brandt KR, Scott CG, Ma L, Mahmoudzadeh AP, Jensen MR, Whaley DH, et al. Comparison of clinical and automated breast density measurements: implications for risk prediction and supplemental screening. Radiology. 2016;279(3):710–719. doi: 10.1148/radiol.2015151261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Veen EM, Brentnall AR, Byers H, Harkness EF, Astley SM, Sampson S, et al. Use of single-nucleotide polymorphisms and mammographic density plus classic risk factors for breast cancer risk prediction. JAMA Oncol. 2018;4(4):476–482. doi: 10.1001/jamaoncol.2017.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eng A, Gallant Z, Shepherd J, McCormack V, Li J, Dowsett M, et al. Digital mammographic density and breast cancer risk: a case-control study of six alternative density assessment methods. Breast Cancer Res. 2014;16(5):439. doi: 10.1186/s13058-014-0439-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michailidou K, Beesley J, Lindstrom S, Canisius S, Dennis J, Lush MJ, et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet. 2015;47(4):373–380. doi: 10.1038/ng.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purrington KS, Slager S, Eccles D, Yannoukakos D, Fasching PA, Miron P, et al. Genome-wide association study identifies 25 known breast cancer susceptibility loci as risk factors for triple-negative breast cancer. Carcinogenesis. 2014;35(5):1012–9. [DOI] [PMC free article] [PubMed]
- 47.Bertrand KA, Scott CG, Tamimi RM, Jensen MR, Pankratz VS, Norman AD, et al. Dense and nondense mammographic area and risk of breast cancer by age and tumor characteristics. Cancer Epidemiol Biomark Prev. 2015;24(5):798–809. doi: 10.1158/1055-9965.EPI-14-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bertrand KA, Tamimi RM, Scott CG, Jensen MR, Pankratz VS, Visscher D, et al. Mammographic density and risk of breast cancer by age and tumor characteristics. Breast Cancer Res. 2013;15(6):R104. doi: 10.1186/bcr3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holm J, Li J, Darabi H, Eklund M, Eriksson M, Humphreys K, et al. Associations of breast cancer risk prediction tools with tumor characteristics and metastasis. J Clin Oncol. 2016;34(3):251–258. doi: 10.1200/JCO.2015.63.0624. [DOI] [PubMed] [Google Scholar]
- 50.Nguyen TL, Aung YK, Evans CF, Dite GS, Stone J, MacInnis RJ, et al. Mammographic density defined by higher than conventional brightness thresholds better predicts breast cancer risk. Int J Epidemiol. 2017;46(2):652–661. doi: 10.1093/ije/dyw212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt DF, Makalic E, Goudey B, et al. Cirrus: an automated mammography-based measure of breast cancer risk based on textural features. J Natl Cancer Inst. 2018; In Press. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Study design and characteristics for eight participating studies. Table S2. Characteristics of cases and non-cases by study (% of non-missing). Table S3. Associations (OR, 95% CI) of adjusted percent density (PD), adjusted dense area (DA) and polygenic risk score (PRS) with breast cancer by study. Adjusted for age and BMI. Table S4. Goodness of fit test P values for joint effect of adjusted* density measures with 77-SNP polygenic risk score (PRS) on breast cancer, based on six population-based studies. PRS and density measures are continuous (per 1 standard deviation, SD). (DOCX 34 kb)
Data Availability Statement
The datasets generated and/or analyzed during the current study are part of the Breast Cancer Association Consortium and would be available with the appropriate permissions, including an application process and appropriate data transfer agreements.