Abstract
Background
Studies carried out with novel 13 strains of Trichoderma, isolated from mangrove sediments (PE, Brazil) using morphophysiological and molecular characterization, followed evaluation of biocontrol using Fusarium strains isolated from Caatinga soil (PE, Brazil). Trichoderma strains were characterized by polyphasic taxonomic approach, and the extracted DNA was amplified with primers ITS 1 and 4, and sequenced. The biocontrol evaluation was conducted at 24 and 48 h of growth intervals by Tukey test, with a significance of 5%. Antibiosis tests were assessed in vitro by dual plate and partition plate techniques against Fusarium strains.
Results
Trichoderma molecular identification, sequences of 500 bp were amplified, deposited into GenBank, and used for phylogenetic analyses. The strains were identified as T. asperellum (10), as T. harzianum (2) and one as T. longibrachiatum. Growth rate presented an average of 0.1207 cm h−1 for Trichoderma and lower growth rate of 0.031 cm h−1 for Fusarium spp., respectively. Antibiosis tests presented the best antagonist level of efficiency for T. asperellum UCP 0149 against F. solani UCP 1395 (82.2%) and F. solani UCP 1075 (70.0%), followed by T. asperellum UCP 0319 against F. solani UCP1083 (73.4%) and T. asperellum UCP 0168 against F. solani UCP1098 (71.5%), respectively.
Conclusions
The data obtained in this study as tool for identification of novel Trichoderma strains serve as basis for development of several sustainable use for biotechnological processes. Those Trichoderma strains found promising for the management antagonistic potential and interaction could aid the conduct of biotechnological biocontrol of contaminants, and improve environmental conditions for the health of plants.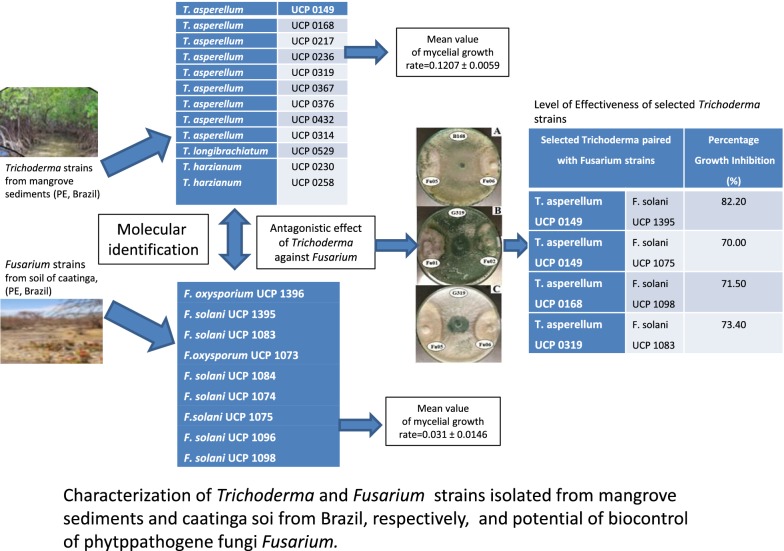
Keywords: Filamentous fungi, Molecular identification, Biocontrol
Background
Brazil possesses 1,225,444 hectares (approximately 12,250 km2) of mangrove. Mangrove are considered important and transitory costal ecosystem between land environments and marine environments. These ecosystems are a dynamic ecotone between terrestrial and marine habitats, located in typical tropical and subtropical regions. They are affected by tides and of great ecological, economic, and social significance [1–3].
Approximately 90% of mangroves are distributed in South-East Asia, America, and Africa. Fungal diversity including lower fungi (oomycetes and thraustochytrids) and higher fungi (ascomycetes and basidiomycetes) are present in mangrove habitats. They are playing an important role in the nutritive cycle and support the mangrove ecosystem Important studies realized with mangrove fungi and suggest promising sources for screening new products considering the adaption of extreme conditions of the environment [4, 5].
Fungi of the genus Trichoderma have been isolated from estuarine environments rich in lignocellulosic materials. Their great capacity for competition and adaptation in these environments is due to: their ability to synthesize biomolecules such as enzymes and antimicrobial metabolites such as antibiotics, which are of great economic and industrial interest; their hyper parasitism; and their competition for space, oxygen, and nutrients, which has been drawing the interest of researchers [6–9].
The assays using these microorganisms, such as the species of the genus Trichoderma, biomolecules producers with the ability to control phytopathogens mainly in the biological and agricultural areas, resulting in formulations of regulated and commercialized products for agricultural use [10, 11]. Among these, T. harzianum is one of the most researched biocontrol species, followed by T. viride, T. koningii, T. hamatum and T. pseudokoningii [12].
Trichoderma spp. are used as biological control agents against soil-borne pathogens, such as Fusarium, Pythium and Rhizoctonia spp., which infect grain cultures like the French beans described in Kariuki et al. [13]. These microorganisms have been used in the management of such pathogens, especially to the export market, because they act producing antibiotics and cell wall degrading enzymes, besides acting through mechanisms of mycoparasitism and competition for nutrients and space, exercising a biological control [14–16].
However, this ability to produce fungitoxic substances can vary between species and among isolates of the same species, where some strains can produce antimicrobial metabolites while others act as promoters of plant development, making necessary a taxonomic identification among species [17, 18].
Taxonomic confirmation of species of the genus Trichoderma, based solely on morphological markers, can be considered limited and of low accuracy due to the plasticity of its characteristics. Therefore, molecular techniques must be combined with the adoption of a variety of parameters for correct identification of species and for phylogenetic comparisons based on target sequences, thus determining precise relationships between isolates from this genus [19, 20].
Currently, pest and disease control measures are carried out on a large scale using agrochemicals. Nonetheless, the use of such substances—besides representing onerous productive costs reflected on the price of the final products is associated to several problems, such as emergence of resistant pathogens, environmental contamination [21, 22]. Biological control stands out as an alternative for a more sustainable agricultural development, besides contributing to environmental conservation. This alternative measure has been more and more widespread on the account of being relatively simple, clean, and less onerous. In vitro tests are the basis for selecting and assessing the potential and feasibility of biocontrol of microorganisms that can prompt the growth or development of phytopathogenic agents [23, 24].
In this context, novel Trichoderma spp. strains were identified by polyphasic taxonomy and molecular characterization, and explored as an effective and sustainable approach for the management of biocontrol of potentially phytopathogenic Fusarium strains.
Results and discussion
Morphological identification and characterization of Trichoderma strains
The genus Trichoderma has gained immense importance since several decades due to its antagonistic ability against wide range of plant pathogens and growth promotion in crop plants. Thirteen of Trichoderma strains were morphologically analyzed, they shown compatible with the description for genus, respectively. They were selected and maintained in periodic cultivation on Potato Dextrose Agar (PDA) medium. Macroscopic morphology was observed in Trichoderma spp. strains and revealed colonies with rapid growth, concentric halos and floccose or compact surface that looked like tufts on the culture medium. The mycelium, initially of a white color, acquired green, yellow shades, or remained white, due to the abundant production of conidia. Microscopically, it was observed abundant sporulation of smooth or rough-appearance conidia, originating from branched and irregularly verticillated conidiophores, and presented conidiogenic (phialides) cells, which generally were ampliform or fusiform and arranged in clusters, according to Gam et al. [25] (Fig. 1).
Fig. 1.
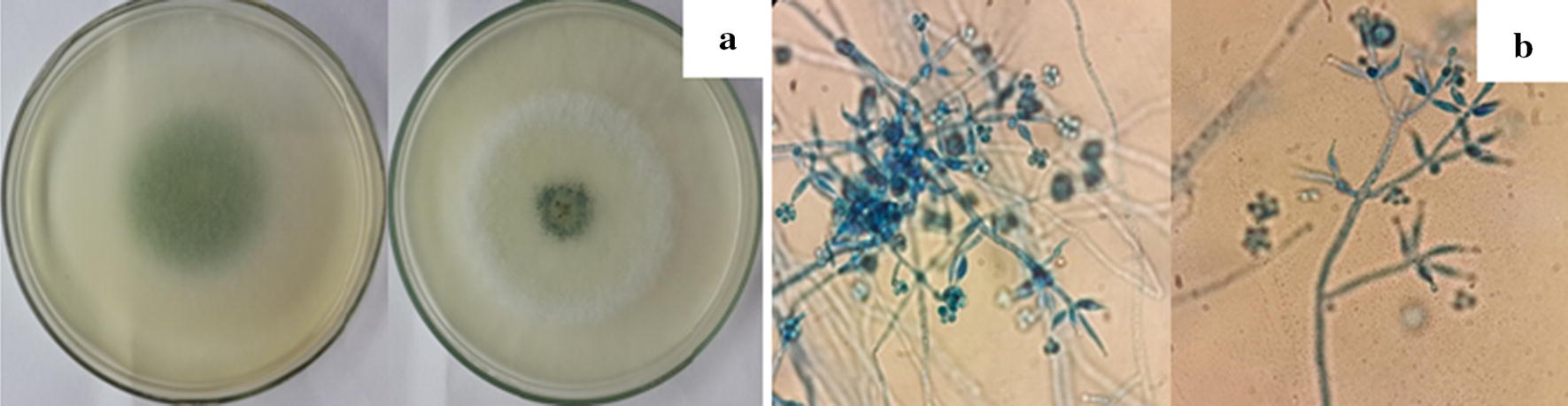
Characterization of Trichoderma strains by macroscopic morphology of the growth pattern of colonies in PDA medium after 5 days: a Optical microscopy arrangements of hyphae; b Conidiophores and conidia. Magnification ×400
Based on these characteristics, the Trichoderma strains could be classified in three groups, according to the identification key described by Gams and Bisset [20], Rifai [26] and Samuels et al. [27] as shown in Table 1.
Table 1.
Trichoderma classification of thirteen strains isolated from mangrove sediments used in this study according to Gams and Bisset [20], Rifai [26] and Samuels et al. [27]
| Groups | Morphology of the colony |
Trichoderma strains Universidade Católica de Pernambuco (UCP) |
|---|---|---|
| Group I | In the verse the colony presented itself dark green in the center and light yellow in the borders, or fully white and reverse with no color change |
Trichoderma sp.UCP 0149 Trichoderma sp. UCP 0217 Trichoderma sp.UCP 0258 Trichoderma sp.UCP 0319 Trichoderma sp.UCP 0367 Trichoderma sp. UCP 0376 Trichoderma sp. UCP 0314 |
| Group II | In the verse the colony presented itself white in the center and yellow or white in the borders, and reverse with no color change |
Trichoderma sp. UCP 0168 Trichoderma sp. UCP 0230 Trichoderma sp. UCP 0236 Trichoderma sp. UCP 0432 Trichoderma sp. UCP 0476 |
| Group III | In the verse the colony presented itself light yellow and reverse with no color change | Trichoderma sp. UCP 0529 |
Trichoderma spp. showed morphological colonies, where the green color of the conidia is interleaved with the white of the mycelium, which is consistent with the characteristics previously described for this fungus using the identification key of Samuels [28] were obtained. For the strains of Trichoderma asperellum showed globous, subglobous to ellipsoid, and some ovoid conidia, ampuliform phialides, and presence of chlamydospores were described Samuels et al. [29].
The results obtained with morphological aspects of Trichoderma strains are corroborated with Samuels et al. [27] to Trichoderma harzianum which species presents subglobous to ellipsoid conidia, ampuliform phialides, and globous and subglobous chlamydospores and the results were corroborated with the findings of the literature [30–32].
Samuels et al. [29] and Rifai [26] describes Trichoderma longibrachiatum possesses subglobous to ovoid conidia and lageniform phialides. Bisset [20], still, reports the presence of yellowish-green pigment in the reverse of some cultures of this species; however, it was not observed in this work. However, the characteristics of the colony morphology serve to identify fungi of this genus Trichoderma, it is insufficient to distinguish the species, and is necessary to confirm those species through molecular methods [33–37]. The authors emphasizes that intraspecific differentiation is more complex as due to the overlapping of several distinctive characters, resulting in a much artificial and subjective key. The identification of the isolates in this study yielded ten species of T. asperellum was the most frequently sampled, two species, of which T. harzianum, and one as T. longibrachiatum. The presence of T. asperellum had already been reported in mangrove sediments from India [38].
Molecular identification and phylogenetic analysis
Alexopoulos et al. [39] states that this complexity observed comes from the morphological concept of species, which is based solely on the similarities and on the characters’ discontinuity. The phylogenetic concept of species considers the genealogical, evolutionary relationships among the organisms of the group. The thirteen Trichoderma strains were identified by molecular level after morphological characterization using primers ITS 1 and ITS 4 amplified sequences of approximately 500 base pairs, observed on the agarose gel (Fig. 2).
Fig. 2.
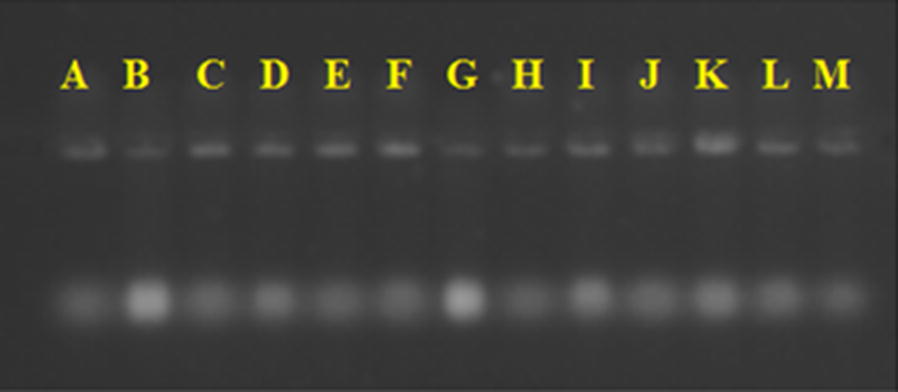
PCR amplification on 1.5% agarose gel with products obtained from the regions of ITS primers of Trichoderma isolates which have distinct lineage strains. Trichoderma isolates as mentioned in Table 1
Advances in the field of molecular biology, mainly in DNA analysis, enhance the way taxonomy has been done, thus justifying the performing of polyphasic approximations in studies of the different fungal groups. Using techniques of molecular identification, Hermosa et al. [40] obtained bands between 560 and 600 bp when they amplified ITS regions in order to identify 17 isolates of Trichoderma spp., among which are T. harzianum, T. atroviride, T. asperellum, T. longibrachiatum, used as biocontrol. Likewise, Menezes et al. [41] identified three isolates of Trichoderma spp. used on the biocontrol of Fusarium. The results described by these authors are similar to those found in this work, where primers ITS 1 and ITS 4 amplified sequences of approximately 500 base pairs for the studied strains of the genus Trichoderma.
The sequences, obtained from the sequencing, were assembled by the use of Staden package [42]. For close species, the sequences were compared to the other available on GenBank, using the BLAST tool available on the National Center for Biotechnology Information (NCBI). Multiples alignments of the sequences were estimated using Muscle algorithm in MEGA v.7 and adjusted manually when necessary. For multilocus analysis, the loci were concatenated by the use of Sequence Matrix v.1.8 [43, 44]. The sequences’ access codes of deposit on GenBank are in Table 2.
Table 2.
Trichoderma strains identification, culture collection number, isolation from, country and number of the genetic sequences of the ITS region
| Taxon | Collection number | Isolation from | Country | Number NCBI/ITS |
|---|---|---|---|---|
| T. asperellum | CBS 433.97c | Soil | USA | AY380912 |
| T. asperellum | IBSD T39 | Soil | India | JX518901 |
| T. asperellum a | UCP 0149 | Mangrove sediments | Brazil | MF974884 |
| T. asperellum a | UCP 0168 | Mangrove sediments | Brazil | MF974875 |
| T. asperellum a | UCP 0217 | Mangrove sediments | Brazil | MF974876 |
| T. asperellum a | UCP 0236 | Mangrove sediments | Brazil | MF974877 |
| T. asperellum a | UCP 0319 | Mangrove sediments | Brazil | MF974878 |
| T. asperellum a | UCP 0367 | Mangrove sediments | Brazil | MF974879 |
| T. asperellum a | UCP 0376 | Mangrove sediments | Brazil | MF974880 |
| T. asperellum a | UCP 0432 | Mangrove sediments | Brazil | MF974881 |
| T. asperellum a | UCP 0314 | Mangrove sediments | Brazil | MF974883 |
| T. asperellum a | UCP 0476 | Mangrove sediments | Brazil | MF974882 |
| T. brunneoviride b | CBS121130c | – | Germany | EU518659 |
| T. longibrachiatum | CBS 816.68c | – | USA | EU401556 |
| T. longibrachiatum | NRRL 54514 | Gloeophyllum trabeum | USA | HQ882796 |
| T. longibrachiatum a | UCP 0529 | Mangrove sediments | Brazil | MF974874 |
| T. harzianum | CBS 226.95c | Soil | UK | AJ222720 |
| T. harzianum | CBS 227.95 | Soil | UK | AJ222721 |
| T. harzianum a | UCP 0230 | Mangrove sediments | Brazil | MF974886 |
| T. harzianum a | UCP 0258 | Mangrove sediments | Brazil | MF974885 |
CBS: Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; IBSD: Institute of Bioresources and Sustainable Development, Manipur, India; NRRL: Agricultural Research Service Culture Collection, Peoria, USA.; UCP: Universidade Católica de Pernambuco
aTaxon in bold were found in this study. ITS: Internal transcribed spacer (ITS1-5.8S-ITS2)
bOutgroup isolate
cType species culture
Among 13 Trichoderma strains analyzed, ten strains were identified as T. asperellum UCP 0149, following codifications UCP 0168, UCP 0217, UCP 0236, UCP 0319, UCP 0367, UCP 0376, UCP0432, UCP 0314 and UCP 0476; two strains as T. harzianum UCP 0230 and UCP 0258, and strain T. longibrachiatum UCP 0529 (Table 2).
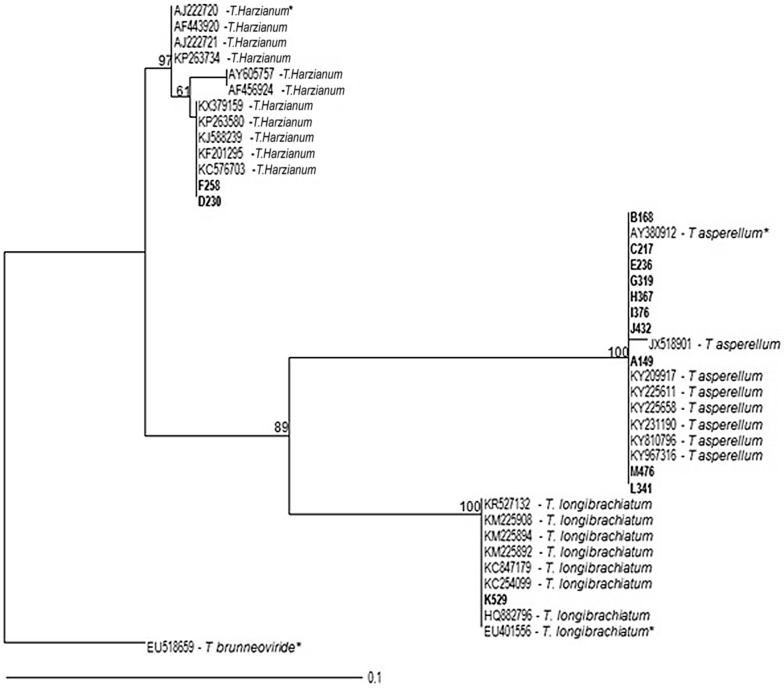
All sequences were used for the formation of the phylogenetic tree, along with sequences of species types. The alignment and phylogenetic analysis of these sequences showed groupings that corroborate the taxonomic identification of the species studied in this work, presenting 602 characters, of which 68 were informative, 94 were variable and 506 were constant. The maximum likelihood analysis generated a consistent topology tree, in which three groups corresponding to the genus Trichoderma were observed, namely: T. harzianum, T. longibrachiatum and T. asperellum, which showed ramifications among the individuals of the other clades, as verified in Fig. 3.
Fig. 3.
Phylogenetic analysis of maximum likelihood tree was constructed by 602 bp aligned with partial sequences of the Internal transcribed spacer—ITS (ITS1, 5.8 S and ITS2), with 19 taxa belonging to the Trichoderma species, compatible with the isolates examined in this research
The species chosen as outgroup was T. brunneoviride, for despite being within the genus, it possesses a nucleotide sequence that diverges on other clades members, occupying a base position and differing from other groupings [45–47].
The isolates UCP 0230 and UCP 0258 were grouped in the T. harzianum section with the England isolate type, forming a clade with bootstrap values of 85%. When related to each other, they formed a clade with bootstrap values of 99% (Fig. 3). In the T. longibrachiatum section, the UCP 0529 isolate formed a clade with bootstrap values of 100% with the isolate type of the United States of America (USA). The T. asperellum section comprised the remaining isolates in two subclades with 100% bootstrap values, where T. asperellum UCP 0376, UCP 0314 and UCP 0432 were grouped with the India rate and T. asperellum UCP 0476, UCP 0319, UCP 0236, UCP 0168, UCP 0367, UCP 0149, UCP 0217 with the USA type rate. However, the clade formed a subgroup with bootstrap values of 90% when compared to the isolate from the T. longibrachiatum section. This indicates a probable speciation event between these two species, thus forming a consistent grouping within the section. Bootstrap values above 70% (indicating the ML/MP ratios) are shown near the nodes. Isolates from this study were written in bold in the highlighted branch; Trichoderma brunneoviride, isolate CBS121130, was used as outgroup, and taxa typed with NCBI access code followed by (*) represents the type of species (Fig. 3).
On the purpose of establishing the relationship among genetic proximity, phylogeny and antagonistic activity against possible phytopathogens of the genus Fusarium, and of eliminating possible duplicities of such isolates, it was carried out a sequencing of the genomic DNA of Trichoderma strains selected for this work. Tests with such goal were also conducted by Colonia and Chagas Junior [48] using ITS region sequencing in the identification of the strains of Trichoderma spp. isolated from Trichoplus JCO fertilizer.
Results from the phylogenetic groupings showed that the strains of Trichoderma used in this work formed a reliable maximum likelihood tree, where support values—verified in each group—and the taxonomic inferences attributed to the individuals—in accordance with the respective grouping sections—are adequate, according to Kubicek et al. [49].
In this study, strains of T. harzianum formed a consistent cluster supported by a bootstrap value of 85%. However, the subdivision observed in this clade, that separates isolates UCP 0230/UCP 0258 from CBS 226.95 and CBS 227.95 (Fig. 3), can be related to recent speciation phenomena or genetic variability present in this species. For Druzhinina et al. [50], T. harzianum is one of the species with most variability and, nowadays, is considered a species complex.
The species chosen as outgroup was T. Brunneoviride because, despite being within the genus, it has a nucleotides sequence that diverges in the other clades individuals. Besides, it occupies a base position that differs from other groups [47, 51].
Studies carried out by Hoyos-Carvajal et al. [32] indicated the predominance of T. asperellum species, followed by T. harzianum, in the biodiversity of the neotropics, what can be explained by the high intraspecific genetic variability, colonization ability and high levels of sporulations in different substrates and carbon sources. In this work, it was also verified the predominance of T. asperellum species, followed by T. harzianum, among the samples of the genus Trichoderma selected and identified by molecular techniques.
Usage of molecular tools for species identification based on DNA sequences has been showing that Trichoderma species structured on classic concepts are, in fact, composed of two or more phylogenetic species (aggregate species), which can exhibit specific or global ecological niches [52]. The species T. harzianum is been recognized as global species, colonizing many substrates and ecological niches [53].
Evaluation of mycelial growth rate of Trichoderma and Fusarium strains
All Trichoderma strains showed a similar in vitro growth pattern, with an average growth rate of 0.1207 cm h−1. However, Fusarium spp. growth rates showed the media of 0.031 cm h−1. This can be considered significant when compared to the mean deviation of 0.0146 cm h−1, which represents a variation of approximately half of this average. The growth rates of Trichoderma spp. were approximately four times greater than the Fusarium strains growth rate after a 72 h period of evaluation (Table 3).
Table 3.
In vitro colony diameter and mycelial growth rate of Trichoderma spp. and Fusarium strains in the 48 h interval
| Trichoderma strains | DG (cm) | MGR | Fusarium strains | DG (cm) | MGR | ||
|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 24 h | 48 h | ||||
| T. asperellum UCP 0149 | 1.9 | 4.7 | 0.11 | F. oxysporum UCP 1396 | 1.1 | 2.4 | 0.05 |
| T. asperellum UCP 0168 | 2.0 | 5.2 | 0.13 | F. oxysporum UCP 1073 | 0.7 | 2.1 | 0.05 |
| T. asperellum UCP 0217 | 2.2 | 5.2 | 0.12 | F. solani UCP 1083 | 0.7 | 1.1 | 0.01 |
| T. harzianum UCP 0230 | 1.5 | 4.0 | 0.10 | F. solani UCP 1395 | 0.8 | 1.3 | 0.02 |
| T. asperellum UCP 0236 | 2.0 | 5.0 | 0.12 | F. solani UCP 1084 | 0.8 | 1.9 | 0.04 |
| T. harzianum UCP 0258 | 2.0 | 5.0 | 0.12 | F. solani UCP 1074 | 0.7 | 1.3 | 0.02 |
| T. asperellum UCP 0319 | 1.7 | 4.8 | 0.12 | F.solani UCP 1075 | 0.6 | 1.3 | 0.02 |
| T. asperellum UCP 0367 | 1.3 | 5.0 | 0.15 | F. solani UCP 1096 | 0.6 | 1.1 | 0.02 |
| T. asperellum UCP 0376 | 1.3 | 4.5 | 0.12 | F. solani UCP 1098 | 0.7 | 2.0 | 0.05 |
| T. asperellum UCP 0432 | 2.0 | 5.0 | 0.12 | 0.031 ± 0.0146a | |||
| T. asperellum UCP 0314 | 1.5 | 4.5 | 0.12 | ||||
| T. asperellum UCP 0476 | 2.0 | 5.0 | 0.12 | ||||
| T. longibrachiatum UCP 0529 | 1.2 | 4.3 | 0.12 | ||||
| 0.1207 ± 0.0059a | |||||||
DG: in vitro colony diameter growth (cm); MGR: mycelial growth rate (cm h−1)
aMean values of mycelial growth rate ± mean deviation values
Phenomenon of antibiosis of Trichoderma against Fusarium strains
The biocontrol potential by pairing test of Trichoderma strains cultures against the strains of Fusarium showed that all the strains of Trichoderma presented biocontrol action, affecting the development pattern of Fusarium spp. colonies. Trichoderma colonies presented a faster growth in the plates of paired cultures, being capable of growing on the potential pathogens and, thus, preventing their mycelial development by nutrient and space competition. Moreover, it was verified the inhibition by way of production of secondary metabolites, which demonstrated the capacity for biocontrol interaction through more than one mechanism [54].
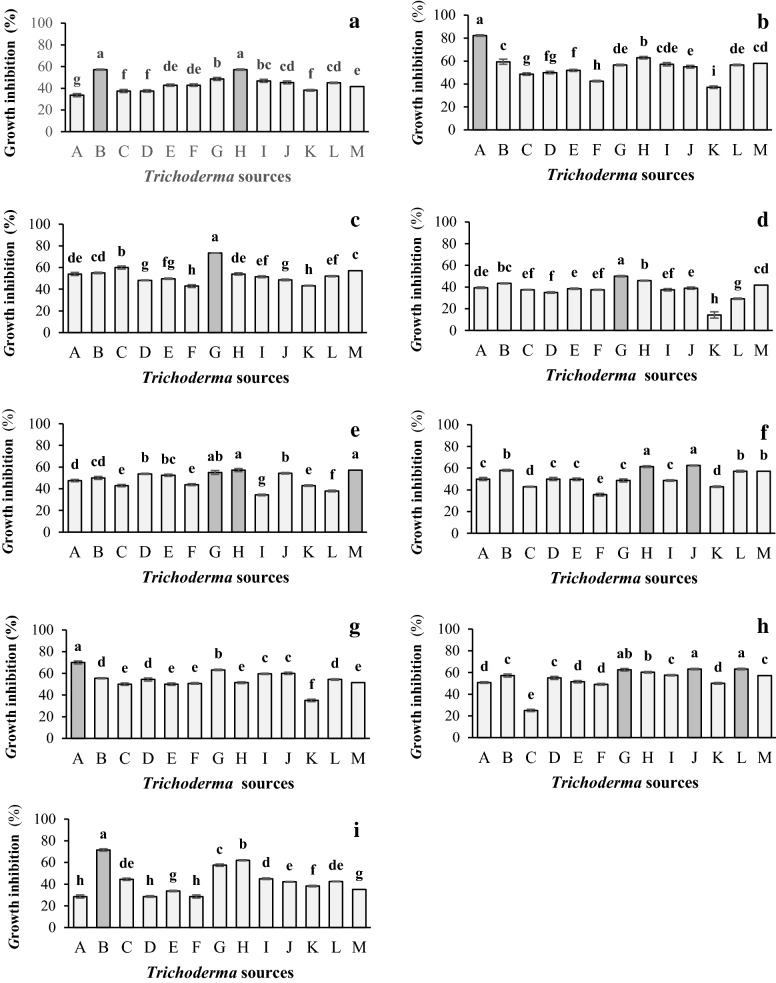
It was verified that six Trichoderma spp. strains reduced down to 50% the growth of Fusarium spp. strains. The strain of T. asperellum UCP 0319 presented two distinct behaviors when paired with different isolates of the genus Fusarium (Fig. 4a, b). The analysis indicated this strain’s effect through mycoparasitism when the antagonist removes nutrients from the parasitical fungi hyphae causing their death, and through competition (Fig. 4a) against the strain of F. oxysporum UCP 1396. The inhibition halo formation can be observed in Fig. 4b against F. solani UCP 1074, as well as against the strains of F. solani UCP 1084, UCP 1395 and UCP 1096. This behavior was also observed in the strains T. asperellum UCP 0217, UCP 0258, UCP 0367 and UCP 0432, and T. harzianum UCP 0230. The analysis of variance, in a factorial arrangement by Tukey’s test of 5% probability, the percentages of inhibition of the possible Fusarium pathogenic strains were significant to different strains of Trichoderma (p ≤ 0.01).
Fig. 4.
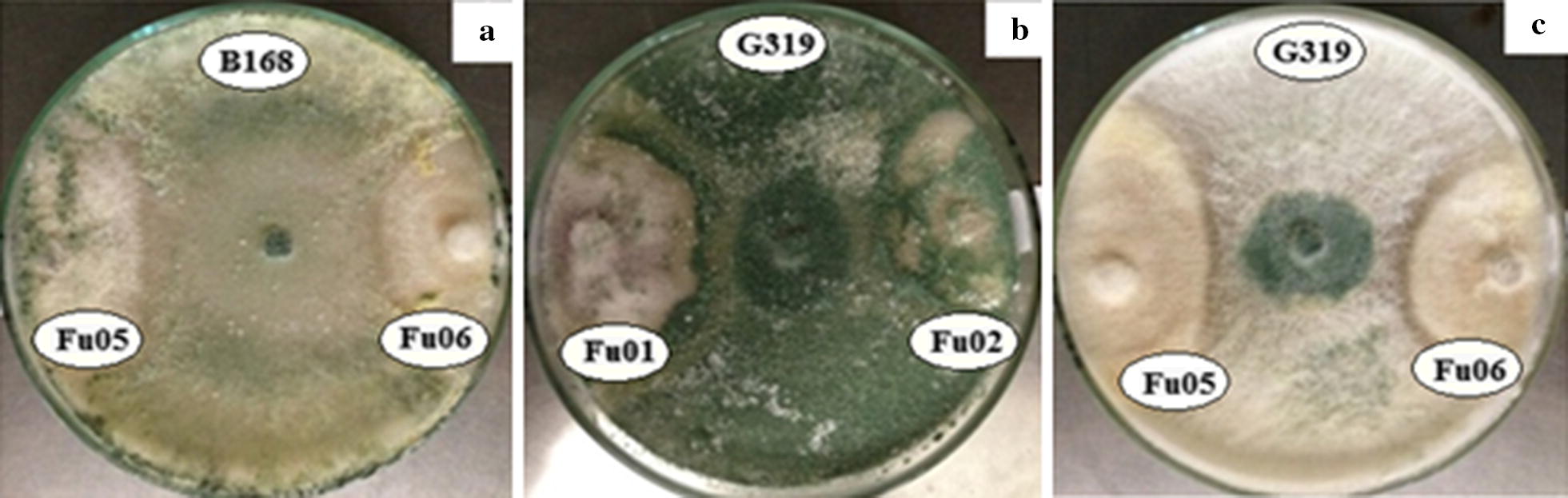
Antagonistic effect through mycoparasitism by the overlapping of colonies (a) and by formation of inhibition halo (b, c) of Trichoderma spp. colonies inhibiting the growth of Fusarium spp. strains in direct confrontation in vitro. Isolates F. oxysporum UCP 1396 and F. solani UCP 1074 under effect of T. asperellum UCP 0319 (a, b). Isolates F. solani UCP 1395 under effect of T. asperellum UCP 0314 (c)
Level of effectiveness of Trichoderma against Fusarium strains
The percentages of mycelial growth inhibition profile (GIP—%) of Trichoderma strains were analyzed by analysis of statistical variance, with significant variation (p ≤ 0.05), when paired were against Fusarium strains variated of 50 to 82.2% of inhibition.
The best antagonist phenomenon of effectiveness was observed to T. asperellum UCP 0149, presenting the highest inhibitory percentage (82.2% inhibition) against Fusarium solani UCP 1395, regarding the others samples analyzed in this work. The statistical analysis showed percentage inhibition of F. solani UCP 1083 by T. asperellum UCP 0319 stood out as the second antagonist effectiveness for this strain, with a percentage inhibition of 73.4%, respectively.
In the light of such results, it can be observed that strains of F. oxysporum are more resistant to the antibiosis action of Trichoderma strains regarding the F. solani. Among the strains of T. asperellum were significantly better antagonists than the strains of T. harzianum and T. longibrachiatum, and showed percentage of inhibition equal or superior to 50% in the most associations. The effective levels of antagonism, according to Sangoyomi [55] and described by Okigbo and Emeka [57], all strains of T. asperellum UCP 0149, UCP 0168, UCP 0432, UCP 0367 and UCP 0319 described in these study, and promoting inhibition over 60% in at least two distinct isolates of Fusarium strains, as well as strains T. asperellum UCP 0314, T. asperellum UCP 0217 and UCP 0376, with the same percentage inhibition in at least one strain of Fusarium (Fig. 5).
Fig. 5.
Antagonistic effect through mycoparasitism due to the overlapping of colonies (a) and due to the formation of inhibition halos (b, c) of Trichoderma spp. Colonies, thus inhibiting the growth of Fusarium spp. strains in direct confrontation in vitro. Isolates F. oxysporium UCP 1396 and F. solani UCP 1074 under effect of T. asperellum UCP 0319 (a, b). Isolates F. solani UCP 1395 under effect of T. asperellum UCP 0314 (c)
The genus Trichoderma is universally recognized by its biotechnological potential in agriculture—as in biocontrol—as well as in industrial applications, in production of enzymes. Therefore, it is a sustainable alternative to chemical pesticides, presenting itself more trustworthy for animal health, ecologically safer, and economically viable [58].
All strains of Trichoderma, isolated from mangrove sediments, reduced the mycelial growth of all potentially pathogenic Fusarium strains, isolated from caatinga soil, tested in this work. The different Trichoderma species showed variable levels of growth rate and antagonism in regards to the Fusarium species in the tests of culture pairing. It was not observed the growth of the probable pathogen on colonies of Trichoderma; most of the isolates were graded 2 according to Bell scale. This fact corroborates the works conducted by Louzada et al. [59], on which the antagonistic potential was tested against S. sclerotiorum and F. solani.
The specificity of biocontrol agents against phytopathogenic agents was evidenced by Vinale et al. [7], Kariuki et al. [13] and Zeilinger et al. [60], recognized Trichoderma spp. as producers of various cell wall degrading enzymes and of secondary metabolites with biocontrol action.
The new strains of T. asperellum UCP 0149, UCP 0168, UCP 0432, UCP 0314, UCP 0319, UCP 0376, UCP 0217 and UCP 0367 identified in this work presented better biocontrol performance, inhibiting the growth of all strains of F. solani UCP 1395, UCP 1075, UCP 1098, UCP 1074, UCP 1096, UCP 1074, UCP 1083 and of F. oxysporum UCP 1396 and UCP 1073, when compared to T. harzianum and T. longibrachiatum. In the present study, it is deduced that the inhibitory actions of the studied strains of Trichoderma are possibly linked to the antibiosis action through the formation of inhibitory halo, hyperparasitism and competition (Tables 2, 3 and 4).
Table 4.
Biocontrol potential of Trichoderma spp. strains against Fusarium spp. strains according to Bell scale [56]
| Trichoderma strains | Fusarium strainsa | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| F1 | F2 | F3 | F4 | F5 | F6 | F7 | F8 | F9 | |
| T. asperellum UCP 0149 | 3.92 | 1.61b | 2.44 | 3.35 | 2.78 | 2.64 | 1.89b | 2.6 | 3.57 |
| T. asperellum UCP 0168 | 2.31 | 2.22b | 2.4 | 3.04 | 2.64 | 2.28 | 2.38 | 2.31 | 1.85b |
| T. asperellum UCP 0217 | 3.52 | 2.72 | 2.2b | 3.52 | 3.08 | 3.08 | 2.64 | 3.75 | 2.97 |
| T. harzianum UCP 0230 | 3.52 | 2.64 | 2.74 | 3.77 | 2.46 | 2.64 | 2.43 | 2.4 | 3.57 |
| T. asperellum UCP 0236 | 3.08 | 2.55 | 2.65 | 3.43 | 2.51 | 2.65 | 2.64 | 2.56 | 3.92 |
| T. harzianum UCP 0258 | 3.08 | 3.11 | 3.08 | 3.52 | 3.01 | 3.71 | 2.6 | 2.69 | 3.57 |
| T. asperellum UCP 0319 | 2.72 | 2.33 | 1.8b | 2.64 | 2.4 | 2.72 | 2.09b | 2.11b | 2.3 |
| T. asperellum UCP 0367 | 2.31 | 2.1b | 2.44 | 2.88 | 2.31 | 2.15b | 2.57 | 2.19b | 2.13b |
| T. asperellum UCP 0376 | 2.82 | 2.31 | 2.56 | 3.52 | 3.85 | 2.72 | 2.22b | 2.3b | 2.93 |
| T. asperellum UCP 0432 | 2.91 | 2.4 | 2.72 | 3.38 | 2.43 | 2.11b | 2.2b | 2.09b | 3.13 |
| T. longibrachiatum UCP 0529 | 3.46 | 3.55 | 3.05 | 4.29 | 3.08 | 3.08 | 3.77 | 2.64 | 3.46 |
| T. asperellum UCP 0314 | 2.93 | 2.33 | 2.54 | 3.54 | 3.48 | 2.31 | 2.43 | 2.09b | 3.11 |
| T. asperellum UCP 0476 | 3.17 | 2.28 | 2.31 | 3.15 | 2.31 | 2.31 | 2.56 | 2.31 | 3.75 |
Okigbo and Emeka [55], Bell et. al. [56] and described by Sangoyomi [57]
aStrains of Fusarium spp. identified on the horizontal axis by the letters: Fusarium oxysporum UCP 1396 (F1); F. solani UCP 1395 (F2); UCP 1083 (F3); F. oxysporum UCP 1073 (F4); UCP 1084 (F5); UCP 1074 (F6); UCP 1075 (F7); UCP 1096 (F8); and UCP 1098 (F9), in joint culture with thirteen isolates of Trichoderma spp. identified on the vertical axis
bEffective inhibition of strains of Fusarium spp
The strains of F. oxysporum presented themselves more resistant to the antibiosis mechanisms when compared to the strains of F. solani. The general average inhibition observed was superior to 50% and the best results were connected to values above 70%, indicating that each species of Trichoderma possesses different abilities to inhibit different strains of Fusarium (Table 4) [1, 58, 61, 62].
Effective results against isolates F. oxysporum in pepper cultivars were also obtained by Hernández Castillo et al. [63] with the use of isolates of Trichoderma asperellum, obtaining an inhibition of 86.5%. According to the authors, the metabolites produced by T. asperellum have potential to reduce the reproductive capacity of F. oxysporum, diminishing sporulation and inhibiting conidia germination.
Different results were observed by Taribuka et al. [64] on selecting strains of Trichoderma that are potentially antagonistic to F. oxysporum. The highest percentages inhibition of growth were provided by T. gamsii (60.61%), followed by T. harzianum.psr-1 (59.08%), T. harzianum.swn-1 (55.80%), T. koningiopsis (55.58%), T. harzianum.swn-2 (54.05%) and T. asperellum (49.67%).
Results obtained in the present study, in antibiosis tests, presented variations regarding the degree of antagonism, where not all isolates of a same species of the genus Trichoderma were able to inhibit with the same effectiveness the same tested Fusarium strain. This fact may be indicative of the existence of specificity between the antagonist and the potential phytopathogen, suggesting the involvement of various genes and genetic factors interacting with the environment.
According to Perello et al. [65], more than one mechanism of action might be simultaneously involved in antagonistic actions. Louzada et al. [59] states that not all isolates of a same species of the biocontrol agent are capable of exercising hyperparasitism, suppressing the pathogen growth through other mechanisms like antibiosis or competition. According to Howell [66], species of Trichoderma have the ability of suppressing the growth of various fungi—in solid culture mediums—possibly linked to antibiosis action, hyperparasitism and competition.
Chen and Zhuang [62] states that species of Trichoderma grow faster because they use the food source in a more efficient way. In this study, the strains of Trichoderma not only presented the highest growth rates in regards to the strains of Fusarium, but they significantly exceeded the values of about 0.33 mm h−1 indicated by Moretto et al. [67]. An agent of biological control can excrete one or more metabolites that slow or inhibits the growth of pathogenic agents in the surrounding area. This phenomenon is called antibiosis [59, 65]. Works developed by Howell [68] prove the production of cell wall degrading enzymes, like chitinases and cellulases, as means of action of Trichoderma spp. against phytopathogenic fungi. According to Bosah et al. [68], the paired culture test is extremely important in the area of biological control of phytopathogens, because a good performance in this test indicates that the antagonist agent is an effective biocontroller.
The strains of T. asperellum UCP 0149, T. asperellum UCP 0168 and T. asperellum UCP 0319 presented the best antagonistic effect on the growth of Fusarium strains, on the observation of the inhibition zone between colonies of both fungi or on the overlapping of the mycelium of Trichoderma in the Fusarium colony (Fig. 4). Observations of such nature are used to evaluate the antagonistic potential of Trichoderma spp. and the obtained results prove that the studied strains have the potential to suppress the growth of Fusarium species, corroborating the findings of Taribuka and Bae [64], respectively.
The species of Trichoderma detect and locate the mycelium of susceptible fungi and grow on its direction, because they respond to the chemical stimuli produced by the host fungus. Besides, competition is one of the main characteristics of Trichoderma isolates due to their high mycelial growth rate [67], as proved in this work. In this context Trichoderma based emerges as an ecologically attractive alternative as biocontrol agent to phytopathogens considering the results obtained suggest the use as commercial products considering the effectiveness level of T. asperellum UCP 0149 against F. solani UCP 1395.
Conclusions
It was shown that, in the mangrove sediment of a Brazilian ecosystem, there are fungi of the novel Trichoderma strains which vary greatly from each other. It is also important to have greater knowledge of the ecology of these species and their responses to environmental or anthropogenic disturbances which may interfere with the equilibrium of these ecosystems. We studied a wide range of morphological and defined phylogenetic lineages based on their morphological characters. The results suggested that T. asperellum, T. harzianum and T. longibrachiatum fungal strains exhibit heterogeneity in genome structure of DNA sequence and similarity of ITS1 and four sequences in most taxa. Trichoderma strains showed a capacity for inhibiting mycelia growth of seven strains of Fusarium solani and two strains of F. oxysporum, all of which were isolated from caatinga soil of Brazil. However, only three selected strains identified as T. asperellum showed the best antagonist results in order to achieve the highest level of effectiveness and the possibility of applying them in an eco-friendly way and at low cost as biological agents against phytopathogenic Fusarium strains, and in addition, as target specific agents when compared with synthetic fungicides. Moreover, studies of those filamentous fungi which have a good potential for antagonistic interaction can both aid the conduct of biotechnological processes, and improve environmental conditions and there the health of plants.
Materials and methods
Microorganism and culture conditions
Thirteen Trichoderma and Nine Fusarium strains were kindly released to the researchers from the UCP (Universidade Católica de Pernambuco) Culture Collection, which is registered in the WFCC (World Federation for Culture Collection). The strains of Fusarium were isolated from Caatinga soil in Serra Talhada, Pernambuco, Brazil; and Trichoderma spp. were obtained from mangrove sediments of Rio Formoso, Pernambuco, Brazil, and maintained on Sabouraud dextrose agar at 5 °C.
Morphological characterization
Morphological identification was accomplished through classification keys of Gams and Bisset [20], Rifai [26] and Samuels et al. [27], taking into consideration macroscopic characteristics like color and texture of the colony surface verse and reverse, presence or absence of pigmentation, and pattern of growth and sporulation. Microscopic characters were analyzed according to morphology, size and disposition of the conidia and the phialides, using slides prepared by the microculture in EMA technique. The material was then dyed with cotton blue so that it could be visualized. Growth rate and morphological characteristics were analyzed in PDA and malt extract (EMA) (Himedia).
Extraction and amplification of genomic DNA
Thirteen samples of Trichoderma spp. underwent species-level molecular identification and phylogenetic analysis. Genomic DNA was extracted from 7 day old mycelial growth at 25 °C by the method adapted from Murray and Thompson [69]. Sequences of internal transcribed spacer (ITS) Regions 1 and 2, including the 5.8S, were amplified using primers ITS 1 (TCC GTA GGT GAA CCT GCG G) and ITS 4 (TCC TCC GCT TAT TGA TAT GC). Each 25 μl of the polymerase chain reaction (PCR) mix included: 13.85 μl ultrapure water, 1 μl template DNA, 1.5 μl of each primer (10 μM, synthesized by Invitrogen-Carlsbad, CA), 2.5 μl of dNTP mix and 4.63 μl of Taq DNA polymerase mix (0.05 μl-1 Taq DNA polymerase, reaction buffer, 4 mM MgCl2, Thermo Scientific, Waltham, USA). PCR reactions were performed in a SimpliAmp™ Thermal Cycler (applied biosystems) at 94 °C for 5 min, followed by 35 cycles at 94 °C for 1 min (denaturation), 57 °C for 1 min.
Analysis of the products obtained by PCR
To verify the efficiency of the PCR reaction, 3 μl of the substances obtained were stained with 3 μl of SYBR® Green dye (Thermo Scientific, Waltham, USA) and analyzed by 1.0% agarose gel electrophoresis in 0.5× TBE buffer (Tris–borate–EDTA 100 mM Tris base and 2.0 mM EDTA solution pH 8.0) and 50 mM of boric acid. Electrophoresis occurred at 75v for a period of 40 min. After the run, the gels were revealed and visualized under an ultraviolet transilluminator to check the amplification and purity, and then were photographed for documentation purposes [69, 70]. The amplicons were purified and sequenced by Macrogen Inc., Korea (http://www.127macrogen.com). Nucleotide sequences obtained were checked and edited using Staden Package 2.0 software packages. Subsequently, the consensus sequences obtained in this study were compared with other using Molecular Evolutionary Genetics Analysis (Mega BLAST) tool and deposited into GenBank databse (http://www.ncbi.nlm.nih.gov).
Molecular identification and phylogeny
Nucleotides sequences obtained were checked and edited with Staden Package 2.0 software packages. Subsequently, the consensus sequences were compared to the Lasiodiplodia ex-type reference sequences, retrieved from GenBank using Mega BLAST, and deposited into GenBank database (http://www.ncbi.nlm.nih.gov). Nineteen taxa belonging to the Trichoderma species, compatible with the isolates of this study, were used to construct the phylogenetic tree, with Trichoderma brunneoviride CBS121130 used as outgroup and taxa written with NCBI access code and followed by (*) representing the type species. The phylogeny was inferred by comparing locus ITS individual phylogenetic groupings through Analysis of Maximum Likelihood Estimates (MLE) on CIPRES portal (http://www.phylo.org). MLE analyses were estimated with RAxML-HPC2 on XSEDE, model GTR GAMMA, and the best topology tree related to the Bootstrap values in 1000 pseudoreplicates. Only sequences that obtained published results—and that were searched on BLASTN—were used, with high score (1000) and e-value equal to zero.
Determination of growth rates Trichoderma spp. and Fusarium strains
In order to evaluate growth rates, each isolate of Trichoderma spp. and Fusarium strains were cultured individually for 5 days at 25 °C in PDA medium. From this culture, disks (0.6 cm in diameter) of fungal structures were deposited in the center of a Petri dish containing PDA. These dishes were incubated at 25 °C with a 12 h photoperiod. After 24 and 48 h of incubation, the diameters (cm h−1) of colonies were measured in two perpendicular directions. Growth rates were determined according to the equation: GR = (G2 − G1)/(T2 − T1); in which: GR = growth rate; G1 = growth after 24 h; G2 = growth after 48 h; T1 = 24 h; T2 = 48 h [67].
Antagonist determination effect of Trichoderma spp. against Fusarium strains
To analyze the antagonistic action of the isolates of Trichoderma spp. on the samples of Fusarium strains, the technique used was direct confrontation in vitro, using the method of culture pairing in Petri dishes. Following Moretto et al. [67], for each Fusarium strains, discs (6 mm diameter) containing fungal structures were deposited at one end of the Petri dish containing PDA medium (approximately 1 cm from the end of the plate). After 72 h, a disk of Trichoderma spp., with 5 days of growth, was deposited 3.5 cm away from the colony of the possible phytopathogen. The design was entirely randomized with four replicates. Control was represented by the Fusarium strains samples without the presence of the antagonist and by the Trichoderma samples without the presence of the probable phytopathogen. The plates were maintained at 25 °C with a 12 h photoperiod for 6 days. After this period, the growth of colonies of Fusarium strains was checked. In accordance with the methodology of Camporota [71], the percentage of colonization (%C) of each antagonist isolate was calculated using the formula: %C = (DT/DE) × 100, where DT is the distance between colonies after mycelial growth stabilizes and DE is initial distance between the two mycelial discs. The inhibition index of Fusarium in relation to Trichoderma was determined by the relation I = 100 − %C. In addition to the %C values, each Fusarium isolate was classified as per the degree of antagonism (G), according to a scale of notes by Bell et al. [40], shown in Table 5. The percent growth inhibition was determined as a guide in selecting the degree of effectiveness in the control of Fusarium spp. samples for the three treatments.
Table 5.
Classification of the degree of antagonism (G), according to the scale of Bell et al. [56]
| Colonization pattern | Degree of antagonism (G) |
|---|---|
| Biocontrol agent grows completely over the pathogen, covering the entire surface of the culture medium | 1 |
| Biocontrol agent grows to at least about 2/3 of the surface of the culture medium | 2 |
| Biocontrol agent and pathogen colonize approximately half the surface of the culture medium (more than 1/3 and less than 2/3) and neither appears dominating the other | 3 |
| Pathogen colonizes at least 2/3 of the surface of the culture medium and exhibits resistance to the biocontrol agent | 4 |
| Pathogen grows completely on the biocontrol agent and occupies the entire surface of the culture medium | 5 |
Statistical analysis
Differences of the antagonistic action of Trichoderma isolates over Fusarium were determined by factorial design in ANOVA and the means were compared by Tukey test at 5% significance using the ASSISTAT® program.
Highlights
We have focused upon Trichoderma-mediated fungal taxonomy, biodiversity and antibiosis against to Fusarium strains.
Significant presence of Trichoderma asperellum, followed T.harzianum and T. longibrachiatum were detected from mangrove sediments.
Trichoderma asperellum strains showed their efficiency as biocontrol agent of phytopathogenes Fusarium solani.
Authors’ contributions
PRBF and GMCT conceived and designed the experiments; PRBF performed the experiments; AFS, ILC and DL analyzed the data; GMCT contributed reagents/materials/analysis tools; PRBF, MACL and GMCT wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
The authors are grateful to the Nucleus of Research in Environmental Sciences and Biotechnology, Catholic University of Pernambuco, Brazil, for the use of laboratories.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the GenBank database, http://www.ncbi.nlm.nih.gov.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was financially supported by CNPq (National Council for Scientific and Technological Development) Process No. 311373/2014-3, FACEPE (Foundation for the Support of Science and Technology of the State of Pernambuco) Process No. APQ-0291-2.12/15, and Coordination Unit for the Improvement of Higher Level Education Personnel (CAPES).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- BOD
biochemical oxygen demand
- CBS
Centraal bureau voor Schimmelcultures, Utrecht, The Netherlands
- DMSO
dimethyl sulfoxide
- DNA
deoxyribonucleic acid
- dNTP
nucleoside triphosphates containing deoxyribose
- EDTA
ethylene diamine tetra acetic acid
- PDA
Potato Dextrose Agar
- GIP
growth inhibition profile
- IBSD
Institute of Bioresources and Sustainable Development, Manipur, India
- ITS
internal transcribed spacer
- MgCl
magnesium chloride
- MLE
Maximum Likelihood Estimates
- NCBI
National Center for Biotechnology Information
- NRRL
Agricultural Research Service Culture Collection, Peoria, USA
- PCR
polymerase chain reaction
- UCP
Universidade Católica de Pernambuco
- UK
United Kingdom
- USA
United States of America
- WFCC
World Federation for Culture Collection
Contributor Information
Patrícia Rego Barros Filizola, Email: patriciarbfilizola@gmail.com.
Marcos Antônio Cavalcanti Luna, Email: marcos.cluna@gmail.com.
Adriana Ferreira de Souza, Email: adrife.souza@gmail.com.
Iwanne Lima Coelho, Email: iwannecoelho@gmail.com.
Delson Laranjeira, Email: delson.laranjeira@ufrpe.br.
Galba Maria Campos-Takaki, Phone: +55-81-2119-4017, Email: galba_takaki@yahoo.com.br.
References
- 1.Schaeffer-Novelli Y, Cintrón-Molero G, Soares MLG, De-Rosa T. Brazilian mangroves. Aquat Ecosyst Health Manag. 2000;3:561–570. [Google Scholar]
- 2.Ghizelini AM, Mendonça-Hagler LCS, Macrae A. Microbial diversity in Brazilian mangrove sediments—a mini review. Braz J Microbiol. 2012;43(4):1242–1254. doi: 10.1590/S1517-83822012000400002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hooper DU, Chapin FS, Ewell JJ, Hector A, Inchausti P, Lavorel S, Lawton JH, Lodge DM, Loreau M, Naeem S, Schmid B, Setälä H, Symstad AJ, Vandermeer J, Wardle DA. Effects of biodiversity on ecosystem functioning: a consensus of current knowledge. Ecol Monogr. 2005;75:3–35. [Google Scholar]
- 4.Gopal B, Chauhan M. Biodiversity and its conservation in the Sundarban mangrove ecosystem. Aquat Sci. 2006;68:338–354. [Google Scholar]
- 5.Thatoi H, Behera BC, Mishra RR. Ecological role and biotechnological potential of mangrove fungi: a review. Mycology. 2013;4:54–71. [Google Scholar]
- 6.Harman GE, Petzoldt R, Comis A, Chen J. Interactions between Trichoderma harzianum strain T22 and maize inbred line Mo17 and effects of these interactions on diseases caused by Pythium ultimum and Colletotrichum graminicola. Phytopathology. 2004;94(2):147–153. doi: 10.1094/PHYTO.2004.94.2.147. [DOI] [PubMed] [Google Scholar]
- 7.Vinale F, Marra R, Scala F, Ghisalberti EL, Lorito M, Sivasithamparam K. Major secondary metabolites produced by two commercial Trichoderma strains active against different phytopathogens. Lett Appl Microbiol. 2006;43:143–148. doi: 10.1111/j.1472-765X.2006.01939.x. [DOI] [PubMed] [Google Scholar]
- 8.Rubio MB, Quijada MN, Pérez E, Domínguez S, Monte E, Hermosa R. Identifying beneficial qualities of Trichoderma parareesei for plants. Appl Environ Microbiol. 2014;80:1864–1873. doi: 10.1128/AEM.03375-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kathiresan K, Saravanakumar K, Anburaj R, Gomathi V, Abirami G, Sahu SK, Anandhan S. Microbial enzyme activity in decomposing leaves of mangroves. Int J Adv Biotechnol Res. 2011;2(3):382–389. [Google Scholar]
- 10.Benítez T, Rincón AM, Limón MC, Codón AC. Biocontrol mechanisms of Trichoderma strains. Int Microbiol. 2004;7:249–260. [PubMed] [Google Scholar]
- 11.Marques E, Irene Martins I, Mello SCM. Antifungal potential of crude extracts of Trichoderma spp. Biota Neotrop. 2018;18(1):1–6. [Google Scholar]
- 12.Keswani C, Mishra S, Sarma BK, Singh SP, Singh SP, Singh HB. Unraveling the efficient applications of secondary metabolites of various Trichoderma spp. Appl Microbiol Biotechnol. 2014;98:533–544. doi: 10.1007/s00253-013-5344-5. [DOI] [PubMed] [Google Scholar]
- 13.Kariuki GM, Kinyua ZM, Amata RL, Otipa MJ, Chege SW. Efficacy TRIAL of trianum-p as a biopesticide for the management of soil-borne diseases of French beans in Kenya. Kenya Agric Res Inst. 2010;1105:129–130. [Google Scholar]
- 14.Verma M, Brara SK, Tyagia RD, Surampalli RY, Valéro JR. Antagonistic fungi, Trichoderma spp.: panoply of biological control. Biochem Eng J. 2007;37(1):1–20. [Google Scholar]
- 15.Harman GE. Myths and dogmas of biocontrol. Changes in perceptions derived from research on Trichoderma harzianum T-22. Plant Dis. 2000;84:377–393. doi: 10.1094/PDIS.2000.84.4.377. [DOI] [PubMed] [Google Scholar]
- 16.Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. Trichoderma species—opportunistic, avirulent plant symbionts. Nat Rev Microbiol. 2004;2:43–56. doi: 10.1038/nrmicro797. [DOI] [PubMed] [Google Scholar]
- 17.Gwa VI, Nwankiti AO. In vitro antagonistic potential of Trichoderma harzianum for biological control of Fusarium moniliforme isolated from Dioscorea rotundata tubers. Virol Mycol. 2017;6(2):2–8. [Google Scholar]
- 18.Contreras-Cornejo HA, Macías-Rodrígues L, Cortés-Penagos C, López-Bucio J. Trichoderma virens, a plant beneficial fungus, enhances biomass production and promotes lateral root growth through an auxin-dependent mechanism in Arabidopsis. Plant Physiol. 2009;149(3):1579–1592. doi: 10.1104/pp.108.130369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoyos-Carvajal L, Orduz S, Bissett J. Growth stimulation in bean (Phaseolus vulgaris L.) by Trichoderma. Biol Control. 2009;5:409–416. [Google Scholar]
- 20.Bissett J, Gams W, Jaklitsch W, Samuels GJ. Accepted Trichoderma names in the year 2015. IMA Fungus. 2015;6(2):263–295. doi: 10.5598/imafungus.2015.06.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Druzhinina IS, Kubicek CP, Komon-Zelazowska M, Mulaw TB, Bissett J. The Trichoderma harzianum demon: complex speciation history resulting in coexistence of hypothetical biological species, recent agamospecies and numerous relict lineages. BMC Evol Biol. 2010;10:1–14. doi: 10.1186/1471-2148-10-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor JW, Geiser DM, Burt A, Koufopanou V. The evolutionary biology and population genetics underlying fungal strain typing. Clin Microbiol Rev. 1999;12:126–146. doi: 10.1128/cmr.12.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El-Abbassi A, Saadaoui N, Kiai H, Raiti J, Hafidi A. Potential applications of olive mill wastewater as biopesticide for crops protection. Sci Total Environ. 2017;576:10–21. doi: 10.1016/j.scitotenv.2016.10.032. [DOI] [PubMed] [Google Scholar]
- 24.Harman GE. Overview of mechanisms and uses of Trichoderma spp. Phytopathology. 2006;96:190–194. doi: 10.1094/PHYTO-96-0190. [DOI] [PubMed] [Google Scholar]
- 25.Gams W, Bisset J. Morphology and identification of Trichoderma. In: Kubicek CP, Harman GE, Ondik KL, editors. Trichoderma and Gliocladium: basic biology, taxonomy, and genetics. London: Taylor & Francis; 2007. pp. 3–74. [Google Scholar]
- 26.Rifai MA. A revision of the genus Trichoderma. Mycol Pap. 1969;116:1–56. [Google Scholar]
- 27.Samuels GJ, Ismaiel A, Mulaw TB, Szakacs G, Druzhinina IS, Kubicek CP, et al. The Longibrachiatum Clade of Trichoderma: a revision with new species. Fungal Divers. 2012;5591:77–108. doi: 10.1007/s13225-012-0152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samuels GJ. Trichoderma: systematics, the sexual state, and ecology. Phytopathology. 2006;96(2):195–206. doi: 10.1094/PHYTO-96-0195. [DOI] [PubMed] [Google Scholar]
- 29.Samuels GJ, Ismaiel A, Bom MC, Respinis S, Petrini O. Trichoderma asperellum sensu lato consists of two cryptic species. Mycologia. 2010;102(4):944–966. doi: 10.3852/09-243. [DOI] [PubMed] [Google Scholar]
- 30.Kullnig CM, Szakacs G, Kubicek CP. Molecular identification of Trichoderma species from Russia, Siberia and the Himalaya. Mycol Res. 2000;104:1117–1125. [Google Scholar]
- 31.Kuhls K, Lieckfeldt E, Samuels GJ, Meyer W, Kubicek CP, Börner T. Revision of Trichoderma sect. Longibrachiatum including related teleomorphs based on analysis of ribosomal DNA internal transcribed spacer sequences. Mycologia. 1997;89(3):442–460. [Google Scholar]
- 32.Hoyos-Carvajal L, Orduz S, Bissett J. Genetic and metabolic biodiversity of Trichoderma from Colombia and adjacent neotropic regions. Fungal Genet Biol. 2009;46:61–631. doi: 10.1016/j.fgb.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 33.Du Plessis IL, Druzhinina IS, Atanasova L, Yarden O, Jacobs K. The diversity of Trichoderma species from soil in South Africa with five new additions. Mycologia. 2018;159:1–25. doi: 10.1080/00275514.2018.1463059. [DOI] [PubMed] [Google Scholar]
- 34.Murray MAS, Thompson WF. Rapid isolation of hight molecular weight DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Özkale E, Demirel R. Molecular characterization of some soil originated Trichoderma Species. Hacettepe J Biol Chem. 2017;45(4):585–591. [Google Scholar]
- 36.Vaidya G, Lohman DJ, Meier R. Sequence Matrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 2011;27(2):171–180. doi: 10.1111/j.1096-0031.2010.00329.x. [DOI] [PubMed] [Google Scholar]
- 37.Mulaw TB, Druzhinina IS, Kubicek CP, Atanasova L. Novel endophytic Trichoderma spp. isolated from healthy Coffea arabica roots are capable of controlling coffee Tracheomycosis. Diversity. 2013;5:750–766. [Google Scholar]
- 38.Druzhinina IS, Kopchinskiy AG, Kubicek CP. The first 100 Trichoderma species characterized by molecular data. Mycoscience. 2006;47:55–64. [Google Scholar]
- 39.Alexopoulos CJ, Mims CW, Blackwell M. Introductory mycology. New York: Wiley; 1996. [Google Scholar]
- 40.Hermosa MR, Keck E, Chamorro I, Rubio B, Sanz L, Vizcaino JA, et al. Genetic diversity shown in Trichoderma biocontrol isolates. Mycol Res. 2013;108:897–906. doi: 10.1017/s0953756204000358. [DOI] [PubMed] [Google Scholar]
- 41.Menezes JP, Lupatini M, Antoniolli ZI, Blume E, Junges E, Manzoni CG. Variabilidade genética na região its do rDNA de isolados de Trichoderma spp. (Biocontrolador) e Fusarium oxysporum f. sp. Chrysanthemi. Ciênc Agrotec. 2010;34(1):132–139. [Google Scholar]
- 42.Staden R, Beal KF, Bonfield JK. The Staden Package, 1998. In: Misener S, Krawetz SA, editors. Bioinformatics methods and protocols. Methods in Molecular Biology™. Totowa: Humana Press; 2000. p. 132. [DOI] [PubMed] [Google Scholar]
- 43.Yoshida R, Fukumizu K, Vogiatzis C. Multilocus phylogenetic analysis with gene tree clustering. Ann Oper Res. 2019;276(1–2):293–313. [Google Scholar]
- 44.Gherbawy YA, Hussein NA, Al-Qurashi AA. Molecular characterization of Trichoderma populations isolated from soil of Taif City, Saudi Arabia. Int J Curr Microbiol App Sci. 2014;3(9):1059–1071. [Google Scholar]
- 45.Nuankaew K, Sotome K, Lumyong S, Boonlue S. Trichoderma polyalthiae sp. nov., an endophytic fungus from Polyalthia debilis. Phytotaxa. 2018;371:5. [Google Scholar]
- 46.Lee SB, Taylor JW. Phylogeny of five fungus-like protoctistan Phytophthora species, inferred from the internal transcribed spacers of ribosomal DNA. Mol Biol Evol. 1992;9(4):636–653. doi: 10.1093/oxfordjournals.molbev.a040750. [DOI] [PubMed] [Google Scholar]
- 47.Hermosa MR, Grondona I, Iturriaga EA, Diaz-Minguez JM, Castro C, Monte E, Garcia-Acha I. Molecular characterization and identification of biocontrol isolates of Trichoderma spp. Appl Environ Microbiol. 2000;66(5):1890–1898. doi: 10.1128/aem.66.5.1890-1898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colonia BSO, Chagas Junior AF. Screening and detection of extra cellular cellulases (Endo- and Exo- glucanases) secreted by filamentous fungi isolated from soils using rapid tests with chromogenic dyes. Afr J Biotechnol. 2014;13(52):4694–4701. [Google Scholar]
- 49.Kubicek CP, Herrera-Estrella A, Seidl-Seiboth V, Martinez DA, Druzhinina IS, Thon M, et al. Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 2011;12(4):40. doi: 10.1186/gb-2011-12-4-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Druzhinina IS, Komoń-Zelazowska M, Atanasova L, Seidl V, Kubicek CP. Evolution and ecophysiology of the industrial producer Hypocrea jecorina (anamorph Trichoderma reesei) and a new sympatric agamospecies related to it. PLoS ONE. 2010;5:9191. doi: 10.1371/journal.pone.0009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kullnig-Gradinger CM, Szakacs G, Kubicek CP. Phylogeny and evolution of the gems Trichoderma: a multigene approach. Mycol Res. 2002;106:757–767. [Google Scholar]
- 52.Jaklitsch WM, Samuels GJ, Dodd SL, Lu BS, Druzhinina IS. Hypocrea rufa/Trichoderma viride: a reassessment, and description of five closely related species with and without warted conidia. Stud Mycol. 2006;56:135–177. doi: 10.3114/sim.2006.56.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chaverri Kubicek CP, Komon-Zelazowska M, Druzhinina IS. Fungal genus Hypocrea/Trichoderma: from barcodes to biodiversity. J Zhejiang Univ Sci B. 2008;9:753–763. doi: 10.1631/jzus.B0860015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hajiehhrari B, Torabi-Giglou M, Mohammadi MR, Davari M. Biocontrol potential of some Iranian Trichoderma isolates in the control of soil-borne plant pathogenic fungi. Afr J Biotechnol. 2008;7:967–972. [Google Scholar]
- 55.Okigbo RN, Emeka AN. Biological control of rot-inducing fungi of water yam (Dioscorea alata) with Trichoderma harzianum, Pseudomonas syringae and Pseudomonas chlororaphis. J Stored Prod Postharvest Res. 2010;1(2):18–23. [Google Scholar]
- 56.Bell DK, Wells HD, Markhabell DK, Wells CR. In vitro antagonism of Trichoderma species against six fungal plant pathogens. Phytopathology. 1982;72:379–382. [Google Scholar]
- 57.Sangoyomi TE, Owoseni AA, Adebayo OS, Omilani OA. Evaluation of some botanicals against bacterial wilt of tomatoes. Int Res J Microbiol. 2011;2(9):365–369. [Google Scholar]
- 58.Waghunde R, Shelake RM, Sabalpara AN. Trichoderma: a significant fungus for agriculture and environment. Afr J Agric Res. 2016;11:1952–1965. [Google Scholar]
- 59.Louzada GAS, Carvalho DDC, Mello SCM, Lobo Júnior M, Martins I, Braúna LM, et al. Potencial antagônico de Trichoderma spp. originários de diferentes ecossistemas contra Sclerotinia scleotiorum e Fusarium solani. Biota Neotrop. 2009;9:145–149. [Google Scholar]
- 60.Zeilinger S, Omann M. Trichoderma biocontrol: signal transduction pathways involved in host sensing and mycoparasitism. Gene Regul Syst Biol. 2007;8:227–234. doi: 10.4137/grsb.s397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gupta BP, Mathur A, Koul A, Shrivastava V. Study on the extraction and purification of metabolites obtained from Trichoderma harzianum. Asian J Pharm Pharmacol. 2019;5(3):547–550. [Google Scholar]
- 62.Chen K, Zhuang WY. Three new soil-inhabiting species of Trichoderma in the Stromaticum clade with test of their antagonism to pathogens. Curr Microbiol. 2017;74:1049. doi: 10.1007/s00284-017-1282-2. [DOI] [PubMed] [Google Scholar]
- 63.Hernández Castillo FD, Flores W, Castillo RF, Morales GG, Castro D, Epifanio A. Antibiosis in vitro of Trichoderma strains metabolic extract on mycelial growth and reproductive capacity of Fusarium oxysporum isolated from pepper plants. Capsicum Br Biotechnol J. 2014;4:387–399. [Google Scholar]
- 64.Taribuka J, Wibowo A, Widyastuti SM, Sumardiyono C. Potency of six isolates of biocontrol agents endophytic Trichoderma against Fusarium wilt on banana. J Degrad Min Land Manage. 2017;4:723–731. [Google Scholar]
- 65.Perelló A, Mónaco C, Simón MR, Sisterna M, Dal Bello G. Biocontrol efficacy of Trichoderma isolates for tan spot of wheat in Argentina. Crop Prot. 2003;22:1099–1106. [Google Scholar]
- 66.Howell CR. Understanding the mechanisms employed by Trichoderma virens to effect biological control of cotton diseases. Phytopathology. 2006;96:178–180. doi: 10.1094/PHYTO-96-0178. [DOI] [PubMed] [Google Scholar]
- 67.Moretto KCK, Gimenes-Fernandes N, Santos JM. Influence of Trichoderma spp. on Colletotrichum acutatum mycelial growth and morphology and on infection of ‘Tahiti’ lime detached flowers. Summa Phytopathol. 2001;27:357–364. [Google Scholar]
- 68.Bosah O, Igeleke CA, Omorusi VI. In vitro microbial control of pathogenic Sclerotium rolfsii. Int J Agric Biol. 2010;12:474–476. [Google Scholar]
- 69.Green MR, Sambrook J. Analysis of DNA by agarose gel electrophoresis. Cold Spring Harb Protoc. 2019 doi: 10.1101/pdb.top100388. [DOI] [PubMed] [Google Scholar]
- 70.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 71.Szekeres A, Leitge B, Kredics L, Manczingera L, Vágvölgyia C. A novel, image analysis-based method for the evaluation of in vitro antagonism. J Microbiol Methods. 2006;65(3):619–622. doi: 10.1016/j.mimet.2005.09.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the GenBank database, http://www.ncbi.nlm.nih.gov.