Abstract
Background
Magnetotactic bacteria (MTB) are ubiquitous in natural aquatic environments. MTB can produce intracellular magnetic particles, navigate along geomagnetic field, and respond to light. However, the potential mechanism by which MTB respond to illumination and their evolutionary relationship with photosynthetic bacteria remain elusive.
Results
We utilized genomes of the well-sequenced genus Magnetospirillum, including the newly sequenced MTB strain Magnetospirillum sp. XM-1 to perform a comprehensive genomic comparison with phototrophic bacteria within the family Rhodospirillaceae regarding the illumination response mechanism. First, photoreceptor genes were identified in the genomes of both MTB and phototrophic bacteria in the Rhodospirillaceae family, but no photosynthesis genes were found in the MTB genomes. Most of the photoreceptor genes in the MTB genomes from this family encode phytochrome-domain photoreceptors that likely induce red/far-red light phototaxis. Second, illumination also causes damage within the cell, and in Rhodospirillaceae, both MTB and phototrophic bacteria possess complex but similar sets of response and repair genes, such as oxidative stress response, iron homeostasis and DNA repair system genes. Lastly, phylogenomic analysis showed that MTB cluster closely with phototrophic bacteria in this family. One photoheterotrophic genus, Phaeospirillum, clustered within and displays high genomic similarity with Magnetospirillum. Moreover, the phylogenetic tree topologies of magnetosome synthesis genes in MTB and photosynthesis genes in phototrophic bacteria from the Rhodospirillaceae family were reasonably congruent with the phylogenomic tree, suggesting that these two traits were most likely vertically transferred during the evolution of their lineages.
Conclusion
Our new genomic data indicate that MTB and phototrophic bacteria within the family Rhodospirillaceae possess diversified photoreceptors that may be responsible for phototaxis. Their genomes also contain comprehensive stress response genes to mediate the negative effects caused by illumination. Based on phylogenetic studies, most of MTB and phototrophic bacteria in the Rhodospirillaceae family evolved vertically with magnetosome synthesis and photosynthesis genes. The ancestor of Rhodospirillaceae was likely a magnetotactic phototrophic bacteria, however, gain or loss of magnetotaxis and phototrophic abilities might have occurred during the evolution of ancestral Rhodospirillaceae lineages.
Electronic supplementary material
The online version of this article (10.1186/s12864-019-5751-9) contains supplementary material, which is available to authorized users.
Keywords: Magnetospirillum, Rhodospirillaceae, Phototrophic, Magnetosome, Oxidative stress
Background
Magnetotactic bacteria (MTB) are a collection of microbes that produce intercellular, nanosized and chain-arranged magnetite (Fe3O4) or greigite (Fe3S4) crystals called magnetosomes [1–3]. Magnetosome biomineralization is a highly organized process under the strict genetic control of a cluster of genes named the magnetosome gene cluster (MGC) [3, 4]. Magnetosomes enable MTB to navigate along the Earth’s magnetic field, usually down to the sediment near the oxic-anoxic transition zone (OATZ), and this ability is known as magnetotaxis [4–8]. However, there is convincing evidence that some MTB are able to actively respond to different wavelengths of light, including the ultraviolet spectrum [9–17]. For example, several MTB, such as multicellular magnetotactic prokaryotes (MMPs) and the marine coccus strain MC-1, showed a negative response to light [9–14]. It has been suggested that the photosensing domain protein genes in uncultured MMP genomes are involved in phototactic movements and that MMPs may avoid damage or lethality caused by long-term irradiation from light and ultraviolet radiation [10, 14]. Moreover, the light wavelength-dependent MMP motility and magnetic sensibility changes have also been discovered [12].
Illumination also influences the growth and magnetosome synthesis of cultured MTB [15–17]. Recent research found that Magnetospirillum magneticum AMB-1, a well-studied MTB strain, was able to not only swim towards visible light [15] but also increase magnetosome synthesis and upregulate stress-related genes [16]. Ultraviolet illumination can delay AMB-1 cell growth and induce both cellular and DNA damage [17]. These phenomena demonstrate an intriguing topic regarding the artificial control of MTB motility and growth by magnetism and photons in bioengineering. However, the mechanism by which MTB respond to illumination and potentially cope with the damage induced by illumination (both visible and ultraviolet) remain unknown.
MTB are distributed in five bacterial phyla, namely, Proteobacteria, Nitrospirae, Omnitrophica, Latescibacteria and Planctomycetes [18, 19]. Magnetospirillum spp. are a group of facultative anaerobic microaerophiles that are members of a well-studied and sequenced genus belonging to the family Rhodospirillaceae in the class Alphaproteobacteria [1, 2, 20].
Rhodospirillaceae, so-called purple nonsulfur bacteria, encompass a total of 34 genera within the order Rhodospirillales and has the type genus Rhodospirillum, which is capable of photosynthesis [20]. Many members of this family can synthesize bacteriochlorophyll a and carotenoids and grow photoheterotrophically under anoxic conditions in light while chemoheterotrophically in darkness. Despite the disparate life styles of magnetotactic Magnetospirillum spp. and phototrophic Rhodospirillum spp., they were closely clustered based on 16S rRNA gene phylogenetic trees [20, 21].
An important discovery was made by Kolinko et al. via the transfer of magnetosome biomineralization genes from Magnetospirillum gryphiswaldense to the photosynthetic model organism Rhodospirillum rubrum, suggesting that both bacteria have the ability to host MGCs in their genomes and to provide a similar intracellular microenvironment for synthesizing magnetosomes [22]. Therefore, the close relationship between these MTB and phototrophic bacteria motivated us to question whether the common ancestor of MTB and phototrophic bacteria within the family Rhodospirillaceae possessed both abilities. Two hypotheses regarding MTB evolution remain hotly debated. One hypothesis is that the common ancestor was able to produce magnetosomes, but some bacteria lost this trait due to environmental differentiation or physiological pressures [21–24]. The other hypothesis is that the magnetosome synthesis genes were inherited via horizontal gene transfer (HGT) [25–27].
To detect the possible relationship between magnetotactic bacteria and other members within the family Rhodospirillaceae, we performed a comprehensive genomic comparison between magnetosome synthesis and photosynthetic bacteria within this family. First, we attempted to reveal potential photoresponse mechanisms via the identification of photosensitive genes in their genomes; second, we compared their gene similarities and differences in stress-related systems that may be induced by illumination; and finally, we discussed the possible evolutionary history of MTB and phototrophic bacteria in Rhodospirillaceae.
Results
Genomic features and magnetosome gene clusters of Magnetospirillum sp. XM-1
Magnetospirillum sp. XM-1 was isolated from Xi’an city moat, China. This strain can synthesize chain-arranged magnetite magnetosomes within the cell [28]. The XM-1 genome comprises one circular chromosome of 4,825,187 bp and one plasmid of 167,290 bp with a GC content of 65.64 and 66.48%, respectively (Additional file 1: Figure S1), and is most closely related to the completely sequenced genome of Magnetospirillum magneticum AMB-1 strain [29]. The 16S rRNA gene of XM-1 shows a high identity (> 99%) with the strain AMB-1, but the whole-genome colinearity analysis reveals that the XM-1 genome displays multiple rearrangements and insertions when compared with the AMB-1 genome (Additional file 1: Figure S2). The average nucleotide identity (ANI) and average amino acid identity (AAI) of the two complete genomes are 86.7 and 73.2%, respectively, which are lower than the defined species cut-off values (> 95–96% for ANI and > 95% for AAI), indicating that XM-1 represents a novel MTB species within the genus Magnetospirillum.
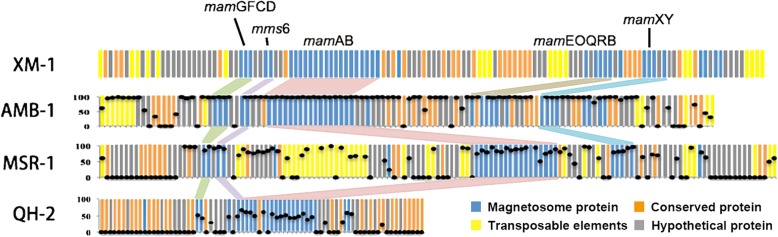
The genomic information of the other three published and completely sequenced MTB strains, i.e., Magnetospirillum magneticum AMB-1 [30], Magnetospirillum gryphiswaldense MSR-1 [31], and Magnetospira sp. QH-2 [32], are listed in Additional file 2: Table S1. The genome sizes of the four strains are approximately 4.02 Mb to 4.97 Mb; however, only XM-1 and QH-2 contain plasmids. All four stains contain MGCs in their chromosomes. The magnetosome synthesis gene operons mamGFCD, mms6, mamAB, mamEOQRB and mamXY are present in their genomic regions, which are also composed of multiple conserved and hypothetical proteins and transposable element genes. All canonical magnetosome genes in the MGC show high identities between XM-1 and the other three MGCs from AMB-1, MSR-1 and QH-2 (Fig. 1). Except for the known magnetosome genes, many genes encoding hypothetical proteins are also conserved in all four MGCs, possibly having critical functions in magnetosome synthesis or iron regulation. Several transposable element genes display high identities among XM-1, AMB-1 and MSR-1, indicating that the entire MGC region may have been inherited vertically from a common ancestor or horizontally transferred from an MTB donor to their ancestral lineage before diversification.
Fig. 1.
Magnetosome gene cluster comparison between Magnetospirillum sp. XM-1 and Magnetospirillum strains AMB-1, MSR-1 and Magnetospira sp. QH-2, respectively. The black dots represent the highest identities between protein sequences from the XM-1 MGC and MGCs from the other three MTB strains using the BLASTp algorithm. The conserved magnetosome synthesis genes are marked above
The Magnetospirillum sp. XM-1 genome displays high similarity with the phototrophic bacteria Phaeospirillum spp. genomes
The genome of Magnetospirillum sp. XM-1 also shares high identity to the genomes of nonmagnetotactic phototrophic bacteria Phaeospirillum molischianum DSM 120 and Phaeospirillum fulvum MGU-K5, with AAIs of 70.82 and 70.8%, respectively. These AAI values are higher than that between XM-1 and MSR-1 (AAI: 67.88%), both of which are from the same genus, Magnetospirillum. These results are consistent with previous phylogenetic analyses based on the 16S rRNA gene, which showed that Phaeospirillum spp. branched within the MTB genus Magnetospirillum [21, 29, 31].
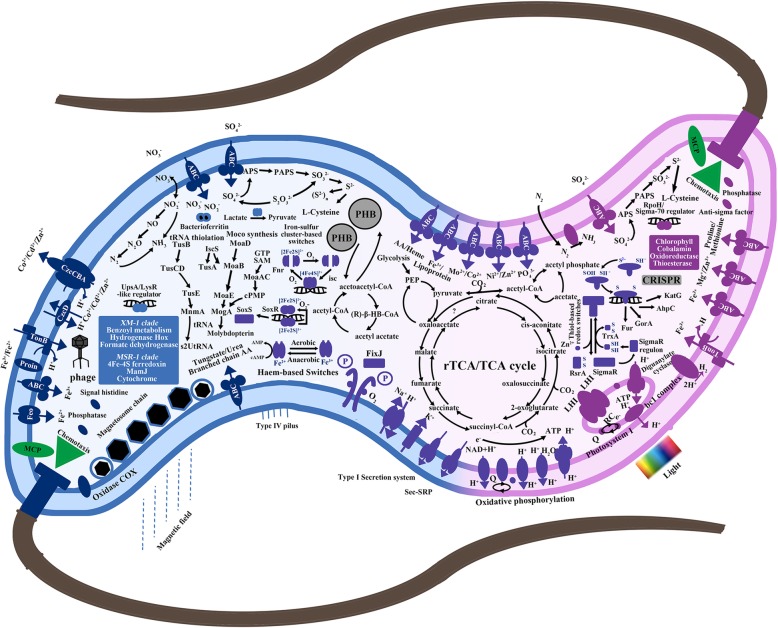
To better understand the possible relationship between MTB and phototrophic bacteria, the metabolic potentials (Fig. 2) of the newly sequenced Magnetospirillum sp. XM-1 genome were compared with the closely related photosynthetic bacteria in the genus Phaeospirillum (Phaeospirillum molischianum DSM 120 and Phaeospirillum fulvum MGU-K5) [33, 34]. The carbon metabolism of Magnetospirillum sp. XM-1 and Phaeospirillum spp. is similar, and both of their genomes possess genes involved in carbohydrate utilization, and this has been demonstrated by the heterotrophic growth of these strains with sole carbon sources such as acetate, fumarate and succinate in microaerobic conditions [28, 35–37]. For autotrophic growth, nearly all Magnetospirillum genomes possess genes of the reductive tricarboxylic acid (rTCA) cycle and the reductive pentose phosphate pathway, i.e., the Calvin–Benson–Bassham (CBB) cycle to fix CO2; however, only strains from the genus Magnetospirillum were experimentally verified to be capable of carbon fixation with the addition of NaHCO3 as a carbon source and Na2S2O3 as an electron donor [28, 38]. In contrast, several autotrophic growth tests on the genus Phaeospirillum showed that both photolithoautotrophy (anaerobic with light, electron donor: Na2S, Na2S2O3 and carbon source: NaHCO3) and chemolithoautotrophy (aerobic in darkness with electron donor: Na2S2O3 and carbon source: NaHCO3) could not be conducted [35, 36]. For nitrogen metabolism, nitrogenase genes have been found in both genera, and atmospheric dinitrogen can act as the sole nitrogen source during growth in multiple experimental tests [28, 29, 33, 34, 37–39]. However, only Magnetospirillum can use other forms of inorganic nitrogen sources such as nitrate, nitrite and ammonium through the denitrification pathway [28, 40], while Phaeospirillum can use ammonium, glutamate and urea as nitrogen sources [35, 36]. For sulfur metabolism, the assimilatory sulfate reduction pathway and dissimilatory sulfite reductase genes (dsr) are present in both Magnetospirillum and Phaeospirillum, which have been tested by the addition of sulfate, sulfite and cysteine as sulfur sources [28, 37, 40].
Fig. 2.
Comparison of metabolic reconstruction maps between Magnetospirillum sp. XM-1 (components colored in blue) and Phaeospirillum spp. (components colored in pink). The components that are shared by both genera are colored in purple. Please see Additional file 4: Table S3 for details
Genomic evidence of the photoresponse mechanism in MTB and phototrophic bacteria in the family Rhodospirillaceae
To make a comprehensive comparison, an additional 7 near-complete MTB genomes from Rhodospirillaceae were chosen for this study, namely, Magnetovibrio blakemorei MV-1 [41], Magnetospirillum caucaseum SO-1 [42], Magnetospirillum magnetotacticum MS-1 [43], Magnetospirillum marisnigri SP-1 [44], Magnetospirillum moscoviense BB-1 [44], Magnetospirillum sp. 64–120 [45] and Terasakiella sp. PR1 [46]. We did not include the analysis of Magnetococcus marinus MC-1 [47], Magnetococcus massalia MO-1 [48] or Magnetofaba australis IT-1 [49] because a recent study has shown that MO-1, together with MC-1 and IT-1, may comprise a new class of Etaproteobacteria and represent the earliest branching lineage in Alphaproteobacteria [48, 50]. Other selected genomes within the family Rhodospirillaceae, including nearly all sequenced phototrophic and nonphototrophic nonmagnetic bacteria, are approximately 3 Mb to 7 Mb in size (Additional file 3: Table S2, downloaded from NCBI prokaryote genomic database before Jan, 2017), with the exception of Endolissoclinum faulkneri L2 and L5 (~ 2 Mb), which are thought to have a parasitic lifestyle. As expected, no magnetosome genes were found in the genomes of phototrophic bacteria, whereas no photosynthesis genes were identified in the MTB genomes. Nevertheless, all bacteria within Rhodospirillaceae contain distinct photoreceptor genes and stress response genes such as oxidative stress genes, iron homeostatic genes and DNA damage repair genes.
Photoreceptors in MTB from the family Rhodospirillaceae
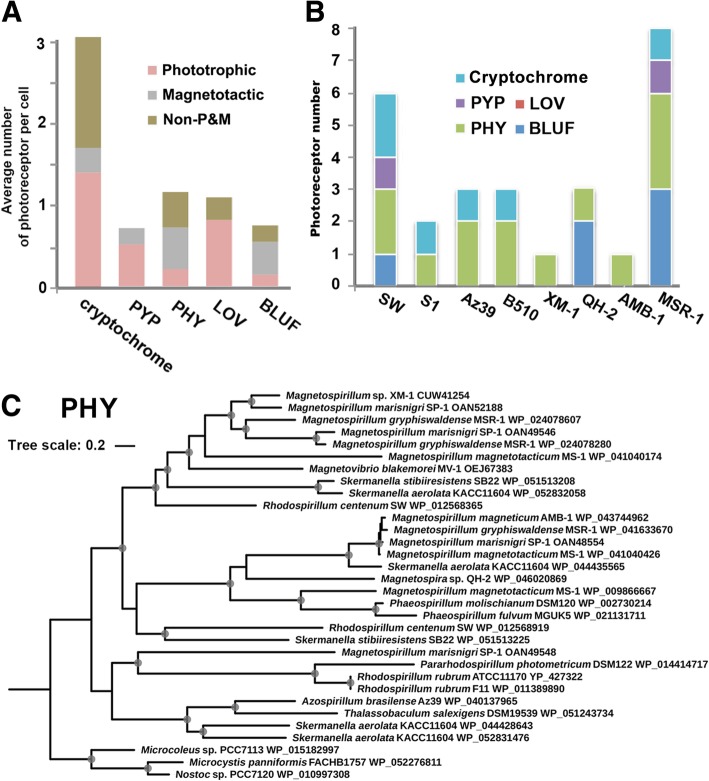
Diverse photosensitive domain-containing proteins, including blue light-sensitive sensors of blue light using FAD (BLUF), light-oxygen-voltage photoreceptor (LOV) domain-containing proteins, red/far red light-sensitive phytochromes (PHYs), photoactive yellow proteins (PYPs) and cryptochrome domain-containing protein genes, were found in MTB and other genomes of members in the Rhodospirillaceae family (Fig. 3). No obvious pattern in the category and number of photoreceptors was found among MTB, phototrophic and nonphototrophic nonmagnetotactic bacteria (Fig. 3a), i.e., different bacterial strains contained different numbers and types of photoreceptors regardless of classification as MTB or phototrophic bacteria (Fig. 3b, here we displayed eight representative strains).
Fig. 3.
(a) Average number of photoreceptors per cell in MTB, phototrophic bacteria and nonphototrophic nonmagnetotactic bacteria within the family Rhodospirillaceae. Photoreceptors were sorted by the following types: cryptochrome, PYP, PHP, LOV and BLUF domain-containing proteins. (Note: the number represents the total number of a certain type of photoreceptor divided by the bacterial genome number). (b) Photoreceptors predicted from the completely sequenced MTB strains XM-1, MSR-1, AMB-1, QH-2, phototrophic bacteria strains S1 and SW and nonphototrophic nonmagnetotactic bacteria Az39 and B510. (c) Phylogenetic trees based on the PHY domain-containing photosensor protein sequences from the genomes of the Rhodospirillaceae family
Specifically, nearly all MTB and Phaeospirillum strains as well as many other phototrophic and nonphototrophic nonmagnetic bacteria within the family Rhodospirillaceae possess one or more PHY domain photoreceptor genes, which is a known sensor for red/far-red light. The MTB strains MSR-1, SP-1 and BB-1 from Magnetospirillum and phototrophic bacteria from the genera Phaeospirillum, Skermanella and Rhodocista possess PYP domain photoreceptor genes. These photosensitive domains usually are coupled with homology of the amino-acid motif GGDEF (Gly-Gly-Asp-Glu-Phe) and conversed residues of the EAL domain proteins, which might be involved in light-dependent gene regulation, although their exact functions in MTB have not yet been verified by laboratory experiments. Some strains in Rhodospirillaceae, such as Magnetospirillum MSR-1 and BB-1 and Magnetospira sp. QH-2, and the genera Azospirillum and Caenispirillum contain BLUF domain protein genes, while no LOV domain protein-coding genes have been identified in MTB. Intriguingly, cryptochrome domain-containing protein-coding genes, which encode deoxyribodipyrimidine photolyase, are present in nearly all of the Rhodospirillaceae non-MTB strains and MTB MSR-1, SP-1 and BB-1 but are absent in other MTB strains.
In general, the results above show that MTB in Rhodospirillaceae contain distinct types of photoreceptor genes that are also shared by phototrophic bacteria and nonphototrophic nonmagnetic bacteria. No preference for a certain photoreceptor was found among the Rhodospirillaceae bacteria. For example, the phylogenetic analysis of the PHY domain photoreceptor, which is most widely spread in MTB and other members in Rhodospirillaceae, indicates that the photoreceptor is not conserved within MTB or phototrophic bacteria. MTB from the same genus even carry different subtypes of the PHY photoreceptor gene, as revealed in the topology of the phylogenetic tree (Fig. 3c). We assume that photoreceptor genes in Rhodospirillaceae were loosely selected or underwent active horizontal gene transfer during their early evolutionary processes. Although most of these bacteria have been experimentally shown to actively or negatively react with illumination, their physiological functions that are induced by the photoresponse still need to be systematically studied.
Stress response pathway comparison within the family Rhodospirillaceae
Since illumination can cause cell damage and even cell death, we specifically focused on oxidative stress, iron homeostasis and DNA damage repair between the MTB and non-MTB within Rhodospirillaceae. The representative completely sequenced genomes of the MTB strains XM-1, AMB-1, MSR-1 and QH-2, phototrophic strains S1 and SW, and nonmagnetic and nonphototrophic strains Az39 and B510 were used to display the general stress response system (the oxidative-related genes of all Rhodospirillaceae members are listed in Additional file 5: Table S4).
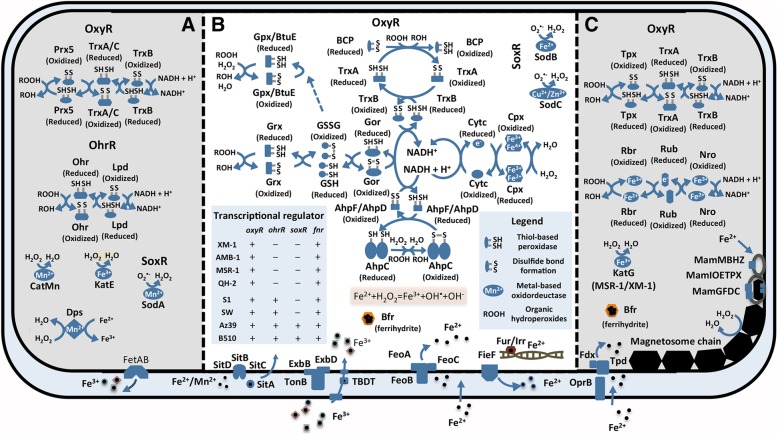
In the Rhodospirillaceae family, MTB and non-MTB contain complex systems to cope with intracellular oxidative stress (Fig. 4). Genes encoding the oxidative stress regulatory protein OxyR and redox-sensitive transcriptional activator SoxR-regulated thiol-based stress response systems, namely, the thioredoxin peroxidase BCP, alkyl hydroperoxide reductase peroxiredoxin AhpC, glutaredoxin Grx and glutathione peroxidase Gpx, together with the metal-based cytochrome C peroxidase Cpx and superoxide dismutase SodB/C, were found in nearly all studied genomes and considered to be the main oxidative balancing mechanisms. Nevertheless, MTB and non-MTB also have some different preferences for genes related to stress response systems; for example, the MTB genomes contain the 2-Cys peroxiredoxin Tpx and rubrerythrin Rbr for peroxide stress, while the non-MTB genomes possess the peroxiredoxin Prx5 system. Compared with non-MTB, nearly all MTB lack the genes encoding the organic hydroperoxide responding Cys-based redox sensor OhrR and SoxR proteins. Most non-MTB genomes contain the catalase CatMn, KatE and DNA protection protein Dps genes to scavenge hydrogen peroxide, while MTBs contain fewer enzymes (catalase KatG) for H2O2 elimination. However, several studies have shown that the magnetosome itself was able to catalyze H2O2 to H2O and O2 [51, 52].
Fig. 4.
Comparison of oxidative stress systems between MTB and non-MTB within the family Rhodospirillaceae. (a), (b) and (c) are the oxidative stress response and elimination processes that are mostly present in the genomes of only non-MTB, both MTB and non-MTB and only MTB, respectively. Iron transport systems from Rhodospirillaceae are displayed in the cell periplasmic space at the bottom of the diagram. Please see Additional file 5: Table S4 for details
Oxidative stress is also closely associated with iron homeostasis because ferrous iron can generate reactive oxygen species through the Fenton reaction (Additional file 1: Figure S3). Most MTB and non-MTB possess genes encoding multiple iron transport systems, including the Feo-dependent ferrous transport system, TonB-dependent ferric transport system, ferrous efflux FieF, iron storage Bfr and iron regulation factor Fur/Irr (Additional file 1: Figure S3). Compared to the non-MTB genomes, the MTB genomes usually contain two or more Feo-dependent ferrous transport system genes, and one feo gene operon has been identified within MGC. Nearly all members of the Rhodospirillaceae family contain genes encoding the TonB-dependent ferric transport system, but these genes are absent in Magnetospira sp. QH-2. The QH-2 genome contains more genes encoding iron export proteins, such as FetAB and FieF, than iron import proteins. The Tpd-like high-affinity ferrous iron transporter genes are present in the MTB genomes (except for QH-2) along with several copies (usually two) of the Btr iron storage protein genes. These iron-specific genes are vital for iron acquisition and magnetosome synthesis.
Illumination can also induce DNA damage through oxidative stresses or directly react with DNA molecules. Nearly all Rhodospirillaceae strains contain genes encoding DNA damage repair regulators such as LexA and RecA, the recombination system RecBFGJNOQR and RuvABC (Additional file 1: Figure S4). Compared to non-MTB, MTB within the Rhodospirillaceae family generally lack genes encoding enzymes involved in direct reversal (AlkB, Dcd) and base excision repair (AlkA, Mug, Nfi). However, most MTB strains contain genes for methylation systems, such as Dam and Dcm, and double copy genes of single-stranded binding protein Ssb, which is essential for DNA replication and repair. Interestingly, the newly isolated strain XM-1 has three copies of the translation error-prone DNA polymerase V UmuCD genes, MSR-1 and AMB-1 possess two and one copy of UmuCD, respectively, while few homologs were found in non-MTB strains. Ultraviolet radiation can also cause pyrimidine dimers, which can be repaired by the nucleotide excision repair (NER) excinuclease system. Nearly all studied strains contain genes of NER excinuclease systems UvrABCD, while only non-MTB and MTB MSR-1, BB-1 and SP-1 possess the deoxyribodipyrimidine photolyase Phr gene.
In general, stress response pathways are universally shared by all members of the Rhodospirillaceae family. They contain complex but similar systems to cope with intracellular oxidative stress and balance iron homeostatic conditions. Although these bacteria also contain comprehensive DNA damage repair genes in their genomes, MTB in the Rhodospirillaceae family overall contain fewer DNA damage repair genes when compared with non-MTB as revealed by the current database (Additional file 1: Figure S5).
Phylogenomic analyses of the relationship between MTB and phototrophic bacteria in the family Rhodospirillaceae
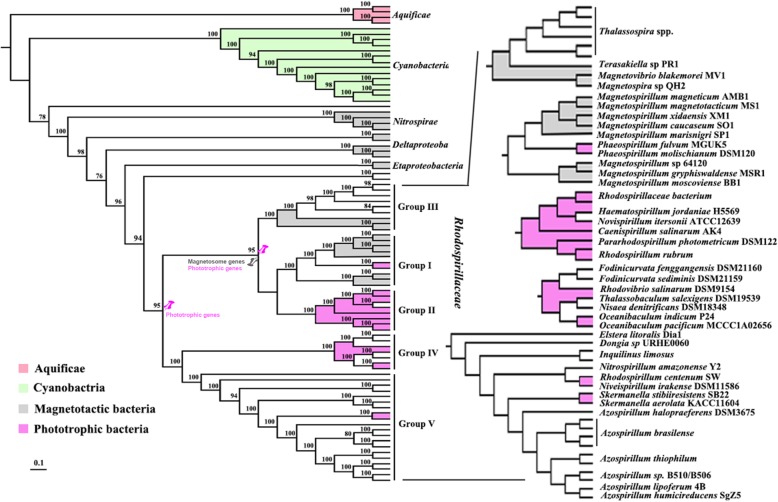
Based on the phylogenomic tree, the members in the family Rhodospirillaceae can be separated into five main clades (Fig. 5): Magnetospirillum and Phaeospirillum (Group I); Rhodospirillum, Pararhodospirillum, Novispirillum, Caenispirillum and Haematospirillum (Group II); Terasakiella, Thalassospira, Magnetospira and Magnetovibrio (Group III); Nisaea, Thalassobaculum, Oceanibaculum, Fodinicurvata and Rhodovibrio (Group IV); and Azospirillum, Niveispirillum, Nitrospirillum, Skermanella, Rhodocista (Rhodospirillum centenum SW), Dongia, Elstera and Inquilinus (Group V). Group I and Group II cluster together with Group III, while Group IV and Group V cluster closer (Fig. 5). Group I mainly consists of MTB, while most of the members from Group II are able to perform photosynthesis. In Group III, nonmagnetotactic Thalassospira clustered together with three MTB strains (genera Terasakiella, Magnetospira and Magnetovibrio). No MTB has been found in Group IV or Group V.
Fig. 5.
Phylogenomic tree of genomes from the family Rhodospirillaceae using the PhyloPhlAn method. Bacterial genomes from Aquificae and Cyanobacteria were also selected as out groups. Different colors represent distinct features within the Rhodospirillaceae family. The gray and pink colors represent magnetotactic and phototrophic bacteria, respectively. Five groups were defined based on the topology of the phylogenomic tree, namely, Magnetospirillum and Phaeospirillum (Group I); Rhodospirillum, Pararhodospirillum, Novispirillum, Caenispirillum and Haematospirillum (Group II); Terasakiella, Thalassospira, Magnetospira and Magnetovibrio (Group III); Nisaea, Thalassobaculum, Oceanibaculum, Fodinicurvata and Rhodovibrio, (Group IV); and Azospirillum, Niveispirillum, Nitrospirillum, Skermanella, Rhodocista (Rhodospirillum centenum SW), Dongia, Elstera and Inquilinus (Group V). The drawing pins in the nodes of Groups I, II and III indicate that these three groups have a common ancestor that contains both magnetosome synthesis (gray pushpin) and photosynthesis genes (pink pushpin)
In the phylogenomic tree, the MTB Magnetospirillum strains and phototrophic bacteria Rhodospirillum strains are closely clustered, and in the photosynthesis branch, the Phaeospirillum genus clusters within the Magnetospirillum genus in Group I using concatenated genomic protein sequence alignment and clustering, which is in line with a previous 16S rRNA gene phylogenetic study [21]. Moreover, core genomic analysis was also conducted, and all predicted proteins from the 84 bacteria genomes described above were clustered into 409,598 total categories based on their protein sequences by OrthoMCL (Additional file 1: Figure S6, Additional file 4: Table S3). A high number of 2290 orthologous genes are shared by all five groups within the family Rhodospirillaceae. Interestingly, ancestor analysis based on the classified groups using Dollo parsimony method in COUNT program (Fig. 5, shown as drawing pins) indicates that both magnetosome synthesis and photosynthesis genes are present at the ancestral nodes of Groups I, II and III, while photosynthesis genes appeared at the ancestral nodes of all Rhodospirillaceae. This result provides a strong indication that both traits are likely vertically inherited rather than inherited through the HGT process.
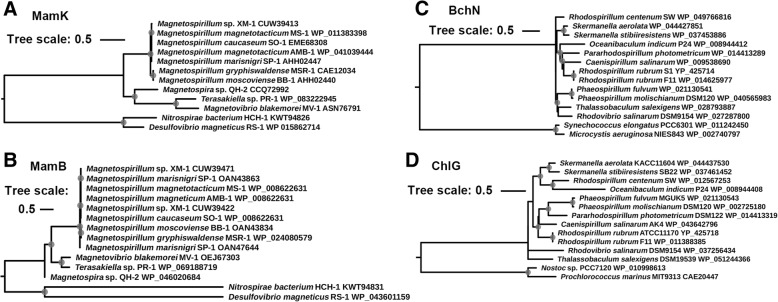
To evaluate the independent phylogenetic relationships of MTB and phototrophic bacteria in Rhodospirillaceae, both conserved magnetosome-associated genes (mamK and mamB) and photosynthesis-related genes (chlG and bchN) were used as representatives for phylogenetic tree construction (Fig. 6). The proteins MamK and MamB are responsible for magnetosome chain formation and iron transportation during magnetosome synthesis, respectively, while the proteins ChlG and BchN are involved in bacteriochlorophyll/chlorophyll a synthesis and light-independent protochlorophyllide synthesis, respectively. These genes are specific to MTB or phototrophic bacteria, i.e., only MTB genomes harbored magnetosome synthesis genes, whereas only phototrophic bacteria genomes harbored photosynthesis genes. The topologies of the phylogenetic trees of both magnetosome synthesis and photosynthesis genes are reasonably consistent with the topology in the phylogenomic tree. These results suggest that MTB closely resemble phototrophic bacteria in this family and again indicate that magnetosome synthesis and photosynthesis abilities were likely vertically transferred during the evolution of the Rhodospirillaceae ancestor.
Fig. 6.
Phylogenetic trees based on MamK (a), MamB (b), BchN (c) and ChlG (d) protein sequences from the genomes of the family Rhodospirillaceae
Discussion
The potential mechanism of MTB response to illumination
Bacterial phototaxis usually involves photosensing proteins and was first discovered in phototrophic organisms. Photosensing proteins function in the regulation of the expression of the photosynthesis machinery and protection of cells from harmful exposure to illumination [53]. Photoreceptor proteins were also found in nonphotosynthetic bacteria and functioned in signal transduction mostly for motility, biofilm forming, life cycle and the induction of carotene for protection against harmful light [54, 55]. The discoveries that MTB were also able to respond to blue and UV light were intriguing. Several photoreceptors were found in the uncultured MMP genomes [14]. It was hypothesized that, in environmental samples, a fraction of MMPs consistently display north-seeking behavior, which contrasts the expected swimming direction in the southern hemisphere, and this behavior was suggested to be regulated by light, stimulating the microorganism to move downwards to find suitable living environments [10].
Nevertheless, in most cases, MTB usually swim along the magnetic field against the oxygen concentration down to the surface sediments to occupy the OATZ [1, 2]. Moreover, Magnetospirillum magneticum AMB-1 was found to migrate towards visible light, while a similar phenomenon was observed in XM-1 swimming towards UVA radiation (Additional file 1: Figure S7) rather than moving away such as MMPs. The different phototaxis behaviors possessed by different MTB strains may be due to their distinct photoreceptors. The members of the Magnetospirillum clade, including the strains XM-1 and AMB-1, contain PHY domain photosensitive proteins that are considered to be red/far-red light-sensing bacteriophytochromes [54]. This photoreceptor may help Magnetospirillum to receive the photon signal for motility or potential physiological changes. XM-1 also contains a putative LOV/PAS-like domain-containing protein-coding gene that may help receive short wavelength light [54]. These Magnetospirillum strains usually live in aquatic environments and exhibit the ability to navigate towards sediment. The migration towards light may help them to move upwards to find better living environments that may contain trace oxygen. However, we cannot rule out that the light-dependent signal may respond to other physiological processes or only the redundant genes that their ancestor left behind. Moreover, other Magnetospirillum strains, such as MSR-1, BB-1 and QH-2, contain multiple photoreceptors, such as PYP, PHY and BLUF domain proteins, that respond to different wavelengths of the spectrum [53–55]. Thus, illumination may influence MTB behaviors or physiology in a more diverse way than previously thought. However, genetic experiments are needed to clearly illustrate the functions of these photoreceptors.
Relationship between MTB and phototrophic bacteria within the Rhodospirillaceae family
In the family Rhodospirillaceae, the phototrophic bacteria Rhodospirillum and Phaeospirillum closely cluster with Magnetospirillum MTB in the phylogenomic tree (Fig. 5), and their central metabolic pathways and stress response systems are highly similar (Figs. 2, 4 and Additional file 1: Figure S4). Moreover, the successful transfer of the Magnetospirillum gryphiswaldense MGC into phototrophic Rhodospirillum rubrum, which enabled its magnetosome production ability [22], further indicates that they have the potential to hold both magnetotactic and phototrophic abilities. This genetically engineered organism was the first report of magnetosome-producing photosynthetic bacteria and consequently posed an interesting topic regarding whether the ancestor of Rhodospirillaceae bacteria was able to photosynthesis while mineralizing magnetic particles for navigation, but lost one or both abilities during evolutionary adaptive radiation in new niches, or whether divergent Rhodospirillaceae bacteria acquired one of these abilities via HGT during evolution. The photosynthesis ability within Rhodospirillaceae was generally believed to diverge from a phototrophic ancestor before the divergence of Proteobacteria [56, 57]. The ancestor analysis by COUNT program and phylogenetic tree of photosynthesis genes also confirmed that these genes might be vertically transferred in the family Rhodospirillaceae (Fig. 5). For the origin of microbial magnetotaxis and magnetosome biomineralization, arguments regarding the mono- or multiple-origin have been addressed by several studies, and more studies tend to support the hypothesis that MTB have only one common ancestor [24, 26, 58–60]. These hypotheses together might support the coexistence of magnetotaxis and photosynthesis within one cell in nature.
Gene loss, which is a mechanism for bacterial adaptation to different ecotypes in heterogeneous environments [61–63], might result in the divergence of this family. The evolutionary loss of traits can occur if traits are selected against or if a trait becomes redundant, and this is typically driven by the weakening or removal of selection pressures that are responsible for maintaining the trait [64, 65]. In this study, most of the MTB strains in the family Rhodospirillaceae contained MGC that was enriched with mobile elements and was easily deleted from the genome, especially in oxygen-rich environments [25]. Moreover, studies on aerobic phototrophic bacteria in eutrophic oceanic areas or rich organic media with prolonged growth could also allow the loss of phototrophic genes [66]. Therefore, if the original Rhodospirillaceae ancestor was equipped with both abilities, the ancestor may have diverged into phototrophic, magnetotactic and nonphototrophic nonmagnetotactic bacteria during niche differentiation.
Geological implication
The possession of both magnetotaxis and phototrophic abilities might be evolutionarily beneficial for aquatic bacteria in early Earth’s history. The negative biological effects of ultraviolet radiation (UVR) in the Archean era have been estimated to be approximately three-fold higher than the present time [67]. Hence, without the ozone layer as a natural protection filtering UVR [68], the photosynthesis bacteria may suffer strong UV-induced damage living in the light penetration zone in the upper layer of the aquatic environment. To avoid deleterious or lethal UVR while still acquiring enough illumination for energy, it is reasonable to assume that these ancient phototrophic microbes may have developed the simple mechanism of navigation, i.e., magnetotaxis. This allowed them to swim downwards away from the strong UVR gradient and helped them find the ultraviolet tolerance photosynthesis zone (UTPZ) instead of the OATZ in the present time (Additional file 1: Figure S8).
Conclusion
With increasing evidence that MTB are able to positively or negatively respond to light, the knowledge of why MTB can react to light, how they mediate damage induced by light, and the evolutionary relationship between MTB and phototrophic bacteria are of great interest to microbiologists. In the current study, by utilizing the sequenced genomes of Magnetospirillum, phototrophic Rhodospirillum and other bacteria within the Rhodospirillaceae family, analyses of their photoreceptors, oxidative stress systems, DNA damage repair abilities and phylogenomics were conducted to answer these questions. Both MTB and phototrophic bacteria contain photosensitive domain proteins. The oxidative stress and DNA damage repair systems of the family Rhodospirillaceae are complex but do not show significant differences. Magnetospirillum species closely cluster with the phototrophic bacteria Rhodospirillum and Phaeospirillum in the phylogenomic tree. The topology of the phylogenomic tree is similar to both phylogenetic tree topologies of the magnetosome synthesis and photosynthesis genes. These results indicate that MTB, phototrophic and nonphototrophic nonmagnetic bacteria from the Rhodospirillaceae family likely evolved vertically with magnetosome synthesis and photosynthesis genes. The ancestor of Rhodospirillaceae was likely a magnetotactic phototrophic bacteria, however, gain or loss of magnetotaxis and phototrophic abilities might have occurred during the evolution of ancestral Rhodospirillaceae lineages.
Methods
The XM-1 genome analysis and the selection of the Rhodospirillaceae genomes
The MTB strain XM-1 was isolated from Xi’an city moat in Northwest China, cultured in optimized XM-C medium [28] and then sequenced by Illumina HiSeq 2000, and the gaps were closed by PCR [29]. The XM-1 genome is available in the NCBI repository under Project Number PRJEB11958, https://www.ncbi.nlm.nih.gov/bioproject/PRJEB11958/. The genome was annotated at MicroScope MaGe [69], and the genomic colinearity of XM-1 and other MTB were analyzed by the program mummer 3.0 [70]. Average nucleotide identity (ANI) and average amino acid identity (AAI) were also calculated with the ANI/AAI calculator [71].
All genomes of cultivated bacteria within the family Rhodospirillaceae were downloaded from NCBI (https://www.ncbi.nlm.nih.gov), and genomes with low quality (contigs > 500) were removed. A total of 84 high-quality genomes (Additional file 2: Table S1) were obtained, and the metabolic capacities were analyzed with the KEGG database [72].
Phylogenetic and phylogenomic trees calculation
The representative 16S rRNA genes of MTB and the Rhodospirillaceae strains were downloaded from NCBI. The phylogenetic tree was constructed using MEGA6.06 [73]. For the phylogenomic tree, MTB genomes outside Rhodospirillaceae, as well as Aquificae and Cyanobacteria genomes, were also downloaded from NCBI as out groups. All genomes within the Rhodospirillaceae family and out groups used in the current study were first aligned by PhyloPhlAn software [74], and the phylogenomic tree was built by the RAxML method [75] with the --auto-prot = bic option using the PROTGAMMAAUTO model with a bootstrap value of 1000.
Comparative genomic analysis of Rhodospirillaceae genomes
The orthologous protein families from the 84 selected genomes were identified using OrthoMCL [76] with a BLASTp E-value threshold of 10− 5, a 50% coverage cutoff with 30% identity and a default MCL inflation parameter of 1.5. For oxidative stress and iron homeostasis analysis, PeroxiBase (http://peroxibase.toulouse.inra.fr/) and one previously reported iron transporter database [77] were modified and used to search for the related genes using BLASTp with an E-value of 10− 20 and 75% coverage with 30% identity as a cutoff. The resulting protein sequences were BLASTp searched against the nonredundant NCBI protein database for confirmation. Only those sequences that reported top hits to the correct functions were considered in our analyses. The ambiguous sequences that contained specific functional domains were checked manually in the NCBI Conserved Domains database. To address the evolution histories of magnetosome synthesis and phototrophic abilities from the family Rhodospirillaceae, ancestral family sizes were inferred using the program COUNT with Dollo parsimony algorithm [78]. This approach strictly prohibits multiple gains of genes and allows reconstructing gene gain and loss events at both observed species and potential ancestors (leaves and nodes on the phylogenetic tree).
Additional files
Figure S1. Circular diagrams of the Magnetospirillum sp. XM-1 chromosome and plasmid show the relevant genome features. Figure S2. Colinearity plot shows the comparison of genome from the Magnetospirillum sp. XM-1 and Magnetospirillum magneticum AMB-1. Figure S3. (A) The comparison of iron related genes. (B) Summary of iron homeostasis features identified in the genomes. (C) Sketch of iron homeostasis systems in the Rhodospirillaceae family. Figure S4. The comparison of DNA damages repair genes. Figure S5. The comparison of the percentage of DNA damage repair gene from MTB and non-MTB. Figure S6. (A) Vann diagram of core, dispensable and specific genes from five groups. (B) Vann diagram of core, dispensable and specific genes. Figure S7. MTB strain XM-1 swam towards the UVA radiation and accumulated at the illuminate side of the quartz bottle. Figure S8. Sketch map shows the strategy of possible photosynthesis magnetotactic bacteria in Archean Eon when surface UV radiation was high. Magnetotaxis could help them to swim down to the ultraviolet tolerance photosynthesis zone (UTPZ) to avoid lethal doses of irradiation while harvest enough light. (PDF 3745 kb)
Table S1. General features of the XM-1 genome compared with other representative MTB genomic sequences from Rhodospirillaceae (PDF 64 kb)
Table S2. The bacteria genomes used in this study (PDF 105 kb)
Table S3 A. The core inventory shared by the family Rhodospirillaceae; Table S3 B. The core genes among MTB, phototrophic bacteria, non-phototrophic non-magnetotactic bacteria within Rhodospirillaceae and other MTB that are not related to the family Rhodospirillaceae (XLSX 178 kb)
Table S4 A. Oxidative related genes from the Rhodospirillaceae members; Table S4 B. Iron related genes from the Rhodospirillaceae members; Table S4 C DNA repair related genes from the Rhodospirillaceae members (XLSX 459 kb)
Acknowledgements
The authors thank Xiang Xiao and Haiwei Luo for useful discussions and Minghua Liu, Mo Huang and Jiahua Wang for kindly helping with computer programming support. The authors also thank three anonymous reviewers for their constructive comments, Artemis Louyakis for improving the English usage.
Funding
This work was funded by the National Natural Science Foundation of China (41621004, 41330104), the CAS Key Research Program of Frontier Sciences. The funding provided by the National Natural Science Foundation of China (41621004, 41330104) and the CAS Key Research Program of Frontier Sciences covered the field trip for sampling, complete genome sequencing and the experiments in the laboratories, as well as language editing and publishing.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the NCBI repository under Project Number PRJEB11958, https://www.ncbi.nlm.nih.gov/bioproject/PRJEB11958/.
Abbreviations
- AAI
Average amino acid identity
- ANI
Average nucleotide identity
- BER
Base pair excision repair
- BLUF
Blue-light using FAD
- CBB cycle
Calvin–Benson–Bassham cycle
- DSR
Dissimilatory sulfite reductase
- GGDEF
Gly-Gly-Asp-Glu-Phe amino-acid motif
- HGT
Horizontal gene transfer
- LOV
Light-oxygen-voltage photoreceptor
- MGC
Magnetosome gene cluster
- MMP
Multicellular magnetotactic prokaryote
- MTB
Magnetotactic bacteria
- NER
Nucleotide excision repair
- OATZ
Oxic-anoxic transition zone
- PHY
Red/far red light sensitive phytochrome
- PYP
Photoactive yellow protein
- ROS
Reactive oxygen species
- rTCA cycle
reductive tricarboxylic acid cycle
- RubisCO
Ribulose-1,5-bisphosphate carboxylase-oxygenase
- UTPZ
Ultraviolet tolerance photosynthesis zone
- UVR
Ultraviolet radiation
Author’s contributions
YW designed the experiments, analyzed data, and wrote the paper; YP initiated the project and wrote the paper; GC helped with the comparative genomics study and improved the paper; and WL, YL and FW wrote the paper and substantively revised it. All authors have read and approved the manuscript.
Ethics approval and consent to participate
No permissions were needed to collect the MTB strain from Xi’an city moat.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yinzhao Wang, Email: wyz@sjtu.edu.cn.
Giorgio Casaburi, Email: giorgiocasaburi@gmail.com.
Wei Lin, Email: weilin0408@gmail.com.
Ying Li, Email: yingli528@vip.sina.com.
Fengping Wang, Email: fengpingw@sjtu.edu.cn.
Yongxin Pan, Email: yxpan@mail.iggcas.ac.cn.
References
- 1.Bazylinski DA, Lefèvre CT, Schüler D. Magnetotactic bacteria. In: Rosenberg E, DeLong FE, Lory S, Stackebrandt E, Thompson F, editors. The prokaryotes. Berlin Heidelberg: Springer; 2013. pp. 453–494. [Google Scholar]
- 2.Uebe R, Schüler D. Magnetosome biogenesis in magnetotactic bacteria. Nat Rev Microbiol. 2016;14:621–637. doi: 10.1038/nrmicro.2016.99. [DOI] [PubMed] [Google Scholar]
- 3.Ali I, Peng C, Khan ZM, Naz I. Yield cultivation of magnetotactic bacteria and magnetosomes: a review. J Basic Microb. 2017;57:643–652. doi: 10.1002/jobm.201700052. [DOI] [PubMed] [Google Scholar]
- 4.Komeili A. Molecular mechanisms of compartmentalization and biomineralization in magnetotactic bacteria. FEMS Microbiol Rev. 2012;36:232–255. doi: 10.1111/j.1574-6976.2011.00315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ali I, Peng C, Khan ZM, Naz I, Sultan M. An overview of heavy metal removal from wastewater using magnetotactic bacteria. J Chem Technol Biotechnol. 2018. 10.1002/jctb.5648.
- 6.Islam T, Peng C, Ali I. Morphological and cellular diversity of magnetotactic bacteria: a review. J Basic Microb. 2017. 10.1002/jobm.201700383. [DOI] [PubMed]
- 7.Bazylinski DA, Frankel RB. Magnetosome formation in prokaryotes. Nat Rev Microbiol. 2004;2:217. doi: 10.1038/nrmicro842. [DOI] [PubMed] [Google Scholar]
- 8.Lin W, Bazylinski DA, Xiao T, Wu LF, Pan Y. Life with compass: diversity and biogeography of magnetotactic bacteria. Environ Microbiol. 2013;16:264658. doi: 10.1111/1462-2920.12313. [DOI] [PubMed] [Google Scholar]
- 9.Frankel RB, Bazylinski DA, Johnson MS, Taylor BL. Magneto-aerotaxis in marine coccoid bacteria. Biophys J. 1997;73:994–1000. doi: 10.1016/S0006-3495(97)78132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shapiro OH, Hatzenpichler R, Buckley DH, Zinder SH, Orphan VJ. Multicellular photo-magnetotactic bacteria. Environ Microbiol Reports. 2011;3:233–238. doi: 10.1111/j.1758-2229.2010.00215.x. [DOI] [PubMed] [Google Scholar]
- 11.Almeida FP, Viana NB, Lins U, Farina M, Keim CN. Swimming behaviour of the multicellular magnetotactic prokaryote ‘Candidatus Magnetoglobus multicellularis’ under applied magnetic fields and ultraviolet light. Antonie Van Leeuwenhoek. 2013;103:845–857. doi: 10.1007/s10482-012-9866-0. [DOI] [PubMed] [Google Scholar]
- 12.de Azevedo LV, de Barros HL, Keim CN, Acosta-Avalos D. Effect of light wavelength on motility and magnetic sensibility of the magnetotactic multicellular prokaryote ‘Candidatus Magnetoglobus multicellularis’. Antonie Van Leeuwenhoek. 2013;104:405–412. doi: 10.1007/s10482-013-9964-7. [DOI] [PubMed] [Google Scholar]
- 13.de Melo RD, Acosta-Avalos D. Light effects on the multicellular magnetotactic prokaryote ‘Candidatus Magnetoglobus multicellularis’ are cancelled by radiofrequency fields: the involvement of radical pair mechanisms. Antonie Van Leeuwenhoek. 2016;110:177–186. doi: 10.1007/s10482-016-0788-0. [DOI] [PubMed] [Google Scholar]
- 14.Abreu F, Morillo V, Nascimento FF, Werneck C, Cantão ME, Ciapina LP, et al. Deciphering unusual uncultured magnetotactic multicellular prokaryotes through genomics. ISME J. 2014;8:1055. doi: 10.1038/ismej.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C, Ma Q, Jiang W, Song T. Phototaxis in the magnetotactic bacterium Magnetospirillum magneticum strain AMB-1 is independent of magnetic fields. Appl Microbiol Biotechnol. 2011;90:269–275. doi: 10.1007/s00253-010-3017-1. [DOI] [PubMed] [Google Scholar]
- 16.Li K, Wang P, Chen C, Chen C, Li L, Song T. Light irradiation helps magnetotactic bacteria eliminate intracellular reactive oxygen species. Environ Microbiol. 2017;19:3638–3648. doi: 10.1111/1462-2920.13864. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Lin W, Li J, Pan Y. Changes of cell growth and magnetosome biomineralization in Magnetospirillum magneticum AMB-1 after ultraviolet-B irradiation. Front Microbiol. 2013;4:397. doi: 10.3389/fmicb.2013.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefevre CT, Bazylinski DA. Ecology, diversity, and evolution of Magnetotactic Bacteria. Microbiol Mol Biol Rev. 2013;77:497–526. doi: 10.1128/MMBR.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin W, Pan Y, Bazylinski DA. Diversity and ecology of and biomineralization by magnetotactic bacteria. Environ Microbiol Rep. 2017;9:345–356. doi: 10.1111/1758-2229.12550. [DOI] [PubMed] [Google Scholar]
- 20.Baldani JI, Videira SS, dos Santos Teixeira KR, Reis VM, de Oliveira ALM, Schwab S, et al. The family Rhodospirillaceae. The prokaryotes: Springer. p. 533–618.
- 21.Lefèvre CT, Schmidt ML, Viloria N, Trubitsyn D, Schüler D, Bazylinski DA. Insight into the evolution of magnetotaxis in Magnetospirillum spp., based on mam gene phylogeny. Appl Environ Microbiol. 2012;78:7238–7248. doi: 10.1128/AEM.01951-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolinko I, Lohße A, Borg S, Raschdorf O, Jogler C, Tu Q, et al. Biosynthesis of magnetic nanostructures in a foreign organism by transfer of bacterial magnetosome gene clusters. Nat Nanotechnol. 2014;9:193. doi: 10.1038/nnano.2014.13. [DOI] [PubMed] [Google Scholar]
- 23.Lefèvre CT, Trubitsyn D, Abreu F, Kolinko S, Almeida LGP, Vasconcelos ATR, et al. Monophyletic origin of magnetotaxis and the first magnetosomes. Environ Microbiol. 2013;15:2267–2274. doi: 10.1111/1462-2920.12097. [DOI] [PubMed] [Google Scholar]
- 24.Lin W, Patersona GA, Zhu Q, Wang Y, Kopylovad E, Li Y, et al. Origin of microbial biomineralization and magnetotaxis during the Archean. P Natl Acad Sci USA. 2017;114:2171–2176. doi: 10.1073/pnas.1614654114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ullrich S, Kube M, Schübbe S, Reinhardt R, Schüler D. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J Bacterial. 2005;187:7176–7184. doi: 10.1128/JB.187.21.7176-7184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abreu F, Cantao ME, Nicolás MF, Barcellos FG, Morillo V, Almeida LG, et al. Common ancestry of iron oxide-and iron-sulfide-based biomineralization in magnetotactic bacteria. ISME J. 2011;5:1634. doi: 10.1038/ismej.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jogler C, Kube M, Schübbe S, Ullrich S, Teeling H, Bazylinski DA, et al. Comparative analysis of magnetosome gene clusters in magnetotactic bacteria provides further evidence for horizontal gene transfer. Environ Microbiol. 2009;11:1267–1277. doi: 10.1111/j.1462-2920.2009.01854.x. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Lin W, Li J, Zhang T, Li Y, Tian J, et al. Characterizing and optimizing magnetosome production of Magnetospirillum sp. XM-1 isolated from Xi'an City moat, China. FEMS Microbiol Lett. 2015;362:fnv167. doi: 10.1093/femsle/fnv167. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Zhang T, Lin W, Zhang B, Cai Y, Yang C, et al. Complete genome sequence of Magnetospirillum sp. strain XM-1, isolated from the Xi’an City moat. China Genome Announ. 2016;4:e01171–e01116. doi: 10.1128/genomeA.01171-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsunaga T, Okamura Y, Fukuda Y, Wahyudi AT, Murase Y, Takeyama H. Complete genome sequence of the facultative anaerobic magnetotactic bacterium Magnetospirillum sp strain AMB-1. DNA Res. 2005;12:157–166. doi: 10.1093/dnares/dsi002. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Wang Q, Zhang W, Wang Y, Li L, Wen T, et al. Complete genome sequence of Magnetospirillum gryphiswaldense MSR-1. Genome Announ. 2014;2:e00171–e00114. doi: 10.1128/genomeA.00171-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji B, Zhang SD, Arnoux P, Rouy Z, Alberto F, Philippe N, et al. Comparative genomic analysis provides insights into the evolution and niche adaptation of marine Magnetospira sp. QH-2 strain. Environ Microbiol. 2013;16:525–544. doi: 10.1111/1462-2920.12180. [DOI] [PubMed] [Google Scholar]
- 33.Duquesne K, Prima V, Ji B, Rouy Z, Médigue C, Talla E, Sturgis JN. Draft genome sequence of the purple photosynthetic bacterium Phaeospirillum molischianum DSM120, a particularly versatile bacterium. J Bacteriol. 2012;194:3559–3560. doi: 10.1128/JB.00605-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivanovsky RN, Keppen OI, Lebedeva NN, Beletsky AV, Mardanov AV, Grouzdev DS. Draft genome sequence of the anoxygenic phototrophic bacterium Phaeospirillum fulvum MGU-K5. Genome Announ. 2017;5:e00895–e00817. doi: 10.1128/genomeA.00895-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raj PS, Chakravarthy SK, Ramaprasad EVV, Sasikala C, Ramana CV. Phaeospirillum tilakii sp. nov., a phototrophic alphaproteobacterium isolated from aquatic sediments. Int J Syst Evol Microbiol. 2012:621069–74. [DOI] [PubMed]
- 36.Lakshmi KVNS, Sasikala C, Takaichi S, Ramana CV. Phaeospirillum oryzae sp. nov., a spheroplast-forming, phototrophic alphaproteobacterium from a paddy soil. Int J Syst Evol Microbiol. 2011;61:1656–1661. doi: 10.1099/ijs.0.025544-0. [DOI] [PubMed] [Google Scholar]
- 37.Imhoff JF. Phaeospirillum. Bergey's manual of systematics of archaea and Bacteria. 2015. pp. 1–7. [Google Scholar]
- 38.Thrash JC, Ahmadi S, Torok T, Coates JD. Magnetospirillum bellicus sp. nov., a novel dissimilatory perchlorate-reducing alphaproteobacterium isolated from a bioelectrical reactor. Appl Environ Microbiol. 2010;76:4730–4737. doi: 10.1128/AEM.00015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bazylinski DA, Dean AJ, Schüler D, Phillips EJ, Lovley DR. N2-dependent growth and nitrogenase activity in the metal-metabolizing bacteria, Geobacter and Magnetospirillum species. Environ Microbiol. 2000;2:266–273. doi: 10.1046/j.1462-2920.2000.00096.x. [DOI] [PubMed] [Google Scholar]
- 40.Bazylinski DA, Blakemore RP. Denitrification and assimilatory nitrate reduction in Aquaspirillum magnetotacticum. Appl Environ Microbiol. 1983;46:1118–1124. doi: 10.1128/aem.46.5.1118-1124.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trubitsyn D, Abreu F, Ward FB, Taylor T, Hattori M, Kondo S, et al. Draft genome sequence of Magnetovibrio blakemorei strain MV-1, a marine vibrioid magnetotactic bacterium. Genome Announ. 2016;4:e01330–e01316. doi: 10.1128/genomeA.01330-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grouzdev DS, Dziuba MV, Sukhacheva MS, Mardanov AV, Beletskiy AV, Kuznetsov BB, Skryabin KG. Draft genome sequence of Magnetospirillum sp. strain SO-1, a freshwater magnetotactic bacterium isolated from the Ol’khovka river, Russia. Genome Announ. 2014;2. [DOI] [PMC free article] [PubMed]
- 43.Richter M, Kube M, Bazylinski DA, Lombardot T, Glöckner FO, Reinhardt R, Schüler D. Comparative genome analysis of four magnetotactic bacteria reveals a complex set of group-specific genes implicated in magnetosome biomineralization and function. J Bacteriol. 2007;189:4899–4910. doi: 10.1128/JB.00119-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koziaeva VV, Dziuba MV, Ivanov TM, Kuznetsov BB, Skryabin KG, Grouzdev DS. Draft genome sequences of two magnetotactic bacteria, Magnetospirillum moscoviense BB-1 and Magnetospirillum marisnigri SP-1. Genome Announ. 2016;(4):e00814–6. [DOI] [PMC free article] [PubMed]
- 45.Kantor RS, van Zyl AW, van Hille RP, Thomas BC, Harrison ST, Banfield JF. Bioreactor microbial ecosystems for thiocyanate and cyanide degradation unravelled with genome-resolved metagenomics. Environ Microbiol. 2015;17:4929–4941. doi: 10.1111/1462-2920.12936. [DOI] [PubMed] [Google Scholar]
- 46.Monteil CL, Perrière G, Menguy N, Ginet N, Alonso B, Waisbord N, et al. Genomic study of a novel magnetotactic Alphaproteobacteria uncovers the multiple ancestry of magnetotaxis. Environ Microbiol. 2018;20:4415–4430. doi: 10.1111/1462-2920.14364. [DOI] [PubMed] [Google Scholar]
- 47.Schuebbe S, Williams TJ, Xie G, Kiss HE, Brettin TS, Martinez D, et al. Complete genome sequence of the chemolithoautotrophic marine magnetotactic coccus strain MC-1. Appl Environ Microbiol. 2009;75:4835–4852. doi: 10.1128/AEM.02874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ji B, Zhang SD, Zhang WJ, Rouy Z, Alberto F, Santini CL, et al. The chimeric nature of the genomes of marine magnetotactic coccoid-ovoid bacteria defines a novel group of Proteobacteria. Environ Microbiol. 2016;19:1103–1119. doi: 10.1111/1462-2920.13637. [DOI] [PubMed] [Google Scholar]
- 49.Araujo ACV, Morillo V, Cypriano J, Teixeira LCRS, Leão P, Lyra S, et al. Combined genomic and structural analyses of a cultured magnetotactic bacterium reveals its niche adaptation to a dynamic environment. BMC Genomics. 2016;17:363. doi: 10.1186/s12864-016-2693-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo H, Moran MA. How do divergent ecological strategies emerge among marine bacterioplankton lineages? Trends Microbiol. 2015;23:577–584. doi: 10.1016/j.tim.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 51.Guo F, Yang W, Jiang W, Geng S, Peng T, Li J. Magnetosomes eliminate intracellular reactive oxygen species in Magnetospirillum gryphiswaldense MSR-1. Environ Microbiol. 2012;14:1722–1729. doi: 10.1111/j.1462-2920.2012.02707.x. [DOI] [PubMed] [Google Scholar]
- 52.Gao Y, Wei Z, Li F, Yang ZM, Chen YM, Zrinyi M, et al. Synthesis of a morphology controllable Fe3O4 nanoparticle/hydrogel magnetic nanocomposite inspired by magnetotactic bacteria and its application in H2O2 detection. Green Chem. 2014;16:1255–1261. doi: 10.1039/C3GC41535J. [DOI] [Google Scholar]
- 53.Elías-Arnanz M, Padmanabhan S, Murillo FJ. Light-dependent gene regulation in nonphototrophic bacteria. Curr Opin Mircobiol. 2011;14:128–135. doi: 10.1016/j.mib.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 54.van der Horst MA, Key J, Hellingwerf KJ. Photosensing in chemotrophic, non-phototrophic bacteria: let there be light sensing too. Trends Microbial. 2007;15:554–562. doi: 10.1016/j.tim.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 55.Nudel CB, Hellingwerf KJ. Photoreceptors in chemotrophic prokaryotes: the case of Acinetobacter spp. revisited. Photochem Photobiol. 2015;91:1012–1020. doi: 10.1111/php.12491. [DOI] [PubMed] [Google Scholar]
- 56.Zheng Q, Zhang R, Fogg PC, Beatty JT, Wang Y, Jiao N. Gain and loss of phototrophic genes revealed by comparison of two Citromicrobium bacterial genomes. PLoS One. 2012;7:e35790. doi: 10.1371/journal.pone.0035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mulkidjanian AY, Galperin MY. A time to scatter genes and a time to gather them: evolution of photosynthesis genes in bacteria. In: Thomas Beatty J editors. Advances in Botanical Research. Academic Press; 2013. pp. 1–35. [Google Scholar]
- 58.Lefèvre CT, Wu LF. Evolution of the bacterial organelle responsible for magnetotaxis. Trends Microbiol. 2013;21:534–543. doi: 10.1016/j.tim.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 59.Wang Y, Pan Y. Ultraviolet-B Radiation Effects on the Community, Physiology, and Mineralization of Magnetotactic Bacteria. In: de Bruijn FJ editor. Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria. John Wiley & Sons, Inc; 2016. p. 532–544.
- 60.Lefèvre CT. Genomic insights into the early-diverging magnetotactic bacteria. Environ Microbiol. 2016;18:1–3. doi: 10.1111/1462-2920.12989. [DOI] [PubMed] [Google Scholar]
- 61.Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 62.Luo H, Löytynoja A, Moran MA. Genome content of uncultivated marine Roseobacters in the surface ocean. Environ Microbial. 2012;14:41–51. doi: 10.1111/j.1462-2920.2011.02528.x. [DOI] [PubMed] [Google Scholar]
- 63.Albalat R, Cañestro C. Evolution by gene loss. Nat Rev Genet. 2016;17:379–391. doi: 10.1038/nrg.2016.39. [DOI] [PubMed] [Google Scholar]
- 64.Maughan H, Callicotte V, Hancock A, Birky CW, Nicholson WL, Masel J. The population genetics of phenotypic deterioration in experimental populations of Bacillus subtilis. Evolution. 2006;60:686–695. doi: 10.1111/j.0014-3820.2006.tb01148.x. [DOI] [PubMed] [Google Scholar]
- 65.Lahti DC, Johnson NA, Ajie BC, Otto SP, Hendry AP, Blumstein DT, et al. Relaxed selection in the wild. Trends Ecol Evol. 2009;24:487–496. doi: 10.1016/j.tree.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 66.Zheng Q, Zhang R, Koblížek M, Boldareva EN, Yurkov V, Yan S, Jiao N. Diverse arrangement of photosynthetic gene clusters in aerobic anoxygenic phototrophic bacteria. PLoS One. 2011;6:e25050. doi: 10.1371/journal.pone.0025050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cockell CS. Ultraviolet radiation and the photobiology of earth's early oceans. Origins Life Evol B. 2000;30:467–500. doi: 10.1023/A:1006765405786. [DOI] [PubMed] [Google Scholar]
- 68.Cockell CS, Raven JA. Ozone and life on the Archaean earth. Philos T R Soc A. 2007;365:1889–1901. doi: 10.1098/rsta.2007.2049. [DOI] [PubMed] [Google Scholar]
- 69.Médigue C, Calteau A, Cruveiller S, Gachet M, Gautreau G, Josso A, et al. MicroScope-an integrated resource for community expertise of gene functions and comparative analysis of microbial genomic and metabolic data. Brief Bioinform. bbx113. [DOI] [PMC free article] [PubMed]
- 70.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, et al. Versatile and open software for comparing large genomes. Genome boil. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee I, Kim YO, Park SC, Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Micr. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 72.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 73.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Segata N, Börnigen D, Morgan XC, Huttenhower C. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun. 2013;4. [DOI] [PMC free article] [PubMed]
- 75.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li M, Toner BM, Baker BJ, Breier JA, Sheik CS, Dick GJ. Microbial iron uptake as a mechanism for dispersing iron from deep-sea hydrothermal vents. Nat Commun. 2014;5:3192. doi: 10.1038/ncomms4192. [DOI] [PubMed] [Google Scholar]
- 78.Csuros M. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics. 2010;26:1910–1912. doi: 10.1093/bioinformatics/btq315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Circular diagrams of the Magnetospirillum sp. XM-1 chromosome and plasmid show the relevant genome features. Figure S2. Colinearity plot shows the comparison of genome from the Magnetospirillum sp. XM-1 and Magnetospirillum magneticum AMB-1. Figure S3. (A) The comparison of iron related genes. (B) Summary of iron homeostasis features identified in the genomes. (C) Sketch of iron homeostasis systems in the Rhodospirillaceae family. Figure S4. The comparison of DNA damages repair genes. Figure S5. The comparison of the percentage of DNA damage repair gene from MTB and non-MTB. Figure S6. (A) Vann diagram of core, dispensable and specific genes from five groups. (B) Vann diagram of core, dispensable and specific genes. Figure S7. MTB strain XM-1 swam towards the UVA radiation and accumulated at the illuminate side of the quartz bottle. Figure S8. Sketch map shows the strategy of possible photosynthesis magnetotactic bacteria in Archean Eon when surface UV radiation was high. Magnetotaxis could help them to swim down to the ultraviolet tolerance photosynthesis zone (UTPZ) to avoid lethal doses of irradiation while harvest enough light. (PDF 3745 kb)
Table S1. General features of the XM-1 genome compared with other representative MTB genomic sequences from Rhodospirillaceae (PDF 64 kb)
Table S2. The bacteria genomes used in this study (PDF 105 kb)
Table S3 A. The core inventory shared by the family Rhodospirillaceae; Table S3 B. The core genes among MTB, phototrophic bacteria, non-phototrophic non-magnetotactic bacteria within Rhodospirillaceae and other MTB that are not related to the family Rhodospirillaceae (XLSX 178 kb)
Table S4 A. Oxidative related genes from the Rhodospirillaceae members; Table S4 B. Iron related genes from the Rhodospirillaceae members; Table S4 C DNA repair related genes from the Rhodospirillaceae members (XLSX 459 kb)
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the NCBI repository under Project Number PRJEB11958, https://www.ncbi.nlm.nih.gov/bioproject/PRJEB11958/.