Abstract
In recent work, Whiteley et al. (2019) define a family of bacterial nucleotidyltransferases (CD-NTases) capable of synthesizing pyrimidine containing cyclic dinucleotides and cyclic trinucleotides. CD-NTases are broadly distributed across bacterial phyla, suggesting that they play important roles in bacterial physiology and modulation of the metazoan host innate immune system.
Much like a symphony with diverse instruments each playing their own individual part to beautifully harmonize, bacteria have a wide diversity of second-messenger molecules that coordinate function to drive adaptation to environmental change. At the turn of the millennium, the importance of a new set of second-messenger “instruments,” the cyclic dinucleotides (CDNs), began to be appreciated. Recent research by Whiteley et al. (2019) adds to this signaling orchestra with the discovery of widely distributed oligonucleotide signaling molecules, including previously uncharacterized pyrimidine-containing CDNs and cyclic trinucleotides (CTN), suggesting that we are just beginning to understand the melody of bacterial second-messenger signaling.
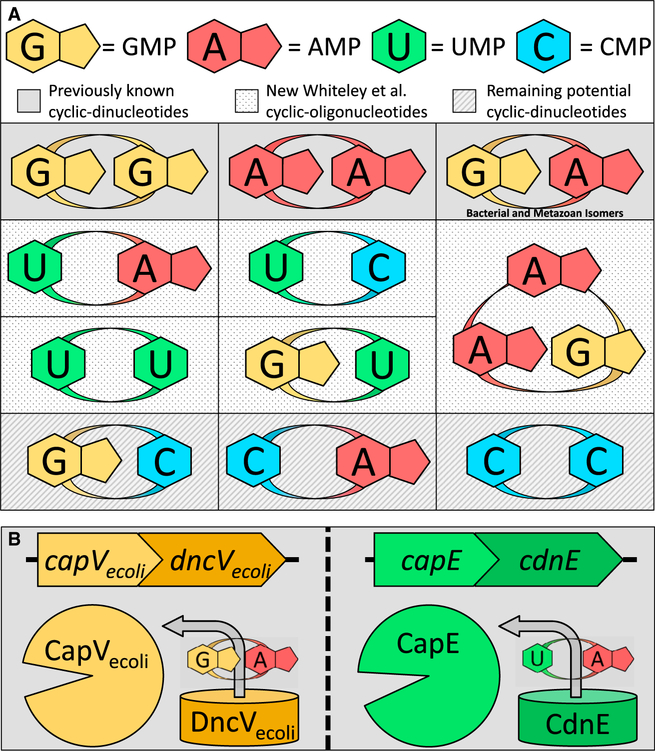
Formerly, the four known CDN signaling molecules were all composed of purine nucleotides: cyclic-di-GMP (c-di-GMP), cyclic-di-AMP (c-di-AMP), and 3′3′ cyclic-GMP-AMP (3′3′-cGAMP) in bacteria and the metazoan 2′3′ cyclic-GMP-AMP (2′3′-cGAMP) (Figure 1A). While each bacterial CDN has been implicated in a myriad of functions, c-di-GMP is most commonly associated with coordinating the transition between biofilm formation and motility (Römling et al., 2013), c-di-AMP with the regulation of osmotic stress (Corrigan & Grundling 2013), and 3′3′-cGAMP with membrane degradation (Severin et al., 2018). In metazoans, the enzyme cGAS synthesizes 2′3′-cGAMP upon recognition of DNA that is inappropriately localized to the cytoplasm (i.e., in case of viral infection or genome instability) (Margolis et al., 2017). The subsequent detection of 2′3′-cGAMP, as well as the bacterial CDNs, by CDN-binding immune modulators stimulates a proinflammatory response (McFarland et al., 2017; Margolis et al., 2017). Pyrimidine-containing CDNs were conspicuously absent, and it was hypothesized that their limited capacity for hydrogen bonding and base stacking interactions made uracil and cytosine intrinsically less favorable substrates for regulating critical signaling networks (Nelson & Breaker 2017).
Figure 1. Expansion of the Cyclic-oligonucleotide Lexicon.
(A) The four previously known CDN molecules were all composed of purine nucleotides (c-di-GMP, c-di-AMP, and both the bacterial 3′3′ cyclic-GMP-AMP and metazoan 2′3′ cyclic-GMP-AMP). With the identification of the bacterial CD-NTase family of enzymes, Whiteley et al. (2019) have expanded this signaling lexicon to include cyclic-UMP-AMP, cyclic-UMP-CMP, cyclic-di-UMP, cyclic-GMP-UMP, and the cyclic trinucleotide cyclic-AMP-AMP-GMP. Three possible cyclic dinucleotides remain to be discovered: cyclic-GMP-CMP, cyclic-CMP-AMP, and cyclic-di-CMP.
(B) Cartoon depicting the genetic duplication of the V. cholerae capV and dncV-like partial operons found in the mammalian commensal strain of E. coli ECOR31. Left: the more conserved pair of enzymes, dncVecoli and capVecoli, predominantly synthesize and respond, respectively, to cyclic-GMP-AMP. Right: the more degenerate pair, cdnE and capE, predominantly synthesize and respond, respectively, to cyclic-UMP-AMP.
Central to the research of Whiteley et al. (2019) was the distribution and conservation of the catalytic activity of the bacterial 3′3′-cGAMP synthase DncV. DncV is structurally similar to cGAS but distinct from the diguanylate cyclase (DGC) and diadenylate cyclase (DAC) domains that synthesize c-di-GMP and c-di-AMP, respectively. In V. cholerae, dncV is encoded on a unique mobile genetic element, VSP-1, and the 3′3′-cGAMP it synthesizes activates the neighboring patatin-like phospholipase CapV (Severin et al., 2018). Whiteley et al. (2019) noticed that an animal commensal strain of Escherichia coli (ECOR31) harbors a duplication of capV and dncV, and they named the more conserved gene pair capVecoli and dncVecoli and the more degenerate copy capE and cdnE (Figure 1B). In vitro evaluation of nucleotide synthase activity revealed that while DncVecoli primarily produced 3′3′-cGAMP, analogous to V. cholerae DncV (Davies et al., 2012), the more divergent CdnE generated the hybrid CDN cyclic-UMP-AMP (cUMP-AMP). Incredibly, the phospholipase activities of CapVecoli and CapE were only activated in vitro by the CDN produced by their neighboring synthase (Figure 1B). This describes a naturally synthesized pyrimidine containing CDN and cUMP-AMP responsive effector protein, and it also demonstrates the insulated signaling capacity of CDN networks originating from homologous synthases.
To understand how CdnE synthesizes cUMP-AMP, Whiteley et al. (2019) determined the crystal structure of a thermophilic CdnE ortholog from Rhodothermus marinus in complex with non-hydrolyzable nucleotide substrates. This analysis revealed that an active site Asparagine (Asn) engages in hydrogen binding with the uracil base. Conversely, the active sites of DncV and cGAS contain an analogous Serine (Ser) residue that interacts with purine bases. Hypothesizing that this active site Asn was a structural determinant of nucleotide discrimination in CdnE, Whiteley et al. (2019) tested the in vitro catalytic active of the CdnEAsn166Ser variant. Amazingly, this single amino acid substitution reprogrammed CdnE from producing predominantly cUMP-AMP to preferentially synthesize c-di-AMP. This was reversibly demonstrated in the CdnE homolog from Elizabethkingia meningoseptica, where substitution of the natively encoded Ser with Asn switched the enzyme from preferentially synthesizing purine derived CDNs to those containing pyrimidines. The authors named this family of enzymes CD-NTases for cGAS/DncV-like nucleotidyltransferases, making this the third family of enzymes—along with DGCs and DACs—that can synthesize CDNs. The flexibility to change the catalytic repertoire of CD-NTases through a single amino acid substitution then raises the questions: what other cyclic-nucleotide species can be synthesized by CD-NTases, and which bacteria harbor them?
Building on the shoulders of a prescient bioinformatic analysis in 2015 by Burroughs et al., (2015), Whiteley et al. (2019) performed an expansive bioinformatic search for CD-NTases and discovered that homologs are widely distributed across nearly all bacterial phyla and encoded in greater than 10% of the available bacterial genomes. Additionally, like DncV and CdnE, many of these enzymes appear to be encoded in conserved operons within mobile genetic elements, suggesting they are not part of the core genomes of bacterial species but rather newly acquired adaptive traits. Eight CD-NTase clades were identified based on sequence alignments of the more than 5,600 unique enzymes identified. To understand the enzymatic repertoire of CD-NTases in these clades, Whiteley et al. (2019) purified 66 candidate enzymes and evaluated their capacity to catalyze nucleotide-based chemistry in vitro. Sixteen of the most active CD-NTases synthesized oligomeric nucleotide-derived products including seven CDN species (such as the CDN cyclic-di-UMP), the CTN cyclic-AMP-AMP-GMP (cAAG), and several nucleotide products of unknown composition and structure (Figure 1A). These observations greatly expand the diversity of naturally synthesized nucleotide-based signaling molecules and suggest that they may regulate undefined biological functions in bacteria and in their metazoan host.
In metazoans, cytoplasmic 2′3′-cGAMP, as well as the three previously known bacterial CDNs, stimulate a proinflammatory response through differing allosteric interactions with the CDN-sensing immune modulators STING (Margolis et al., 2017) and RECON (McFarland et al., 2017). Hypothesizing that some of these cyclic oligonucleotides might also regulate innate immunity, Whiteley et al. (2019) incubated cUMP-AMP and cAAG with STING and RECON. Interestingly, neither of these signals activated STING, but both were able to bind RECON and alter its catalytic activity. The co-crystal structure of cAAG in complex with mouse RECON revealed that binding interactions between the two molecules extend beyond the known CDN binding site to include additional conserved residues found in other RECON homologs. This raises the possibility that the cyclic oligonucleotides described in this study may have a role in mediating commensal and pathogenic interactions between bacteria and their host.
By defining the CD-NTase family of diverse cyclic dinucleotide synthases, Whiteley et al. (2019) greatly expand our knowledge of the cyclic-oligonucleotide chemical lexicon. These observations raise exciting questions and prospects for future research: what biological processes do these signals regulate, and when are they synthesized in vivo? How are these signals degraded once they are no longer needed? Why are the signaling networks packaged in mobile genetic elements and not part of core genomes? CD-NTases are commonly found in bacteria that have close association with plant and animal hosts, and the interaction of the signals they synthesize with STING and RECON suggests that they mediate interactions with the innate immune system. The research in this paper ushers in a new era of CDN and now CTN signaling, and it is going to be fascinating to see what music they make.
ACKNOWLEDGMENTS
C.M.W. acknowledges support from NIH grants GM109259, GM110444, and AI130554. We appreciate Nicolas Fernandez for comments on the manuscript.
REFERENCES
- Burroughs AM, Zhang D, Schäffer DE, Iyer LM, and Aravind L (2015). Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res. 43, 10633–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, and Gründling A (2013). Cyclic di-AMP: another second messenger enters the fray. Nat. Rev. Microbiol 11, 513–524. [DOI] [PubMed] [Google Scholar]
- Davies BW, Bogard RW, Young TS, and Mekalanos JJ (2012). Coordinated regulation of accessory genetic elements produces cyclic dinucleotides for V. cholerae virulence. Cell 149, 358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis SR, Wilson SC, and Vance RE (2017). Evolutionary origins of cGAS-STING signaling. Trends Immunol. 38, 733–743. [DOI] [PubMed] [Google Scholar]
- McFarland AP, Luo S, Ahmed-Qadri F, Zuck M, Thayer EF, Goo YA, Hybiske K, Tong L, and Woodward JJ (2017). Sensing of bacterial cyclic dinucleotides by the oxidoreductase RECON promotes NF-kB activation and shapes a proinflammatory antibacterial state. Immunity 46, 433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, and Breaker RR (2017). The lost language of the RNA World. Sci. Signal 10, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U, Galperin MY, and Gomelsky M (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev 77, 1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin GB, Ramliden MS, Hawver LA, Wang K, Pell ME, Kieninger AK, Khataokar A, O’Hara BJ, Behrmann LV, Neiditch MB, et al. (2018). Direct activation of a phospholipase by cyclic GMP-AMP in El Tor Vibrio cholerae. Proc. Natl. Acad. Sci. USA 115, E6048–E6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley AT, Eaglesham JB, de Oliveira Mann CC, Morehouse BR, Lowey B, Nieminen EA, Danilchanka O, King DS, Lee ASY, Mekalanos JJ, and Kranzusch PJ (2019). Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 567, 194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]