Abstract
Understanding the number of times a trait has evolved is a necessary foundation for comprehending its potential relationships with selective regimes, developmental constraints and evolutionary diversification. Rodents make up over 40% of extant mammalian species, and their ecological and evolutionary success has been partially attributed to the increase in biting efficiency that resulted from a forward shift of one or two portions of the masseter muscle from the zygomatic arch onto the rostrum. This forward shift has occurred in three discrete ways, but the number of times it has occurred has never been explicitly quantified. We estimated an ultrametric phylogeny, the first to include all rodent families, using thousands of ultraconserved elements. We examined support for evolutionary relationships among the five rodent suborders and then incorporated relevant fossils, fitted models of character evolution, and used stochastic character mapping to determine that a portion of the masseter muscle has moved forward onto the rostrum at least seven times (with one reversal) during the approximately 70 Myr history of rodents. Combined, the repeated evolution of this key innovation, its increasing prevalence through time, and the species diversity of clades with this character underscores the adaptive value of improved biting efficiency and the relative ease with which some advantageous traits arise.
Keywords: convergence, hystricomorphy, parallel evolution, Rodentia, stochastic character mapping, ultraconserved elements
1. Introduction
The acquisition and mechanical processing of foods are critical tasks that undoubtedly result in strong selective pressure on the rostrum and masticatory apparatus of vertebrates [1]. The strength of this selective pressure is manifested in the repeated evolution of morphotypes in taxa that use the same foods, but are not each other's closest relatives. Examples of rostral morphotypes that have evolved repeatedly include pharyngeal jaws specialized for grinding shrimp in African cichlids [2], elongated rostra and reduced dentitions in ant-eating mammals [3] and decurved bills in nectivorous tanagers [4]. Diprotodonty, the spatial and functional separation of the incisors and cheek teeth, is another rostral morphotype that has evolved independently in multiple mammal clades (e.g. some marsupials, multituberculates, aye-ayes and rodents) [5]. In rodents, this arrangement allows engagement of either the incisors or the cheek teeth, but never both simultaneously. Thus, rodents gnaw with their incisors (e.g. chiselling, cropping, bark removal) while the molars are disengaged [6], and at other times, they grind food items with their cheek teeth while the incisors are disengaged [7].
Modifications to diprotodonty in rodents have increased the efficiency of gnawing and chewing, almost certainly contributing to their substantial evolutionary and ecological success. Gnawing and chewing are enabled by the masseter muscle, which pulls the jaw forward. It originates on the zygomatic arch in the earliest rodents and the living mountain beaver (protrogomorphy, figure 1). However, these feeding activities are more efficient when portions of the masseter have a more anterior origin on the rostrum [8–10]. Over the course of rodent evolution, the origin of different portions of the masseter has moved forward onto the rostrum in three general ways: (i) the deep masseter extends underneath the zygomatic arch on to the rostrum and no muscle passes through a small infraorbital foramen (sciuromorphy); (ii) part of the zygomatico-mandibularis (part of the masseter) originates on the rostrum and passes through a greatly enlarged infraorbital foramen (hystricomorphy); or (iii) the characteristics of sciuromorphy and hystricomorphy are combined with the deep masseter extended on to the rostrum underneath the zygomatic arch and the infraorbital portion of the zygomatico-mandibularis passes through a moderately sized infraorbital foramen (myomorphy) [11] (figure 1). Cox et al. [12] suggested that bite efficiency differs among these three masseteric systems, with sciuromorphy improving gnawing efficiency, hystricomorphy improving chewing efficiency and myomorphy maximizing the efficiency of both gnawing and chewing.
Figure 1.
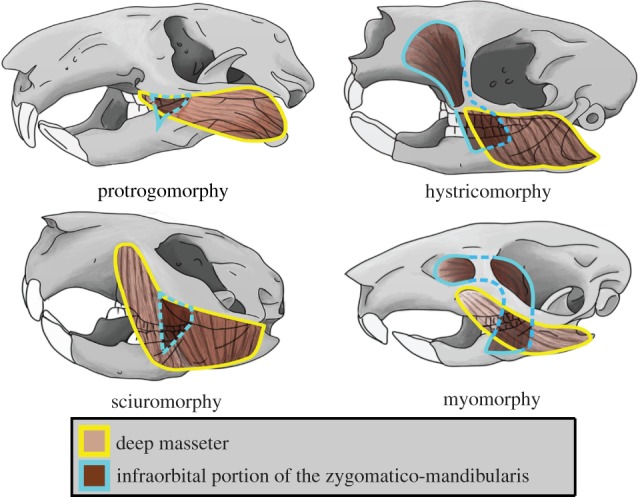
Illustrations of discrete masseter character states.
Although some variation on the four masseteric systems exists [13,14], the largely discrete differences between forms led early taxonomists (prior to recognition of protrogomorphy) to organize rodent diversity into Sciuromorpha, Hystricomorpha and Myomorpha [8,15,16]. However, convergent evolution of the sciuromorphous, hystricomorphous and myomorphous masseter architectures from a protrogomorphous ancestor has been suspected for decades [11], and has been confirmed by molecular phylogenetic studies [17–24]. These molecular phylogenies, built from limited numbers of loci, indicate that Rodentia comprises the monophyletic suborders Ctenohystrica (gundis, mole-rats and guinea pig-related species), Sciuromorpha (dormice, the mountain beaver and squirrels), Anomaluromorpha (springhares and scaly-tailed flying squirrels), Castorimorpha (beavers and pocket gophers) and Myomorpha (mice), with the last three forming a ‘mouse-related clade’. Relationships among Ctenohystrica, Sciuromorpha and the mouse-related clade, as well as among the three suborders within the mouse-related clade, however, remain debated, and their resolution is necessary for a clearer image of how masseter morphology has evolved. Short branches around these nodes, indicating rapid divergence, have resulted in low support and conflicting topologies. A plurality of studies prefer a sister relationship between Ctenohystrica and the mouse-related clade and to a lesser extent a sister relationship between Anomaluromorpha and Myomorpha [25]. Nevertheless, confidence has been placed in the Ctenohystrica and mouse-related clade sister relationship because it is supported by SINEs, which are argued to be homoplasy-free [20,24].
Several authors have used hundreds to thousands of nuclear loci to examine topology at the rodent root and recovered all three possible topologies. However, these studies have found low statistical support [26–28] or neglected to closely evaluate evidence at this node [29,30]. The latter is problematic as large molecular datasets often have inflated bootstrap values [31], while small numbers of anomalous loci can drive phylogenetic signal [32]. Additionally, no phylogenomic study of rodent relationships has included samples from more than nine of 32 families, or the taxa necessary to discern the topology of suborders within the mouse-related clade. Poor taxon sampling can lead to inaccurate topological inferences [33,34].
Uncertainty in the relationships among rodent suborders has prevented objective estimates of the number of times the hystricomorphous and sciuromorphous masseter architectures have evolved. Estimates of how many times a portion of the zygomatico-mandibularis has moved its origin forward through the infraorbital foramen onto the rostrum vary widely, and include, for instance, four origins across crown rodents [35], four origins in Ctenohystrica alone [36] and 11 origins of sciuromorphy and hystricomorphy combined [11]. An improved understanding of how many times these modifications have arisen would provide a foundation for understanding the genetic and developmental pathways that underlie these adaptations, as well as the traits' effect on the evolutionary and ecological success of rodents. In this study, we infer relationships among rodent suborders using ultraconserved element (UCE) sequences from at least one representative of each rodent family. We then incorporate relevant fossil taxa, scored for masseter morphology, within an ultrametric tree and use stochastic character mapping to estimate the number of transitions among masseter character states.
2. Methods
We collected UCE loci from 17 species using in silico alignment of a 5060 UCE probe set [37] to genomes available from NCBI, followed by extraction of the matched region +300 bp of flanking nucleotides on each side. To improve taxon sampling, we enriched and sequenced UCEs in 34 additional species in order to have representatives from each family within Rodentia and as many subfamilies as possible. We extracted genomic DNA from tissue samples using a Qiagen DNeasy Blood and Tissue Kit. As there is no fresh tissue of Platacanthomys lasiurus available, we extracted genomic DNA using a phenol-chloroform protocol on a piece of dried skin taken from a museum study skin (http://protocols.faircloth-lab.org/en/latest/protocols-lab/extraction/phenol-chloroform-extraction.html).
After quantifying DNA in extracts with a Qubit 2.0 fluorometer, we cleaned 1000 ng aliquots with 3×the volume of Sera-Mag Carboxylate-modified SpeedBeads [38] and eluted DNA into 30 µl of TE buffer. We mechanically sheared DNA in 2.5 min increments at 17 milliamps with an Epigentek Episonic sonicator until the average fragment size was about 500 bp, as assessed by eye on an electrophoretic gel. Our sample of P. lasiurus was excluded from this step.
We prepared, enriched and sequenced DNA libraries as described by Esselstyn et al. [27] and used the Python package PHYLUCE [39] for subsequent data processing. We assembled cleaned reads into contigs using Trinity v. r2013.08.14 [40] and extracted contigs for each taxon that matched UCE loci. We assembled an incomplete dataset containing UCE loci that were present in at least 39 of the 51 taxa (76%). We aligned each locus with MAFFT [41], and we trimmed resulting alignments to allow missing nucleotides at the flanks of each alignment only if at least 65% of taxa contained data, which is the default in PHYLUCE. We further trimmed uncertain alignment regions using Gblocks [42] with default parameters except for the minimum number of sequences for a flank position, which we set at 65%. We created a concatenated file of all loci [43] using PHYLUCE. Our alignments contain two lagomorphs as an outgroup (Oryctolagus cuniculus and Ochotona princeps) and 49 ingroup species representing all 32 rodent families [44].
We generated a concatenated tree using RAxML-HPC2 with a GTR + Γ model and 100 bootstrap replicates. We reconstructed gene trees for each locus with RAxML 8.2.9 [45] using the GTR + Γ model, with 10 maximum-likelihood searches performed per locus. We then used PHYLUCE to generate 100 multi-locus bootstraps of the data [46] and performed gene tree inference on these replicates in RAxML for use in approximate multi-species coalescent methods to estimate the species tree in ASTRAL v. 4.10.12 [47,48] and ASTRID v. 1.4 [49]. We summarized bootstrap values and annotated trees from each analysis using DendroPy 3.12.0 [50].
To understand how many individual gene trees support each possible topology at our nodes of interest, we calculated the proportion of gene trees containing particular bipartitions using a custom Python script (http://github.com/carloliveros/mammals [29]). We removed Platacanthomys lasiurus from this analysis because it was an outlier for average locus length (electronic supplementary material, table S1). We also determined the strength of support of individual loci by comparing the difference in per-locus log-likelihood values [29,31] between our concatenated topology and each possible alternative topology at our nodes of interest.
We transformed our concatenated topology into a dated, ultrametric tree using the penalized likelihood approach of TreePL [51]. We used nine fossil node calibrations to set minimum dates across the tree (table 1), but excluded two commonly used fossils (Spurimus and Douglassciurus jeffersoni) because of doubt over their phylogenetic relationships [60–62]. We set maximum dates for these calibrations to the beginning of the geological epoch prior to each fossil's occurrence, except for the Mus–Rattus maximum divergence date, which is from Kimura et al. [59].
Table 1.
Fossil constraints corresponding to nodes in figure 1 that were used in dating analysis with TreePL.
| constraint | constrained clade | fossil species | minimum age (Mya) | maximum age (Mya) | minimum age citations |
|---|---|---|---|---|---|
| A | Rodentia | Paramys atavus | 56 | 93.9 | [52] |
| B | Castorimorpha | Mattimys | 52.4 | 66 | [53] |
| C | Sciuromorpha | Eogliravus | 52.5 | 66 | [54] |
| D | Myomorpha | Pappocricetodon | 41 | 66 | [54] |
| E | Caviomorpha–Phiomorpha | Cachiyacuy contamanensis | 40.9 | 66 | [55,56] |
| F | Anomaluromorpha | Pondaungimys | 37.1 | 66 | [57] |
| G | Chinchilloidea–Octodontoidea | Eoviscaccia frassinettii | 31.3 | 56 | [56] |
| H | Diatomyidae–Ctenodactylidae | Fallomus | 28.3 | 56 | [58] |
| I | Rattus–Mus | Karnimata | 11.1 | 16 | [59] |
To include fossil taxa in our reconstruction of masseter evolution in rodents, we added fossil tips to the dated tree from Marivaux et al. [54]. Only fossils with confident sister relationships and masseter morphology states that differ from their extant sister group were grafted to the tree. These fossils were added using the R function fossil.graft (https://github.com/evolucionario [63]).
Masseter morphology character states for extant taxa were assigned based largely on primary sources involving dissections [35,36,64–83] (electronic supplementary material, table S1), summaries provided by Rodrigues [84] and Montgelard et al. [18], and by examination of skull specimens of each sampled family (except Diatomyidae). We chose to assign the four classic masseter states despite known variation within states. Our goals were to be objective, to keep the number of states manageable, and to provide the overarching framework needed for future investigations of morphological variation within each of these categories. Character states for fossil taxa were assigned based on Marivaux et al. [54].
We fit models of character evolution in which all transition rates are equal (ER), forward and reverse transitions occur at equal rates (SYM) and all possible transitions can occur at different rates (ARD) using the fitMk function in the R package phytools [63,85]. Since myomorphy is defined by the attachment of two separate muscles originating on the rostrum and protrogomorphy has no rostral attachments, we considered direct transitions between these two states to be highly unlikely. We therefore also fit partially ordered models in which transitions are possible between all states except directly between protrogomorphy and myomorphy. We used the model with the best weighted AIC score to generate 1000 character histories using the make.simmap function in phytools [85]. We then summarized these histories to estimate the number of transitions between masseter states in the dated tree.
3. Results
Our alignment consists of 2213 loci from 49 ingroup and two outgroup taxa, while allowing up to 12 missing taxa per locus. Individual loci are 121–947 () bp long with 0–288 () informative sites.
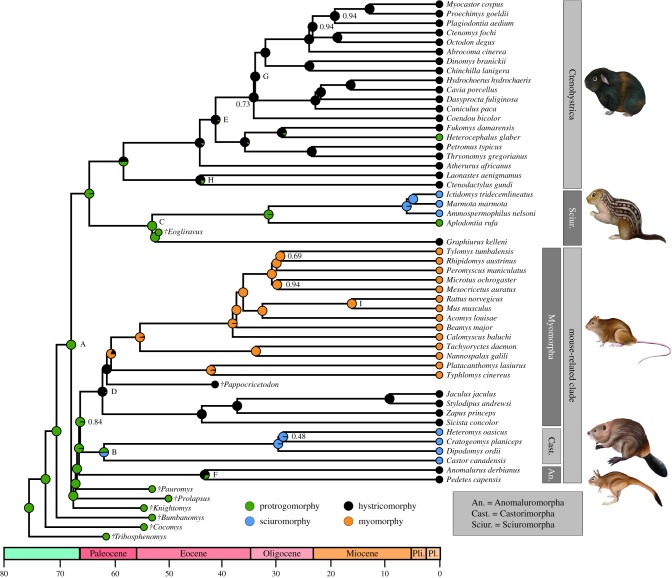
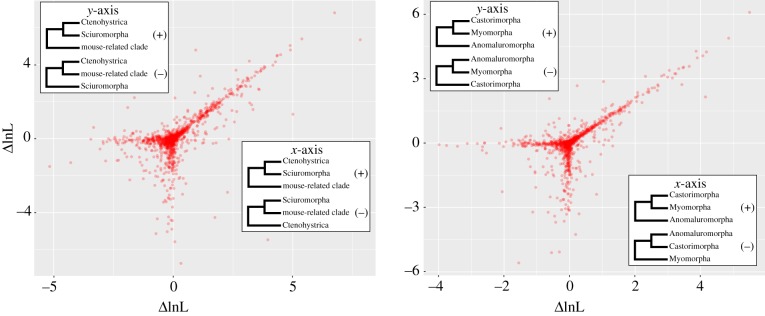
Our concatenated, ASTRAL and ASTRID trees all recovered a sister relationship between Sciuromorpha and Ctenohystrica, but with varying bootstrap support (BS: 100, 79, 66, respectively; figure 2; electronic supplementary material, figure S1). Further supporting this conclusion, our bipartition frequency analysis revealed that an order of magnitude more gene trees were consistent with a sister relationship between Sciuromorpha and Ctenohystrica versus the two alternative topologies (table 2). For relationships within the mouse-related clade, the concatenated and ASTRAL trees identify a sister relationship between Castorimorpha and Myomorpha (BS = 84, 99, respectively), while ASTRID supports Castorimorpha and Anomaluromorpha as sisters (BS = 78). More gene trees were topologically consistent with the latter than the former (table 2). Importantly, none of our analyses recovered a sister relationship between the two mouse-related suborders with hystricomorphous taxa (Anomaluromorpha and Myomorpha). Our per-locus site-likelihood analysis resulted in low magnitude ΔlnL values and did not show a pattern of a few loci or clusters of loci driving support for particular relationships at either node of interest (figure 3).
Figure 2.
A time-calibrated, family-level phylogeny of rodents, showing the masseter muscle character states of extant and fossil taxa as well as the probability of each state at ancestral nodes based on the ordered ARD model. Character states correspond to masseter architecture shown in figure 1. Numbers at nodes indicate bootstrap support less than 100 and letters at nodes correspond to fossil constraints from table 1. ‘Pli.’ denotes the Pliocene epoch and ‘Pl’ denotes the Pleistocene epoch on the timeline. Outgroups Ochotona princeps and Oryctolagus cuniculus were pruned out prior to stochastic character mapping analysis.
Table 2.
Percentages of gene tree topologies consistent with each phylogenetic hypothesis for the crown rodent and mouse-related clade nodes.
| node | hypothesized clade | gene tree frequency (%) |
|---|---|---|
| crown rodent | Sciuromorpha + Ctenohystrica | 3.208 |
| crown rodent | Sciuromorpha + mouse-related | 0.2259 |
| crown rodent | Ctenohystrica + mouse-related | 0.09038 |
| mouse-related clade | Anomaluromorpha + Castorimorpha | 3.299 |
| mouse-related clade | Anomaluromorpha + Myomorpha | 0.2259 |
| mouse-related clade | Castorimorpha + Myomorpha | 0.3163 |
Figure 3.
Plots of per-locus ΔlnL values for the comparison of figure 2 topology (supported by RAxML concatenation and ASTRAL) with alternative topologies at two nodes of interest showing low magnitude values and no outlying clusters of loci. Positive values indicate support for figure 2 topology while negative values indicate support for an alternative hypothesis. (Online version in colour.)
TreePL produced an ultrametric tree with divergence dates broadly comparable with previous studies [86,87] and inferred a divergence date at an unconstrained node for Aplodontidae and Sciuridae at 31 Mya, consistent with the appearance of the first definitive squirrel [88]. We therefore consider this temporal framework suitable for stochastic character mapping. The ARD model had the highest AIC score in both ordered and unordered comparisons (table 3). When we compared the unordered and ordered ARD models, the ordered model had substantially higher support (table 3). Stochastic character mapping using the ordered ARD model estimated an average of: 5.9 transitions from protrogomorphy to hystricomorphy and 1.2 transitions from hystricomorphy back to protrogomorphy, 2.5 transitions from protrogomorphy to sciuromorphy and 1.2 transitions from hystricomorphy to myomorphy, but none in the reverse of these directions; and no transition between hystricomorphy and sciuromorphy or sciuromorphy and myomorphy.
Table 3.
Comparison of ordered and unordered models of masseter muscle evolution in which all transition rates are equal (ER), forward and reverse transitions occur at equal rates (SYM), and all possible transitions occur at different rates (ARD).
| comparison | model | AIC | delta AIC | weighted AIC |
|---|---|---|---|---|
| unordered | ER | 83.502 | 5.402 | 0.047 |
| SYM | 80.125 | 2.043 | 0.252 | |
| ARD | 78.081 | 0.000 | 0.701 | |
| ordered | ER | 81.725 | 4.974 | 0.052 |
| SYM | 78.125 | 1.374 | 0.317 | |
| ARD | 76.751 | 0.000 | 0.630 | |
| ARD | unordered | 78.08 | 1.33 | 0.34 |
| ordered | 76.75 | 0.00 | 0.66 |
4. Discussion
Our well-supported inference of a Ctenohystrica–Sciuromorpha clade is concordant with previous work which removed the fastest evolving sites from a combination of exonic, mitochondrial and intronic loci [18] as well as Meredith et al.'s [21] combination of exons and untranslated regions. However, it contradicts most previous studies, including phylogenomic analyses of exons [26–28] and retroposons [20,24], which most often recover a sister relationship between Ctenohystrica and the mouse-related clade.
Examination of phylogenomic conflict among studies of avian relationships revealed that the use of coding or non-coding genes has a greater topological effect than dataset size or taxon sampling [89]. Alignments of non-coding genes produced more reliable inferences because modelling sequence evolution may be more challenging for exons. Inferences from SINEs have also recovered a sister relationship between the Ctenohystrica and mouse-related clade, in conflict with the Ctenohystrica–Sciuromorpha clade we recovered. Although Alu elements in primates are nearly homoplasy-free [90], other types of SINEs are arguably subject to higher rates of homoplasy [91]. Indeed, Churakov et al. [20] found two shared insertion events between Ctenohystrica and Sciuromorpha and attributed this to ancient hybridization, but it is also consistent with homoplasy.
Relationships among suborders in the mouse-related-clade have been difficult to disentangle but our concatenation and one coalescent analysis (ASTRAL) both recovered a sister relationship between Castorimorpha and Myomorpha, which is congruent with work which used either mitochondrial or a combination of nuclear and mitochondrial genes in a Bayesian framework [23,81]. None of our analyses supported a sister relationship between the two clades with hystricomorphous members, Anomaluromorpha and Myomorpha, as has been recovered with limited statistical support by most previous studies [17,19,21,22].
Fitting models of character evolution, followed by stochastic character mapping provided refreshing clarity regarding the pattern of masseter evolution. Support for the ordered ARD model in which direct transitions between protrogomorphy and myomorphy are not possible is consistent with classic conjecture and paleontological evidence that myomorphous species can only evolve from those which already have one masseter muscle attached on the rostrum [11,14]. The ordered ARD model suggests protrogomorphy is the ancestral state for crown rodents, in accordance with the abundance of protrogomorphous rodents known from the Palaeocene and early Eocene [11,54]. Subsequent transitions from protrogomorphy to states with one portion of the masseter muscle moved forward onto the rostrum (hystricomorphy and sciuromorphy) are common and may be overestimated, but reversals are rare (figure 2). We infer independent origins of hystricomorphy to have occurred in Anomaluromorpha, Ctenohystrica, Gliridae (represented by Graphiurus) and Myomorpha while the only possible reversal from hystricomorphy to protrogomorphy occurred in the Heterocephalidae (naked mole-rats). A fifth origin of hystricomorphy is assumed in the Theridomyidae, a family of middle Eocene to early Miocene European rodents with uncertain relationships to extant taxa [54,92] excluded from our analysis.
While the improved gnawing and chewing efficiency resulting from myomorphy has been credited with the evolutionary success of Myomorpha [12], none of these masseter morphology character transitions occur at the same nodes as diversification rate shifts detected by more recent species rich phylogenetic analyses of Rodentia [22,93]. This suggests that other factors (e.g. tooth morphology, dispersal ability), perhaps in concert with masseter morphology, have fostered the ecological and evolutionary success of Cricetidae and Muridae [22]. Alternatively, current estimates of diversification rate shifts in Rodentia could have been confounded by failure of current methods to accurately estimate extinction rates [94].
Our inferences of character state evolution benefited from incorporation of fossil taxa at the rodent root, the root of the mouse-related clade, the protrogomorphous glirid Eogliravus and the hystricomorphous cricetid Pappocricetodon. We repeated our stochastic character mapping analyses without the fossil taxa and this erroneously resulted in the rodent root as hystricomorphous as well as no transitions from protrogomorphy to hystricomorphy. A hystricomorphous ancestor conflicts with the fossil record, which shows early and abundant protrogomorphs [9,65]. Relying solely on extant taxa for the ancestral state reconstruction would have led to a distorted and incoherent pattern of rodent masseter evolution, as has been shown for systems with a general trend in the direction of character state evolution, such as body symmetry in echinoderms and the evolution of feathers and flight in birds [94–96].
Intra-family variation in masseter architecture is known from two families. In Gliridae, we sampled the hystricomorphous Graphiurus while the remaining taxa are myomorphous. The presence of myomorphy in both the Sciuromorpha and Myomorpha suborders is consistent with paleontological and developmental work suggesting myomorphy has independently evolved twice [35,97,98]. In addition, members of the Bathyergidae are known to exhibit varying degrees of hystricomorphy and protrogomorphy over the course of their development [36,99]. Detailed anatomical study and elucidation of phylogenetic relationships within Gliridae and Bathyergidae are necessary to improve our understanding of masseter evolution in these families.
Morphological variation also exists within each of the classically defined masseter states that we modelled. For instance, within hystricomorphy, the degree to which the zygomatico-mandibularis attaches to the rostrum through the infraorbital foramen varies across taxa. More specifically, within the Ctenohystrica, differences in infraorbital foramen result in characteristic profiles for each family [100]. Future work should focus on whether morphological variation among descendants of each independent origin of the classic morphotypes has also resulted in biomechanical convergence.
Masseter architecture and the increased gnawing efficiency resulting from attachment of this muscle at the rostrum have been critical components of rodent systematics and biology for nearly 200 years, yet the forces that selected for sciuromorphy, hystricomorphy and myomorphy remain unknown. Our inference of five independent origins of hystricomorphy, one reversal to protrogomorphy and two origins of sciuromorphy provides a valuable framework that can be integrated with anatomical, biomechanical, dietary and fossil data to elucidate the relative roles of developmental constraint, selective regimes and stochasticity in the evolution of masseter architecture diversity. The repeated forward movement of the masseter muscle onto the rostrum and the increasing prevalence of this trait through time shows that convergence has played a critical role in rodents' extraordinary evolutionary and ecological success.
Supplementary Material
Supplementary Material
Acknowledgements
We are grateful to the following institutions for providing tissue samples and/or access to skull specimens: Field Museum of Natural History, Australian National Wildlife Collection, Louisiana State University Museum of Natural Science, Royal Ontario Museum, American Museum of Natural History, Sam Noble Museum, Museum of Vertebrate Zoology, Harvard Museum of Comparative Zoology and Museum of Southwestern Biology. Figures 1–2 illustrations are by Subir Shakya. Discussions with Pierre-Henri Fabre, Lionel Hautier, Ryan Norris, Nate Upham, Suyin Ting, Chuankuei Li, Jin Meng, Monique Vianey-Liaud, Zachary Rodriguez, Laura Lagomarsino and Jonathan Nations greatly improved the quality of this manuscript. We thank Philip Cox and two anonymous reviewers for their helpful suggestions.
Data accessibility
Raw reads and DNA sequences are available on the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) and GenBank, accessible through NCBI BioProject PRJNA513335. BioSample accessions are listed in the electronic supplementary material. The concatenated alignment, individual locus alignments and trees can be accessed from the Dryad Digital Repository at https://doi.org/10.5061/dryad.3f83707 [43].
Authors' contributions
M.T.S. conceived the study and collected data, M.T.S. and C.H.O. conducted analyses, J.A.E. obtained funding and M.T.S. drafted the manuscript, with editorial input from C.H.O. and J.A.E.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by NSF through DEB-1441634 and DEB-1754393.
References
- 1.Liem KF, Bemis WE, Walker WF, Grande L. 2001. Functional anatomy of the vertebrates: an evolutionary perspective, 3rd edn Belmont, CA: Brooks/Cole. [Google Scholar]
- 2.Muschick M, Indermaur A, Salzburger W. 2012. Convergent evolution within an adaptive radiation of cichlid fishes. Curr. Biol. 22, 2362–2368. ( 10.1016/j.cub.2012.10.048) [DOI] [PubMed] [Google Scholar]
- 3.Feldhamer GA, Drickamer LC, Vessey SH, Merritt JF. 2004. Mammalogy: adaptation, diversity, and ecology, 2nd edn New York: NY: McGraw-Hill. [Google Scholar]
- 4.Burns KJ, Hackett SJ, Klein NK. 2003. Phylogenetic relationships of Neotropical honeycreepers and the evolution of feeding morphology. J. Avian Biol. 34, 360–370. ( 10.1111/j.0908-8857.2003.03171.x) [DOI] [Google Scholar]
- 5.Morris PJR, Cobb SNF, Cox PG. 2018. Convergent evolution in the Euarchontoglires. Biol. Lett. 14, 9–12. ( 10.1098/rsbl.2018.0366) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landry SO. 1970. The Rodentia as omnivores. Q. Rev. Biol. 45, 351–372. ( 10.1086/406647) [DOI] [PubMed] [Google Scholar]
- 7.Vaughan TA, Ryan JM, Czaplewski NJ. 2015. Mammalogy, 6th edn Burlington, MA: Jones and Bartlett Learning. [Google Scholar]
- 8.Wood AE. 1955. A revised classification of the rodents. J. Mammal. 36, 165–187. ( 10.1644/859.1.Key) [DOI] [Google Scholar]
- 9.Wood AE. 1959. Eocene radiation and phylogeny of the Rodents. Evolution 13, 354–361. ( 10.2307/2406112) [DOI] [Google Scholar]
- 10.Druzinsky RE. 2010. Functional anatomy of incisal biting in Aplodontia rufa and sciuromorph rodents, part 2: sciuromorphy is efficacious for production of force at the incisors. Cells Tissues Organs 192, 50–63. ( 10.1159/000284930) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood AE. 1965. Grades and clades among rodents. Evolution 19, 115–130. ( 10.1111/j.1558-5646.1965.tb01696.x) [DOI] [Google Scholar]
- 12.Cox PG, Rayfield EJ, Fagan MJ, Herrel A, Pataky TC, Jeffery N. 2012. Functional evolution of the feeding system in rodents. PLoS ONE 7, e36299 ( 10.1371/journal.pone.0036299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korth WW. 1994. The tertiary record of rodents in North America. New York, NY: Plenum Press. [Google Scholar]
- 14.Lindsay EH. 2007. Cricetidae. In Evolution of tertiary mammals of North America, vol. 2 (eds Janis CM, Gunnell GF, Uhen MD), pp. 456–479. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 15.Brandt JF. 1855. Beitrage zur nahern Kenntniss der Saugethiere Russlands. Buchdruckerei der Kais. Akad. der Wissenschaften 6, 1–375. [Google Scholar]
- 16.Simpson GG. 1945. The principles of classification and a classification of mammals. Bull. Am. Museum Nat. Hist. 85, 1–350. [Google Scholar]
- 17.Huchon D, Chevret P, Jordan U, Kilpatrick CW, Ranwez V, Jenkins PD, Brosius J, Schmitz J. 2007. Multiple molecular evidences for a living mammalian fossil. Proc. Natl Acad. Sci. USA 104, 7495–7499. ( 10.1073/pnas.0701289104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montgelard C, Forty E, Arnal V, Matthee CA. 2008. Suprafamilial relationships among Rodentia and the phylogenetic effect of removing fast-evolving nucleotides in mitochondrial, exon and intron fragments. BMC Evol. Biol. 8, 321 ( 10.1186/1471-2148-8-321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanga-Kanfi S, Miranda H, Penn O, Pupko T, DeBry RW, Huchon D. 2009. Rodent phylogeny revised: analysis of six nuclear genes from all major rodent clades. BMC Evol. Biol. 9, 71 ( 10.1186/1471-2148-9-71) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Churakov G, Sadasivuni MK, Rosenbloom KR, Huchon D, Brosius J, Schmitz J. 2010. Rodent evolution: back to the root. Mol. Biol. Evol. 27, 1315–1326. ( 10.1093/molbev/msq019) [DOI] [PubMed] [Google Scholar]
- 21.Meredith RW, et al. 2011. Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334, 521–524. ( 10.1126/science.1211028) [DOI] [PubMed] [Google Scholar]
- 22.Fabre P-H, Hautier L, Dimitrov D, Douzery EJ. 2012. A glimpse on the pattern of rodent diversification: a phylogenetic approach. BMC Evol. Biol. 12, 88 ( 10.1186/1471-2148-12-88) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heritage S, Fernández D, Sallam HM, Cronin DT, Esara Echube JM, Seiffert ER. 2016. Ancient phylogenetic divergence of the enigmatic African rodent Zenkerella and the origin of anomalurid gliding. PeerJ 4, e2320 ( 10.7717/peerj.2320) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doronina L, Matzke A, Churakov G, Stoll M, Huge A, Schmitz J. 2017. The beaver's phylogenetic lineage illuminated by retroposon reads. Sci. Rep. 7, 43562 ( 10.1038/srep43562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fabre P-H, Hautier L, Douzery PEJ. 2015. Synopsis of rodent molecular phylogenetics, systematics and biogeography. In Evolution of the rodents: advances in phylogeny, functional morphology and development (eds PG Cox, L Hautier), pp. 19–69. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 26.Song S, Liu L, Edwards SV, Wu S. 2015. Correction for Song et al., resolving conflict in eutherian mammal phylogeny using phylogenomics and the multispecies coalescent model. Proc. Natl Acad. Sci. USA 112, E6079 ( 10.1073/pnas.1518753112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feijoo M, Parada A. 2017. Macrosystematics of eutherian mammals combining HTS data to expand taxon coverage. Mol. Phylogenet. Evol. 113, 76–83. ( 10.1016/j.ympev.2017.05.004) [DOI] [PubMed] [Google Scholar]
- 28.Scornavacca C, Galtier N. 2017. Incomplete lineage sorting in mammalian phylogenomics. Syst. Biol. 66, 112–120. ( 10.1093/sysbio/syw082) [DOI] [PubMed] [Google Scholar]
- 29.Esselstyn JA, Oliveros CH, Swanson MT, Faircloth BC. 2017. Investigating difficult nodes in the placental mammal tree with expanded taxon sampling and thousands of ultraconserved elements. Genome Biol. Evol. 9, 2308–2321. ( 10.1093/gbe/evx168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu L, et al. 2017. Genomic evidence reveals a radiation of placental mammals uninterrupted by the KPg boundary. Proc. Natl Acad. Sci. USA 114, E7282–E7290. ( 10.1073/pnas.1616744114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen X-X, Hittinger CT, Rokas A. 2017. Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nat. Ecol. Evol. 1, 1–10. ( 10.1038/s41559-017-0126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown JM, Thomson RC. 2017. Bayes factors unmask highly variable information content, bias, and extreme influence in phylogenomic analyses. Syst. Biol. 66, 517–530. ( 10.1093/sysbio/syw101) [DOI] [PubMed] [Google Scholar]
- 33.Zwickl DJ, Hillis DM. 2002. Increased taxon sampling greatly reduces phylogenetic error. Syst. Biol. 51, 588–598. ( 10.1080/10635150290102339) [DOI] [PubMed] [Google Scholar]
- 34.Heath T, Hedtke S, Hillis D. 2008. Taxon sampling and the accuracy of phylogenetic analyses. J. Syst. Evol. 46, 239–257. [Google Scholar]
- 35.Hautier L, Michaux J, Marivaux L, Vianey-Liaud M. 2008. Evolution of the zygomasseteric construction in Rodentia, as revealed by a geometric morphometric analysis of the mandible of Graphiurus (Rodentia, Gliridae). Zool. J. Linn. Soc. 154, 807–821. ( 10.1111/j.1096-3642.2008.00453.x) [DOI] [Google Scholar]
- 36.Cox PG, Faulkes CG. 2014. Digital dissection of the masticatory muscles of the naked mole-rat, Heterocephalus glaber (Mammalia, Rodentia). PeerJ 2, e448 ( 10.7717/peerj.448) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faircloth BC, McCormack JE, Crawford NG, Harvey MG, Brumfield RT, Glenn TC. 2012. Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Syst. Biol. 61, 717–726. ( 10.1093/sysbio/sys004) [DOI] [PubMed] [Google Scholar]
- 38.Rohland N, Reich D. 2012. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 22, 939–946. ( 10.1101/gr.128124.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faircloth BC. 2016. PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics 32, 786–788. ( 10.1093/bioinformatics/btv646) [DOI] [PubMed] [Google Scholar]
- 40.Grabherr MG, et al. 2013. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29, 644–652. ( 10.1038/nbt.1883.Trinity) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. ( 10.1093/oxfordjournals.molbev.a026334) [DOI] [PubMed] [Google Scholar]
- 43.Swanson MT, Oliveros CH, Esselstyn JA. 2019. Data from: A phylogenomic rodent tree reveals the repeated evolution of masseter architectures Dryad Digital Repository. ( 10.5061/dryad.3f83707) [DOI] [PMC free article] [PubMed]
- 44.Wilson DE, Reeder DM (eds). 2005. Mammal species of the world, 3rd edn Baltimore, MD: Johns Hopkins University Press. [Google Scholar]
- 45.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. ( 10.1093/bioinformatics/btu033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seo TK. 2008. Calculating bootstrap probabilities of phylogeny using multilocus sequence data. Mol. Biol. Evol. 25, 960–971. ( 10.1093/molbev/msn043) [DOI] [PubMed] [Google Scholar]
- 47.Mirarab S, Reaz R, Bayzid MS, Zimmermann T, Swenson M, Warnow T. 2014. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics 30, 541–548. ( 10.1093/bioinformatics/btu462) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mirarab S, Warnow T. 2015. ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics 31, i44–i52. ( 10.1093/bioinformatics/btv234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vachaspati P, Warnow T. 2015. ASTRID: accurate species TRees from internode distances. BMC Genomics 16(Suppl 1), S3 ( 10.1186/1471-2164-16-S10-S3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sukumaran J, Holder MT. 2010. DendroPy: a Python library for phylogenetic computing. Bioinformatics 26, 1569–1571. ( 10.1093/bioinformatics/btq228) [DOI] [PubMed] [Google Scholar]
- 51.Smith SA, O'Meara BC. 2012. TreePL: divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 28, 2689–2690. ( 10.1093/bioinformatics/bts492) [DOI] [PubMed] [Google Scholar]
- 52.Benton MJ, Donoghue PCJ, Asher RJ, Friedman M, Near TJ, Vinther J. 2015. Constraints on the timescale of animal evolutionary history. Palaeontol. Electron. 18, 1–107. ( 10.26879/424) [DOI] [Google Scholar]
- 53.Flynn LJ, Jacobs LL. 2008. Castoroidea. In Evolution of tertiary mammals of North America, vol. 2 (eds Janis CM, Gunnell GF, Uhen MD), pp. 391–405. New York, NY: Cambridge University Press. [Google Scholar]
- 54.Marivaux L, Vianey-Liaud M, Jaeger J-J. 2004. High-level phylogeny of early tertiary rodents: dental evidence. Zool. J. Linn. Soc. 142, 105–134. ( 10.1111/j.1096-3642.2004.00131.x) [DOI] [Google Scholar]
- 55.Phillips MJ. 2014. Four mammal fossil calibrations: balancing competing palaeontological and molecular considerations. Palaeontol. Electron. 18.1.5FC, 1–16. [Google Scholar]
- 56.Antoine PO, et al. 2012. Middle eocene rodents from peruvian amazonia reveal the pattern and timing of caviomorph origins and biogeography. Proc. R. Soc. B 279, 1319–1326. ( 10.1098/rspb.2011.1732) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marivaux L, Ducrocq S, Jaeger J-J, Marandat B, Sudre J, Chaimanee Y, Tun ST, Htoon W, Soe AN. 2005. New remains of Pondaungimys anomaluropsis (Rodentia, Anomaluroidea) from the Latest Middle Eocene Pond Aung Formation of Central Myanmar. J. Vertebr. Paleontol. 25, 214–227. ( 10.1671/0272-4634(2005)025[0214:NROPAR]2.0.CO;2) [DOI] [Google Scholar]
- 58.Dawson MR. 2006. Laonastes and the ‘Lazarus Effect’ in recent mammals. Science 311, 1456–1458. ( 10.1126/science.1124187) [DOI] [PubMed] [Google Scholar]
- 59.Kimura Y, Hawkins MTR, McDonough MM, Jacobs LL, Flynn LJ. 2015. Corrected placement of Mus-Rattus fossil calibration forces precision in the molecular tree of rodents. Sci. Rep. 5, 1–9. ( 10.1038/srep14444) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heissig K. 2003. Origin and early dispersal of the squirrels and their relatives. Deinsea 10, 277–286. [Google Scholar]
- 61.Rose KD. 2006. The beginning of the Age of mammals. Baltimore, MD: The Johns Hopkins University Press. [Google Scholar]
- 62.Phillips MJ, Fruciano C. 2018. The soft explosive model of placental mammal evolution. BMC Evol. Biol. 18, 1–13. ( 10.1186/s12862-018-1218-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.R Core Team. 2016. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 64.Woods CA. 1972. Comparative myology of jaw, hyoid, and pectoral appendicular regions of new and old world hystricomorph rodents. Bull. Am. Museum Nat. Hist. 147, 115–198. [Google Scholar]
- 65.Wood AE. 1985. The relationships, origin and dispersal of the hystricognathous rodents. In Evolutionary relationships among rodents: A multidisciplinary analysis (eds Luckett WP, Hartenberger J-L), pp. 475–514. New York, NY: Plenum Press. [Google Scholar]
- 66.Woods CA, Hermanson JW. 1985. Myology of hystricognath rodents: An analysis of form, function, and phylogeny. In Evolutionary relationships among rodents: A multidisciplinary analysis (eds Luckett WP, Hartenberger J-L), pp. 515–548. New York, NY: Plenum Press. [Google Scholar]
- 67.Van Daele PAAG, Herrel A, Adriaens D.. 2008. Biting performance in teeth-digging African Mole-Rats (Fukomys, Bathyergidae, Rodentia). Physiol. Biochem. Zool. 82, 40–50. ( 10.1086/594379) [DOI] [PubMed] [Google Scholar]
- 68.Turnbull WD. 1970. Mammalian masticatory apparatus. Fieldiana Geol. 18, 153–356. [Google Scholar]
- 69.Hautier L, Saksiri S. 2009. Masticatory muscle architecture in the Laotian rock rat Laonastes aenigmamus (Mammalia, Rodentia): new insights into the evolution of hystricognathy. J. Anat. 215, 401–410. ( 10.1111/j.1469-7580.2009.01130.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hautier L. 2010. Masticatory muscle architecture in the gundi Ctenodactylus vali (Mammalia, Rodentia). Mammalia 74, 153–162. ( 10.1515/MAMM.2010.025) [DOI] [Google Scholar]
- 71.Druzinsky RE. 2010. Functional anatomy of incisal biting in Aplodontia rufa and sciuromorph rodents, part 1: masticatory muscles, skull shape and digging. Cells Tissues Organs 191, 510–522. ( 10.1159/000284931) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arata AA. 1967. Muroid, gliroid, and dipodoid rodents. In Recent mammals of the world: a synopsis of families (eds Anderson S, Jones JK), pp. 226–253. New York, NY: Ronald Press Company. [Google Scholar]
- 73.Satoh K, Iwaku F. 2009. Structure and direction of jaw adductor muscles as herbivorous adaptations in Neotoma mexicana (Muridae, Rodentia). Zoomorphology 128, 339–348. ( 10.1007/s00435-009-0094-8) [DOI] [Google Scholar]
- 74.Greene EC. 1935. Anatomy of the rat. New York, NY: Hafner Publishing Company. [Google Scholar]
- 75.Bekele A. 1983. The comparative functional morphology of some head muscles of the rodents Tachyoryctes splendens and Rattus rattus. I. Mammalia 47, 395–419. ( 10.1515/mamm.1983.47.4.549) [DOI] [Google Scholar]
- 76.Klingener D. 1964. The comparative myology of four dipodoid rodents (genera zapus, napaeozapus, sicista, and jaculus) Misc. Publ. Museum Zool. Univ. Michigan 124, 1–100. [Google Scholar]
- 77.Fahlbusch V. 1985. Origin and evolutionary relationships among geomyoids. In Evolutionary relationships among rodents: a multidisciplinary analysis (eds Luckett WP, Hartenberger J-L), pp. 617–629. New York, NY: Plenum Press. [Google Scholar]
- 78.Brylski P. 1993. The Evolutionary Morphology of Heteromyids. In Biology of the heteromyidae (eds Genoways HH, Brown JH), pp. 357–385. Washington, DC: American Society of Mammalogists. [Google Scholar]
- 79.Cox PG, Baverstock H. 2016. Masticatory muscle anatomy and feeding efficiency of the American Beaver, Castor canadensis (Rodentia, Castoridae) . J. Mamm. Evol. 23, 191–200. ( 10.1007/s10914-015-9306-9) [DOI] [Google Scholar]
- 80.Jaeger J-J, Denys C, Coiffait B. 1985. New Phiomorpha and Anomaluridae from the late Eocene of north-west Africa: Phylogenetic implications. In Evolutionary relationships among rodents: A multidisciplinary analysis, pp. 567–588. New York, NY: Plenum Press. [Google Scholar]
- 81.Fabre PH, Tilak MK, Denys C, Gaubert P, Nicolas V, Douzery EJP, Marivaux L. 2018. Flightless scaly-tailed squirrels never learned how to fly: a reappraisal of Anomaluridae phylogeny. Zool. Scr. 47, 404–417. ( 10.1111/zsc.12286) [DOI] [Google Scholar]
- 82.Cox PG. 2017. The jaw is a second-class lever in Pedetes capensis (Rodentia: Pedetidae). PeerJ 5, e3741 ( 10.7717/peerj.3741) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krystufek B, Vohralik V. 2009. Mammals of Turkey and Cyprus, vol. 3: Rodentia II. Koper, Slovenia: Annales Majora. [Google Scholar]
- 84.Rodrigues HG. 2015. The great variety of dental structures and dynamics in rodents: new insights into their ecological diversity. In Evolution of the rodents. Advances in phylogeny, functional morphology and development (eds Cox PG, Hautier L), pp. 424–447. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 85.Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. ( 10.1111/j.2041-210X.2011.00169.x) [DOI] [Google Scholar]
- 86.Wu S, Wu W, Zhang F, Ye J, Ni X, Sun J, Edwards SV, Meng J, Organ CL. 2012. Molecular and paleontological evidence for a post-Cretaceous origin of rodents. PLoS ONE 7, e46445 ( 10.1371/journal.pone.0046445) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Patterson BD, Upham NS. 2014. A newly recognized family from the Horn of Africa, the Heterocephalidae (Rodentia: Ctenohystrica). Zool. J. Linn. Soc. 172, 942–963. ( 10.1111/zoj.12201) [DOI] [Google Scholar]
- 88.Vianey-Liaud M. 1974. Palaeosciurus goti nov. sp., écureuil terrestre de l'Oligocène moyen du Quercy. Données nouvelles sur l'apparition des sciuridés en Europe. Ann. Paleontol. 60, 101–124. [Google Scholar]
- 89.Reddy S, et al. 2017. Why do phylogenomic data sets yield conflicting trees? Data type influences the avian tree of life more than taxon sampling. Syst. Biol. 66, 857–879. ( 10.1093/sysbio/syx041) [DOI] [PubMed] [Google Scholar]
- 90.Doronina L, Reising O, Clawson H, Ray DA, Schmitz J. 2019. True homoplasy of retrotransposon insertions in primates. Syst. Biol. 68, 482–493. ( 10.1093/sysbio/syy076) [DOI] [PubMed] [Google Scholar]
- 91.Han KL, et al. 2011. Are transposable element insertions homoplasy free? An examination using the avian tree of life. Syst. Biol. 60, 375–386. ( 10.1093/sysbio/syq100) [DOI] [PubMed] [Google Scholar]
- 92.Vianey-Liaud M, Marivaux L. 2016. Autopsie d'une radiation adaptative: Phylogénie des Theridomorpha, rongeurs endémiques du Paléogène d'Europe—histoire, dynamique évolutive et intérêt biochronologique. Palaeovertebrata 40, e1 ( 10.18563/pv.40.3.e1) [DOI] [Google Scholar]
- 93.Upham N, Esselstyn JA, Jetz W.2019. Ecological causes of uneven diversification and richness in the mammal tree of life. bioRxiv, 504803. (doi:10.1101/504803)
- 94.Marshall CR. 2017. Five palaeobiological laws needed to understand the evolution of the living biota. Nat. Ecol. Evol. 1, 1–6. ( 10.1038/s41559-017-0165) [DOI] [PubMed] [Google Scholar]
- 95.Rozhnov S. 2014. Symmetry of echinoderms: from initial bilaterally-asymmetric metamerism to pentaradiality. Nat. Sci. 06, 171–183. ( 10.4236/ns.2014.64021) [DOI] [Google Scholar]
- 96.Brusatte SL, O'Connor JK, Jarvis ED. 2015. The origin and diversification of birds. Curr. Biol. 25, R888–R898. ( 10.1016/j.cub.2015.08.003) [DOI] [PubMed] [Google Scholar]
- 97.Vianey-Liaud M. 1985. Possible evolutionary relationships among Eocene and Lower Oligocene of rodents from Asia, Europe and North America. In Evolutionary relationships among rodents: a multidisciplinary analysis (eds Luckett WP, Hartenberger J-L), pp. 277–309. New York, NY: Plenum Press. [Google Scholar]
- 98.Maier W, Klingler P, Ruf I. 2002. Ontogeny of the medial masseter muscle, pseudo-myomorphy, and the systematic position of the Gliridae (Rodentia, Mammalia). J. Mamm. Evol. 9, 253–269. ( 10.1023/A:1023979212759) [DOI] [Google Scholar]
- 99.Maier W, Schrenk F. 1987. The hystricomorphy of the Bathyergidae, as determined from ontogenetic evidence. Zeitschrift fur Saugetierkd. 52, 156–164. [Google Scholar]
- 100.Hautier L, Cox PG, Lebrun R. 2015. Grades and clades among rodents: the promise of geometric morphometrics. In Evolution of the rodents: advances in phylogeny, functional morphology and development (eds Cox PG, Hautier L), pp. 277–299. Cambridge, UK: Cambridge University Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Swanson MT, Oliveros CH, Esselstyn JA. 2019. Data from: A phylogenomic rodent tree reveals the repeated evolution of masseter architectures Dryad Digital Repository. ( 10.5061/dryad.3f83707) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Raw reads and DNA sequences are available on the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) and GenBank, accessible through NCBI BioProject PRJNA513335. BioSample accessions are listed in the electronic supplementary material. The concatenated alignment, individual locus alignments and trees can be accessed from the Dryad Digital Repository at https://doi.org/10.5061/dryad.3f83707 [43].