Abstract
North African history and populations have exerted a pivotal influence on surrounding geographical regions, although scant genetic studies have addressed this issue. Our aim is to understand human historical migrations in the coastal surroundings of North Africa. We built a refined genome-wide dataset of North African populations to unearth the fine-scale genetic structure of the region, using haplotype information. The results suggest that the gene-flow from North Africa into the European Mediterranean coast (Tuscany and the Iberian Peninsula) arrived mainly from the Mediterranean coast of North Africa. In Tuscany, this North African admixture date estimate suggests the movement of peoples during the fall of the Roman Empire around the fourth century. In the Iberian Peninsula, the North African component probably reflects the impact of the Arab expansion since the seventh century and the subsequent expansion of the Christian Kingdoms. By contrast, the North African component in the Canary Islands has a source genetically related to present-day people from the Atlantic North African coast. We also find sub-Saharan gene-flow from the Senegambia region in the Canary Islands. Specifically, we detect a complex signal of admixture involving Atlantic, Senegambian and European sources intermixing around the fifteenth century, soon after the Castilian conquest. Our results highlight the differential genetic influence of North Africa into the surrounding coast and show that specific historical events have not only had a socio-cultural impact but additionally modified the gene pool of the populations.
Keywords: North Africa, Canary Islands, Iberian Peninsula, gene-flow, haplotype-based methods, fineStructure
1. Introduction
North Africa is a genetically diverse region from a human population perspective. North African populations show a complex and heterogeneous genetic structure that has been described as an amalgam of at least four different ancestral components: Middle Eastern, sub-Saharan African, European and autochthonous North African [1]. Most of the genetic studies about North Africa have focused on the inner relationships among populations, or the gene-flow from nearby populations [1,2]. However, there are scant studies that have focused on North African gene-flow into neighbouring regions [3]. It is well known that the surrounding coast has been historically influenced by North African peoples [4,5]; however, the demographic impact of those contacts has not been properly addressed. Our aim is to assess the North African demographic and genetic influence in nearby regions outside the African continent by assessing the gene-flow in three geographical neighbouring regions with documented contacts with North Africa: the Canary Islands, the Iberian Peninsula and Tuscany.
The Canary Islands, located in the Atlantic coast of North Africa, have been inhabited since approximately 1000 BCE [5,6]. The islands were known by the Phoenicians, Greeks and Romans; however, it is thought that there was no contact with the autochthonous settlers of the islands since the fourth century until the Castilian conquest in the fifteenth century [7]. By the time of this European conquest of the Islands, the aboriginal population size has been estimated around 100 000 individuals [8]. A northwest African origin of the first settlers of the islands is consistent with patterns of uniparental and classical genetic markers in modern and ancient samples [9,10]. In particular, the presence of haplogroups in the Canary Islands that are only found in individuals of North African descent, such as mitochondrial (mtDNA) haplogroup U6 [11] and Y-chromosome haplogroup M81 [12], among some others considered founder lineages, support the North African origin of the islanders. The frequencies of these haplogroups in the extant population of the Canary Islands show a clear sexual bias: the percentage of the maternal North African component estimated through the analysis of mtDNA lineages is high, between 42 and 74% [10]; while the paternal component analysed through the study of Y-chromosome lineages is lower, between 5 and 16% [9]. Additionally, Botigué et al. [3], analysing genome-wide data, showed a higher identity by descent sharing between individuals from the Canary Islands and North Africa compared to individuals from continental Europe, suggesting a higher gene-flow from the African continent to the Islands. Finally, genome-wide analysis with ancient DNA from the Canary Islands has corroborated the North African origin of the autochthonous component and its presence in current Canary Islanders [13]. However, the exact dates of admixture from Europe and the precise geographical origin of the North African component in the Islands have not been addressed.
The most well-documented contact between North Africa and the Iberian Peninsula is the Arab expansion, which crossed the Mediterranean and arrived in Gibraltar in the eighth century. However, genetic studies based on uniparental markers, together with archaeological and anthropological evidence, have suggested previous contacts across the Gibraltar Strait that date back to prehistoric times [4,5,11,14,15]. Recently, ancient DNA studies have supported prehistoric migrations from North Africa into the Iberian Peninsula since around 4000 ya (years ago) [16–18]. Moreover, mtDNA, Y-chromosome and short tandem repeat studies [4,19–22], as well as genome-wide and ancient DNA analyses [3,17,23], have also shown gene-flow in historical times. Moorjani et al. [23] dated African gene-flow into southern Europe around 55 generations ago, with the highest proportions in Iberia: 3.2 ± 0.3% in Portugal, and 2.4 ± 0.3% in Spain, which was related to a demographic impact either in Roman or Arab periods. Botigué et al. [3] showed that the inclusion of North African populations in their analyses increased those estimated percentages of gene-flow, suggesting a higher North African gene-flow in Iberia, and that the sub-Saharan gene-flow detected entered with the North African wave, challenging the interpretation of a direct sub-Saharan influence in southern Europe. Additionally, the North African gene-flow in the Iberian Peninsula was dated to 6–10 generations ago, although previous gene-flow was not discarded. In a large study of human populations admixture, Hellenthal et al. [24] described a complex scenario with continuous gene-flow during the past 2000 years in Iberia with North and sub-Saharan Africans. In sum, although all studies agree on the genetic influence of North Africa in Iberia, there is no clear consensus in the pattern of gene-flow and the estimated dates of the North African admixture.
The presence of the Etruscans, in what is nowadays referred to as the Tuscany territory in the Italian Peninsula, has been largely documented. However, the genetic footprint of the Etruscans in current populations has been only claimed in some isolated populations, but not in the Tuscan general population [25]. Moreover, although a Middle Eastern or Anatolian origin has been hypothesized for the Etruscans [26], recent studies analysing mtDNA have rejected an origin outside Italy [25,27]. Recently, an exhaustive genome-wide study of the Italian population [28] has dated different admixture events in Italy coming from different sources, including old events dated around 3000 ya that involved Caucasus, Middle Eastern and central Italian populations; whereas other more recent admixture processes involved gene-flow from north-central Europe around the collapse of the Roman Empire, a period which has been associated with extensive human movements. This continuous gene-flow in multiple directions at different times has yielded a complex genetic structure in the Italian Peninsula shown in both uniparental and genome-wide analyses [28,29], and traces of North African influences have also been detected, although the amount and timing of such contributions to Italy have not been assessed [28,30].
Our aim is to assess the impact of gene-flow from North Africa to surrounding populations for which there is documented evidence of contact between the populations, in particular the Canary Islands, Iberia and Tuscany. Previous North African–European gene-flow analyses [3,23] were limited by the scant geographical distribution of North African samples available. In order to overcome these issues, we use the largest genome-wide dataset of North African samples available, including different Berber groups that have not been included in previous studies of North African gene-flow, which allows us to describe detailed and complete genetic scenarios for North African admixture into the surrounding areas. The application of haplotype-based methods to a large dataset of samples and autosomal markers might refine our knowledge on the (i) estimated dates of the admixture events; (ii) the specific geographical sources of the gene-flow; and (iii) the quantification of the amount of gene-flow in the three targeted populations.
2. Material and methods
(a). Building the dataset
We built a dataset of more than 1200 samples that includes European and sub-Saharan African samples from the 1000 genomes project [31]; Iberian, Basque and Canary Islands populations from Botigué et al. [3]; and a large and diverse dataset of North African populations (which includes both Arab and Berbers and covers a wide geographical extension) from Henn et al. [1] and Arauna et al. [2] (see the electronic supplementary material, table S1 and figure S1). For some of the Iberian samples from the 1000 genomes project, no geographical coordinates are available; and, therefore, for some analyses they were assigned as ‘Iberian’ without specifying the location. Both Plink versions 1.07 and 1.9 have been used depending on the analyses [32,33]. Single nucleotide polymorphisms (SNPs) missing in more than 10% of the individuals, those that failed Hardy–Weinberg test at 0.01 significance threshold, and those with a minor allele frequency below 0.05 were discarded. After filters, 267 475 SNPs remained for analyses. Individuals sharing more than 85% of their genome identity by state were removed, and remaining individuals with more than 10% of missing SNPs were also excluded. For the analyses that required linkage equilibrium, SNPs were pruned using a pairwise linkage disequilibrium maximum threshold of 0.5 using a windows size of 50 a shift step of 5, remaining 149 956 SNPs.
(b). Haplotype-based methods
(i). Phasing
The phasing of SNPs was performed with Shapeit [34], using the population-averaged genetic map from the HapMap phase II [35] and the 1000 genomes dataset as a reference panel [31]. This phasing step was performed after an alignment with the reference panel and the removal of SNPs that did not align.
(ii). ChromoPainter
ChromoPainter [36] was run to infer the genome-wide number and proportion of haplotype segments for which each individual shared with every other individual, without population specification (i.e. using all sampled individuals as both recipients and donors, -a mode). We followed the protocol analogous to that outlined in Hellenthal et al. [24]. In particular, first the global mutation probability and the switch rate parameters were estimated using the expectation-maximization algorithm implemented in ChromoPainter with the following parameters: -i 10 -in -iM, in chromosomes 1, 7, 14 and 20 for all individuals. The mutation probability and the switch rate parameters estimated were averaged across these four chromosomes, weighting by the number of SNPs per chromosome. The average weighted values were 0.00017 and 208.30557 for the global mutation and switch rate, respectively. ChromoPainter was run afterwards for all chromosomes using these fixed global mutation and switch rates values. The final co-ancestry matrices (i.e. *.chuncklengths.out and *.chunckcounts.out files) were summed across chromosomes.
(iii). fineStructure
We used ChromoCombine to estimate the fineStructure C parameter (c = 0.264). Then, following Leslie et al. [37], fineStructure v.2.0.4 [36] was run using 2 million iterations of Markov chain Monte Carlo, sampling values every 10 000 iterations following 1 million ‘burn-in’ iterations (i.e. -x 1 000 000 -y 2000000-z 10000). Finally, the fineStructure tree was inferred using default parameters (i.e. –m T). Three seeds were estimated in order to check robustness of the analyses. Based on the fineStructure results, we established genetic clusters to use as ‘populations' for subsequent Globetrotter analyses.
(iv). Globetrotter
We applied Globetrotter to identify and date admixture events in each of our target populations using genome-wide linkage disequilibrium decay patterns, under a model that assumes instantaneous admixture involving two or more groups at 1 or 2 times in the past, followed by random mating among individuals from the admixed population. To do so, we followed the protocol of Hellenthal et al. [24]. In particular, after defining genetic clusters based on fineStructure results (see fineStructure clusters in figure 2 and electronic supplementary material, table S1), we performed a separate run of ChromoPainter painting each cluster using all other clusters as donors (i.e. disallowing ‘self-copying’ from other members of the own cluster). The clusters are assigned geographical names in order to facilitate the comprehension, however the detailed information of the distribution of the samples contributing to each cluster can be found in the electronic supplementary material, table S4 and figures S2 and S4. Then, we ran Globetrotter [24] using the copy vectors (i.e. *chunklength.out file) from the first ChromoPainter run used in fineStructure (i.e. that painted each individual using all other sampled individuals) and the painting (i.e. *samples.out files) from the second ChromoPainter run (i.e. that painted each individual using all other individuals outside of their cluster). The null.ind parameter was set to 1 for all the Globetrotter analyses, as recommended, to account for decay in linkage disequilibrium that may not be attributable to genuine admixture signals. Four different target groups were tested separately for admixture: Canary_Islands, Iberian_Peninsula, Tuscany and Basque. For each of the four targets, other clusters were used as surrogates, except that the Canary_Islands cluster was not included as a surrogate when testing the Iberian_Peninsula cluster for admixture, owing to the relatively high genetic similarity between Iberian_Peninsula and the Canary Islands. We performed 100 bootstrap iterations to infer confidence intervals for date estimates, for both one- and two-date models of admixture. As a result, for each target cluster we have 200 estimates of the fit of the model, combined across one and two dates of admixture, and the estimated dates for each bootstrap. We assumed a generation time of 25 years [38].
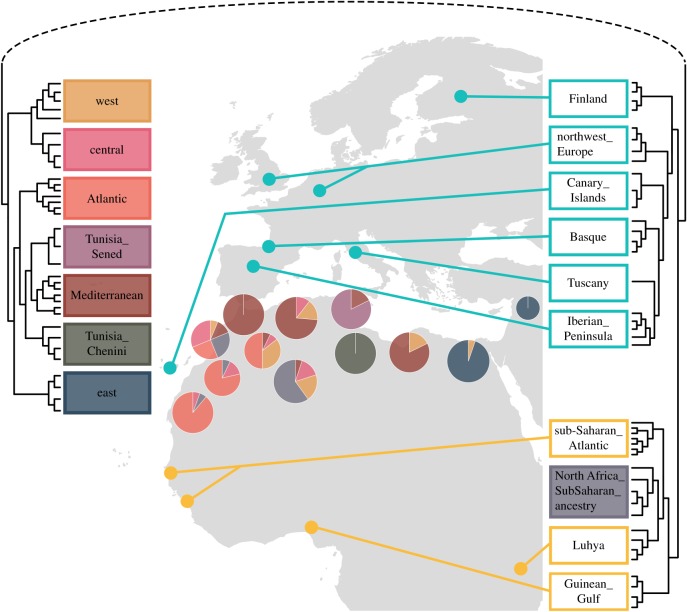
Figure 2.
fineStructure clustering shown as a dendrogram and its correspondence in a map. The filled rectangles are the North African samples, and the proportion of individuals from each of the clusters in each geographical sampled population is shown in pie-charts. Clusters containing European and sub-Saharan African individuals are denoted by non-filled rectangles coloured blue and yellow, respectively, and are labelled primarily according to geography.
Admixture between more than two sources at a given time is inferred by Globetrotter as multiway admixture, and described as two events that each involve two sources (where each such source may comprise some unknown mixture of the genuine admixing groups). To better interpret these events, in these multiway cases we manually reviewed the co-ancestry curves generated for each pair of surrogate populations to establish the sources participating in the admixture process, as illustrated in the electronic supplementary material, figure S7. In all these cases, we found evidence for three distinct sources intermixing. In particular, we assumed the three surrogates (or groups of surrogates) demonstrating the patterns in electronic supplementary material, figure S7 represented three distinct admixing sources. We represent the genetic make-up of each of these three sources in the electronic supplementary material, table S3 by decomposing the Globetrotter proportions estimation considering only two sources and recalculating those proportions considering the three manually inferred sources.
3. Results
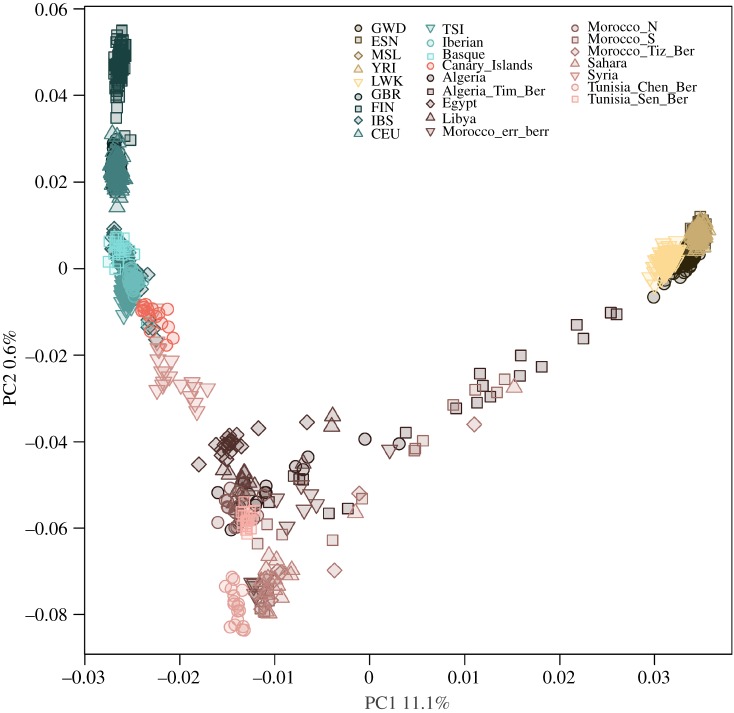
We have compiled a dataset of more than 1200 samples that includes a large and diverse dataset of North African populations to study the influence of North African gene-flow in neighbouring populations. Principal components analysis (PCA) of all populations in the dataset differentiates sub-Saharan and European populations along the first PC (PC1) (figure 1). The North African samples are widely spread along the first PC reflecting high heterogeneity, in accordance with the previously described differential admixture of the subpopulations [1,2]. PC2 further differentiates North African samples and highlights the genetic diversity within North Africa. On the first two PCs, the Canary Islands samples are placed close to the Iberian samples but shifted towards the Middle East and North Africa. When focusing on the European samples, three largely non-overlapping clusters can be observed: the Finns; northern and western Europeans (Great Britain and Utah residents with northern and western European ancestry (CEU)); and southern Europeans (Tuscany, Iberia, Basque Country and also the Canary Islands).
Figure 1.
Principal component analysis (PCA) of the samples included in the present study. See material and methods section for details.
We used haplotype-based methods to dissect the genetic structure of the studied populations and understand their genetic relationships. We performed fineStructure analyses (figure 2) and identified three major splits separating our data: North Africa, Europe and sub-Saharan Africa. Within these major geographical clusters, several subclusters can be identified that suggest a finer resolution of genetic structure. For example, within the European cluster, six sub-clusters are found that correlate with geography: Iberian Peninsula, Tuscany, Basque, Canary Islands, northwest Europe and Finland (electronic supplementary material, figure S1). Thirteen Syrian samples clustered together with the Canary Islands populations and were removed from further analyses. Similarly, within sub-Saharan African samples we find four sub-clusters that correspond closely with sampling locations: Luhya (from Kenya), sub-Saharan Atlantic (GWD and MSL), Guinean Gulf (YRI and ESN) and North Africa_sub-Saharan_ancestry, which is composed of North African samples with substantial sub-Saharan admixture (as previously described in [2]). By contrast, sub-clusters within North Africa do not show as precise a correlation with geography, with several sub-clusters containing individuals that span broad geographical areas: east, west, central, Atlantic, Mediterranean, Tunisia Chenini and Tunisa Sened (the last two have been already described as drifted populations that show high levels of relatedness [1,2] (figure 2 and electronic supplementary material, figure S2). Finally, a dissection of the Iberian Peninsula sub-cluster shows four minor clusters: NorthWest_Iberian, South_Iberian and two clusters without clear geographical structure (Iberian_Peninsula1 and Iberian_Peninsula2; electronic supplementary material, figure S3). One Iberian individual was an outlier (did not cluster), and therefore this individual was not included in further analyses.
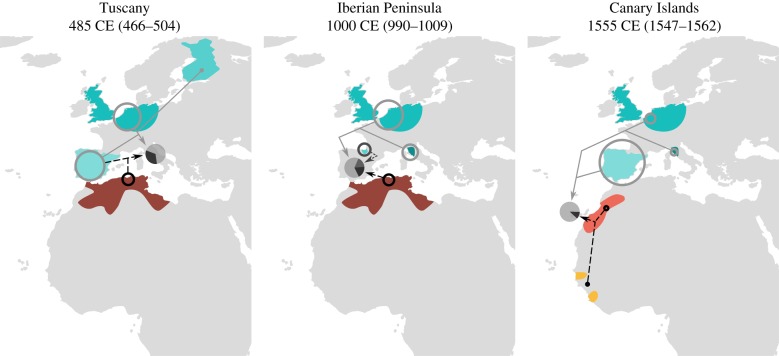
We identified and dated admixture events with Globetrotter using the clusters defined in figure 2 (figure 3). We focused on Tuscany, Iberia and the Canary Islands, three populations that surround North Africa for which there is documented contact with North Africa [28,29,39–42], in order to dissect possible admixture events between these geographical areas. We also tested admixture in the Basque population, but no admixture was detected. Assuming a single date of admixture per group, different times of admixture were inferred for the three populations: in Tuscany, the mean estimated admixture time after 100 bootstrap iterations was 485 ± 19 Current Era (CE); in the Iberian Peninsula the estimated gene-flow was dated to 1000 ± 9 CE; and, finally, in the Canary Islands the estimated date of admixture with North Africa was 1555 ± 7 CE (electronic supplementary material, figure S4 and table S2). However, while the data strongly supports a single event of North African admixture in Tuscany; in the Canary Islands and the Iberian Peninsula a history of multiple episodes of gene-flow cannot be ruled out, according to the goodness-of-fit test for two admixture events (electronic supplementary material, figure S5). The Globetrotter manual notes that the program concludes ‘multiple dates’ of admixture when its goodness-of-fit score for two dates relative to the fit of one date is above 0.35 which are based on simulation results [24]. In our dataset, 7% and 3.5% of the bootstraps exceed 0.30 for the Canary Islands and Iberian Peninsula, respectively (electronic supplementary material, figure S5).
Figure 3.
Globetrotter admixture results for the three geographical regions analysed (Tuscany, Iberia and the Canary Islands). The mean admixture date and confidence intervals for each admixture event are shown above the graphs. The geographical locations of surrogates that contribute more than 2.5% are coloured in the maps, with circle sizes showing the proportion of contribution. Coloured areas boundaries are defined by the genetic clusters' geographical distribution. Each different shade of grey corresponds to a different admixing source group, with the surrogates representing that source group linked via a continuous or dashed line. The pie in each graph shows the proportion inferred from each admixing source for the given target population (Tuscany, Iberia or the Canary Islands, respectively).
The sources inferred in the admixture events are also different in each of these three populations. In Tuscany, Globetrotter concludes a simple admixture event between two sources (figure 3). The major source is inferred to be related to present-day European groups, with the largest component being Iberian-like but with an additional northwestern European-like component. The minor contributing source inferred for Tuscany relates genetically to individuals from the Mediterranean shore of North Africa, though this minor source also contains an Iberian component. By contrast, in the Iberian Peninsula we detected a more complex pattern of gene-flow of a three-way admixture between a North African-like source from the Mediterranean shore, a Basque-like source and a European-like source with northwest and south (Tuscany) components, possibly at different times as noted above. Finally, in the Canary Islands, admixture is detected between a European-like source, mainly related to people from the Iberian Peninsula but with some relatedness to northwest Europeans and Tuscans, and a second source of admixture representing a composite of present-day North Africans from the Atlantic and sub-Saharan Africans from the Senegambia region.
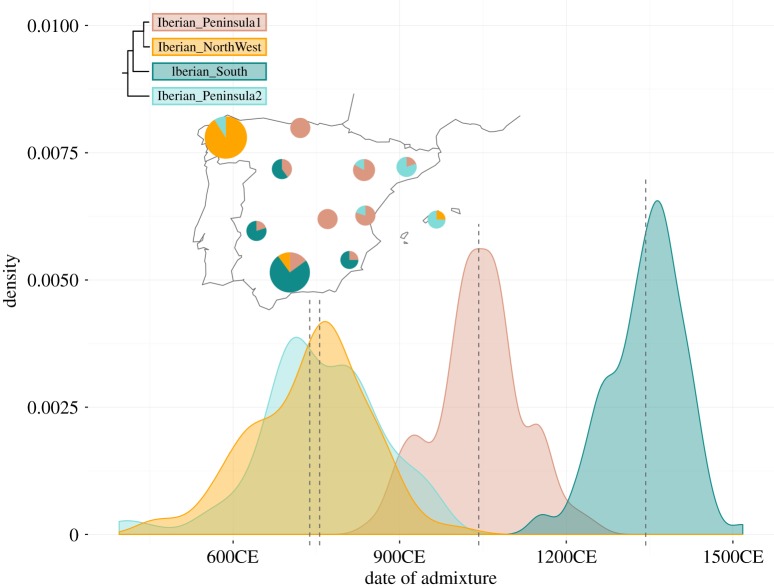
Since the Iberian Peninsula analysis showed a complex pattern of gene-flow that could be attributed to the presence of genetic substructure, we analysed the genetic subclusters within Iberia. Four different minor genetic clusters could be identified, as described above. The analysis of these four minor clusters allowed us to dissect the sources and dates of admixture within the Iberian Peninsula (figure 4). Globetrotter infers a single pulse of admixture for each of the Iberian_Northwest and Iberian_Peninsula2 minor clusters, with overlapping dates of gene-flow related to North African sources occurring around the eighth century (717–759 CE and 734–778 CE, respectively, 95% confidence interval (CI)). In the Iberian_Peninsula1 minor cluster, the inferred date of North African related admixture is around the eleventh century (1027–1058, 95% CI), while for the Iberian_South minor cluster, Globetrotter dates admixture to the second half of the fourteenth century (1330–1356, 95% CI). However, in the last two cases, again multiple episodes of gene-flow cannot be ruled out (electronic supplementary material, figure S6), and thus figure 4 may reflect dates of more recent gene-flow and mask older gene-flow. In all Iberian clusters, Globetrotter infers a North African-like source that mainly relates to our Mediterranean cluster. However, Iberian_Northwest and Iberian_Peninsula2 (which are the clusters for which Globetrotter infers older, single pulses of admixture), also show a North African west-like component (electronic supplementary material, table S3).
Figure 4.
Density plot for the admixture dates estimates after 100 bootstrap iterations of Globetrotter. The x-axis shows the date of admixture in years. On the top left the fineStructure dendrogram and the geographical distribution of minor clusters for the Iberian samples are shown, with each pie showing the proportion of individuals from that sampling location that were assigned to each of the four minor clusters (colours). The size of each circle corresponds to the number of sampled individuals. One cluster was formed by only one individual and therefore is not considered.
In summary, the North African gene-flow detected in the three geographical areas analysed (Tuscany, Iberia and the Canary Islands) differ not only in the estimated dates of admixture, but also in the sources of admixture and amount of DNA inherited for each source. In particular, Tuscany and Iberia show admixture from a Mediterranean-like source, while the Canary Islands show admixture from an Atlantic North African-like source (figure 2; electronic supplementary material, table S2).
4. Discussion
The aim of our study was to dissect gene-flow from North Africa to three surrounding coastal areas that have been documented to have had historical contact with North Africans: Tuscany, Iberia and the Canary Islands. We applied haplotype-based methods on a large sample set using genome-wide markers in order to refine our knowledge of the gene-flow between these geographical areas, focusing on the following: (i) the estimated dates of the admixture, (ii) the geographical origins of the sources of the admixture events, and (iii) the proportions of the gene-flow. The extensive dataset and the use of haplotype-based methods allowed us to estimate precise and narrow CIs for admixture dates which we correlated with historical processes. Different estimated times, sources and proportions of admixture were detected in each of the three populations analysed.
While all three populations show evidence of admixture between European-like and North African-like source groups, the geographical characterization of the North African source varies across populations. In particular, the North African source in the Canary Islands is more genetically similar to populations along the Atlantic coast, while the North African source in Iberia and Tuscany is more genetically similar to populations along the Mediterranean Coast.
In the Canary Islands, our date of admixture corresponds to the time of the Castilian conquest (fifteenth century). The European contribution is mainly Iberian, but it also shows a small amount of northwest European genetic influence, which might be related to the presence of Normans involved in the first steps of the conquest [43]. The African source shows both a North African component from the Atlantic and a sub-Saharan component from Senegambia.
The mixture of the Atlantic and Senegambia components in the Canary Islands could be explained by admixture at different times prior to European contact. Our data suggest that the initial settlers of the Islands may have already been a composite of these two components. This scenario is supported by the presence of sub-Saharan mitochondrial lineages (i.e. L haplogroups) [10,44–46] in ancient Canary samples. Alternatively, admixture between the Atlantic and the Senegambia components could have occurred by gene-flow from Senegambia at different times after the initial settlement of the Islands and before their admixture with Europeans. However, the sub-Saharan gene-flow into North Africa is high and has been continuous through time, which makes it difficult to discern whether the Senegambia component was already present in North Africa before the first colonization of the Islands or whether it arrived later on. Moreover, the initial colonization of the Islands was very recent, making it difficult to ascertain how much of the North African component may be attributable to the initial settlers versus potential gene-flow from North Africa after the initial colonization. Future studies including ancient DNA from North Africa could help resolve these issues.
Both the dates and the origin of the gene-flow from the North African Mediterranean coast suggest a genetic impact of the Arab expansion in the Iberian Peninsula. The northwest of the Iberian Peninsula shows our oldest estimated date of North African admixture and is consistent with a single pulse of admixture around the time of the early arrival and conquest of Iberia by the Arabs. By contrast, our results suggest that the south of the Iberian Peninsula experienced more recent admixture and perhaps continuous gene-flow. In this case, the admixture is dated to the last periods of the Arab rule in the Peninsula in the second half of the fourteenth century. In 1212, the Christian Kingdoms became allies in the Battle of Navas de Tolosa and conquered all southern territories except the Nasrid Kingdom of Granada, which was conquered at the end of the fifteenth century. The inferred continuous gene-flow suggests that contact between the Arab and southern Iberian populations was not limited to that time period, and the estimated dates represent an upper bound on centuries of admixture (figure 4; electronic supplementary material, figures S5 and S6). Collectively, we can identify at least two different gene-flow events in the Iberian Peninsula for which the inferred dates correlate with Arab rule in the territory: an early concentrated event in the northwest of the Peninsula, and a continuous and more recent event in the south. Moreover, the North African populations that settled in the Peninsula during the Arab conquest may have had different origins (both in time and in geography), which could be indicative of different migration waves (electronic supplementary material, table S3).
In three of the four minor genetic clusters identified for the Iberian Peninsula (Iberian_Peninsula1, Iberian_Peninsula2 and Iberian_South), three-way admixture was detected between European-like (mainly Iberian), North African-like and Basque-like sources. Alternatively, in the case of the other minor cluster, Iberian_NorthWest, only two sources of admixture (North African-like and Iberian-like) were detected. This is in agreement with different admixture events occurring at different moments and in which different populations were involved. The fact that in the northwest of Iberia the admixture does not involved a Basque-like component, while it participated in the admixture events detected in the rest of the Iberian Peninsula, suggests different Iberian populations participated in geographically separated admixture events. This may reflect different waves of the Christian Kingdoms expansion.
The genome-wide study of Fiorito et al. [28] performed admixture analyses in a large-scale Italian dataset, and highlighted more complex events of admixture than the one described herein in Tuscany. Specifically, they described continuous gene-flow from different sources since 3000 ya, which could be the result of their more geographically diverse sample set relative to our geographically localized sample of Tuscany. Perhaps because of this, we infer only a single pulse of admixture which coincides with the movement of people during the fall of the Roman Empire, which was just one of the multiple events detected by Fiorito et al. [28]. Nonetheless, our focus on North African populations has allowed us to propose a more precise origin for the North African gene-flow into Tuscany, with our best surrogate group being comprised present-day people living on the Mediterranean shores of North Africa.
Our study highlights the importance of including an extensive and diverse North African dataset in genetic studies. North Africa is a very heterogeneous region, with ample sociological, historical and genetic diversity. Our use of an extensive dataset and the use of population clusters based on genetic homogeneity allowed us to detect and describe events of admixture with more precision than previous studies investigation the influence of North African gene-flow into surrounding regions. Recent methods based on haplotype information, such as those presented here, will illuminate the finer structure and genetic history of Iberian populations, particularly as sampling increases both in terms of numbers and geographical regions encompassed [47]. In the case of the Canary Islands, ancient DNA studies might also help to better understand the origin of the first settlers of the islands and identify its influence in modern populations [48].
Supplementary Material
Acknowledgements
We thank Francesc Calafell and Alex Mas-Sandoval for helpful discussion. We would like to thank all participants for collaborating in the present study.
Data accessibility
This article has no additional data.
Authors' contributions
L.R.A. participated in data analysis, design of the study and manuscript writing. D.C. and G.H. contributed to manuscript writing and design of the study. All authors gave final approval for publication.
Competing interests
The authors declare no competing interest.
Funding
This work was supported by the Spanish MINECO grant nos. CGL2013-44351-P and CGL2016-75389-P (MINECO/FEDER, UE) and the ‘María de Maeztu’ Programme for Units of Excellence in R&D (MDM-2014-0370); and the Generalitat de Catalunya grant no. 2014SGR866. G.H. is supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (grant no. 098386/Z/12/Z) and supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre.
References
- 1.Henn BM, et al. 2012. Genomic ancestry of North Africans supports back-to-Africa migrations. PLoS Genet. 8, e1002397 ( 10.1371/journal.pgen.1002397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arauna LR, et al. 2017. Recent historical migrations have shaped the gene pool of Arabs and Berbers in North Africa. Mol. Biol. Evol. 34, 318–329. ( 10.1093/molbev/msw218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Botigué LR, et al. 2013. Gene flow from North Africa contributes to differential human genetic diversity in southern Europe. Proc. Natl Acad. Sci. USA 110, 11 791–11 796. ( 10.1073/pnas.1306223110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plaza S, Calafell F, Helal A, Bouzerna N, Lefranc G, Bertranpetit J, Comas D. 2003. Joining the Pillars of Hercules: mtDNA sequences show multidirectional gene flow in the western Mediterranean. Ann. Hum. Genet. 67, 312–328. ( 10.1046/j.1469-1809.2003.00039.x) [DOI] [PubMed] [Google Scholar]
- 5.Secher B, Fregel R, Larruga JM, Cabrera VM, Endicott P, Pestano JJ, González AM. 2014. The history of the North African mitochondrial DNA haplogroup U6 gene flow into the African, Eurasian and American continents. BMC Evol. Biol. 14, 109 ( 10.1186/1471-2148-14-109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mederos JF Navarro. 1997. Arqueología de las Islas Canarias. Espac. Tiempo y Forma, Ser. I, Prehist. y Arqueol. 10, 447–478. [Google Scholar]
- 7.Peña PA. 2013. Consideraciones en relación con la colonización protohistórica de las Islas Canarias. Any. Estud. Atlánticos 59, 519–562. [Google Scholar]
- 8.Rodríguez-Martin C, Martín-Oval M. 2009. Guanches, una historia bioantropológica (Museo Arqueológico de Tenerife).
- 9.Fregel R, Gomes V, Gusmão L, González AM, Cabrera VM, Amorim A, Larruga JM. 2009a. Demographic history of Canary Islands male gene-pool: replacement of native lineages by European. BMC Evol. Biol. 9, 181 ( 10.1186/1471-2148-9-181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maca-Meyer N, Arnay M, Rando JC, Flores C, González AM, Cabrera VM, Larruga JM. 2004. Ancient mtDNA analysis and the origin of the Guanches. Eur. J. Hum. Genet. 12, 155–162. ( 10.1038/sj.ejhg.5201075) [DOI] [PubMed] [Google Scholar]
- 11.Maca-Meyer N, González AM, Pestano J, Flores C, Larruga JM, Cabrera VM. 2003. Mitochondrial DNA transit between West Asia and North Africa inferred from U6 phylogeography. BMC Genet. 4, 15 ( 10.1186/1471-2156-4-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solé-Morata N, García-Fernández C, Urasin V, Bekada A, Fadhlaoui-Zid K, Zalloua P, Comas D, Calafell F. 2017. Whole Y-chromosome sequences reveal an extremely recent origin of the most common North African paternal lineage E-M183 (M81). Sci. Rep. 7, 15941 ( 10.1038/s41598-017-16271-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodríguez-Varela R, et al. 2017. Genomic analyses of pre-European conquest human remains from the Canary Islands reveal close affinity to modern North Africans. Curr. Biol. 27, 3396–3402. ( 10.1016/j.cub.2017.09.059) [DOI] [PubMed] [Google Scholar]
- 14.Bosch E, Calafell F, Comas D, Oefner PJ, Underhill PA, Bertranpetit J. 2001. High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between northwestern Africa and the Iberian Peninsula. Am. J. Hum. Genet. 68, 1019–1029. ( 10.1086/319521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Currat M, Poloni ES, Sanchez-Mazas A. 2010. Human genetic differentiation across the Strait of Gibraltar. BMC Evol. Biol. 10, 237 ( 10.1186/1471-2148-10-237) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González-Fortes G, et al. 2019. A western route of prehistoric human migration from Africa into the Iberian Peninsula. Proc. R. Soc. B 286, 20182288 ( 10.1098/rspb.2018.2288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olalde I, et al. 2019. The genomic history of the Iberian Peninsula over the past 8000 years. Science 363, 1230–1234. ( 10.1126/science.aav4040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valdiosera C, et al. 2018. Four millennia of Iberian biomolecular prehistory illustrate the impact of prehistoric migrations at the far end of Eurasia. Proc. Natl Acad. Sci. USA 115, 3428–3433. ( 10.1073/pnas.1717762115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams SM, et al. 2008. The genetic legacy of religious diversity and intolerance: paternal lineages of Christians, Jews, and Muslims in the Iberian Peninsula. Am. J. Hum. Genet. 83, 725–736. ( 10.1016/j.ajhg.2008.11.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capelli C, et al. 2009. Moors and Saracens in Europe: estimating the medieval North African male legacy in southern Europe. Eur. J. Hum. Genet. 17, 848–852. ( 10.1038/ejhg.2008.258) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casas MJ, Hagelberg E, Fregel R, Larruga JM, González AM. 2006. Human mitochondrial DNA diversity in an archaeological site in al-Andalus: genetic impact of migrations from North Africa in Medieval Spain. Am. J. Phys. Anthropol. 131, 539–551. ( 10.1002/ajpa.20463) [DOI] [PubMed] [Google Scholar]
- 22.Regueiro M, Garcia-Bertrand R, Fadhlaoui-Zid K, Álvarez J, Herrera RJ. 2015. From Arabia to Iberia: a Y chromosome perspective. Gene 564, 141–152. ( 10.1016/j.gene.2015.02.042) [DOI] [PubMed] [Google Scholar]
- 23.Moorjani P, et al. 2011. The history of African gene flow into southern Europeans, Levantines, Jews. PLoS Genet. 7, e1001373 ( 10.1371/journal.pgen.1001373) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hellenthal G, Busby G.BJ, Band G, Wilson JF, Capelli C, Falush D, Myers S. 2014. A genetic atlas of human admixture history. Science 343, 747–751. ( 10.1126/science.1243518) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghirotto S, et al. 2013. Origins and evolution of the Etruscans' mtDNA. PLoS ONE 8, e55519 ( 10.1371/journal.pone.0055519) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Achilli A, et al. 2007. Mitochondrial DNA variation of modern Tuscans supports the near eastern origin of Etruscans. Am. J. Hum. Genet. 80, 759–768. ( 10.1086/512822) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belle EMS, Ramakrishnan U, Mountain JL, Barbujani G. 2006. Serial coalescent simulations suggest a weak genealogical relationship between Etruscans and modern Tuscans. Proc. Natl Acad. Sci. USA 103, 8012–8017. ( 10.1073/pnas.0509718103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fiorito G, Di Gaetano C, Guarrera S, Rosa F, Feldman MW, Piazza A, Matullo G. 2016. The Italian genome reflects the history of Europe and the Mediterranean basin. Eur. J. Hum. Genet. 24, 1056–1062. ( 10.1038/ejhg.2015.233) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boattini A, et al. 2013. Uniparental markers in Italy reveal a sex-biased genetic structure and different historical strata. PLoS ONE 8, e65441 ( 10.1371/journal.pone.0065441) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Busby GBJ, et al. 2015. The role of recent admixture in forming the contemporary west Eurasian genomic landscape. Curr. Biol. 25, 2518–2526. ( 10.1016/j.cub.2015.08.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526, 68–74. ( 10.1038/nature15393) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 ( 10.1186/s13742-015-0047-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell S, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. ( 10.1086/519795) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Connell J, et al. 2014. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 10, e1004234 ( 10.1371/journal.pgen.1004234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The International HapMap Consortium. 2003. The International HapMap Project. Nature 426, 789–796. ( 10.1038/nature02168) [DOI] [PubMed] [Google Scholar]
- 36.Lawson DJ, Hellenthal G, Myers S, Falush D. 2012. Inference of population structure using dense haplotype data. PLoS Genet. 8, e1002453 ( 10.1371/journal.pgen.1002453) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leslie S, et al. 2015. The fine-scale genetic structure of the British population. Nature 519, 309–314. ( 10.1038/nature14230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laval G, Patin E, Barreiro LB, Quintana-Murci L. 2010. Formulating a historical and demographic model of recent human evolution based on resequencing data from noncoding regions. PLoS ONE 5, e10284 ( 10.1371/journal.pone.0010284) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brett M, Fentress E. 1997. The Empire and the Other: Romans and Berbers. In The Berbers, pp. 50–80. Oxford, UK: Blackwell Publishers Ltd. [Google Scholar]
- 40.Camps G.1995. Les Berbères : mémoire et identité (Errance) See https://upfinder.upf.edu/iii/encore/record/_C_Rb1160244_Sgabriel%20camps_Orightresult_U_X4?lang=spi&suite=def .
- 41.Camps G, Vela i Aulesa C.1998. Los bereberes: de la orilla del Mediterráneo al límite meridional del Sáhara (Icaria) See https://upfinder.upf.edu/iii/encore/record/_C_Rb1192860_Sgabriel%20camps_Orightresult_U_X4?lang=spi&suite=def .
- 42.Naylor PC. 2009. North Africa: a history from antiquity to the present. Austin, TX: University of Texas Press. [Google Scholar]
- 43.Reverón BB. 1944. Las canarias y la conquista Franco-Normanda: Juan de Bethencourt See https://mdc.ulpgc.es/utils/getfile/collection/MDC/id/44156/filename/80639.pdf.
- 44.Fregel R, Pestano J, Arnay M, Cabrera VM, Larruga JM, González AM. 2009b. The maternal aborigine colonization of La Palma (Canary Islands). Eur. J. Hum. Genet. 17, 1314–1324. ( 10.1038/ejhg.2009.46) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fregel R, Cabrera VM, Larruga JM, Hernández JC, Gámez A, Pestano JJ, Arnay M, González AM. 2015. Isolation and prominent aboriginal maternal legacy in the present-day population of La Gomera (Canary Islands). Eur. J. Hum. Genet. 23, 1236–1243. ( 10.1038/ejhg.2014.251) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ordóñez AC, Fregel R, Trujillo-Mederos A, Hervella M, de-la-Rúa C, Arnay-de-la-Rosa M. 2017. Genetic studies on the prehispanic population buried in Punta Azul Cave (El Hierro, Canary Islands). J. Archaeol. Sci. 78, 20–28. ( 10.1016/j.jas.2016.11.004) [DOI] [Google Scholar]
- 47.Bycroft C, Fernández-Rozadilla C, Ruiz-Ponte C, Quintela I, Carracedo Á, Donnelly P, Myers S. 2019. Patterns of genetic differentiation and the footprints of historical migrations in the Iberian Peninsula. Nat. commun. 10, 551 ( 10.1038/s41467-018-08272-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fregel R, et al. 2019. Mitogenomes illuminate the origin and migration patterns of the indigenous people of the Canary Islands. PLoS ONE 14, e0209125 ( 10.1371/journal.pone.0209125) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.